Abstract
Inhibitor of apoptosis protein (IAP)-like protein-1 (ILP-1) (also known as X-linked IAP [XIAP] and mammalian IAP homolog A [MIHA]) is a potent inhibitor of apoptosis and exerts its effects, at least in part, by the direct association with and inhibition of specific caspases. Here, we describe the molecular cloning and characterization of a human gene related to ILP-1, termed ILP-2. Despite high homology to ILP-1, ILP-2 is encoded by a distinct gene, which in normal tissues is expressed solely in testis. In contrast to ILP-1, overexpression of ILP-2 had no protective effect on apoptosis mediated by Fas (also known as CD95) or tumor necrosis factor. However, ILP-2 potently inhibited apoptosis induced by overexpression of Bax or by coexpression of caspase 9 with Apaf-1, and preincubation of cytosolic extracts with ILP-2 abrogated caspase activation in vitro. A processed form of caspase 9 could be coprecipitated with ILP-2 from cells, suggesting a physical interaction between ILP-2 and caspase 9. Thus, ILP-2 is a novel IAP family member with restricted specificity for caspase 9.
Apoptosis is a genetically determined, biochemically ordered process in which cells are induced to initiate a cellular suicide program in response to physiologic signals, cellular damage, or virus infection (24, 46, 47). The principal effectors are a family of cysteine proteases known as caspases (35, 42). These proteases, which cleave with a high degree of specificity after aspartate residues, are initially produced in the cell as inactive precursors whose activation is controlled by a variety of regulatory factors and events. Numerous regulators of apoptosis have been identified and have been shown to exert their effects ultimately by affecting levels of caspase activity. For example, the Bcl-2 family of proteins regulates apoptosis through the control of mitochondrial integrity. Proapoptotic signals have been shown to result in the release of cytochrome c from the mitochondria into the cytosol (4, 6, 14, 25, 34, 44, 48). Released cytochrome c is thought to stimulate the formation of an active holoenzyme containing the CED-4-like protein Apaf-1 together with caspase 9 (1, 5, 31), and this holoenzyme induces the activation of the downstream effector caspase, caspase 3. Overexpression of prosurvival members of the Bcl-2 family has been shown to maintain mitochondrial membrane integrity, preventing cytochrome c release and thereby inhibiting formation of the Apaf-1:caspase 9 holoenzyme (21).
In contrast to the mechanisms employed by Bcl-2-related proteins, several members of the inhibitor of apoptosis (IAP) family of proteins are thought to bind directly to caspases and inhibit their enzymatic activity (10, 11). Three IAPs, ILP-1 (also known as X-linked IAP [XIAP] and MIHA), c-IAP1, and c-IAP2, are widely expressed in mammalian tissues and are broad inhibitors of apoptosis (15, 26, 32, 43). Their predicted open reading frames share several common features: all three IAPs have three tandem repeats of an approximately 70-residue domain known as the BIR (baculovirus iap repeat), followed by an amphipathic spacer region of unknown function and a single carboxy-terminal RING finger domain (2, 22). In vitro evidence suggests that c-IAP1, c-IAP2, and ILP-1 inhibit cell death by interacting with and inhibiting specific caspases, namely caspases 3, 7, and 9 (11, 12, 33). Studies of individual BIRs suggest that these are the domains responsible for caspase interaction and inhibition (40, 41). The RING finger domain of the IAPs has recently been shown to play a role in regulating their signal-induced autoubiquitination and resulting degradation (49).
Here, we report the identification of ILP-2, a novel member of the IAP family that is most closely related in sequence to ILP-1. ILP-2 encodes a single amino-terminal BIR domain followed by a spacer region and a carboxy-terminal RING finger domain. In normal tissues, expression of the ILP-2 gene was detected only in testis, but ILP-2 was also detected in lymphoblastoid cells. Human ILP-2 potently inhibited apoptosis induced by overexpression of Bax or by caspase 9 and Apaf-1, but unlike ILP-1, ILP-2 had no protective effect on apoptosis mediated by Fas (also known as CD95). In a cell-free system utilizing cytosolic extracts, ATP-dependent caspase activation was abrogated when recombinant ILP-2 protein was added prior to ATP. However, once active, caspases in the cell-free system were not inhibited by ILP-2. In contrast, ILP-1 blocked both caspase activation and activity in the cell-free system. In transient-transfection studies, a processed form of caspase 9 was coprecipitated with ILP-2, suggesting a direct physical interaction between ILP-2 and caspase 9. Thus, ILP-2 is a novel member of the IAP family with restricted apoptotic inhibitory functions.
MATERIALS AND METHODS
Molecular cloning and characterization of ILP-2.
Combinations of oligonucleotides which collectively span the ILP-1 gene were tested in PCRs using human genomic DNA and cDNA prepared from peripheral blood leukocyte RNA. A PCR product was obtained from genomic DNA with two primers, 5′-CCTGGCGCGAAAAGGTGGACAAGTC-3′ and 5′-TGTTCACATCACACATTCAATCAGGG-3′, and using the following PCR conditions: 45 s at 94°C, 45 s at 60°C, and 120 s at 72°C for 30 cycles. This product was found to encode a previously unidentified sequence, which was designated ILP-2.
Based on this novel sequence, two PCR primers that specifically recognize ILP-2 and that do not cross-react with ILP-1 were designed: 5′-ACTTGAGGGAGCTCTGGTACAAAC-3′ and 5′-AGTGACCAGATGTCCACAAGG-3′. These primers, which amplify a ∼300-bp product, were used for PCR-mediated isolation of a bacterial artificial chromosome (BAC) genomic clone (Genome Systems, Westchester, Pa.) by using the same PCR conditions described above.
Genomic DNA from the primates indicated in the text was prepared from buffy coats isolated from peripheral blood (Buckshire Corporation, Perkasie, Pa.) by Lofstrand Laboratories (Gaithersburg, Md.). Cloning of primate genomic ILP-2 was performed by PCR under standard conditions using the ILP-2-specific primers 5′-TGAATCTGATGTTGCGAGTTCTG-3′ and 5′-GTTGAGTCACATCACACATTTAATC-3′. PCR products were either sequenced directly or subcloned into pCR2.1 (Invitrogen, Carlsbad, Calif.) prior to sequencing.
RT-PCR expression analysis.
Reverse transcriptase-PCR (RT-PCR) was performed on total RNA of testis, heart, and lung (Clontech, Palo Alto, Calif.) as follows. RNase-free DNase I (1 μl; Promega) was added to 4 μl of total RNA and was incubated at 37°C for 30 min and then at 75°C for 5 min. RT-PCR was performed according to the manufacturers' instructions using a cDNA cycle kit (Invitrogen) and Advantage polymerase (Clontech). Controls without reverse transcriptase were included in every reaction. The human ILP-2-specific PCR primers (5′-ACTTGAGGGAGCTCTGGTACAAAC-3′ and 5′-AGTGACCAGATGTCCACAAGG-3′) were used for detection of ILP-2 transcripts. For internal controls, GAPDH-specific primers 5′-ACCACCATGGAGAAGGCTGG-3′ and 5′-CTCAGTGTAGCCCAGGATGC-3′ were used in parallel reactions that produced a 500-bp product.
Plasmids.
The pEBB expression vector and the pEBG mammalian glutathione S-transferase (GST) expression vector were kindly provided by B. Mayer. The pEBB-FLAG-ILP-1 and pEBG-ILP-1 plasmids have been described previously (14). The Apaf-1 expression vector was a gift of R. Armstrong. The P1 artificial chromosome (PAC) genomic clone of ILP-1 was a gift of the Sanger Centre (library RPCI6, clone dA30A23). The Bax expression vector was kindly provided by S. Korsmeyer. The Fas expression vector was a kind gift of R. Siegel. Expression vectors encoding wild-type and dominant-negative caspase 9 (20, 25) were kindly provided by C. Zacharchuk. Site-directed mutagenesis (QuikChange; Stratagene, La Jolla, Calif.) was performed on the caspase 9 plasmid to generate two mutations: Asp-315 to an Ala codon and Asp-315 to the TGA stop codon. The pEBG-ILP-1 (Δ2BIR) vector encodes residues 262 to 497 of ILP-1.
The open reading frames of ILP-1 and ILP-2 were subcloned into pEBB-FLAG and pEBG for expression in mammalian cells or into the bacterial expression vector pGEX4T-1 (Amersham Pharmacia, Piscataway, N.J.) for expression in Escherichia coli cells.
Southern and Northern blot analyses.
BAC and PAC DNA (5 ng) and genomic DNA (10 μg) were digested overnight with EcoRI (New England Biolabs, Beverly, Mass.). For Southern blot analysis, the restriction fragments were separated by 1% agarose gel electrophoresis at 40 V, transferred to a Hybond-N+ nylon membrane (Amersham Pharmacia), and immobilized by UV cross-linking. The DNA probe was obtained by isolating a fragment of human ILP-2 from a cDNA expression vector using the restriction enzymes BamHI and HindIII. 32P-labeling was performed as described (19) using Klenow enzyme (New England Biolabs). The DNA probe was separated from unincorporated nucleotides on a G-25 spin column (Amersham Pharmacia). Hybridization and washing were performed with Hybrisol II (Intergen, Gaithersburg, Md.) according to the manufacturer's instructions.
Northern blot hybridization was performed similarly using Human Multiple Tissue Northern blots (Clontech), and a 32P-labeled DNA fragment from ILP-2 spanning the entire open reading frame was prepared exactly as described above. Blots were washed at high stringency following the manufacturer's instructions.
Cells, transfections, and viability assays.
Human embryonic kidney 293 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 2 mM glutamine at 37°C in 5% CO2. Six-well plates were seeded with 2 × 105 cells per well in 2 ml of medium. Cells were transfected the following day by the calcium phosphate procedure as described previously (13). For viability experiments, 293 cells were transfected with 0.5 μg of green fluorescent protein (GFP) (pEGFP-N1; Clontech) together with plasmids encoding Bax, Fas, caspase 9, and Apaf-1 as indicated in the text. Cells were stained with 4′,6′-diamidino-2-phenylindole (DAPI; Molecular Probes, Eugene, Oreg.) 24 h after transfection and were examined for apoptosis by fluorescence microscopy as previously described (16).
Protein expression and purification.
pGEX-CD1 (29), pGEX-ILP-1, or pGEX-ILP-2 was expressed in BL-21(DE3) E. coli cells (Stratagene). Bacterial cultures (500 ml) were grown overnight at 37°C before induction with 1 mM isopropyl-1-β-d-thiogalactopyranoside (IPTG) at 30°C for 3 h and then were purified by standard glutathione-Sepharose affinity purification methods. Briefly, bacteria were lysed in 50 ml of GST lysis buffer (50 mM Tris, pH 7.5, 1 mM EDTA, 100 mM NaCl, 1% Triton X-100), sonicated five times for 10 s, and centrifuged for 15 min at 10,000 × g at 4°C. The supernatant was applied to a column containing 1 ml of a 50% slurry of glutathione-Sepharose (Amersham Pharmacia) and washed with 10 ml of GST lysis buffer. Protein fractions were eluted with elution buffer (10 mM glutathione and 100 mM Tris, pH 7.5) and dialyzed against phosphate-buffered saline. Expression of proteins was analyzed by gradient sodium dodecyl sulfate (SDS)–4 to 12% polyacrylamide gel electrophoresis (PAGE; Invitrogen) followed by Coomassie blue staining, and protein concentrations were measured by the method described by Bradford (3) using a commercial kit (Bio-Rad, Hercules, Calif.).
Extract preparation and caspase assays.
Cytosolic extracts from 293 cells were prepared essentially as previously described (17). Cells were washed and the pellet was resuspended in 10 volumes of extract preparation buffer (50 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] at pH 7.0, 50 mM KCl, 5 mM EGTA, 2mM MgCl2, 1 mM dithiothreitol [DTT]) supplemented with 10 μg of cytochalasin B/ml and protease inhibitor cocktail (0.1 mM phenylmethylsulfonyl fluoride and 2 μg [each] of chymostatin, pepstatin, leupeptin, and antipain/ml; Sigma, St. Louis, Mo.). Cells were immediately pelleted and lysed by three cycles of freezing and thawing. The lysate was centrifuged at 100,000 × g for 1 h to produce the cell extract. The final concentration of extract was adjusted to 15 μg/μl in modified extract preparation buffer containing 10 mM KCl.
Recombinant GST, GST–ILP-1 or GST–ILP-2 (100 ng each) was added to 293 cytosolic extract (150 μg) either before or after extract activation with ATP, in a total volume of 20 μl, as detailed in the text. Following incubation for the indicated periods of time at 37°C, 2 μl of the reaction mixture was incubated with 200 μl of assay buffer (50 mM PIPES at pH 7.0, 0.1 mM EDTA, 10% glycerol, 1 mM DTT) containing 20 μM Ac-DEVD-AFC (N-acetyl-Asp-Glu-Val-Asp-AFC [7-amino-4-trifluoromethylcoumarin]) or 50 μM Ac-LEHD-AFC (N-acetyl-Leu-Glu-His-Asp-AFC) (Biomol Research Laboratories, Plymouth Meeting, Pa.) for 10 min. The release of free AFC was measured with a Cytofluor 4000 fluorescence plate reader (Perseptive Biosystems, Framingham, Mass.).
Coprecipitation and immunoblot analysis.
Human embryonic kidney 293 cells were transiently transfected in six-well plates with the indicated plasmids. After 15 h, cells were washed once with phosphate-buffered saline and lysed for 15 min in 0.3 ml of 1% Triton X-100 buffer containing 25 mM HEPES (pH 7.9), 100 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM phenylmethyl sulfonyl fluoride 1 mM DTT, and 1 protease inhibitor cocktail tablet (Roche Diagnostics, Indianapolis, Ind.) per 10 ml of lysis buffer. For coprecipitation, 150-μl aliquots of lysates were incubated with 20 μl of glutathione-Sepharose beads (Amersham Pharmacia) at 4°C for 1 h. Beads were washed four times with 1 ml of lysis buffer. The bead-associated proteins along with aliquots of total lysates were resolved by a 4 to 12% gradient SDS-PAGE (Invitrogen) and transferred to nitrocellulose membranes (Invitrogen). After blocking the membranes in 5% dry milk in Tris-buffered saline with 0.2% Tween, the membranes were incubated with either anti-GST antibody (Santa Cruz Laboratories, Santa Cruz, Calif.), anti-Apaf-1 antibody (a kind gift of Y. Lazebnik, Cold Spring Harbor, N.Y.), or anti-caspase 9 antibodies (Pharmingen, San Diego, Calif. or Upstate Biotechnology, Lake Placid, N.Y.) as indicated in the figure legends and were visualized by enhanced chemiluminescence (ECL; Amersham Pharmacia).
Gel filtration chromatography.
Chromatography was carried out using an ÄKTA fast-performance liquid chromatograph (FPLC) (Amersham Pharmacia) at 4°C. Samples (0.2 ml, 1.5 mg of protein) were applied to an HR 10/30 Superose 6 column (Amersham Pharmacia) equilibrated in 25 mM HEPES (pH 7.0), 10 mM KCl, 50 mM NaCl, 5 mM EGTA, 1 mM MgCl2, 5% sucrose, 0.1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propane-sulfonate}, and 1 mM DTT. Proteins were eluted at 0.3 ml/min, and 0.5-ml fractions were collected. An aliquot of each fraction was immediately assayed for caspase activity or snap frozen for later analysis by immunoblotting. The Superose 6 column had previously been calibrated using blue dextran, thyroglobulin, ferritin, catalase, bovine serum albumin, and ovalbumin as standards (Amersham Pharmacia).
RESULTS
Cloning and genetic analysis of human ILP-2.
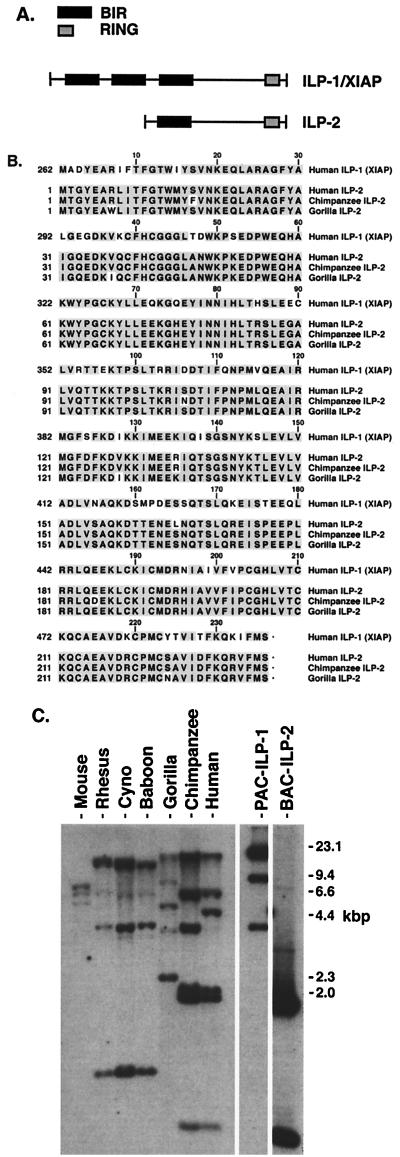
To search for human homologs of ILP-1, combinations of PCR primers which spanned the entire open reading frame of human ILP were designed. Human genomic DNA and cDNA prepared from peripheral blood leukocyte RNA were used as templates for PCR. Sequencing of the PCR products revealed novel ILP-related sequences in addition to that of ILP-1. The full-length genomic clone of one of these sequences was isolated and termed ILP-2. The predicted open reading frame of ILP-2 lacks the first two BIR domains and is comprised of a single amino-terminal BIR followed by a spacer domain and a carboxy-terminal RING domain (Fig. 1A). The coding sequence of ILP-2 is very similar to that of ILP-1, with 80% identity (95% homology) at the amino acid level to the corresponding region of ILP-1 (Fig. 1B).
FIG. 1.
ILP-2 is conserved during the evolution of great apes. (A) Schematic representation of ILP-1 and ILP-2. (B) Sequence comparison of the ILP-1 and ILP-2 proteins. The predicted amino acid sequences of the corresponding region of human ILP-1 (residues 262 to 497) are shown aligned with the full coding sequence of human ILP-2, chimpanzee ILP-2, and gorilla ILP-2. The GenBank accession number of human ILP-2 is AF164682, and the accession numbers of chimpanzee and gorilla ILP-2 are AY030052 and AY030053, respectively. (C) Southern blot analysis of EcoRI-digested genomic DNA from humans, great apes (chimpanzee and gorilla), Old World monkeys (baboon, cynomolgus monkey, and rhesus monkey), and mouse (C57BL6) was obtained by using a probe which detects both ILP-1 and ILP-2. Included in the experiment were the human genomic clones of ILP-1 (PAC–ILP-1) and ILP-2 (BAC–ILP-2). The gorilla ILP-2 gene does not contain an EcoRI site in its coding sequence, accounting for a profile that differs slightly from those of the human and chimpanzee genes.
The chromosomal localization of ILP-2 was determined by fluorescence in situ hybridization and radiation hybrid analysis and was found to map to a location distinct from that of ILP-1. ILP-2 mapped to 19q 13.3-q13.4 (S. S. Mir and C. S. Duckett, unpublished observations), whereas ILP-1 has been localized to Xq25 (30). Unlike ILP-1, ILP-2 lacks introns. Analysis of the genomic sequence of ILP-2 revealed the presence of a poly(A) tail downstream of its 3′ untranslated region, which suggests that ILP-2 is a processed gene derived from ILP-1 by a retrotransposition event.
ILP-2 is conserved in primates.
To determine whether ILP-2 has been conserved throughout evolution, the ILP-2 genes of several primates were characterized. The open reading frame of ILP-2 in both chimpanzee and gorilla was conserved (Fig. 1B), suggesting that the selective pressure on ILP-2 has been retained. Interestingly, no ILP-2 sequences could be isolated from any species other than primates. ILP-2 was detected only in humans and great apes (chimpanzee and gorilla; Fig. 1B) and could not be isolated from genomic DNA prepared from Old World monkeys (baboon, cynomolgus monkey, and rhesus monkey). This observation was examined further by performing Southern analysis of genomic DNA from primates as well as mouse by using a radiolabeled probe prepared from ILP-2 but which identifies both ILP-1 and ILP-2. The human genomic clones of ILP-1 and ILP-2 were included in order to identify bands seen in primate samples. Analysis revealed a similar pattern of bands present in genomic DNA from the Old World monkeys (Fig. 1C). Consistent with results obtained by PCR experiments, this pattern is more complex in genomic DNA from the great apes, suggesting the presence of additional ILP-1-related genes that are not present in Old World monkeys. Bands in genomic DNA from great apes, but not that from Old World monkeys, migrated closely with the two major bands representing the human genomic clone of ILP-2, suggesting that ILP-2 emerged after their divergence. The pattern of bands seen in genomic DNA from primates was not present in mouse genomic DNA. These data suggest that the retrotransposition event which gave rise to ILP-2 occurred during the evolution of great apes.
Expression of ILP-2.
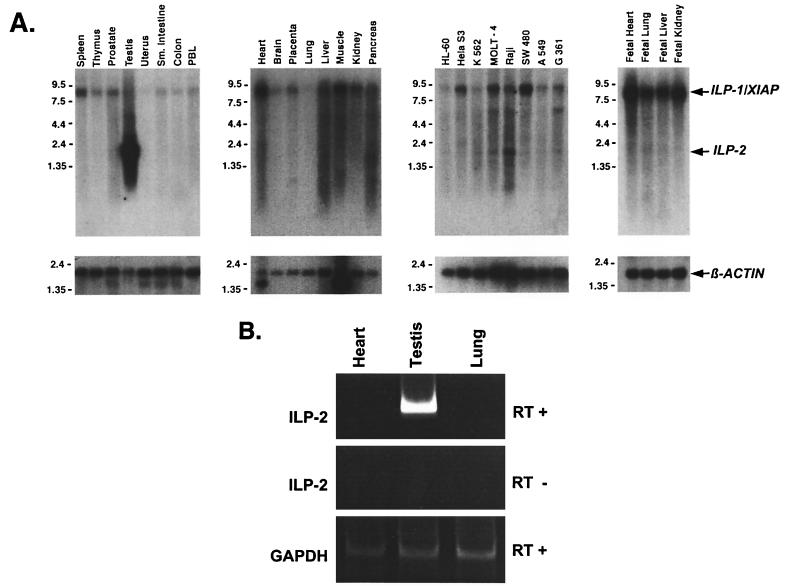
The expression pattern of ILP-2 was examined by Northern analysis of both fetal and adult human tissues, as well as of a panel of transformed cell types. In primary cells, a ∼2-kb transcript in human testis was detected with a radiolabeled ILP-2 fragment (Fig. 2A), indicating a testis-restricted pattern of expression. In the Northern blot panel of selected tumor lines (Fig. 2A), a band of the same size was also detected in the Raji B-lymphoblastoid cell line, suggesting that deregulated expression of ILP-2 might occur in a subset of transformed cells.
FIG. 2.
Expression of ILP-2. (A) Northern blot analysis of human multiple-tissue blots and cell lines (Clontech) was performed using high-stringency conditions and a 32P-labeled cDNA fragment of full-length ILP-2. The ∼8.5-kb band present in all lanes corresponds to ILP-1, which cross-reacts with the ILP-2 probe. As a loading control, the blot was probed for β-actin (below). Sizes (in kilobase) are indicated. (B) RT-PCR of total RNA from human tissues of heart, testis, and lung using ILP-2-specific oligonucleotide primers and reverse transcriptase (top). As a negative control, PCRs were performed without reverse transcriptase (middle). A control PCR using GAPDH-specific primers and reverse transcriptase amplified equal amounts of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) PCR fragment of the RNA of each tissue (bottom).
To confirm that ILP-2 expression is restricted to testis, RT-PCR analysis was performed using PCR primers to a region of ILP-2 which is divergent from ILP-1. These primers were identical to those used to isolate full-length ILP-2 clones and do not cross-react with ILP-1. Using RNA prepared from human tissues of testis, heart, and lung, ILP-2 expression was detected only in human testis (Fig. 2B).
ILP-2 inhibits apoptosis induced by Bax or coexpression of caspase 9 and Apaf-1.
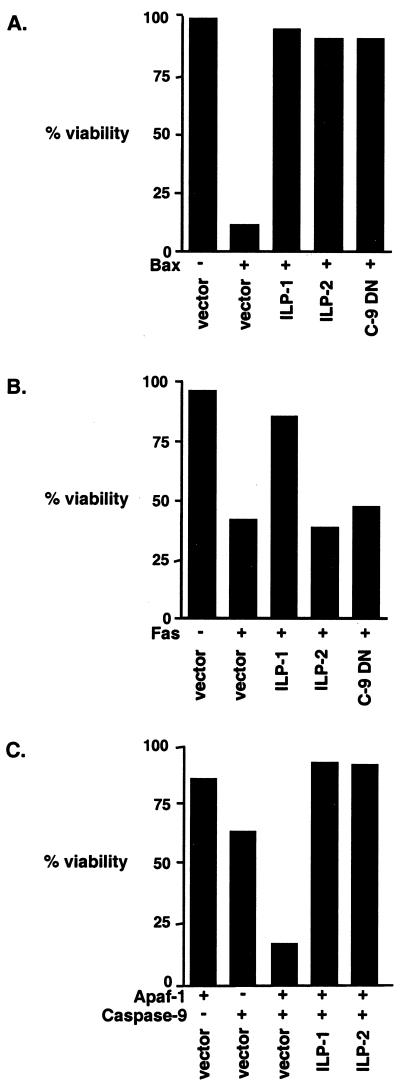
Since ILP-2 is highly homologous to ILP-1, the two proteins were compared for their ability to regulate apoptosis. Expression vectors encoding several proapoptotic stimuli were coexpressed in 293 cells with vectors encoding ILP-1, ILP-2, or control vectors (Fig. 3). Apoptosis induced by ectopic expression of Bax was profoundly inhibited by ILP-2 at levels which were essentially identical to those observed with ILP-1 (Fig. 3A). However, expression of ILP-2 had no inhibitory effect on apoptosis induced by Fas (Fig. 3B). Induction of apoptosis by Bax is thought to occur through the insertion of Bax into the mitochondrial outer membrane. This event is then followed by perturbation of the integrity of the mitochondrial membrane, the release of cytochrome c, and the activation of a complex which contains the apoptosis activating factor Apaf-1 and caspase 9 (4, 31). In contrast, the induction of apoptosis by overexpression of Fas in 293 cells occurs through a caspase 9-independent pathway (9), since it is not inhibited by dominant-negative caspase 9 (Fig. 3B). These data suggested that ILP-2 might inhibit Bax-induced death by suppression of caspase 9 activity. Therefore, the ability of ILP-2 to inhibit caspase 9-induced death was assessed. Consistent with previous reports (7), ectopic expression of caspase 9 alone did not induce apoptosis of 293 cells, although coexpression of caspase 9 with Apaf-1 strongly induced the apoptosis of transfected cells (Fig. 3C). ILP-2 potently inhibited cell death induced by caspase 9 plus Apaf-1 (Fig. 3C), suggesting that ILP-2 suppresses Bax-induced apoptosis through inhibition of caspase 9–Apaf-1 activity.
FIG. 3.
ILP-2 inhibits apoptosis induced by Bax and caspase 9 plus Apaf-1 but not by Fas. Human embryonic kidney 293 cells were transfected with a GFP plasmid (0.5 μg/well) in combination with either a Bax expression plasmid (0.25 μg/well) (A), Fas expression plasmid (2 μg/well) (B), or Apaf-1 (2 μg/well) and caspase 9 (0.5 μg/well) plasmids, as indicated (C). Cells were cotransfected with a plasmid encoding GFP as a marker of transfection, together with a control plasmid or plasmids expressing ILP-1, ILP-2, or dominant-negative caspase 9 (C-9 DN) (2 μg/well) as indicated. Cells were fixed and stained with DAPI, and transfected wells were scored for viability based on nuclear morphology as described in Materials and Methods. Data shown are representative of at least three independent experiments.
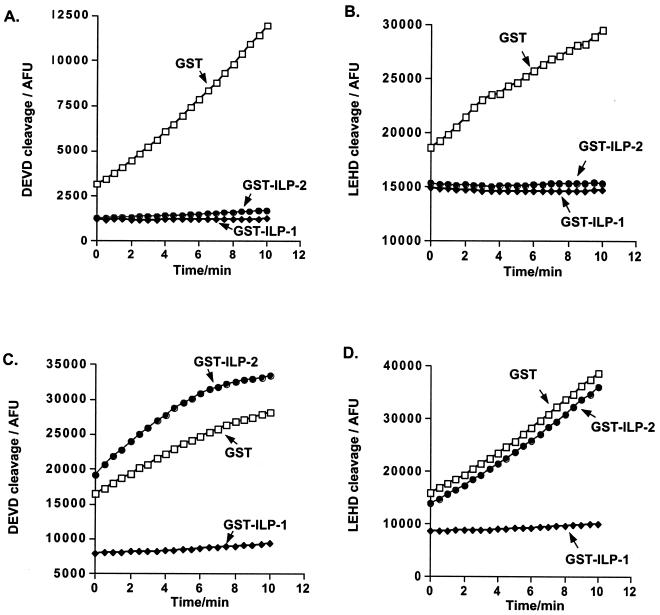
Binding of Apaf-1 to cytochrome c in the presence of ATP or dATP results in the recruitment and processing of caspase 9 (25). These events can be observed in a cell-free system by incubating cytosolic extracts with ATP (1, 18, 27). The addition of ATP leads to the processing of caspase 9, which in turn has been shown to cleave and activate caspase 3. ILP-1 has been reported to inhibit both caspase 9-dependent activation of extracts as well as caspase 3-dependent activity of extracts following activation by ATP (9). To determine whether ILP-2 can inhibit this process, cytosolic extracts were preincubated with GST, GST–ILP-1, or GST–ILP-2 proteins prior to the addition of ATP. Under these conditions, both ILP-1 and ILP-2 abrogated ATP-induced caspase activation. This inhibition was detected using either a caspase 3 substrate (DEVD-AFC; Fig. 4A) or a caspase 9 substrate (LEHD-AFC; Fig. 4B).
FIG. 4.
ILP-2 inhibits caspase activation in a cell-free system. (A, B) Cytosolic extracts (150 μg) from 293 cells were preincubated with GST–ILP-1, GST–ILP-2, or GST control proteins (100 ng each) as indicated for 20 min, at which time ATP (1 mM final concentration) was added and the reaction mixture was incubated for a further 30 min. (C, D) Cytosolic extracts were incubated with ATP for 30 min, after which time the recombinant GST–ILP-1, GST–ILP-2, or GST control proteins were added and the reaction mixture was incubated for a further 20 min. Caspase activity was measured by cleavage of the caspase 3-specific fluorogenic substrate DEVD-AFC (A, C) or the caspase 9-specific substrate LEHD-AFC (B, D), as described in Materials and Methods.
The effects of ILP-1 and ILP-2 were next examined on extracts which had already been activated. Cytosolic extracts were activated by the addition of ATP for 30 min, and recombinant GST–ILP-1, GST–ILP-2, or GST control proteins were subsequently added. Under these conditions, ILP-2 had no inhibitory effect, while ILP-1 completely blocked caspase activity. Similar results were obtained using either a caspase 3-specific substrate (Fig. 4C) or a caspase 9-specific substrate (Fig. 4D). Thus, data from both cell-based and cell-free systems suggested that ILP-2 acts through inhibition of caspase 9 activity.
ILP-2 binds caspase 9.
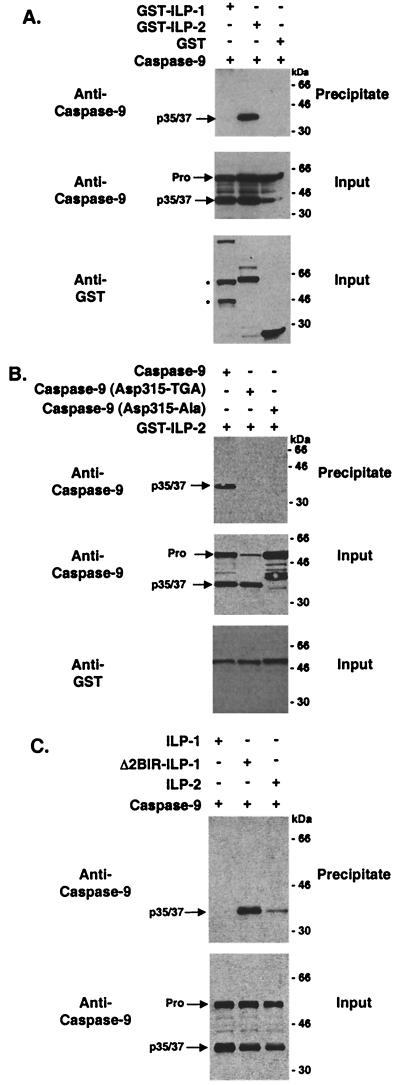
To examine whether ILP-2 interacts with caspase 9 in cells, mammalian expression vectors encoding ILP-1 or ILP-2 fused to GST were cotransfected with a caspase 9 expression vector into 293 cells. Cell lysates were precipitated with glutathione-Sepharose beads and analyzed with an anti-caspase 9 antibody. A processed form of caspase 9 was found to coprecipitate with ILP-2 but not with ILP-1 (Fig. 5A) with an approximate molecular mass of 35 kDa.
FIG. 5.
ILP-1 (Δ2BIR) and ILP-2 interact with processed caspase 9 in cells. 293 cells were transfected with expression vectors encoding GST, GST–ILP-1, GST–ILP-2, and/or caspase 9 as indicated (2 μg/well) (A), GST–ILP-2, caspase 9, and/or caspase 9 mutants as indi- cated (B), and GST–ILP-1, GST–ILP-1 (Δ2BIR), GST–ILP-2, and/or caspase 9 as indicated (C). Cell lysates were coprecipitated with glutathione-Sepharose beads, washed, resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with an anti-caspase 9 antibody (Pharmingen). The full-length (Pro) and processed (p35/37) forms of caspase 9 are indicated. Cell lysates were also examined by immunoblot analysis to confirm transfection efficiency and loading (input). The bands marked with asterisks are found only when the GST–ILP-1 vector is coexpressed with caspase 9 and presumably represent the caspase 9-induced cleavage products which have been reported previously (9, 23). Molecular masses (in kilodaltons) are shown.
While full-length caspase 9 has been shown to be enzymatically active (38), the processed form is thought to possess a higher specific activity (37, 38). Recent studies have demonstrated that an early step in the processing of full-length caspase 9 occurs at Asp-315 (37). To determine whether the caspase 9 species observed to interact with ILP-2 in cells represents caspase 9 that has been cleaved at Asp-315, mutants of caspase 9 were generated, in which the Asp-315 codon was mutated to an Ala codon or to the TGA stop codon. The truncated caspase 9 mutant (Asp-315 codon to TGA) comigrated with processed caspase 9, while mutation of caspase 9 (Asp-315 codon to Ala codon) inhibited cleavage of caspase 9 to its processed form (Fig. 5B, right). Surprisingly, neither of the caspase 9 mutants interacted with ILP-2, raising the possibility that in cells the interaction between ILP-2 and caspase 9 may require a conformational difference which is present only in the active tetrameric caspase.
Cleaved ILP-1 interacts with processed caspase 9 in cells.
Recent in vitro studies suggest that ILP-1 is cleaved by caspase 9 at Asp-242 (9). This cleavage event generates a form of ILP-1 which lacks its first two BIR domains (Δ2BIR) and is therefore analogous to ILP-2. To determine whether cleaved ILP-1 interacts with caspase 9 in cells, a GST-tagged deletion construct of ILP-1 was generated (Δ2BIR) which consists of the third BIR, spacer, and RING domains of ILP-1. Expression vectors encoding ILP-1, ILP-1 (Δ2BIR), or ILP-2 were cotransfected with a caspase 9 expression vector into 293 cells. Cell lysates were precipitated with glutathione-Sepharose beads and analyzed with a caspase 9 antibody. Processed caspase 9 was found to coprecipitate with ILP-1 (Δ2BIR) as well as ILP-2 but not with full-length ILP-1 (Fig. 5C). These studies suggest that cleaved but not full-length ILP-1 interacts with processed caspase 9 in cells.
ILP-2 inhibits processing of caspase 9 in cell-free system.
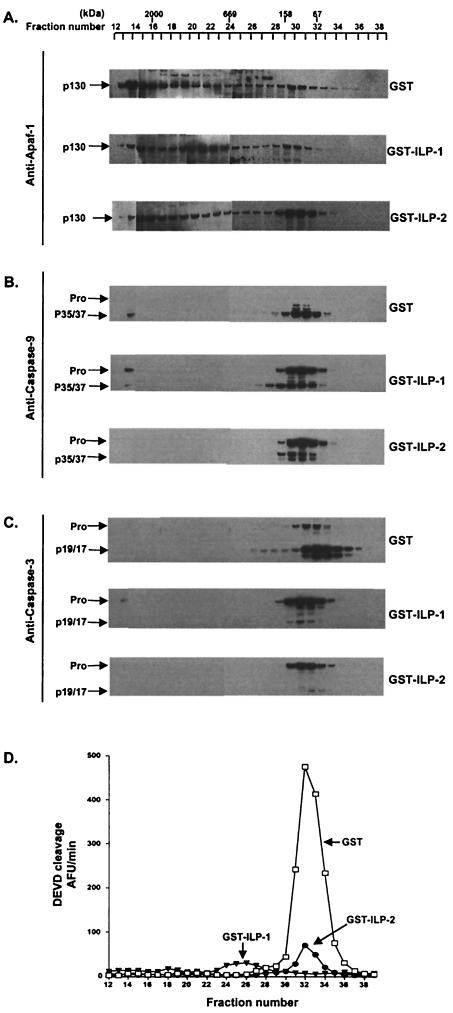
The data shown in Fig. 4 and 5 suggest that the ability of ILP-2 to impede the activation of caspase 3 is indirect, perhaps functioning at the level of caspase 9. While the coprecipitation studies shown in Fig. 5 revealed that ILP-2 can coassociate with processed caspase 9, the possibility remained that ILP-2 might also function to impair either holoenzyme formation or the processing of full-length caspase 9. To test this possibility, cytosolic extracts from 293 cells were preincubated with recombinant, purified GST–ILP-1, GST–ILP-2, or control GST proteins prior to the addition of ATP and were subsequently resolved by gel filtration chromatography using a Superose 6 column. Fractions eluted from the column were evaluated for DEVDase activity, which measures downstream caspase 3 activity, and in parallel, aliquots were immunoblotted and probed with antibodies to Apaf-1, caspase 9, and caspase 3 (Fig. 6). No significant DEVDase activity was detected in higher-molecular-mass fractions corresponding to the predicted molecular mass (∼700 to 1,400 kDa) of the apoptosome (5). However, oligomeric Apaf-1, necessary for the formation of the holoenzymatic complex, was detected and was slightly inhibited in extracts incubated with GST–ILP-1 or GST–ILP-2 (Fig. 6A). In GST-incubated extracts, processed caspase 9 was faintly detected in a high-molecular-weight fraction (fraction 14) which presumably corresponds to the Apaf-1:caspase 9 holoenzyme, while full-length caspase 9 was detected in the equivalent fraction in ILP-1-incubated extracts. Caspase 9 was not detected in the corresponding fraction in ILP-2-incubated extracts (Fig. 6B). Caspases 3 and 9 could readily be detected in fractions which, by comparison with molecular mass standards, would be predicted to be uncomplexed with Apaf-1 (Fig. 6B and C). The fractions containing these free caspases corresponded exactly to the fractions with the peak enzymatic activity, as measured by the DEVDase assay (Fig. 6D). As expected, the DEVDase activities of corresponding fractions from extracts preincubated with either GST–ILP-1 or GST–ILP-2 were greatly reduced. These fractions from GST–ILP-1 and GST–ILP-2-incubated extracts were found to contain caspase 3 predominantly in its full-length form. Also as predicted, the majority of caspase 3 in the GST-incubated extracts was found to be in the active p19/17 form (Fig. 6C). In the case of ILP-2, this is presumably due to the suppression of caspase 9 activity, thereby impeding the ability of caspase 9 to cleave full-length caspase 9. Surprisingly, however, immunoblot analysis of fractions from the ILP-2-incubated extracts revealed that a large proportion of caspase 9 was also found in its full-length form.
FIG. 6.
ILP-1 and ILP-2 inhibit processing of caspases 9 and 3 in a cell-free system. 293 cell extracts (1.5 mg) were incubated with recombinant, purified GST–ILP-1, GST–ILP-2 or control GST proteins (1 μg) for 30 min, activated with ATP for 30 min, and resolved by gel filtration chromatography on a Superose 6 column (Amersham Pharmacia) using an ÄKTA FPLC system (Amersham Pharmacia). Aliquots of the fractions eluted from the column were separated by SDS-PAGE, immunoblotted onto nitrocellulose, and probed with the indicated antibodies as follows. (A) Apaf-1 (Y. Lazebnik, Cold Spring Harbor Laboratories, N.Y.). The calculated molecular mass of Apaf-1 was consistent with the expected molecular mass of 130 kDa and is indicated (p130). (B) Caspase 9 (Upstate Biotechnology). The full-length (Pro) and processed 35- and 37-kDa species (p35/37) are indi- cated. (C) Caspase 3 (Pharmingen). The inactive precursor (Pro) and the 19- and 17-kDa processed, active forms (p19/17) are indicated. (D) Aliquots of the fractions were also evaluated for DEVDase activity, using DEVD-AFC (20 μM) as substrate as described in Materials and Methods. Molecular masses (in kilodaltons) are shown.
DISCUSSION
ILP-1 is a potent inhibitor of apoptosis which exerts its effects, at least in part, through the direct binding and inhibition of specific caspases. For example, ILP-1 can bind active caspase 3 with nanomolar affinity and is a powerful inhibitor of its enzymatic activity (10). Interestingly, ILP-1 is able to bind only the active, heterotetrameric form of caspase 3 and does not associate with its inactive precursor (12). An important consequence of the high affinity of ILP-1 for caspase 3 is that ILP-1 is capable of suppressing a wide variety of apoptotic signals, including signaling through death receptors as well as DNA-damaging agents, such as irradiation and chemotoxic drugs. While these stimuli have been shown to initiate distinct apoptotic pathways involving the sequential activation of different caspases, they are thought to converge at the downstream effector caspases, particularly caspase 3, and this is thought to account for the wide-ranging protective effects of ILP-1 (10).
In this report, we describe the cloning and characterization of a novel IAP gene, ILP-2. Examination of the genomic structure of the ILP-2 locus revealed the absence of introns as well as an autosomal location. Most gene families are thought to have arisen during evolution by duplication of an ancestral gene, often followed by sequence divergence and ultimately to distinct biological functions. In this case, the general intron-exon structure of the gene family should be conserved. The lack of introns in ILP-2 strongly suggests that it was generated by a retroprocessing event (45), that is, by the integration into the genome of a fully spliced ILP-1 cDNA generated by reverse transcription. In some cases, retrotransposons have been found to encode functional proteins (28). One example, which has striking similarity to the situation of ILP-1 and ILP-2, is that of the X-linked gene XAP-5 and its autosomal retrotransposon-derived counterpart, X5L (36). Like ILP-2, X5L has an intronless open reading frame which is highly conserved, encodes a functional protein, and is preferentially expressed in testis. While the functions of XAP-5 and X5L are unknown, it has been proposed that the testis-specific expression of X5L serves to compensate for the absence of XAP-5 in the Y-containing gamete during spermatogenesis (36). In this regard, ILP-2 is highly analogous to X5L, since it is autosomal (Mir and Duckett, unpublished) and has a conserved open reading frame and in primary tissues is expressed exclusively in the testis (Fig. 2). However, the retroposition event which resulted in ILP-2 most likely occurred relatively recently in primate evolution, while the X5L retroposition is thought to have taken place before the radiation of eutherian mammals (36, 39). The human ILP-1 and ILP-2 DNA sequences are significantly closer to each other than they are to murine Xiap. This, combined with Southern blot analysis data (Fig. 1C), suggests that a murine counterpart of ILP-2 might not exist, and therefore a direct test of the biological significance of ILP-2 by gene disruption cannot be performed. However, since ILP-1 can be proteolytically processed to a form which closely resembles ILP-2, it will be of particular interest to examine the tissue expression and functional properties of processed ILP-1.
Analysis of the ILP-2 open reading frame reveals a close similarity to ILP-1. The most significant difference between ILP-2 and ILP-1 is that ILP-2 lacks the two amino-terminal BIR domains which are present in ILP-1. Given the high primary sequence homology between the two proteins, it is more likely that this major structural difference, rather than the small degree of sequence variation, is the main determinant of the differences in biological activity between the two proteins. In support of this hypothesis, two recent reports have described the existence of a proteolytically processed form of ILP-1 (9, 23) which shares many structural and functional properties with ILP-2.
Despite its similarity to ILP-1, ILP-2 was found to function in a far more restricted manner. Overexpression of ILP-2 failed to inhibit apoptosis induced by signals through death domain-containing receptors but potently suppressed apoptosis induced by Bax or by ectopic expression of Apaf-1 and caspase 9. In addition, cytosolic extracts preincubated with either recombinant ILP-1 or ILP-2 before activation with ATP were found to suppress caspase activity. However, ILP-2 was unable to inhibit caspase activity when added after the extracts had been activated with ATP, while ILP-1 retained its inhibitory function under these conditions.
In many respects, the properties of ILP-2 appear to be very similar to a recently described cleaved form of ILP-1 which lacks the first two amino-terminal BIR domains (9, 23). Both proteins were capable of suppressing apoptosis induced by Bax or by Apaf-1 and caspase 9, and neither ILP-2 nor the equivalent version of ILP-1 was able to inhibit Fas-mediated apoptosis. However, in transfected cells, processed caspase 9, but not full-length caspase 9, was found to coprecipitate with ILP-2 or its ILP-1 equivalent, and in contrast to a previous report (11), caspase 9 did not coprecipitate with full-length ILP-1. A recent study by Datta et al. (8) reported that ILP-1/XIAP could coprecipitate from cells with processed caspase 9, but not full-length caspase 9, and this would appear to be consistent with our data. One possibility is that different forms of ILP-1 and caspase 9 may exist in distinct cellular compartments. For example, processed caspase 9 of the form observed in Fig. 6 is thought to be generated in the Apaf-1-containing apoptosome (1, 31, 34), and so it is formally possible that, in a cell, ILP-1 and ILP-2 might specifically be targeted to this enzyme complex.
Of particular interest was the observation that preincubation of extracts with GST–ILP-2 resulted in fractions containing a large proportion of full-length caspase 9 (Fig. 6B), while in cotransfection studies, processed caspase 9, but not full-length caspase 9, coprecipitated with ILP-2 (Fig. 5A). While we currently have no definitive explanation for these apparently contradictory findings, one possibility that would reconcile the data would be if Apaf-1, following caspase 9 recruitment, processing, and release, could recruit further full-length caspase 9 molecules. If ILP-2 were targeted to the Apaf-1-containing apoptosome, its binding to processed caspase 9 might affect the release of processed caspase 9 from Apaf-1 and the subsequent recruitment of other caspase 9 molecules. Thus, the observed result would be an abundance of full-length caspase 9, even though ILP-2 was functioning specifically on processed caspase 9, and this is consistent with our findings. While this is an attractive possibility, other models can be proposed, and further studies will be required to determine the precise mode of ILP-2 action.
These studies reinforce the notion that IAP proteins can function to suppress apoptosis at multiple points in the cell death pathway. The high degree of specificity of ILP-2 may be very useful in better understanding the apoptotic machinery, and this may also be important as other Apaf-1-independent pathways are identified. Finally, ILP-2 or the equivalent ILP-1 fragment may represent a novel therapeutic target, since disruption of these proteins from the holoenzyme would be predicted to specifically sensitize tumor cells to chemotherapy-induced apoptosis.
ACKNOWLEDGMENTS
Bettina W. M. Richter and Samy S. Mir contributed equally to this study.
We thank R. Armstrong, B. Mayer, S. Korsmeyer, R. Siegel, and C. Zacharchuk for plasmids, Y. Lazebnik for Apaf-1 antibody, the Sanger Centre for the PAC genomic clone of ILP-1, and J. Ashwell for helpful advice and critical reading of the manuscript. S.S.M. is an HHMI-NIH Research Scholar.
Footnotes
This paper is dedicated to the memory of Lois K. Miller. This is manuscript no. 45 of the Genoma 2000/ITBA Project funded by Cariplo.
REFERENCES
- 1.Beere H M, Wolf B B, Cain K, Mosser D D, Mahboubi A, Kuwana T, Tailor P, Morimoto R I, Cohen G M, Green D R. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat Cell Biol. 2000;2:469–475. doi: 10.1038/35019501. [DOI] [PubMed] [Google Scholar]
- 2.Borden K L, Freemont P S. The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Budihardjo I, Oliver H, Lutter M, Luo X, Wang X. Biochemical pathways of caspase activation during apoptosis. Annu Rev Cell Dev Biol. 1999;15:269–290. doi: 10.1146/annurev.cellbio.15.1.269. [DOI] [PubMed] [Google Scholar]
- 5.Cain K, Bratton S B, Langlais C, Walker G, Brown D G, Sun X M, Cohen G M. Apaf-1 oligomerizes into biologically active approximately 700-kDa and inactive approximately 1.4-MDa apoptosome complexes. J Biol Chem. 2000;275:6067–6070. doi: 10.1074/jbc.275.9.6067. [DOI] [PubMed] [Google Scholar]
- 6.Cain K, Brown D G, Langlais C, Cohen G M. Caspase activation involves the formation of the aposome, a large (approximately 700 kDa) caspase-activating complex. J Biol Chem. 1999;274:22686–22692. doi: 10.1074/jbc.274.32.22686. [DOI] [PubMed] [Google Scholar]
- 7.Chau B N, Cheng E H, Kerr D A, Hardwick J M. Aven, a novel inhibitor of caspase activation, binds Bcl-xL and Apaf-1. Mol Cell. 2000;6:31–40. [PubMed] [Google Scholar]
- 8.Datta R, Oki E, Endo K, Biedermann V, Ren J, Kufe D. XIAP regulates DNA damage-induced apoptosis downstream of caspase-9 cleavage. J Biol Chem. 2000;275:31733–31738. doi: 10.1074/jbc.M910231199. [DOI] [PubMed] [Google Scholar]
- 9.Deveraux Q L, Leo E, Stennicke H R, Welsh K, Salvesen G S, Reed J C. Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. EMBO J. 1999;18:5242–5251. doi: 10.1093/emboj/18.19.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deveraux Q L, Reed J C. IAP family proteins—suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 11.Deveraux Q L, Roy N, Stennicke H R, Van Arsdale T, Zhou Q, Srinivasula S M, Alnemri E S, Salvesen G S, Reed J C. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devereaux Q L, Takahashi R, Salvesen G S, Reed J C. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 13.Duckett C S, Gedrich R W, Gilfillan M C, Thompson C B. Induction of nuclear factor κB by the CD30 receptor is mediated by TRAF1 and TRAF2. Mol Cell Biol. 1997;17:1535–1542. doi: 10.1128/mcb.17.3.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duckett C S, Li F, Wang Y, Tomaselli K J, Thompson C B, Armstrong R C. Human IAP-like protein regulates programmed cell death downstream of Bcl-xL and cytochrome c. Mol Cell Biol. 1998;18:608–615. doi: 10.1128/mcb.18.1.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duckett C S, Nava V E, Gedrich R W, Clem R J, Van Dongen J L, Gilfillan M C, Shiels H, Hardwick J M, Thompson C B. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J. 1996;15:2685–2694. [PMC free article] [PubMed] [Google Scholar]
- 16.Duckett C S, Thompson C B. CD30-dependent degradation of TRAF2: implications for negative regulation of TRAF signaling and the control of cell survival. Genes Dev. 1997;11:2810–2821. doi: 10.1101/gad.11.21.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fearnhead H O, McCurrach M E, O'Neill J, Zhang K, Lowe S W, Lazebnik Y A. Oncogene-dependent apoptosis in extracts from drug-resistant cells. Genes Dev. 1997;11:1266–1276. doi: 10.1101/gad.11.10.1266. [DOI] [PubMed] [Google Scholar]
- 18.Fearnhead H O, Rodriguez J, Govek E E, Guo W, Kobayashi R, Hannon G, Lazebnik Y A. Oncogene-dependent apoptosis is mediated by caspase-9. Proc Natl Acad Sci USA. 1998;95:13664–13669. doi: 10.1073/pnas.95.23.13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 20.Hegde R, Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Alnemri E S. Blk, a BH3-containing mouse protein that interacts with Bcl-2 and Bcl-xL, is a potent death agonist. J Biol Chem. 1998;273:7783–7786. doi: 10.1074/jbc.273.14.7783. [DOI] [PubMed] [Google Scholar]
- 21.Hengartner M O. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 22.Joazeiro C A, Weissman A M. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 23.Johnson D E, Gastman B R, Wieckowski E, Wang G Q, Amoscato A, Delach S M, Rabinowich H. Inhibitor of apoptosis protein hILP undergoes caspase-mediated cleavage during T lymphocyte apoptosis. Cancer Res. 2000;60:1818–1823. [PubMed] [Google Scholar]
- 24.Kerr J F, Wyllie A H, Currie A R. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 26.Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda J-E, MacKenzie A, Korneluk R G. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature. 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Kim C N, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 28.Maestre J, Tchenio T, Dhellin O, Heidmann T. mRNA retroposition in human cells: processed pseudogene formation. EMBO J. 1995;14:6333–6338. doi: 10.1002/j.1460-2075.1995.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perkins N D, Edwards N L, Duckett C S, Agranoff A B, Schmid R M, Nabel G J. A cooperative interaction between NF-κB and Sp1 is required for HIV-1 enhancer activation. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajcan-Separovic E, Liston P, Lefebvre C, Korneluk R G. Assignment of human inhibitor of apoptosis protein (IAP) genes xiap, hiap-1, and hiap-2 to chromosomes Xq25 and 11q22–q23 by fluorescence in situ hybridization. Genomics. 1996;37:404–406. doi: 10.1006/geno.1996.0579. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez J, Lazebnik Y. Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 1999;13:3179–3184. doi: 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothe M, Pan M-G, Henzel W J, Ayres T M, Goeddel D V. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 33.Roy N, Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saleh A, Srinivasula S M, Balkir L, Robbins P D, Alnemri E S. Negative regulation of the Apaf-1 apoptosome by Hsp70. Nat Cell Biol. 2000;2:476–483. doi: 10.1038/35019510. [DOI] [PubMed] [Google Scholar]
- 35.Salvesen G S, Dixit V M. Caspases: intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 36.Sedlacek Z, Munstermann E, Dhorne-Pollet S, Otto C, Bock D, Schutz G, Poustka A. Human and mouse XAP-5 and XAP-5-like (X5L) genes: identification of an ancient functional retroposon differentially expressed in testis. Genomics. 1999;61:125–132. doi: 10.1006/geno.1999.5931. [DOI] [PubMed] [Google Scholar]
- 37.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Alnemri E S. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 38.Stennicke H R, Deveraux Q L, Humke E W, Reed J C, Dixit V M, Salvesen G S. Caspase-9 can be activated without proteolytic processing. J Biol Chem. 1999;274:8359–8362. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- 39.Stewart C B, Disotell T R. Primate evolution—in and out of Africa. Curr Biol. 1998;8:R582–R588. doi: 10.1016/s0960-9822(07)00367-3. [DOI] [PubMed] [Google Scholar]
- 40.Sun C, Cai M, Gunasekera A H, Meadows R P, Wang H, Chen J, Zhang H, Wu W, Xu N, Ng S C, Fesik S W. NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature. 1999;401:818–822. doi: 10.1038/44617. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi R, Deveraux Q, Tamm I, Welsh K, Assa-Munt N, Salvesen G S, Reed J C. A single BIR domain of XIAP sufficient for inhibiting caspases. J Biol Chem. 1998;273:7787–7790. doi: 10.1074/jbc.273.14.7787. [DOI] [PubMed] [Google Scholar]
- 42.Thornberry N A, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 43.Uren A, Pakusch M, Hawkins C, Puls K L, Vaux D L. Cloning and expression of apoptosis inhibitory protein homologs that function to inhibit apoptosis and/or bind tumor necrosis factor receptor-associated factors. Proc Natl Acad Sci USA. 1996;93:4974–4978. doi: 10.1073/pnas.93.10.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vander Heiden M G, Chandel N S, Williamson E K, Schumacker P T, Thompson C B. Bcl-xL regulates the membrane potential and volume homeostasis of mitochondria. Cell. 1997;91:627–637. doi: 10.1016/s0092-8674(00)80450-x. [DOI] [PubMed] [Google Scholar]
- 45.Vanin E F. Processed pseudogenes: characteristics and evolution. Annu Rev Genet. 1985;19:253–272. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- 46.Vaux D L, Haecker G, Strasser A. An evolutionary perspective on apoptosis. Cell. 1994;76:777–779. doi: 10.1016/0092-8674(94)90350-6. [DOI] [PubMed] [Google Scholar]
- 47.White E. Death-defying acts: a meeting review on apoptosis. Genes Dev. 1993;7:2277–2284. doi: 10.1101/gad.7.12a.2277. [DOI] [PubMed] [Google Scholar]
- 48.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Cai J, Peng T-I, Jones D P, Wang X. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 49.Yang Y, Fang S, Jensen J P, Weissman A M, Ashwell J D. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;28:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]