Abstract
RAG-1 and RAG-2 initiate V(D)J recombination by binding to specific recognition sequences (RSS) and then cleave the DNA in two steps: nicking and hairpin formation. Recent work has established that a dimer of RAG-1 and either one or two monomers of RAG-2 bind to a single RSS, but the enzymatic contributions of the RAG molecules within this nucleoprotein complex and its functional organization have not been elucidated. Using heterodimeric protein preparations containing both wild-type and catalytically deficient RAG-1 molecules, we found that one active monomer is sufficient for both nicking and hairpin formation at a single RSS, demonstrating that a single active site can carry out both cleavage steps. Furthermore, the mutant heterodimers efficiently cleaved both RSS in a synaptic complex. These results strongly suggest that two RAG-1 dimers are responsible for RSS cleavage in a synaptic complex, with one monomer of each dimer catalyzing both nicking and hairpin formation at each RSS.
V(D)J recombination assembles separate antigen receptor gene segments into an exon encoding the antigen binding domain of immunoglobulin and T-cell receptor proteins. The V(D)J recombinase consists of two proteins, RAG-1 and RAG-2, which bind to recognition sequences (RSS) located adjacent to the V, D, and J coding segments. Recombination is initiated by the RAG proteins, which introduce a nick precisely between the RSS and the coding segment, creating a free 3′ OH which is then used as a nucleophile to attack the opposite strand, resulting in a blunt, 5′-phosphorylated signal end and a hairpin coding end (17, 22).
Although nicking can occur at a single RSS, efficient hairpin formation normally requires a pair of different RSS, one with a 12- and one with a 23-nucleotide spacer (known, respectively, as the 12-RSS and the 23-RSS), with the two being assembled into a synaptic complex (9, 30). This restriction, known as the 12/23 rule, prevents immunologically irrelevant recombination events from scrambling the immune receptor loci. The 12/23 rule can, however, be bypassed under certain conditions, such as when Mn2+ is substituted for the physiological divalent cation, Mg2+ (30).
In order to understand the mechanism of cleavage and the molecular basis of the 12/23 rule, it is necessary to delineate the organization of the nucleoprotein complexes that carry out cleavage at a single RSS and at a 12/23 RSS pair. Recent work has shown that a single molecule of DNA containing an RSS is bound by a RAG-1 dimer, with one or two monomers of RAG-2, and that this nucleoprotein complex is competent for cleavage (3, 21, 29). This single RSS complex is quite stable; it is resistant to incubation with a 250-fold excess of specific competitor at 37°C for at least 1 h (8). Furthermore, purified RAG-1 dimers are stable in solution, even in the absence of DNA, remaining associated after overnight incubation at 4°C (21). Additional evidence for the stability of RAG-1 dimers is provided by the observation that heterodimer formation requires coexpression of two different RAG-1 molecules: mixing of individually purified RAG-1 homodimers did not result in detectable heterodimer formation (29).
Although the stoichiometry of RAG-1 molecules bound to a single RSS has been established, our understanding of the anatomy of a functional RAG-RSS complex remains incomplete. A question of particular mechanistic importance is whether the two cleavage steps, nicking and hairpin formation, utilize one or both active sites present in the RAG-1 dimer bound at a single RSS. Precedents exist for both possibilities. The Tn7 transposase utilizes two distinct active sites, in two different transposase proteins, to catalyze cleavage of the top and bottom strands (24). In contrast, the transposases of bacteriophage Mu, Tn10, and Tn5 each use a single active site to carry out cleavage and strand transfer (4, 11, 18, 19, 32).
To address these questions, we have taken advantage of catalytically deficient RAG-1 mutants. We recently identified three catalytic amino acids (D600, D708, and E962) in RAG-1 that are thought to be involved in coordinating a divalent metal ion that is essential for cleavage (14). There is evidence that two of these amino acids, D708 and D600, directly contact the metal ion (7, 12, 14). Importantly, mutant proteins bearing a single-amino-acid substitution at any of these three positions exhibit essentially wild-type RSS binding and 12/23 synaptic complex formation but are completely unable to carry out either nicking or hairpin formation (7, 12, 14). Because metal ions are required for DNA binding (2, 23), it is likely that these catalytically deficient mutants can bind metal ions but fail to position them properly for catalysis. Moreover, pairwise combinations of these mutant proteins do not restore enzymatic activity when assayed either in vivo or in vitro, demonstrating that these three catalytic amino acids are not contributed by different monomers of RAG-1 (unpublished observations). We have used purified mutant heterodimers of RAG-1 that contain one wild-type monomer and one catalytically deficient monomer to determine that a single RAG-1 monomer performs both nicking and hairpin formation at a single RSS. Furthermore, analysis of cleavage of paired 12/23 complexes suggests that the synaptic complex contains two RAG-1 dimers.
MATERIALS AND METHODS
Protein purification.
Baculovirus transfer vectors encoding the mutant RAG proteins or their wild-type counterparts were made by subcloning into the pFastBac transfer vector (GIBCO/BRL). Sf9 cells were coinfected with one recombinant baculovirus encoding a RAG-1 molecule with an N-terminal maltose binding protein (MBP) fusion and three copies of a C-terminal Myc tag (MBP–RAG-1[384-1008]–Myc3, ∼130 kDa, referred to as mR1) and a second recombinant baculovirus encoding a RAG-1 molecule lacking the N-terminal MBP fusion but containing a C-terminal polyhistidine and three copies of the Myc tag (RAG-1[384-1008]–His9–Myc3, ∼85 kDa, referred to as R1h) (2, 13, 17). Typically, 500-ml volumes of Sf9 cells at a density of 2 × 106 cells/ml were infected with high-titer virus stocks. Cells were harvested ∼60 h postinfection and lysed in 30 ml of lysis buffer (20 mM Tris-Cl, pH 7.9, at 4°C; 0.5 M NaCl; 20% glycerol; 2 mM β-mercaptoethanol) plus 10 mM imidazole by dounce homogenization (20 strokes, tight pestle). The resulting lysate was centrifuged at 100,000 × g for 30 min at 4°C. The supernatant was loaded onto a 0.5-ml metal chelating Sepharose column (Pharmacia) charged with NiSO4. The column was washed with 10 ml of lysis buffer containing 60 mM imidazole and eluted with 5 ml of lysis buffer containing 250 mM imidazole. The eluate was diluted with 8 ml of amylose A (25 mM sodium phosphate, pH 7.2; 0.5 M NaCl; 10% glycerol; 1 mM dithiothreitol [DTT]) and loaded onto a 0.5-ml amylose resin column. The column was washed with 10 ml of amylose A, and the protein was eluted with amylose A plus 10 mM maltose. Fractions containing the RAG proteins were dialyzed against 1,000 volumes of storage buffer (25 mM K-HEPES, pH 7.5; 150 mM potassium glutamate; 20% glycerol; 2 mM DTT) for 3 h at 4°C. The protein was aliquotted, flash-frozen in liquid nitrogen, and stored at −80°C. Glutathione S-transferase (GST)-tagged RAG-2(1-383) was purified as described previously (25, 26).
Electrophoretic mobility shift assays.
Purified RAG-1 and RAG-2 proteins (100 ng of each, as measured by Coomassie blue-stained gels) were incubated with 25 fmol of the annealed oligonucleotide substrate DAR39/40 (17) in 10 μl of reaction buffer (37.8 mM HEPES-KOH, pH 7.5; 51 mM potassium glutamate; 10% glycerol; 3 mM DTT; 2.5 pmol of the nonspecific competitor oligonucleotide FM117 [17]; 1 mM MgCl2; 60 μg of bovine serum albumin [BSA] per ml; 0.006% NP-40; 20% dimethyl sulfoxide). The incubations were at 30°C for 30 min, and they were cross-linked by the addition of glutaraldehyde (to 0.1%), with an additional incubation for 10 min at 37°C, as described previously (2, 8). DNA binding was analyzed by nondenaturing electrophoresis through a 4% polyacrylamide gel run in 1× Tris-borate-EDTA (TBE) (80 mA · h at 4°C). Dried gels were visualized by autoradiography and/or with a phosphorimager.
Oligonucleotide cleavage assays.
Oligonucleotide cleavage assays were performed as described previously (13). RAG-1 and RAG-2 (100 ng of each; approximately 400 fmol of RAG-1 dimer) were incubated with 250 fmol of annealed DAR39/40 (17) (DAR39 was 5′ 32P end labeled) in a 10-μl reaction mixture (40 mM HEPES-KOH, pH 7.5; 60 mM potassium glutamate; 5; 60 mM potassium glutamate; 10% glycerol; 3 mM DTT; 1 mM MnCl2; 60 μg of BSA per ml; 0.006% NP-40) at 37°C for 45 min unless otherwise stated. The reactions were stopped by adding an equal volume of a solution containing 94% formamide, 20 mM EDTA, and 0.05% bromophenol blue. The reaction products were separated by electrophoresis through a 10% acrylamide gel containing 30% formamide, 0.67× TBE, 7 M urea, and 12.5 mM HEPES-KOH, pH 7.5, for 2 h at 75 W. Wet gels were visualized by autoradiography or with a phosphorimager.
Plasmid cleavage assays.
RAG-1 and RAG-2 were incubated with 20 ng of pJH290 for 3 h at 30°C (50 mM HEPES-KOH, pH 8.0; 11 mM KCl; 4 mM NaCl; 15 mM KGlu; 5 mM MgCl2; 4% glycerol; 1 mM DTT; 100 ng of BSA per μl; 20 ng of HMG-1 per μl). The cleavage reactions were terminated by the addition of 100 μl of stop buffer (100 mM Tris-Cl, pH 8.0; 0.2% sodium dodecyl sulfate; 0.35 mg of proteinase K per ml; 10 mM EDTA) and incubated at 55°C for 1 h. The deproteinized cleavage products were phenol-chloroform extracted, ethyl alcohol precipitated, resuspended in 20 μl of Tris-EDTA, and digested with PvuII (0.1 U) for 30 min at 37°C. Half of the digested reaction products were then separated by electrophoresis through a 4.5% acrylamide gel, transferred to a solid support, and hybridized with a randomly primed 693-bp PvuII fragment from pJH290 that is complementary to all cleavage products.
RESULTS AND DISCUSSION
Expression and purification of heterodimeric RAG-1 protein.
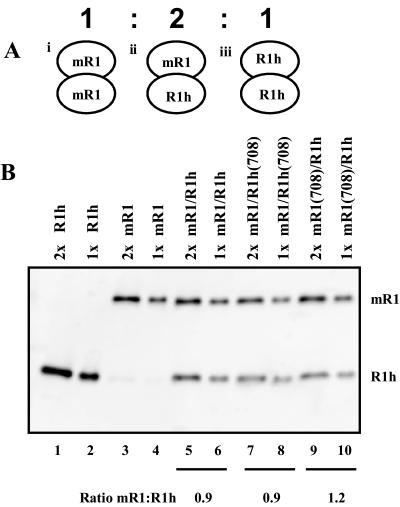
To purify heterodimers containing one wild-type monomer and one catalytically deficient monomer (D708N, D600N, or E962Q), we coinfected insect cells with recombinant baculoviruses encoding two different versions of RAG-1, one with an N-terminal MBP fusion (referred to as mR1) and one with a C-terminal polyhistidine tag (referred to as R1h). Three different dimers can be formed within a cell infected by both viruses, as diagrammed in Fig. 1A: an mR1-mR1 homodimer, an mR1-R1h heterodimer, and an R1h-R1h homodimer. Assuming random association, these three species should be formed in a 1:2:1 ratio. Heterodimers (mR1-R1h) were purified by sequential affinity chromatography using an Ni2+ chelating resin (which binds the polyhistidine tag) followed by an amylose A resin (which binds the MBP tag). This sequential chromatography scheme purifies heterodimers away from contaminating homodimers (purified heterodimer preparations contain less than 5% homodimer [see below]).
FIG. 1.
Heterodimeric RAG-1 protein preparations containing equal amounts of the two RAG-1 monomers. (A) Three possible combinations (i, ii, and iii, respectively) of RAG-1 dimers resulting from random assortment during coinfection. Form ii is the only dimer retained after sequential affinity chromatography. (B) Representative Western blot analysis of heterodimeric RAG-1 protein preparations using the anti-c-Myc antibody (clone 9E10). The quantitation of band intensity was carried out by fluorimager analysis. The average molar ratios (fluorimager units for mR1 divided by fluorimager units for R1h for both amounts loaded [1x and 2x]) were calculated from three independent protein preparations of each heterodimer.
We prepared two types of mutant heterodimers, MBP-tagged wild-type–histidine-tagged mutant [mR1-R1h(708)] and MBP-tagged mutant–histidine-tagged wild-type [mR1(708)-R1h]. As expected, both preparations yielded similar results. To determine the molar ratio of mR1 to R1h in each preparation, we performed Western blot analysis using an anti-c-Myc antibody which recognizes a C-terminal Myc epitope tag present on both proteins (Fig. 1B). Each determination was made at two different protein concentrations to ensure that the values fell within the linear range of the assay. The resulting data show that the different heterodimer preparations contained equal amounts of RAG-1 protein. The control wild-type heterodimer (mR1-R1h) showed an average mR1/R1h molar ratio of 0.9, as determined by fluorimager analysis of three different protein preparations (representative data are shown in Fig. 1B, lanes 5 and 6). The two mutant heterodimers [mR1-R1h(708) (Fig. 1B, lanes 7 and 8) and mR1(708)-R1h (Fig. 1B, lanes 9 and 10)] yielded average molar ratios of 0.9 and 1.2, respectively, demonstrating that the purified heterodimers contained equal proportions of wild-type and mutant monomers.
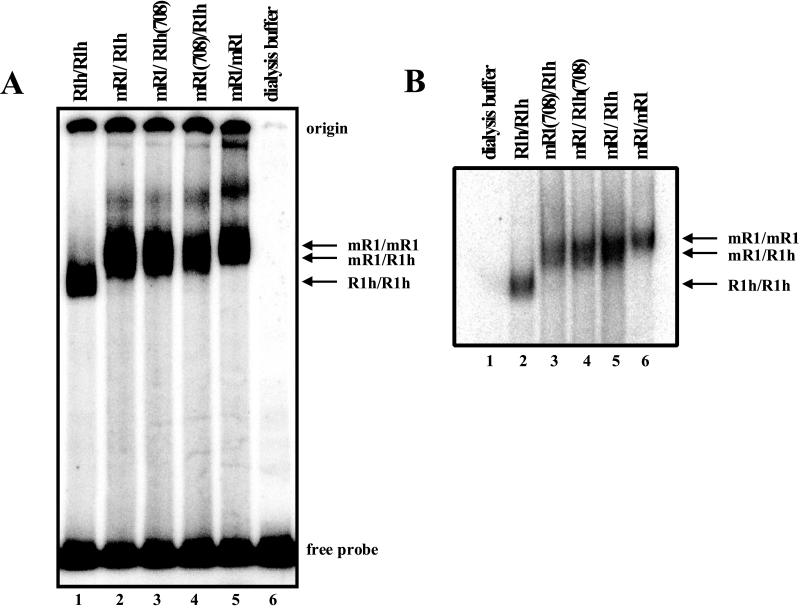
The 1:1 molar ratio of the recovered proteins suggests that the purified heterodimer preparations were not significantly contaminated by homodimers. To verify this and to directly detect the heterodimers, we assayed the ability of purified heterodimers to bind to an RSS-containing oligonucleotide using a standard electrophoretic mobility shift assay (8). Mobility shift assays were performed in the presence of independently purified GST-tagged RAG-2, as both RAG proteins are required for stable RSS binding (2, 8, 29). R1h-R1h homodimers produce a shifted species with substantially faster mobility than the mR1-mR1homodimers (Fig. 2A, compare lanes 1 and 5; Fig. 2B, lanes 2 and 6). As expected, the purified wild-type and mutant heterodimers, mR1-R1h, mR1-R1h(708), and mR1(708)-R1h, migrated to an intermediate position (Fig. 2A, lanes 2 to 4), providing direct evidence that the purified species are indeed heterodimers. (The heterodimers were more clearly separated from the mR1-mR1homodimers after extended periods of electrophoresis, as shown in Fig. 2B [compare lanes 3 through 5 with lane 6].) Note that both mutant heterodimers bound to the RSS at essentially wild-type levels, verifying that the D708N mutation does not significantly affect DNA binding, as shown previously for D708N homodimers (14).
FIG. 2.
Electrophoretic mobility shift analysis of homodimers and heterodimers. Electrophoretic mobility shift analysis of a 12-RSS-containing oligonucleotide substrate was performed with either homodimeric or heterodimeric RAG-1 and GST-tagged RAG-2. All reaction mixtures contain GST-tagged RAG-2, but only RAG-1 is listed above the lanes for simplicity. Dialysis buffer used for RAG-1 purification was used for no RAG-1 control. The heterodimers shift the probe to an intermediate size relative to the large and small homodimers. In panel B, samples were run for a longer time.
This assay can also be used to detect the presence of contaminating R1h homodimers in the purified heterodimer preparations, taking advantage of the substantial difference in the mobilities of the R1h-R1h and the mR1-R1h species. Unfortunately, the difference between the mobilities of the mR1-R1h and mR1-mR1 complexes is not as great, which makes it difficult to detect small amounts of contaminating mR1-mR1 homodimers, if present. Nevertheless, all experiments were done with both mR1(708)-R1h and mR1-R1h(708) heterodimer preparations, which yielded the same results (see below); contamination of the mR1(708)-R1h heterodimer by small amounts of catalytically inactive mR1(708)-mR1(708) mutant homodimers should not affect the results. No R1h-R1h homodimers were detected in the mR1-R1h, mR1-R1h(708), or mR1(708)-R1h heterodimer preparations (Fig. 2), even with long autoradiographic exposures (data not shown). Based on electrophoretic mobility shift analysis of a protein titration (data not shown), we conclude that the purified heterodimer preparations contained <5% homodimer, in agreement with the results of Western blot analysis shown in Fig. 1. These results also show that the RAG-1 heterodimers are stable and did not reassort (to generate a homodimer from two heterodimers) during the 40-min incubation at 37°C prior to gel electrophoresis. Similar results were obtained with mR1(600)-R1h and mR1(962)-R1h (data not shown). While these data do not rule out the possibility that DNA-protein complexes could rearrange without complete dissociation, they are consistent with the results of previous studies in which the reassortment of heterodimers was not detected (29).
One functional RAG-1 active site is sufficient for both nicking and hairpin formation in Mn2+.
The catalytically competent RAG–12-RSS complex (also called the 12-SC) contains a dimer of RAG-1, one 12-RSS, and either one or two RAG-2 molecules (3, 21, 29). How many functional active sites within the RAG-1 dimer bound to a single RSS are necessary for nicking and hairpin formation? The simplest possibility is either that (i) one RAG-1 monomer nicks and the other monomer is responsible for hairpin formation or that (ii) a single monomer's active site performs both catalytic steps. These two models can be tested using the purified mutant heterodimers which contain one wild-type RAG-1 monomer and one mutant RAG-1 monomer that is incapable of either nicking or hairpin formation. Model 1 predicts that a standard substrate would be nicked, without hairpin formation (since nicking must precede hairpin formation); use of a prenicked substrate would allow hairpin formation. According to model 2, both nicking and hairpin formation should be observed with the use of standard substrates.
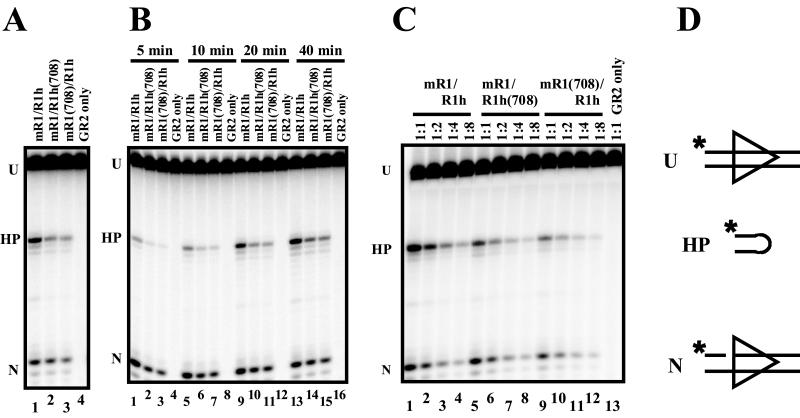
To address this question, standard single-RSS cleavage reactions were performed using a 12-RSS, either wild-type or mutant heterodimeric RAG-1, and wild-type GST-tagged RAG-2. The results of a typical experiment are shown in Fig. 3A. In the presence of Mn2+, which allows efficient cleavage of a single RSS, both wild-type and mutant heterodimers carried out both nicking and hairpin formation (Fig. 3A, lanes 1 to 3). (The 50% reduction in both nicking and hairpin formation by the mutant heterodimers is discussed below.) As expected, no cleavage was observed using RAG-2 only (Fig. 3A, lane 4). Two other mutant heterodimers [mR1(600)-R1h and mR1(962)-R1h] were also able to carry out nicking and hairpin formation under these conditions (data not shown), in agreement with the predictions of model 2.
FIG. 3.
One functional RAG-1 monomer is capable of nicking and hairpin formation at a single RSS. Abbreviations: U, uncut substrate; HP, hairpin product; N, nicked intermediate. (A) Standard cleavage reactions using a radiolabeled 12-RSS-containing oligonucleotide substrate and Mn2+ were performed. (B) Cleavage was allowed to proceed for the times noted. (C) RAG-1 and RAG-2 proteins were diluted with dialysis buffer prior to incubation, and cleavage was allowed to proceed for 45 min. (D) Schematic of substrate and cleavage products. ∗, radiolabel.
The results described above indicate that a single RAG-1 molecule is capable of carrying out both cleavage steps. It is important, however, to rule out two alternative scenarios in which more than one active RAG-1 monomer might contact the RSS. First, the cleavage events detected in our experiments might occur in higher-order complexes or aggregates containing more than one heterodimer of RAG-1. Gel filtration analysis of the mutant heterodimer [mR1(708)-R1h] provides strong evidence against this possibility, because the fractions with the highest cleavage activity migrated with an apparent molecular mass that corresponds to dimers of RAG-1 (data not shown), in agreement with the results of previous gel filtration analysis of wild-type RAG-1 dimers (3, 21). Second, we considered the possibility that cleavage by the mutant heterodimers might occur by a bind-and-release mechanism (nicking, release of the nicked substrate, rebinding, and hairpin formation), although this mechanism is not utilized by wild-type RAG proteins (8, 15). To address this possibility, we examined the kinetics of nicking and hairpin formation by the mutant heterodimers. The time course experiment results shown in Fig. 3B demonstrate that nicking and hairpin formation by the wild-type and both mutant heterodimers follow similar kinetics. Nicking, along with a low level of hairpin formation, is readily apparent at the first time point (5 min) (Fig. 3B, lanes 1 to 3). Hairpin formation continued to increase up to the last time point (40 min) (Fig. 3B, lanes 13 to 15). The parallel kinetic profiles of wild-type and mutant heterodimers argue against the possibility that hairpin formation by the mutant heterodimers occurs by a bind-and-release model (additional evidence is presented below).
Another prediction of model 2 is that the overall level of cleavage by the mutant heterodimers should be decreased twofold, since both the wild-type and mutant monomers in each heterodimer should have an equal probability of binding an RSS. The results shown in Fig. 3A and B are in agreement with this prediction, since at all time points of examination the nicks and hairpins generated by the mutant heterodimers were approximately twofold less abundant (by phosphorimager analysis) than the products generated by wild-type heterodimers. To confirm this measurement, we performed a protein titration, testing a series of twofold dilutions (Fig. 3C). This analysis again demonstrated that the reactions performed by both undiluted mutant heterodimers (Fig. 3C, lanes 5 and 9) most closely mimic the activity observed with the 1:2 diluted wild-type heterodimer (Fig. 3C, lane 2). Together, these data suggest that one functional RAG-1 active site is sufficient for both nicking and hairpin formation at a single RSS in Mn2+. Furthermore, these results are inconsistent with the bind-and-release model, which predicts that cleavage by the heterodimers should yield a level of hairpin formation no more than 25% of the wild-type level and that this yield should be even lower if release and rebinding are not 100% efficient. (According to this model, only 50% of the initial RAG-RSS complexes should be competent for nicking. Hairpin formation requires release of the RAG dimers; only 50% of the rebound dimers would be in the proper position to catalyze hairpin formation, resulting in a theoretical maximum efficiency of 25%.)
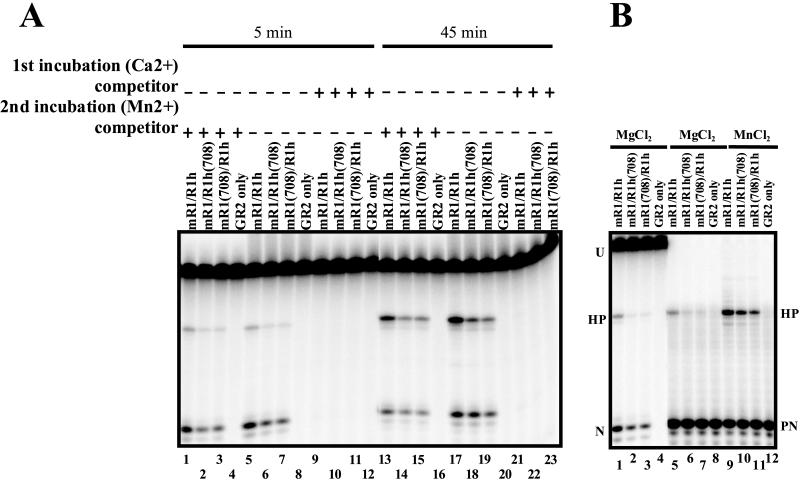
As a final test of the bind-and-release model, we performed competition experiments. This model predicts that hairpin formation by the mutant heterodimers occurs only after release and rebinding of the RAG proteins. Therefore, addition of excess unlabeled competitor RSS at the onset of catalysis should specifically block the appearance of radiolabeled hairpin products. RAG-RSS complexes were allowed to form during a 60-min preincubation in the presence of a radiolabeled 12-RSS and Ca2+ (which supports DNA binding but not catalysis). After the preincubation, Mn2+ was added either with or without a 100-fold molar excess of unlabeled 12-RSS competitor and cleavage was allowed to occur for either 5 or 45 min (Fig. 4A, lanes 1 through 12 or 13 through 23, respectively). The levels of nick and hairpin products were not significantly affected by the presence of competitor RSS in the second (Mn2+) incubation (Fig. 4A, lanes 1 through 4 and 13 through 16). In contrast, when added to the first incubation (with Ca2+), the same amount of competitor effectively abolished both nicking activity and hairpin formation (Fig. 4A, lanes 8 through 12 and 21 through 23). These data are inconsistent with the bind-and-release model, which predicts that competitor DNA should not affect nicking but should abolish hairpin formation. Based on these results and the data described above, we conclude that the mutant RAG heterodimers remain stably associated with the RSS substrate during catalysis. Therefore, the ability of the mutant heterodimers to both nick and form a hairpin at a single RSS strongly suggests that one functional active site within the context of a RAG-1 dimer performs both catalytic steps.
FIG. 4.
Mutant heterodimer cleavage activity is competitor resistant. Abbreviations: U, uncut substrate; HP, hairpin product; N, nicked intermediate; PN, prenicked substrate. (A) This assay was staged, using two incubations. In the first incubation, RAGs and substrate were incubated in Ca2+ to allow complex formation in either the presence or the absence of specific cold competitor. In the second incubation, Mn2+ was added to allow catalysis in either the presence or the absence of specific cold competitor. (B) Cleavage in the presence of Mg2+ or Mn2+ was tested on an uncut substrate or a prenicked substrate.
One RAG-1 active site is also sufficient for both cleavage steps at a single RSS in the presence of Mg2+.
The experiments described above were performed in the presence of Mn2+, which is known to relax the specificity of many nucleases (31), including the RAG proteins (17). Furthermore, Mn2+ nonspecifically rescues catalytic activities of several mutant RAG proteins (but not the DDE mutants) by unknown mechanisms (14). Therefore, it was important to examine single-RSS cleavage in the presence of Mg2+ (hairpin formation was inefficient but detectable under these conditions). As we observed in experiments with Mn2+, both mutant heterodimer preparations were capable of nicking and hairpin formation in the presence of Mg2+, yielding levels of these products that were approximately twofold lower than those observed with wild-type heterodimers (Fig. 4B, lanes 1 to 3). The mutant heterodimers were also capable of catalyzing hairpin formation using a prenicked substrate, again approximately 50% less efficiently than the wild type both in the presence of Mg2+ (Fig. 4B, lanes 5 through 7) and in the presence of Mn2+ (Fig. 4B, lanes 9 through 11). Thus, a single functional RAG-1 active site in the RAG-RSS complex was sufficient for nicking and hairpin formation in the presence of both Mn2+ and Mg2+. The ratios of nick formation to hairpin formation for wild-type and mutant heterodimers were closely matched at early time points. In some experiments, after prolonged incubation the mutant heterodimers showed somewhat less hairpin formation than wild-type heterodimers. The mechanistic basis for this behavior is unclear. It is important to note, however, that cleavage at a single RSS is not necessarily a physiologic reaction (analysis of cleavage at a 12/23 RSS pair is presented below).
Our data demonstrate that the V(D)J recombinase can utilize a single RAG-1 active site to perform both catalytic steps of cleavage at a single RSS. Recent experiments, published while this report was in preparation (27), also support this conclusion. The 50% reduction in cleavage efficiency observed with the mutant heterodimers further indicates that the active monomer must be bound in a specific orientation. Given that the Hin homology domain of RAG-1 (amino acids 389 to 446) is responsible for binding to the nonamer (5, 26, 28), it is possible that one RAG-1 monomer binds to the nonamer and positions the second monomer at the cleavage site—the junction between the heptamer and the coding flank. In fact, recent experiments indicate that cleavage at a single RSS is mediated by the RAG-1 monomer that is not bound to the nonamer (27).
The Tn10 transposase uses a single active site to perform all four reactions necessary for transposition: nicking, hairpin formation, hairpin opening, and strand transfer (4, 11). We have shown that a single RAG-1 active site can be used for both nicking and hairpin formation. Since the RAG proteins are capable of transposition (1, 10), it will be interesting to determine whether the same RAG-1 monomer catalyzes all three steps: nicking, hairpin formation, and transposition.
Cleavage of a 12/23 synaptic complex requires two RAG-1 dimers.
Under physiological conditions, hairpin formation is substantially stimulated by the formation of a synaptic complex in which the RAG proteins are bound to both a 12- and a 23-RSS (6, 9, 30). The stoichiometry of this complex with respect to RAG-1 has not been established. Two models have been proposed for the formation of these complexes (3, 27, 29). A RAG complex containing a RAG-1 dimer associated with a single RSS could simply capture a second RSS, leading to a synaptic complex containing only one dimer of RAG-1. Alternatively, the synaptic complex could form by collision of two RAG-RSS complexes, leading to a complex containing two RAG-1 dimers. Examples of both dimeric (19) and tetrameric (18, 32) configurations are provided by bacterial transposases. In both cases, only one active monomer at each end of the transposable element is necessary for all catalytic activities (4, 11, 18, 19, 32).
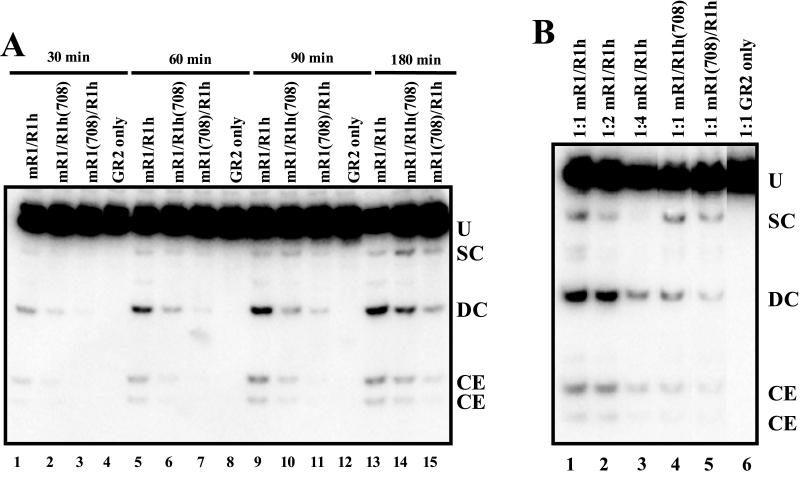
To probe the organization of the synaptic complex, we assessed the ability of the mutant heterodimers to perform coupled cleavage of a plasmid substrate, pJH290, which contains a 12/23 RSS pair (16). Using this substrate, cleavage at a single RSS can be readily distinguished from coupled cleavage at a 12/23 RSS pair. Thus, if the synaptic complex contains a single RAG-1 heterodimer, cleavage should occur only at a single RSS, because only one active site is present in the complex. If, however, the synaptic complex contains two heterodimers, cleavage can occur at both RSS, producing a doubly cleaved fragment.
We performed an analysis of the kinetics of plasmid cleavage using wild-type and mutant heterodimers (Fig. 5A). The mutant heterodimers produced appreciable levels of the doubly cut species (Fig. 5A, lanes 2 and 3 and lanes 6 and 7, as well as lanes 10 and 11 and lanes 14 and 15), indicating the presence of two functional RAG-1 active sites and suggesting that the synaptic complex contained two RAG-1 dimers. Nevertheless, we considered two alternative scenarios in which a synaptic complex containing a single RAG-1 dimer could generate doubly cleaved products. First, one active site might nick and form a hairpin(s) efficiently at both RSS (although this would presumably require major structural rearrangements within the synaptic complex), which would result in wild-type cleavage at both RSS if no particular orientation was required or a 50% reduction if the active monomer had to bind either the 12- or the 23-RSS. Second, the doubly cleaved products could be derived from sequential single RSS cleavage events. Our kinetics analysis argues strongly against the latter possibility, because singly cleaved intermediates should precede the appearance of doubly cleaved products. Double cleavage by both mutant heterodimers was observed at the earliest time point (30 min) (Fig. 5A, lanes 2 and 3); no singly cleaved species were detected until the 60-min time point (Fig. 5A, lanes 6 and 7). Note that the earlier time point is shorter than the incubation time used for the mobility shift experiments (40 min), in which no detectable rearrangement of protein subunits within the RAG-1 dimer was observed. Therefore, it is unlikely that doubly cleaved species arise from successive single-RSS cleavage events, since they appear before the singly cleaved species accumulates. Additional support for this interpretation is provided by competition experiments, which showed that cleavage of plasmid substrates is resistant to a 100-fold molar excess of specific RSS competitor DNA (data not shown).
FIG. 5.
Two dimers of RAG-1 exist in a synaptic complex. Abbreviations: U, uncut substrate; SC, cleavage at one RSS; DC, cleavage at both RSS; CE, coding end. (A) Cleavage was allowed to proceed for the times noted; (B) RAG-1 and RAG-2 proteins were diluted with dialysis buffer prior to incubation and cleavage was allowed to proceed for 30 min.
The two-dimer model is further supported by quantitative analysis of the ratio of single- to double-cleavage events. This model predicts that the mutant heterodimer should generate singly and doubly cleaved products in a ratio of 2:1, because single-cleavage events can occur whenever either RSS has a catalytically competent monomer in the proper configuration (two out of four possible combinations), whereas double cleavage requires both monomers to be in the correct position (one out of four possible combinations). In agreement with these expectations, Fig. 5A demonstrates an increased ratio of singly cut to doubly cut species derived from the mutant heterodimers (relative to wild-type) and a level of doubly cleaved products that was decreased compared to that observed with the wild-type heterodimer (Fig. 5A, compare lanes 13 through 15). To confirm these results, we performed a protein titration (Fig. 5B) which showed that the level of doubly cleaved products produced by the undiluted mutant heterodimers (Fig. 5B, lanes 4 and 5) was equal to that generated by a 1:4 dilution of the wild-type heterodimer (Fig. 5B, lane 3), as predicted by the two-dimer model. These results are inconsistent with the single-dimer model in which only one functional monomer is required for cleavage at both RSS, because this model predicts either no decrease (if the functional monomer can be bound to either RSS) or a twofold decrease (if the functional monomer must bind in a particular orientation) in doubly cut products. Our data clearly show a fourfold reduction, consistent with the two-dimer model. Furthermore, the ratio of single-cut to double-cut products generated by the mutant heterodimers was higher than it was in the case of wild-type homodimers at all dilutions tested, in agreement with the two-dimer model. Together, these data strongly suggest that the synaptic complex contains two dimers of RAG-1 and that one monomer of RAG-1 at each RSS is sufficient for cleavage.
Several well-characterized transposases perform cleavage in trans: the active monomer responsible for cleaving at one end of the transposon is donated by a transposase monomer bound to the other end (4, 11, 18, 19, 32). Hairpin formation by the RAG proteins (in Mg2+) is strongly stimulated by the presence of a 12/23 RSS pair (6, 9, 30). Catalysis of this step in trans would provide a simple way to ensure that double-strand break formation requires synaptic complex assembly, and would help to enforce coupled cleavage. Our data show that under conditions that bypass the requirement for a 12/23 RSS pair (in Mn2+), hairpin formation can be carried out in cis, by the same RAG-1 monomer that is responsible for nicking. We obtained similar results in Mg2+, under conditions that support inefficient hairpin formation at a single RSS. The substantial stimulation of hairpin formation observed in the presence of a 12/23 RSS pair in Mg2+ may reflect a conformational change induced by the addition of the second dimer of RAG-1 in the synaptic complex. Future experiments are required to determine the organization of the RAG monomers in the synaptic complex and to determine whether the individual cleavage steps are catalyzed in cis or in trans.
Another unanswered question is the role of RAG-2 in catalysis. Although it is not clear whether RAG-2 contributes amino acids to the active site, we have recently identified a RAG-2 mutant that specifically inhibits hairpin formation (20). The use of this mutant in experiments similar to those described here should facilitate determination of the organization of RAG-2 monomers in the synaptic complex, providing a more comprehensive understanding of the architecture of the DNA-protein complex that carries out V(D)J recombination.
ACKNOWLEDGMENTS
Monica Calicchio and Wei-han Kan provided technical assistance, and we thank Suzanne Robertson and Denise Guzman for secretarial support. We are grateful to Vicky Brandt for editorial help and to Tania Baker and Ilana Goldhaber-Gordon for helpful discussions. Mary Purugganan, Heather Schultz, Leslie Huye, Sundeep Shah, and Matt Neiditch provided critical suggestions on the manuscript.
This work was supported by a grant from the National Institutes of Health (AI-36420). M.A.L. was supported by a National Institutes of Health Predoctoral Fellowship (T32-AI07495). D.B.R. is an Assistant Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Agrawal A, Eastman Q M, Schatz D G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 2.Akamatsu Y, Oettinger M A. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailin T, Mo X, Sadofsky M J. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol Cell Biol. 1999;19:4664–4671. doi: 10.1128/mcb.19.7.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolland S, Kleckner N. The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell. 1996;84:223–233. doi: 10.1016/s0092-8674(00)80977-0. [DOI] [PubMed] [Google Scholar]
- 5.Difilippantonio M J, McMahan C J, Eastman Q M, Spanopoulou E, Schatz D G. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 6.Eastman Q M, Leu T M J, Schatz D G. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 7.Fugmann S D, Villey I J, Ptaszek L M, Schatz D G. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- 8.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 9.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 10.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy A K, Haniford D B, Mizuuchi K. Single active site catalysis of the successive phosphoryl transfer steps by DNA transposases: insights from phosphorothioate stereoselectivity. Cell. 2000;101:295–305. doi: 10.1016/s0092-8674(00)80839-9. [DOI] [PubMed] [Google Scholar]
- 12.Kim D R, Dai Y, Mundy C L, Yang W, Oettinger M A. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D R, Oettinger M A. Functional analysis of coordinated cleavage in V(D)J recombination. Mol Cell Biol. 1998;18:4679–4688. doi: 10.1128/mcb.18.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landree M A, Wibbenmeyer J A, Roth D B. Mutational analysis of RAG-1 and RAG-2 identifies three active site amino acids in RAG-1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Swanson P, Desiderio S. RAG-1 and RAG-2-dependent assembly of functional complexes with V(D)J recombination substrates in solution. Mol Cell Biol. 1997;17:6932–6939. doi: 10.1128/mcb.17.12.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieber M R, Hesse J E, Lewis S, Bosma G C, Rosenberg N, Mizuuchi K, Bosma M J, Gellert M. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell. 1988;55:7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- 17.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 18.Namgoong S-Y, Harshey R M. The same two monomers within a MuA tetramer provide the DDE domains for the strand cleavage and strand transfer steps of transposition. EMBO J. 1998;17:3775–3785. doi: 10.1093/emboj/17.13.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naumann T A, Reznikoff W S. Trans catalysis in Tn5 transposition. Proc Natl Acad Sci USA. 2000;97:8944–8949. doi: 10.1073/pnas.160107997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu J, Kale S B, Schultz H Y, Roth D B. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol Cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 21.Rodgers K K, Villey I J, Ptaszek L, Corbett E, Schatz D G, Coleman J E. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Res. 1999;27:2938–2946. doi: 10.1093/nar/27.14.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roth D B, Zhu C, Gellert M. Characterization of broken DNA molecules associated with V(D)J recombination. Proc Natl Acad Sci USA. 1993;90:10788–10792. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santagata S, Aidinis V, Spanopoulou E. The effect of Me2+ cofactors at the initial stages of V(D)J recombination. J Biol Chem. 1998;273:16325–16331. doi: 10.1074/jbc.273.26.16325. [DOI] [PubMed] [Google Scholar]
- 24.Sarnovsky R J, May E W, Craig N L. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 1996;15:6348–6361. [PMC free article] [PubMed] [Google Scholar]
- 25.Sawchuk D J, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig M C, Cortes P. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J Exp Med. 1997;185:2025–2031. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spanopoulou E, Zaitseva F, Wang F-H, Santagata S, Baltimore D, Panayotou G. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 27.Swanson P C. The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol Cell Biol. 2001;21:449–458. doi: 10.1128/MCB.21.2.449-458.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swanson P C, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 29.Swanson P C, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol Cell Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Gent D C, Ramsden D A, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 31.Vermote C L M, Halford S E. EcoRV restriction endonuclease: communication between catalytic metal ions and DNA recognition. Biochemistry. 1992;31:6082–6089. doi: 10.1021/bi00141a018. [DOI] [PubMed] [Google Scholar]
- 32.Williams T L, Jackson E L, Carritte A, Baker T A. Organization and dynamics of the Mu transpososome: recombination by communication between two active sites. Genes Dev. 1999;13:2725–2737. doi: 10.1101/gad.13.20.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]