Abstract
Bop1 is a novel nucleolar protein involved in rRNA processing and ribosome assembly. We have previously shown that expression of Bop1Δ, an amino-terminally truncated Bop1 that acts as a dominant negative mutant in mouse cells, results in inhibition of 28S and 5.8S rRNA formation and deficiency of newly synthesized 60S ribosomal subunits (Z. Strezoska, D. G. Pestov, and L. F. Lau, Mol. Cell. Biol. 20:5516–5528, 2000). Perturbation of Bop1 activities by Bop1Δ also induces a powerful yet reversible cell cycle arrest in 3T3 fibroblasts. In the present study, we show that asynchronously growing cells are arrested by Bop1Δ in a highly concerted fashion in the G1 phase. Kinase activities of the G1-specific Cdk2 and Cdk4 complexes were downregulated in cells expressing Bop1Δ, whereas levels of the Cdk inhibitors p21 and p27 were concomitantly increased. The cells also displayed lack of hyperphosphorylation of retinoblastoma protein (pRb) and decreased expression of cyclin A, indicating their inability to progress through the restriction point. Inactivation of functional p53 abrogated this Bop1Δ-induced cell cycle arrest but did not restore normal rRNA processing. These findings show that deficiencies in ribosome synthesis can be uncoupled from cell cycle arrest and reveal a new role for the p53 pathway as a mediator of the signaling link between ribosome biogenesis and the cell cycle. We propose that aberrant rRNA processing and/or ribosome biogenesis may cause “nucleolar stress,” leading to cell cycle arrest in a p53-dependent manner.
Proliferating cells can delay or block cell cycle transitions in response to a variety of extracellular regulatory signals as well as to perturbations in intracellular processes. Several types of stress, such as DNA damage, defects in replication and chromosome segregation, and accumulation of misfolded proteins in the endoplasmic reticulum are now known to elicit checkpoint responses that prevent progression through the cell cycle (16, 25, 69). These responses are often altered in neoplastic cells, suggesting that the regulatory mechanisms involved play important roles in tumor development (24).
In a previous study, we applied a genetic selection procedure to search for sequences in a cDNA library that can cause reversible arrest of the cell cycle (45). One cDNA clone (Bop1Δ) that induced a particularly strong inhibition of DNA synthesis in NIH 3T3 fibroblasts encoded an amino-terminally truncated form of a novel WD40 repeat protein, named Bop1 (block of proliferation). Expression of Bop1Δ interfered with the functions of the endogenous Bop1 in a dominant manner, which likely accounted for the strong growth-inhibitory potential of this clone.
Subsequent studies revealed that Bop1 was predominantly localized to the nucleolus and cofractionated with preribosomal particles (58). Bop1Δ exhibited a similar localization but lacked some of the critical functions of the wild-type protein, leading to a dominant negative phenotype. Expression of this mutant form of Bop1 in LAP3 cells completely blocked formation of the mature 28S and 5.8S rRNAs and resulted in reduced levels of 60S ribosome subunits in the cytoplasm, while synthesis of 18S rRNA and production of 40S subunits were unaffected (58). Analysis of pre-rRNA processing revealed that conversion of the 36S precursor to the 32S pre-rRNA was reduced and that the 32S precursor was not processed to the 28S and 12S/5.8S rRNAs but instead was degraded (58).
Although these findings indicated the role of Bop1 in processing of the 28S and 5.8S rRNAs and 60S ribosome assembly, it remained unclear how expression of Bop1Δ might exert an antiproliferative effect. In this study, we show that the cell cycle arrest caused by Bop1Δ-mediated perturbation of Bop1 function exhibits features of a G1 checkpoint associated with upregulation of the Cdk inhibitors (CKIs) p21 and p27 and downregulation of the G1-specific Cdk2 and Cdk4 activities. Inactivation of p53 alleviated Bop1Δ-induced cell cycle arrest. These findings show, for the first time, a p53-dependent cross-talk between ribosome biogenesis and cell cycle progression. We propose a model in which p53 senses nucleolar stress as a result of rRNA processing errors and induces cell cycle arrest as a consequence.
MATERIALS AND METHODS
Cells and expression constructs.
LAP3 is a cell line derived from NIH 3T3 fibroblasts that supports isopropyl-β-d-thiogalactopyranoside (IPTG) inducible expression from pX vectors (46). Bop1Δ is a mutant of mouse Bop1 lacking 231 amino acids from the amino terminus cloned in pX11 (previously named B5-35). Cell lines obtained by transfection of LAP3 cells with either the empty vector pX11 (LAP3/1) or Bop1Δ (Bop1Δ/2 and Bop1Δ/6) have been characterized previously (45); they were referred to as pX11/1, B5-35/2, and B5-35/6, respectively. pJ4Ω16E6 and pJ4Ω16E6Δ111–115 express wild-type E6 and mutant E6 defective in p53 binding, respectively (11). The retroviral vector pBabe-puro-GSE56 expresses a fragment of p53 that acts as a genetic suppressor element that effectively antagonizes wild-type p53 function (43). Cells were cultured as described previously (45).
Analysis of DNA, RNA, and protein synthesis.
To measure the rate of DNA synthesis, cells were seeded at 3 × 104 cells per well in a 24-well plate, and IPTG was added to 1 mM the next day. After incubation for the indicated durations, cells were labeled with 5 μCi of [methyl-3H]thymidine (ICN) per ml for 35 min, then chilled on ice, washed twice with phosphate-buffered saline (PBS) and twice for 5 min each with 10% trichloroacetic acid, and lysed in 200 μl of 1% sodium dodecyl sulfate (SDS)–0.1 M NaOH. For analysis of rRNA processing, cells were pulse-labeled with 2.5 μCi of [3H]uridine (New England Nuclear) per ml for 30 min, washed once with medium, then chased with nonradioactive medium for 2 h, and lysed with Trizol (Gibco-BRL). Purified RNA samples were normalized by scintillation counting, separated on a formaldehyde-agarose gel, and transferred to a nylon membrane, which was treated with En3Hance (New England Nuclear) and exposed to film. To measure the rate of protein synthesis, cells were incubated for 30 min in medium with 0.1× the normal methionine concentration and labeled with 2.5 μCi of [35S]methionine (New England Nuclear) per ml for 1 h. Cells were washed with PBS and trichloroacetic acid as described above and dissolved in 0.5% SDS. Cell number was determined by the fluorometric DNA assay with Hoechst 33258 (48).
Flow cytometry.
Cells were harvested by trypsinization, washed in PBS, pelleted, resuspended in PBS containing 0.2% NP-40 and 0.5% bovine serum albumin plus 100 μg of RNase A and 25 μg of propidium iodide per ml, and incubated for 2 to 3 h at 4°C in the dark. Data were collected on a FACSort cytometer (Becton Dickinson). Aggregates were gated out, and histograms were drawn for presentation using WINMDI software; cell cycle analysis was done using ModFit LT (Verity).
Immunoblot and kinase assays.
Cell lysates prepared as described before (37) were normalized by protein content using the Bio-Rad DC protein assay reagents and bovine serum albumin as a standard. Antibodies against Cdk2 (M2), cyclins D1 (72-13G), E (M-20), and A (H-432), p21 (C-19), and p27(C-19) were from Santa Cruz; anti-Cdk4 (DCS-35) was from NeoMarkers; antibodies against phosphorylated Ser780 of pRb were from New England Biolabs; and anti-retinoblastomal protein (anti-pRb; G3-245) was from PharMingen. Secondary horseradish peroxidase-coupled antibodies and a chemiluminescence detection kit were from Amersham Pharmacia. Cdk2 assays were performed according to a published protocol (12). Cdk4 assays were done as described previously (37), with the following changes: lysates containing 400 μg of protein were immunoprecipitated for 2.5 h with 2 μg of antibody; reactions were carried out in 30 μl of 50 mM HEPES-NaOH (pH 7.5)–10 mM KCl–10 mM MgCl2–1 mM EGTA–1 mM dithiothreitol–10 mM β-glycerophosphate–1 mM ATP containing 2 μg of maltose-binding protein–pRb(701–928) fusion protein (New England Biolabs) as a substrate, and stopped after 10 min at 30°C, when they were in the linear range. Analysis of pRb phosphorylation was done as described previously (45).
RESULTS
Expression of Bop1Δ arrests cell cycle progression at G1.
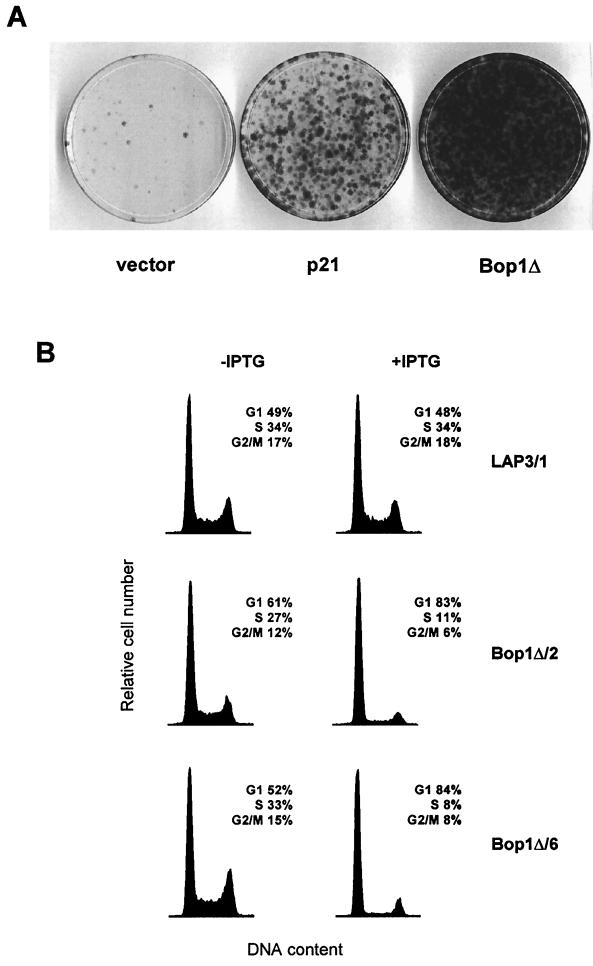
Bop1Δ, an N-terminally truncated form of Bop1, acts in a dominant negative manner to perturb Bop1 activity (58). Expression of Bop1Δ induces a potent but reversible cell cycle arrest in NIH 3T3-derived LAP3 cells (45). This effect can be demonstrated in a cell suicide assay in which proliferating cells that incorporate bromodeoxyuridine (BrdU) into newly synthesized DNA are killed by the combined action of the drug and visible light. Cells that are induced to express a stably transfected gene that blocks DNA synthesis are protected from the lethal effects of BrdU and light and can form colonies after removal of BrdU and repression of the transfected gene (46). As shown in Fig. 1A, Bop1Δ induced a reversible block of DNA synthesis in LAP3 cells with very high efficiency, allowing the survival of a large number of cells. By comparison, expression of the CKI p21 in this system also induced reversible cell cycle arrest, but not as efficiently as Bop1Δ (Fig. 1A). Notably, many cells displayed morphological abnormalities after transient overexpression of p21 and failed to form colonies, whereas Bop1Δ-arrested cells rapidly reverted to normal morphology and resumed proliferation after its repression (unpublished observations). Thus, Bop1Δ can elicit a potent cell cycle-inhibitory response in LAP3 cells without significantly affecting cell viability.
FIG. 1.
Expression of Bop1Δ in asynchronously growing cells induces reversible G1 growth arrest. (A) LAP3 cells were cotransfected with pPGK-puro, which confers resistance to puromycin, and either the empty IPTG-inducible vector (pX11) or constructs for inducible expression of the Cdk inhibitor p21 (pX11-p21) or Bop1Δ (pX11-Bop1Δ). Equal numbers of stably transfected, puromycin-resistant cells were treated in parallel with IPTG for 24 h to induce expression and then with IPTG and BrdU for 48 h to selectively kill proliferating cells, as previously described (46). Cells that did not replicate DNA during this period and therefore survived the treatment were rescued by removal of the BrdU and IPTG, grown for 8 days, and stained with crystal violet. The number of colonies thus reflects the number of cells that were reversibly cell cycle arrested while under IPTG induction of the transfected gene. (B) Bop1Δ causes accumulation of cells in G1. Parallel cultures of asynchronously growing cells were left untreated (−IPTG) or treated with inducer for 24 h (+IPTG) and subjected to flow cytometry analysis. Two independent clonal lines that inducibly express Bop1Δ (Bop1Δ/2 and Bop1Δ/6) and a control clonal line (LAP3/1) transfected with the empty vector were analyzed. Histograms of the cellular DNA content and the calculated distributions of cell populations in different phases of the cell cycle are shown.
To assess whether Bop1Δ arrests cells at a specific point of the cell cycle, we generated clonal lines of LAP3 cells in which Bop1Δ was expressed under the control of an IPTG-inducible promoter (45, 58). We incubated asynchronously growing cells with IPTG for 24 h and analyzed their DNA content by flow cytometry, using untreated cells as a control. Induction of Bop1Δ resulted in a significant accumulation of cells with a G1 DNA content and a concomitant decrease in the number of cells with an S and G2/M content in independently derived clonal lines, whereas no effect of IPTG was observed in control cells (Fig. 1B). For example, in clonal line Bop1Δ/6, the fraction of cells in G1 increased from 52 to 84%, while S-phase cells decreased from 33 to 8% after 24 h of Bop1Δ expression (Fig. 1B). These data suggest that expression of Bop1Δ in asynchronously growing cells leads to cell cycle arrest in the G1 phase.
Effects of Bop1Δ on cyclins, CKIs, and pRb phosphorylation.
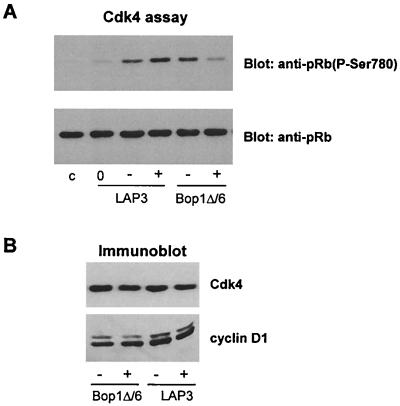
Cyclins D, E, and A, acting in concert with their associated Cdk catalytic subunits, are key regulators of G1 progression and S-phase entry (53). We investigated whether expression of Bop1Δ might affect these cyclin-Cdk complexes. First, we examined the activity of Cdk4, a major cyclin D-dependent kinase in NIH 3T3 cells. We immunoprecipitated cell extracts from the clonal line Bop1Δ/6 with anti-Cdk4 antibody and analyzed the immune complexes for kinase activity towards a recombinant Rb protein. In this kinase assay, we used detection of reaction products with antibodies specific for phosphorylated serine 780 (phospho-Ser780) of pRb, a site efficiently phosphorylated by cyclin D1-Cdk4 in vitro (29). The Cdk4-associated activity in Bop1Δ/6 cells treated with IPTG was substantially decreased compared with that in untreated cells (Fig. 2A). In control samples, IPTG treatment of the parental LAP3 cells did not reduce Cdk4 activity, while withdrawal of serum led to a significant reduction, confirming the specificity of the assay (Fig. 2A). Protein immunoblots showed that the amounts of Cdk4 and its catalytic counterpart cyclin D1 were similar in cells that were induced to express Bop1Δ and untreated cells (Fig. 2B). Thus, Bop1Δ induction in LAP3 cells inhibits Cdk4 activity, although it does not downregulate expression of either cyclin D1 or Cdk4.
FIG. 2.
Bop1Δ inhibits Cdk4-associated activity but not the abundance of Cdk4 or cyclin D1. (A) pRb kinase activity in Cdk4 immune complexes. Cdk4-specific phosphorylation of recombinant pRb as a substrate (see Materials and Methods) was determined by immunoblotting of reaction products with antibodies specific to phosphorylated Ser780 of pRb [anti-pRb(P-Ser780)] (top), and then the same blot was reprobed with anti-pRb antibody to show equal amounts of the substrate in each lane (bottom). Assays were performed with immunoprecipitates from Bop1Δ/6 and parental LAP3 cells that were untreated (−) or treated with IPTG for 24 h (+). Lane 0, LAP3 cells serum starved for 48 h to determine basal kinase activity; lane c, no-cell-lysate control. (B) Cdk4 and cyclin D1 levels in the cell lysates shown in panel A were determined by immunoblotting of cell lysates with the indicated antibodies.
Cyclin D1 protein is metabolically short-lived, and its abundance is responsive to mitogenic signals (2, 4, 13, 72). The similar levels of cyclin D1 before and after Bop1Δ induction suggest that growth factor-sensing signaling pathways are not affected by Bop1Δ expression. In support of this conclusion, Bop1Δ did not inhibit mitogen-dependent transcriptional activation in early G1 of the immediate-early genes c-fos or c-myc or phosphorylation of ERK1 after serum stimulation of G0 cells (data not shown).
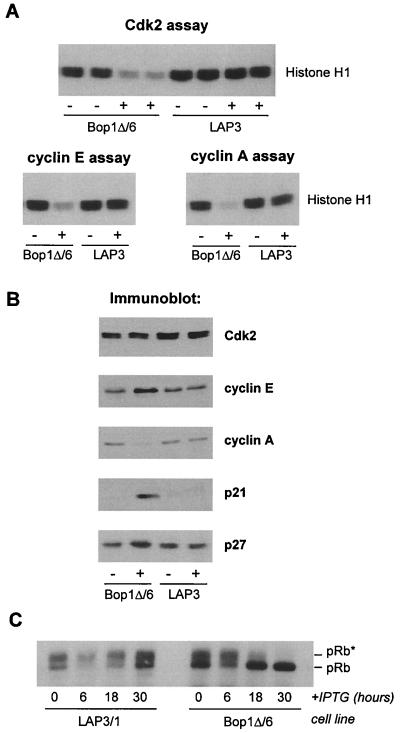
Next, we investigated Cdk2 and its cyclin partners cyclins E and A. Cdk2 complexes were immunoprecipitated from induced and noninduced Bop1Δ/6 cells and analyzed for catalytic activity in vitro using histone H1 as a substrate. While high levels of Cdk2-associated kinase activity were present in control cell lines as well as in untreated Bop1Δ/6 cells, this activity was dramatically decreased in immunoprecipitates from induced Bop1Δ/6 cells (Fig. 3A). When a similar assay was performed with cyclin E immune complexes recovered from IPTG-treated Bop1Δ/6 cells, their kinase activity was also significantly reduced compared to that in uninduced cells and control cell lines (Fig. 3A), consistent with a low activity displayed by the immunoprecipitates of Cdk2. The lack of cyclin E-Cdk2 kinase activity was not due to an absence of either cyclin or Cdk subunits. Immunoblotting of protein lysates showed that the amounts of Cdk2 and cyclin E were not decreased after Bop1Δ induction; in fact, cyclin E levels were elevated (Fig. 3B). This increase in cyclin E protein level was highly reproducible and is likely due to its stabilization in inactive cyclin E-Cdk2 complexes; activation of these complexes has been shown to promote cyclin E degradation by causing its autophosphorylation (9, 73).
FIG. 3.
Effects of Bop1Δ on cyclin E/A-Cdk2, CKIs, and pRb phosphorylation. (A) Histone H1 kinase activity of Cdk2 immunoprecipitates (top; duplicate assays shown) and cyclin E and cyclin A immunoprecipitates (bottom) were determined in the presence of [γ-32P]ATP. Reaction products were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and detected by autoradiography. (B) Lysates from cells that were untreated (−) or treated with IPTG for 24 h (+) were resolved by SDS-PAGE and immunoblotted with antibodies against the indicated proteins. (C) At the indicated times after IPTG addition to growing cultures of Bop1Δ/6 and control LAP3/1 cells, pRb was immunoprecipitated from cell lysates, separated on a 7.5% polyacrylamide gel, and detected by immunoblotting with anti-pRb antibody. pRb∗, hyperphosphorylated pRb.
The cyclin A-associated histone H1 kinase activity was also substantially decreased in IPTG-induced Bop1Δ/6 cells (Fig. 3A). In contrast to cyclin E, only a small amount of cyclin A was detectable in protein lysates (Fig. 3B), likely contributing to the absence of the associated kinase activity. The low level of cyclin A protein is consistent with the inactivity of cyclin E-Cdk2, since transcription of the cyclin A gene is normally induced in late G1 after inactivation of pRb family members by cyclin E-Cdk2 (23, 27, 42). RNA blot analysis confirmed that cyclin A mRNA levels were significantly reduced in Bop1Δ-arrested cells (data not shown).
The activity of cyclin-Cdk complexes can be modulated by the binding of CKIs. We examined the levels of two CKIs, p21 and p27, both of which are implicated in negative regulation of cyclin D-, E-, and A-associated kinases (reviewed in reference 54). Immunoblot analysis of protein extracts showed strong induction of p21 in Bop1Δ/6 cells at 24 h after IPTG treament and a relatively small but detectable increase in the levels of p27; neither protein was induced by IPTG treatment in control cells (Fig. 3B). The increase in p27 concentration is consistent with the observed inactivity of cyclin E/A-Cdk2 complexes. p27 is a binding partner and substrate of cyclin E/A-Cdk2 that is capable of inhibiting its catalytic activity (7, 50), while activated cyclin E-Cdk2 in turn phosphorylates p27 and promotes its degradation (40, 52, 63). Increased amounts of p27 in Bop1Δ-expressing cells therefore confirm that cyclin E-Cdk2 is in the inactive state, which may be stabilized by p27. Both p21 and p27 are known to play a role in inhibiting the activities of different G1-specific cyclin-Cdks in response to various antiproliferative signals. The accumulation of these CKIs may be an important mechanism contributing to inhibition of Cdks and cell cycle arrest following Bop1Δ induction.
The above results indicate that Bop1Δ prevents activation of two key Cdks in G1, Cdk2 and Cdk4. One important target of these kinases is proteins of the Rb family. The inhibition of G1 cyclin-Cdk activities can inhibit pRb hyperphosphorylation, a critical event associated with a transit through the restriction point in mid- to late G1 (5, 53, 68). Consistent with the observed inhibition of Cdk activities, treatment of growing Bop1Δ/6 cells with IPTG led to the disappearance of hyperphosphorylated pRb forms and accumulation of growth-inhibitory, hypophosphorylated pRb (Fig. 3C). Collectively, these data provide a strong indication that Bop1Δ does not arrest cells randomly but in a uniform fashion by inhibiting key regulators of the G1 phase and thus preventing entry into S phase.
Kinetics of Bop1Δ effects on rRNA processing and DNA and protein synthesis.
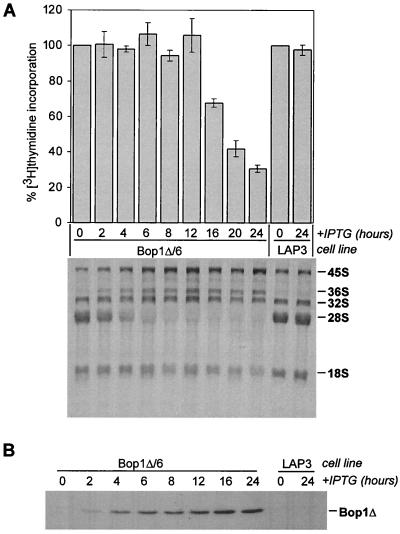
We were interested to determine the temporal relationship between the cell cycle effects of Bop1Δ and its inhibition of rRNA processing. To assess the kinetics of these two events, growing cells of the inducible line Bop1Δ/6 were treated with IPTG for different periods of time and analyzed for processing of rRNA labeled with [3H]uridine and DNA synthesis by measuring [3H]thymidine incorporation. The electrophoretic analysis of labeled rRNA showed that induction of Bop1Δ disrupted normal processing of 28S rRNA almost immediately. When low levels of Bop1Δ protein started to be detectable by immunoblot analysis 2 h after IPTG addition, an increase in the levels of the 36S rRNA precursor appeared (Fig. 4). This increase indicates inhibition of rRNA processing because normally this precursor is short-lived and does not accumulate. Between 6 and 8 h after Bop1Δ induction, formation of the mature 28S rRNA was completely blocked, whereas the 32S and 36S precursors accumulated to high levels. A detailed description of these effects of Bop1Δ on rRNA processing is provided elsewhere (58). Although rRNA production was clearly disrupted, no detectable decrease in DNA synthesis, as measured by incorporation of [3H]thymidine, occurred until 12 h after IPTG addition, at which point the DNA synthesis rate started to drop precipitously (Fig. 4). These data indicate that the primary effect of Bop1Δ is on rRNA biosynthesis, preceding its effects on the cell cycle by >6 h.
FIG. 4.
Time course of Bop1Δ induction and its effects on DNA synthesis and rRNA processing. (A) The Bop1Δ and LAP3 cell lines were treated with IPTG for the indicated times, and their rates of DNA synthesis (top) were determined by measuring incorporation of [3H]thymidine. Histograms show the average counts per minute in quadruplicate samples, expressed as a percentage of that in untreated cultures; error bars indicate standard deviation. For analysis of rRNA processing (bottom), RNA from cells undergoing parallel treatment and labeled with [3H]uridine was separated on an agarose gel, transferred to a nylon membrane, and visualized by fluorography. Positions of mature 28S and 18S rRNAs and major precursors are marked. (B) Bop1Δ detected by immunoblotting with anti-Bop1 antibodies at various times after IPTG induction in Bop1Δ/6 cells. Parental LAP3 cells were used as a control.
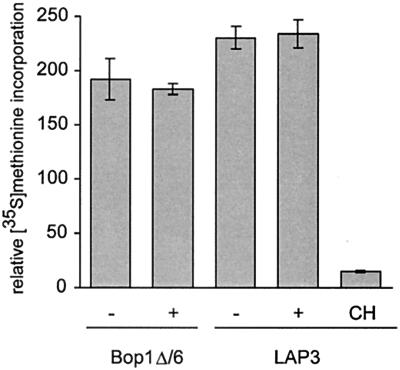
What is the mechanism of Bop1Δ-induced inhibition of the cell cycle? Since this protein inhibits production of 60S ribosome subunits, we first considered the possibility that cells could become arrested due to the lack of ribosomes and repression of translation. To test this idea, we analyzed the rate of protein synthesis by measuring [35S]methionine incorporation in Bop1Δ/6 cells. At 24 h after Bop1Δ induction, when the cells arrested in G1, methionine incorporation into newly synthesized proteins did not change significantly (Fig. 5). Other observations also indicate that cell cycle arrest caused by Bop1Δ occurs prior to depletion of the ribosome pool that might lead to a general inhibition of translation. Analysis of polysome profiles in Bop1Δ/6 cells as late as 32 h after Bop1Δ induction showed no significant effect on polysome integrity, indicating that translation was ongoing and active (58). Inhibition of translation was reported to cause rapid depletion of cyclin D1 (3); however, there is no decline in cyclin D1 levels when cells become arrested after Bop1Δ induction (Fig. 2B). The absence of immediate effects on translation after Bop1Δ induction is not surprising given the large number of existing ribosomes in the cell, which can be efficiently recycled into new translating polysomes. Indeed, prior studies in different experimental systems showed that blocking production of ribosomes may not immediately affect overall protein synthesis rates (10, 28, 65). We conclude that general repression of protein synthesis is not sufficient to explain the cell cycle arrest brought about by Bop1Δ.
FIG. 5.
Rate of global protein synthesis is unaffected by Bop1Δ at the time of cell cycle arrest. [35S]methionine incorporation was measured in triplicate cultures of Bop1Δ/6 or LAP3 cells that were either untreated (−) or treated with IPTG for 24 h (+). Histograms show average incorporation normalized to cell number, and error bars show standard deviation. The background incorporation was determined in cells treated with cycloheximide (CH) (10 μg/ml) for 30 min to block protein synthesis.
Bop1Δ-induced cell cycle arrest is mediated through p53.
We hypothesized that Bop1Δ-induced G1 arrest might represent a form of stress response caused by faulty rRNA synthesis, analogous to responses to DNA damage, heat shock, and other insults. In animal cells, a variety of stress-induced cell cycle arrest responses are mediated through the p53 pathway (31, 34, 57), prompting us to investigate whether p53 might be involved in Bop1Δ-induced cell cycle arrest.
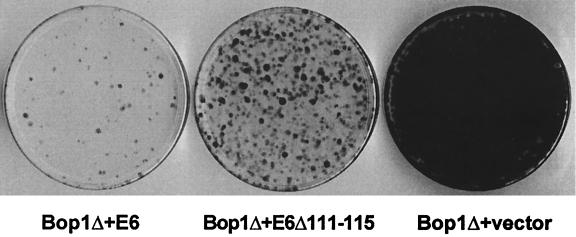
To assess the possible involvement of p53, we introduced the inducible pX11-Bop1Δ construct into LAP3 cells together with a vector expressing human papillomavirus type 16 E6 protein, which targets p53 for degradation (51). As a control, we used mutant E6Δ111–115, which retains the transactivation activities of wild-type E6 but is impaired for p53 binding and degradation (11). When equal numbers of cells from the pools of stable clones were subjected to the BrdU-light assay to test their ability to undergo reversible cell cycle arrest (as described above for Fig. 1), survival of cells expressing E6 was dramatically reduced, suggesting that these cells continued replicating their DNA after Bop1Δ induction (Fig. 6). The E6Δ111–115 mutant also relieved Bop1Δ-induced arrest in this assay, but to a lesser extent than wild-type E6. The simplest interpretation of the difference between p53-binding E6 and nonbinding E6Δ111–115 is that efficient cell cycle arrest in Bop1Δ-expressing cells requires p53 activity, although the partial effect of E6Δ111–115 implies that other molecular pathways in addition to p53 may also be involved.
FIG. 6.
Expression of human papillomavirus type 16 E6 protein alleviates Bop1Δ-induced cell cycle arrest. LAP3 cells were cotransfected with pX11-Bop1Δ, the selection marker pPGK-puro, and either pJ4Ω16E6 or pJ4Ω16E6Δ111–115, which drive expression of wild-type E6 and mutant E6 defective in p53 binding, respectively (11), or vector DNA. Pools of stably transfected clones were obtained by puromycin selection, and equal numbers of cells from each pool were subjected to BrdU and light treatment as described in the legend to Fig. 1.
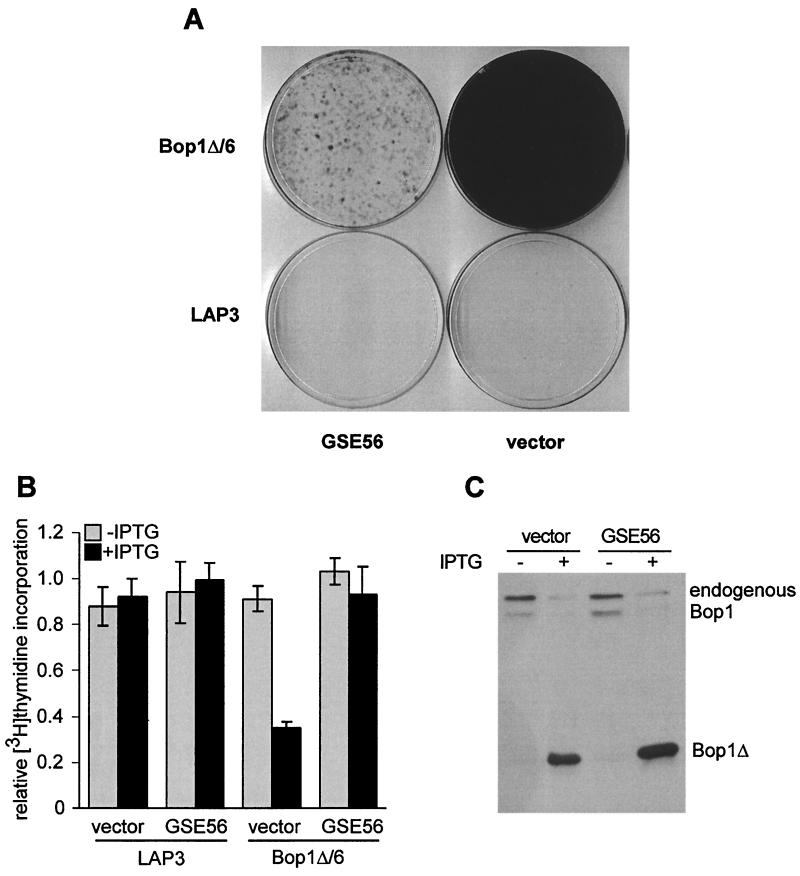
To clarify the role of p53, we sought to inhibit its function by an independent and more specific means. A short fragment of p53, termed GSE56, was identified in a genetic screen as a powerful dominant inhibitor of wild-type p53 function (43). We cloned the GSE56 coding sequence in the retrovirus vector pBabe-puro, infected the Bop1Δ/6 cell line and parental LAP3 cells with either the GSE56-expressing retrovirus or the control empty vector, and selected cells for puromycin resistance. The ability of these infected cells to undergo cell cycle arrest upon Bop1Δ induction was first examined using the BrdU-light assay. Bop1Δ/6 cells infected with the GSE56-expressing virus survived the BrdU-light treatment very poorly, whereas infection with the vector alone did not significantly affect Bop1Δ-induced arrest (Fig. 7A). These results indicate that inactivation of p53 by GSE56 interferes with the cell cycle effects of Bop1Δ. Consistent with these observations, Bop1Δ/6 cells expressing GSE56 displayed no significant reduction in the rate of DNA synthesis after 20 h of IPTG treatment, in contrast to cells infected with the vector (Fig. 7B). Control experiments showed that expression of GSE56 in these cell lines did not decrease accumulation of Bop1Δ after IPTG induction (Fig. 7C). Cells infected with the GSE56 virus continued to proliferate despite expression of Bop1Δ after several days of incubation with IPTG, although at a slower rate than untreated cells (data not shown). Collectively, these results strongly indicate that disruption of p53 function abolishes the stringent cell cycle block imposed by Bop1Δ.
FIG. 7.
Bop1Δ-induced cell cycle arrest is p53 dependent. (A) Bop1Δ/6 cells were infected with pBabe-puro-GSE56, which antagonizes p53 function (43), or pBabe-puro vector. BrdU-light treatment with puromycin-selected pools was performed as described in the legend to Fig. 1 except that BrdU was added at 15 h after IPTG induction. Parental LAP3 cells infected with the same viruses and treated in parallel are shown for comparison. (B) [3H]thymidine incorporation was measured in Bop1Δ/6 and control LAP3 cells carrying retrovirus transduced GSE56 or empty pBabe-puro vector that were either untreated or treated with IPTG for 20 h. Histograms show average label incorporation normalized to cell number in triplicate cultures, and error bars indicate standard deviation. (C) Induction of Bop1Δ is unaffected by GSE56. Whole-cell lysates prepared from Bop1Δ/6 cells used for the above experiments and normalized to protein content were analyzed by immunoblotting with antibodies against Bop1.
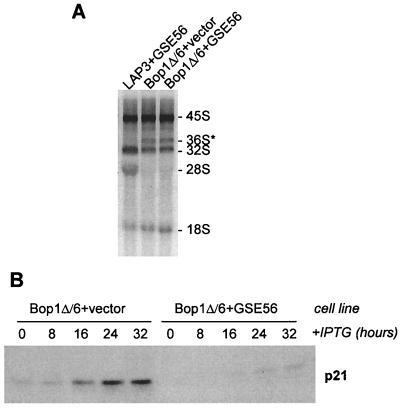
To address whether expression of GSE56 might somehow interfere with the effects of Bop1Δ on rRNA processing, we analyzed the rRNA synthesized in these cells. Expression of GSE56 did not affect the ability of Bop1Δ to block maturation of the 28S rRNA (Fig. 8A), even though it relieved the Bop1Δ-induced cell cycle block (Fig. 7A and B). Thus, the rRNA processing block and the cell cycle arrest caused by Bop1Δ can be uncoupled by inactivation of p53.
FIG. 8.
Functional inactivation of p53 in Bop1Δ-expressing cells does not affect rRNA processing block but decreases p21 induction. (A) Synthesis of 28S rRNA is impaired in Bop1Δ/6 cells expressing GSE56. rRNA processing was analyzed by [3H]uridine labeling as in Fig. 4; note the lack of 28S rRNA labeling and aberrant accumulation of the 36S precursor. (B) Expression of Bop1Δ was induced in puromycin-selected Bop1Δ/6 cell populations infected with pBabe-puro or pBabe-puro-GSE56. Cell lysates were prepared at different times after induction, normalized to protein content, and analyzed by immunoblotting with antibodies against p21.
One of the principal mechanisms by which p53 suppresses cell proliferation is the induction of the CKI p21 (15, 66). Consistent with this notion, we have found that cells arrested by Bop1Δ display elevated levels of p21 (see Fig. 3B). We reasoned that if the cell cycle-inhibitory response in Bop1Δ-expressing cells depended on p53 function, inactivation of p53 should negatively affect p21 induction in these cells. Indeed, infection of Bop1Δ/6 cells with the GSE56 virus led to a striking decrease in p21 accumulation after Bop1Δ induction (Fig. 8B).
Other molecular features of the Bop1Δ-induced cell cycle arrest, such as marked repression of cyclin E-Cdk2 activity, hypophosphorylation of pRB, and low levels of cyclin A with no decrease in cyclin E and D1 levels (Fig. 2 and 3), are similar to those observed previously for p53-mediated cell cycle inhibition (14). We note that although these features are also found when p53 is induced by genetic damage, it is unlikely that such damage occurs in Bop1Δ-expressing cells. First, these cells remain viable and rapidly resume normal proliferation when Bop1Δ is repressed (see above). Second, DNA damage results in highly increased p53 levels in LAP3 cells, but only a small increase in p53 is detectable after Bop1Δ induction (data not shown). Taken together, these findings strongly suggest that expression of Bop1Δ leads to activation of a checkpoint mechanism that blocks G1/S transition in a p53-dependent manner.
DISCUSSION
Bop1 is a newly characterized nucleolar protein essential for the processing of 28S and 5.8S rRNAs and 60S ribosome biosynthesis (58). Expression of a dominant negative mutant of Bop1, Bop1Δ, in asynchronous, logarithmically growing cells blocks synthesis of the 60S ribosome subunits and brings about a strong G1 arrest. In the present study, we have characterized the nature of the cell cycle arrest conferred by Bop1Δ and shown that this arrest is dependent on functional p53. These observations implicate the p53 pathway in a heretofore unknown function as a monitor of ribosome biogenesis and provide important insight into the molecular mechanisms linking ribosome biogenesis and cell proliferation in mammalian cells.
Induction of the Bop1 dominant negative mutant in LAP3 cells elicits changes in cell cycle regulators consistent with a G1 checkpoint response. Progression through G1 requires the activity of several cyclin-Cdk complexes (53, 54). Our results indicate that expression of Bop1Δ causes inhibition of both Cdk4 and Cdk2 activities: (i) their immunoprecipitated complexes display low kinase activity in vitro, and (ii) the lack of hyperphosporylation of pRb suggests that inhibition of the corresponding holoenzyme activities takes place in the cell as well. The Rb family proteins are important targets of cyclin D-Cdk4 and cyclin E/A-Cdk2 (5, 39, 53). Hyperphosphorylation of pRb is associated with, and partially controls, passage through the restriction point in late G1, which marks commitment to DNA synthesis (68). The absence of hyperphosphorylated forms of pRb in Bop1Δ-arrested cells indicates that this critical event does not occur.
The idea that cell cycle progression may depend on some aspect of ribosome biogenesis was first proposed in early studies on the cell cycle (6, 55), although the nature of this connection has remained unknown. Growth-inhibitory stimuli have been shown to repress RNA polymerase I transcription, suggesting that alterations in ribosome production may thus modulate the potential for cellular proliferation (8, 22, 64). We infer that ribosome depletion per se is unlikely the only determinant of cell cycle inhibition connected with ribosome biogenesis. When cells are devoid of functional p53, they can progress through G1 to the S phase in spite of the absence of 60S subunit synthesis caused by expression of Bop1Δ (Fig. 7 and 8). Conversely, cells bearing functional p53 become arrested in response to Bop1Δ when they possess enough ribosomes for virtually unabated protein synthesis (Fig. 5). Thus, defects in ribosome synthesis and inhibition of the cell cycle can be dissociated—when p53 function is impaired, Bop1Δ expression leads to rRNA processing defects but does not induce strong cell cycle arrest (Fig. 7 and 8).
Although we cannot rule out the possibility that Bop1Δ directly activates the p53-mediated cell cycle arrest pathway, we favor the idea that expression of Bop1Δ triggers an arrest response indirectly by causing perturbations in rRNA and/or ribosome biogenesis. This model is supported by the observation that DNA synthesis inhibition is delayed compared to inhibition of 28S rRNA maturation (Fig. 4), suggesting that the primary effect of Bop1Δ is on rRNA biosynthesis and cell cycle arrest is likely a secondary effect. In addition, previous studies with mammalian cells indicate that interference with ribosome biogenesis by other means can also inhibit cell proliferation. For example, the ts422E temperature-sensitive mutant of the Syrian hamster cell line BHK21 that is unable to produce mature 28S rRNA and 60S ribosome subunits was rapidly growth arrested at the nonpermissive temperature (38, 61); the nature of this growth defect, however, has not been clearly defined (20, 41). Antisense-mediated inhibition of the nucleolar protein p120, a human homolog of the yeast Nop2p that is implicated in biosynthesis of the large ribosome subunit, arrested human lymphocytes in G1 (17). Recently, conditional deletion of the ribosomal S6 gene was shown to inhibit cell proliferation in the livers of mice (65). Deletion of S6 using the Cre/LoxP system in this study abrogated production of 40S subunits in liver cells and inhibited their entry into S phase, leading to the suggestion that control mechanisms may have evolved in the cells to recognize lesions in ribosome biogenesis. The similarity of antiproliferative effects observed in our study and these distinctly different experimental systems strongly argues that various defects in ribosome biogenesis may trigger a cell cycle-inhibitory response.
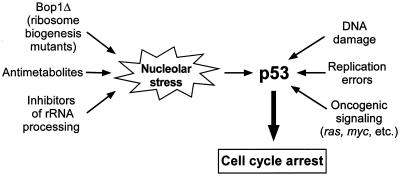
We propose a hypothesis that the signal transmitted to the cell cycle machinery is generated by a mechanism that monitors some aspects of ribosome production in the nucleolus. The most immediate effect of Bop1Δ expression is disruption of several steps in rRNA processing, which prevents formation of mature 60S ribosome subunits (58). In recent years, it has become apparent that many complex processes in the cell are monitored by checkpoint systems that generate interruptions of the cell cycle when various types of stress are detected (16, 25, 69). It is conceivable that cells possess a similar checkpoint mechanism to respond to nucleolar stress—perturbations in the nucleolar biosynthetic machinery that produces ribosomes (Fig. 9). This idea appears especially appealing because ribosome biogenesis is a complex process that is highly sensitive to various disturbances in cellular metabolism, including various chemical inhibitors (1, 10, 30, 56, 70, 71), reduced protein synthesis (21, 44), and starvation (35, 62). Hence, detection of anomalies in ribosome biogenesis could potentially provide integration of a variety of inputs indicating unfavorable or toxic environmental conditions. In this line of reasoning, the inhibitory effect of nucleolar stress on the cell cycle may represent a built-in protective mechanism to prevent DNA replication under suboptimal metabolic conditions.
FIG. 9.
Nucleolar stress model of cell cycle arrest due to perturbation in ribosome biogenesis. Expression of mutant proteins such as Bop1Δ, exposure to chemical inhibitors of synthesis, and maturation of rRNA and other ribosome components in mammalian cells induce nucleolar stress, causing cell cycle arrest by triggering activation of the p53 pathway. p53 is required for sensing cellular stress caused by a variety of conditions, including DNA damage, replication defects, and gratuitous oncogene expression.
The nucleolar stress model is consistent with our finding that p53 plays a role in mediating Bop1Δ-induced cell cycle arrest. The tumor suppressor p53 participates in responses to numerous extra- and intracellular stresses (31, 34, 57). Intriguingly, several components of the p53 pathway are localized, at least transiently, to the nucleolus, including p53 itself (49), Mdm2 (60, 67), and p19Arf (32, 74). Mdm2 and p53 were also detected in complexes containing ribosomal protein L5 and 5S and 5.8S rRNAs (18, 36). The significance of the connection of p53 with the nucleolus is unclear but has been suggested to involve sequestration from the nucleus, nuclear export, and colocalization with sites of RNA synthesis. Herein we show, for the first time, a functional link between p53 activity and a protein directly involved in rRNA processing and ribosome biogenesis. The association of components of the p53 pathway with the nucleolus may thus reflect a previously unrecognized role of this pathway in monitoring nucleolar function.
One interesting feature of the nucleolar stress model is that it provides a possible mechanism for the antiproliferative effects of a diverse group of metabolic inhibitors, including many clinically important anticancer drugs, that strongly inhibit the rRNA synthesis and processing machinery (1, 10, 56, 70, 71). At present, the contribution of impaired RNA synthesis and processing to their action is poorly understood. Nevertheless, there is extensive evidence that the antiproliferative properties of 5-fluorouridine and 5-fluorouracil, commonly used in cancer chemotherapy, depend strongly on their effects on RNA metabolism, which can be experimentally separated from other metabolic effects (19, 26, 47, 59). A large group of ribonucleotide biosynthesis inhibitors were also shown to induce p53-dependent G1 arrest, leading to the suggestion that this effect might be mediated by inhibition of synthesis of some specific RNA molecules (33). Thus, it would be of particular interest to determine whether molecular mechanisms linked to rRNA processing and ribosome production might mediate cell cycle effects of different chemotherapeutic agents that target RNA.
ACKNOWLEDGMENTS
We thank Marina Polonskaia for help with FACS analysis and Andrei Gudkov, Pradip Raychaudhuri, and Karen Vousden for plasmids.
This work was supported by a grant from the National Institutes of Health (CA52220).
REFERENCES
- 1.Abe T, Fukamachi Y, Kanazawa Y, Furukawa H, Shimizu K, Hirano T, Kasai H, Kashimura M, Higashi K. Inhibition of nucleolar function and morphological change by adriamycin associated with heat shock protein 70 accumulation. Jpn J Cancer Res. 1996;87:945–951. doi: 10.1111/j.1349-7006.1996.tb02124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aktas H, Cai H, Cooper G M. Ras links growth factor signaling to the cell cycle machinery via regulation of cyclin D1 and the Cdk inhibitor p27KIP1. Mol Cell Biol. 1997;17:3850–3857. doi: 10.1128/mcb.17.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aktas H, Fluckiger R, Acosta J A, Savage J M, Palakurthi S S, Halperin J A. Depletion of intracellular Ca2+ stores, phosphorylation of elF2alpha, and sustained inhibition of translation initiation mediate the anticancer effects of clotrimazole. Proc Natl Acad Sci USA. 1998;95:8280–8285. doi: 10.1073/pnas.95.14.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldin V, Lukas J, Marcote M J, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 5.Bartek J, Bartkova J, Lukas J. The retinoblastoma protein pathway in cell cycle control and cancer. Exp Cell Res. 1997;237:1–6. doi: 10.1006/excr.1997.3776. [DOI] [PubMed] [Google Scholar]
- 6.Baserga R, Estensen R D, Petersen R O. Inhibition of DNA synthesis in Ehrlich ascites cells by actinomycin D. II. The presynthetic block in the cell cycle. Proc Natl Acad Sci USA. 1965;54:1141–1148. doi: 10.1073/pnas.54.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blain S W, Montalvo E, Massague J. Differential interaction of the cyclin-dependent kinase (Cdk) inhibitor p27Kip1 with cyclin A-Cdk2 and cyclin D2-Cdk4. J Biol Chem. 1997;272:25863–25872. doi: 10.1074/jbc.272.41.25863. [DOI] [PubMed] [Google Scholar]
- 8.Cavanaugh A H, Hempel W M, Taylor L J, Rogalsky V, Todorov G, Rothblum L I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 9.Clurman B E, Sheaff R J, Thress K, Groudine M, Roberts J M. Turnover of cyclin E by the ubiquitin-proteasome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes Dev. 1996;10:1979–1990. doi: 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]
- 10.Cohen M B, Glazer R I. Cytotoxicity and the inhibition of ribosomal RNA processing in human colon carcinoma cells. Mol Pharmacol. 1985;27:308–313. [PubMed] [Google Scholar]
- 11.Crook T, Tidy J A, Vousden K H. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell. 1991;67:547–556. doi: 10.1016/0092-8674(91)90529-8. [DOI] [PubMed] [Google Scholar]
- 12.DeGregori J, Leone G, Ohtani K, Miron A, Nevins J R. E2F-1 accumulation bypasses a G1 arrest resulting from the inhibition of G1 cyclin-dependent kinase activity. Genes Dev. 1995;9:2873–2887. doi: 10.1101/gad.9.23.2873. [DOI] [PubMed] [Google Scholar]
- 13.Diehl J A, Cheng M, Roussel M F, Sherr C J. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dulic V, Kaufmann W K, Wilson S J, Tlsty T D, Lees E, Harper J W, Elledge S J, Reed S I. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 15.el-Deiry W S, Harper J W, O'Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 16.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 17.Fonagy A, Swiderski C, Dunn M, Freeman J W. Antisense-mediated specific inhibition of P120 protein expression prevents G1- to S-phase transition. Cancer Res. 1992;52:5250–5256. [PubMed] [Google Scholar]
- 18.Funtoura B M, Sorokina E A, David E, Carroll R B. p53 is covalently linked to 5.8S rRNA. Mol Cell Biol. 1992;12:5145–5151. doi: 10.1128/mcb.12.11.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghoshal K, Jacob S T. An alternative molecular mechanism of action of 5-fluorouracil, a potent anticancer drug. Biochem Pharmacol. 1997;53:1569–1575. doi: 10.1016/s0006-2952(97)00040-3. [DOI] [PubMed] [Google Scholar]
- 20.Grummt F, Grummt I, Mayer E. Ribosome biosynthesis is not necessary for initiation of DNA replication. Eur J Biochem. 1979;97:37–42. doi: 10.1111/j.1432-1033.1979.tb13083.x. [DOI] [PubMed] [Google Scholar]
- 21.Hadjiolov A A. The nucleolus and ribosome biogenesis. New York, N.Y: Springer-Verlag; 1985. [Google Scholar]
- 22.Hannan K M, Kennedy B K, Cavanaugh A H, Hannan R D, Hirschler-Laszkiewicz I, Jefferson L S, Rothblum L I. RNA polymerase I transcription in confluent cells: Rb downregulates rDNA transcription during confluence-induced cell cycle arrest. Oncogene. 2000;19:3487–3497. doi: 10.1038/sj.onc.1203690. [DOI] [PubMed] [Google Scholar]
- 23.Harbour J W, Luo R X, Dei S A, Postigo A A, Dean D C. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 24.Hartwell L H, Kastan M B. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 25.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 26.Heimer R, Sartorelli A C. RNA-directed actions of 5-fluorouridine in hemin stimulated K-562 erythroleukemia cells. Cancer Biochem Biophys. 1992;12:221–239. [PubMed] [Google Scholar]
- 27.Huet X, Rech J, Plet A, Vie A, Blanchard J M. Cyclin A expression is under negative transcriptional control during the cell cycle. Mol Cell Biol. 1996;16:3789–3798. doi: 10.1128/mcb.16.7.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston G C, Singer R A. RNA synthesis and control of cell division in the yeast S. cerevisiae. Cell. 1978;14:951–958. doi: 10.1016/0092-8674(78)90349-5. [DOI] [PubMed] [Google Scholar]
- 29.Kitagawa M, Higashi H, Jung H K, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J, Segawa K, Yoshida E, Nishimura S, Taya Y. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar A, Wu R S. Role of ribosomal RNA transcription in ribosome processing in HeLa cells. J Mol Biol. 1973;80:265–276. doi: 10.1016/0022-2836(73)90172-1. [DOI] [PubMed] [Google Scholar]
- 31.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 32.Lindstrom M S, Klangby U, Inoue R, Pisa P, Wiman K G, Asker C E. Immunolocalization of human p14(ARF) to the granular component of the interphase nucleolus. Exp Cell Res. 2000;256:400–410. doi: 10.1006/excr.2000.4854. [DOI] [PubMed] [Google Scholar]
- 33.Linke S P, Clarkin K C, Di Leonardo A, Tsou A, Wahl G M. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 34.Ljungman M. Dial 9-1-1 for p53: mechanisms of p53 activation by cellular stress. Neoplasia. 2000;2:208–225. doi: 10.1038/sj.neo.7900073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maden B E, Vaughan M H, Warner J R, Darnell J E. Effects of valine deprivation on ribosome formation in HeLa cells. J Mol Biol. 1969;45:265–275. doi: 10.1016/0022-2836(69)90104-1. [DOI] [PubMed] [Google Scholar]
- 36.Marechal V, Elenbaas B, Piette J, Nicolas J C, Levine A J. The ribosomal L5 protein is associated with Mdm-2 and Mdm-2-p53 complexes. Mol Cell Biol. 1994;14:7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J Y. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meiss H K, Basilico C. Temperature sensitive mutants of BHK 21 cells. Nat New Biol. 1972;239:66–68. doi: 10.1038/newbio239066a0. [DOI] [PubMed] [Google Scholar]
- 39.Mittnacht S. Control of pRB phosphorylation. Curr Opin Genet Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 40.Montagnoli A, Fiore F, Eytan E, Carrano A C, Draetta G F, Hershko A, Pagano M. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mora M, Darzynkiewicz Z, Baserga R. DNA synthesis and cell division in a mammalian cell mutant temperature sensitive for the processing of ribosomal RNA. Exp Cell Res. 1980;125:241–249. doi: 10.1016/0014-4827(80)90208-6. [DOI] [PubMed] [Google Scholar]
- 42.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S-phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ossovskaya V S, Mazo I A, Chernov M V, Chernova O B, Strezoska Z, Kondratov R, Stark G R, Chumakov P M, Gudkov A V. Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc Natl Acad Sci USA. 1996;93:10309–10314. doi: 10.1073/pnas.93.19.10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pederson T, Kumar A. Relationship between protein synthesis and ribosome assembly in HeLa cells. J Mol Biol. 1971;61:655–668. doi: 10.1016/0022-2836(71)90070-2. [DOI] [PubMed] [Google Scholar]
- 45.Pestov D G, Grzeszkiewicz T M, Lau L F. Isolation of growth suppressors from a cDNA expression library. Oncogene. 1998;17:3187–3197. doi: 10.1038/sj.onc.1202260. [DOI] [PubMed] [Google Scholar]
- 46.Pestov D G, Lau L F. Genetic selection of growth-inhibitory sequences in mammalian cells. Proc Natl Acad Sci USA. 1994;91:12549–12553. doi: 10.1073/pnas.91.26.12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pritchard D M, Watson A J, Potten C S, Jackman A L, Hickman J A. Inhibition by uridine but not thymidine of p53-dependent intestinal apoptosis initiated by 5-fluorouracil: evidence for the involvement of RNA perturbation. Proc Natl Acad Sci USA. 1997;94:1795–1799. doi: 10.1073/pnas.94.5.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rago R, Mitchen J, Wilding G. DNA fluorometric assay in 96-well tissue culture plates using Hoechst 33258 after cell lysis by freezing in distilled water. Anal Biochem. 1990;191:31–34. doi: 10.1016/0003-2697(90)90382-j. [DOI] [PubMed] [Google Scholar]
- 49.Rubbi C P, Milner J. Non-activated p53 co-localizes with sites of transcription within both the nucleoplasm and the nucleolus. Oncogene. 2000;19:85–96. doi: 10.1038/sj.onc.1203378. [DOI] [PubMed] [Google Scholar]
- 50.Russo A A, Jeffrey P D, Patten A K, Massague J, Pavletich N P. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 51.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 52.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 53.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 54.Sherr C J, Roberts J M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 55.Singer R A, Johnston G C. Transcription of rRNA genes and cell cycle regulation in the yeast Saccharomyces cerevisiae. In: Padilla G M, McCarty K S, editors. Genetic expression and the cell cycle. New York, N.Y: Academic Press; 1982. pp. 181–198. [Google Scholar]
- 56.Snyder A L, Kann H E J, Kohn K W. Inhibition of the processing of ribosomal precursor RNA by intercalating agents. J Mol Biol. 1971;58:555–565. doi: 10.1016/0022-2836(71)90371-8. [DOI] [PubMed] [Google Scholar]
- 57.Somasundaram K, el-Deiry W S. Tumor suppressor p53: regulation and function. Front Biosci. 2000;5:D424–D437. doi: 10.2741/somasund. [DOI] [PubMed] [Google Scholar]
- 58.Strezoska Z, Pestov D G, Lau L F. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol Cell Biol. 2000;20:5516–5528. doi: 10.1128/mcb.20.15.5516-5528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takimoto C H, Tan Y Y, Cadman E C, Armstrong R D. Correlation between ribosomal RNA production and RNA-directed fluoropyrimidine cytotoxicity. Biochem Pharmacol. 1987;36:3243–3248. doi: 10.1016/0006-2952(87)90640-x. [DOI] [PubMed] [Google Scholar]
- 60.Tao W, Levine A J. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toniolo D, Basilico C. Processing of ribosomal RNA in a temperature sensitive mutant of BHK cells. Biochim Biophys Acta. 1976;425:409–418. doi: 10.1016/0005-2787(76)90005-8. [DOI] [PubMed] [Google Scholar]
- 62.Vaughan M H J, Soeiro R, Warner J R, Darnell J E. The effects of methionine deprivation on ribosome synthesis in HeLa cells. Proc Natl Acad Sci USA. 1967;58:1527–1534. doi: 10.1073/pnas.58.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vlach J, Hennecke S, Amati B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voit R, Hoffmann M, Grummt I. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 1999;18:1891–1899. doi: 10.1093/emboj/18.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volarevic S, Stewart M J, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma S C, Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 66.Waldman T, Kinzler K W, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 67.Weber J D, Taylor L J, Roussel M F, Sherr C J, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 68.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 69.Weinert T. DNA damage and checkpoint pathways: molecular anatomy and interactions with repair. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- 70.Weiss J W, Pitot H C. Inhibition of ribosomal RNA maturation in Novikoff hepatoma cells by toyocamycin, tubercidin, and 6-thioguanosine. Cancer Res. 1974;34:581–587. [PubMed] [Google Scholar]
- 71.Wilkinson D S, Tlsty T D, Hanas R J. The inhibition of ribosomal RNA synthesis and maturation in Novikoff hepatoma cells by 5-fluorouridine. Cancer Res. 1975;35:3014–3020. [PubMed] [Google Scholar]
- 72.Winston J T, Pledger W J. Growth factor regulation of cyclin D1 mRNA expression through protein synthesis-dependent and -independent mechanisms. Mol Biol Cell. 1993;4:1133–1144. doi: 10.1091/mbc.4.11.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Won K A, Reed S I. Activation of cyclin E/CDK2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. EMBO J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, Xiong Y. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]