Abstract
The discovery that the V(D)J recombinase functions as a transposase in vitro suggests that transposition by this system might be a potent source of genomic instability. To gain insight into the mechanisms that regulate transposition, we investigated a phenomenon termed target commitment that reflects a functional association between the RAG transposase and the target DNA. We found that the V(D)J recombinase is quite promiscuous, forming productive complexes with target DNA both before and after donor cleavage, and our data indicate that the rate-limiting step for transposition occurs after target capture. Formation of stable target capture complexes depends upon the presence of active-site metal binding residues (the DDE motif), suggesting that active-site amino acids in RAG-1 are critical for target capture. The ability of the RAG transposase to commit to target prior to cleavage may result in a preference for transposition into nearby targets, such as immunoglobulin and T-cell receptor loci. This could bias transposition toward relatively “safe” regions of the genome. A preference for localized transposition may also have influenced the evolution of the antigen receptor loci.
Jawed vertebrates create a diverse repertoire of antigen receptors through a series of programmed DNA rearrangements during lymphocyte differentiation. This process, V(D)J recombination, is catalyzed by a multisubunit recombinase that contains two lymphoid-cell-specific proteins, RAG1 and RAG2. These proteins recognize recombination signal sequences (RSS) that flank all T- and B-cell antigen receptor gene segments (hereafter referred to as coding segments). Double-stranded DNA breaks are introduced at these sites via a two-step mechanism. First, a nick is introduced between the RSS and the adjacent coding segment; second, the newly generated 3′OH attacks the corresponding phosphodiester bond on the opposite strand, creating a covalently sealed (hairpin) coding end and a 5′-phosphorylated blunt signal end. The broken ends are repaired with the participation of several double-stranded break repair proteins, thus creating a diverse array of rearranged antigen receptor genes (reviewed in references 10 and 22).
There are many similarities between the mechanism of V(D)J recombination and the movements of transposable elements (reviewed in reference 32). In light of these similarities and the unusual organization of the RAG genes, it has been suggested that the V(D)J recombination system evolved from an ancient cut-and-paste-type transposon (29, 35, 39). This hypothesis is supported by the observation that RAG proteins, together with the nonspecific DNA-bending protein HMG1, can form an active transposase in vitro, capable of efficiently inserting DNA molecules terminating in signal ends into target DNA molecules without significant target site specificity (1, 14).
The discovery that the V(D)J recombinase can function as a transposase in vitro raised the possibility that this system might be a potent source of genomic instability in developing lymphocytes that are actively undergoing V(D)J recombination (14). The presence of germ line antigen receptor rearrangements in sharks and skates suggests that RAG-mediated transposition could contribute to genome rearrangements in nonlymphoid cells as well (23, 31). It is, therefore, of great interest to understand the mechanism of transposition and its regulation.
Linear DNA molecules terminating in signal ends—the substrates for transposition—are quite abundant and apparently long-lived in developing lymphocytes (30, 33). These signal ends are thought to remain bound to the RAG proteins (2, 13). In view of the observation that such precleaved signal ends complexed to the RAG proteins efficiently undergo transposition in vitro, it would seem that V(D)J recombination in vivo is a rather risky enterprise. However, RAG-mediated transposition has not yet been documented in vivo, and it is likely that regulatory mechanisms are in place to limit the frequency of transposition in lymphocytes (1, 10, 14, 26, 32).
Selection of the target DNA molecule seems a reasonable step by which to regulate transposition specifically without affecting V(D)J recombination. Some transposons, such as Tn7, show a strong preference for specific target sites (8). In fact, cleavage at Tn7 ends does not proceed until a proper target site has been identified (6). Some mobile elements with much less stringent target sequence requirements, such as the bacteriophage Mu transposase, can interact with the target DNA both prior to and after donor cleavage (7, 27). In contrast, the Tn10 transposase, whose behavior closely parallels that of the V(D)J recombinase in other respects (17), does not stably interact with its target until after the transposon ends have been liberated from the flanking DNA (34). Cleavage may be required to expose the target DNA binding pocket of the transposase (34). Recent experiments have established that the RAG proteins contact the coding flank DNA (9), and it has been suggested that like Tn10, the RAG proteins may interact with the target DNA only after cleavage (1).
We investigated interactions between the RAG transposase and target DNA. In addition to identifying a RAG-donor-target complex (termed a target capture complex), we examined a phenomenon termed target commitment, a functional association between the RAG-RSS complex and target DNA that is resistant to addition of a competitor target. Target commitment can be distinct from target capture: in the case of Tn10 only a subset of target capture complexes exhibit commitment, which is viewed as a more functionally significant interaction (34). We found that the RAG proteins indeed exhibit target commitment. Furthermore, our data demonstrate that, contrary to expectation, the V(D)J recombinase is a promiscuous transposase that can form productive complexes with target DNA both before and after RSS cleavage. The ability of the RAG transposase to commit to target prior to cleavage may result in a preference for transposition into nearby targets, such as immunoglobulin and T-cell receptor loci. Functional and evolutionary implications of this newly discovered property of the RAG proteins are discussed.
MATERIALS AND METHODS
DNA.
Uncleaved 16-bp coding flank 12- and 23-RSS were created by annealing the DAR39/40 and DG61/62 (25) oligonucleotides, respectively. Uncleaved 41-bp coding flank 12- and 23-RSS oligonucleotides were created by annealing SK42 (5′-CTGCAGGTACAGGACGAGTTCTACAGATCTGGCCTGTCTGCCACAGTGCTACAGACTGGAACAAAAACCCTGCAG) to its complement, SK43, and MBN21 (5′-CTGCAGGTACAGGACGAGTTCTACAGATCTGGCCTGTCTGCCACAGTGGTAGTACTCCACTGTCTGGCTGTACAAAAAC CCTGCAG) to its complement, MBN22. Precleaved 12- and 23-RSS oligonucleotides were created by annealing DG10 and DG4 to their complements (25). The target used for physical detection of target capture complexes was the oligonucleotide mm30b (5′-ATCGAGGACGCAGTTACGTTCCCGGAGATC) annealed to its complement, mm30t. All oligonucleotides were purchased from GIBCO Life Technologies and were gel purified before use. pUC19 and pcDNA1/AMP were used as the short and long plasmid targets, respectively. The linearized target size standards were created by digesting the appropriate target plasmids with BamHI and by 5′ end labeling with T4 polynucleotide kinase.
Proteins.
Recombinant truncated (core) RAG proteins (amino acids 384 to 1008 of RAG1 and 1 to 387 of RAG2) were purified from baculovirus-infected insect cells as previously described (3, 19, 25, 41). Both proteins contain a carboxy-terminal nine-histidine tag, three human c-myc epitope tags, and an amino-terminal maltose-binding protein fusion. The target capture assay and DDE mutant RAG experiments were performed using RAG-1 and RAG-2 glutathione S-transferase (GST) fusions copurified from Chinese hamster ovary (RMP41) cells as previously described (36–38). Both types of protein preparations were capable of target commitment; however, the GST epitope-tagged proteins were more active.
Target commitment.
Unless otherwise noted, 10-μl preincubation mixtures contained 32.7 mM K+-HEPES (pH 7.5), 2.6 mM dithiothreitol, 19.4 mM potassium glutamate, 5.1 mM CaCl2, 6% glycerol, 60 μg of bovine serum albumin/μl, 0.006% NP-40, 150 ng each of MR1 and MR2, 1.5 ng of HMG1/μl, and donor and target DNA as indicated. Following preincubation at the indicated time and temperature, reaction mixtures were spiked with mixtures of MgCl2 or other Me2+ as indicated (3 mM, final concentration), polyethylene glycol 8000 (10%, final wt/vol ratio), and the indicated target or Tris-EDTA (TE) for a final reaction volume of 15 μl and were incubated at 37°C for 30 min (except in the kinetic analyses). Fifteen microliters of stop buffer (100 mM Tris [pH 8.0], 10 mM EDTA, 0.2% sodium dodecyl sulfate [SDS], 0.35 mg of proteinase K/ml) was added, and incubation continued at 37°C for >1.5 h. All reactions were subjected to electrophoresis through 1% SeaKem GTG agarose (FMC BioProducts) 25-cm gels containing 0.2 μg of ethidium bromide/ml at 180 V for ∼200 min in 1× Tris-borate-EDTA. Dried gels were visualized with a PhosphorImager.
Target commitment assays.
Uncleaved donor reactions were performed as indicated above with 0.02 pmol of 5′ 32P-end-labeled 12-RSS donor, 0.02 pmol of 23-RSS donor, and 0.04 pmol of pcDNA1/AMP and/or pUC19 target. Precleaved donor reactions were performed as indicated above, using 0.12 pmol of 5′ 32P-end-labeled 12-RSS donor, 0.12 pmol of 23-RSS donor, and 0.03 pmol of pcDNA1/AMP and/or pUC19. DDE mutant experiments were performed as indicated above with 0.02 pmol of 5′ 32P-end-labeled 12-RSS donor, 0.02 pmol of 23-RSS donor, and 0.04 pmol of pcDNA1/AMP and/or pUC19 target.
Target capture assays.
Preincubations (6.5 μl) were carried out at 37°C for 10 min, in mixtures containing 25 mM K-morpholinepropanesulfonic acid (MOPS) (pH 7.0), 4 mM dithiothreitol, 75 mM potassium glutamate, 5.4 mM CaCl2, 100 μg of bovine serum albumin/ml, 150 ng each of GST-R1 and GST-R2, 20 ng of HMG1, and 0.13 pmol each of uncleaved 12- and 23-RSS donor. The reaction mixtures were spiked with mixtures of MgCl (5 mM, final concentration), dimethyl sulfoxide (DMSO) (10%, final wt/vol ratio), and 0.65 pmol of annealed oligonucleotide target for a final reaction volume of 10 μl and were incubated at 37°C for 15 min. These reactions were also performed without the addition of DMSO. Target capture was found to be completely independent of the presence of DMSO. Two microliters of 50% glycerol was added to all reaction mixtures except for the stop buffer-treated reaction mixture, which received 10 μl of stop buffer and was incubated for 15 min at 37°C prior to the addition of 4 μl of 50% glycerol. Entire reaction mixtures were then loaded directly onto a 4 to 20% gradient nondenaturing polyacrylamide gel and run at 120 V for 110 min in 1× Tris-borate-EDTA at 4°C. Dried gels were visualized by PhosphorImager analysis. Electrophoretic mobility shifts of labeled donor were conducted as previously described (21), except that glutaraldehyde was omitted.
RESULTS
Target commitment prior to donor cleavage.
The ability of the Tn10 transposase to interact with its target has been analyzed using staged reactions in which the transposase (along with appropriate transposon end sequences) is first incubated with target DNA in Ca2+, which promotes assembly of protein-DNA complexes but does not support transposition. This preincubation is followed by the addition of a second distinguishable target along with Mg2+, a divalent metal ion that allows transposition. A functionally significant interaction between the transposase-end complex and the target (commitment) is inferred if preferential integration into the first target is observed (34).
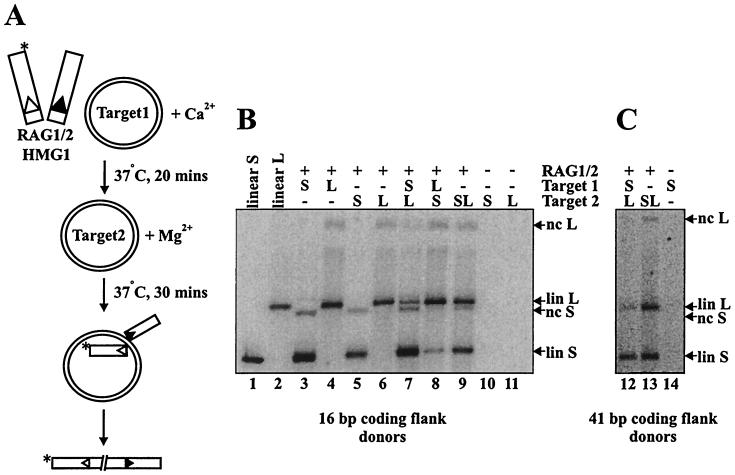
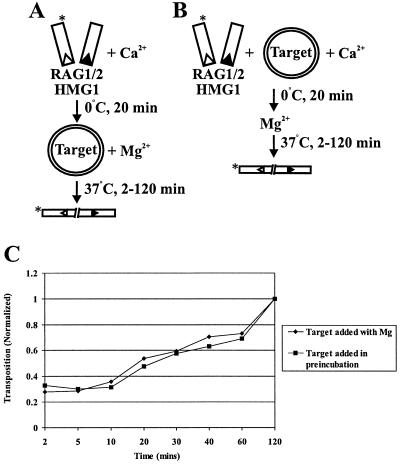
We adopted a similar strategy to study the interactions of the RAG transposase with target DNA. We performed staged reactions in which uncleaved 12- and 23-spacer RSS-containing oligonucleotides were preincubated with the RAG proteins and HMG1 under conditions that support assembly but not transposition (generally in Ca2+). Donor molecules containing either 16 or 41 bp of flanking DNA were tested, yielding similar results (Fig. 1B and C, respectively). Target plasmids of two distinct sizes (2.7 and 4.8 kb) were used (illustrated schematically in Fig. 1A). Target 1 was added in the preincubation, and target 2 was added, along with Mg2+, in the second incubation. If the RAG transposase captures the target prior to cleavage and remains committed to this target, then target 1 should be preferred over an equimolar amount of target 2. If, on the other hand, the RAG proteins do not commit to the first target prior to cleavage, then there should be no preference for transposition into either target.
FIG. 1.
The RAG transposase can commit to target prior to donor cleavage. (A) Schematic of target commitment assay with uncleaved donor. The asterisk indicates the location of radiolabel. (B) Transposition products are either linearized (lin) or nicked circular (nc) target species resulting from concerted or single integration of donor molecules, respectively (single events are not pictured in the schematic). Linear products of concerted transposition events migrate slightly more slowly than the linearized standards due to the presence of the integrated donor molecules. − indicates that dialysis buffer was substituted for RAG protein or that TE was substituted for DNA in indicated reactions. S and L refer to short and long targets, respectively.
As shown in Fig. 1B, incubation of the RAG proteins with either target 1 or 2 alone yielded predominantly double-ended transposition events, resulting in linearization of the target plasmid (Fig. 1B, lanes 3 and 4; compare with linearized plasmid standards in lanes 1 and 2), in agreement with previous studies (1, 14). Consistent with earlier reports (1, 14), approximately 1% of the radiolabeled donor was integrated into the plasmid targets. As shown in Fig. 1, the staged reactions reveal a clear preference for the first target, regardless of whether the smaller or larger plasmid served as target 1 (Fig. 1B, lanes 7 and 8, and C, lane 12). The preference for target 1 was at the expense of target 2 utilization (compare Fig. 1B, lanes 7 and 8, with lanes 6 and 5, respectively). As controls, both plasmids were added together during the preincubation (data not shown) or at the time of Mg2+ addition (Fig. 1B, lane 9, and C, lane 13); as expected, no target preference was observed. Thus, the RAG proteins, unlike the Tn10 transposase, interact with and commit to the target prior to cleavage.
Target commitment after donor cleavage.
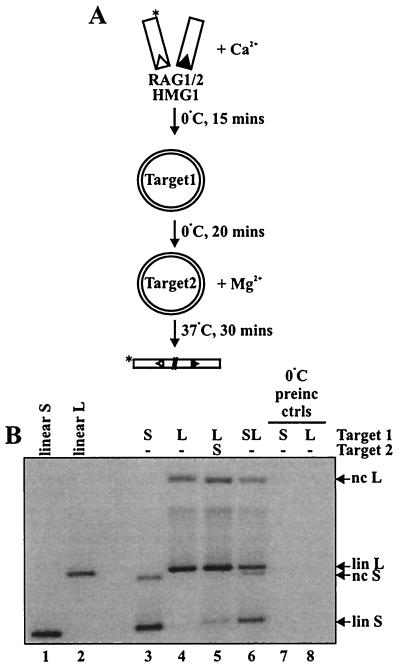
We next examined the behavior of precleaved donor DNA molecules. Staged reactions were performed as described above, except that the preincubation step was performed at 0°C to prevent transposition during the preincubation (Fig. 2A), because precleaved donors are active for transposition in Ca2+ (14). To verify that transposition did not occur at 0°C, additional reactions with target present during the Ca2+ preincubation phase were performed in parallel without a Mg2+ addition step; no transposition was observed in multiple experiments (for example, see Fig. 2B, lanes 7 and 8). No preferential use of either target was observed when both targets were present in the preincubation (Fig. 2B, lane 6) or in the second, Mg2+-containing incubation (data not shown). The staged reactions, however, revealed a preferential use of target 1 (Fig. 2B, lane 5; see also Fig. 6B, lane 6), indicating that precleaved complexes support target commitment.
FIG. 2.
The RAG transposase can commit to target after donor cleavage. (A) Schematic of RAG transposase target commitment assay with precleaved donor end molecules. Asterisks indicate the position of radiolabel. (B) Agarose gel of transposition products. −, no addition of target; preinc, preincubation; ctrls, controls; lin; linear; nc, nicked circular; S, short; L, long.
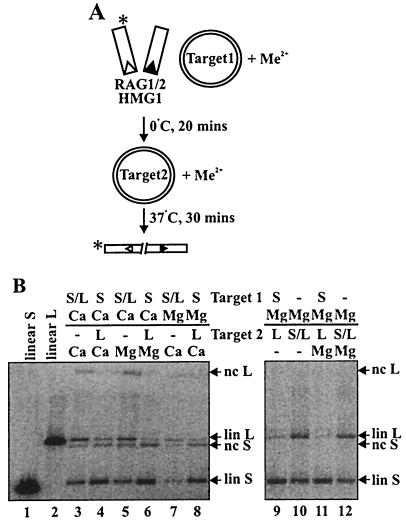
FIG. 6.
The RAG transposase commits to target in the presence of both Ca2+ and Mg2+. (A) Schematic of assay employed to test the contributions of Ca2+ and Mg2+ to target commitment. Preincubation mixtures contained 5.1 mM concentrations of the indicated divalent metal ion; the indicated divalent metal ion was added with target 2 or TE to a final concentration of 3 mM. Asterisks indicate the position of radiolabel. (B) Truncated RAG proteins (lacking the N-terminal maltose-binding protein fusion) were used in the experiment shown. Commitment to target 1 was observed in Ca2+/Ca2+, Ca2+/Mg2+, Mg2+/Ca2+, and Mg2+/Mg2+ reactions. lin, linear; nc, nicked circular; S, short; L, long, −, no addition of target.
Preincubation with target does not accelerate the rate of strand transfer.
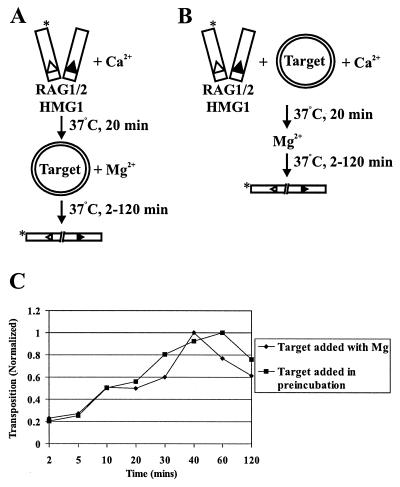
Target commitment could simply reflect accelerated kinetics of transposition, as the formation of a target capture complex could be a rate-limiting step. Indeed, in the case of Tn10, preincubation of target with transposition complexes containing precleaved donor DNA molecules substantially accelerates the rate of strand transfer in staged reactions (34). We therefore examined the effects of preincubation with uncleaved donor DNA on the kinetics of strand transfer, following the protocols schematized in Fig. 3A and B. We added target either simultaneously with Mg2+ (Fig. 3A) or during the preincubation (Fig. 3B). The rate of strand transfer was not significantly accelerated by preincubating with target (Fig. 3C). As expected, preincubation with target did not affect the rate of donor cleavage (data not shown). Transposition decreased after approximately 40 to 60 min, perhaps as a result of disintegration (26).
FIG. 3.
Preincubation with target DNA does not accelerate the kinetics of strand transfer of uncleaved donor ends. Panels A and B show assays utilizing uncleaved donor molecules in which target was added either with Mg2+ or during preincubation. Asterisks indicate the position of radiolabel. (C) The amount of transposition is plotted as a function of the incubation time with Mg2+ at 37°C. Target was added either with Mg2+ or during preincubation.
We next studied the kinetic effects of target preincubation on transposition by precleaved donor molecules, as depicted schematically in Fig. 4A and B. Target was added at the time of Mg2+ addition (Fig. 4A) or during the preincubation (Fig. 4B). As with the uncleaved donor, the time course of transposition of precleaved donor molecules was not significantly affected by preincubation with target (Fig. 4C). These data demonstrate that preincubation with target DNA molecules does not significantly increase the rate of transposition, suggesting that a reaction step subsequent to target capture is rate limiting. In fact, varying the time of target 1 preincubation revealed that target commitment occurred very quickly, in less than 1 min (the earliest time point; data not shown).
FIG. 4.
Preincubation with target DNA does not accelerate the kinetics of strand transfer of precleaved donor ends. Panels A and B picture assays utilizing precleaved donor molecules in which target was added either with Mg2+ or during the preincubation. Asterisks indicate position of radiolabel. (C) The amount of transposition observed is plotted as a function of the incubation time with Mg2+ at 37°C. Target was added either with Mg2+ or during preincubation.
Transposase-target interactions are reversible in Ca2+.
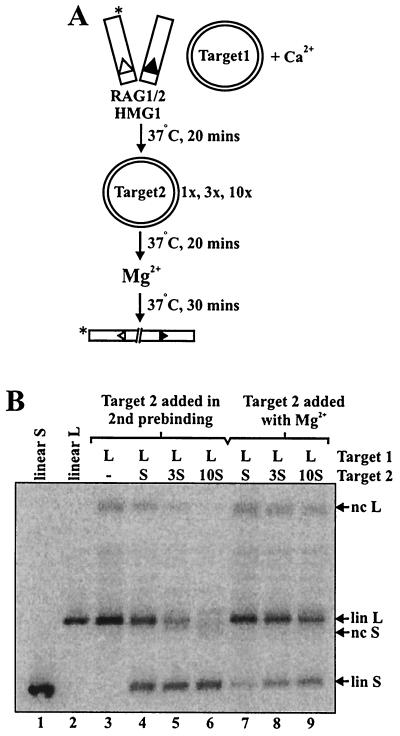
The target commitment experiments described above demonstrate that transposase-target associations formed during preincubation in Ca2+ are resistant to competitor target added at the time of Mg2+ addition. Since Ca2+ supports binding, but not catalysis, we wondered whether the target-transposase association would also be resistant to competitor added in Ca2+. To examine this question, samples preincubated with uncleaved donor and target 1 in Ca2+ were subjected to a second 20-min preincubation in Ca2+ with various amounts of target 2 as competitor (illustrated schematically in Fig. 5A). Even an equimolar amount of target 2 abolished the preference for target 1 (Fig. 5B, lane 4); 3- and 10-fold molar excesses of target 2 progressively diminished the use of target 1 (Fig. 5B, lanes 5 and 6). Note that when high concentrations of competitor target were added in the Ca2+ preincubation, the efficiency of transposition was somewhat diminished (Fig. 5B, compare lanes 3 and 6). This is because the large amounts of plasmid DNA act as a nonspecific competitor, inhibiting synaptic complex formation (12; M. B. Neiditch and D. B. Roth, unpublished data).
FIG. 5.
Transposase-target interactions leading to commitment are reversible in Ca2+. (A) Schematic of assay utilized to determine the stability of target commitment under preincubation and catalytic conditions. The asterisk indicates the position of radiolabel. (B) Preincubations were performed for 20 min, with 0.02 pmol of 12- and 23-spacer RSS donors and 1× (0.04 pmol) pcDNA1/AMP (target 1). 1×, 3×, or 10× molar amounts of pUC19 (target 2) were added as indicated (lanes 4 to 6), and preincubations were continued for 20 min. Reaction mixtures were supplemented with MgCl2 and polyethylene glycol, bringing the final reaction conditions to 3 mM MgCl2 and 10% polyethylene glycol 8000 in 15 μl. These reactions were compared to similar reactions in which 1×, 3×, or 10× target 2 was added at the time of Mg2+ addition (lanes 7 to 9). nc, nicked circular; lin, linear; S, short; L, long; −, no addition of target 2.
These data demonstrate that the transposase-target interactions leading to commitment are reversible in Ca2+. Identical results were obtained with precleaved donor molecules (data not shown), indicating that the target capture complex is not stabilized by the absence of flanking donor DNA. In contrast, commitment to target 1 was resistant to even a 10-fold molar excess of target 2 added during the Mg2+ addition step (Fig. 5B, lane 9). This competitor resistance was also observed when the 2.7-kb target was used as target 1 and when the 4.8-kb target was utilized as target 2 (data not shown). The relative inability of target 2 to compete with target 1 in Mg2+ implies that the RAG-RSS-target complexes are fairly stable in Mg2+ (see below for further discussion).
Effects of divalent metal ions on target commitment.
The lack of stable target commitment in Ca2+ indicates that the transposase-target interactions responsible for commitment are readily reversible under these conditions. Because target commitment is observed upon addition of Mg2+, we wondered whether Mg2+ might increase the stability of transposase-target interactions, locking the transposase onto the target. Alternatively, the establishment of conditions that support catalysis (rather than the presence of Mg2+ per se) might promote an alteration of the transposase-donor-target interaction that induces target commitment.
To test these hypotheses, we made use of the fact that precleaved donor DNA molecules are competent for transposition in both Ca2+ and Mg2+. We performed staged reactions in which the second step was carried out in Ca2+ at 37°C, allowing catalysis to be initiated in the absence of Mg2+ (Fig. 6A). Reactions in which both incubations were carried out in Ca2+ exhibited target commitment that was indistinguishable from reactions performed in the standard fashion, in which Ca2+ is followed by Mg2+ (a representative experiment is shown in Fig. 6B; compare lanes 4 and 6). Target commitment was also observed when target 1 was preincubated in Mg2+ and target 2 was added with Ca2+ (Fig. 6B, lane 8) and when target 1 was preincubated in Mg2+ and target 2 was added with (Fig. 6B, lanes 11 and 12) or without (Fig. 6B, lanes 9 and 10) additional Mg2+. Thus, it is the switch to catalytic conditions, rather than the presence of a specific metal ion, that induces target commitment.
Physical analysis of target capture complexes.
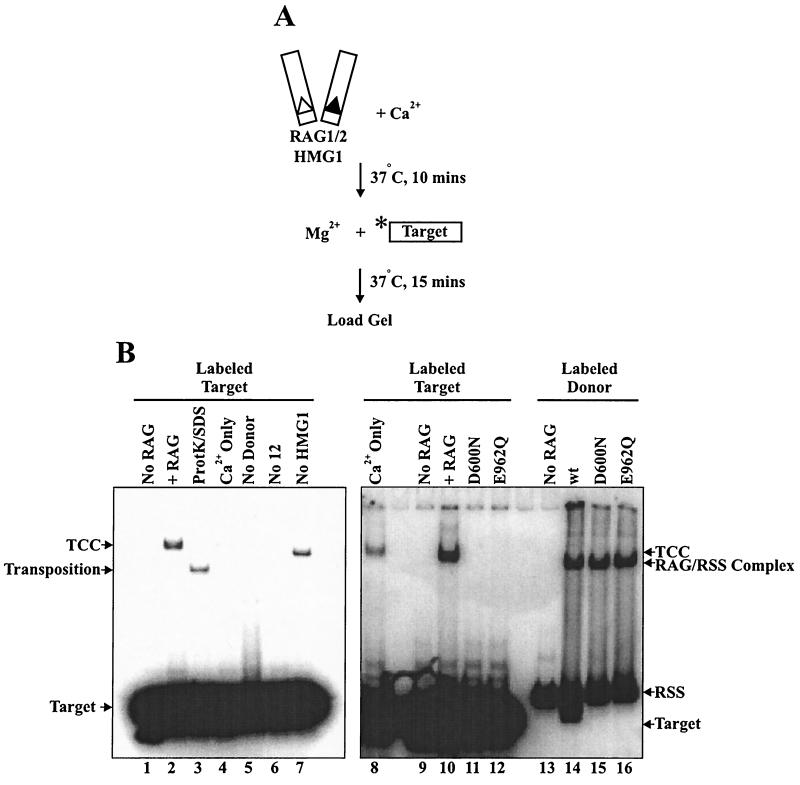
The ability of the RAG transposase to display target commitment implies a stable association between the RAG-RSS complex and the target. To detect such target capture complexes directly, we performed electrophoretic mobility shift assays using a radiolabeled oligonucleotide. No protein cross-linking reagents (such as glutaraldehyde) were used. We observed target capture complexes (Fig. 7B, lane 2) that were dependent on the presence of both RAG proteins (Fig. 7B, lane 1) and donor RSS (Fig. 7B, lane 5). As expected, efficient target capture complex formation required the presence of both a 12- and a 23-RSS (Fig. 7B, lane 6, and data not shown). Ca2+ alone supported inefficient but detectable target capture (Fig. 7B, lanes 4 and 8). Some target capture complex formation occurred in the absence of HMG1, yielding a complex with faster electrophoretic mobility (Fig. 7B, lane 7) that was not observed in standard reactions.
FIG. 7.
Physical detection of RAG target capture complexes. (A) Schematic of assay employed to detect noncovalently associated RAG-donor-target complexes. The asterisk indicates the position of radiolabel. (B) Lanes marked + RAG contain RAG proteins, HMG1, 12- and 23-donor RSS, target, and Mg2+. TCC denotes the position of target capture complexes. All reaction mixturess were loaded directly to gel without extraction. ProtK, proteinase K; wt, wild type.
To determine whether the DNA-protein complexes contained transposition products, reaction mixtures were deproteinized by treatment with SDS and proteinase K and loaded directly to the gel without extraction or precipitation. The target capture complex band was completely ablated upon SDS-proteinase K treatment, and a new band of altered mobility was detected, representing products resistant to SDS-proteinase K treatment. This indicates a covalent association between donor and target DNA, which was further verified by denaturing gel electrophoresis (data not shown). In multiple experiments, however, we noted that only approximately 50% of the target capture complexes were resistant to SDS-proteinase K treatment, indicating that a substantial fraction of target capture complexes had not undergone strand transfer. This observation is consistent with the ability of target capture complexes to form in Ca2+, a condition that does not support strand transfer of uncleaved donor RSS. These data demonstrate the presence of RAG-donor-target complexes (some containing substrates and some containing completed transposition products) that are sufficiently stable to withstand gel electrophoresis.
Analysis of catalytically deficient RAG mutants.
The Tn10 transposase active site has been implicated in transposase-target interactions (16). To gain insight into the mode of target capture employed by the RAG transposase, we examined catalytically deficient RAG mutants bearing point mutations in acidic amino acids important for coordination of divalent metal ions (the so-called DDE motif). DDE mutants are defective for both the hydrolysis and transesterification steps of RSS cleavage and fail to carry out transposition of either uncleaved or precleaved donor RSS in Mg2+ (11, 18, 21).
We used the target capture assay to examine the ability of D600N and E962Q to capture target in Mg2+. Both mutants (as well as wild-type RAG-1) bound RSS in the presence of wild-type RAG-2, yielding a RAG-RSS complex that has a somewhat faster mobility than the RAG-RSS-target complex (Fig. 7B, lanes 14 to 16). The mutants, however, failed to form stable target capture complexes (Fig. 7B, lanes 11 and 12). These data indicate that target capture involves active-site residues in RAG-1.
The D600 and E962 mutants are capable of catalyzing inefficient but detectable transposition of precleaved donor molecules in the presence of Mn2+ (18; Neiditch and Roth, unpublished data). We took advantage of this fact to examine the effects of the D600N and E962Q mutants on target commitment using precleaved donor RSS. Although transposition was quite inefficient, both mutants exhibited target commitment under these conditions (data not shown). These data suggest that under these conditions (in the presence of Mn2+, which bypasses the effects of the DDE mutants on the strand transfer step), the mutant proteins can bind to target DNA. It is difficult to draw firm conclusions about the roles of D600 and E962 in target commitment from these data, however, because it is not clear why Mn2+ rescues the strand transfer activity of the mutant proteins.
DISCUSSION
The interaction between the transposase and target DNA is a critical step in RAG-mediated transposition. Here we show that the RAG-RSS complex stably associates with target DNA. To examine the functional significance of these complexes, we employed staged reactions in which a RAG-RSS complex was preincubated with target DNA under conditions that allow binding but not strand transfer and was then incubated under catalytic conditions. Transposition into the first target was favored even when a large excess of a second target was added in the second incubation. Target commitment was observed with both uncleaved and precleaved donor DNA molecules, contrary to expectation (1). In this respect, the RAG proteins resemble the Mu transposase, which can capture target both before and after donor cleavage (27), and are quite distinct from Tn7 and Tn10, which capture target DNA only before and after cleavage, respectively (5, 6, 34). The RAG transposase is also different from Tn10 in that it does not show an increased rate of strand transfer if preincubated with target. This suggests that the rate-limiting step for RAG-mediated transposition occurs after target capture.
What is the basis of target commitment?
Preincubation in Ca2+ presumably favors the formation of productive RAG-RSS-target complexes. Our data indicate that these complexes are rapidly exchangeable under the preincubation conditions but rapidly become committed upon shift to catalytic conditions, resisting even large molar excesses of a second target. How do catalytic conditions facilitate target commitment? We considered the possibility that commitment could simply reflect efficient strand transfer upon shift to conditions that support catalysis, but two lines of evidence argue against this model. First, strand transfer is slow relative to the rate of target capture complex dissociation: whereas formation of significant amounts of strand transfer products takes at least 10 min (Fig. 3 and 4), under preincubation conditions target capture complexes are readily dissociated by addition of a second target within 2 min (Neiditch and Roth, unpublished observations). Second, target commitment can be established within 20 min at 0°C in the absence of detectable strand transfer (Fig. 2). These data demonstrate that target commitment and strand transfer are distinct steps. Furthermore, the observation that target commitment precedes strand transfer suggests that the RAG proteins may not be irreversibly committed to transposition after capture of the target. In the in vivo situation, signal joint formation may compete with transposition even after the transposase has captured (and possibly committed to) a target DNA molecule. Further experiments are required to examine this possibility.
Others have shown that target capture by Tn10 requires removal of IHF, a DNA-bending protein that is important for assembly of the paired-end complex (34). Preincubation with plasmid DNA titrates IHF away from the Tn10-donor complex, causing a conformational change in the transposase that facilitates target capture. We have observed two distinct forms of the RAG target capture complex, with different electrophoretic mobilities, formed with and without HMG1. These data suggest that target capture can occur in the absence of HMG1 (although the presence of low levels of HMG1 in our RAG protein preparations has not been rigorously excluded). These two forms of the target capture complex may have different properties with respect to target binding and target commitment. Ongoing experiments are examining this possibility.
We hypothesize that commitment reflects a change in the mode of target binding, perhaps resulting from a conformational change in the active site of the transposase so that the target is bound more stably. Precedents for this model are provided by DNA binding proteins that search nonspecifically for target sequences using a readily reversible binding mode and then lock on using a different binding mode upon identifying specific recognition sequences (for example, see reference 15). A similar two-step binding model has been suggested for target capture by the Tn10 transposase (34).
How do the RAG proteins bind to target DNA?
The regions of the RAG transposase responsible for binding to target DNA remain unknown. The observation that the Tn10 transposase exhibits target commitment only with precleaved donor DNA molecules suggests that the same binding site that contacts the flanking donor DNA also binds to the target (34). Our data do not support a similar model for the RAG transposase: we observed target commitment with both uncleaved and precleaved donor DNA molecules. Nevertheless, the stoichiometry of the RAG-RSS-target complex is not known, and the possibility remains that RAG monomers not involved in donor cleavage mediate important interactions with target DNA.
Taking into account studies of Tn10 that indicate that the active-site amino acids responsible for coordinating catalytic metal ions play a role in target binding (16), we hypothesized that the more stable binding mode observed under catalytic conditions involves the acidic active-site residues recently discovered in RAG1 (11, 18, 21). Indeed, our finding that the D600N and E962Q mutants are severely defective for target capture in Mg2+ suggests that these active-site amino acids (which may function to coordinate catalytic divalent metal ions) play an important role in target binding. This is consistent with our finding that stable target commitment is observed only under catalytic conditions. Exactly how metal coordinating residues are involved in target capture remains unclear. These amino acids may be directly involved in target DNA binding; alternatively, the mutations may affect target binding indirectly by altering the conformation of the active site (16).
Biological considerations.
Once produced, signal ends have two possible fates: joining to form a signal joint (the normal pathway) or transposition. RAG-mediated transposition in vivo is likely to have deleterious consequences, for both the cell and the organism, including oncogenic chromosome translocations (14, 32). The fact that transposition has not yet been detected in vivo suggests that this reaction may indeed be carefully controlled (10). The ability of the RAG transposase to select a target DNA molecule prior to cleavage may have important biological ramifications. Target commitment may represent a critical decision point, and our data suggest that the decision to transpose actually precedes RSS cleavage.
The ability to commit to a target prior to cleavage may influence the choice of target sites. Donor cleavage removes a physical constraint and may allow the transposon to diffuse freely throughout the nucleus. If the target is chosen before the transposon is excised, transposition might preferentially occur into targets that are physically close to the relevant antigen receptor loci. Recent evidence demonstrates that genetic rearrangements preferentially involve DNA segments that are spatially juxtaposed (20, 28).
Thus, RAG-mediated transposition may favor targets that lie close to the donor sites, such as the antigen receptor loci themselves (which are in an accessible chromatin configuration at the time of rearrangement). Indeed, both the Ac plant transposable element and Drosophila P elements demonstrate a proclivity for transposition to local target sites (4, 24, 40), suggesting that these transposases also capture target before donor cleavage. A bias toward local transposition by the RAG proteins could serve to channel transposition events into relatively safe regions of the genome. Moreover, localized RAG-mediated transposition may have influenced the evolution of the antigen receptor loci. Evidence suggests that these loci have evolved via repeated rounds of local transposition into the germ line by an ancient transposon (39); this process could have been assisted by the ability of the RAG proteins to commit to target prior to cleavage.
ACKNOWLEDGMENTS
We thank Tania Baker and Ilana Goldhaber-Gordon for many helpful discussions; Vicky Brandt for editorial assistance; Meni Melek for providing physical assay conditions; Ilana Goldhaber-Gordon, S. Kale, J. Qiu, L. E. Huye, and H. Yarnall-Schultz for valuable comments on the manuscript; and L. E. Huye for HMG1 protein. S. Robertson and D. Guzman provided secretarial assistance. We also thank anonymous referees for insightful suggestions.
D.B.R. is an Assistant Investigator of the Howard Hughes Medical Institute. M.A.L. was supported by a predoctoral fellowship from the National Institutes of Health (T32-AI07495). This work was supported by a grant from the National Institutes of Health (AI-36420).
REFERENCES
- 1.Agrawal A, Eastman Q M, Schatz D G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal A, Schatz D G. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 3.Akamatsu Y, Oettinger M A. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Athma P, Grotewold E, Peterson T. Insertional mutagenesis of the maize P gene by intragenic transposition of Ac. Genetics. 1992;131:199–209. doi: 10.1093/genetics/131.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bainton R, Gamas P, Craig N L. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell. 1991;65:805–816. doi: 10.1016/0092-8674(91)90388-f. [DOI] [PubMed] [Google Scholar]
- 6.Bainton R J, Kubo K M, Feng J N, Craig N L. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993;72:931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- 7.Baker T A, Mizuuchi M, Mizuuchi K. MuB protein allosterically activates strand transfer by the transposase of phage Mu. Cell. 1991;65:1003–1013. doi: 10.1016/0092-8674(91)90552-a. [DOI] [PubMed] [Google Scholar]
- 8.Craig N L. Target site selection in transposition. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 9.Eastman Q M, Villey I J, Schatz D G. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol Cell Biol. 1999;19:3788–3797. doi: 10.1128/mcb.19.5.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fugmann S D, Lee A I, Shockett P E, Villey I J, Schatz D G. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 11.Fugmann S D, Villey I J, Ptaszek L M, Schatz D G. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- 12.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 13.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;7:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 14.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 15.Jack W E, Terry B J, Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc Natl Acad Sci USA. 1982;79:4010–4014. doi: 10.1073/pnas.79.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Junop M S, Haniford D B. Factors responsible for target site selection in Tn10 transposition: a role for the DDE motif in target DNA capture. EMBO J. 1997;16:2646–2655. doi: 10.1093/emboj/16.10.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy A K, Guhathakurta A, Kleckner N, Haniford D B. Tn10 transposition via a DNA hairpin intermediate. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 18.Kim D R, Dai Y, Mundy C L, Yang W, Oettinger M A. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D R, Oettinger M A. Functional analysis of coordinated cleavage in V(D)J recombination. Mol Cell Biol. 1998;18:4679–4688. doi: 10.1128/mcb.18.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozubek S, Lukasova E, Ryznar L, Kozubek M, Liskova A, Govorun R D, Krasavin E A, Horneck G. Distribution of ABL and BCR genes in cell nuclei of normal and irradiated lymphocytes. Blood. 1997;89:4537–4545. [PubMed] [Google Scholar]
- 21.Landree M A, Wibbenmeyer J A, Roth D B. Mutational analysis of RAG-1 and RAG-2 identifies three active site amino acids in RAG-1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis S M. The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 23.Lewis S M, Wu G E. The old and the restless. J Exp Med. 2000;191:1631–1636. doi: 10.1084/jem.191.10.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machida C, Onouchi H, Koizumi J, Hamada S, Semiarti E, Torikai S, Machida Y. Characterization of the transposition pattern of the Ac element in Arabidopsis thaliana using endonuclease I-SceI. Proc Natl Acad Sci USA. 1997;94:8675–8680. doi: 10.1073/pnas.94.16.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 26.Melek M, Gellert M. RAG1/2-mediated resolution of transposition intermediates: two pathways and possible consequences. Cell. 2000;101:625–633. doi: 10.1016/s0092-8674(00)80874-0. [DOI] [PubMed] [Google Scholar]
- 27.Naigamwalla D Z, Chaconas G. A new set of Mu DNA transposition intermediates: alternate pathways of target capture preceding strand transfer. EMBO J. 1997;16:5227–5234. doi: 10.1093/emboj/16.17.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikiforova M N, Stringer J R, Blough R, Medvedovic M, Fagin J A, Nikiforov Y E. Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science. 2000;290:138–141. doi: 10.1126/science.290.5489.138. [DOI] [PubMed] [Google Scholar]
- 29.Oettinger M A, Schatz D G, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 30.Ramsden D A, Gellert M. Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev. 1995;9:2409–2420. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- 31.Roth D B. From lymphocytes to sharks: V(D)J recombinase moves to the germline. Genome Biol. 2000;1:1014.1–1014.4. doi: 10.1186/gb-2000-1-2-reviews1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roth D B, Craig N L. VDJ recombination: a transposase goes to work. Cell. 1998;94:411–414. doi: 10.1016/s0092-8674(00)81580-9. [DOI] [PubMed] [Google Scholar]
- 33.Roth D B, Nakajima P B, Menetski J P, Bosma M J, Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor δ rearrangement signals. Cell. 1992;69:41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- 34.Sakai J, Kleckner N. The Tn10 synaptic complex can capture a target DNA only after transposon excision. Cell. 1997;89:205–214. doi: 10.1016/s0092-8674(00)80200-7. [DOI] [PubMed] [Google Scholar]
- 35.Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light chain genes. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- 36.Sawchuk D J, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig M C, Cortes P. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J Exp Med. 1997;185:2025–2031. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz H Y, Landree M A, Kale S B, Roth D B. Joining-deficient RAG-1 mutants block V(D)J recombination in vivo. Mol Cell. 2001;7:65. doi: 10.1016/s1097-2765(01)00155-1. [DOI] [PubMed] [Google Scholar]
- 38.Spanopoulou E, Zaitseva F, Wang F-H, Santagata S, Baltimore D, Panayotou G. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 39.Thompson C B. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995;3:531–539. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 40.Tower J, Karpen G H, Craig N, Spradling A C. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]