Abstract
The receptor tyrosine kinase RET functions as the signal transducing receptor for the GDNF (for “glial cell-derived neurotrophic factors”) family of ligands. Mutations in the RET gene were implicated in Hirschsprung disease (HSCR), multiple endocrine neoplasia type 2 (MEN 2), and thyroid carcinomas. In this report we demonstrate that the docking protein FRS2 is tyrosine phosphorylated by ligand-stimulated and by constitutively activated oncogenic forms of RET. Complex formation between RET and FRS2 is mediated by binding of the phosphotyrosine-binding domain of FRS2 to pY1062, a residue in RET that also functions as a binding site for Shc. However, overexpression of FRS2 but not Shc potentiates mitogen-activated protein (MAP) kinase activation by RET oncoproteins. We demonstrate that oncogenic RET-PTC proteins are associated with FRS2 constitutively, leading to tyrosine phosphorylation of FRS2, MAP kinase stimulation, and cell proliferation. However, loss-of-function HSCR-associated RET mutants exhibit impaired FRS2 binding and reduced MAP kinase activation. These experiments demonstrate that FRS2 couples both ligand-regulated and oncogenic forms of RET, with the MAP kinase signaling cascade as part of the response of RET under normal biological conditions and pathological conditions, such as MEN 2 and papillary thyroid carcinomas.
The GDNF family of neurotrophins consists of four members collectively designated GFLs (GDNF family ligands): glial cell-derived neurotrophic factor (GDNF), neurturin, persephin, and artemin. These growth factors play a crucial role in regulating the survival of neurons of the peripheral and central nervous system. The GDNF proteins signal through a multicomponent receptor complex consisting of a ligand-binding GDNF family receptor (GFR), designated the α subunit (GFRα), and the proto-oncogenic receptor tyrosine kinase, RET, which forms the β subunit. The four GFRα receptors, GFRα1, -2, -3, and -4, are linked to the cell membrane via glycosyl-phosphatidylinositol anchors. It was shown that GFRα1, -2, -3, and -4 predominantly bind GDNF, neurturin, artemin, and persephin, respectively. RET functions as a common intracellular signal transducing component in conjunction with each of the GFRα subunits (1). However, it was recently proposed that the GFRα receptors may signal autonomously in the absence of RET (49) and that they target RET to lipid rafts on the cell membrane (47). The GFRα and RET receptors are expressed in both distinct and overlapping regions of the developing embryo and the adult mouse. Mice deficient in either GDNF, RET, or GFRα1 fail to develop the ureteric bud or undergo metanephric development, demonstrating that this ligand-receptor complex plays a role in kidney morphogenesis. Moreover, the mutant mice lack enteric neurons throughout their digestive tracts (39). The striking similarities in the developmental defects observed in the mutant mice is consistent with the notion that GDNF acts primarily through the GFRα1-RET receptor complex (39).
RET has been associated with several human diseases and genetic syndromes. Loss-of-function mutations in RET, designated Hirschsprung disease (HSCR), cause defective intestinal innervation and congenital megacolon (8, 32). Gene rearrangements leading to fusion of the kinase domain of RET with heterologous proteins containing dimerization motifs result in constitutively activated RET proteins. Such fusion proteins are expressed in papillary thyroid carcinomas (PTC) and have been termed RET-PTC (35). Several different RET-PTC genes have been identified in thyroid carcinomas differing in the RET fusion partner. Finally, germ line point mutations in RET result in inherited multiple endocrine neoplasia types 2A and 2B (MEN 2A and MEN 2B) and familial medullary thyroid carcinoma (13). It has been shown that the oncogenic activity of RET in MEN and familial medullary thyroid carcinoma is caused by mutations leading to formation of unpaired cysteines in the extracellular domain which facilitate receptor dimerization and activation (40).
Three RET protein variants (RET9, RET43, and RET51) have been shown to be generated by alternative RNA splicing, leading to the expression of RET isoforms with identical primary structures up to amino acid 1063, followed by unique carboxy-terminal sequences (27). RET51 contains two additional tyrosines (23), one of which, Y1096, functions as a binding site of Grb2 (2). It has been reported that rather than being involved in Ras–mitogen-activated protein (MAP) kinase activation, Y1096 plays an important role in recruiting Gab2–phosphatidylinositol 3-kinase complexes to RET (6). Phosphorylated Y905, Y1015, and Y1062, amino acids common to the three RET protein forms, have been characterized as docking sites for several signaling molecules. pY905 mediates the recruitment of the SH2 domain-containing adapter proteins Grb7 and Grb10 (29, 30). Phospholipase Cγ binds to activated RET via pY1015 (7), and Shc is recruited to activated RET by means of binding of its phosphotyrosine-binding (PTB) domain to pY1062, a tyrosine residue located within a canonical NKLpY (single-letter amino acid code) PTB motif (3, 4, 24). In addition, the PDZ and LIM domain-containing protein Enigma binds to RET via Y1062 in a phosphorylation-independent manner (11, 12). It was demonstrated that Y1062 is essential for the mitogenic and transforming activity of RET-MEN 2A and RET-PTC alleles (4, 46) and for the survival-promoting signal induced by RET-MEN 2A (10).
It is now well established that the Ras-MAP kinase (MAPK) signaling cascade plays a pivotal role in cell mitogenesis and transformation (25) and that the Grb2-Sos complex serves as a link between activated receptor tyrosine kinases (RTKs) and Ras. The Grb2-Sos complex may be recruited to the plasma membrane by direct interaction with activated RTKs or indirectly through membrane-linked docking proteins that are tyrosine phosphorylated in response to RTK stimulation (33, 45). Direct recruitment of the Grb2-Sos complex occurs through the binding of the SH2 domain of Grb2 to the pYXN motif on activated RTKs, as shown for the epidermal growth factor (EGF) receptor (22). However, receptors lacking the pYXN motif, such as the insulin or fibroblast growth factor (FGF) receptors, utilize the docking proteins IRS1 or FRS2, respectively, for recruitment of Grb2-Sos complexes (45). Thus, docking proteins play a dominant role in recruiting the Grb2-Sos complex to the plasma membrane to mediate Ras activation by RTKs which do not bind Grb2 directly (47). In addition, docking proteins play a role in the targeting of signaling molecules to the plasma membrane and in the expansion of the repertoire of signaling pathways activated by RTKs.
In this report, we show that FRS2 proteins are tyrosine phosphorylated in response to activation of the RET receptor. Our data show that the PTB domain of FRS2α binds to pY1062 of RET, an autophosphorylation site which also serves as the binding site for Shc. Accordingly, two natural HSCR-associated mutants of the NKLpY(1062) motif of RET show defective binding to FRS2α. FRS2α is constitutively associated with the chimeric RET-PTC oncoproteins, both in a human tumor cell line and in transfected cells. Moreover, overexpression of FRS2 leads to potentiation of MAPK activation by oncogenic RET mutants. These experiments reveal an important role for FRS2 in the normal function of RET and in diseases caused by gain-of-function or loss-of-function RET mutations.
MATERIALS AND METHODS
Expression plasmids.
Expression plasmids for FRS2α and its nonmyristylated mutant (G2A) were described previously (21). All RET constructs used in this work, unless otherwise specified, were performed in a background of a RET9 isoform. The RET-PTC3 and RET-PTC3 (Y588F) cDNAs were cloned in the pBABE retroviral vector and in the pCDNA3(Myc-His) vector (Invitrogen, Groningen, The Netherlands) fused in frame at the C terminus with a myc epitope or a His tag (15, 41). The RET plasmids coding for the MEN 2A-associated changes, C634Y and C634R, as well as those coding for the HSCR-associated changes, L1061P and ΔN1059, were previously described (14, 40). The chimeric EGF receptor-RET (E-R) receptor was reported previously (42). The hemagglutinin (HA) epitope-tagged MAPK (HA-ERK2) and myc epitope-tagged p52shc constructs were described previously (14, 16). The RET (C634Y, Y905F), RET (C634Y, Y1062F), RET (C634Y, K758 M), RET (C634Y, Y1015F), and RET (C634Y)-51 (expressing the RET51 form of the MEN 2A-associated C634Y RET mutant) constructs were generated by site-directed mutagenesis using a QuickChange mutagenesis kit (Stratagene, La Jolla, Calif.). The mutations were confirmed by DNA sequencing.
Antibodies and recombinant proteins.
Polyclonal anti-RET antibodies were raised against the recombinant kinase domain of the RET protein (42). Anti-FRS2 antibodies have been described previously (21). Anti-phosphotyrosine antibodies (4G10) were from Upstate Biotechnology Inc. (Lake Placid, N.Y.). Rabbit polyclonal anti-MAPK (#9101) and anti-phospho-MAPK (#9102) antibodies were from New England Biolabs (Beverley, Mass.). Anti-Grb2, anti-glutathione S-transferase (GST), anti-tag, and secondary antibodies coupled to horseradish peroxidase were from Santa Cruz Biotechnology (Santa Cruz, Calif.).
The GST fusion protein of the PTB domains of FRS2α and Shc were expressed in Escherichia coli and purified with glutathione-conjugated agarose beads (Sigma, St. Louis, Mo.) by standard procedures.
Cell culture and transfection.
NIH 3T3 fibroblasts were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum and 100 U (each) of penicillin and streptomycin (Gibco BRL, Paisley, Pa.)/ml. NIH 3T3 cells expressing the E-R chimeric receptor have been described previously (42). For EGF stimulation experiments, E–R-expressing cells were serum starved for 36 h prior to stimulation (50 ng/ml, 5 min). Human 293 cells were grown in DMEM supplemented with 10% fetal calf serum and antibiotics. Transient transfections in 293 cells were carried out using LipofectAMINE (Gibco BRL) according to the protocol recommended by the manufacturer. The papillary thyroid carcinoma cells (PTC) and human thyroid cells (HTC) were cultured as described (9). PC Cl 3 is a thyroid epithelial cell line derived from 18-month-old Fischer rats. These cells were cultured in Coon's modified F12 medium supplemented with 5% calf serum (Gibco BRL). For both stable and transient transfections, approximately 5 × 105 PC Cl 3 cells were plated 48 h before transfection. Three hours before transfection the medium was changed to DMEM containing 5% calf serum. Calcium phosphate-DNA precipitates were prepared by standard procedures and were incubated with the cells for 1 h. Then DNA precipitates were removed and cells were incubated in 15% glycerol for 2 min. Finally, the cells were washed with DMEM and incubated for 48 h in Coon's modified F12 medium containing 5% calf serum. PC-PTC3 and PC-PTC3 (Y588F) cells were obtained by expressing the wild type or the Y588F RET-PTC3 mutant in PC Cl 3 cells. Representative mass populations of several hundred clones were obtained by puromycin selection. PC-PTC3 cells transfected with FRS2α were selected with 400 μg of Geneticin (G418) per ml. PC–E-R cells were obtained by transfecting PC Cl 3 cells with the E-R construct (42). PC-PTC3 and PC-PTC3 (Y588F) cells were subjected to transient transfection with FRS2α or p52Shc expression vectors and then processed by cytofluorimetric analysis. Trace amounts of enhanced green fluorescent protein (EGFP) were added to identify transfected cells. For cytofluorimetric analysis cells were fixed in methanol for 1 h at −20°C, rehydrated in phosphate-buffered saline (PBS) for an additional hour at 4°C, and then treated with RNase A (50 μg/ml) for 30 min. Propidium iodide (25 μg/ml) was added to the cells, and samples were analyzed with a FACScan flow cytometer (Becton Dickinson, San Jose, Calif.) interfaced with an Hewlett Packard computer (Palo Alto, Calif.). The percentages of cells in the G0/G1, S, and G2/M phases were determined. The average values from three independent experiments were obtained. [3H]thymidine incorporation assays were performed as described previously (42). Briefly, cells were grown to confluence in 24-well plates (Costar, Acton, Mass.) and then maintained for 24 h in Coon's modified F12 medium containing 1% serum in the presence of 4 μCi of [3H]thymidine (Amersham, Buck, United Kingdom) per ml. [3H]thymidine incorporation was measured in triplicate. The average values from three independent experiments were obtained.
Immunoprecipitation and peptide binding experiments.
Cells were solubilized in lysis buffer containing 50 mM HEPES (pH 7.5), 1% (vol/vol) Triton X-100, 150 mM NaCl, 5 mM EGTA, 50 mM NaF, 20 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 2 mM phenylmethylsulfonyl fluoride, and 0.2 μg (each) of aprotinin and leupeptin per ml. Cell lysates were subjected to immunoprecipitation with different antibodies or subjected to pull-down binding assays with purified recombinant proteins immobilized on agarose beads. The protein complexes were washed with the lysis buffer, eluted, and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Immunoblotting with specific antibodies and enhanced chemiluminescence (ECL; Amersham) were employed for immunodetection of proteins in complexes. Peptides dissolved in the appropriate solvent were added to the cell lysates. Incubation conditions for binding and washing of protein complexes, SDS-PAGE separation, and detection by immunoblotting were performed as detailed above. The peptides used in this study include STWIENKLYGRIS (p1062) and STWIENKLpYGRIS (pp1062), spanning the RET sequence around Y1062, and QISLDNPDpYQQDF (pEGFR), spanning the EGFR sequence that functions as the binding site for the PTB domain of Shc (pY1148) (5). These peptides were synthesized by Neosystem Laboratoire (Strasbourg, France). Subcellular fractionation was performed according to published procedures (8). Cells were homogenized in a hypotonic buffer containing 20 mM HEPES (pH 7.5), 5 mM EGTA, 50 mM NaF, 20 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 2 mM phenylmethylsulfonyl fluoride, and 0.2 μg (each) of aprotinin and leupeptin per ml. The nuclear pellet was removed after centrifugation at 1,000 × g for 5 min at 4°C. The postnuclear fraction was centrifuged at 100,000 × g for 45 min to separate the soluble and particulate fractions.
RESULTS
Ligand-stimulated E-R chimeric receptor induces tyrosine phosphorylation of FRS2.
Although a large body of evidence has implicated RET in oncogenesis, the underlying mechanisms of signaling through activated RET remain poorly understood. In order to study intracellular signaling through RET, we have generated NIH 3T3 cells which express a chimeric E-R receptor, whereby the extracellular ligand-binding domain of RET was replaced with that of the EGF receptor (42) (Fig. 1A). The parental NIH 3T3 cells express negligible amounts of endogenous EGFRs and no RET receptors. The use of this chimeric receptor allowed the analysis of signal transduction through RET independently of the GFRα coreceptors, which have been shown to be capable of autonomous signaling in response to ligand binding (49). Stimulation of NIH 3T3 cells expressing the E-R receptors with EGF has been shown to result in mitogenesis and cell transformation (42). Moreover, stimulation of RET results in tyrosine phosphorylation of Shc and complex formation with Grb2-Sos (3, 4, 24).
FIG. 1.
FRS2 coimmunoprecipitates with the activated RET RTK. (A) A schematic representation of RET and the chimeric EGFR-RET receptor. CAD, cadherin homologous domain; CYS, cysteine-rich sequence; TM, transmembrane domain. (B) Quiescent NIH 3T3 cells expressing E-R receptors were left unstimulated or were stimulated with EGF (50 ng/ml, 5 min), and lysates from these cells were immunoprecipitated (IP) with anti-Grb2 antibodies, followed by SDS-PAGE and immunoblotting (IB) with anti-pTyr antibodies. The migration of E-R, Shc, and FRS2 as revealed by immunoblotting with specific antibodies is marked. pY, pY1062. (C) Quiescent parental NIH 3T3 (NIH) and NIH–E-R cells were left unstimulated or were stimulated with EGF (50 ng/ml, 5 min). Lysates from these cells were immunoprecipitated with anti-FRS2 antibodies followed by SDS-PAGE and immunoblotting with anti-pTyr antibodies. (D) Quiescent parental PC Cl 3 (PC) and PC–E-R cells were left unstimulated or were stimulated with EGF (50 ng/ml, 5 min). Lysates were immunoprecipitated with anti-FRS2 antibodies followed by SDS-PAGE and immunoblotting with anti-pTyr antibodies.
To gain further insight into signaling via RET, we have identified proteins that become tyrosine phosphorylated and associated with Grb2 in cells in response to RET activation. Quiescent NIH 3T3 cells expressing the E-R receptors were stimulated with EGF, and lysates from stimulated or unstimulated cells were subjected to immunoprecipitation with anti-Grb2 antibodies. The immunocomplexes were resolved by SDS-PAGE followed by immunoblotting with anti-pTyr antibodies. As shown in Fig. 1B, four major tyrosine-phosphorylated proteins, with apparent molecular sizes of 160, 90, 52, and 46 kDa, coimmunoprecipitated with Grb2 upon RET activation. The 160-kDa protein and the 52- and 46-kDa proteins were identified by immunoblotting with specific antibodies as the E-R chimeric receptor and isoforms of Shc, respectively (data not shown). The 90-kDa species (marked by an asterisk in Fig. 1B) was of particular interest. We examined whether this 90-kDa protein corresponded to the docking protein FRS2. Serum-starved NIH 3T3 cells expressing the E-R receptors were stimulated with EGF, and lysates from stimulated or unstimulated cell were subjected to immunoprecipitation with anti-FRS2 antibodies. Parental NIH 3T3 cells were used as a control. The immunoprecipitates were resolved by SDS-PAGE followed by immunoblotting with anti-pTyr antibodies. As shown in Fig. 1C, FRS2 is tyrosine phosphorylated in response to E-R activation. Moreover, the activated E-R chimeric receptor was coimmunoprecipitated with FRS2, demonstrating that FRS2 forms a complex with activated RET. As RET has been shown to be oncogenically activated in thyroid cells, we have expressed the E-R receptor in these cells and studied signaling via RET following EGF stimulation (42). Rat thyroid epithelial PC Cl 3 cells were transfected with E-R, and marker-selected mass populations (PC–E-R) of these cells were further explored. Serum-starved PC–E-R or parental untransfected cells were stimulated with EGF, and lysates prepared from these cells were subjected to immunoprecipitation with anti-FRS2 antibodies followed by immunoblotting with anti-pTyr or anti-RET antibodies. The experiment presented in Fig. 1D shows EGF-induced tyrosine phosphorylation of FRS2 and complex formation between FRS2α and activated E-R in thyroid cells.
The PTB domain of FRS2 binds to pY1062 of RET.
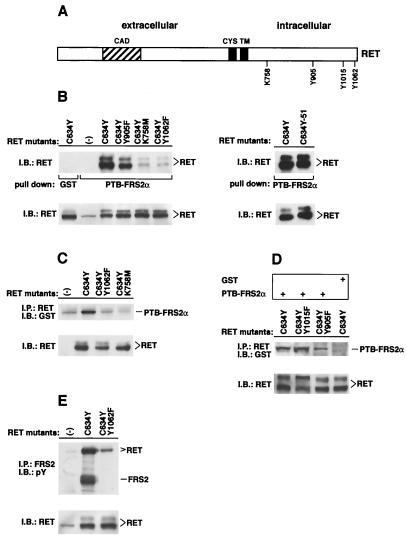
We proceeded to investigate the nature of the interaction between FRS2 and RET. To this end, we made use of a constitutively activated RET mutant responsible for the MEN 2A cancer syndrome, RET (C634Y). The use of this mutant rather than the wild-type receptor circumvents the necessity to cotransfect the GFRα coreceptors and confines the analysis to RET-mediated signaling. It has been shown that the substitution of C634 with different amino acids (such as Y or R) induces receptor activation through disulfide-linked RET dimerization (40). We also constructed three additional RET mutants in the C634Y background. These include RET (C634Y, Y905F), a mutant with active but reduced kinase activity (18), RET (C634Y, K758 M) (11), a kinase-negative mutant, and RET (C634Y, Y1062F), a mutant unable to recruit Shc (3, 4, 24) (Fig. 2A). RET (C634Y), RET (C634Y, Y905F), RET (C634Y, K758 M), and RET (C634Y, Y1062F) were transiently expressed in human 293 cells, and lysates prepared from these cells were subjected to a pull-down binding assay with the PTB domain of FRS2α expressed as a GST fusion protein and immobilized on agarose beads. GST alone was used as a control. Specifically bound proteins were eluted, resolved by SDS-PAGE, and electroblotted onto nitrocellulose membranes. The binding of RET proteins to the PTB domain of FRS2α was detected by immunoblotting with anti-RET antibodies. This experiment shows that the PTB domain binds to phosphorylated Y1062 on RET (Fig. 2B, left). Alternative splicing at the RET carboxyl terminus generates three protein variants (RET9, RET43, and RET51) with different C termini. Y1062 and residues N terminal to this amino acid are common to RET9, RET51, and RET43 (27). We have examined the possibility of whether the different C termini of the different isoforms influenced complex formation between RET and FRS2. RET (C634Y) and RET (C634Y)-51, expressing the C634Y MEN 2A mutant in the RET9 or RET51 background, respectively, were expressed in 293 cells, and lysates were subjected to a pull-down assay with the PTB domain of FRS2α. The two forms of RET (C634Y) showed similar abilities to bind the PTB domain of FRS2α (Fig. 2B, right). These findings are consistent with earlier studies demonstrating that amino acids N terminal to the pTyr residue are essential for specific binding of PTB domains to target sequences (20, 26, 45).
FIG. 2.
The PTB domain of FRS2α binds to pY1062 of RET. (A) A schematic diagram of the RET protein denoting residues that were mutated. CAD, cadherin homologous domain; CYS, cysteine-rich sequence; TM, transmembrane domain. (B) 293 cells were transfected with the expression plasmids encoding RET (C634Y), RET (C634Y, Y905F), RET (C634Y, K758 M), RET (C634Y, 1062F) (left-side gel), or RET (C634Y)-51 (right-side gel). The lysates (500 μg) were subjected to a pull-down binding assay with the GST fusion protein of the PTB domain of FRS2α or with GST alone as a control. The protein complexes were resolved by SDS-PAGE followed by immunoblotting (IB) with anti-RET antibodies (upper gel). Equal amounts of total cell lysates from the lysate sample of each mutant were resolved by SDS-PAGE followed by immunoblotting with anti-RET antibodies to demonstrate the comparable expression of the different mutants (lower gel). (C and D) 293 cells were transfected with the expression plasmids encoding RET (C634Y), RET (C634Y, Y1062F), RET (C634Y, K758 M), RET (C634Y, Y1015F), or RET (C634Y, Y905F). One milligram of each lysate sample was immunoprecipitated (IP) with anti-RET antibodies. The immunoprecipitates were recovered with protein A-Sepharose beads and were washed. The beads were then incubated with a purified GST fusion protein of the PTB domain of FRS2α or with GST alone as a control (top section of panel D). The protein complexes were washed and analyzed by SDS-PAGE followed by immunoblotting with anti-GST antibodies (upper gel of panel C and upper gel of panel D). Equal amounts of total cell lysates were stained with anti-RET antibodies to evaluate the expression of the different mutants (lower gels of each panel). (E) 293 cells were transfected with the expression plasmids encoding RET (C634Y) or RET (C634Y, Y1062F). One milligram of each lysate sample was immunoprecipitated with anti-FRS2 antibodies. The immunoprecipitates were resolved by SDS-PAGE followed by immunoblotting with anti-pTyr antibodies (upper gel). Equal amounts of total cell lysates from the lysate sample of each mutant were resolved by SDS-PAGE followed by immunoblotting with anti-RET antibodies to determine the expression of the different mutants (lower gel). pY, pY1062.
To confirm that the association between the PTB domain of FRS2α and RET requires the phosphorylation of Y1062, a series of RET mutants were expressed in 293 cells and subjected to immunoprecipitation with anti-RET antibodies. The immunoprecipitates were washed, left immobilized on protein A-agarose beads, and then incubated with the purified GST fusion protein of the PTB domain of FRS2α. The protein complex bound to the protein A-agarose beads was further washed, eluted, and subjected to SDS-PAGE followed by immunoblotting with anti-GST antibodies. As shown in Fig. 2C, the PTB domain of FRS2α bound specifically to the activated RET mutant (C634Y). The kinase-defective Y905F mutant exhibited reduced binding towards the PTB domain (Fig. 2D). A point mutant, Y1015F, which fails to bind to the SH2 domain of phospholipase Cγ (7), bound normally to the FRS2 PTB domain, and no binding was observed with GST alone (Fig. 2D).
We proceeded to verify whether the interaction of the FRS2 proteins with RET was dependent on pY1062 in intact cells. RET (C634Y) or RET (C634Y, Y1062F) was transiently expressed in 293 cells, and lysates were subjected to immunoprecipitation with anti-FRS2 antibodies followed by SDS-PAGE and immunoblotting with anti-pTyr or anti-RET antibodies. As shown in Fig. 2E, while the activated RET (C634Y) mutant formed a complex with tyrosine-phosphorylated FRS2, RET (C634Y, Y1062F) did not. Collectively, these experiments demonstrate that FRS2 binds to pY1062 of RET through its PTB domain.
HSCR-associated RET mutations impair interaction of RET with FRS2.
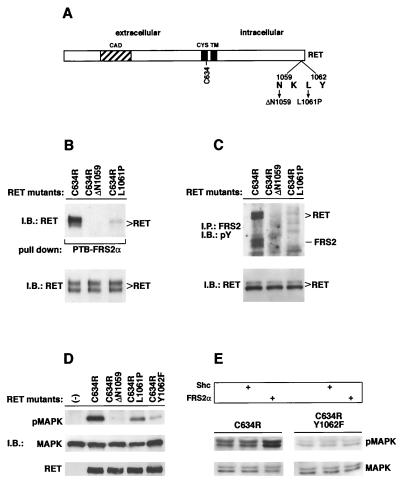
Many HSCR cases are associated with naturally occurring loss-of-function mutations of the RET RTK. HSCR mutations include deletions, insertions, frameshifts, and nonsense and missense mutations dispersed throughout the RET coding sequence (37). Recently, two distinct mutations of RET were mapped in the vicinity of the binding site for the PTB domains of Shc and FRS2: deletion of Asn 1059 (ΔN1059) and replacement of Leu 1061 by a Pro (L1061P) (14). To evaluate their effects on the interaction between RET and FRS2, we introduced these changes into the constitutively activated MEN 2A-associated RET (C634R) mutant (as schematically shown in Fig. 3A) and analyzed the binding of these mutants with FRS2 in vitro and in living cells. RET (C634R) or the HSCR mutants were expressed in 293 cells, and lysates prepared from the cells were subjected to a pull-down binding assay with the PTB domain of FRS2α expressed as a GST fusion protein and immobilized on agarose beads. The binding of the RET proteins was detected by immunoblotting with anti-RET antibodies. The experiment presented in Fig. 3B demonstrates that the binding of both the HSCR mutants to the PTB domain of FRS2α was significantly impaired, although the binding of the L1061P mutant was reduced to a lesser extent than the binding of the ΔN1059 mutant. Thus, the binding of RET to the PTB domain of FRS2α requires Asn and Leu residues at positions −3 and −1 relative to pY1062, respectively.
FIG. 3.
HSCR-associated RET gene mutations impair the binding of FRS2 to activated RET. (A) A schematic diagram of the RET protein denoting residues which have been mutated. CAD, cadherin homologous domain; CYS, cysteine-rich sequence; TM, transmembrane domain. (B) RET (C634R) alone and RET (C634R) bearing the HSCR-associated changes, ΔN1059 or L1061P, were transiently expressed in 293 cells. Cell lysates were subjected to a pull-down binding assay, with the GST fusion protein of the PTB domain of FRS2α immobilized on beads. The protein complexes were eluted, resolved by SDS-PAGE, and immunoblotted (IB) with anti-RET antibodies (upper gel). Equal amounts of total cell lysates were resolved by SDS-PAGE followed by immunoblotting with anti-RET antibodies to evaluate the expression of the different mutants (lower gel). (C) 293 cells were transfected with plasmids encoding RET (C634R) or the mutants bearing the HSCR-associated changes, ΔN1059 or L1061P. One milligram of each lysate sample was immunoprecipitated (IP) with anti-FRS2 antibodies. The immunoprecipitates were eluted and resolved by SDS-PAGE followed by immunoblotting with anti-pTyr antibodies (upper gel). Equal amounts of total cell lysates were resolved by SDS-PAGE, followed by immunoblotting with anti-RET antibodies to evaluate the expression of the different mutants (lower gel). pY, pY1062. (D) 293 cells were transfected with plasmids coding for an epitope-tagged MAPK construct (HA-ERK2) together with RET (C634R) or the HSCR changes, ΔN1059 and L1061P. Each sample (50 μg) was resolved by SDS-PAGE followed by immunoblotting with anti-RET (upper gel) or anti-pMAPK (middle gel) or with the anti-HA epitope antibodies for normalization (MAPK, lower gel). −, 293 cells transfected with HA-ERK2 alone. (E) 293 cells were transfected with plasmids encoding RET (C634R) or RET (C634R, Y1062F) in the presence of p52Shc or FRS2α-encoding plasmids as indicated (top section). Each lysate sample (50 μg) was resolved by SDS-PAGE followed by immunoblotting with anti-pMAPK (middle section) or with anti-MAPK antibodies for normalization (bottom section). RET, FRS2, and Shc expression was confirmed by immunoblotting (not shown).
To analyze the effect of the HSCR mutants on the interaction between RET and FRS2 in living cells, we expressed RET (C634R) or the HSCR mutants in 293 cells and subjected the cell lysates to immunoprecipitation with anti-FRS2 antibodies. The immunocomplexes were eluted and resolved by SDS-PAGE followed by immunoblotting with anti-pTyr antibodies (Fig. 3C). Consistent with the data from the in vitro binding assay, endogenous FRS2 was not tyrosine phosphorylated and did not form a complex with the HSCR mutants.
It has been shown that tyrosine-phosphorylated FRS2 recruits both Grb2 and Shp2 molecules, events involved in MAPK activation (16). The phosphorylation of Shc (3, 4, 24) and of FRS2 (this report) is dependent on their binding to activated RET at the NKLpY motif. It thus follows that the NKLpY motif should be essential for RET-mediated MAPK activation and that the Y1062F, ΔN1059, and L1061P RET mutants should be impaired in stimulating MAPK activity. To test this hypothesis, we coexpressed an epitope-tagged MAPK (HA-ERK2) with RET (C634R) or the HSCR mutants in 293 cells and analyzed MAPK activation in these cells using antibodies which specifically recognize activated phospho-MAPK. The experiment presented in Fig. 3D shows that the HSCR mutants induced a weak MAPK response. Since the L1061P mutant retains residual FRS2 and Shc binding, it is possible that this mutant can still weakly activate MAPK stimulation.
To distinguish between the contributions of Shc and FRS2 in mediating MAPK response to activated RET, we compared MAPK activation in 293 cells transiently transfected with the active C634R or the defective C634R, Y1062F RET constructs in the presence of expression vectors for either the p52 form of Shc or FRS2α. The level of ectopically expressed Shc or FRS2α over the expression levels of the two endogenous proteins has been shown to be approximately fivefold (data not shown). Activation of endogenous MAPK was determined by analyzing the samples with phospho-MAPK antibodies. Figure 3E shows that overexpression of FRS2α potentiates by approximately threefold MAPK activation as compared to that of cells transfected with RET (C634R) alone (left panel) or of cells transfected with RET (C634R) together with Shc. However, overexpression of FRS2α with the RET (C634R, Y1062F) mutant did not activate MAPK response (Fig. 3E, right panel).
To investigate the contribution of the individual amino acids in the vicinity of Y1062 to the interaction of RET with FRS2α, peptide competition assays were performed. RET (C634Y) was expressed in 293 cells by transient transfection, and cell lysates were subjected to a pull-down binding assay with the GST fusion protein of the PTB domain of FRS2α or Shc immobilized on beads. A phosphopeptide containing the pY1062 sequence from RET9 (pp1062, STWIENKLpY1062GRIS) or the nonphosphorylated version of the same sequence (p1062) was added to the binding reaction at 1, 10, or 50 μM (Fig. 4A). While the phosphopeptide (pp1062) strongly inhibited the interaction between RET (C634Y) and the PTB domains of both FRS2α and Shc, the nonphosphorylated peptide (p1062) did not. Interestingly, the phosphopeptide encompassing the Shc binding site on the EGF receptor (pEGFR, QISLDNPDpYQQDF) inhibited the binding of the PTB domain of Shc to RET (C634Y) efficiently but did not affect the binding of the PTB domain of FRS2α to RET (C634Y) (Fig. 4B). These results show that the amino acids in the vicinity of Y1062 of RET significantly affect the interaction of the receptor with the PTB domains of Shc and FRS2α. Furthermore, while the NKLpY motif is critical for binding of both docking proteins, the surrounding amino acids determine the specificity of binding of the PTB domains of Shc and FRS2 to pY1062.
FIG. 4.
The PTB domain of FRS2α binds to pY1062 of RET. The RET (C634Y) or, as a control, RET (C634Y, Y1062F) proteins were transiently expressed in 293 cells. Equal amounts of the lysate were incubated with the GST fusion proteins of the PTB domains of FRS2α or Shc in the presence of 1, 10, or 50 μM concentrations of the pp1062, p1062 (A), or pEGFR (B) peptides. pp1062 is a phosphorylated RET peptide (STWIENKLpY1062GRIS), and p1062 is the corresponding nonphosphorylated peptide. pEGFR is an EGFR-derived phosphopeptide (QISDNPDpY1148QQDF) which functions as a binding site for the PTB domain of Shc. Bound proteins were eluted and resolved by SDS-PAGE followed by immunoblotting (IB) with anti-RET antibodies. (C) Equal amounts of total cell lysates from the lysate sample of RET (C634Y) or RET (C634Y, Y1062F) were resolved by SDS-PAGE followed by immunoblotting with anti-RET antibodies to evaluate the expression of the different mutants.
Differential effects of FRS2 and Shc overexpression on RET–PTC-induced MAPK response, cell cycle progression, and membrane targeting.
PTC is the most common form of thyroid malignancy in humans. The predominant molecular lesion that is associated with these tumors is the fusion of RET with heterologous proteins. The fusion proteins, termed RET-PTC, are expressed in thyroid carcinomas, are cytosolic, and contain coiled-coil domains that are responsible for RET-PTC dimerization and activation (48). Several oncogenic RET-PTC fusion proteins have been characterized (15, 41). The RET-PTC1 and RET-PTC3 oncoproteins result from the fusion of the kinase domain of RET with the coiled-coil motifs of the H4 protein and the RFG gene product, respectively (Fig. 5A) (16, 44). It has been shown that transgenic mice expressing either RET-PTC1 or RET-PTC3 develop thyroid carcinomas (19, 38, 43).
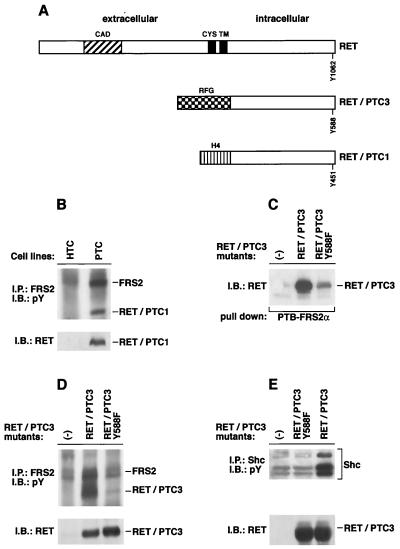
FIG. 5.
RET-PTC oncoproteins interact with FRS2. (A) A schematic representation of the wild-type RET protein and the chimeric RET-PTC1 and RET-PTC3 transforming proteins. Tyrosine 1062 of RET and the corresponding tyrosines in RET-PTCs are indicated. CAD, cadherin homologous domain; CYS, cysteine-rich sequence; TM, transmembrane domain. (B) Lysates from normal thyroid epithelial cells (HTC) and papillary thyroid carcinoma cells (PTC) were immunoprecipitated (IP) with anti-FRS2 antibodies followed by SDS-PAGE and immunoblotting (IB) with anti-pTyr antibodies (upper gel). An immunoblot with anti-RET antibodies was performed to show the presence of RET-PTC1 protein in PTC but not HTC cells (lower gel). pY, pY1062. (C) Expression vectors for RET-PTC3 and RET-PTC3 (Y588F) were expressed in 293 cells, and a pull-down assay with GST fusion protein of the PTB domains of FRS2α was performed; untransfected 293 cells (−) were used as a control. (D) 293 cells were transfected with expression vectors encoding RET-PTC3 and RET-PTC3 (Y588F). One milligram of each lysate sample was immunoprecipitated with anti-FRS2 antibodies. The immunoprecipitates were eluted and resolved by SDS-PAGE followed by immunoblotting with anti-pTyr antibodies (upper gel). Equal amounts of total cell lysates were resolved by SDS-PAGE followed by immunoblotting with anti-RET antibodies to evaluate the expression of the different mutants (lower gel). (E) 293 cells were transfected with expression vectors encoding RET-PTC3 or RET-PTC3 (Y588F). One milligram of each lysate sample was immunoprecipitated with anti-Shc antibodies. The immunoprecipitates were eluted and resolved by SDS-PAGE followed by immunoblotting with anti-pTyr antibodies (upper gel). Equal amounts of total cell lysates were resolved by SDS-PAGE followed by immunoblotting with anti-RET antibodies to evaluate the expression of the different mutants (lower panel).
We proceeded to examine the interaction between RET-PTC fusion proteins and FRS2 by using the PTC human thyroid papillary carcinoma cell line which expresses RET-PTC1 (17) or by using 293 cells transiently transfected with RET-PTC3. Lysates from PTC cells or from normal human thyroid cells were subjected to immunoprecipitation with anti-FRS2 antibodies followed by SDS-PAGE and immunoblotting with anti-pTyr or anti-RET antibodies (Fig. 5B). The experiment presented in Fig. 5B shows that tyrosine-phosphorylated FRS2 forms a complex with RET-PTC1 in lysates of these cells.
To analyze in more detail the nature of the interaction between FRS2 and RET-PTC3, expression vectors for RET-PTC3 or RET-PTC3 (Y588F) were transiently expressed in 293 cells. It is of note that Y588 in RET-PTC3 corresponds to Y1062 in RET. The lysates from RET-PTC3- or RET-PTC3 (Y588F)-expressing cells were subjected to a pull-down binding assay with the GST fusion protein of the PTB domain of FRS2α immobilized on agarose beads. Bound proteins were eluted, resolved by SDS-PAGE, and electroblotted on a nitrocellulose membrane followed by immunoblotting with anti-RET antibodies. As shown in Fig. 5C, while RET-PTC3 formed a complex with the PTB domain of FRS2α, the association of RET-PTC3 (Y588F) with FRS2α was very weak. We proceeded to verify the interaction of RET-PTC3 or the RET-PTC3 (Y588F) mutant with endogenous FRS2 in intact cells. In this experiment 293 cells were transfected with RET-PTC3 or RET-PTC3 (Y588F), and cell lysates were subjected to immunoprecipitation with anti-FRS2 antibodies followed by SDS-PAGE and immunoblotting with anti-pTyr or anti-RET antibodies. Consistent with the results from the in vitro binding assay, RET-PTC3 but not RET-PTC3 (Y588F) was found to be capable of binding to and tyrosine phosphorylation of endogenous FRS2 in intact cells (Fig. 5D). Previous studies have shown that Shc is tyrosine phosphorylated in PTC cells as a consequence of binding to RET via pY1062 (34). We therefore analyzed tyrosine phosphorylation of Shc in 293 cells transiently expressing RET-PTC3 or the RET-PTC3 (Y588F) mutant. Endogenous Shc was immunoprecipitated with anti-Shc antibodies and resolved by SDS-PAGE followed by immunoblotting with anti-pTyr antibodies. This experiment shows an increase in tyrosine phosphorylation of Shc in transfected RET-PTC3 cells but not in RET-PTC3 (Y588F) cells (Fig. 5E), thus demonstrating that Y1062 in RET (corresponding to Y588 in RET-PTC3) is indeed required for tyrosine phosphorylation of Shc as shown in this report for tyrosine phosphorylation of FRS2.
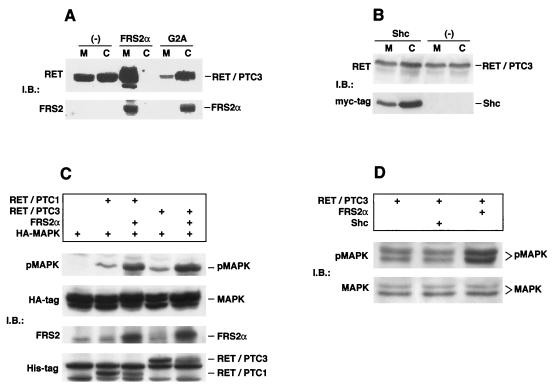
Since FRS2 is linked to the cell membrane, we have examined the possibility that FRS2 plays a role in targeting RET-PTC to the cell membranes. In this experiment 293 cells were transfected with expression vector for RET-PTC3 alone or together with expression vector for FRS2α or the G2A mutant, a nonmyristylated mutant that fails to interact with the cell membrane (21). The cells were homogenized and fractionated into soluble or particulate fractions (a fraction containing a mixture of plasma membrane, mitochondria, Golgi, lysosomes, and vesicles). The fractions were resolved by SDS-PAGE followed by immunoblotting with anti-RET or anti-FRS2 antibodies to analyze the relative distribution of the RET-PTC3 and FRS2α proteins in the two fractions. As shown in Fig. 6A, RET-PTC3 is localized primarily in the particulate fraction when coexpressed with wild-type FRS2α and in the soluble fraction when coexpressed with the nonmyristylated mutant of FRS2α. When highly overexpressed alone, RET-PTC3 was found in both the soluble and particulate fractions. On the basis of this experiment, we propose that the binding of RET-PTC to FRS2α may facilitate translocation of the protein to the cell membrane. However, similar experiments performed with Shc demonstrated that overexpression of Shc did not influence the distribution of RET-PTC3 between the soluble and particulate fractions (Fig. 6B).
FIG. 6.
FRS2 promotes localization of RET-PTC3 protein to particulate fraction and potentiates RET-PTC-mediated MAPK activation. (A) 293 cells were transfected with RET-PTC3 alone or together with wild-type FRS2α or the G2A nonmyristylated mutant. Cells were homogenized and separated into the postnuclear soluble (C) and particulate membrane (M) fractions. After removal of cell nuclei, about 30% of the total proteins were found in the membrane fraction and 70% were in the cytosolic fraction. Each fraction (50 μg) was subjected to SDS-PAGE and immunoblotted (IB) with anti-RET antibodies or anti-FRS2 antibodies. The efficiency of fractionation was determined by using EGFR and eps15 as specific markers for the membrane and cytosolic fractions, respectively (data not shown). (B) 293 cells were transfected with RET-PTC3 alone or together with a myc-tagged p52Shc expression vector. Cells were homogenized and separated into the postnuclear soluble and particulate membrane fractions. Each fraction (50 μg) was subjected to SDS-PAGE and immunoblotted with anti-RET or anti-myc antibodies. The efficiency of fractionation was determined by using EGFR and eps15 as specific markers for the membrane and cytosolic fractions, respectively (data not shown). (C) 293 cells were transfected with expression vectors encoding epitope-tagged MAPK (HA-ERK2) together with expression vectors for myc- and His-tagged RET-PTC1 or RET-PTC3 alone or in combination with FRS2α as indicated. Each lysate sample (50 μg) was resolved by SDS-PAGE followed by immunoblotting with (i) pMAPK antibodies, (ii) anti-HA antibodies (MAPK), and (iii) anti-His-tagged antibodies (RET-PTC1 and RET-PTC3). FRS2α expression was detected by immunoprecipitation of 200 μg of each sample followed by SDS-PAGE and immunoblotting with anti-FRS2 antibodies. (D) 293 cells were transfected with expression vectors for myc- and His-tagged RET-PTC3 alone or in combination with FRS2α or p52Shc as indicated. Each lysate sample (50 μg) was resolved by SDS-PAGE followed by immunoblotting with pMAPK or MAPK antibodies. RET-PTC3, FRS2, and Shc expression was confirmed by immunoblotting (data not shown).
It was previously demonstrated that tyrosine-phosphorylated FRS2α mediates receptor-induced MAPK activation by means of recruitment of Grb2-Sos complexes, directly and indirectly through Shp2 (16, 21). In this report we show that the MAPK response of RET mutants defective in the recruitment of FRS2 and Shc is severely impaired. To test the role of FRS2 and Shc in RET–PTC-induced MAPK stimulation, we have examined the effect of overexpression of FRS2 or Shc on RET–PTC-induced MAPK response. In this experiment a HA-ERK2 construct was coexpressed together with His-tagged constructs of RET-PTC1 or RET-PTC3 in 293 cells and the cell lysates were subjected to immunoblotting with antibodies specific for activated phospho-MAPK. When singly transfected, both RET-PTC1 and RET-PTC3 induce a modest stimulation of MAPK (Fig. 6C). However, when either of the two RET-PTC proteins was coexpressed with FRS2α the resulting MAPK response was strongly potentiated 5- to 10-fold (Fig. 6C). To determine whether Shc had a similar effect, we compared MAPK stimulation in 293 cells transiently transfected with RET-PTC3 in the presence of expression vectors for p52Shc or FRS2α. While overexpression of FRS2α potentiated MAPK activation by approximately fivefold in cells transfected with RET-PTC3, overexpression of Shc together with RET-PTC3 did not induce a similar stimulation of MAPK (Fig. 6D).
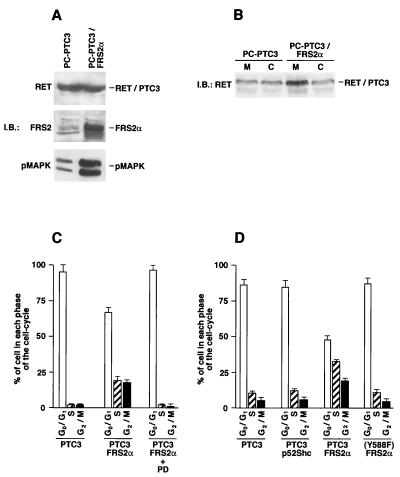
To further evaluate whether FRS2 potentiates biological effects of the RET-PTC proteins, we used thyroid epithelial cells expressing RET-PTC3. The PC Cl 3 rat thyroid epithelial cell line expressing RET-PTC3 (PC-PTC3 cells) has modest phenotypic effects, such as the loss of thyroid-differentiated functions and thyrotropin dependency (44; R. M. Melillo and M. Santoro, unpublished observations). PC-PTC3 cells were transfected with an expression vector for FRS2α, and mass populations of several hundred clones were obtained and expanded. One representative population was used for further studies. PC-PTC3–FRS2α showed high levels of expression of FRS2α (Fig. 7A) and an increased MAPK response compared to MAPK stimulation detected in the parental PC-PTC3 cells (Fig. 7A). We also compared the effect of FRS2 expression on the subcellular distribution of RET-PTC3 in these cells. Parental PC-PTC3 and PC-PTC3–FRS2α cells were homogenized and fractionated into soluble (C) or particulate (M) fractions followed by immunoblotting with anti-RET antibodies. The experiment presented in Fig. 7B shows increased RET-PTC3 protein in the particulate fraction in cells expressing FRS2α (Fig. 7B), similar to the results obtained with 293 cells.
FIG. 7.
Potentiation of RET-PTC3 induced stimulation of S-phase entry of PC Cl 3 thyroid cells by overexpression of FRS2. (A) PC-PTC3 cells (PC Cl 3 cells expressing the RET-PTC3 oncogene) were transfected with the FRS2α expression vector. A representative cell population was designated PC-PTC3–FRS2α. Aliquots of 100 μg of each lysate sample from the PC-PTC3 and PC-PTC3–FRS2α cells were resolved by SDS-PAGE, followed by immunoblotting (IB) with anti-RET, anti-FRS2, or anti-pMAPK antibody. (B) PC-PTC3 and PC-PTC3–FRS2α cells were homogenized and separated into the postnuclear soluble (C) and particulate membrane (M) fractions. Each fraction (100 μg) was subjected to SDS-PAGE and immunoblotted with anti-RET antibodies. The efficiency of fractionation was determined by using anti-EGFR and anti-eps15 antibodies as specific markers for the membrane and cytosolic fractions, respectively (data not shown). (C) PC-PTC3 and FRS2α-expressing PC-PTC3 cells were harvested after 96 h of serum deprivation and analyzed by flow cytometry. When indicated, PC-PTC3–FRS2α cells were pretreated with 20 μM PD98059 (PD). The percentage of cells in each phase of the cell cycle is depicted in a bar graph. The results are the averages of three independent experiments. (D) PC-PTC3 or PC-PTC3 (Y588F) cells were transiently transfected with expression vectors for FRS2α or p52Shc together with an expression vector for EGFP to identify transfected cells. On average, approximately 70% of transfected cells were EGFP positive (not shown). Cells were harvested after 48 h of serum deprivation (1% serum), and EGFP-positive cells were analyzed by flow cytometry. The results are the averages of three independent experiments.
It has been shown that PC-PTC3 cells arrested in the G1 phase of the cell cycle are unable to proceed to the S phase when serum is removed from the culture medium (43). We have examined the possibility of whether overexpression of FRS2α could promote G1/S transition in G1-arrested PC-PTC3 cells. Cell cycle kinetics of parental and FRS2α-expressing cells was examined upon serum deprivation (96 h). Under these conditions, approximately 45% of the cells that express FRS2α (PC-PTC3–FRS2α) were found in the S and G2/M phases, while parental cells (PC-PTC3) were found to be almost entirely arrested in the G0/G1 phase of the cell cycle (Fig. 7C). The FRS2-dependent progression of cell cycle appears to be dependent on stimulation of the MAPK cascade, as treatment of the PC-PTC3–FRS2α cells with 20 μM PD98059, a specific inhibitor of MEK1, arrested the treated cells in the G0/G1 phase (Fig. 7C). However, expression of FRS2α alone in PC Cl 3 cells did not have a significant effect on MAPK stimulation or on serum-independent cell cycle progression (data not shown). As a more direct measure of DNA synthesis, we next compared stimulation of [3H]thymidine incorporation in the two cell populations. The cells were maintained for 24 h in 1% serum in the presence of 4 μCi of [3H]thymidine/ml, and [3H]thymidine incorporation was measured in triplicate. PC-PTC3–FRS2α showed a (2.2 ± 0.3)-fold (average of three different experiments) increase in thymidine incorporation compared to thymidine incorporation measured for PC-PTC3 cells.
We have also compared the ability of FRS2 and Shc to promote cell cycle progression of PC-PTC3 cells by using flow cytometry. PC-PTC3 and PC-PTC3 (Y588F) cells were transiently transfected with expression vectors for FRS2α or Shc or with an empty vector as a control. A plasmid coding for EGFP was added to the transfection mixture to allow the identification of transfected cells by flow cytometry or fluorescence microscopy to demonstrate that approximately 70% of the cells were transfected (data not shown). The transfected cells were maintained in low serum (1%) for 48 h then fixed and stained with propidium iodide followed by analysis of the EGFP-positive cells by flow cytometry. The results presented in Fig. 7D show that expression of FRS2 but not Shc induced the progression of PC-PTC3 cells into the S and G2/M phases. However, cell cycle progression was not induced in cells expressing the RET-PTC3 (Y588F) mutant.
Taken together, these experiments show that both FRS2 and Shc are recruited by RET by means of pY1062. However, FRS2 appears to play a more prominent role than Shc in membrane targeting, MAPK stimulation, and cell cycle progression induced by RET and by its oncogenic forms.
DISCUSSION
The FRS2 docking proteins are major substrates of the FGF and NGF receptors (16, 21). The two members of the FRS2 family (FRS2α and FRS2β) are structurally similar, comprising an N-terminal myristylation sequence, a PTB domain, and a C-terminal fragment that contains multiple tyrosine residues that, when phosphorylated, constitute recognition motifs for the SH2 domains of Grb2 and Shp2, leading to the recruitment of Grb2-Sos complexes to the cell membrane (16, 21, 50). The binding of Shp2 to FRS2 results in its own tyrosine phosphorylation followed by complex formation with Grb2. Thus, upon tyrosine phosphorylation, the FRS2 proteins mediate the recruitment of multiple Grb2-Sos complexes both directly and indirectly through Shp2. Compared to other docking proteins, FRS2α and FRS2β show several unique properties. While most docking proteins, such as IRS1, -2, -3, and –4, interact with multiple signaling molecules, such as Grb2, Shp2, Nck, and phosphatidylinositol-3 kinase, the FRS2 proteins mainly appear to be involved in the recruitment of Grb2 and Shp2. In addition, IRS, Dok, and Shc are translocated from the cytosol to the plasma membrane in response to receptor activation, while FRS2 proteins are permanently linked to the plasma membrane via a myrstyl anchor (45).
FRS2 proteins bind directly to the FGF and NGF receptors and, as demonstrated in this report, to RET through their PTB domains. However, while the interaction between the FRS2 proteins and the FGF receptor does not depend on tyrosine phosphorylation, their binding to the NGF receptor or RET requires tyrosine autophosphorylation of these receptors. FRS2 proteins bind specifically to a highly conserved region in the juxtamembrane region of FGFR1, recognizing the sequence KSIPLRRQVTVS (28, 50). In contrast, the binding of FRS2 proteins to Trk (26, 28) or RET is mediated through a canonical PTB domain recognition sequence comprising the NXXpY motif. Accordingly, in this report we show that the PTB domain of FRS2α binds to RET through an NKLpY1062 motif at the C-terminal tail of the receptor. Another docking protein, Shc, also binds to activated Trk and RET receptors at pY490 and pY1062, respectively, by means of its PTB domain. From the characterization of interactions between FRS2 proteins and the FGF, Trk, and RET receptors, it appears that the PTB domains of FRS2 proteins are capable of recognizing relatively diverse sequences in different receptors. Although the β-turn structure formed by the NPXpY motif in several binding sites of PTB domains has been shown to be required for high-affinity interaction with PTB domains, both the recognition sites for the PTB domains of FRS2α and FRS2β on FGFR1 and RET lack the β-turn-forming motifs. Thus, the PTB domains of FRS2 proteins exhibit ligand-binding specificities that are not restricted to recognition of phosphotyrosine-containing or β-turn-forming sequences.
In addition to the recruitment of Shc and FRS2 proteins, it has been shown that Y1062 in RET is required for the binding of Enigma, a protein composed of a PDZ domain followed by three LIM domains. Enigma has been shown to associate directly with RET or the RET-PTC fusion proteins constitutively through its second LIM domain (11, 12). Several studies have demonstrated that Y1062 of RET plays a critical role in mediating the mitogenic effects of wild-type RET as well as the transforming potential of the RET-PTC oncoproteins (4, 11, 12, 46). Recently, two mutations of RET in the vicinity of Y1062, resulting in the deletion of Asn1059 (ΔN1059) or the replacement of Leu1061 by Pro (L1061P), respectively, were identified in HSCR patients (14). Biochemical analysis provided evidence that the recruitment of Shc to the ΔN1059 or L1061P mutants of the receptor was impaired (14). In this report, we demonstrate that the ΔN1059 or L1061P mutations also interfere with the binding of FRS2 to the RET protein. Intriguingly, while most HSCR mutations are heterozygous, the L1061P mutation, which causes only a partial defect in Shc and FRS2 binding and partial attenuation of MAPK response, has been found in the homozygous state in HSCR patients (14).
We have shown that a phosphopeptide corresponding to the binding site of the PTB domain of Shc on the EGF receptor effectively inhibits the binding of the PTB domain of Shc to RET but does not interfere with the binding of FRS2α to the same receptor. This result suggests that the PTB domains of Shc and FRS2α bind to overlapping but not identical sites on RET within the context of pY1062. It would be of interest to identify loss-of-function mutations in RET that show differential binding towards Shc, FRS2, and perhaps also Enigma. The characterization of the binding of Shc, FRS2, and Enigma to such mutants may provide insights into the molecular mechanisms of interaction of these proteins with RET in the vicinity of Y1062.
Gain-of-function mutations in RET genes give rise to constitutively activated receptors, resulting in the MEN 2 class of dominantly inherited cancer syndromes (13). Here, we report that two of the naturally occurring activating point mutations in the MEN 2A syndrome (C634Y and C634R) are constitutively associated with FRS2α. As shown previously, Shc is also constitutively associated with and tyrosine phosphorylated by the MEN 2 form of RET. Tyrosine-phosphorylated FRS2 and Shc proteins mediate the recruitment of the Grb2-Sos complexes to the plasma membrane to activate Ras, resulting in activation of the MAPK cascade (45). Thus, FRS2 and Shc may be dominant targets of the activated forms of RET, acting as mediators of RET-induced cell transformation. However, the roles played by FRS2 and Shc in RET signaling and oncogenesis do not appear to be identical. Here we show that overexpression of FRS2, but not Shc, promotes MAPK activation induced by RET-MEN 2A and RET-PTC3. These findings indicate that FRS2 is particularly efficient in coupling RET to the MAPK pathway. This can be due to the ability of FRS2 to recruit multiple Grb2-Sos complexes, the localization of FRS2 at the cell membrane, and its ability to recruit directly Shp2, which also appears to play a role in MAPK stimulation. These findings do not exclude a role for Shc in this process; rather, they indicate that FRS2 plays a more prominent role in mediating cellular signaling of oncogenic RET proteins.
Somatic events that lead to the activation of RET have been extensively documented in human PTC (35). In these cases, chromosomal translocations occur resulting in the fusion of the RET locus with other genes. These rearrangements, which give rise to the RET-PTC oncoproteins, may play a causative role in thyroid cancer. However, the mechanism(s) by which these proteins cause cell transformation is poorly understood. Here we show that FRS2 is recruited by RET-PTC proteins and that FRS2 overexpression potentiates MAP activation and mitogenesis induced by these oncogenic proteins. Biochemical fractionation experiments demonstrate that overexpression of FRS2 leads to accumulation of RET-PTC3 in the particulate fraction. It is likely that the FRS2–RET-PTC complex is localized, at least in part, at the cell membrane due to interactions with FRS2, as FRS2 is linked to the cell membrane by myristylation (21). FRS2-mediated plasma membrane recruitment of RET-PTC proteins is likely to contribute to the activation of signaling cascades that are initiated at the plasma membrane. By contrast, experiments presented in this report show that Shc is unable to recruit RET-PTC3 protein to the particulate cell fraction.
Our findings demonstrate that FRS2 couples RET with the Ras-MAPK signaling cascade. Potentiation of this central signaling cascade can be involved in neoplastic diseases associated with gain-of-function mutations of RET genes. On the other hand, impairment of the link between RET and MAPK takes place in congenital megacolon (HSCR) (14, 39). Accordingly, De Vita et al. demonstrated that the survival signal induced by RET is dependent upon activation of Akt and MAPK (10). It has been shown that enteric neural crest cells undergo apoptosis in the foregut of embryos lacking the RET receptor (39), indicating that impairment in cell survival pathways may be one of the major consequences of RET inactivation in HSCR. Indeed, we have recently demonstrated that FRS2 plays an important role in the control of a cell survival pathway mediated by Gab1 and phosphatidylinositol-3 kinase (29). On the basis of the experiments described in this report revealing a role for FRS2 in signaling via RET, we propose that FRS2 proteins play a pivotal role in coupling RET with the MAPK signaling cascade under normal physiological conditions and by the oncogenic forms of RET.
ACKNOWLEDGMENTS
We thank G. Viglietto for the cytofluorimetric analyses and C. Monaco for the RET-PTC molecular constructs. We are grateful to J. Gilder for editing the text.
This study was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC), by E. C. grant BMH4-CT96–0814, by Programma Biotecnologie legge 95/95 (MURST 5%), and by the Ligue Nationale contre le Cancer grant to M.B. M.S. was supported by a fellowship from FIRC (Italian Foundation for Cancer Research).
REFERENCES
- 1.Airaksinen M S, Titievsky A, Saarma M. GDNF family neurotrophic factor signaling: four masters, one servant? Mol Cell Neurosci. 1999;13:313–325. doi: 10.1006/mcne.1999.0754. [DOI] [PubMed] [Google Scholar]
- 2.Alberti L, Borrello M G, Ghizzoni S, Torriti F, Rizzetti M G, Pierotti M A. Grb2 binding to the different isoforms of Ret tyrosine kinase. Oncogene. 1998;17:1079–1087. doi: 10.1038/sj.onc.1202046. [DOI] [PubMed] [Google Scholar]
- 3.Arighi E, Alberti L, Torriti F, Ghizzoni S, Rizzetti M G, Pelicci G, Pasini B, Bongarzone I, Piutti C, Pierotti M A, Borrello M G. Identification of Shc docking site on Ret tyrosine kinase. Oncogene. 1997;14:773–782. doi: 10.1038/sj.onc.1200896. [DOI] [PubMed] [Google Scholar]
- 4.Asai N, Murakami H, Iwashita T, Takahashi M. A mutation at tyrosine 1062 in MEN2A-Ret and MEN2B-Ret impairs their transforming activity and association with shc adaptor proteins. J Biol Chem. 1996;271:17644–17649. doi: 10.1074/jbc.271.30.17644. [DOI] [PubMed] [Google Scholar]
- 5.Batzer A G, Blaikie P, Nelson K, Schlessinger J, Margolis B. The phosphotyrosine interaction domain of Shc binds an LXNPXY motif on the epidermal growth factor receptor. Mol Cell Biol. 1995;15:4403–4409. doi: 10.1128/mcb.15.8.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besset V, Scott R P, Ibanez C F. Signaling complexes and protein-protein interactions involved in the activation of the Ras and PI3K pathways by the c-Ret receptor tyrosine kinase. J Biol Chem. 2000;275:39159–39166. doi: 10.1074/jbc.M006908200. [DOI] [PubMed] [Google Scholar]
- 7.Borrello M G, Alberti L, Arighi E, Bongarzone I, Battistini C, Bardelli A, Pasini B, Piutti C, Rizzetti M G, Mondellini P, Radice M T, Pierotti M A. The full oncogenic activity of Ret/ptc2 depends on tyrosine 539, a docking site for phospholipase C gamma. Mol Cell Biol. 1996;16:2151–2163. doi: 10.1128/mcb.16.5.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlomagno F, De Vita G, Berlingieri M T, de Franciscis V, Melillo R M, Colantuoni V, Kraus M H, Di Fiore P P, Fusco A, Santoro M. Molecular heterogeneity of RET loss of function in Hirschsprung's disease. EMBO J. 1996;15:2717–2225. [PMC free article] [PubMed] [Google Scholar]
- 9.de Nigris F, Visconti R, Cerutti J, Califano D, Mineo A, Santoro M, Santelli G, Fusco A. Overexpression of the HIP gene coding for a heparin/heparan sulfate-binding protein in human thyroid carcinomas. Cancer Res. 1998;58:4745–4751. [PubMed] [Google Scholar]
- 10.De Vita G, Melillo R M, Carlomagno F, Visconti R, Castellone M D, Bellacosa A, Billaud M, Fusco A, Tsichlis P N, Santoro M. Tyrosine 1062 of RET-MEN2A mediates activation of Akt (protein kinase B) and mitogen-activated protein kinase pathways leading to PC12 cell survival. Cancer Res. 2000;60:3727–3731. [PubMed] [Google Scholar]
- 11.Durick K, Gill G N, Taylor S S. Shc and Enigma are both required for mitogenic signaling by Ret/ptc2. Mol Cell Biol. 1998;18:2298–2308. doi: 10.1128/mcb.18.4.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durick K, Wu R Y, Gill G N, Taylor S S. Mitogenic signaling by Ret/ptc2 requires association with Enigma via a LIM domain. J Biol Chem. 1996;271:12691–12694. doi: 10.1074/jbc.271.22.12691. [DOI] [PubMed] [Google Scholar]
- 13.Eng C, Clayton D, Schuffenecker I, Lenoir G, Cote G, Gagel R F, van Amstel H K, Lips C J, Nishisho I, Takai S I, Marsh D J, Robinson B G, Frank-Raue K, Raue F, Xue F, Noll W W, Romei C, Pacini F, Fink M, Niederle B, Zedenius J, Nordenskjold M, Komminoth P, Hendy G N L M, Mulligan L M, et al. The relationship between specific RET proto-oncogene mutations and disease phenotype in multiple endocrine neoplasia type 2. International RET mutation consortium analysis. JAMA. 1996;276:1575–1579. [PubMed] [Google Scholar]
- 14.Geneste O, Bidaud C, Vita G D, Hofstra R M, Tartare-Deckert S, Buys C H, Lenoir G M, Santoro M, Billaud M. Two distinct mutations of the RET receptor causing Hirschsprung's disease impair the binding of signaling effectors to a multifunctional docking site. Hum Mol Genet. 1999;8:1989–1999. doi: 10.1093/hmg/8.11.1989. [DOI] [PubMed] [Google Scholar]
- 15.Grieco M, Santoro M, Berlingieri M T, Melillo R M, Donghi R, Bongarzone I, Pierotti M A, Della Porta G, Fusco A, Vecchio G. PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell. 1990;60:557–563. doi: 10.1016/0092-8674(90)90659-3. [DOI] [PubMed] [Google Scholar]
- 16.Hadari Y R, Kouhara H, Lax I, Schlessinger J. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol Cell Biol. 1998;18:3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishizaka Y, Ushijima T, Sugimura T, Nagao M. cDNA cloning and characterization of ret activated in a human papillary thyroid carcinoma cell line. Biochem Biophys Res Commun. 1990;168:402–408. doi: 10.1016/0006-291x(90)92335-w. [DOI] [PubMed] [Google Scholar]
- 18.Iwashita T, Asai N, Murakami H, Matsuyama M, Takahashi M. Identification of tyrosine residues that are essential for transforming activity of the ret proto-oncogene with MEN2A or MEN2B mutation. Oncogene. 1996;12:481–487. [PubMed] [Google Scholar]
- 19.Jhiang S M, Sagartz J E, Tong Q, Parker-Thornburg J, Capen C C, Cho J Y, Xing S, Ledent C. Targeted expression of the ret/PTC1 oncogene induces papillary thyroid carcinomas. Endocrinology. 1996;137:375–378. doi: 10.1210/endo.137.1.8536638. [DOI] [PubMed] [Google Scholar]
- 20.Kavanaugh W M, Williams L T. An alternative to SH2 domains for binding tyrosine-phosphorylated proteins. Science. 1994;266:1862–1865. doi: 10.1126/science.7527937. [DOI] [PubMed] [Google Scholar]
- 21.Kouhara H, Hadari Y R, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 22.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature. 1993;363:85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Vega Q C, Decker R A, Pandey A, Worby C A, Dixon J E. Oncogenic RET receptors display different autophosphorylation sites and substrate binding specificities. J Biol Chem. 1996;271:5309–5312. doi: 10.1074/jbc.271.10.5309. [DOI] [PubMed] [Google Scholar]
- 24.Lorenzo M J, Gish G D, Houghton C, Stonehouse T J, Pawson T, Ponder B A, Smith D P. RET alternate splicing influences the interaction of activated RET with the SH2 and PTB domains of Shc, and the SH2 domain of Grb2. Oncogene. 1997;14:763–771. doi: 10.1038/sj.onc.1200894. [DOI] [PubMed] [Google Scholar]
- 25.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 26.Meakin S O, MacDonald J I, Gryz E A, Kubu C J, Verdi J M. The signaling adapter FRS-2 competes with Shc for binding to the nerve growth factor receptor TrkA. A model for discriminating proliferation and differentiation. J Biol Chem. 1999;274:9861–9870. doi: 10.1074/jbc.274.14.9861. [DOI] [PubMed] [Google Scholar]
- 27.Myers S M, Eng C, Ponder B A, Mulligan L M. Characterization of RET proto-oncogene 3′ splicing variants and polyadenylation sites: a novel C-terminus for RET. Oncogene. 1995;11:2039–2045. [PubMed] [Google Scholar]
- 28.Ong S H, Guy G R, Hadari Y R, Laks S, Gotoh N, Schlessinger J, Lax I. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol Cell Biol. 2000;20:979–989. doi: 10.1128/mcb.20.3.979-989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ong, S. H., Y. R. Hadari, N. Gotoh, G. R. Guy, J. Schlessinger, and I. Lax. Stimulation of PI-3 kinase by FGF receptors is mediated by coordinated recruitment of multiple docking proteins. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 30.Pandey A, Duan H, Di Fiore P P, Dixit V M. The Ret receptor protein tyrosine kinase associates with the SH2-containing adapter protein Grb10. J Biol Chem. 1995;270:21461–21463. doi: 10.1074/jbc.270.37.21461. [DOI] [PubMed] [Google Scholar]
- 31.Pandey A, Liu X, Dixon J E, Di Fiore P P, Dixit V M. Direct association between the Ret receptor tyrosine kinase and the Src homology 2-containing adapter protein Grb7. J Biol Chem. 1996;271:10607–10610. doi: 10.1074/jbc.271.18.10607. [DOI] [PubMed] [Google Scholar]
- 32.Pasini B, Borrello M G, Greco A, Bongarzone I, Luo Y, Mondellini P, Alberti L, Miranda C, Arighi E R, Bocciardi R, et al. Loss of function effect of RET mutations causing Hirschsprung disease. Nat Genet. 1995;10:35–40. doi: 10.1038/ng0595-35. [DOI] [PubMed] [Google Scholar]
- 33.Pawson T, Scott J D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 34.Pelicci G, Lanfrancone L, Salcini A E, Romano A, Mele S, Borrello M G, Segatto O, Di Fiore P P, Pelicci P G. Constitutive phosphorylation of Shc proteins in human tumors. Oncogene. 1995;11:899–907. [PubMed] [Google Scholar]
- 35.Pierotti M A, Bongarzone I, Borello M G, Greco A, Pilotti S, Sozzi G. Cytogenetics and molecular genetics of carcinomas arising from thyroid epithelial follicular cells. Genes Chromosomes Cancer. 1996;16:1–14. doi: 10.1002/(SICI)1098-2264(199605)16:1<1::AID-GCC1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 36.Plyte S, Majolini M B, Pacini S, Scarpini F, Bianchini C, Lanfrancone L, Pelicci P, Baldari C T. Constitutive activation of the Ras/MAP kinase pathway and enhanced TCR signaling by targeting the Shc adaptor to membrane rafts. Oncogene. 2000;19:1529–1537. doi: 10.1038/sj.onc.1203451. [DOI] [PubMed] [Google Scholar]
- 37.Ponder B A. The phenotypes associated with ret mutations in the multiple endocrine neoplasia type 2 syndrome. Cancer Res. 1999;59:1736–1741. [PubMed] [Google Scholar]
- 38.Powell D J, Jr, Russell J, Nibu K, Li G, Rhee E, Liao M, Goldstein M, Keane W M, Santoro M, Fusco A, Rothstein J L. The RET/PTC3 oncogene: metastatic solid-type papillary carcinomas in murine thyroids. Cancer Res. 1998;58:5523–5528. [PubMed] [Google Scholar]
- 39.Rosenthal A. The GDNF protein family: gene ablation studies reveal what they really do and how. Neuron. 1999;22:201–203. doi: 10.1016/s0896-6273(00)81077-6. [DOI] [PubMed] [Google Scholar]
- 40.Santoro M, Carlomagno F, Romano A, Bottaro D P, Dathan N A, Grieco M, Fusco A, Vecchio G, Matoskova B, Kraus M H, et al. Activation of RET as a dominant transforming gene by germline mutations of MEN2A and MEN2B. Science. 1995;267:381–383. doi: 10.1126/science.7824936. [DOI] [PubMed] [Google Scholar]
- 41.Santoro M, Dathan N A, Berlingieri M T, Bongarzone I, Paulin C, Grieco M, Pierotti M A, Vecchio G, Fusco A. Molecular characterization of RET/PTC3; a novel rearranged version of the RET proto-oncogene in a human thyroid papillary carcinoma. Oncogene. 1994;9:509–516. [PubMed] [Google Scholar]
- 42.Santoro M, Wong W T, Aroca P, Santos E, Matoskova B, Grieco M, Fusco A, di Fiore P P. An epidermal growth factor receptor/ret chimera generates mitogenic and transforming signals: evidence for a ret-specific signaling pathway. Mol Cell Biol. 1994;14:663–675. doi: 10.1128/mcb.14.1.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santoro M, Chiappetta G, Cerrato A, Salvatore D, Zhang L, Manzo G, Picone A, Portella G, Santelli G, Vecchio G, Fusco A. Development of thyroid papillary carcinomas secondary to tissue-specific expression of the RET/PTC1 oncogene in transgenic mice. Oncogene. 1996;12:1821–1826. [PubMed] [Google Scholar]
- 44.Santoro M, Melillo R M, Grieco M, Berlingieri M T, Vecchio G, Fusco A. The TRK and RET tyrosine kinase oncogenes cooperate with ras in the neoplastic transformation of a rat thyroid epithelial cell line. Cell Growth Differ. 1993;4:77–84. [PubMed] [Google Scholar]
- 45.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 46.Segouffin-Cariou C, Billaud M. Transforming ability of MEN2A-RET requires activation of the phosphatidylinositol 3-kinase/AKT signaling pathway. J Biol Chem. 2000;275:3568–3576. doi: 10.1074/jbc.275.5.3568. [DOI] [PubMed] [Google Scholar]
- 47.Tansey M G, Baloh R H, Milbrandt J, Johnson E M., Jr GFR alpha-mediated localization of RET to lipid rafts is required for effective downstream signaling, differentiation, and neuronal survival. Neuron. 2000;25:611–623. doi: 10.1016/s0896-6273(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 48.Tong Q, Xing S, Jhiang S M. Leucine zipper-mediated dimerization is essential for the PTC1 oncogenic activity. J Biol Chem. 1997;272:9043–9047. doi: 10.1074/jbc.272.14.9043. [DOI] [PubMed] [Google Scholar]
- 49.Trupp M, Scott R, Whittemore S R, Ibanez C F. Ret-dependent and -independent mechanisms of glial cell line-derived neurotrophic factor signaling in neuronal cells. J Biol Chem. 1999;274:20885–20894. doi: 10.1074/jbc.274.30.20885. [DOI] [PubMed] [Google Scholar]
- 50.Xu H, Lee K W, Goldfarb M. Novel recognition motif on fibroblast growth factor receptor mediates direct association and activation of SNT adapter proteins. J Biol Chem. 1998;273:17987–17990. doi: 10.1074/jbc.273.29.17987. [DOI] [PubMed] [Google Scholar]