Abstract
Polycomb-group (Pc-G) genes are required for the stable repression of the homeotic selector genes and other developmentally regulated genes, presumably through the modulation of chromatin domains. Among the Drosophila Pc-G genes, Enhancer of zeste [E(z)] merits special consideration since it represents one of the Pc-G genes most conserved through evolution. In addition, the E(Z) protein family contains the SET domain, which has recently been linked with histone methyltransferase (HMTase) activity. Although E(Z)-related proteins have not (yet) been directly associated with HMTase activity, mammalian Ezh2 is a member of a histone deacetylase complex. To investigate its in vivo function, we generated mice deficient for Ezh2. The Ezh2 null mutation results in lethality at early stages of mouse development. Ezh2 mutant mice either cease developing after implantation or initiate but fail to complete gastrulation. Moreover, Ezh2-deficient blastocysts display an impaired potential for outgrowth, preventing the establishment of Ezh2-null embryonic stem cells. Interestingly, Ezh2 is up-regulated upon fertilization and remains highly expressed at the preimplantation stages of mouse development. Together, these data suggest an essential role for Ezh2 during early mouse development and genetically link Ezh2 with eed and YY1, the only other early-acting Pc-G genes.
Polycomb-group (Pc-G) genes are required for the stable repression of the homeotic selector genes and other developmentally regulated genes, thereby providing differentiation programs with a “transcriptional memory” (reviewed in reference 25). Among the Drosophila Pc-G genes, Enhancer of zeste [E(z)] (19, 20) and extra sex combs (esc) (12) are exceptional, since both genes are required early during development, in contrast to other Drosophila Pc-G genes that appear to have later functions. For example, E(z) is involved in the repression of some of the early-acting segmentation-gap genes (24, 26). E(z) and esc are the Pc-G genes most highly conserved throughout evolution, being the only Pc-G genes found in the Caenorhabditis elegans genome (17, 21). E(z) function also influences chromosome integrity during the rapid nuclear divisions in the first 2 h of development (19). Moreover, E(z) null alleles have been shown to reduce immunostaining of several Pc-G proteins in polytene chromosomes and to result in a general decondensation of chromatin structure (28) that is reflected by increased chromosome breakage and a low mitotic index (11). Thus, E(z) appears to be a pleiotropic Pc-G gene with functions in epigenetic gene regulation, chromosome architecture, and growth control.
Mammalian homologues of E(z) have been isolated and characterized and are encoded by two loci in the mouse, Ezh1 and Ezh2 (Enhancer of zeste homologues) (15, 22). Alignments of Ezh1 and Ezh2 proteins with Drosophila E(Z) reveal four conserved regions of homology, including domain I, domain II, and a cysteine-rich amino acid stretch which precedes the C-terminal SET domain (22). The primary difference between the Ezh loci appears to lie in their expression profiles, with Ezh2 being predominantly expressed during embryonic development, whereas Ezh1 is more abundant in adult tissues (22).
Genetic and biochemical evidence from both Drosophila and the mouse support the existence of two distinct Pc-G complexes, with eed-Ezh2 defining one complex (33, 37) and Pc-G proteins such as Bmi-1 and Polycomb constituting the other (2). eed is the murine homologue of the Drosophila Pc-G gene esc (32) and is required for gastrulation in the mouse (8). By contrast, gene targeting approaches in the mouse revealed later developmental functions for members of the Bmi-1–Polycomb complex (reviewed in reference 36). Another Pc-G protein, YY1, was identified through homology to pleiohomeotic (3) and was subsequently shown to reside within the Bmi-1–Polycomb complex (10) and also to interact with eed (31). YY1 is unique among the Pc-G proteins in that it displays sequence-specific DNA binding activity (3), with disruption of YY1 resulting in peri-implantation lethality (7).
E(Z) is an original member of Drosophila chromatin regulators which defined the evolutionarily conserved SET domain (reviewed in reference 18). The SET domain of mammalian SU(VAR)3-9 homologues has recently been shown to harbor an intrinsic histone methyltransferase activity (29). Although E(Z)-related proteins have not so far been associated with histone methyltransferase activity, histone deacetylases (HDACs) 1 and 2 are present in the eed-Ezh2 complex (35), and transcriptional repression of this complex is mediated via histone deacetylation (35). Ezh proteins interact with eed through domain II (33, 37) but do not by themselves recruit HDACs (35). The SET domain of E(Z)-related proteins has been shown to be a target for mammalian Sbf-1 (5), a SET domain binding antiphosphatase, providing a link for Ezh in the control of proliferation and differentiation (5). In addition, domain II of Ezh2 interacts with the product of the proto-oncogene Vav (14), suggesting that Ezh2 may also be involved in signal-dependent T-cell activation. A potential role for Ezh2 in B- and T-cell development and proliferation is complemented by its abundant expression in the thymus and in organs of the developing immune system (15, 22).
With strong evidence linking Ezh2 to conserved mechanisms of eukaryotic gene silencing (22) and a possible role for Ezh2 in mouse development, growth control, and B- and T-cell proliferation, we decided to address the in vivo function of Ezh2 by generating an Ezh2 null mutation in the mouse germ line.
MATERIALS AND METHODS
Generation and genotyping of Ezh2 mutant mice.
Ezh2 maps to mouse chromosome 6 (23). A partial genomic clone of Ezh2 (23) was used to generate short (1.4 kb) and long (4.2 kb) arms of homology, in a strategy to produce an in-frame fusion of the first 200 amino acids of Ezh2 with β-galactosidase (LacZ) modified with a nuclear localization signal. The diphtheria toxin A (DTA) gene under the control of the MCI promoter (provided by T. Kallunki and M. Karin, San Diego, Calif.) was used to select against random integration of the targeting construct and was inserted 3′ of the long arm of homology. A pGNA-derivative targeting construct comprising a neomycin gene for positive selection and two polyadenylation sites (see Fig. 1A) was linearized with NotI and electroporated into feeder-independent CCE and feeder-dependent embryonic day 14.1 (E14.1) embryonic stem (ES) cells.
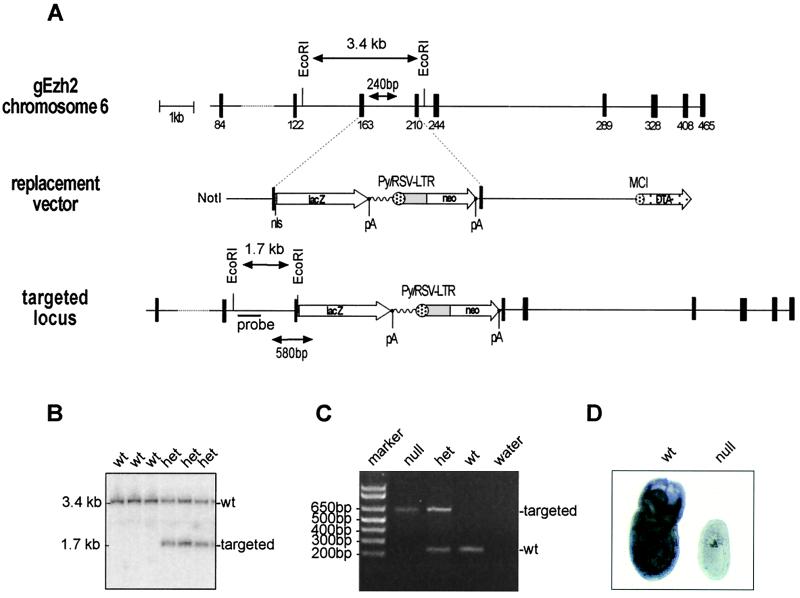
FIG. 1.
Targeting and genotyping of Ezh2-deficient mice. (A) Diagrammatic representation of the Ezh2 genomic locus, the replacement vector, and the targeted Ezh2 allele. Exons are indicated by black boxes with numbers referring to the starting amino acid positions of the respective exons (23). Also shown are the diagnostic EcoRI restriction sites and the internal probe used for Southern blot analysis, as are the primer pairs (arrowheads) for PCR genotyping. pA indicates polyadenylation signals. (B) Southern blot analysis of EcoRI-restricted DNA isolated from offspring of heterozygous (het) intercrosses. wt, wild type. (C) PCR genotyping of genomic DNA isolated from embryos of heterozygous intercrosses. (D) Whole-mount RNA in situ hybridization with day 7.5 wild-type and Ezh2 null embryos with an Ezh2-specific antisense probe.
ES cells were put under G418 selection, and homologous recombination was screened by PCR, using a nested reaction with primers external to the short arm (Ezh2 PCR1, 5′-GTTCTGGATCAGGATGGCAC; Ezh2 PCR2n, 5′-GTTGCCATGACAGTGGCAGCTC) and primers in the lacZ gene (lacZ PCR1, 5′-AACCCGTCGGATTCTCCGTGGGAAC; lacZ PCR2, 5′-CTCAGGAAGATCGCACTCCAGCC). Targeting was confirmed by Southern blot analysis of EcoRI-digested ES cell DNA with a 360-bp internal probe, generated with the primers Ezh2 probe 4f (5′-TCTGTTTATGGGGCATGTGC) and Ezh2 probe 4r (5′-ATCTGGAAGAATAGTGAAGGC). This probe detects a 3.4-kb fragment from the wild-type allele and a 1.7-kb fragment from the targeted allele (see Fig. 1A and B).
E14.1 targeted ES cells were used to generate chimeric mice. Heterozygous mice were interbred to obtain Ezh2-deficient embryos, which were genotyped by PCR using the following primers: g33 (5′-GTGCTGATAGGCCTTCAC) and lacZ PCR1 (above) to amplify a 580-bp fragment from the targeted allele, and Ezh2 900r (5′-CAGAGCACCTGGGAGCTGCTG) and g3 (5′-GAGGTTGATTCTTGTTTCCTATGC) to amplify a 240-bp fragment from the wild-type allele (see Fig. 1A and C).
Embryological and histological techniques.
Basic embryological and histological methods were performed as described elsewhere (16). Pregnant mice at defined times postcoitus were sacrificed by cervical dislocation, the deciduae were isolated, and the embryos were dissected.
Whole-mount RNA in situ hybridization analysis.
A 275-bp fragment of the EZH2 cDNA (22), encoding amino acids 330 to 455, was cloned into the pGEM-3Zf vector (Promega) to derive sense and antisense riboprobes. This human EZH2 probe contains 11 base pair mismatches (95% identity) with the corresponding portion of the murine Ezh2 gene and can be used to detect Ezh2 transcripts. Whole-mount RNA in situ hybridization of embryos was performed as described elsewhere (4).
Blastocyst outgrowth and ES cell derivation.
Blastocyst outgrowth and ES cell derivation procedures were performed as described elsewhere (1).
RT-PCR to detect Ezh1 and Ezh2 transcripts.
Total RNA was prepared from pools of oocytes, fertilized oocytes, morulae, and blastocysts using glycogen as a carrier. Reverse transcription (RT) was performed with random hexamers (G. Schaffner, IMP) on the above-described RNA preparations and on 1 μg of total spleen RNA. Primer pairs that are specific for Ezh1 (E1f, 5′-TGAAATCTGAGTATATGCGGC; E1r, 5′-AGATATCCTGGCTGTCGAAC) or Ezh2 (E2f, 5′-GCCAGACTGGGAAGAAATCTG; E2r, 5′-TGTGCTGGAAAATCCAAGTCA) were used in a subsequent PCR to generate a 236-bp product for Ezh1 and a 270-bp product for Ezh2. The mouse Gapdh (30) primers (G1, 5′-CGGAGTCAACGGATTTGGTCGTAT; G2, 5′-AGCCTTCTCCATGGTGGTGAAGAC) were used to control for the quality and amount of the reverse-transcribed RNA.
RESULTS
Targeting the Ezh2 locus.
A conventional targeting approach was used to inactivate the Ezh2 locus on mouse chromosome 6 (23). The Ezh2 gene was disrupted by homologous recombination in ES cells, replacing parts of exons five and six with a lacZ gene and a neo gene (Fig. 1A). This targeting strategy produces an in-frame fusion of the first 200 amino acids of Ezh2 with LacZ. Domain I, the eed interaction region (33, 37), is present in the fusion protein, but domain II and the C-terminal SET domain will not be expressed upon targeting (Fig. 1D). The Ezh2-LacZ fusion protein could not be used as a marker of Ezh2 expression because it did not retain β-galactosidase activity (data not shown). The frequency of homologous recombination in feeder-dependent ES cells was 11%. Injection of three independently targeted clones produced several high chimeras from two clones. Chimeras derived from one targeted clone passed the mutation through the germ line. EcoRI-restricted DNA from heterozygous mice produces a 3.4-kb fragment from the wild-type locus and a diagnostic 1.7-kb fragment from the targeted allele (Fig. 1B). Homozygous mutation of the Ezh2 gene results in early embryonic lethality (see below). In order to genotype Ezh2 mutant embryos, a PCR-based approach to detect both the wild-type and mutant alleles was developed. The locations of the respective primer pairs are indicated in Fig. 1A. PCR amplification produces a fragment of 240 bp for the endogenous allele and a fragment of 580 bp for the targeted allele (Fig. 1C). To demonstrate the generation of a loss-of-function allele, RNA in situ hybridization was performed on wild-type and Ezh2-deficient day 7.5 embryos, using an Ezh2-specific antisense probe; Ezh2 transcripts hybridizing to this antisense probe were not detectable in mutant embryos (Fig. 1D).
Ezh2 is required for mouse development.
Genotyping of offspring by Southern blot analysis from Ezh2 heterozygous intercrosses revealed the absence of mice homozygous for the Ezh2 mutation (Table 1), indicating that Ezh2 is required for embryonic development. In order to determine when the Ezh2 mutation produces a lethal phenotype, timed matings were set up and embryos were obtained at day 7.5 and day 10.5. Genotyping of day 10.5 embryos by Southern blot analysis revealed that no Ezh2 mutant mice were identified (Table 1). We next analyzed day 7.5 embryos. Genotyping by PCR indicated the presence of Ezh2 mutants, albeit just under Mendelian ratios. All mutant embryos identified by PCR were significantly smaller than littermates and fell into two distinct categories. The first category comprised embryos which were extremely growth retarded and resembled in size a day 5.5 embryo. The second category of embryos comprised those that were growth retarded and displayed increased amounts of extraembryonic tissue. Indeed, the ratio between embryonic and extraembryonic tissues was visibly skewed, with only a small part of the embryo consisting of embryonic regions. Sub-Mendelian numbers of Ezh2 heterozygous animals were observed at birth (Table 1). Genotyping of day 10.5 embryos derived from either heterozygous intercrosses or wild-type-to-heterozygous crosses revealed the presence of Mendelian ratios of heterozygous animals (Table 1 and data not shown). This suggests that a fraction of Ezh2 heterozygous mice develop problems during mid- to late gestation. However, the nature of these heterozygous defects was not further investigated.
TABLE 1.
Genotypes of live births and embryos from Ezh2 heterozygous intercrossesa
| Category | Total no. | No. wt | No. het | No. null | No. resorbed | ND |
|---|---|---|---|---|---|---|
| Live births | 82 | 45 | 37 | 0 | ||
| Embryos (E10.5) | 19 | 4 | 6 | 0 | 9 | |
| Embryos (E7.5) | 41 | 11 | 19 | 6 | 2 | 3 |
Numbers and genotypes of life births and day 10.5 and day 7.5 embryos from Ezh2 heterozygous intercrosses. ND, not determined; wt, wild-type; het, heterozygous.
Arrested development and gastrulation failure in embryos from Ezh2 heterozygous intercrosses.
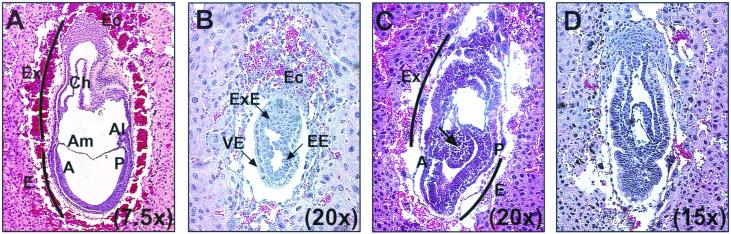
We then examined day 7.0 to day 8.5 embryos derived from heterozygous intercrosses histologically. Entire deciduae were isolated, fixed, and stained with hematoxylin-eosin. Preparations were sectioned, and embryos were scored for being either of wild-type (Fig. 2A) morphology, abnormal, or resorbed. At day 8.5, no abnormal embryos were identified, although a significant number of resorptions were observed (Table 2). By contrast, at day 7.0 to day 7.5, 11 abnormal embryos and 3 resorptions were detected. These 11 presumptive Ezh2-deficient mice fell into two distinct categories, similar to the analysis with whole-mount preparations (see above). The most severely growth-retarded embryos ceased developing shortly after implantation and resembled a day 5.5 embryo (Fig. 2B). This category of mutants accounted for 45% (5 of 11) of the abnormal embryos. The other mutants appeared to initiate but not complete gastrulation (Fig. 2C and D). The initiation of gastrulation was confirmed by the presence of mesoderm cells and verified by RNA in situ analysis of the mesoderm marker brachyury (13) in whole-mount null embryos (data not shown). However, the majority of the mesoderm-like cells appear to migrate into extraembryonic regions and form a bulge of cells which pushes on the embryonic portion of the embryo (Fig. 2C). The embryo illustrated in Fig. 2D consists almost completely of extraembryonic tissues and represents a mutant which continued the aberrant cell migration shown in Fig. 2C. The mutant embryos represented in Fig. 2C and D each accounted for 27% (3 of 11) of abnormal embryos. This later-onset phenotype resembles that of the eed mutation, where embryos fail to complete gastrulation due to defects in morphogenetic movements (9).
FIG. 2.
Arrested development and gastrulation failure in presumptive Ezh2-deficient embryos. Deciduae at day 7.0 to day 7.5 were isolated, sectioned at 5-μm thickness, stained with hematoxylin-eosin, and scored for being normal or abnormal based on size and morphology. (A) An almost-sagittal section through a wild-type embryo at day 7.5. (B to D) Sections of presumptive Ezh2 mutants isolated at days 7.0 to 7.5. The arrow in panel C highlights excessive amounts of mesoderm aberrantly accumulating onto the abnormal embryo. The different magnifications of each photograph are indicated. Abbreviations: A, anterior; Al, allantois; Am, amnion; E, embryonic; EE, embryonic ectoderm; Ec, ectoplacental cone; Ex, extraembryonic; ExE, extraembryonic ectoderm; P, posterior; Ch, chorion; VE, visceral endoderm.
TABLE 2.
In utero histology of embryos from Ezh2 heterozygous intercrossesa
| Age | No. of embryos | No. abnormal | No. resorbed | Total no. (A + R)b |
|---|---|---|---|---|
| E8.5 | 22 | 5 | 5 | |
| E7.0–E7.5 | 57 | 11 | 3 | 14 |
Statistical analysis of perturbed morphologies in day 8.5 embryos and day 7.0 to day 7.5 embryos from Ezh2 heterozygous intercrosses. For examples of abnormal embryos, see Fig. 2.
A + R, no. abnormal plus no. resorbed.
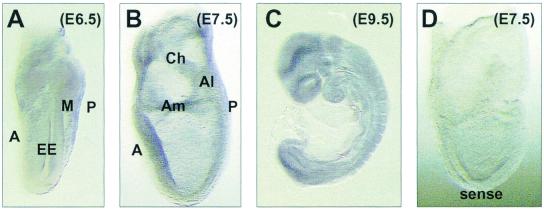
We investigated the spatial expression profiles of Ezh2 by whole-mount RNA in situ hybridizations with day 6.5 to day 9.5 mouse embryos. Whereas no staining was observed with the control sense probe (Fig. 3D), the Ezh2 antisense probe revealed broad expression of Ezh2 throughout the gastrulating mouse embryo (day 6.5; Fig. 3A). This expression profile indicates a role for Ezh2 during gastrulation and would be consistent with the defects of the presumptive Ezh2-deficient embryos described above. However, Ezh2 was also expressed broadly at later stages of mouse development (E7.5 to E9.5; Fig. 3B and C), suggesting additional functions for Ezh2 in postgastrulation development.
FIG. 3.
Ezh2 is broadly expressed during early mouse embryogenesis. Whole-mount RNA in situ hybridization analysis of wild-type day 6.5 (A), day 7.5 (B), and day 9.5 (C) embryos with an Ezh2-specific antisense probe or, as a control, with an Ezh2-specific sense probe (D) is shown. Abbreviations: A, anterior; Al, allantois; Am, amnion; Ch, chorion; EE, embryonic ectoderm; M, mesoderm; P, posterior.
Ezh2 is required for ES cell derivation.
To further analyze the Ezh2 null phenotype, we decided to generate Ezh2-deficient ES cells, which would allow us to gain insight into Ezh2 function through cellular assays and chimera studies. We derived ES cells from blastocysts obtained from heterozygous intercrosses. The inner cell mass (ICM) of expanded blastocysts was disaggregated and seeded under ES cell conditions, while the trophoblast layer was used for PCR genotyping. From 62 blastocysts processed for ES cell derivation, 33 ES cell lines were established, and none of these lines was homozygous for the Ezh2 mutation (Table 3). A representative wild-type ES cell clone is shown in Fig. 4A. Attempts to derive ES cells from null blastocysts resulted in non-ES-like cells that failed to proliferate, acquired large vacuoles, and subsequently died (Fig. 4B). ES cell lines were established from both wild-type and heterozygous blastocysts with an approximate efficiency of 62% (Table 3). With this degree of efficiency, we would have expected 6 null ES cell lines from 10 null blastocysts; we therefore conclude that Ezh2 is required for the derivation of ES cells.
TABLE 3.
Derivation of ES cell lines from blastocystsa
| Group | Total no. | No. with genotype
|
||
|---|---|---|---|---|
| Wt | Het | Null | ||
| Blastocysts | 62 | 25 | 27 | 10 |
| ES cell lines | 33 | 16 | 17 | 0 |
Statistical analysis to derive ES cell lines from day 3.5 blastocysts of the indicated genotypes (see also Fig. 4A and B). Wt, wild-type; Het, heterozygous.
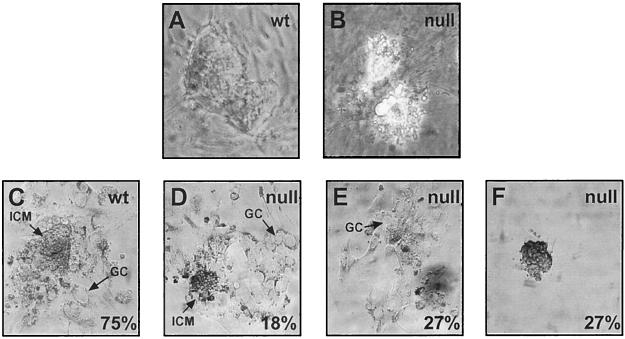
FIG. 4.
Impaired ES cell derivation and outgrowth potential of Ezh2-deficient blastocysts. Blastocysts were isolated from heterozygous intercrosses at day 3.5 and processed for either ES cell derivation (A and B) or outgrowth experiments (C to F). In panel A, a wild-type (wt) ES cell colony is shown, whereas panel B depicts the highly vacuoled and short-lived mutant cells derived from an Ezh2 null blastocyst. (C to F) Blastocysts were cultured in vitro for 7 days and photographed. Panels C and D show successful outgrowths from a wild-type blastocyst and an Ezh2 null blastocyst. Arrows point to the ICM and to the trophoblast giant cells (GC). Each percentage indicates the proportion of blastocysts (n = 68) displaying the depicted phenotype (see Table 4).
Impaired outgrowth potential of Ezh2-deficient blastocysts.
We next examined the outgrowth potential of blastocysts. Blastocysts were cultured under conditions where the ICM should expand and outgrow, while the trophoblast layer should become adherent and differentiate into giant cells (Fig. 4C). Cells were kept in culture for 7 days after seeding, photographed for documentation, and subsequently genotyped by PCR. Seventy-five percent of wild-type blastocysts and 51% of heterozygous blastocysts underwent normal outgrowth, compared to only 18% of null blastocysts (Table 4; Fig. 4D). The null blastocysts, which failed to expand normally, displayed three main phenotypes. One subset of null blastocysts did not show growth of the ICM, while the trophoblast layer attached and differentiated into giant cells (termed “no ICM” in Table 4; Fig. 4E). Another group of null blastocysts did not show any growth of the ICM or of the trophoblast (termed “no outgrowth” in Table 4; Fig. 4F). The final group of null blastocysts was nonviable and died after a brief time in culture. These data demonstrate that blastocyst outgrowth in vitro is impaired in the absence of Ezh2, which appears to be primarily attributable to the failure of the growth of the ICM, since the trophoblast layer of Ezh2-deficient blastocysts has the potential to adhere and differentiate into giant cells.
TABLE 4.
Blastocyst outgrowth experimentsa
| Ezh2 genotype | No. total | No. (%) with phenotype
|
|||
|---|---|---|---|---|---|
| Normal | No ICM | No outgrowth | Nonviable | ||
| Wt | 28 | 21 (75) | 2 (7) | 4 (14) | 1 (3) |
| Het | 29 | 15 (51) | 6 (2) | 6 (21) | 2 (7) |
| Null | 11 | 2 (18) | 3 (27) | 3 (27) | 3 (27) |
Statistical analysis of the blastocyst outgrowth experiments (n = 68). Successful outgrowths (normal) are illustrated in Fig. 4C and D, with 75% of wild-type (wt) but only 18% of null blastocysts being capable of expanding the ICM. No ICM, blastocysts failed to expand the ICM while the trophoblast layer attached and differentiated into giant cells (Fig. 4E). No outgrowth, blastocysts did not outgrow ICM or trophoblast cells (Fig. 4F). Nonviable, blastocysts died after a brief time in culture. Het, heterozygous.
Ezh2 is up-regulated in fertilized oocytes and preimplantation embryos.
Finally, we determined the preimplantation expression profile of Ezh2 by RT-PCR on total RNA prepared from oocytes, fertilized oocytes, and preimplantation embryos. To control for the relative abundance of Ezh2 transcripts, we included RT-PCR analyses for the second murine Ezh gene, Ezh1 (22), and for the ubiquitously expressed Gapdh gene. Whereas Ezh1 is expressed in fertilized oocytes, Ezh1 transcripts are not detectable in morulae or blastocysts and are also not present in unfertilized oocytes (Fig. 5, upper panel). By contrast, Ezh2 is also up-regulated upon fertilization but remains abundantly expressed at all stages of preimplantation (Fig. 5, middle panel). Splenic RNA was used as a positive control for the RT-PCR, since both Ezh loci are similarly expressed in this tissue (22). These data indicate persistent expression of Ezh2, but not of Ezh1, in mouse preimplantation embryos.
FIG. 5.
Preimplantation expression of Ezh1 and Ezh2 in the early mouse embryo. RT-PCR was used to detect Ezh1, Ezh2, and Gapdh transcripts in total RNA preparations from wild-type oocytes, fertilized oocytes, morulae, and blastocysts. As a control, RT-PCR was also performed on total RNA from spleen tissue.
DISCUSSION
Ezh2 is an early-acting Pc-G gene during mouse preimplantation.
Based on the above analysis, Ezh2 joins YY1 as the only other described Pc-G gene (7) with expression at preimplantation stages of mouse development (Fig. 5). By contrast, although eed and Ezh2 physically interact (33, 37), zygotic eed expression does not appear to start until shortly after implantation at day 5.5 (32). These data suggest additional and earlier roles for Ezh2 outside the eed-Ezh2 complex. Ezh2 could function during preimplantation development to prepare the embryo for rapid cell proliferations and the initiation and maintenance of complex gene expression programs. Since Ezh2 is a SET domain-containing protein, it is intriguing to infer that some of these early functions may impinge on a more regional modification of the underlying chromatin structure, presumably by enhancing or interfering with the methylation of a yet-unknown substrate.
Alternatively, Ezh2 may be recruited to sites in the genome of the preimplantation embryo which require heritable regulation upon implantation. During implantation and concomitant with the onset of eed expression, Ezh2 would interact with eed to target HDACs (35) to these genomic sites. Indeed, this hierarchy is supported by this and other (G. Lagger, D. O'Carroll, T. Jenuwein, and C. Seiser, unpublished data) genetic studies with the mouse. Ezh2-deficient blastocysts display an impaired potential for outgrowth (Table 4 and Fig. 4D to F), and a subset of Ezh2 mutants (45%) cease to progress after implantation (Fig. 2B). The later-developing Ezh2 mutants bear a striking resemblance to those with the eed phenotype, which is required for morphogenetic movements during gastrulation, with eed mutants dying around day 8.5 (8, 9). Similarly, a portion of Ezh2 mutants (55%) initiate but fail to complete gastrulation and also display an apparent defect in mesoderm migration (Fig. 2C and D).
Ezh2: a regulator of proliferation?
The Ezh2 null mutation results in early embryonic lethality, and Ezh2-deficient mouse embryos die during the transition from pre- to postimplantation development (Tables 1 and 2 and Fig. 2). This transition is accompanied by elevated rates of cell division, and around gastrulation these divisions are characterized by very short cell cycles (4 to 6 h for some cell types) (16). The broad expression profile of Ezh2 during pre- and postimplantation stages of mouse development (Fig. 5 and Fig. 3, respectively) would be consistent with Ezh2 having a more global role in the control and/or progression of proliferation. Although Ezh2-deficient blastocysts maintain the potential to outgrow both the ICM and the trophoblast cells, most null blastocysts fail to expand their ICM in vitro (Tables 3 and 4 and Fig. 4). Similarly, Ezh2-deficient embryos show arrested development after implantation (Fig. 2).
There are many additional lines of evidence linking Ezh2 function to proliferation in cell types that are more specialized than those present in the preimplantation mouse embryo. For example, Ezh2 was also identified through an interaction with the gene product of the proto-oncogene Vav (14), which is required as a signaling molecule for stimulated lymphocyte proliferation (34). Resting splenic B or T cells do not express Ezh2, but upon mitogen stimulation, Ezh2 is rapidly up-regulated (D. O'Carroll and T. Jenuwein, unpublished data). In addition, the transition of resting mantle B cells to proliferating follicular centroblasts coincides with the appearance of both Ezh2 and eed expression (27). In general, SET domain proteins appear to be important chromatin-associated targets for signaling pathways involved in regulating growth control. Forced expression of Sbf-1, a SET domain binding factor resembling an antiphosphatase (5), results in oncogenic transformation of fibroblasts or enhanced proliferation of primary B-cell cultures (6). Sbf-1 was shown to interact with the SET domain of E(Z)-related proteins (5).
In summary, we have shown that the Pc-G gene Ezh2 is required for early mouse development. The severity of the phenotype distinguishes Ezh2 from most other Pc-G genes and defines Ezh2 as an early-acting gene along with YY1 and eed (7, 32). Our genetic analysis thus indicates a functional overlap for these Pc-G genes during mouse preimplantation and extends the recent observation that YY1 can physically interact with the eed-Ezh2 protein complex (31). Although Ezh2 represents one of the most early-acting Pc-G genes, it is also broadly expressed in the postimplantation embryo and in lymphoid organs (15, 22). Based on the data presented here, we propose that Ezh2 is an important regulator for growth control whose disruption at later stages of mouse development would severely impair the overall proliferation potential of Ezh2-mutated cells.
ACKNOWLEDGMENTS
We thank Gotthold Schaffner for sequence analysis and oligonucleotide synthesis, Hans-Christian Theussl for blastocyst injection of ES cell clones, Terry Magnuson and Elizabeth M. Morin-Kensicki (Case Western Reserve University, Cleveland, Ohio) for kind teaching of early embryological techniques, Annette Neubüser for advice on Ezh2 mutant morphology, and Stephen Rea for critical reading of the manuscript.
Research in T.J.'s laboratory is supported by the IMP through Boehringer Ingelheim, the Austrian Research Promotion Fund, and the Vienna Economy Promotion Fund. Research in M.A.S.'s laboratory is supported by a Wellcome Trust Grant (036481), and S.E. holds a Ph.D. scholarship from Boehringer Ingelheim.
REFERENCES
- 1.Abbondanzo S J, Gadi I, Stewart C L. Derivation of embryonic stem cell lines. Methods Enzymol. 1993;225:803–823. doi: 10.1016/0076-6879(93)25052-4. [DOI] [PubMed] [Google Scholar]
- 2.Alkema M J, Bronk M, Verhoeven E, Otte A, van't Veer L J, Berns A, van Lohuizen M. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian Polycomb complex. Genes Dev. 1997;11:226–240. doi: 10.1101/gad.11.2.226. [DOI] [PubMed] [Google Scholar]
- 3.Brown J L, Mucci D, Whiteley M, Dirksen M L, Kassis J A. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 4.Conlon R A, Herrmann B G. Detection of messenger RNA by in situ hybridization to postimplantation embryo whole mounts. Methods Enzymol. 1993;225:373–383. doi: 10.1016/0076-6879(93)25026-x. [DOI] [PubMed] [Google Scholar]
- 5.Cui X, De Vivo I, Slany R, Miyamoto A, Firestein R, Cleary M L. Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet. 1998;18:331–337. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- 6.De Vivo I, Cui X, Domen J, Cleary M L. Growth stimulation of primary B cell precursors by the anti-phosphatase Sbf1. Proc Natl Acad Sci USA. 1998;95:9471–9476. doi: 10.1073/pnas.95.16.9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donohoe M E, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faust C, Schumacher A, Holdener B, Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development. 1995;121:273–285. doi: 10.1242/dev.121.2.273. [DOI] [PubMed] [Google Scholar]
- 9.Faust C, Lawson K A, Schork N J, Thiel B, Magnuson T. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development. 1998;125:4495–4506. doi: 10.1242/dev.125.22.4495. [DOI] [PubMed] [Google Scholar]
- 10.Garcia E, Marcos-Gutierrez C, del Mar Lorente M, Moreno J C, Vidal M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999;18:3404–3418. doi: 10.1093/emboj/18.12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatti M, Baker B S. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 1989;3:438–453. doi: 10.1101/gad.3.4.438. [DOI] [PubMed] [Google Scholar]
- 12.Gutjahr T, Frei E, Spicer C, Baumgartner S, White R A H, Noll M. The Polycomb-group gene, extra sex combs, encodes a nuclear member of the WD-40 repeat family. EMBO J. 1995;14:4296–4306. doi: 10.1002/j.1460-2075.1995.tb00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrmann B G, Labeit S, Poustka A, King T R, Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- 14.Hobert O, Jallal B, Ullrich A. Interaction of Vav with ENX-1, a putative transcriptional regulator of homeobox gene expression. Mol Cell Biol. 1996;16:3066–3073. doi: 10.1128/mcb.16.6.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobert O, Sures I, Ciossek T, Fuchs M, Ullrich A. Isolation and developmental expression analysis of Enx-1, a novel mouse Polycomb group gene. Mech Dev. 1996;55:171–184. doi: 10.1016/0925-4773(96)00499-6. [DOI] [PubMed] [Google Scholar]
- 16.Hogan B, Beddinton R, Costantini F, Lacy E. Manipulating the mouse embryo. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 17.Holdeman R, Nehrt S, Strome S. MES-2, a maternal protein essential for viability of the germline in Caenorhabditis elegans, is homologous to a Drosophila Polycomb group protein. Development. 1998;125:2457–2467. doi: 10.1242/dev.125.13.2457. [DOI] [PubMed] [Google Scholar]
- 18.Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones R S, Gelbart W M. Genetic analysis of the Enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics. 1990;126:185–199. doi: 10.1093/genetics/126.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones R S, Gelbart W M. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol Cell Biol. 1993;13:6357–6366. doi: 10.1128/mcb.13.10.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korf I, Fan Y, Strome S. The Polycomb group in Caenorhabditis elegans and maternal control of germline development. Development. 1998;125:2469–2478. doi: 10.1242/dev.125.13.2469. [DOI] [PubMed] [Google Scholar]
- 22.Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laible G, Haynes A R, Lebersorger A, O'Carroll D, Mattei M G, Denny P, Brown S D, Jenuwein T. The murine Polycomb-group genes Ezh1 and Ezh2 map close to Hox gene clusters on mouse chromosomes 11 and 6. Mamm Genome. 1999;10:311–314. doi: 10.1007/s003359900993. [DOI] [PubMed] [Google Scholar]
- 24.Moazed D, O'Farrell P H. Maintenance of the engrailed expression pattern by Polycomb group genes in Drosophila. Development. 1992;116:805–810. doi: 10.1242/dev.116.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paro R, Strutt H, Cavalli G. Heritable chromatin states induced by the Polycomb and trithorax group genes. Novartis Found Symp. 1998;214:51–61. doi: 10.1002/9780470515501.ch4. [DOI] [PubMed] [Google Scholar]
- 26.Pelegri F, Lehmann R. A role of Polycomb group genes in the regulation of gap gene expression in Drosophila. Genetics. 1994;136:1341–1353. doi: 10.1093/genetics/136.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raaphorst F M, van Kemenade F J, Fieret E, Hamer K M, Satijn D P, Otte A P, Meijer C J. Cutting edge: Polycomb gene expression patterns reflect distinct B cell differentiation stages in human germinal centers. J Immunol. 2000;164:1–4. doi: 10.4049/jimmunol.164.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Rastelli L, Chan C S, Pirrotta V. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rea S, Eisenhaber F, O'Carroll D, Strahl B D, Sun Z W, Schmid M, Opravil S, Mechtler K, Ponting C P, Allis C D, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 30.Sabath D E, Broome H E, Prystowski M B. Glyceraldehyde-3-phosphate dehydrogenase mRNA is a major interleukin 2-induced transcript in a cloned T-helper lymphocyte. Gene. 1990;91:185–191. doi: 10.1016/0378-1119(90)90087-8. [DOI] [PubMed] [Google Scholar]
- 31.Satijn D P E, Hamer K M, den Blaauwen J, Otte A P. The Polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol Cell Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;383:250–253. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- 33.Sewalt R G, van der Vlag J, Gunster M J, Hamer K M, den Blaauwen J L, Satijn D P, Hendrix T, van Driel R, Otte A P. Characterization of interactions between the mammalian Polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian Polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–3595. doi: 10.1128/mcb.18.6.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarakhovsky A, Turner M, Schaal S, Mee P J, Duddy L P, Rajewsky K, Tybulewicz V L. Defective antigen receptor-mediated proliferation of B and T cells in the absence of Vav. Nature. 1995;374:467–470. doi: 10.1038/374467a0. [DOI] [PubMed] [Google Scholar]
- 35.van der Vlag J, Otte A P. Transcriptional repression mediated by the human Polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 36.van Lohuizen M. Functional analysis of mouse Polycomb group genes. Cell Mol Life Sci. 1998;54:71–79. doi: 10.1007/s000180050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Lohuizen M, Tijms M, Voncken J W, Schumacher A, Magnuson T, Wientjens E. Interaction of mouse Polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol Cell Biol. 1998;18:3572–3579. doi: 10.1128/mcb.18.6.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]