Abstract
Ubiquitination is used to target both normal proteins for specific regulated degradation and misfolded proteins for purposes of quality control destruction. Ubiquitin ligases, or E3 proteins, promote ubiquitination by effecting the specific transfer of ubiquitin from the correct ubiquitin-conjugating enzyme, or E2 protein, to the target substrate. Substrate specificity is usually determined by specific sequence determinants, or degrons, in the target substrate that are recognized by the ubiquitin ligase. In quality control, however, a potentially vast collection of proteins with characteristic hallmarks of misfolding or misassembly are targeted with high specificity despite the lack of any sequence similarity between substrates. In order to understand the mechanisms of quality control ubiquitination, we have focused our attention on the first characterized quality control ubiquitin ligase, the HRD complex, which is responsible for the endoplasmic reticulum (ER)-associated degradation (ERAD) of numerous ER-resident proteins. Using an in vivo cross-linking assay, we directly examined the association of the separate HRD complex components with various ERAD substrates. We have discovered that the HRD ubiquitin ligase complex associates with both ERAD substrates and stable proteins, but only mediates ubiquitin-conjugating enzyme association with ERAD substrates. Our studies with the sterol pathway-regulated ERAD substrate Hmg2p, an isozyme of the yeast cholesterol biosynthetic enzyme HMG-coenzyme A reductase (HMGR), indicated that the HRD complex discerns between a degradation-competent “misfolded” state and a stable, tightly folded state. Thus, it appears that the physiologically regulated, HRD-dependent degradation of HMGR is effected by a programmed structural transition from a stable protein to a quality control substrate.
Ubiquitin-mediated, proteasome-dependent degradation is often employed by the cell to destroy both normal and misfolded proteins for purposes of regulation and quality control (39, 51). In general, proteins destined for degradation by the proteasome are covalently modified with the small protein ubiquitin through the collective action of a hierarchy of enzymes (21, 40, 46). A single ubiquitin-activating enzyme, or E1 protein, activates ubiquitin for transfer to a small number of ubiquitin-conjugating enzymes, or E2 proteins, which are divergent in their targeting function and underlie the first level of specificity for substrate selection. The ubiquitin-conjugating enzymes then covalently attach the ubiquitin moiety to the side chain of an internal lysine residue within their target substrates (11, 14, 79), in a reaction requiring additional specificity factors referred to as ubiquitin ligases, or E3 proteins, which constitute the largest class of proteins in the ubiquitination hierarchy.
Currently known ubiquitin ligases have catalytic subunits that fall into two distinct classes: HECT domain proteins, such as E6-AP and Rsp5p (35, 43, 74), and RING-H2 domain proteins, such as Rbx1p and Hrd1p (3, 32, 76, 78). HECT domain ubiquitin ligase action involves transient covalent linkage of ubiquitin to the ubiquitin ligase itself, followed by transfer of the attached ubiquitin moiety to the target substrate (75). RING-H2 domain ubiquitin ligase action appears to involve promotion of direct ubiquitin transfer between the ubiquitin-conjugating enzyme and the substrate. In addition to catalyzing substrate ubiquitination, many ubiquitin ligases also catalyze their own ubiquitination (3, 18, 27, 57, 61, 67).
Current models suggest that RING-H2 ubiquitin ligases function by providing specific binding sites for both the target substrate and the relevant ubiquitin-conjugating enzyme to effect transfer of ubiquitin from the conjugating enzyme to the target. In fact, the RING-H2 domain of cCbl mediates direct interaction with UbcH7 (92). The simultaneous binding of the substrate and ubiquitin-conjugating enzyme to the RING-H2 ubiquitin ligase to stimulate substrate ubiquitination can be referred to as a mutual binding mechanism. The specific binding of target substrates may be brought about directly by the ubiquitin ligase itself or by proteins associated with the ubiquitin ligase. For example, in the SCF ubiquitin ligase complex, the RING-H2 protein Hrt1p (also called Rbx1p or Roc1p) employs a scaffold complex containing an effector protein that specifically binds to the target substrate (19, 62, 76–78). Different substrate-binding proteins are incorporated into the scaffold complex to change the complex's substrate specificity (19, 23, 63, 77). Similarly, the APC ubiquitin ligase complex, containing the RING-H2 protein Apc11p (53, 62, 76, 90), also utilizes a scaffold complex to bind the target substrate (52, 64, 89, 90). In contrast, the N-end rule ubiquitin ligase contains multiple substrate recognition sites within the RING-H2 protein itself (1, 2, 30, 70), allowing the same protein to bind multiple substrates and related allosteric regulators. Other ubiquitin ligases that also directly bind their specific substrate targets include cCbl (47, 54) and Mdm2 (41, 42). In each case, the specificity of the ubiquitin ligase complex is a consequence of its ability to specifically bind the target protein.
Ubiquitin-mediated degradation is employed in cellular quality control to remove misfolded and misassembled proteins (51, 84). For quality control degradation to be effective, the ubiquitination machinery must recognize common structural hallmarks of damage or misfolding in a diverse group of proteins with little or no primary sequence homology. Recently, we and others have identified a RING-H2 ubiquitin ligase complex, referred to as the HRD complex below, that is responsible for degradation of numerous quality control substrates (3, 6, 27, 32, 67). Presumably, these and other quality control ubiquitin ligases also function by promoting association between the substrate and the ubiquitin-conjugating enzyme. However, it is not clear if the mutual binding model of ubiquitin ligase function is applicable to the large and varied set of possible quality control substrates with no sequence similarity. It may be that the quality control ubiquitin ligases recognize appropriate substrates by evaluation of the folding state rather than specific sequence motifs within the target, although no evidence has yet been presented for such a model.
A significant component of cellular quality control degradation occurs by endoplasmic reticulum (ER)-associated degradation (ERAD). Mutant lumenal or integral membrane proteins incapable of attaining correct structure or proper assembly are destroyed by ERAD (4, 20, 32, 36, 45, 66, 68, 82, 85, 87, 88, 93). ERAD is not restricted to aberrant proteins, as a significant fraction of newly made, normal proteins are destroyed by ERAD as part of normal ER physiology (55, 80, 83). Furthermore, ERAD is employed in the feedback regulation of HMG-coenzyme (CoA) reductase (HMGR), so that signals from the sterol synthesis pathway control HMGR stability (12, 17, 25, 31, 34, 60).
In Saccharomyces cerevisiae, ERAD is mediated in large part by the action of a RING-H2 ubiquitin ligase complex composed of the integral ER membrane proteins Hrd1p (Der3p) and Hrd3p (3, 6, 27, 32, 66, 67). Hrd1p contains an N-terminal, multispanning membrane anchor and a C-terminal cytosolic domain (27), which contains a RING-H2 motif homologous to several characterized ubiquitin ligases (18, 47, 49, 57). Consistent with this homology, Hrd1p functions both in vitro and in vivo as a ubiquitin ligase (3), catalyzing the processive transfer of ubiquitin to itself and other proteins. In vitro, Hrd1p shows preference for a misfolded protein as a multiubiquitination substrate (3) and thus may directly participate in some aspects of quality control recognition. In vivo, Hrd1p is rate limiting for ubiquitination and degradation of numerous ERAD substrates (3, 27, 67) and specifically employs only Ubc7p and Ubc1p in its action (3), with Ubc7p being quantitatively more important in the ERAD pathway. Hrd3p is a single-spanning ER membrane protein, with the majority of its sequence residing in the ER lumen (27, 32, 67, 71). The Hrd3p lumenal regions are both necessary and sufficient for Hrd3p action in ERAD (27). One function of the Hrd3p lumenal domain is to regulate the activity and stability of the cytosolic Hrd1p RING-H2 domain through transmembrane communication mediated by the Hrd1p membrane anchor (27). However, Hrd3p has ERAD functions independent of Hrd1p stabilization (27).
Because Hrd1p and Hrd3p form an ER membrane-associated ubiquitin ligase complex required for quality control ERAD, it is likely that the complex recognizes common structural features among its diverse substrates. Here, we directly tested the model that in vivo HRD complex promotes association between ERAD substrates and the ER ubiquitin-conjugating enzyme Ubc7p. From these studies, it is clear that the HRD complex does indeed mediate fruitful proximity between ERAD substrates and the appropriate ubiquitin-conjugating enzyme. Additionally, it appears that the HRD complex scans ER proteins for their degradation status and, upon encountering a protein that appears to be misfolded, promotes access of the appropriate ubiquitin-conjugating enzyme to the substrate, allowing robust yet selective ubiquitination of the desired target. According to this model, and in contrast to regulatory ubiquitin ligases, the HRD complex does not promote specificity by restricting its interactions only to substrates. Rather, the HRD complex functions in ubiquitination by discerning substrate properties subsequent to interaction. This model also suggests a mechanism for regulated ERAD of HMGR, which provides a significant component of eukaryotic regulation of the sterol pathway. Regulated HMGR ERAD appears to occur through a controlled structural transition of the HMGR transmembrane domain that allows it to be recognized as a quality control substrate by the HRD machinery. These studies have mechanistic implications for understanding both the specificity of quality control degradation and the clinically relevant axis of sterol regulation that employs ERAD to control the cellular synthesis of cholesterol (29).
MATERIALS AND METHODS
Materials and reagents.
All enzymes were obtained from New England Biolabs (Beverly, Mass.). Chemical reagents were obtained from Sigma Chemical (St. Louis, Mo.). Dithiosuccinimidylproprionate (DSP) was obtained from Pierce (Rockford, Ill.). Protein A-Sepharose CL-4B was obtained from Amersham Pharmacia (Piscataway, N.J.). Lovastatin, L-659,699, and zaragozic acid were generously donated by Merck (Rahway, N.J.). ECL enhanced chemiluminescence immunodetection reagents were from Amersham Corp. (Arlington Heights, Ill.). Anti-Myc 9E10 antibody was used as a cell culture supernatant obtained by growing the 9E10 hybridoma (ATCC CRL 1729) in RPMI 1640 culture medium (Life Technologies, Grand Island, N.Y.) with 10% fetal calf serum. Antihemagglutinin (HA) antibody was an ascites fluid obtained from Babco (Berkeley, Calif.). Affinity-purified goat anti-mouse immunoglobulin-horseradish peroxidase (HRP) conjugate was obtained from Sigma Chemical.
Recombinant DNA and molecular cloning.
The splicing-by-overlap-extension method was use to make epitope-tagged versions of each gene (38). PCR was performed as previously described (24). A list of primers used is available upon request. All epitope-tagged plasmids were verified for their ability to completely complement a null mutation in their respective genomic copy.
The plasmid that expressed a single, myc epitope-tagged version of Hmg1p (1myc-Hmg1p) was constructed as follows. Partial complementary primers that encoded a single myc epitope sequence (EQKLISEEDL) were used to amplify a fragment of HMG1 from pRH144-2 (31), which resulted in a single myc epitope coding sequence inserted between codons 621 and 622 of HMG1. The PCR fragment was cloned between the BsrGI and PflMI sites in pRH144-2. The resulting plasmid was named pRH945.
The plasmid that expressed a double, HA epitope-tagged version of Ubc7p (2HA-Ubc7p) was constructed as follows. The UBC7 gene was amplified from RHY623 genomic DNA (25). A DNA fragment that encoded two tandem HA epitope tags was amplified from pGTEP (obtained from B. Futcher). The two PCR fragments were spliced together by PCR, and the resulting DNA fragment was cloned between the PstI and SalI sites in pRH423 (34), which resulted in pRH685. The 1.3-kb PvuII-SphI fragment from pRH685, which contained the 2HA-UBC7 gene, was cloned between the Ecl136II and SphI sites in pRH507. The resulting TRP1 2HA-UBC7-containing plasmid was called pRH373.
The plasmid that expressed a triple HA epitope-tagged version of Ubc6p (3HA-Ubc6p) was made as follows. The UBC6 gene was amplified from RHY623 genomic DNA and cloned between the PstI and NheI sites in pRH442 (25). The resulting plasmid was called pRH1149. A DNA fragment encoding a triple HA epitope tag was amplified from pGTEP and cloned between the NheI and SalI sites in pRH1149, which resulted in pRH1151. A 1.6-kb fragment which contained the 3HA-UBC6 gene was cloned between the PstI and KpnI sites in pRH507 (85). The resulting TRP1 3HA-UBC6-containing plasmid was called pRH1217.
The plasmid used to make hrd3Δ::LEU2 was constructed as follows. A 3.1-kb XhoI-SpeI fragment from pRH508 which contained the HRD3 gene (32) was inserted between the BamHI and EcoRI sites in pBluescript KSII, resulting in pRH1175. The LEU2 gene was PCR amplified from pRS405, digested with XbaI, and inserted between the BsaBI and NheI sites in pRH1175. The resulting hrd3Δ::LEU2 plasmid was named pRH1185.
The plasmid used to make ubc7Δ::LEU2 was constructed as follows. A 650-bp fragment containing the coding region for the UBC7 gene was PCR amplified from pRH685 and inserted between the PstI and SalI sites in pBluescript KSII, which resulted in pRH1176. pRH1176 contained an HpaI and a BsrGI site with the UBC7 sequence. A 1.5-kb HpaI-BsrGI fragment from pRS405 containing the LEU2 gene was inserted between the HpaI and BsrGI sites in pRH1176. The resulting ubc7Δ::LEU2 plasmid was named pRH1186.
Strains and media.
Escherichia coli DH5α strains were grown at 37°C in Luria-Bertani medium with ampicillin (100 μg/ml). Yeast strains were grown at 30°C in minimal medium supplemented with glucose and the appropriate amino acids, as described (24). The lithium acetate method was used to transform yeast cells with plasmid DNA (44).
Yeast strains RHY1914 (MATα his3Δ200 lys2-801 ade2-101 leu2Δ ura3-52::1MYC-HMG2::URA3 trp1::HISG met2 hmg1::LYS2 HMG2) and RHY1915 (MATα his3Δ200 lys2-801 ade2-101 leu2Δ ura3-52::1MYC-HMG1::URA3 trp1::HISG met2 hmg1::LYS2 HMG2) were used as parent strains. The plasmid containing 3HA-Hrd1p, pRH1196, was digested with BglII and integrated at the HRD1 genomic locus. The plasmid containing 3HA-Hrd3p, pRH1263, was digested with BsrGI and integrated at the HRD3 genomic locus. The plasmid containing 2HA-Ubc7p, pRH373, was digested with BsgI and integrated at the TRP1 genomic locus. The plasmid containing 3HA-Ubc6p, pRH1217, was digested with BsrGI and integrated at the UBC6 genomic locus. Transformants were selected for TRP+ prototrophy. The plasmid containing hrd1Δ::LEU2, pRH1184 (85), was digested with XhoI and BamHI and integrated at the HRD1 genomic locus. The plasmid containing hrd3Δ::LEU2, pRH1185, was digested with XhoI and NdeI and integrated at the HRD3 genomic locus. The plasmid containing ubc7Δ::LEU2, pRH1186, was digested with PstI and SalI and integrated at the UBC7 genomic locus. The plasmid containing cue1::LEU2, pTX118 (5), was digested with ApaI and SacI and integrated at the CUE1 genomic locus. Transformants were selected for LEU+ prototrophy.
Ubiquitination assays.
Ubiquitination assays were performed as previously described (3).
Degradation assay.
Cycloheximide-chase assays were performed as previously described (24).
Cross-linking assays.
Cross-linking assays were done as previously described (27), except that 30 μl of anti-HMGR antiserum was used for the immunoprecipitation. In a given set of experiments, samples were always run on a single gel so that transfer, immunoreactivity, and exposure times were kept constant between samples that were to be compared.
Flow cytometry.
Flow cytometry was performed as previously described (24), using a FACSscan (Beckton Dickinson, Palo Alto, Calif.) analytical flow microfluorimeter with settings for fluorescein-labeled antibody analysis. To examine the effects of lovastatin and zaragozic acid on Hmg2p-green fluorescent protein (GFP) steady-state levels, the drugs were added to early-log-phase cultures (optical density at 600 nm [OD600] of <0.2) to the desired final concentrations, and the cells were grown for an additional 4 h (two doublings). In some cases, cells were grown to log phase, with 10% glycerol initially added to the medium or 10% glycerol added at the time of drug addition.
TLC.
Thin-layer chromatography (TLC) was performed as previously described (28).
Protease protection assays.
Microsomes were isolated as previously described (27). Protease protection assays were performed as previously described (27) except that anti-Hmg2p antiserum was used to detect Hmg2p.
RESULTS
ERAD of numerous ER lumenal and membrane proteins requires the Hrd1p/Hrd3p ubiquitin ligase complex (4, 6, 66, 67), including the regulated ERAD of the yeast HMGR isozyme Hmg2p (3, 27, 32). In addition to Hrd1p and Hrd3p, ERAD requires the ubiquitin-conjugating enzyme Ubc7p (4, 34, 37, 66) and its membrane association factor Cue1p (5). From this, it seems likely that some or all of the HRD complex proteins and their associated factors could directly interact with substrates to target them for ubiquitination.
To test this hypothesis, we required an assay that would allow detection of the transient interactions that might exist between the ER membrane-bound HRD ubiquitin ligase complex components and the targeted degradation substrates. Furthermore, the substrates and HRD complex under study are, for the most part, integral membrane proteins, and the assay had to be amenable to the physical constraints inherent with such molecules. Finally, to study the participation of the HRD complex in regulation of Hmg2p, the assay had to allow for the presence and action of small signaling molecules from the mevalonate pathway that our studies show are at low cellular abundance and transiently produced (25, 28, 31). Assays normally used for interaction studies are the yeast two-hybrid assay, coimmunoprecipitation, and chemical cross-linking. To measure transient interactions, however, the two-hybrid assay was not a viable option because by nature, the transient interactions would likely be of low affinity, nor is there a transparent adaptation of this technique for studying interactions between membrane proteins. Similarly, because the HRD complex components and some of its substrates are integral membrane proteins (27, 31, 32), a cross-linking approach was required in lieu of the more usual coimmunoprecipitation studies of ubiquitin ligase activity and substrate association (47, 49, 78, 86). Coimmunoprecipitation assays were not applicable to our system, since the solubilization methods required to extract the proteins from the ER membrane would likely disrupt the interactions between substrates and membrane-bound HRD proteins and would most certainly dilute or remove the transient signaling molecules that control ERAD of Hmg2p. In contrast, chemical cross-linking has been used with great success to determine transient interactions between components of the ER membrane-bound Sec61p translocon and its translocation substrates (48, 58, 59, 65, 72). Additionally, we developed an in vivo modification of the cross-linking assay (27) which allows the signaling molecules that control Hmg2p ERAD to remain and function at normal levels and location. Although cross-linking between two proteins indicates their close proximity and is therefore informative of potential protein-protein interactions, the absence of cross-linking does not necessarily preclude an interaction. With this caveat, we proceeded with the cross-linking studies to test interactions between the HRD complex and its substrates.
To analyze substrate-HRD complex interactions, we used the in vivo chemical cross-linking assay previously developed to examine interactions between the subunits of the HRD complex itself (27). For ease of detection, we used functional, HA epitope-tagged versions of each protein in the HRD complex, which have been described previously (3, 27, 71). In addition, we employed a uniquely available set of ERAD substrates from our studies of regulated degradation of yeast HMGR (26, 32), each with an added myc epitope tag. These included the mevalonate pathway-regulated Hmg2p, the homologous but constitutively stable Hmg1p, and the constitutively degraded 6myc-Hmg2p.
Specific cross-linking of Ubc7p with ERAD substrates.
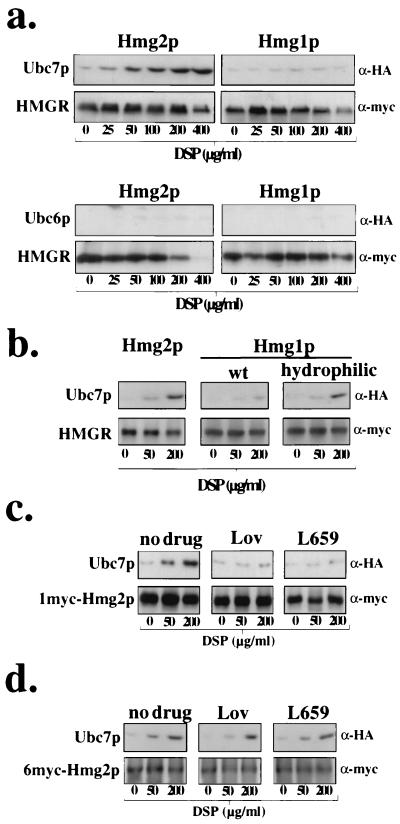
The current model for RING-H2 ubiquitin ligase function is that the ligase promotes proximity between the ubiquitin-conjugating enzyme and the target substrate through their mutual binding, which leads to direct ubiquitination of the substrate. However, ubiquitin ligase-mediated proximity of ubiquitin- conjugating enzyme and its substrate has never been demonstrated in vivo. We first used the cross-linking assay to examine proximity between the regulated ERAD substrate Hmg2p and Ubc7p, the principal ubiquitin-conjugating enzyme employed in its degradation (3, 34). When Hmg2p was immunoprecipitated from cells incubated with increasing concentrations of the cross-linker DSP (56), Ubc7p coimmunoprecipitated with Hmg2p in a cross-linker concentration-dependent fashion (Fig. 1a). Another ER-associated ubiquitin-conjugating enzyme, Ubc6p, which does not significantly participate in the ubiquitination and degradation of Hmg2p (3, 34), did not cross-link to Hmg2p (Fig. 1a). In contrast to Hmg2p, Ubc7p did not demonstrate any significant cross-linking to Hmg1p (Fig. 1a), the exceedingly stable isozyme of HMGR (26, 31). Thus, only a ubiquitin-conjugating enzyme required for Hmg2p degradation cross-linked to Hmg2p, and this cross-linking was in large part restricted to degradation substrate and not a similar, stable protein.
FIG. 1.
Ubc7p cross-linked to degraded Hmg2p in a mevalonate pathway-regulated manner but not to stable Hmg1p. Interaction of 2HA-Ubc7p with 1myc-HMGR was assessed by the in vivo cross-linking assay. Cells were grown to mid-log phase and removed to amine-free medium. DSP was added at the indicated concentrations to separate aliquots of cells, and cross-linking proceeded at 30°C for 30 min. Lysates were prepared, and 1myc-Hmg2p was immunoprecipitated with antiserum raised against its catalytic domain. Immunoprecipitates were denatured under reducing conditions and analyzed by immunoblotting with either the 9E10 antibody (α-myc) to detect 1myc-Hmg2p or the 12CA5 antibody (α-HA) to detect 2HA-Ubc7p. (a) Ubc7p cross-linked to Hmg2p but did not cross-link to Hmg1p. Cross-linking assay was done with strains expressing either 1myc-Hmg2p or 1myc-Hmg1p and coexpressing either 2HA-Ubc7p or 3HA-Ubc6p. The reduced immunoreactivity of 1myc-HMGR with DSP at 400 μg/ml is due to modification of the lysine residue in the myc epitope sequence, not reduced immunoprecipitation of HMGR (unpublished observations). The small amount of 2HA-Ubc7p that is present with DSP at 0 μg/ml in this and the following figures is similar to that observed when preimmune serum is used instead of HMGR-specific antiserum (unpublished observations), indicating that it is nonspecific immunoprecipitation. (b) Ubc7p cross-linked to a degraded version of Hmg1p; cross-linking assay with strains expressing either 1myc-Hmg2p, 1myc-Hmg1p, or hydrophilic-1myc-Hmg1p and coexpressing 2HA-Ubc7p. (c) Addition of either lovastatin or L-659,699 decreased Ubc7p cross-linking. Regulated Ubc7p cross-linking to Hmg2p was observed by preincubating the cells with either no or 50 μg of lovastatin (Lov) or 10 μg of L-659,599 (L659) per ml for 2 h prior to addition of DSP. Cross-linking assay was performed as in Fig. 1. (d) Cross-linking of Ubc7p to 6myc-Hmg2p was not regulated by the mevalonate pathway. Ubc7p cross-linking to 6myc-Hmg2p was observed by preincubating the cells with either 0 or 50 μg lovastatin or 10 μg of L-659,599 per ml for 2 h prior to addition of DSP.
In order for two proteins to be cross-linked by DSP, free and accessible lysine residues within a target protein must be within a short distance of lysine residues within the bait protein. As lysine residues are generally the sites for attachment of ubiquitin (21, 46), it may be that the stable Hmg1p has limited access to its lysine residues for Ubc7p. Accordingly, perhaps Ubc7p did not cross-link with Hmg1p because there were no accessible lysine residues within Hmg1p in close proximity to Ubc7p.
To test if Ubc7p could be cross-linked to Hmg1p, we performed the cross-linking assay on mutant and degraded versions of Hmg1p. From our previous mutagenic analyses that dissected the in cis determinants for Hmg2p degradation (26), we constructed a number of Hmg1p mutants that were no longer stable but were subject to Ubc7p-dependent degradation (unpublished observations). One of these degradation-competent Hmg1p mutants, termed hydrophilic-Hmg1p, was the result of a limited number of substitutions that converted the hydrophobic residues in the first 26 residues to hydrophilic ones but did not change the number of lysine residues within Hmg1p, similar to hydrophilic-Hmg2p previously described (26). When hydrophilic-Hmg1p was immunoprecipitated from cells treated with DSP, Ubc7p now increasingly coimmunoprecipitated with the mutant Hmg1p in a cross-linker concentration-dependent manner (Fig. 1b, lanes hydrophilic versus wt). Thus, it appeared that the difference in Ubc7p cross-linking to normal Hmg1p was a physiological result of its stability rather than an intrinsic inability of Hmg1p to cross-link to Ubc7p.
Hmg2p is unique as an ERAD substrate in that its ubiquitination and degradation rates are feedback regulated by downstream signals from the mevalonate pathway (25, 31). Reduced production of downstream products of the mevalonate pathway by inhibition or downregulation of pathway enzymes results in the stabilization of Hmg2p (25, 31). We determined if Ubc7p cross-linking to Hmg2p was altered in accord with the mevalonate pathway regulation of the Hmg2p degradation rate. To examine if cross-linking between Ubc7p and Hmg2p was affected by this axis of regulation, we tested the effects of drugs that result in stabilization of Hmg2p. Incubation of cells with inhibitors of early pathway enzymes, such as the HMGR inhibitor lovastatin and the upstream HMG-CoA synthase inhibitor L-659,699, blocks Hmg2p ubiquitination and degradation (25, 31, 34). Incubation of cells with either of these drugs drastically reduced Ubc7p cross-linking to Hmg2p (Fig. 1c). The same drug treatments had no effect on Ubc7p cross-linking to 6myc-Hmg2pΔ63–219 (Fig. 1d), a grossly misfolded, epitope-tagged mutant of Hmg2p missing 157 residues of the native Hmg2p sequence, including a putative transmembrane span that undergoes constitutive, unregulated Ubc7p-dependent degradation (25, 32).
In vivo action of the HRD ubiquitin ligase complex.
Thus, Ubc7p cross-linking to ER proteins was directly correlated with their degradation status. Proteins that are naturally stable or are stabilized by physiological conditions within the cell did not cross-link to Ubc7p. Ubc7p-dependent degradation substrates did cross-link to Ubc7p, suggesting that Ubc7p directly associated with ERAD substrates in a highly specific and selective manner. In the case of regulated Hmg2p, the cross-linking between Hmg2p and Ubc7p was physiologically regulated in a manner entirely consistent with the regulation of Hmg2p ubiquitination and degradation by mevalonate pathway production.
HRD complex mediated Ubc7p cross-linking to substrates.
We next investigated the role of the Hrd1p/Hrd3p ubiquitin ligase complex in promoting the proximity of Ubc7p to Hmg2p. To do so, we tested the effects of null alleles of the genes encoding these proteins on Hmg2p-Ubc7p cross-linking. As expected, when the individual null alleles were introduced into the strain that coexpressed the epitope-tagged versions of Hmg2p and Ubc7p, the presence of either the hrd1Δ or the hrd3Δ allele resulted in complete stabilization of Hmg2p through reduced ubiquitination (Fig. 2a and b, respectively). Consistent with their effect on Hmg2p ubiquitination and degradation, the presence of either the hrd1Δ or the hrd3Δ allele resulted in almost complete loss of Ubc7p cross-linking to Hmg2p (Fig. 2c). Importantly, the effects of the hrd1Δ and hrd3Δ alleles were not explained by altered steady-state levels or cellular distribution of Ubc7p. In both of these mutants, Ubc7p stability, steady-state levels, and microsomal association were all similar to those in wild-type cells (Fig. 2d and e). Similar to the hrd1Δ and hrd3Δ alleles, introduction of the cue1Δ allele, which results in loss of the Ubc7p ER membrane anchor Cue1p (5), also stabilized Hmg2p through reduced ubiquitination (Fig. 2a and b). The presence of the cue1Δ allele similarly reduced Ubc7p cross-linking to Hmg2p (Fig. 2c). However, loss of Cue1p prevented Ubc7p from associating with the ER membrane and resulted in rapid degradation of the subsequently soluble Ubc7p protein (Fig. 2d and e). Thus, reduced Ubc7p cross-linking to Hmg2p in cue1Δ cells was likely due to the mislocalization and reduced steady-state levels of Ubc7p. In contrast to the effects of the hrd1Δ and hrd3Δ alleles, introduction of the hrd2-1 allele, which stabilizes Hmg2p by altering proteasome activity but not Hmg2p ubiquitination (32) (Fig. 2a and b), had no effect on Ubc7p cross-linking to Hmg2p (Fig. 2c). From these combined results, the Hrd1p/Hrd3p complex was required to promote a functional proximity between Ubc7p and the target ERAD substrate Hmg2p, consistent with its proposed function as a RING-H2 ubiquitin ligase complex.
FIG. 2.
HRD gene-encoded proteins required for Ubc7p-Hmg2p cross-linking. (a) The appropriate null alleles of each gene required for Hmg2p degradation were introduced into the strain coexpressing 1myc-Hmg2p and 2HA-Ubc7p. Correct introduction of each null allele was determined by observation of 1myc-Hmg2p stabilization in a cycloheximide-chase assay, performed as in Fig. 5 (left panels). Lysates from each indicated time point after addition of cycloheximide were prepared and immunoblotted to determine the level of 1myc-Hmg2p. (b) The presence of cue1Δ, hrd1Δ, and hrd3Δ blocked Hmg2p-regulated ubiquitination. Ubiquitination assays of cells carrying the indicated null allele were performed in the presence of no drug or zaragozic acid (ZA, 10 μg/ml). Upper panels are the result of antiubiquitin (α-Ub) immunoblotting for covalently linked Ub-Hmg2p conjugates. Lower panels are the result of parallel immunoblotting of an aliquot (1/8 total volume) of the same immunoprecipitates with the 9E10 anti-myc antibody (α-myc) to assess total immunoprecipitated Hmg2p. (c) The appropriate null alleles of each gene required for Hmg2p degradation were introduced into the strain coexpressing 1myc-Hmg2p and 2HA-Ubc7p. Cross-linking assay was performed as in Fig. 1. (d) 2HA-Ubc7p levels and degradation were unaffected in hrd1Δ and hrd3Δ cells, but 2HA-Ubc7p was rapidly degraded in cue1Δ cells. Cells expressing 2HA-Ubc7p and the indicated hrd allele were grown to log phase. Lysates from each indicated time point after addition of cycloheximide were prepared and immunoblotted to determine the level of 2HA-Ubc7p. (e) Membrane fractionation of the cells from panel d was performed by osmotically lysing cells and preparing a crude microsomal fraction. The ability of 2HA-Ubc7p to remain membrane bound was assayed under conditions of buffer, 2.5 M NaCl, 2.5 M urea, 0.8 M potassium acetate (KOAc, pH 11.6), or 1% Triton X-100. Lanes S, supernatant fractions. Lanes P, pellet fractions.
Cross-linking of HRD ubiquitin ligase complex to substrates.
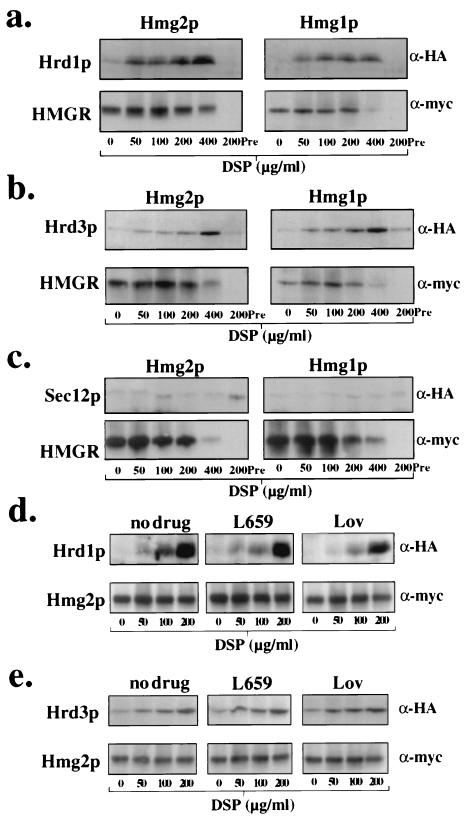
The Hrd1p/Hrd3p ubiquitin ligase complex is required for the specific ubiquitination of ERAD substrates by Ubc7p and appeared to promote a functional association between Ubc7p and ERAD substrates. This suggested that the HRD complex itself associated with target substrates. Accordingly, we evaluated cross-linking of the HRD complex components Hrd1p and Hrd3p with ERAD substrates. Because Hmg2p degradation is regulated by the mevalonate pathway (25, 31), we were particularly interested in the role that potential HRD complex-substrate interactions played in substrate selection and physiological regulation.
To examine such interactions, we used strains that coexpressed either 3HA-Hrd1p or 3HA-Hrd3p and a relevant 1myc-HMGR protein. Using the same in vivo cross-linking assay, we observed that both Hrd1p and Hrd3p cross-linked to Hmg2p (Fig. 3a and b, respectively). Surprisingly, and unlike Ubc7p cross-linking, both Hrd1p and Hrd3p also cross-linked to the stable HMGR isozyme Hmg1p (Fig. 3a and b, respectively). Furthermore, and consistent with this observation, cross-linking of either Hrd1p or Hrd3p to Hmg2p was completely unaffected by incubation of cells with lovastatin or L-659,699 (Fig. 3d and e, respectively), which stabilized Hmg2p (31, 34) and blocked Ubc7p cross-linking to Hmg2p (Fig. 1c). Thus, the Hrd1p/Hrd3p complex appeared to associate with both stable and degraded proteins alike and association with Hmg2p was not regulated. The cross-linking of Hrd1p and Hrd3p to both Hmg1p and Hmg2p did not appear to be due simply to nonspecific cross-linking of all ER proteins to Hmg1p or Hmg2p, as other ER proteins not involved in HRD-dependent degradation did not cross-link to either Hmg1p or Hmg2p, such as Ubc6p (Fig. 1a) and Sec12p (Fig. 3c).
FIG. 3.
Hrd1p/Hrd3p ubiquitin ligase complex cross-linked with both ERAD substrates and stable proteins. (a) Hrd1p cross-linked to both the degraded Hmg2p and the stable Hmg1p. Strains expressing either 1myc-Hmg2p or 1myc-Hmg1p and coexpressing 3HA-Hrd1p were subjected to the cross-linking assay as described for Fig. 1. 1myc-Hmg2p and 1myc-Hmg1p were detected using the 9E10 antibody (α-myc), and 3HA-Hrd1p was detected using the 12CA5 antibody (α-HA). The lower immunoreactivity of 1myc-HMGR with DSP at 400 μg/ml lane is due to modification of the lysine residue in the myc epitope sequence, not reduced immunoprecipitation of HMGR (data not shown). Preimmune serum substituted for anti-Hmg2p antiserum in the indicated sample is indicated as Pre. (b) Hrd3p cross-linked to both the degraded Hmg2p and the stable Hmg1p. Strains expressing either 1myc-Hmg2p or 1myc-Hmg1p and coexpressing 3HA-Hrd3p were subjected to the cross-linking assay as described for panel a. (c) Sec12p did not cross-link to Hmg2p. Strains expressing either 1myc-Hmg2p or 1myc-Hmg1p and coexpressing 3HA-Sec12p were subjected to the cross-linking assay. (d) Hrd1p cross-linking to Hmg2p was unregulated by the mevalonate pathway. Lack of regulated Hrd1p cross-linking to Hmg2p was observed by preincubating the cells with either no drug, lovastatin (Lov) (50 μg/ml), or L-659,599 (L659) (10 μg/ml) for 2 h prior to addition of DSP. (e) Hrd3p cross-linking to Hmg2p was unregulated by the mevalonate pathway. Lack of regulated Hrd3p cross-linking to Hmg2p was observed by preincubating the cells as in panel d.
We next examined cross-linking of each HRD protein to Hmg2p and Hmg1p in a variety of null mutants to evaluate functional relationships. Hrd3p cross-linking to Hmg2p or Hmg1p was equivalent in wild-type cells and cells carrying either the hrd1Δ, ubc7Δ, or cue1Δ allele (Fig. 4a and unpublished observations), indicating that Hrd3p association with ER proteins was independent of Hrd1p, Ubc7p, or Cue1p expression. Hrd1p cross-linking to Hmg2p or Hmg1p was equivalent in wild-type cells and cells carrying the ubc7Δ or cue1Δ allele, but was strongly diminished by the presence of the hrd3Δ allele (Fig. 4b and unpublished observations). However, Hrd1p steady-state levels are eightfold lower in hrd3Δ cells due to its rapid degradation (27) (Fig. 4d).
FIG. 4.
Hrd1p/Hrd3p cross-linking to Hmg2p did not require the presence of Ubc7p or Cue1p. (a) Hrd3p cross-linking to Hmg2p in the presence of either the hrd1Δ, ubc7Δ, or cue1Δ allele. The appropriate null alleles of each gene required for Hmg2p degradation were introduced into the strain coexpressing 1myc-Hmg2p and 3HA-Hrd3p. Cross-linking assay was performed as in Fig. 1. (b) Hrd1p cross-linking to Hmg2p in the presence of either the hrd1Δ, ubc7Δ, or cue1Δ allele. The appropriate null alleles of each gene required for Hmg2p degradation were introduced into the strain coexpressing 1myc-Hmg2p and 3HA-Hrd1p, and the cross-linking assay was performed. (c) Hrd3p function was required for Hrd1p cross-linking to Hmg2p under normal Hrd1p expression levels. Cells expressing 1myc-Hmg2p and 3HA-Hrd1p from its native promoter and coexpressing either the wild-type HRD3 allele (wt), the hrd3Δ allele, or the truncated hrd3 allele (hrd3357–833) were subjected to the cross-linking assay. (d) Expression of Hrd3p357–833 as the only source of Hrd3p allowed stabilization of Hrd1p, but did not allow ERAD. Cycloheximide-chase assays of strains expressing 1myc-Hmg2p and either 3HA-Hrd1p or 2HA-Ubc7p and containing either the wild-type HRD3 allele, the hrd3Δ allele, or the truncated hrd3 allele. Cells were grown to mid-log phase (OD600 of 0.5), and cycloheximide was added to a final concentration of 50 μg/ml. Lysates were prepared at the indicated time points. The 9E10 anti-myc antibody was used to detect 1myc-Hmg2p. An anti-HA ascites fluid was used to detect 3HA-Hrd1p and 2HA-Ubc7p. (e) Hrd3p function was required for Ubc7p cross-linking to Hmg2p under normal Hrd1p expression levels. Cells expressing 1myc-Hmg2p, 2HA-Ubc7p, and Hrd1p from its native promoter and coexpressing either the wild-type HRD3 allele, the hrd3Δ allele, or the truncated hrd3 allele, were subjected to the cross-linking assay as in Fig. 1. (f) The N-terminal region of Hrd3p was required for Hrd3p cross-linking to Hmg2p. Cells expressing 1myc-Hmg2p and carrying the wild-type HRD3 allele, the hrd3Δ allele, or the hrd3357–833 allele were subjected to the cross-linking assay as in Fig. 3.
To determine if Hrd3p was necessary for Hrd1p cross-linking to ER proteins, we required a mutant of Hrd3p that allowed normal Hrd1p stabilization but was deficient for ERAD. Previously, we demonstrated that deletion of the first 356 residues of Hrd3p, resulting in the truncation mutant Hrd3p357–833, allowed normal Hrd1p stabilization but did not allow ERAD (27). Use of this mutant allowed us to assess the role of Hrd3p in HRD complex cross-linking to ER proteins independent of its function in Hrd1p stabilization. In fact, Hrd1p cross-linking to Hmg2p was reduced in cells that expressed the truncation mutant Hrd3p357–833 as the only source of Hrd3p (Fig. 4c), despite the normal steady-state levels of wild-type Hrd1p (27) (Fig. 4d). We also tested the effect of this ERAD-deficient hrd3 truncation allele on Ubc7p cross-linking with substrate. Despite the presence of normal Ubc7p and Hrd1p levels (Fig. 4d), Ubc7p cross-linking to Hmg2p was reduced in cells expressing only the truncation mutant Hrd3p357–833 as the only source of Hrd3p (Fig. 4e), and the effect was identical to that of the hrd3Δ allele. Furthermore, loss of the N-terminal region also resulted in greatly reduced Hrd3p cross-linking with substrate (Fig. 4f). Thus, the Hrd3p N-terminal region was required for Hrd3p-substrate association and appeared to be critical in controlling Hrd1p-Ubc7p cross-linking with substrate under normal Hrd1p steady-state levels. Accordingly, Hrd3p functions both in the maintenance of Hrd1p levels and in a separate ERAD function required to target Ubc7p and Hrd1p to the substrate. Importantly, functional disruption of Hrd1p and Hrd3p cross-linking to ERAD substrates indicated that both Hrd1p and Hrd3p cross-linking was physiologically relevant.
Hmg2p is recognized for degradation by a regulated structural change.
Both Hrd1p and Hrd3p in the HRD ubiquitin ligase complex appeared to associate indiscriminately with both stable proteins and ERAD substrates. However, the complex specifically mediated functional Ubc7p cross-linking with proteins slated for either constitutive or regulated ERAD. Thus, the Hrd1p/Hrd3p ubiquitin ligase complex discriminated ERAD substrates from stable proteins at the level of Ubc7p recruitment through some feature of the targeted protein that distinguishes it as an ERAD substrate. Considering the quality control function of the HRD complex, a reasonable criterion for ERAD substrates would be molecular hallmarks consistent with misfolded or incompletely folded proteins.
According to this model, constitutively degraded substrates would always cross-link with Ubc7p, but physiologically regulated substrates would only cross-link with Ubc7p when degradation signals were high. In fact, both the Hmg2p degradation rate and HRD-dependent Ubc7p cross-linking to Hmg2p are altered accordingly by a feedback regulatory signal generated from the conditions of cellular mevalonate pathway production (25, 31) (Fig. 1c). Because the HRD complex targets misfolded proteins for degradation, we investigated if a mevalonate pathway-regulated alteration in the Hmg2p folding state programmed its controlled ERAD by the quality control HRD complex.
We used a class of compounds known as chemical chaperones to examine the role of the Hmg2p folding state in HRD complex recognition, Ubc7p interaction, and subsequent degradation of Hmg2p. Such agents have been demonstrated to enhance the folding of various mutant and normal proteins in cell and whole-animal studies (8, 9, 73). Importantly, addition of such chemical chaperones to cells has been shown to block substrate-specific ERAD by enhancing the folding of a number of mutant, misfolded proteins. For example, addition of the chemical chaperone glycerol to mammalian cells enhances the folding of the cystic fibrosis transmembrane conductance regulator (CFTR) ΔF508 mutant, which is normally retained in the ER and destroyed by ERAD (82), thereby allowing passage of the mutant CFTR through the secretory pathway, restoring normal CFTR-mediated chloride ion transport into the cell (8, 73). Similarly, addition of the chemical chaperone 4-phenylbutyric acid to mammalian cells enhances folding and secretion of the PiZ isoform of α1-antitrypsin (10), which is also normally retained in the ER and subject to ERAD (13).
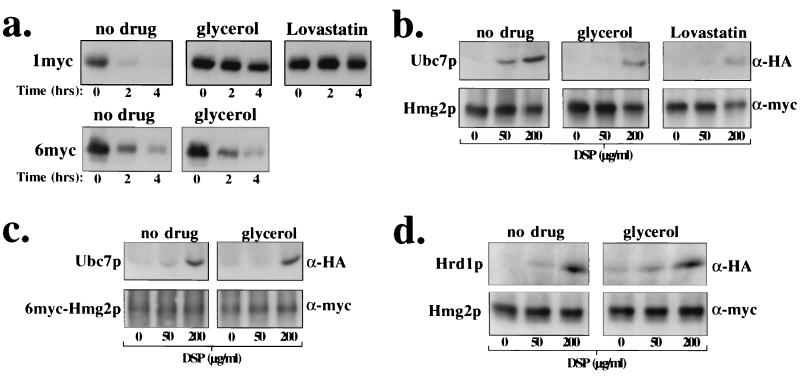
We first determined if addition of the chemical chaperone glycerol to yeast cells could stabilize the normally regulated fluorescence reporter protein Hmg2p-GFP (15, 25, 33). When cells expressing Hmg2p-GFP were grown in 10% glycerol, the Hmg2p-GFP steady-state level was significantly elevated, which was observed as a rightward shift in the fluorescence histogram of glycerol-treated cells compared to untreated cells (Fig. 5a). Such an increase in Hmg2p-GFP steady-state level is normally indicative of Hmg2p-GFP stabilization (15, 25, 26, 33). In contrast, identical treatment of a strain expressing the extremely stable Hmg1p-GFP resulted in only a minor alteration in cellular fluorescence in either case (Fig. 6a). The increase in Hmg2p-GFP steady-state levels in glycerol-treated cells was identical to that in cells treated with lovastatin (Fig. 5a). Importantly, glycerol did not alter production of downstream mevalonate pathway products (Fig. 5b). Furthermore, addition of lovastatin to glycerol-treated cells had no added effect on the glycerol-induced increase in Hmg2p-GFP steady-state levels (unpublished observations). Not only did incubation of cells with glycerol increase Hmg2p-GFP steady-state levels, but the normal, zaragozic acid-induced reduction in Hmg2p-GFP steady-state levels was also completely blocked by addition of glycerol at the same time as zaragozic acid (Fig. 5c). Zaragozic acid is an inhibitor of the downstream enzyme squalene synthase and results in enhanced Hmg2p ubiquitination and degradation through build-up of the pathway molecule farnesyl diphosphate (25, 34).
FIG. 5.
Chemical chaperone glycerol increased the steady-state level of Hmg2p and blocked the degradation-enhancing effect of zaragozic acid. (a) Incubation of cells with glycerol increased the steady-state levels of Hmg2p-GFP. Cells expressing the indicated HMGR-GFP variant were grown in minimal medium to mid-log phase in the presence or absence of glycerol (final concentration, 10%). Lovastatin (final concentration, 25 μg/ml) was added to the indicated sample, and all samples were incubated at 30°C for an additional 4 h. Steady-state GFP fluorescence was analyzed by flow cytometry. (b) Glycerol did not affect mevalonate pathway production. TLC analysis of the nonsaponifiable lipid fraction from yeast cells treated with no drug (nd), 10% glycerol (G), or 50 μg of lovastatin (L) per ml. Cells were incubated with [14C]acetate in the presence of the indicated drug for 6 h. Lipids were extracted and gently saponified, and the nonsaponifiable lipid fraction was extracted and applied to a TLC plate. Migration of standards is marked on the right. (c) Glycerol blunted the degradation-enhancing effect of zaragozic acid. Cells expressing Hmg2p-GFP were grown in minimal medium to mid-log phase. Zaragozic acid (ZA; final concentration, 10 μg/ml) and/or glycerol (final concentration, 10%) was added to the indicated samples, which were incubated at 30°C for an additional 4 h. Steady-state GFP fluorescence was analyzed by flow cytometry.
FIG. 6.
Chemical chaperone glycerol stabilized Hmg2p and reduced Ubc7p cross-linking to Hmg2p. (a) Glycerol stabilized 1myc-Hmg2p but not 6myc-Hmg2p. Cells expressing either 1myc-Hmg2p or 6myc-Hmg2p were grown in minimal medium to mid-log phase. Glycerol (final concentration, 15%) or lovastatin (lovastatin; final concentration, 25 μg/ml) was added to the indicated samples, and the cells were incubated at 30°C for 4 h. Cells were then subjected to a cycloheximide-chase assay. Lysates were prepared at the indicated time points after cycloheximide addition and immunoblotted with the 9E10 antibody to assess the levels of myc-HMGR. (b and c) Glycerol blocked Ubc7p cross-linking to Hmg2p but not 6myc-Hmg2p. Cells coexpressing either 1myc-Hmg2p or 6myc-Hmg2p and 2HA-Ubc7p were grown in minimal medium to mid-log phase. Cells were treated with either no drug, 15% glycerol, or lovastatin (25 mg/ml) for 2 h at 30°C. A similar cross-linking assay was performed as in Fig. 1. (d) Glycerol did not block Hrd1p cross-linking to Hmg2p. Cells coexpressing 1myc-Hmg2p and 3HA-Hrd1p were grown in minimal media to mid-log phase. Cells were treated with either no drug or 15% glycerol for 2 h at 30°C. A similar cross-linking assay was performed as in Fig. 4.
Glycerol is most effective at stabilizing proteins with point mutations that result in misfolding of the mutant protein. Therefore, we did not expect glycerol to have much effect on 6myc-Hmg2pΔ63–219, a grossly misfolded mutant of Hmg2p that is missing 157 residues of the native Hmg2p sequence (32), including a putative transmembrane span. This large deletion of Hmg2p sequence would prevent 6myc-Hmg2pΔ63–219 from ever achieving a normally folded sequence, and consistent with this, addition of glycerol did not significantly affect the steady-state level of 6myc-Hmg2pΔ63–219 (Fig. 5a, right panel).
It was likely that glycerol's effect occurred through stabilization of Hmg2p, possibly by enhancing Hmg2p folding. Therefore, we directly determined the effect of glycerol on the degradation of regulated 1myc-Hmg2p and grossly misfolded 6myc-Hmg2pΔ63–219 by the cycloheximide-chase assay. Incubation of cells in glycerol resulted in stabilization of 1myc-Hmg2p to a similar extent as incubation of cells with lovastatin (Fig. 6a). In a similar experiment, the constitutive degradation of 6myc-Hmg2pΔ63–219 was unaltered by incubation of cells with glycerol (Fig. 6a), consistent with its severely perturbed structure (26, 32).
Finally, we examined the effect of glycerol in the cross-linking between Ubc7p and Hmg2p. When cells were incubated with glycerol, Ubc7p cross-linking to Hmg2p was reduced, and the effect was similar to incubation of cells with lovastatin (Fig. 6b). This was not the result of decreased cross-linking efficiency for Ubc7p, as Ubc7p cross-linked similarly to 6myc-Hmg2pΔ63–219 in both the presence and absence of glycerol (Fig. 6c). This was also not a result of decreased Ubc7p stability or membrane association (unpublished observations), nor was it a result of decreased Hrd1p cross-linking to substrate, as Hrd1p cross-linked to Hmg2p identically in the presence and absence of glycerol (Fig. 6d). Thus, enhanced folding of Hmg2p specifically blocked functional HRD-mediated proximity between Ubc7p and Hmg2p and was without effect on HRD complex association with substrate.
These results supported a model in which the HRD complex detects proteins with appropriate structural features and promotes functional proximity between Ubc7p and the substrate to initiate ERAD. The glycerol experiments implied that the Hmg2p structure was different between the stable state and the degradation-competent state, with the stable state having a more completely folded structure. To determine if Hmg2p did have a dynamic structure, we examined the sensitivity of the Hmg2p structure to exogenously added proteases under different mevalonate pathway conditions. A similar assay has been employed successfully to evaluate the structural differences between the wild-type form of CFTR and the rapidly degraded CFTR ΔF508 mutant (91). We probed the protease sensitivity of Hmg2p in isolated microsomes from yeast cells treated with either no drug, stabilizing lovastatin, or the degradation-enhancing zaragozic acid.
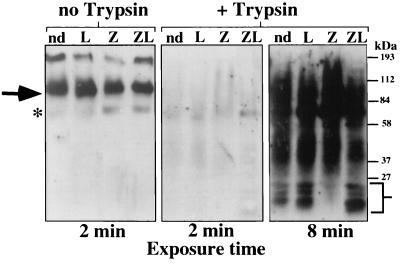
A noticeable and reproducible difference was observed in the proteolytic sensitivity of Hmg2p from the zaragozic acid-treated cells compared to the no-drug cells and the lovastatin-treated cells (Fig. 7, right panels, lanes Z versus nd and L). The complete absence of lower-molecular-weight fragments in the zaragozic acid-treated cells suggested that Hmg2p from zaragozic acid-treated cells had a different structure that altered its sensitivity to exogenously added proteases. Furthermore, the reduced presence of these fragments in the untreated sample correlated with its intermediate rate of degradation. The change in Hmg2p proteolytic sensitivity seen with the zaragozic acid-treated cells was blocked by incubation of these cells in lovastatin prior to addition of zaragozic acid (Fig. 7, right panels, lane ZL), indicating that the effect was regulated by upstream blockade of the mevalonate pathway. Thus, in a manner consistent with the mevalonate pathway's regulation of Hmg2p ubiquitination and degradation, Hmg2p had a differential sensitivity to exogenously added proteases that depended on production of downstream molecules of the mevalonate pathway. These results, and those with the chemical chaperone, strongly suggest that Hmg2p undergoes a mevalonate pathway-regulated structural transition that allows it to be recognized by the HRD complex as a degradation substrate.
FIG. 7.
Hmg2p transmembrane domain underwent structural transition from stable to degradation-competent state. Limited proteolytic analysis of intact microsomes containing 1myc-Hmg2p revealed an altered sensitivity to trypsin when cells were incubated with zaragozic acid. Cells expressing 1myc-Hmg2p were grown to mid-log phase in minimal medium. The cells were treated with either no drug (nd), lovastatin (L; 100 μg/ml, final concentration) for 30 min, zaragozic acid (Z; 10 μg/ml, final concentration) for 15 min, or both lovastatin and zaragozic acid (ZL) for 30 min at 30°C. These treatments did not appreciably alter the steady-state level of Hmg2p during the period of treatment (unpublished observations). Intact, isolated microsomes were prepared from these cells. Separate aliquots of isolated microsomes were treated with trypsin (0.5 μg/ml) for 30 min at 0°C in the absence of detergent. The degree of protease digestion was observed by immunoblotting with a polyclonal antiserum specific for Hmg2p sequences located in both the lumen and cytosol. All samples had similar amounts of undigested Hmg2p in the microsomes prior to treatment with trypsin (no-trypsin lanes, left panel). All total-protein loads were identical in each lane, as assessed from India ink staining of the Western blot (data not shown). Full-length Hmg2p is indicated with an arrow. Lower-molecular-weight bands are indicated with the brackets. The asterisk indicates a nonspecific band that cross-reacts with the anti-Hmg2p antiserum.
DISCUSSION
The ER-associated proteins Hrd1p and Hrd3p function as a RING-H2 ubiquitin ligase complex required for the targeting and ubiquitination of ER-associated, proteasome-dependent degradation substrates (3, 27), such as the ER lumenal protein CPY* (6, 67) and the ER membrane protein Hmg2p (27, 32). A current model for RING-H2 ubiquitin ligase action is that the ubiquitin ligase mediates direct association between the substrate and the appropriate ubiquitin-conjugating enzyme, allowing the specific transfer of ubiquitin to the target protein. From our studies, we demonstrate that the HRD complex does mediate functional association between the ER-associated ubiquitin-conjugating enzyme Ubc7p and ERAD substrates through a unique substrate-targeting mechanism, which may act in an inspection process to target ERAD substrates for ubiquitination and subsequent proteasome-dependent degradation.
Mechanism of substrate selection: a scanning mechanism for the HRD complex.
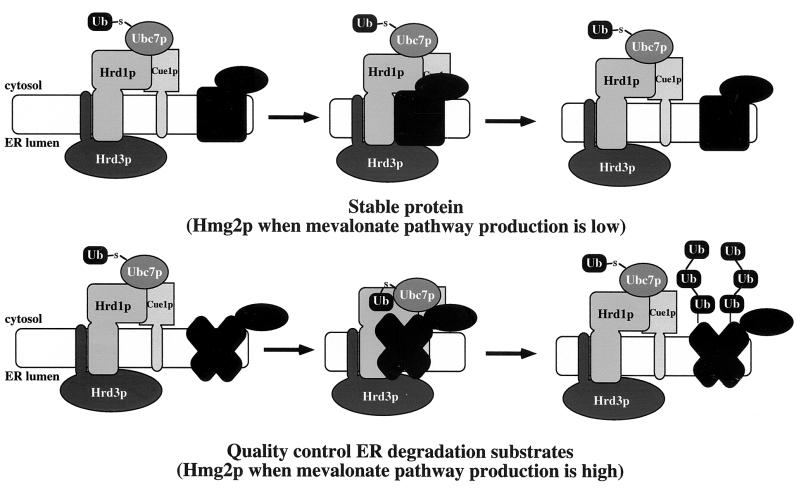
Ubiquitin ligases must interact with substrates as part of the targeting mechanism. In the cases of other characterized ubiquitin ligases, specific sequence motifs in the substrate serve as key specificity determinants for ubiquitin ligase binding and subsequent ubiquitination (1, 2, 19, 23, 41, 42, 47, 53, 54, 57, 62, 63, 76, 77, 90). Unlike other ubiquitin ligase proteins and associated components, however, both Hrd1p and Hrd3p associated with stable proteins in addition to degradation substrates (Fig. 8). Thus, we suggest that the Hrd1p/Hrd3p complex “scans” the ER for candidate substrates, querying all accessible proteins and associating with stable proteins and degradation substrates alike. When a bound protein meets the criteria for ERAD, the HRD complex mediates a functional association between the substrate and the ER-associated ubiquitin conjugating enzyme Ubc7p, thereby allowing direct transfer of ubiquitin from Ubc7p to the substrate (Fig. 8). Substrate specificity lies not in substrate-HRD complex association, but in the subsequent HRD-dependent functional association of the ubiquitin-conjugating enzyme with the substrate. A reasonable criterion for the HRD complex to proceed with functional association of Ubc7p with a substrate to allow substrate ubiquitination would be recognition of hallmarks of misfolding. By this model, the difference between a degradation substrate and a stable protein would be the exposure of such hallmarks by the degradation substrate but not the stable protein. In this regard, the soluble Hrd1p C-terminal domain, which contains the RING-H2 motif, does preferentially program the ubiquitination of a misfolded protein in an in vitro ubiquitination assay (3), indicating that Hrd1p might participate directly in some aspect of quality control recognition.
FIG. 8.
Model for HRD ubiquitin ligase complex association with Hmg2p and mechanism of mevalonate pathway-regulated ERAD. (a) Stable proteins associate with Hrd1p/Hrd3p core complex, but the structure of the stable protein does not allow functional association with Ubc7p. Hmg2p stabilized as a result of low mevalonate pathway production would have similar interaction kinetics with the HRD ubiquitin ligase complex. (b) ERAD substrates associate with Hrd1p/Hrd3p core complex, and their misfolded or mutant structures allow functional association with Ubc7p mediated by the Hrd1p/Hrd3p complex. A structural change within Hmg2p as a result of abundant production of downstream molecules in the mevalonate pathway would result in Hmg2p's acquisition of a similar “misfolded” state as other ERAD substrates.
Our model of HRD ubiquitin ligase action also allowed for a simple mechanism for the sterol pathway regulation of HMGR degradation by the apparent quality control HRD complex. Whereas misfolded degraded substrates always expose quality control hallmarks and are constitutively recognized by the HRD complex, HMGR would only expose similar hallmarks when degradation signals from the sterol pathway were high. Thus, regulated degradation of HMGR would occur through a controlled structural transition from a stable protein to a quality control substrate (Fig. 8). Consistent with this, the chemical chaperone glycerol, which stabilizes some ER-associated misfolded proteins such as CFTR (73), blocked the degradation of Hmg2p but not the degradation of the grossly misfolded 6myc-Hmg2p. Furthermore, glycerol blocked Ubc7p cross-linking to Hmg2p but not 6myc-Hmg2p. Glycerol addition had no effect on HRD complex association with Hmg2p, nor did any of the mevalonate pathway inhibitors that stabilized Hmg2p. In fact, Hmg2p did appear to undergo a mevalonate pathway structural transition, as measured by protease sensitivity. Though this structural transition could be the prelude to exposure of specific degrons, we do not think that is likely based on the fact that exhaustive mutational analysis of the entire Hmg2p transmembrane sequence did not reveal any signature sequences indicative of a degron, but did demonstrate that structural information was required for Hmg2p degradation (26). Taken together, these observations imply that regulation of Hmg2p degradation was not due to altered association of Hmg2p with the HRD complex. Rather, HRD-dependent Hmg2p degradation occurred through a regulated structural transition of the Hmg2p transmembrane domain from a stable state to one that resembled a quality control substrate, allowing the HRD complex to proceed with functional Ubc7p-Hmg2p association and subsequent Hmg2p ubiquitination.
It is quite possible that this substrate-scanning mechanism for the HRD ubiquitin ligase complex is restricted to ER membrane proteins and a different mechanism is employed for lumenal ERAD substrates, as our experiments were limited to membrane-associated ERAD substrates. Consistent with this, the protein Der1p is required for ERAD of the lumenal CPY* (50), but is not utilized for ERAD of the membrane protein Hmg2p (unpublished observations). Thus, a different targeting mechanism for lumenal substrates by the HRD complex may be employed, and interactions between the various HRD complex components and lumenal ERAD substrates should be examined. However, a unique set of lumenal substrates similar to that which we have employed in this study—a regulated ERAD substrate, a constitutive ERAD substrate, and a stable protein not involved in ER quality control—is not available for lumenal proteins. A well-characterized, constitutively degraded, lumenal ERAD substrate does exist in CPY* (6, 20, 37, 67), and we are initiating similar studies to determine CPY*-HRD complex interactions. Without a regulated lumenal ERAD substrate and a stable lumenal protein not involved in ER quality control, it will be difficult to extend the studies that we have reported here.
Mechanism(s) of Ubc7p recruitment.
The fact that Ubc7p cross-linked only to degradation substrates and not to stable proteins leads to a few possible models for Ubc7p recruitment to the substrate by the Hrd1p/Hrd3p complex. First, Ubc7p may not be constitutively associated with the Hrd1p/Hrd3p core complex and may only be recruited to join the complex when a degradation substrate is bound by Hrd1p and Hrd3p, possibly as the result of an allosteric alteration in the Hrd1p/Hrd3p complex induced only by degradation substrate binding. Alternatively, Ubc7p may be constitutively associated with the core Hrd1p/Hrd3p complex, but is not in the correct position to effect ubiquitination of a bound protein. Again, only upon the binding of a degradation substrate by the Hrd1p and Hrd3p would the complex undergo an allosteric alteration that subsequently brings Ubc7p into a correct orientation with the substrate to effect substrate ubiquitination. Lastly, Ubc7p may be constitutively associated with the Hrd1p/Hrd3p core complex in a fixed position conducive to effect substrate ubiquitination so that the complex need not undergo any structural alteration. Instead, the complex distinguishes degradation substrates from stable proteins by the misfolded structure of the degradation substrate that allows Ubc7p direct access to the bound protein's lysine residues, which are used as the sites for the covalent attachment of ubiquitin (21, 46). Only those proteins to be ubiquitinated and degraded would have lysine residues accessible to Ubc7p.
In this regard, our studies delineating the sequence determinants required for Hmg2p-regulated degradation may be instructive. HRD-dependent ubiquitination of Hmg2p requires two distinct lysine residues, Lys6 and Lys357, which appear to be specifically involved in regulation of Hmg2p stability and may serve as the ubiquitination sites within Hmg2p (26). Perhaps the regulated Hmg2p structural transition brings about repositioning of Lys6 and Lys357, allowing for their close proximity to Ubc7p when Hmg2p is engaged by the HRD complex, which would result in their subsequent ubiquitination. Alternatively, Lys6 and Lys357 may act directly to promote global structural transitions of Hmg2p in response to signals from the mevalonate pathway. In this case, Lys6 and Lys357 would not be the recipients of Ubc7p-dependent ubiquitination, but together would be intimately involved in the structural changes that bring other lysine residues in close proximity to Ubc7p when Hmg2p is engaged by the HRD complex. Resolving the details of the regulated Hmg2p structural transition and the general ERAD scanning mechanism is a central concern of our ongoing work and may provide the key to understanding the mode of HRD complex-dependent Ubc7p recruitment to the substrate.
In addition to Ubc7p, Ubc1p has recently been shown to be directed by Hrd1p in vivo to program the ubiquitination of both CPY* and Hmg2p (3, 22). This suggests that the HRD complex also mediates an interaction between ERAD substrates and Ubc1p, although this has yet to be tested.
Role of each component of the complex in substrate binding.
Consistent with its role as a ubiquitin ligase (3), Hrd1p was required for Ubc7p cross-linking to ERAD substrates. The cytosolic RING-H2 domain of Hrd1p mediates direct recruitment of Ubc7p, as this domain is required for direct, in vivo and in vitro interaction between Hrd1p and Ubc7p (3, 16). The RING-H2 motif similarly binds ubiquitin-conjugating enzymes in other homologous ubiquitin ligases (47, 57), and the crystal structure between cCbl and UbcH7 demonstrated that the cCbl RING-H2 motif mediates direct interaction between the ubiquitin ligase and the ubiquitin-conjugating enzyme (92). Hrd1p directly binds Ubc7p (3, 16) and directs Ubc7p proximity to substrates, so it is likely that Hrd1p contains some substrate-binding activity. Because Hrd1p directs ubiquitination of only ERAD substrates, it must have a way of distinguishing between the degradation substrates and the stable proteins that it interacts with. It is possible that Hrd1p may engage all proteins with its transmembrane domain but only binds ERAD substrates with its RING-H2 domain, thus bringing Ubc7p in proximity to the degradation substrate. In fact, the cytosolic domain of Hrd1p has a preference for misfolded proteins in an in vitro ubiquitination assay (3), possibly indicating that the Hrd1p cytosolic domain possesses a component of substrate recognition. Hrd1p functions in in vivo ERAD in the absence of Hrd3p when expressed at sufficiently high levels (27, 67), further supporting the idea of a substrate recognition component within Hrd1p.
To interact with such a diverse array of proteins, the complex must have a general binding mechanism. We cannot rule out that the complex recognizes specific sequences in each target degradation substrate, such as degrons (81), but no consensus sequence has been revealed through our mutagenic or computer analyses (26; unpublished observations). In fact, our recent analysis of the Hmg2p sequence (26) and our studies here indicate that the complex recognizes structural cues within Hmg2p rather than specific sequences. To allow recognition of common structural features among a variety of proteins, perhaps a protein in the complex has a substrate-binding site with binding affinities similar to those of a chaperone. It is quite possible that the initial substrate-scanning site is in the lumen of the ER, where both lumenal proteins and ER membrane proteins would exist. Hrd3p is the only protein of the established ER ubiquitin ligase complex that is almost entirely lumenal (27, 67, 71) and, from our cross-linking data, the only protein that interacted with all tested substrates in vivo in the absence of the other complex components when present at normal steady-state levels. These observations suggest that Hrd3p may serve a primary role in initial substrate recognition.
Hrd3p may also function directly to control Hrd1p RING-H2 ubiquitin ligase activity (27), which promotes proximity between Ubc7p and the substrate, thereby indirectly controlling Ubc7p ubiquitin-conjugating activity. In support of this, a completely lumenal version of Hrd3p functions normally in ERAD (27), indicating that the action of Hrd3p to program the cytosolic ubiquitination of substrates occurs through a lumen-to-cytosol signaling process. Because Hrd3p interacts directly with the Hrd1p transmembrane domain (27), Hrd3p action to allow cytosolic ERAD events is likely transduced through the Hrd1p transmembrane domain. Thus, a defunct signaling process, through either elimination of Hrd1p or loss of Hrd3p function, may prevent Ubc7p association with substrates. In fact, elimination of Hrd1p did prevent Ubc7p cross-linking to substrates. Loss of Hrd3p function, by expression of only the Hrd3p N-terminal truncation mutant, also prevented Ubc7p cross-linking to substrates despite maintenance of normal Hrd1p levels (27). Additionally, although Hrd1p was maintained at normal levels by the Hrd3p truncation mutant, the Hrd3p truncation mutant did not allow Hrd1p cross-linking to substrates. This was most likely the result of an inability of the Hrd3p truncation mutant itself to associate with substrates and is consistent with the Hrd3p N-terminal region's functioning in the initial substrate recognition of the complex. Perhaps the initial binding of substrate with Hrd3p induces allosteric changes in the complex that result in the activation of Hrd1p ubiquitin ligase function. These allosteric changes would initiate correct temporal and spatial activation of Hrd1p RING-H2 ubiquitin ligase function and would promote physical association between ERAD substrates and Hrd1p and Ubc7p, leading to ubiquitination of the bound substrate. It may be that the truncated Hrd3p maintains Hrd1p in an inactive conformation, preventing any functional association of Hrd1p and Ubc7p with substrate. However, the deletion of such a large portion of Hrd3p may result in an aberrant structure of the remaining part of Hrd3p, possibly confounding this interpretation.
The action of the HRD ubiquitin ligase complex involves coordination of processes and information on both sides of the ER membrane (27). One likely model for this coordination is that degradation proceeds by interaction of the Hrd1p/Hrd3p complex with a substrate engaged by the Sec61p retrotranslocation complex, which is required for ER degradation (94). The combined interaction of both Hrd3p and Hrd1p with the Sec61p-engaged substrate would allow the Hrd1p RING-H2 domain to promote Ubc7p association with substrate, resulting in subsequent ubiquitination of the substrate. Further exploration of the mechanism by which the HRD ubiquitin ligase complex operates on both sides of a membrane will be important in understanding specific aspects of ERAD and processes that employ ubiquitination as a mode of transmembrane signal transduction.
Implications of Hmg2p interaction with the HRD complex.
The structural transition mechanism for Hmg2p-regulated degradation has several important implications. It has recently been demonstrated that regulated degradation of mammalian HMGR is mediated by ubiquitination (69), in a manner very similar to Hmg2p (34). Although some differences exist in the molecular details of the two systems, it is very likely that the same mechanism of signal-induced transition to a quality control substrate underlies this important axis of sterol synthesis regulation. At present, it is not clear if the ability to undergo this structural transition is an autonomous feature of the HMGR molecule or if it is mediated by ancillary machinery that brings the transition about. In either case, however, it is possible that small molecules could be designed or discovered to drive the transition in a specific and clinically useful manner. More generally, as quality control degradation systems likely exist in the cytosol, and quite possibly the nucleus or mitochondria, it may be that entry of substrates into these systems occurs by similar mechanisms and could be harnessed in much the same fashion as the ER quality control degradation apparatus. The observation of a regulated transition to a quality control substrate allows the framing of both basic and clinical lines of inquiry to examine the utility and biological generality of this mode of protein regulation.
ACKNOWLEDGMENTS
We thank Merck for the generous gifts of zaragozic acid, L-659,699, and lovastatin. R.G.G. thanks C. Melissa Morelli for essential technical support required to achieve strong signal intensity despite limited application of reagents. R.Y.H. thanks J. Theriot for opening unexpected conceptual doors.
This work was supported by NIH grant DK5199601 (R.Y.H.) and a Searle Scholarship (R.Y.H.).
REFERENCES
- 1.Baker R T, Varshavsky A. Inhibition of the N-end rule pathway in living cells. Proc Natl Acad Sci USA. 1991;88:1090–1094. doi: 10.1073/pnas.88.4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel B, Wünning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bays N W, Gardner R G, Seelig L P, Joazeiro C A, Hampton R Y. Hrd1p is a membrane-anchored ubiquitin ligase required for endoplasmic reticulum-associated degradation. Nat Cell Biol. 2000;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 4.Biederer T, Volkwein C, Sommer T. Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J. 1996;15:2069–2076. [PMC free article] [PubMed] [Google Scholar]
- 5.Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 6.Bordallo J, Plemper R K, Finger A, Wolf D H. Der3p-Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bordallo J, Wolf D H. A RING-H2 finger motif is essential for the function of Der3/Hrd1 in endoplasmic reticulum associated protein degradation in the yeast Saccharomyces cerevisiae. FEBS Lett. 1999;448:244–248. doi: 10.1016/s0014-5793(99)00362-2. [DOI] [PubMed] [Google Scholar]
- 8.Brown C R, Hong-Brown L Q, Biwersi J, Verkman A S, Welch W J. Chemical chaperones correct the mutant phenotype of the delta F508 cystic fibrosis transmembrane conductance regulator protein. Cell Stress Chaperones. 1996;1:117–125. doi: 10.1379/1466-1268(1996)001<0117:ccctmp>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown C R, Hong-Brown L Q, Welch W J. Correcting temperature-sensitive protein folding defects. J Clin Investig. 1997;99:1432–1444. doi: 10.1172/JCI119302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burrows J A, Willis L K, Perlmutter D H. Chemical chaperones mediate increased secretion of mutant alpha 1-antitrypsin (alpha 1-AT) Z: a potential pharmacological strategy for prevention of liver injury and emphysema in alpha 1-AT deficiency. Proc Natl Acad Sci USA. 2000;97:1796–1801. doi: 10.1073/pnas.97.4.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chau V, Tobias J W, Bachmair A, Marriott D, Ecker D J, Gonda D K, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 12.Chun K T, Bar-Nun S, Simoni R D. The regulated degradation of 3-hydroxy-3-methylglutaryl-CoA reductase requires a short-lived protein and occurs in the endoplasmic reticulum. J Biol Chem. 1990;265:22004–22010. [PubMed] [Google Scholar]
- 13.Ciccarelli E, Alonso M A, Cresteil D, Bollen A, Jacobs P, Alvarez F. Intracellular retention and degradation of human mutant variant of A alpha-1 antitrypsin in stably transfected Chinese hamster ovary cell lines. Eur J Biochem. 1993;213:271–276. doi: 10.1111/j.1432-1033.1993.tb17759.x. [DOI] [PubMed] [Google Scholar]
- 14.Cook W J, Jeffrey L C, Kasperek E, Pickart C M. Structure of tetraubiquitin shows how multiubiquitin chains can be formed. J Mol Biol. 1994;236:601–609. doi: 10.1006/jmbi.1994.1169. [DOI] [PubMed] [Google Scholar]
- 15.Cronin S R, Khoury A, Ferry D K, Hampton R Y. Regulation of HMG-CoA reductase degradation requires the P-type ATPase Cod1p/Spf1p. J Cell Biol. 2000;148:915–924. doi: 10.1083/jcb.148.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deak P M, Wolf D H. Membrane topology and function of der3/hrd1p as a ubiquitin-protein ligase (e3) involved in endoplasmic reticulum degradation. J Biol Chem. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- 17.Edwards P A, Lan S F, Tanaka R D, Fogelman A M. Mevalonolactone inhibits the rate of synthesis and enhances the rate of degradation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in rat hepatocytes. J Biol Chem. 1983;258:7272–7275. [PubMed] [Google Scholar]
- 18.Fang S, Jensen J P, Ludwig R L, Vousden K H, Weissman A M. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 19.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p-Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 20.Finger A, Knop M, Wolf D H. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- 21.Finley D, Chau V. Ubiquitination. Annu Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- 22.Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol. 2000;2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- 23.Galan J M, Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci USA. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner R, Cronin S, Leader B, Rine J, Hampton R. Sequence determinants for regulated degradation of yeast 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1998;9:2611–2626. doi: 10.1091/mbc.9.9.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardner R G, Hampton R Y. A highly conserved signal controls degradation of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase in eukaryotes. J Biol Chem. 1999;274:31671–31678. doi: 10.1074/jbc.274.44.31671. [DOI] [PubMed] [Google Scholar]
- 26.Gardner R G, Hampton R Y. A ‘distributed degron’ allows regulated entry into the ER degradation pathway. EMBO J. 1999;18:5994–6004. doi: 10.1093/emboj/18.21.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner R G, Swarbrick G M, Bays N W, Cronin S, Wilhovsky S, Seelig L, Kim C, Hampton R Y. Endoplasmic reticulum degradation requires lumen to cytosol signaling: transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardner R G, Hampton R Y. An oxysterol-derived signal for 3-hydroxy-3-methylglutaryl CoA reductase degradation in yeast. J Biol Chem. 2000;276:8681–8694. doi: 10.1074/jbc.M007888200. [DOI] [PubMed] [Google Scholar]
- 29.Goldstein J L, Brown M S. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 30.Gonda D K, Bachmair A, Wünning I, Tobias J W, Lane W S, Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- 31.Hampton R Y, Rine J. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J Cell Biol. 1994;125:299–312. doi: 10.1083/jcb.125.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hampton R Y, Gardner R G, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hampton R Y, Koning A, Wright R, Rine J. In vivo examination of membrane protein localization and degradation with green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:828–833. doi: 10.1073/pnas.93.2.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hampton R Y, Bhakta H. Ubiquitin-mediated regulation of 3-hydroxy-3-methylglutaryl-CoA reductase. Proc Natl Acad Sci USA. 1997;94:12944–12948. doi: 10.1073/pnas.94.24.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hein C, Springael J Y, Volland C, Haguenauer-Tsapis R, André B. NP11, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 36.Hill K, Cooper A A. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 2000;19:550–561. doi: 10.1093/emboj/19.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiller M M, Finger A, Schweiger M, Wolf D H. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 38.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 39.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 40.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 41.Honda R, Yasuda H. Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, towards p53 or itself is dependent on the RING finger domain of the ligase. Oncogene. 2000;19:1473–1476. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- 43.Huibregtse J M, Scheffner M, Howley P M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A L, Riordan J R. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 46.Jentsch S. The ubiquitin-conjugation system. Annu Rev Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- 47.Joazeiro C A P, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y. The tyrosine kinase negative regulator c-CBL as a RING-type E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 48.Kalies K U, Rapoport T A, Hartmann E. The beta subunit of the Sec61 complex facilitates cotranslational protein transport and interacts with the signal peptidase during translocation. J Cell Biol. 1998;141:887–894. doi: 10.1083/jcb.141.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamura T, Koepp D M, Conrad M N, Skowyra D, Moreland R J, Iliopoulos O, Lane W S, Kaelin W G, Jr, Elledge S J, Conaway R C, Harper J W, Conaway J W. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 50.Knop M, Finger A, Braun T, Hellmuth K, Wolf D H. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 1996;15:753–763. [PMC free article] [PubMed] [Google Scholar]
- 51.Kopito R R. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 52.Kramer K M, Fesquet D, Johnson A L, Johnston L H. Budding yeast RSI1-APC2, a novel gene necessary for initiation of anaphase, encodes an APC subunit. EMBO J. 1998;17:498–506. doi: 10.1093/emboj/17.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leverson J D, Joazeiro C A P, Page A M, Huang H, Hieter P, Hunter T. The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol Biol Cell. 2000;11:2315–2325. doi: 10.1091/mbc.11.7.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levkowitz G, Waterman H, Ettenberg S A, Katz M, Tsygankov A Y, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 55.Liao W, Yeung S-C, Chan L. Proteasome-mediated degradation of apolipoprotein B targets both nascent peptides cotranslationally before translocation and full-length apolipoprotein B after translocation into the endoplasmic reticulum. J Biol Chem. 1998;273:27225–27230. doi: 10.1074/jbc.273.42.27225. [DOI] [PubMed] [Google Scholar]
- 56.Lomant A J, Fairbanks G. Chemical probes of extended biological structures: synthesis and properties of the cleavable protein crosslinking reagent [35S]dithiobis(succinimidyl proprionate) J Mol Biol. 1976;104:243–261. doi: 10.1016/0022-2836(76)90011-5. [DOI] [PubMed] [Google Scholar]
- 57.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mothes W, Prehn S, Rapoport T A. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO J. 1994;13:3973–3982. doi: 10.1002/j.1460-2075.1994.tb06713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Müsch A, Wiedmann M, Rapoport T A. Yeast Sec proteins interact with polypeptides traversing the endoplasmic reticulum membrane. Cell. 1992;69:343–352. doi: 10.1016/0092-8674(92)90414-8. [DOI] [PubMed] [Google Scholar]
- 60.Nakanishi M, Goldstein J L, Brown M S. Multivalent control of 3-hydroxy-3-methylglutaryl coenzyme A reductase: mevalonate-derived product inhibits translation of mRNA and accelerates degradation of enzyme. J Biol Chem. 1988;263:8929–8937. [PubMed] [Google Scholar]
- 61.Nuber U, Schwarz S E, Scheffner M. The ubiquitin-protein ligase E6-associated protein (E6-AP) serves as its own substrate. Eur J Biochem. 1998;254:643–649. doi: 10.1046/j.1432-1327.1998.2540643.x. [DOI] [PubMed] [Google Scholar]
- 62.Ohta T, Michel J J, Schottelius A J, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 63.Patton E E, Willems A R, Sa D, Kuras L, Thomas D, Craig K L, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peters J M. Subunits and substrates of the anaphase-promoting complex. Exp Cell Res. 1999;248:339–349. doi: 10.1006/excr.1999.4443. [DOI] [PubMed] [Google Scholar]
- 65.Pilon M, Römisch K, Quach D, Schekman R. Sec61p serves multiple roles in secretory precursor binding and translocation into the endoplasmic reticulum membrane. Mol Biol Cell. 1998;9:3455–3473. doi: 10.1091/mbc.9.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Plemper R K, Egner R, Kuchler K, Wolf D H. Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J Biol Chem. 1998;273:32848–32856. doi: 10.1074/jbc.273.49.32848. [DOI] [PubMed] [Google Scholar]
- 67.Plemper R K, Bordallo J, Deak P M, Taxis C, Hitt R, Wolf D H. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J Cell Sci. 1999;112:4123–4134. doi: 10.1242/jcs.112.22.4123. [DOI] [PubMed] [Google Scholar]
- 68.Qu D, Teckman J H, Omura S, Perlmutter D H. Degradation of a mutant secretory protein, alpha-1-antitrypsin Z, in the endoplasmic reticulum requires proteasome activity. J Biol Chem. 1996;271:22791–22795. doi: 10.1074/jbc.271.37.22791. [DOI] [PubMed] [Google Scholar]
- 69.Ravid T, Doolman R, Avner R, Harats D, Roitelman J. The ubiquitin-proteasome pathway mediates the regulated degradation of mammalian HMG-CoA reductase. J Biol Chem. 2000;275:35840–35847. doi: 10.1074/jbc.M004793200. [DOI] [PubMed] [Google Scholar]
- 70.Reiss Y, Kaim D, Hershko A. Specificity of binding of NH2-terminal residue of proteins to ubiquitin-protein ligase. Use of amino acid derivatives to characterize specific binding sites. J Biol Chem. 1988;263:2693–2698. [PubMed] [Google Scholar]
- 71.Saito Y, Yamanushi T, Oka T, Nakano A. Identification of SEC12, SED4, truncated SEC16, and EKS1/HRD3 as multicopy suppressors of ts mutants of Sar1 GTPase. J Biochem. 1999;125:130–137. doi: 10.1093/oxfordjournals.jbchem.a022249. [DOI] [PubMed] [Google Scholar]
- 72.Sanders S L, Whitfield K M, Vogel J P, Rose M D, Schekman R W. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell. 1992;69:353–365. doi: 10.1016/0092-8674(92)90415-9. [DOI] [PubMed] [Google Scholar]
- 73.Sato S, Ward C L, Krouse M E, Wine J J, Kopito R R. Glycerol reverses the misfolding phenotype of the most common cystic fibrosis mutation. J Biol Chem. 1996;271:635–638. doi: 10.1074/jbc.271.2.635. [DOI] [PubMed] [Google Scholar]
- 74.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 75.Scheffner M, Nuber U, Huibregtse J M. Protein ubiquitination involving an E1–E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 76.Seol J H, Feldman R M, Zachariae W, Shevchenko A, Correll C C, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies R J. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 78.Skowyra D, Koepp D M, Kamura T, Conrad M N, Conaway R C, Conaway J W, Elledge S J, Harper J W. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 79.Thrower J S, Hoffman L, Rechsteiner M, Pickart C M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Travers K J, Patil C K, Wodicka L, Lockhart D J, Weissman J S, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 81.Varshavsky A. Naming a targeting signal. Cell. 1991;64:13–15. doi: 10.1016/0092-8674(91)90202-a. [DOI] [PubMed] [Google Scholar]
- 82.Ward C L, Omura S, Kopito R R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 83.Wei X, Eisman R, Xu J, Harsch A D, Mulberg A E, Bevins C L, Glick M C, Scanlin T F. Turnover of the cystic fibrosis transmembrane conductance regulator (CFTR): slow degradation of wild-type and delta F508 CFTR in surface membrane preparations of immortalized airway epithelial cells. J Cell Physiol. 1996;168:373–384. doi: 10.1002/(SICI)1097-4652(199608)168:2<373::AID-JCP16>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 84.Wickner S, Maurizi M R, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 85.Wilhovsky S, Gardner R, Hampton R. HRD gene dependence of ER-associated degradation. Mol Biol Cell. 2000;11:1697–1708. doi: 10.1091/mbc.11.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xie Y, Varshavsky A. The E2–E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 1999;18:6832–6844. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang M, Omura S, Bonifacino J S, Weissman A M. Novel aspects of degradation of T cell receptor subunits from the endoplasmic reticulum (ER) in T cells: importance of oligosaccharide processing, ubiquitination, and proteasome-dependent removal from ER membranes. J Exp Med. 1998;187:835–846. doi: 10.1084/jem.187.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu H, Kaung G, Kobayashi S, Kopito R R. Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- 89.Yu H, Peters J M, King R W, Page A M, Hieter P, Kirschner M W. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- 90.Zachariae W, Shevchenko A, Andrews P D, Ciosk R, Galova M, Stark M J, Nasmyth K. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- 91.Zheng F, Kartner N, Lukacs G L. Limited proteolysis as a probe for arrested conformational maturation of DELTA-F508 CFTR. Nat Struct Biol. 1998;5:180–183. doi: 10.1038/nsb0398-180. [DOI] [PubMed] [Google Scholar]
- 92.Zheng N, Wang P, Jeffrey P D, Pavletich N P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 93.Zhou M, Fisher E A, Ginsberg H N. Regulated co-translational ubiquitination of apolipoprotein B100. A new paradigm for proteasomal degradation of a secretory protein. J Biol Chem. 1998;273:24649–24653. doi: 10.1074/jbc.273.38.24649. [DOI] [PubMed] [Google Scholar]
- 94.Zhou M, Schekman R. The engagement of Sec61p in the ER dislocation process. Mol Cell. 1999;4:925–934. doi: 10.1016/s1097-2765(00)80222-1. [DOI] [PubMed] [Google Scholar]