Abstract
Snf-Swi, the prototypical ATP-dependent nucleosome-remodeling complex, regulates transcription of a subset of yeast genes. With the exception of Snf2p, the ATPase subunit, the functions of the other components are unknown. We have investigated the role of the conserved Snf-Swi core subunit Snf5p through characterization of two conditional snf5 mutants. The mutants contain single amino acid alterations of invariant or conserved residues that abolish Snf-Swi-dependent transcription by distinct mechanisms. One mutation impairs Snf-Swi assembly and, consequently, its stable association with a target promoter. The other blocks a postrecruitment catalytic remodeling step. These findings suggest that Snf5p coordinates the assembly and nucleosome-remodeling activities of Snf-Swi.
Chromatin structure inhibits gene transcription by blocking access of components of the transcriptional machinery and gene-specific activator proteins to DNA recognition sequences (18, 22, 33). The structural changes in chromatin that accompany transcriptional activation of a gene often require multiprotein factors that remodel nucleosomes. The conserved Saccharomyces cerevisiae Snf-Swi complex is the prototype of one class of eukaryotic factors that restructures chromatin by a process requiring ATP hydrolysis (7, 31, 46, 66). Histone acetyltransferases represent a second class of multiprotein complexes that modulates nucleosome structure by acetylating the amino-terminal tails of the core histones (2, 21, 58, 70). Genetic and biochemical experiments suggest that these two classes of remodeling factors function interdependently or in parallel in transcription at certain promoters (3, 12, 32, 37, 49, 51, 59, 61, 67). ATP-utilizing factors can also function with histone deacetylases in transcriptional repression (see reference 63).
Other Snf-Swi family complexes have been isolated from human (hSWI/SNF), Drosophila melanogaster (brm), and yeast (RSC) cells (34, 70), indicating the evolutionary importance of the nucleosome-remodeling activity. hSWI/SNF appears to regulate cell cycle progression (73) and cellular differentiation (6). RSC is essential for progression through the mitotic cell cycle (9, 10, 16, 62). Despite functional and compositional differences, several subunit polypeptides of the Snf-Swi complexes are highly conserved. Moreover, three conserved human Snf-Swi members, Snf2p (also known as BRG1 or hBrm), Snf5p (also known as INI1), and Swi3p (also known as BAF155 or BAF170), constitute a core set of Snf-Swi factors (see reference 48). Importantly, INI1 may function as a tumor suppressor (65).
Snf-Swi complexes disrupt histone-DNA contacts in mononucleosomes and nucleosome arrays in reactions requiring ATP hydrolysis in vitro (see reference 66). Possible remodeling mechanisms include the generation of superhelical torsion, the creation of chromatin loops, the generation of activated nucleosome intermediates, and the sliding or transfer of histone octamers (26, 66).
The yeast SNF and SWI genes, first identified as mutants defective in the expression of the SUC2 (snf, for sucrose nonfermenting) and HO (swi, for mating-type switching) genes (43, 56), have now been shown to affect the transcription of many other genes (28, 35, 60). A functional link to chromatin was established by the identification of mutations in genes encoding histones and other chromatin assembly factors as snf and swi suppressors (27, 36, 44, 50, 51). The Snf-Swi proteins are assembled into a large 2-MDa complex comprised of 11 polypeptides that is important but not essential for mitotic growth of cells (8, 13, 45).
In vivo analysis of the chromatin structures of the SUC2 and PHO8 promoters provided evidence that Snf-Swi-dependent chromatin-remodeling activity is required for transcriptional initiation (19, 20, 27, 39, 71). Snf-Swi associates directly with target promoters (12, 14), and both transcriptional activators and repressors have been implicated in the targeting mechanism(s) (see reference 47).
With the exception of the conserved Snf2p-Swi2p ATPase subunit required for nucleosome perturbation, the mechanism of action of other Snf-Swi component proteins has not been addressed. In this study, we have investigated the in vivo roles of Snf5p, a core subunit of Snf-Swi, through genetic and biochemical characterization of two conditional snf5-ts regulatory mutants. We present evidence that Snf5p is involved in maintaining Snf-Swi integrity and in postrecruitment chromatin remodeling in vivo.
MATERIALS AND METHODS
Yeast strains and genetic methods.
Yeast strains BLY1 (MATα his3-Δ200 lys2-801 ura3-52 SUC2), BLY3 (MATα snf5-Δ2 his3-Δ200 ura3-52 ade2-101), BLY35 (MATα snf2-Δ2::URA3 his3-Δ200 ura3-52 ade2-101), BLY61 (MATα snf5-51ts his3-Δ200 ura3-52 ade2-101), and BLY169 (MATα snf5-83bts his3-Δ200 ura3-52 ade2-101) are isogenic derivatives of S288C, and BLY54 (MATa leu2 ura3-1 trp1-1 his3 6lexAOp-LEU2 GAL+) (17) (gift of E. Golemis) is derived from W303. The following media were used: YPD medium (YEP [1% yeast extract and 2% peptone] containing 2% dextrose), YPR medium (YEP supplemented with 2% raffinose [1 μg of antimycin A/ml]), and YPGal medium (YEP supplemented with galactose). Synthetic complete (SC) medium is SC medium containing yeast nitrogen base supplemented with 2% sugars (dextrose or raffinose) and a drop-out mixture of amino acids and bases (52). SD-inositol medium contains inositol-free yeast nitrogen base (Bio-101) and 2% dextrose. Glucose-repressed cultures were grown in YPD at 30°C. Glucose-derepressed cells were grown first to mid-logarithmic phase in YPD, washed twice with water, and transferred to YEP plus 0.05% glucose for the times indicated at either 30 or 37°C. Standard genetic procedures were followed (52).
Plasmids.
LexA hybrid plasmids are derivatives of pSH2-1 (24) and express, from the constitutive ADH1 promoter, the amino-terminal 87 residues of the LexA protein fused to the indicated Snf5p residues. pLY50 carries the wild-type SNF5 gene and was created by cloning the 4.8-kb EcoRI-BamHI fragment of pJW34 (1) into pRS316. Site-directed mutagenesis of SFH1 was carried out with pIN18, a derivative of pRS316 (CEN6 URA3) carrying the 1.9-kb XhoI-AseI fragment of pYC5H (SFH1) (10). Oligonucleotide sequences and plasmid construction details will be provided upon request.
Enzyme assays.
β-Galactosidase activity was assayed in permeabilized cells (23, 40). Secreted invertase activity was assayed in whole cells as previously described (64).
Isolation of snf5 temperature-sensitive alleles.
pLY50 (SNF5 CEN6 URA3) was mutagenized in vitro as previously described (16), and the mutagenized plasmid DNA was used to transform strain BLY3 (snf5Δ) to uracil prototrophy. Approximately 4,500 Ura+ transformants were patched onto YPD plates and then replicated to two sets of YPR plates. One set was incubated at 30°C and the other at 37°C. Plasmid DNA was recovered from those colonies that grew at 30°C but not at 37°C. Mutations located within SNF5 were confirmed by restriction fragment swapping. Five temperature-sensitive mutants were identified, and the alleles were sequenced. The nucleotide changes (and predicted amino acid changes) of the three alleles described here are as follows. snf5-51ts and snf5-65ts (snf5-51ts and snf5-65ts are identical and are hereafter referred to as snf5-51ts) contained a nucleotide change of G-1744 to A (E582K), and snf5-83ts contained nucleotide changes of G-1066 to A (E356K), C-1085 to T (P362L), and G-1423 to A (D475N) and contained a silent mutation at Y360.
To determine the relative contributions of the three snf5-83ts point substitutions to the temperature sensitivity phenotype, we constructed two pRS306-derived integrating plasmids, psnf5-83a, which carries the E356K and P362L mutations, and psnf5-83b, which carries the D475N substitution, and used these to replace the snf5Δ locus. Following integration, snf5-83a cells showed wild-type growth (data not shown), whereas the temperature sensitivity phenotype of the snf5-83bts cells was indistinguishable from that of the original snf5-83ts mutant. All subsequent experiments were carried out with the snf5-83bts allele (D475N). psnf5-51 (E582K) was also constructed and used to replace the snf5Δ locus.
Immunological procedures.
Recombinant Snf5 (amino acids 1 to 193) and Snf2 (amino acids 1256 to 1703) proteins fused to glutathione S-transferase in pGEX-3X and pGEX-2T (Pharmacia), respectively, were purified from Escherichia coli cells and used to immunize rabbits as previously described (25). Immunoblot analysis was performed as described previously (10) using anti-Snf5p (1:2,000), anti-Snf2p (1:1,000), anti-Swi3p (1:1,000; gift of C. L. Peterson), and anti-LexA (1:2,000; gift of R. Brent) polyclonal antibodies and developed with the nitroblue tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate) color reagents except where noted.
Transcriptional activation of lexA-LEU2
Growth of cells expressing the wild-type and mutant Snf5 LexA fusion proteins was first compared on SC-His plates containing 0, 100, 300, or 900 μg of leucine. Mutant Snf5 proteins exhibited temperature-sensitive growth only on plates containing 300 μg of leucine (limiting leucine). Therefore, the transcriptional activation assays were carried out on leucine (900 μg) and limiting leucine (300 μg) plates at 30 and 37°C.
RNA analysis.
Total RNA was prepared as previously described (38), and SUC2 and U6 (as a control) RNA transcript levels were measured by primer extension analysis using specific primers as previously described (50).
Gel filtration of Snf-Swi complex.
Log-phase cultures of the snf5-ts or SNF5 (BLY1) cells grown in YPD were derepressed at either 30 or 37°C for 2 h. Whole-cell protein extracts were prepared and analyzed on a fast protein liquid chromatography Superose 6 gel filtration column (Pharmacia) as previously described (45). Proteins were trichloroacetic acid precipitated, separated on sodium dodecyl sulfate–6% polyacrylamide gels, and analyzed by Western blot analysis.
Protein immunoprecipitation.
Volumes containing 1.2 mg of protein from whole-cell lysates (51) were incubated with 1 μl of anti-Snf5p antibody for 2 h at 4°C in 1.0 ml of immunoprecipitation buffer (51). Immune complexes were collected using protein A-coupled agarose beads and washed as previously described (51).
Chromatin structure analysis.
Nuclei were isolated according to methods described by Roth and Simpson (53) from cells grown under glucose-repressing or -derepressing conditions at 30 or 37°C for 2 h. Micrococcal nuclease (Sigma) digestion, isolation of DNA from nuclei, and primer extension analysis were carried out as previously described (19). Primer extension analysis at the SUC2 locus was performed with Taq DNA polymerase (Fisher Scientific) and oligonucleotide primer F1 (a gift from R. T. Simpson and I. Gavin), which corresponds to SUC2 base pairs −784 to −755 (19).
Chromatin immunoprecipitations.
Approximately 3 × 108 cells were fixed in 1% formaldehyde for 15 min at room temperature. Cross-linked cells were lysed by glass bead breakage in lysis buffer as previously discussed (57). Chromatin was solubilized by sonication to an average DNA fragment size of 0.4 kb. For immunoprecipitation, 1 μl of anti-Snf5p or anti-Snf2p antibody was incubated with 0.8 mg of extract in 1.0 ml of lysis buffer overnight at 4°C (57). Immune complexes were collected and washed as previously described (57), and PCR was performed on extracted DNA with SUC2 gene and reference primer pairs. PCR products were separated on 8% polyacrylamide gels, and photoprocessing was carried out using a Foto Eclipse (Fotodyne) digital imaging system.
RESULTS
The functional domain of Snf5p contains two repeat motifs and is conserved throughout eukaryotes.
Snf5p is an essential and highly conserved component of the multiprotein yeast Snf-Swi complex. The sequences between Snf5p amino acids 455 and 676 are 32 to 46% identical to proteins encoded by open reading frames in human (30, 42), Drosophila (15), Danio rerio (zebrafish) (EMBL accession no. AJ249795.1), Caenorhabditis elegans (EMBL accession no. R07E5.3), Schizosaccharomyces pombe (EMBL accession no. C2F7.08C), S. cerevisiae (10), Arabidopsis thaliana (4), and Tetraodon fluviatilis (puffer fish) (72) cells, suggesting that an activity essential for basic cellular processes has been conserved during evolution. Notably, the highly conserved domain of the indispensable yeast Sfh1p protein, the sole yeast homologue of Snf5p, is sufficient for wild-type SFH1 function (10). In addition, truncating mutations of the conserved domain of human Snf5p are associated with oncogenesis (65).
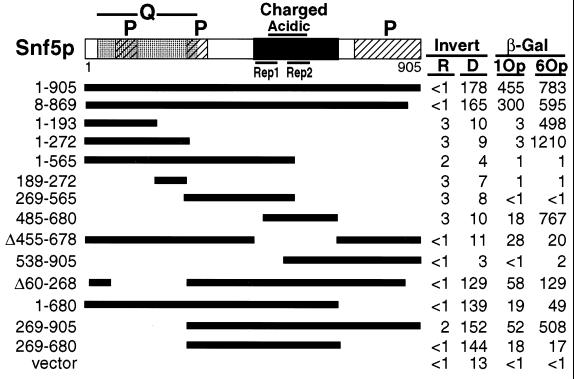
To determine the functional region(s) of Snf5p, a series of lexA-SNF5 fusion and deletion plasmids was constructed and tested for the complementation of an snf5 null mutation for SUC2 invertase activity and for the activation of transcription of a lexAop-GAL1-lacZ target gene. All fusion proteins were expressed at levels comparable to that of LexA-Snf51-905, as determined by immunoblot analysis (data not shown). Plasmids expressing, minimally, amino acids 269 through 680, which encompass the central charged region, including the conserved imperfect direct repeat motifs Rep1 and Rep2 (42), restored wild-type invertase activity (Fig. 1). Moreover, pLexA-Snf5Δ455-678, which lacks sequences encoding the repeats, failed to complement the invertase defects of snf5Δ, providing further evidence for the functional importance of this region. Hybrid proteins containing Snf5p amino acids 1 to 193 or 485 to 680 activated transcription of the GAL1-lacZ reporter gene to levels comparable to that of LexA-Snf51-905 (Fig. 1). Amino acids 1 to 193 may contain a cryptic activation domain, as these sequences are dispensable for SNF5 function at SUC2. Alternatively, activation by this region may reflect an additional, redundant activation function. In contrast, amino acids 485 to 680 lie entirely within the complementing region. The lower levels of GAL1-lacZ activation by LexA-Snf5269-680 could be explained by improper folding of the protein. We conclude that Snf5p amino acids 269 through 680, including the evolutionarily conserved repeat motifs Rep1 and Rep2, are necessary and sufficient for Snf5p function, at least at SUC2.
FIG. 1.
The functional domain of Snf5p. The first and last amino acids of Snf5p fused to LexA, or the amino acids deleted (Δ), are indicated. Invertase activity was measured in strain BLY3 cells (snf5Δ) carrying the indicated plasmids grown under glucose-repressing (R) and glucose-derepressing (D) conditions and is expressed as micromoles of glucose released per minute per 100 mg (dry weight) of cells. β-Galactosidase (β-Gal) activity was measured in strain BLY1 cells (SNF5) carrying the indicated lexA-SNF5 plasmids and target plasmids with a single (1Op) or six overlapping (6Op) lexA operators upstream of the GAL1-lacZ reporter gene, and the results are expressed in Miller units. The glutamine-rich N terminus (Q) is stippled, three proline-rich regions (P) are hatched, a highly charged central region is filled, and Rep1 and Rep2 are the direct imperfect repeats at positions 457 to 498 and 541 to 601, respectively. Invertase and β-galactosidase values represent the averages of four independent isolates. Errors were <12% for values >2.
Conditional temperature-sensitive alleles of SNF5 alter amino acids within evolutionarily conserved repeat motifs.
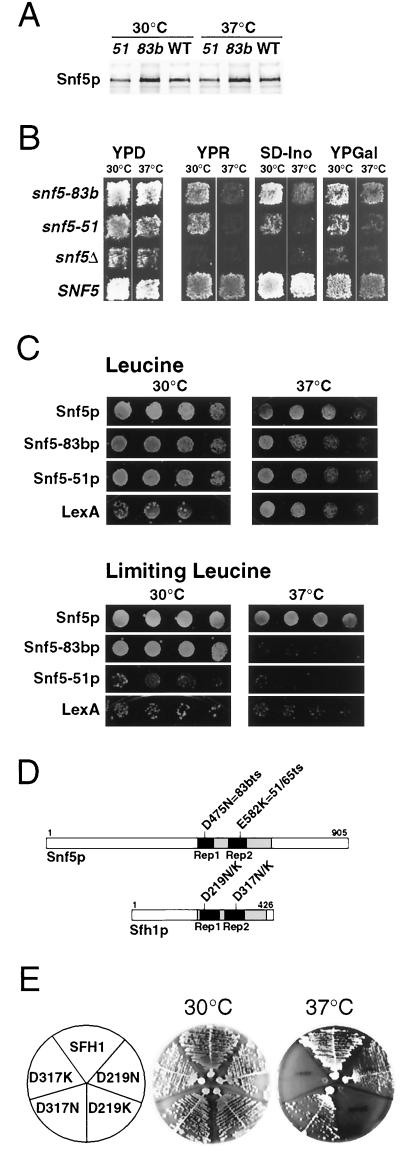
For a more detailed investigation of the role of Snf-Swi in transcriptional regulation in vivo, a genetic screen was initiated to isolate conditional loss-of-function mutations in SNF5. Temperature-sensitive mutations in SNF5 would permit a temporal examination of the physiological impact of loss of Snf-Swi activity. Conditional mutants were identified on the basis of thermolabile growth on raffinose media. Each of the alleles to be described, snf5-51ts and snf5-83bts, is recessive and was complemented by plasmids containing the wild-type SNF5 gene. Snf5 protein levels were comparable in the snf5-ts mutants and SNF5 cells grown at permissive (30°C) or nonpermissive (37°C) temperatures (Fig. 2A).
FIG. 2.
Characterization of snf5-ts mutations. (A) Snf5 protein expression levels are unaltered in the snf5-ts mutants. Log-phase cultures of BLY1 (SNF5 [WT]), BLY61 (snf5-51ts), and BLY169 (snf5-83bts) cells were derepressed for 2 h at 30 or 37°C. Proteins from whole-cell lysates (10) were immunoblotted with anti-Snf5p antiserum. (B) The snf5-ts mutants show distinct growth phenotypes at nonpermissive temperature. The same strains used in panel A and BLY3 (snf5Δ2) were grown in patches on a YPD plate and then replica plated onto YPD, YPR, SD-inositol, or YPGal media and incubated for 2 days at 30 or 37°C. (YPD patches were taken from a different region of the same master plate.) (C) The LexA-Snf5ts fusion proteins fail to activate transcription in vivo. Threefold serial dilutions of log-phase BLY54 (lexAop-LEU2) cells expressing LexA-Snf5p, LexA–Snf5-83bp, LexA–Snf5-51p, or LexA were spotted onto SC (leucine) or SC limiting leucine plates and incubated for 2 to 4 days at 30 or 37°C. (D) The snf5 temperature-sensitive mutations alter highly conserved amino acids. Schemes of Snf5p and Sfh1p are shown drawn to scale, with the conserved domains of each being shaded and Rep1 and Rep2 shown as filled boxes. The positions of mutations and the amino acid changes are indicated. (E) The corresponding mutations in SFH1 confer a temperature-sensitive phenotype. Cells of strains BLY210 (SFH1), BLY213 (sfh1-D219N), BLY214 (sfh1-D219K), BLY215 (sfh1-D317N), and BLY217 (sfh1-D317K), which carry the indicated SFH1 allele on plasmids in the sfh1Δ background, were streaked onto YPD plates and incubated for 2 days at 30 or 37°C.
Transcription through the SUC2, INO1, and GAL1/GAL10 Snf-Swi-dependent promoters (69) was compared in SNF5, snf5Δ, and snf5-ts strains by assaying growth on YPR, SD-inositol, and YPGal media at 30 and 37°C. snf5-51ts and snf5-83bts mutants supported wild-type or nearly wild-type growth on these media at 30°C but not at 37°C, and neither of the snf5-ts mutants showed temperature-sensitive growth defects on glucose (Fig. 2B). In contrast, snf5Δ cells failed to grow on these media at either temperature. We infer from these results that the snf5-ts mutations confer temperature-sensitive transcriptional defects at several Snf-Swi-dependent promoters in vivo.
To explore further the broader transcriptional consequences of Snf5p inactivation, we compared the ability of wild-type and mutant LexA-Snf5 fusion proteins to activate transcription of a chromosomally integrated lexAop-LEU2 target promoter (in which the upstream activation sequence [UAS] is replaced by lexA operator sequences) (Fig. 2C). Wild-type and mutant fusion proteins migrated with the expected apparent molecular weights and were expressed at levels comparable to those of wild-type LexA-Snf5p at 30 and 37°C, as determined by immunoblot analysis (data not shown). Snf5p and several other Snf-Swi proteins, when artificially tethered to DNA, activate transcription of target genes in vivo in an Snf-Swi-dependent manner (38). On leucine plates, cells expressing wild-type Snf5p, mutant Snf5 proteins, or LexA alone grew equally well at 30 and 37°C. On limiting leucine plates, the growth of cells expressing wild-type LexA-Snf5p at both temperatures was comparable. In contrast, although cells expressing LexA-Snf5-83bp grew as well as wild-type Snf5p at 30°C, these cells failed to grow at 37°C. Cells expressing LexA-Snf5-51p were incapable of growth at 37°C, and these cells also exhibited poor growth at 30°C. Thus, both snf5-ts alleles abolished the ability of Snf5p to activate transcription of the LEU2 target gene in vivo, suggesting general transcriptional defects in Snf-Swi function.
The snf5-ts mutations alter highly conserved amino acids in the functional domain of Snf5p (Fig. 2D). snf5-83bts contains an asparagine substituted for Asp475, which is located within Rep1 and is one of two invariant amino acids present in all Snf5p family proteins. snf5-51ts contains a lysine substituted for Glu582, part of the consensus sequence for Rep2 (41).
Sfh1p, the only other yeast protein homologous to Snf5p, is a component of RSC, a multiprotein ATP-dependent nucleosome-remodeling complex related to Snf-Swi (9). The corresponding substitutions in Sfh1p also conferred temperature-sensitive growth on cells (Fig. 2D and E). Asparagine replacements caused slight temperature-sensitive phenotypes while lysine replacements conferred severe temperature-sensitive growth (Fig. 2D and E). The phenotypes conferred by the two snf5 mutations and the corresponding sfh1 mutations highlight the functional importance of this conserved region for the family of ATP-dependent chromatin-remodeling complexes.
snf5-ts mutations regulate transcription of SUC2
To study the consequences of the conditional inactivation of SNF5 at a target promoter in more detail, invertase activity and SUC2 RNA levels were compared in SNF5, snf5Δ, and snf5-ts cells under derepressing conditions at 30 and 37°C (Table 1; Fig. 3). SUC2 is a glucose-repressible gene whose expression is repressed in the presence of high glucose and induced 100-fold by growth in low glucose (29).
TABLE 1.
snf5-ts mutations regulate SUC2 expression in vivo
| Relevant genotype | Invertase activitya
|
||||||
|---|---|---|---|---|---|---|---|
| R | D30°C
|
D37°C
|
|||||
| 1 h | 2 h | 4 h | 1 h | 2 h | 4 h | ||
| SNF5 | 2 | 19 | 108 | 189 | 84 | 170 | 220 |
| snf5Δ | 1 | 5 | 8 | 16 | NDb | ND | ND |
| snf5-5lts | 2 | 8 | 40 | 90 | 12 | 19 | 21 |
| snf5-83bts | 2 | 12 | 77 | 101 | 28 | 51 | 56 |
Units are as follows: micromoles of glucose released per minute per 100 mg (dry weight) of cells. Repressed cells (R) or cells derepressed at 30°C (D30°C) and 37°C (D37°C) for 1, 2, or 4 h were assayed for invertase activity as previously described (64). Data are the averages of those from at least two different cultures. Errors were less than 15%.
ND, not determined.
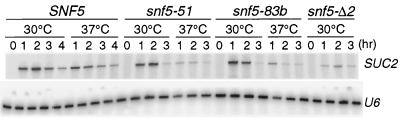
FIG. 3.
snf5-ts mutations regulate SUC2 transcription. Primer extension analysis was carried out with total RNA prepared from repressed cells (0 h) or cells derepressed at 30 or 37°C for the indicated times. Primers specific for SUC2 and U6 transcripts were used for each reaction.
Invertase levels in derepressed SNF5 cells increased steadily from 1 to 4 h at both 30 and 37°C. In contrast, even after 4 h of derepression, the snf5Δ strain was incapable of inducing expression of SUC2. At 30°C, the mutants expressed substantial invertase activity (within twofold of that of the wild type). However, at 37°C after 2 h of derepression, no further accumulation of invertase was observed in the mutants. Thus, invertase expression was tightly controlled in a temperature-sensitive manner in each of the snf5-ts strains.
SUC2 RNA levels in induced SNF5 cells at 30 or 37°C peaked during the first 2 h and then decreased (Fig. 3); a similar pattern was observed for cells of a different genetic background (51). SUC2 transcript levels in both snf5-ts mutants at 30°C were comparable to those of the wild type in the first 2 h, although the subsequent drop in RNA levels at 3 h was more precipitous. In contrast, at 37°C, SUC2 transcript levels in the snf5-ts mutants at 1 h were only slightly higher than those in the snf5 deletion mutant, and by 2 h they were indistinguishable.
When combined, the invertase and RNA analyses show that following a shift to derepressing media at nonpermissive temperature, SUC2 transcription in the snf5-ts mutants was first turned on for a short time and then shut off as Snf-Swi function was lost. These results support recent findings that Snf-Swi is needed continuously for SUC2 transcription in vivo (3, 59).
Snf-Swi assembly is perturbed in the snf5-51ts mutant and only moderately altered in the snf5-83bts mutant.
The transcriptional defects in the snf5-ts mutants could be explained by disassembled Snf-Swi complexes or by assembled but functionally inactive complexes. Therefore, the integrity of Snf-Swi in the snf5-ts mutants was examined by gel filtration (Fig. 4A). Whole-cell extracts were applied to a Sepharose 6 gel filtration column, and elution of Snf-Swi polypeptides was monitored by immunoblot analysis. The fractionation of SNF5 whole-cell extracts showed coelution of the Snf2p, Swi3p, and Snf5p polypeptides as peaks in fraction 19, suggesting an assembled Snf-Swi complex as shown previously (45), and this pattern was unaffected at 37°C. In contrast, in the fractionation of snf5Δ cells, the elution of Snf2p and Swi3p was altered significantly (Fig. 4A) as shown previously (45), suggesting partial disassembly of Snf-Swi. The elution pattern of proteins upon fractionation of snf5-51ts extracts indicated that assembly of Snf-Swi was perturbed at both 30 and 37°C. In contrast, fractionation of whole-cell extracts derived from snf5-83bts cells suggested that the integrity of Snf-Swi was only moderately affected. At 30°C, Snf2p, Swi3p, and Snf5p coeluted exactly as they eluted in SNF5 cells. At 37°C, significant amounts of these polypeptides coeluted and peaked in fraction 19 (Fig. 4A), an elution pattern resembling that of extracts prepared from the Snf-Swi ATPase mutant swi2K798A (45).
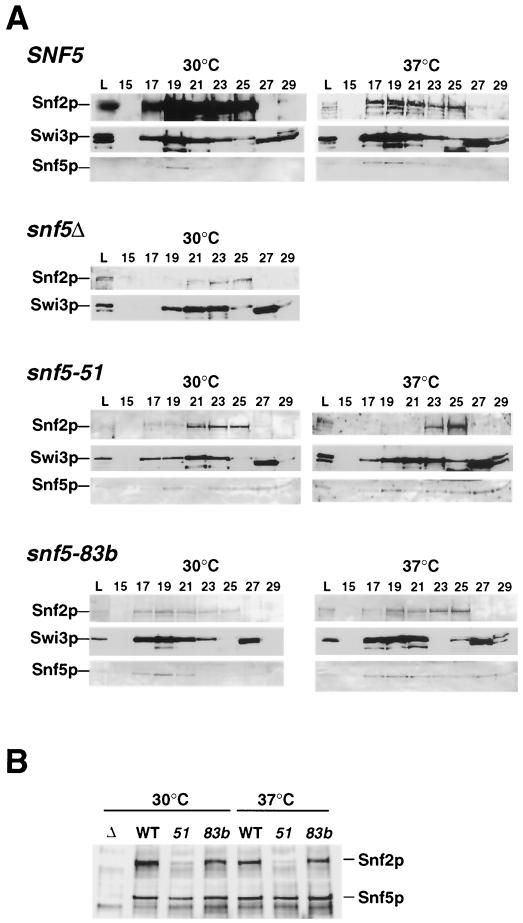
FIG. 4.
Assembly of the Snf-Swi complex is aberrant in the snf5-51ts mutant but only mildly altered in the snf5-83bts mutant. (A) Whole-cell protein extracts prepared from the same strains used in Fig. 2 grown under derepressing conditions for 2 h at 30 or 37°C were fractionated on Superose 6, and fractions were assayed for Snf2p, Swi3p, and Snf5p by immunoblot analysis. Swi3p immunoblots and the Snf2p immunoblot prepared from wild-type cells at 30°C were followed by chemiluminescence; all other immunoblots were followed colorimetrically. Proteins with molecular masses of 669 and 443 kDa should have eluted in fractions 25 and 28, respectively (45). L, load. (B) Immunoprecipitations were carried out as described in Materials and Methods with whole-cell lysates prepared from the same strains described for panel A under the same growth conditions. Anti-Snf5p-precipitated proteins were separated on sodium dodecyl sulfate–4 to 15% polyacrylamide gels and probed with anti-Snf5p or anti-Snf2p antibodies.
Next, the physical association of Snf5p and Snf2p was measured in the two snf5-ts mutants by coimmunoprecipitation assays using anti-Snf5p antibodies (Fig. 4B). Levels of both Snf5p and Snf2p precipitated from snf5-83bts and wild-type whole-cell extracts from cells grown at 30 and 37°C were comparable, consistent with the apparent integrity of Snf-Swi. In contrast, the snf5-51ts mutation dramatically affected the association of Snf5p and Snf2p, even at permissive temperature (30°C). These two lines of evidence suggest that the snf5-51ts mutation severely destabilizes Snf-Swi protein-protein interactions, preventing functional assembly of Snf-Swi. The absence of a dramatic effect on Snf-Swi assembly in snf5-83bts cells suggests that the assembled complex is functionally impaired.
snf5-ts mutants fail to remodel chromatin structure of SUC2 promoter.
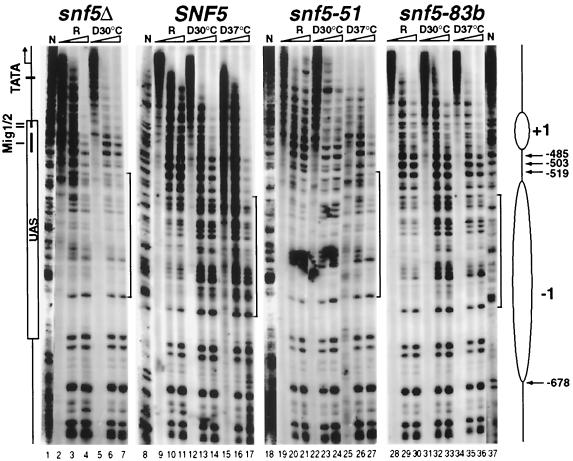
The chromatin structure of the SUC2 promoter changes dramatically upon derepression in an Snf-Swi-dependent, transcription-independent manner (19, 27). To study whether the snf5-ts mutant Snf-Swi complexes are competent for nucleosome remodeling, we analyzed the chromatin organization of a region of the SUC2 promoter sensitive to mutations in SNF/SWI, −678 to −519 (19, 71), by primer extension methodology (Fig. 5).
FIG. 5.
The nucleosome structure of the SUC2 UAS region is not remodeled in the snf5-ts mutants. Primer extension analysis (using the F1 primer) was carried out with chromatin isolated from cells grown under repressing (R) or derepressing conditions at 30°C (D30°C) or 37°C (D37°C) digested previously with increasing amounts of micrococcal nuclease. Schematic features of the SUC2 upstream region are shown on the left (19). The presumed positions of the nucleosomes are indicated by ellipses. Brackets denote regions containing derepression-sensitive nuclease cutting sites. Arrows mark the major sites of digestion in the repressed SUC2 promoter; numbers indicate the distance from the initiating A residue. Triangles indicate increasing concentrations of micrococcal nuclease as previously described (19). The naked DNA samples (N) were digested with micrococcal nuclease as a control. Slight differences in the mobility of DNA fragments in the four strains are due to differences in gel electrophoresis. Strains used were the same as those in Fig. 4.
A dramatic change in chromatin structure accompanied SUC2 derepression in the wild-type cells at both 30 and 37°C (Fig. 5, compare lanes 10 and 11 with lanes 13 and 14 and with lanes 16 and 17), and this change did not occur in the snf5 deletion strain (Fig. 5, compare lanes 3 and 4 with lanes 6 and 7). The enhanced cleavage by micrococcal nuclease in the wild-type cells indicates the disruption of nucleosome −1. For the snf5-51ts mutant, at neither temperature did the micrococcal nuclease digestion pattern differ appreciably under derepressing conditions from that under repressing conditions (Fig. 5, compare lanes 23, 24, 26, and 27 with lanes 20 and 21). In contrast, a significant increase in nuclease cutting was detected in the snf5-83bts mutant at 30°C, similar to that in SNF5 cells (Fig. 5, compare lanes 28 to 33 with lanes 9 to 14). However, at 37°C, nuclease cutting was diminished (Fig. 5, compare lanes 35 and 36 with lanes 32 and 33). These results indicate that the snf5-51ts mutation severely compromises Snf-Swi remodeling activity even at permissive temperature. In contrast, snf5-83bts interferes with chromatin-remodeling activity conditionally.
Snf-Swi occupancy at SUC2 promoter.
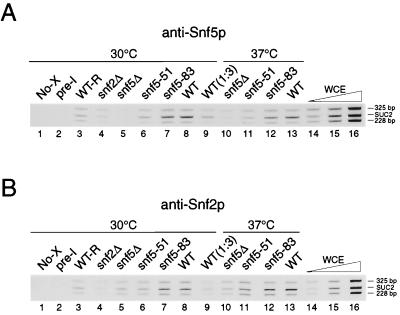
The inability of the snf5-51ts and snf5-83bts mutant Snf-Swi complexes to remodel nucleosomes or activate transcription at target chromosomal loci suggests that these complexes are compromised either in their association with chromatin or in a subsequent chromatin-remodeling step(s). To test these possibilities, we measured the presence of Snf5p and Snf2p in vivo at the SUC2 promoter in wild-type (derepressed and repressed) and mutant cells by chromatin immunoprecipitation. At the same time, chromatin immunoprecipitation assays carried out with extracts prepared from wild-type glucose-repressed and glucose-derepressed cells affords a test of the model that transcription of SUC2 is controlled by promoter recruitment of Snf-Swi. Polyclonal anti-Snf5p and anti-Snf2p antibodies were incubated with formaldehyde cross-linked chromatin, and the immune complexes were collected by binding to protein A-Sepharose beads. Levels of SUC2 promoter DNA and two reference DNAs in the immunoprecipitates were compared using PCR. None of the DNAs was precipitated in the absence of Snf5p or Snf2p, in mock immunoprecipitations using preimmune sera or in immunoprecipitations with non-cross-linked chromatin (Fig. 6A and B).
FIG. 6.
In vivo association of Snf5p and Snf2p with the SUC2 promoter. Chromatin obtained from 2-h glucose-derepressed (except as noted) strains BLY1 (WT), BLY3 (snf5Δ), BLY35 (snf2Δ), BLY61 (snf5-51ts), and BLY169 (snf5-83bts) was immunoprecipitated with anti-Snf5p, anti-Snf2p, or preimmune sera (pre-I). DNA isolated from immunoprecipitates or from whole-cell extracts (WCE) was amplified by PCR with primers specific for the SUC2 UAS (SUC2; 250 bp) combined with two pairs of reference primers for promoter regions of flanking genes 1.7 kb downstream (228-bp product) and 2.4 kb upstream (325-bp product) of SUC2, simultaneously. The PCR products were separated on an 8% polyacrylamide gel. (A) Association of Snf5p with the SUC2 UAS. Lane 1, anti-Snf5p immunoprecipitation with non-cross-linked SNF5 chromatin (No-X); lane 2, preimmune serum immunoprecipitation of SNF5 chromatin; lane 3, anti-Snf5p immunoprecipitation of chromatin from glucose-repressed SNF5 (WT-R) cells; lanes 4 to 9, anti-Snf5p immunoprecipitation of snf2Δ, snf5Δ, snf5-51ts, snf5-83bts, and wild-type chromatin (lane 8, undiluted wild type; lane 9, 1:3 dilution) from derepressed cells at 30°C; lanes 10 to 13, anti-Snf5p immunoprecipitation of snf5Δ, snf5-51ts, snf5-83bts, and wild-type chromatin prepared from derepressed cells at 37°C; lanes 14 to 16, threefold serial dilutions of total input DNA. (B) Association of Snf2p with the SUC2 UAS. The same chromatin solutions used in panel A were immunoprecipitated with anti-Snf2p antibody or preimmune serum.
In wild-type derepressed cells, SUC2 UAS sequences were preferentially precipitated, indicating specific binding of Snf5p and Snf2p proteins to SUC2, whereas under repressing conditions, the selective association of Snf5p and Snf2p proteins was lost (Fig. 6A and B, compare lanes 8 and 13 with lane 3). In addition, we infer from the interdependent binding of Snf5p and Snf2p to SUC2 (Fig. 6A, compare lanes 4 and 8; Fig. 6B, compare lanes 5 and 8) that binding was by the Snf-Swi complex. Together, these results demonstrate for the first time that Snf-Swi is targeted to SUC2 upon derepression.
We next investigated whether binding of Snf5p or Snf2p to chromatin was affected by the snf5-ts mutations. In derepressed snf5-51ts cells at 30 or 37°C, little SUC2 DNA (comparable to that in repressed wild-type cells; Fig. 6A and B, compare lanes 6 and 11 with lane 3) was selectively precipitated by either antibody. In contrast, in snf5-83bts cells, both Snf5p and Snf2p associated with the SUC2 promoter as well as in wild-type at both temperatures (Fig. 6A and B, compare lanes 7 and 8 and lanes 12 and 13), suggesting that this mutation does not regulate Snf-Swi binding to specific target sites in chromatin. Thus, at 37°C, the snf5-83bts Snf-Swi complex is present at SUC2 despite the lack of chromatin-remodeling activity and transcription.
DISCUSSION
Here, we present genetic and biochemical evidence that supports distinct roles for the conserved core subunit, Snf5p, in Snf-Swi assembly and nucleosome-remodeling activities. These data provide new insights into in vivo mechanisms of chromatin targeting and remodeling by Snf-Swi complexes.
In this work, critical roles of Snf5p in Snf-Swi function have been revealed by characterization of two snf5-ts mutants harboring single alterations of invariant or conserved amino acids that reside within two repeat sequences, Rep1 and Rep2. The possibility that these repeats carry out distinct functions is suggested by the finding that the corresponding hSNF5 repeat motifs show differential properties in binding human immunodeficiency virus integrase and c-MYC (11, 41). We found that, under nonpermissive conditions, both mutations in the Snf5p repeat motifs compromise Snf-Swi chromatin remodeling and transcriptional activation but through distinct mechanisms. The snf5-51ts mutant demonstrates that Snf5p is essential for the assembly and promoter targeting of Snf-Swi. In contrast, the snf5-83bts mutant uncovers a critical role for Snf5p in one or more postrecruitment remodeling functions.
The snf5-51ts mutation (an E582K substitution in Rep2) severely perturbs Snf-Swi assembly, even at permissive temperature, and further dissociates Snf5p from Snf2p at nonpermissive temperature (Fig. 4). This result extends the previous whole-cell extract fractionation studies, which showed that Snf-Swi polypeptides no longer copurify in the absence of Snf5p (45), by demonstrating that a single amino acid substitution within Rep2 abolishes the architectural function of Snf5p. Furthermore, in the snf5-51ts mutant at both temperatures, association of Snf5p and Snf2p with the SUC2 promoter is impaired, arguing that an intact Snf-Swi complex is necessary for promoter recruitment.
We observed substantial derepression of the SUC2 gene at 30°C in the snf5-51ts cells despite defective nucleosomal remodeling (compare Table 1 and Fig. 3 to Fig. 4). One possibility is that transient chromatin remodeling at earlier time points was missed, since Snf-Swi recruitment and chromatin-remodeling assays were carried out only at the 2-h time point. However, our preliminary results showed that the chromatin organization of the SUC2 UAS region in snf5-51ts cells derepressed for 30 min is no different than that at 2 h (data not shown). Therefore, we favor the idea that the low amount of Snf-Swi complex present at the UAS region in derepressed snf5-51ts cells (comparable to that in repressed SNF5 cells) is sufficient to allow efficient transcription but fails to stably support transcription (Fig. 3). Snf-Swi could activate transcription by disrupting higher-order chromatin structure, facilitating histone acetylation (3, 35, 51, 59), or interacting with the RNA polymerase holoenzyme (68), any of which might not lead to detectable changes in the remodeling assay.
An alternative possibility is that the snf5-51ts mutant Snf-Swi remodels other regions of the SUC2 promoter, distinct from the UAS, to permit transcription. Although the SUC2 UAS region is essential for gene derepression (5, 54, 55), other SUC2 promoter sequences are also remodeled in an Snf-Swi-dependent manner (5, 19, 27, 39, 71). In addition, Snf-Swi can be differentially recruited to distinct regions of the HO promoter (12). Therefore, it is tempting to speculate that the snf5-51ts mutation affects Snf-Swi remodeling activity at distinct promoter elements (such as the UAS and TATA regions) differently. Experiments are under way to resolve these important issues.
In contrast to the snf5-51ts mutation, the snf5-83bts mutation (a D475N substitution in Rep1) affects neither Snf-Swi assembly nor its recruitment to SUC2, indicating that the remodeling and transcriptional defects at nonpermissive temperature are caused by blockage of one or more postrecruitment Snf-Swi remodeling step(s). Interestingly, hSNF5 has been shown to moderately stimulate the nucleosome-remodeling activity of the human Snf2p homologue, BRG1, in an in vitro reconstitution experiment (48). Snf5p could have a similar stimulatory effect on remodeling, although the effect is expected to be more robust in vivo. Another possibility is that Snf5p plays a role in the coordination of Snf-Swi and other remodeling factors required for chromatin remodeling (20, 61).
The activity of Snf-Swi in vivo is controlled by both chromosomal targeting (see reference 47) and postrecruitment events (20, 61). Our results reveal essential roles for Snf5p in both processes. We propose that Snf5p integrates important protein-protein interactions for Snf-Swi assembly and coordinates promoter recruitment and chromatin remodeling.
ACKNOWLEDGMENTS
Fuqiang Geng and Yixue Cao contributed equally to this work.
We thank Igor Gavin and Robert Simpson for generous advice in the chromatin structure analysis, Laurie Boyer and Craig Peterson for helpful assistance with gel filtration analysis, and Pamela Meluh for CHIP instruction. Mary Ann Osley, Camilo Parada, and the members of the Laurent laboratory are thanked for comments on the manuscript. We also thank Craig Peterson and Roger Brent for gifts of antisera and Irem Nasir for constructing pIN18.
This work was supported in part by an American Cancer Society grant (NP-871), the March of Dimes Birth Defects Foundation (Basil O'Connor Starter Scholar Research Award), and a Public Health Service grant (GM56700) to B.C.L.
REFERENCES
- 1.Abrams E, Neigeborn L, Carlson M. Molecular analysis of SNF2 and SNF5, genes required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:3643–3651. doi: 10.1128/mcb.6.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger S L. Gene activation by histone and factor acetyltransferases. Curr Opin Cell Biol. 1999;11:336–341. doi: 10.1016/S0955-0674(99)80046-5. [DOI] [PubMed] [Google Scholar]
- 3.Biggar S R, Crabtree G R. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 1999;18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brzeski J, Podstolski W, Olczak K, Jerzmanowski A. Identification and analysis of the Arabidopsis thaliana BSH gene, a member of the SNF5 gene family. Nucleic Acids Res. 1999;27:2393–2399. doi: 10.1093/nar/27.11.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bu Y, Schmidt M C. Identification of cis-acting elements in the SUC2 promoter of Saccharomyces cerevisiae required for activation of transcription. Nucleic Acids Res. 1998;26:1002–1009. doi: 10.1093/nar/26.4.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 7.Cairns B R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- 8.Cairns B R, Kim Y-J, Sayre M H, Laurent B C, Kornberg R D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 10.Cao Y, Cairns B R, Kornberg R D, Laurent B C. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol Cell Biol. 1997;17:3323–3334. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng S W, Davies K P, Yung E, Beltran R J, Yu J, Kalpana G V. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- 12.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 13.Côté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 14.Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley M A. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol Cell. 1999;4:75–83. doi: 10.1016/s1097-2765(00)80189-6. [DOI] [PubMed] [Google Scholar]
- 15.Dingwall A K, Beek S J, McCallum C M, Tamkun J W, Kalpana G V, Goff S P, Scott M P. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol Biol Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du J, Nasir I, Benton B K, Kladde M P, Laurent B C. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estojak J, Brent R, Golemis E A. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 19.Gavin I M, Simpson R T. Interplay of yeast global transcriptional regulators Ssn6p-Tup1p and Swi-Snf and their effect on chromatin structure. EMBO J. 1997;16:6263–6271. doi: 10.1093/emboj/16.20.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory P D, Schmid A, Zavari M, Munsterkotter M, Horz W. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 1999;18:6407–6414. doi: 10.1093/emboj/18.22.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 22.Grunstein M. Nucleosomes: regulators of transcription. Trends Genet. 1990;6:395–400. doi: 10.1016/0168-9525(90)90299-l. [DOI] [PubMed] [Google Scholar]
- 23.Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 24.Hanes S D, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 25.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 26.Havas K, Flaus A, Phelan M, Kingston R, Wade P A, Lilley D M J, Owen-Hughes T. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell. 2000;103:1133–1142. doi: 10.1016/s0092-8674(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 27.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 28.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 29.Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 30.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 31.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 32.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 33.Kornberg R D, Lorch Y. Chromatin structure and transcription. Annu Rev Cell Biol. 1992;8:563–587. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- 34.Kornberg R D, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 35.Krebs J E, Fry C J, Samuels M L, Peterson C L. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell. 2000;102:587–598. doi: 10.1016/s0092-8674(00)00081-7. [DOI] [PubMed] [Google Scholar]
- 36.Kruger W, Peterson C L, Sil A, Coburn C, Arents G, Moudrianakis E N, Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 37.Kuo M-H, vom Baur E, Struhl K, Allis C D. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol Cell. 2000;6:1309–1320. doi: 10.1016/s1097-2765(00)00129-5. [DOI] [PubMed] [Google Scholar]
- 38.Laurent B C, Treitel M A, Carlson M. The SNF5 protein of Saccharomyces cerevisiae is a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol Cell Biol. 1990;10:5616–5625. doi: 10.1128/mcb.10.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matallana E, Franco L, Perez-Ortin J E. Chromatin structure of the yeast SUC2 promoter in regulatory mutants. Mol Gen Genet. 1992;231:395–400. doi: 10.1007/BF00292708. [DOI] [PubMed] [Google Scholar]
- 40.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 41.Morozov A, Yung E, Kalpana G V. Structure-function analysis of integrase interactor 1/hSNF5L1 reveals differential properties of two repeat motifs present in the highly conserved region. Proc Natl Acad Sci USA. 1998;95:1120–1125. doi: 10.1073/pnas.95.3.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. A human protein with homology to Saccharomyces cerevisiae SNF5 interacts with the potential helicase hbrm. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neigeborn L, Celenza J L, Carlson M. SSN20 is an essential gene with mutant alleles that suppress defects in SUC2 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:672–678. doi: 10.1128/mcb.7.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peterson C L, Dingwall A, Scott M P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peterson C L, Tamkun J W. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 47.Peterson C L, Workman J L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 48.Phelan M L, Sif S, Narlikar G J, Kingston R E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 49.Pollard K J, Peterson C L. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prelich G, Winston F. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics. 1993;135:665–676. doi: 10.1093/genetics/135.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Recht J, Osley M A. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi/Snf in yeast. EMBO J. 1999;18:101–113. doi: 10.1093/emboj/18.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose M D, Winston F, Hieter P. Methods in yeast genetics, a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1990. [Google Scholar]
- 53.Roth S Y, Simpson R T. Yeast minichromosomes. Methods Cell Biol. 1991;35:289–314. [PubMed] [Google Scholar]
- 54.Sarokin L, Carlson M. Upstream region of the SUC2 gene confers regulated expression to a heterologous gene in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:2521–2526. doi: 10.1128/mcb.5.10.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarokin L, Carlson M. Upstream region required for regulated expression of the glucose-repressible SUC2 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2750–2757. doi: 10.1128/mcb.4.12.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stern M J, Jensen R E, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 57.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 58.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 59.Sudarsanam P, Cao Y, Wu L, Laurent B C, Winston F. The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J. 1999;18:3101–3106. doi: 10.1093/emboj/18.11.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sudarsanam P, Iyer V R, Brown P O, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Syntichaki P, Topalidou I, Thireos G. The Gcn5 bromodomain co-ordinates nucleosome remodeling. Nature. 2000;404:414–417. doi: 10.1038/35006136. [DOI] [PubMed] [Google Scholar]
- 62.Tsuchiya E, Hosotani T, Miyakawa T. A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res. 1998;26:3286–3292. doi: 10.1093/nar/26.13.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tyler J K, Kadonaga J T. The “dark side” of chromatin remodeling: repressive effects on transcription. Cell. 1999;99:443–446. doi: 10.1016/s0092-8674(00)81530-5. [DOI] [PubMed] [Google Scholar]
- 64.Vallier L G, Carlson M. Synergistic release from glucose repression by mig1 and ssn mutations in Saccharomyces cerevisiae. Genetics. 1994;137:49–54. doi: 10.1093/genetics/137.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Versteege I, Sevenet N, Lange J, Rousseau-Merck M-F, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 66.Vignali M, Hassan A H, Neely K E, Workman J L. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 68.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 69.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 70.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 71.Wu L, Winston F. Evidence that Snf-Swi controls chromatin structure over both the TATA and UAS regions of the SUC2 promoter in Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4230–4234. doi: 10.1093/nar/25.21.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao C W, Leu J H, Chin C, Chou C K, Huang C J. Round-spotted pufferfish (Tetraodon fluviatilis) snf5 gene is oriented in a tail-to-tail manner with the set gene which encodes an inhibitor of protein phosphatase 2A. DNA Cell Biol. 1998;17:69–82. doi: 10.1089/dna.1998.17.69. [DOI] [PubMed] [Google Scholar]
- 73.Zhang H S, Gavin M, Dahiya A, Postigo A A, Ma D, Luo R X, Harbour J W, Dean D C. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]