Abstract
Mastermind (Mam) has been implicated as an important positive regulator of the Notch signaling pathway by genetic studies using Drosophila melanogaster. Here we describe a biochemical mechanism of action of Mam within the Notch signaling pathway. Expression of a human sequence related to Drosophila Mam (hMam-1) in mammalian cells augments induction of Hairy Enhancer of split (HES) promoters by Notch signaling. hMam-1 stabilizes and participates in the DNA binding complex of the intracellular domain of human Notch1 and a CSL protein. Truncated versions of hMam-1 that can maintain an association with the complex behave in a dominant negative fashion and depress transactivation. Furthermore, Drosophila Mam forms a similar complex with the intracellular domain of Drosophila Notch and Drosophila CSL protein during activation of Enhancer of split, the Drosophila counterpart of HES. These results indicate that Mam is an essential component of the transcriptional apparatus of Notch signaling.
Notch signaling is an evolutionarily conserved mechanism that mediates cell-cell communications required for cell fate decisions in metazoans (2). For some of these processes, transactivation of the primary target genes of this signaling, Enhancer of split [E(spl)] in Drosophila melanogaster or Hairy Enhancer of split-1 (HES-1) and HES-5 in mammals, has been shown to be essential (2, 22). Activation of these genes involves ligand-induced intracellular (IC) shedding of the Notch receptor, translocation of the IC domain to the nucleus, and association of the IC domain with promoter elements through CSL DNA binding proteins (CBF-1, Suppressor of Hairless [Su(H)], and Lag-1) (12, 21, 28).
Notch receptors are first synthesized as large single-pass transmembrane proteins. During maturation, the Notch receptor is cleaved once in the extracellular region (site 1) and noncovalently reattached (35). Upon binding to ligands that are present on adjacent cells, Notch is cleaved sequentially at an extracellular juxtamembrane region (site 2) and in or near its transmembrane domain (site 3) (35). It has been postulated that the IC domain of the receptor is liberated from the membrane and transported to the nucleus (15, 28, 31), where it participates in transcriptional activation (12, 28). Recently, it has been shown that mice homozygous for the Notch1 allele, which is deficient in the processing of site 3, exhibit embryonic death, resembling mice homozygous for the null allele (11). This result supports the nuclear Notch model, at least in early mammalian development.
Mastermind (Mam), along with several other components of the Notch pathway, is a member of the original group of neurogenic loci of Drosophila (16). Loss-of-function neurogenic phenotypes and strong genetic interactions with other pathway mutations have implicated Mam as an important positive regulator of the Notch signaling pathway (1, 8). The Mam protein has been shown to associate with specific polytene chromosome sites in vivo, and it is often coincident with RNA polymerase II, implying a role in transcriptional regulation (3). However, until very recently (25, 37) Mam had been identified only in the genus Drosophila (20, 30), and its biochemical activity has remained elusive.
We show here that a possible human counterpart (hMam-1) of Drosophila Mam (DMam) stabilizes and participates in the DNA binding complex of the IC domain of human Notch1 and a CSL protein to activate HES promoters. Furthermore, DMam forms a similar complex with the IC domain of Drosophila Notch (DNotch) and Drosophila CSL protein during activation of E(spl). These data confirm and extend recent reports on Mam function (25, 37), further supporting the idea that Mam is an essential component of the transcriptional apparatus of Notch signaling.
MATERIALS AND METHODS
Plasmid constructions.
To produce cDNA for hMam-1 with a hemagglutinin tag attached to the portion of the coding region, corresponding to the N terminus (hMam-1 N-HA), a synthetic double-stranded oligonucleotide that has Kozak's optimal sequence for initiation of translation and a coding sequence for an HA tag (NH2-MGYPYDVPDYASLGPGM, where the final M is the initiator methionine of KIAA0200) was inserted into the KIAA0200 cDNA (19). Deletion mutants of hMam-1 were constructed by digestion with appropriate restriction endonucleases, filling in by T4 DNA polymerases, ligation to synthetic oligonucleotide linkers containing in-frame termination codons, and cloning into plasmid vectors. The various hMam-1 cDNAs and RBP-J cDNA (17) were cloned into pEF-BOS (18) or pMx (23) for expression in mammalian cells. Full-length Notch1 cDNA within pcDNA1/Amp (Invitrogen) was as described previously (4). An expression vector for the IC domain of Notch1 (Notch1IC) was constructed by replacing the portion of the Notch1 cDNA encoding the extracellular and transmembrane domains with a synthetic oligonucleotide encoding the Kozak sequence and the initiator methionine. The resultant vector expresses amino acids 1760 to 2556 of Notch1 following the initiator methionine and glycine. cDNA encoding the IC domain of DNotch (36) was constructed by a similar method and cloned in pMT under the control of the metallothionein promoter. The resultant vector expresses amino acids 1762 (methionine) to 2703. DMam cDNA (30) was cloned in pMT and pAct (under the control of the actin 5C distal promoter). Su(H) tagged with the c-Myc epitope and cloned in pCaSpeR-hs (under the control of the heat shock protein 70 promoter) was as described previously (6). pMTNotch(ICN) (where ICN is a designation for IC Notch), pMTSu(H), and mγ Luc were obtained from L. Grimm (lab of S. Artavanis-Tsakonas, Harvard University).
Immunocytochemistry.
293T cells seeded in Lab-Tek chamber slides (Nalge Nunc) were transfected with plasmids by the calcium phosphate method. Forty-eight hours after transfection, cells were fixed with the IntraStain kit (DAKO) followed by staining with an anti-HA antibody (12CA5) and a fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G. The stained cells were analyzed with a laser-scanning microscope (Zeiss).
Transcriptional activation assays.
NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% calf serum. The cells were seeded in six-well dishes (1.5 × 105 cells/well) and cotransfected with 0.6 μg of pHES-5luc reporter plasmid (21), 0.8 μg of pcDNA1/Amp with or without Notch cDNA, 0.6 μg of pEF-BOS (for full-length proteins) or pMx (for truncations) with or without various hMam-1 cDNAs, and 20 ng of Renilla luciferase internal control plasmid (pRL-CMV; Promega). The transient transfection was done using Lipofectamine (Gibco-BRL). Forty-eight hours after transfection, firefly and Renilla luciferase activities were determined using the Dual Luciferase assay kit (Promega) and a Turner Designs TD20/20 dual luminometer. Firefly luciferase activities were normalized with the Renilla luciferase control activities.
Drosophila S2 cells were grown in serum-free media (Hyclone) supplemented with 50 μg of penicillin and streptomycin (Gibco-BRL)/ml. The cells were seeded in 12-well plates at 23°C and cultured to 40 to 60% confluence. Transfections (Lipofectin; Gibco-BRL) contained 0.2 μg of each expression plasmid, 0.4 μg of reporter, and 0.1 μg of control plasmid and pMT to maintain constant amounts of DNA. After transfection, the cells were incubated in 0.7 mM CuSO4 for 17 to 24 h to induce the MT promoter. Enzyme activity was measured as described above.
Electrophoretic mobility shift assays (EMSA).
293T cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. The cells (2 × 106 cells/10-cm dish) were transfected by the calcium phosphate method with 5 μg of each of the expression vectors for RBP-J, Notch1IC, and various Mam constructs or their empty counterparts. The total amount of plasmid DNA was kept constant (15 μg). Forty-eight hours after transfection, the cells were harvested, washed with phosphate-buffered saline, and suspended in ice-cold 20 mM HEPES-NaOH (pH 7.9) buffer containing 0.5% NP-40, 15% glycerol, 300 mM NaCl, 1 mM EDTA, 10 mM NaF, 1 mM dithiothreitol, 1 mM sodium orthovanadate, 0.5 mM phenylmethylsulfonyl fluoride, 50 μM calpain inhibitor-1, 1 μg of leupeptin/ml, 1 μg of pepstatin/ml, and 1 μg of aprotinin/ml. After 30 min of gentle agitation at 4°C, the supernatants were collected as whole-cell extracts by centrifugation. DNA-protein binding reactions (15 μl) were performed by incubation of the whole-cell extracts (20 μg equivalent as protein amount) in a solution containing 13 mM HEPES-NaOH (pH 7.9) buffer containing 8% glycerol, 50 mM NaCl, 0.4 mM MgCl2, 0.5 mM dithiothreitol, 66.6 μg of poly(dI-dC) · poly(dI-dC)/ml, and 33.3 μg of salmon sperm DNA/ml for 15 min on ice, followed by an additional 30-min incubation with 32P-end-labeled synthetic double-stranded oligonucleotide probe (0.1 to 0.2 ng, 5 to 20 nCi) at room temperature. Half of the mixture was loaded on to polyacrylamide gels (5%) in 0.5 × Tris-borate-EDTA buffer to separate DNA-protein complexes. The complexes were detected by exposing the dried gels to X-ray films. To test the sensitivities of the binding activities to antibodies (supershift experiments), antibodies were added to the binding reactions following the radiolabeled probe (32). The sequences of the oligonucleotides for the probe (−91 to −56 of the mouse HES-1 gene) (12, 33) were 5′-GATCGTTACTGTGGGAAAGAAAGTTTGGGAAGTTTCACAC-3′ and 5′-GATCGTGTGAAACTTCCCAAACTTTCTTTCCCACAGTAAC-3′. The antibodies used for supershift experiments were purified immunoglobulin fractions of anti-RBP-J (0.5 μg; K0043) (27), anti-Notch1 (0.2 μg, sc-6014; Santa Cruz), anti-HA (0.6 μg; 12CA5) and anti-CD44 (1.0 μg; Hermes3).
Immunoprecipitation and immunoblotting.
Whole-cell extracts from 293T cells were prepared as described above. Drosophila S2 cells were maintained in M3 medium containing 10% fetal bovine serum. The cells (4 × 106 cells/10-cm dish) were transfected with CellFECTIN (Gibco-BRL) with 1.7 μg of each of the plasmid vectors for Su(H), DNotchIC, and DMam or their empty control vectors to keep the total amount constant (5.1 μg). Twenty-four hours after transfection, CuSO4 (final concentration, 0.7 mM) was added to the medium to induce the MT promoter. Beginning 48 and 72 h after transfection, the cells were incubated twice at 37°C for 1 h at 30-min intervals to induce the heat shock promoter (6). The cells were harvested 2 h after the final heat shock. Preparation of whole-cell extracts was done as for 293T cells. The extracts were immunoprecipitated with anti-Notch1 (sc-6014) or anti-c-Myc (9E10) with protein G-Sepharose (Pharmacia). Proteins separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis were electrophoretically transferred onto polyvinylidene difluoride membranes (Bio-Rad). Primary antibodies used for blotting were anti-HA (12CA5), anti-Notch1 (sc-6014), anti-RBP-J (T6719) (27), anti-c-Myc (9E10), anti-DNotch (9C6), and anti-DMam (3). They were visualized with appropriate secondary antibodies conjugated with horseradish peroxidase and SuperSignal chemiluminescent substrate (Pierce). In some cases, the membranes were stripped and reblotted.
RESULTS
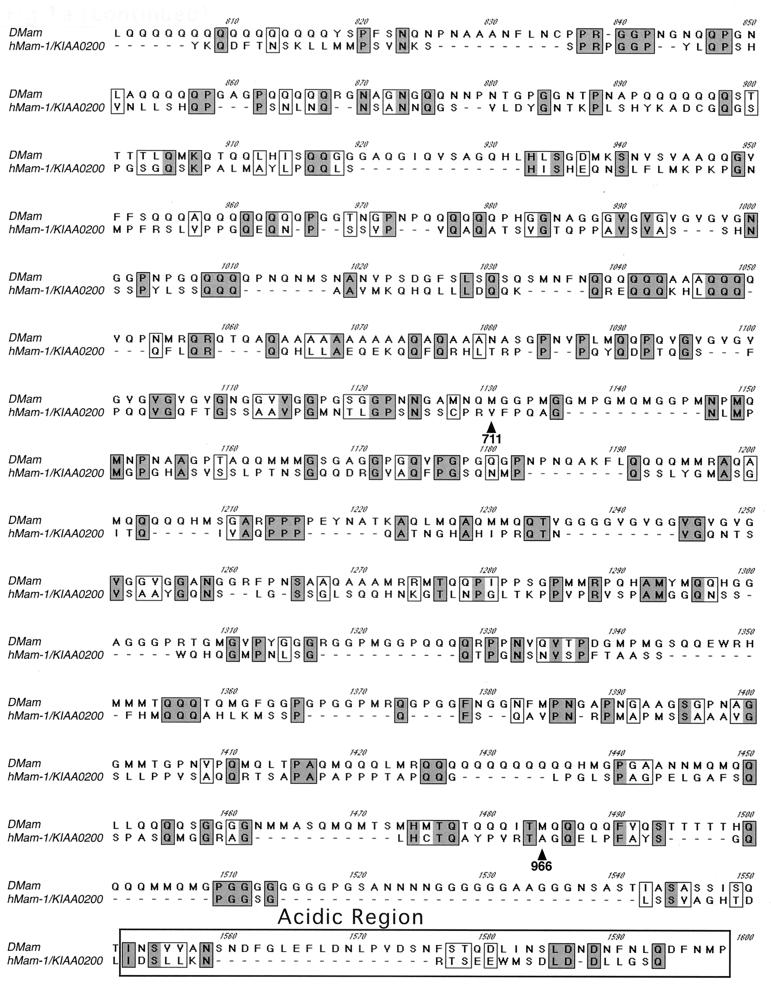
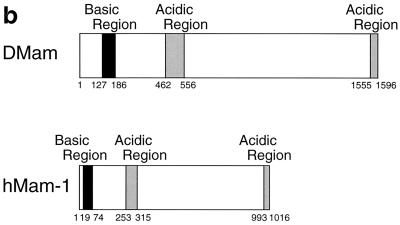
In order to investigate potential roles for Mam in Notch signaling, we searched sequence databases for related proteins. A human cDNA sequence that shows significant similarity to DMam (30) was identified (Fig. 1a). This cDNA, KIAA0200, was isolated during a project to clone long cDNAs (19). No function for this sequence has been reported. The arrangements of basic and acidic amino acid clusters in the protein and DMam are well conserved, implying that their higher-order structures are related (Fig. 1b). Based on our observations reported below, we have tentatively named KIAA0200 hMam-1.
FIG. 1.
Primary structure of Mam proteins. (a) Primary sequences of DMam (GenBank accession number X54251) and hMam-1 (D83785). Sequences were aligned using the ClustalW algorithm. Identical amino acids are in shaded boxes. Similar amino acids are in open boxes. The basic and acidic regions are in labeled boxes. The residues converted in the deletion mutants of hMam-1 are indicated by arrowheads. The overall identity of hMam-1 and DMam is 22%. Identity over the most highly conserved basic region is 38%. (b) Arrangement of basic and acidic regions of the DMam and hMam-1 proteins.
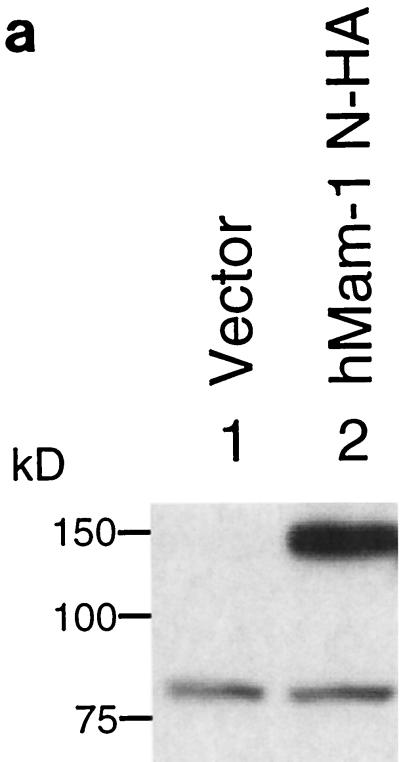
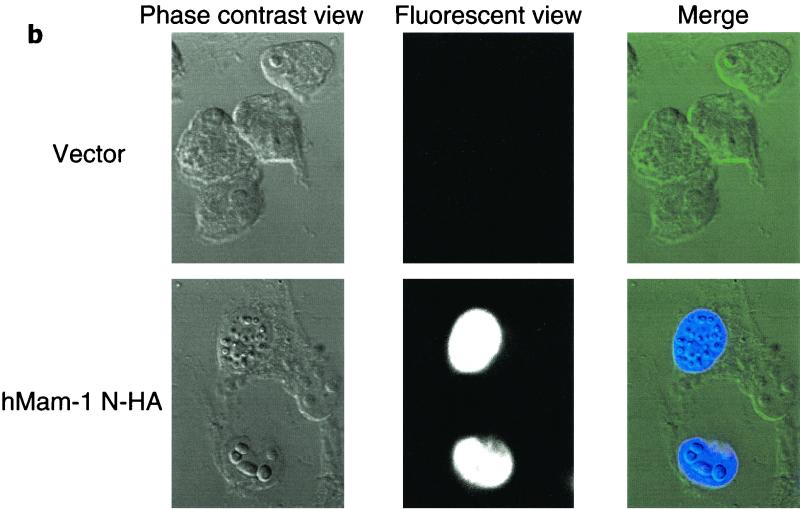
To identify the product of a full-length cDNA of hMam-1, we attached an HA tag to the portion of the coding region corresponding to the N terminus and cloned it into a mammalian expression vector. When this vector was transfected to 293T cells, a single protein with an apparent molecular mass of 140 kDa was detected by Western blotting analysis with an anti-HA antibody (Fig. 2a). Immunocytochemistry with the same antibody showed that this antigen is in the nuclei of the transfected cells (Fig. 2b), as previously reported for DMam (3, 30).
FIG. 2.
Identification and nuclear localization of the hMam-1 protein. (a) Identification of the hMam-1 protein. 293T cells were transfected with an expression vector for hMam-1 N-HA or the empty control vector (pEF-BOS). Whole-cell extracts prepared from these cells were analyzed by immunoblotting using an anti-HA antibody. (b) Nuclear localization of the hMam-1 protein. 293T cells transfected with the expression vector for hMam-1 N-HA or the empty control vector (pEF-BOS) were stained with the anti-HA antibody and a fluorescein isothiocyanate-conjugated secondary antibody. Phase-contrast, fluorescent, and merged views are shown.
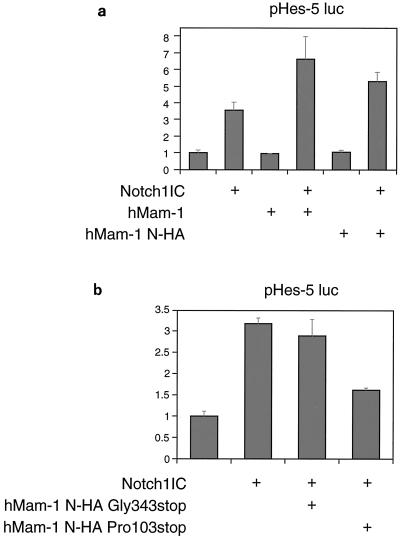
If hMam-1 is a homolog or ortholog of DMam, its expression should have effects on the mammalian Notch signaling pathway. Ligand binding to Notch on cell surfaces induces cleavage of the receptor in or near its transmembrane domain (site 3), releasing the IC domain of the receptor from the membrane (15, 28, 31). The Notch1-mediated activation of the HES-1 promoter has been shown to require this cleavage (28), and this can be mimicked experimentally by expression of the IC domain of Notch (12, 21). We examined whether hMam-1 acts synergistically with Notch1IC in this system by cotransfecting the expression vector of hMam-1 with that of the IC domain of Notch1 into NIH 3T3 cells. As shown in Fig. 3a, the HES-5 promoter was activated by the expression of Notch1IC alone but not by the expression of hMam-1 alone. Coexpression of Notch1IC and hMam-1 augmented the activation of the HES-5 promoter. More-modest effects were observed using the HES-1 promoter (data not shown).
FIG. 3.
Effects of full-length and truncated forms of hMam-1 on transactivation of the HES-5 promoter. NIH 3T3 cells were transfected with expression vectors for Notch and Mam or their empty counterparts as controls. The transfection also contains the pHES-5 luciferase reporter and an internal control for transfection. Vertical axes represent normalized luciferase activity relative to the mean activity of empty vector-transfected cells. Error bars indicate standard deviations (n = 3). (a) hMam-1 and Notch1IC synergistically activate the target promoter. (b) hMam-1 truncated at proline residue 103 depresses the activity induced by Notch1IC.
The mRNA of hMam-1 is expressed ubiquitously in human tissues (19) and many cell lines (Y. Konuma, K. Harigaya, and M. Kitagawa, unpublished data). Thus, we assumed that the murine version of the Mam-1 protein is expressed in NIH 3T3 cells and contributes to the activation of the HES promoters observed in the absence of the transfection of hMam-1. In Drosophila, it has been shown that overexpression of C-terminally truncated versions of DMam disrupts Notch pathway function (8). We made similar truncations of hMam-1 and examined the effects of their expression on the transactivation of the HES promoters in the transactivation assay system. Coexpression of the shortest truncated form of hMam-1 reduced the Notch1IC-induced activation of HES-5 (Fig. 3b) and HES-1 (data not shown). Additionally, expression of Notch2IC in combination with full-length and truncated hMam-1 elicited similar effects on the activation of HES promoters (data not shown). These data suggest that hMam-1 may act like DMam does in the insect system, as a positive regulator of Notch signaling and HES promoter activity. They suggest further that hMam-1 may act in concert with the IC domain of Notch.
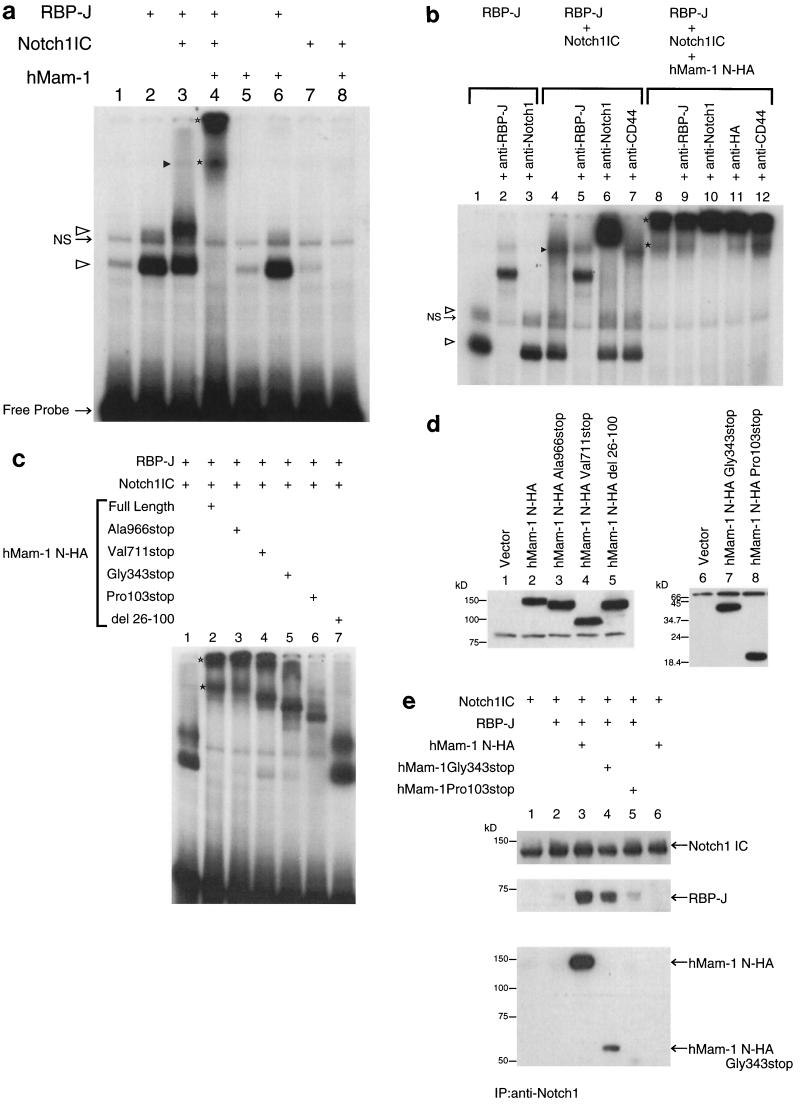
The Notch IC domain has been shown to translocate to the nucleus and associate with the sequence-specific DNA binding protein CSL (9, 12, 29). Therefore, we tested for physical interaction of the hMam-1 protein with this complex. For this purpose, we employed an EMSA using an oligonucleotide probe derived from the HES-1 promoter. This sequence is essential for activation by the Notch1IC domain (12, 21). As shown in Fig. 4a, lane 2, an extract from cells transfected with a major form of mammalian CSL proteins (RBP-J, also known as CBF-1) (7) exhibited two specific bands. Cotransfection of Notch1IC with RBP-J induced another complex that migrates more slowly than the bands mentioned above (Fig. 4a, lane 3). Interestingly, coexpression of hMam-1 (either tagged or not) with these two proteins abolished all the complexes and induced two novel bands that migrate more slowly (Fig. 4a, lane 4, and b, lane 8). The complexes from the extract of RBP-J-transfected cells were supershifted by incubation with an antibody against the RBP-J protein (Fig. 4b, lanes 1 to 3), verifying involvement of RBP-J (34). In the extract cotransfected with RBP-J and Notch, two faster-migrating bands that show apparent similarity in their mobility to the RBP-J complex could be supershifted by the anti-RBP-J antibody but not by the anti-Notch1 antibody (Fig. 4b, lanes 4 to 7). The more slowly migrating complex could be supershifted by the anti-Notch1 antibody and diminished or cleared by the anti-RBP-J antibody, indicating its identity as the Notch1IC–RBP-J complex. The amount of the complex supershifted by the anti-Notch1 antibody (Fig. 4b, lane 6) is relatively large compared to the Notch1IC–RBP-J band. This phenomenon is reproducible and might be due to stabilization of the complex by binding to the antibody. Cotransfection of Notch1IC with RBP-J also makes the slower-migrating form of the RBP-J-specific complexes stronger (Fig. 4a, lane 3, and b, lane 4). This might be due to the induction of modification of RBP-J by the presence of Notch1IC. In the extract transfected with RBP-J, Notch1IC, and the tagged hMam-1, the anti-Notch1 antibody shifted the faster band (Fig. 4b, lane 10). The appearance of this supershift by the anti-Notch1 antibody is very different from that of the Notch1IC–RBP-J band by the same antibody (compare lanes 6 and 10 of Fig. 4b), indicating that the faster band that lies just above the Notch1IC–RBP-J band is indeed distinct from it. The anti-HA antibody did produce a minor shift (Fig. 4b, lane 11); however, there is no direct evidence of the involvement of RBP-J in these complexes, as available antibody against RBP-J does not react with them (Fig. 4b, lane 9). Stereospecific inhibition might inhibit access of the antibodies to the antigen in this particular context. We did observe that transfection of hMam-1 and Notch1IC without RBP-J greatly reduced the quantities of these complexes (Fig. 4a, lane 8) and that RBP-J is coimmunoprecipitated with hMam-1 and Notch1IC (Fig. 4e; also see below), which is strong evidence that RBP-J is present. Expression of Notch2IC with hMam-1 and RBP-J induced slow-migrating complexes that look similar to those found in the case of Notch1IC expression (data not shown). Expression of hMam-1 without Notch1IC does not significantly alter the DNA binding activity of RBP-J or endogenous RBP-J-like factor (Fig. 4a, lanes 1, 2, 5, and 6).
FIG. 4.
Formation of the Mam-Notch-CSL complex on its target promoter elements. (a) Extracts of transfected 293T cells exhibit activities that bind to the RBP-J element of the HES-1 promoter. Cells were transfected with the expression vectors for the indicated proteins. NS designates a nonspecific complex, open arrowheads mark RBP-J-specific bands, the closed arrowhead marks the RBP-J–Notch1IC-specific band, and stars mark hMam-1–RBP-J–Notch1IC-specific bands. The closed star marks the hMam-1–RBP-J–Notch1IC band that could be supershifted by the anti-Notch1 and anti-HA antibodies (see panel b). (b) Effects of antibodies on binding activities. Complexes formed in the presence of RBP-J, RBP-J–Notch1IC, or hMam-1–RBP-J–Notch1IC were tested for supershifts after incubation with the indicated antibodies. Anti-CD44 was included as a control. NS, arrowheads, and stars are as described for panel a. (c) hMam-1 and its truncations containing the basic charge cluster can associate with RBP-J and Notch1IC within ternary complexes. The hMam-1 truncations, characterized below, were tested for the ability to form the ternary complex that is observed with full-length hMam-1. 293T cells were transfected with the indicated combinations of the expression vectors, and their extracts were analyzed for binding to the RBP-J element. The binding complexes migrate at a rate proportional to the length of the truncated form of hMam-1. Stars are as described for panel a. (d) Expression of the truncated forms of the hMam-1 protein. Extracts of 293T cells transfected with the vectors for indicated proteins were blotted and stained with anti-HA antibody. (e) Expression of hMam-1 enhances the physical association of RBP-J with Notch1IC. 293T cell extracts transfected with indicated vectors were immunoprecipitated by anti-Notch1 antibody. The precipitates were separated on an SDS gel, blotted onto a membrane, and then stained sequentially with anti-Notch1, anti-RBP-J, and anti-HA antibodies. The hMam-1 N-HA Pro103stop protein was too small to be resolved on this gel.
To map the domain(s) of hMam-1 required for these physical associations, we examined the effects of C-terminal truncations of hMam-1 on the DNA binding complexes involving RBP-J and Notch1IC. The truncations include those exhibiting dominant negative effects on the transcription activation assay. As shown in lanes 3 to 6 of Fig. 4c, all the truncations up to amino acid 103 greatly diminished the complexes involving RBP-J only. Furthermore, each of these truncations induced more slowly migrating complexes whose mobility correlates with the molecular mass of each truncation (Fig. 4d). These results support the idea that both of the slowly migrating complexes contain the hMam-1 protein. The faster-migrating complex involving shorter truncations of hMam-1 migrates more rapidly than the Notch1IC–RBP-J complex (Fig. 4a and b). This could be due to a change in complex stoichiometry or composition. Furthermore, approximately 100 amino acids from the N terminus are sufficient to alter the mobility of the DNA binding complexes, and a deletion construct lacking amino acids 26 to 100 exhibits virtually no activity on the complexes (Fig. 4c, lane 7). Thus, the N-terminal region of hMam-1 is necessary and sufficient to mediate the physical association.
Coimmunoprecipitation assays revealed that hMam-1 associates with Notch1IC and RBP-J even in the absence of the binding site of DNA (Fig. 4e, lane 3). This assay also revealed that hMam-1 associates with Notch1IC only in the presence of RBP-J (Fig. 4e, lanes 3 and 6). In EMSA, expression of hMam-1 without Notch1IC does not significantly alter the DNA binding activity of RBP-J (Fig. 4a, lanes 2 and 6), indicating that hMam-1 associates with RBP-J only in the presence of Notch1IC. These results suggest that hMam-1 associates with the complex of the two proteins but not with the single protein species. The coimmunoprecipitation assays further reveal that expression of hMam-1 enhances the physical association of Notch1IC and RBP-J (12, 28) (Fig. 4e, lanes 2 and 3). The two shortest truncations could also be coimmunoprecipitated with Notch1 with RBP-J, enhancing the association of these two proteins in varying degrees (Fig. 4e, lanes 4 and 5). More carboxyl portions of the hMam-1 protein presumably contain a domain(s) necessary for transcriptional activation, as overexpression of the N-terminal region hampers the transactivation induced by Notch signaling (Fig. 3b).
There is substantial genetic evidence implicating DMam as a positive effector in Notch signaling (1, 8). In contrast, no genetic information is yet available for hMam-1, and additionally, hMam-1 has diverged significantly from the DMam sequence outside the charged amino acid clusters (Fig. 1a). Therefore, additional evidence that linked the functions of these two proteins was sought. We investigated whether DMam can form a complex with DNotch and Su(H) (Drosophila CSL) proteins in Drosophila cells.
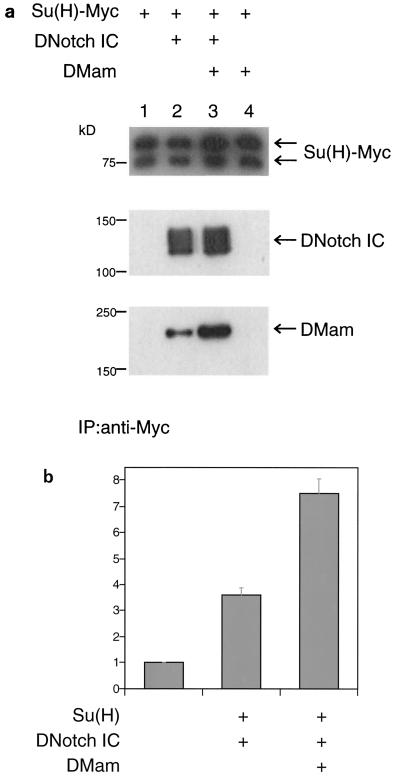
Drosophila S2 cells endogenously express DMam. S2 cells were cotransfected with either Myc epitope-tagged Su(H) [Su(H)-Myc] and DNotch1IC or Su(H)-Myc, DNotchIC, and DMam. Coimmunoprecipitations showed that endogenous DMam (Fig. 5a, lane 2) or transfected DMam (Fig. 5a, lane 3) exists in a complex with Su(H) and DNotchIC. These data also revealed that DNotchIC is required for DMam's association with the complex (Fig. 5a, lanes 1 and 4). As reported earlier (5), cotransfection of a Notch IC domain and Su(H) activates expression of an E(spl) reporter (Fig. 5b). Consistent with the hMam-1 data, we observed that cotransfection of DMam augments the activation levels of the E(spl) reporter (Fig. 5b).
FIG. 5.
DMam function in cell culture. (a) Physical association of DMam with DNotchIC and Su(H). S2 cell extracts were immunoprecipitated with an anti-c-Myc antibody. The precipitates were resolved on an SDS gel, transferred to a membrane, and stained sequentially with anti-c-Myc, anti-Notch, and anti-DMam antibodies. Lane 2 shows that DMam is expressed endogenously in S2 cells. (b) DMam augments the activation of an E(spl) mγ reporter. The vertical axis represents normalized relative luciferase activity. The cotransfection of DMam with DNotch ICN (DNotchIC) and Su(H) leads to a doubling of reporter activity. Five experiments were performed, and all data was obtained in triplicate. Error bars indicate standard errors of the means.
DISCUSSION
We believe that hMam-1 is the human version of the Notch pathway component Mam for the following reasons. The expression of full-length hMam-1 augments Notch pathway-mediated activation of HES targets, while the expression of truncated forms depresses HES activation. hMam-1 contributes to the formation of a ternary complex along with RBP-J and the IC domain of Notch, and this complex can associate with an HES promoter sequence. The hMam-1 basic charge cluster appears to mediate the association with the ternary complex. Finally, DMam forms a ternary complex with the IC domain of DNotch and Su(H) that appears to function similarly to the mammalian complex. Association with this ternary complex appears to be a conserved feature of Mam proteins.
The results presented here suggest that Mam is an element involved with orchestrating the formation of transactivating complexes on the promoters of the target genes. In the absence of Notch receptor activation, the CSL protein associates with histone deacetylases (10, 13) and binds the promoter, effecting transcription repression (9). Under these conditions, Mam may not be associated with the complex. After Notch signaling is evoked, Mam can interact with the nuclear forms of Notch and CSL, thereby contributing to an activation complex. This complex likely recruits histone acetylases (14).
It is difficult to detect NotchIC in vivo or in response to ligand stimulation in vitro. It is thought that this is because the ligand-induced release of NotchIC from the cell membrane occurs in a very small fraction of Notch receptors (15, 28, 31). Our results may provide support for this nuclear Notch model.
Recent reports by other groups (25, 37) have suggested that Mam proteins form a ternary complex with CSL and NotchIC to activate transcription. The results presented here are consistent with this hypothesis and further show that full-length species of Mam, CSL, and NotchIC can form DNA binding complexes on physiological promoter elements from both humans and Drosophila. We also presented evidence that Mam proteins can stabilize the DNA binding complex made by NotchIC and CSL.
Recently a new component of Notch signaling, LAG-3, has been identified in Caenorhabditis elegans (24). LAG-3 has properties similar to Mam in that it forms a complex with the IC domain of Notch (GLP-1 and LIN-12 in the species) and CSL (LAG-1) (24). LAG-3 has no significant sequence similarity with Mam besides polyglutamine stretches, and there is no Mam-like sequence in the C. elegans genome (26). It appears that C. elegans has evolved a distinct protein which performs a role similar to that of Mam.
ACKNOWLEDGMENTS
We thank T. Nagase for KIAA0200 cDNA; S. Artavanis-Tsakonas and R. Mann for Notch1 and Notch2 cDNA; T. Kitamura for pMx; R. Kageyama for pHES-1luc and pHES-5luc; T. Honjo for the cDNA and antibodies for RBP-J; M. Tomizawa and M. Higashi for computer software; K. Azuma for antibodies; T. Umemiya for immunocytochemistry; N. Yumoto for the expression vector for Notch1IC; M. Ito, K. Nakao-Sawai, and H. Okano for DNotchIC and S2 cells; L. Grimm for pMTNotch (ICN), pMTSu(H), and mγ Luc; M. Masada, N. Terada, and S. Masuda for support; T. Hiwasa for advice; and colleagues of the Department of Pathology, Chiba University School of Medicine, for discussions and encouragement. cDNA for RBP-J was supplied by from RIKEN DNA bank.
This work was supported by grants in aid from the Ministry of Education, Science, Sports, and Culture of Japan to M.K. (grant no. 11680671) and K.H. (grant no. 12215018); a grant from the Smoking Research Foundation to K.H.; grants from the Naito Foundation and the Sumitomo Foundation to K.M.; and a grant from the National Science Foundation (grant no. IBN 9904411) to B.Y.
REFERENCES
- 1.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 3.Bettler D, Pearson S, Yedvobnick B. The nuclear protein encoded by the Drosophila neurogenic gene mastermind is widely expressed and associates with specific chromosomal regions. Genetics. 1996;143:859–875. doi: 10.1093/genetics/143.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capobianco A J, Zagouras P, Blaumueller C M, Artavanis-Tsakonas S, Bishop J M. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eastman D S, Slee R, Skoufos E, Bangalore L, Bray S, Delidakis C. Synergy between Suppressor of Hairless and Notch in regulation of Enhancer of split mγ and mδ expression. Mol Cell Biol. 1997;17:5620–5628. doi: 10.1128/mcb.17.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fortini M E, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 7.Furukawa T, Maruyama S, Kawaichi M, Honjo T. The Drosophila homolog of the immunoglobulin recombination signal-binding protein regulates peripheral nervous system development. Cell. 1992;69:1191–1197. doi: 10.1016/0092-8674(92)90640-x. [DOI] [PubMed] [Google Scholar]
- 8.Helms W, Lee H, Ammerman M, Parks A L, Muskavitch M A, Yedvobnick B. Engineered truncations in the Drosophila mastermind protein disrupt Notch pathway function. Dev Biol. 1999;215:358–374. doi: 10.1006/dbio.1999.9477. [DOI] [PubMed] [Google Scholar]
- 9.Hsieh J J-D, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–995. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh J J-D, Zhou S, Chen L, Young D B, Hayward S D. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huppert S S, Le A, Schroeter E H, Mumm J S, Saxena M T, Milner L A, Kopan R. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature. 2000;405:966–970. doi: 10.1038/35016111. [DOI] [PubMed] [Google Scholar]
- 12.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 13.Kao H Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurooka H, Honjo T. Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem. 2000;275:17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 15.Lecourtois M, Schweisguth F. Indirect evidence for Delta-dependent intracellular processing of notch in Drosophila embryos. Curr Biol. 1998;8:771–774. doi: 10.1016/s0960-9822(98)70300-8. [DOI] [PubMed] [Google Scholar]
- 16.Lehman R F, Jimenez W, Dietrich U, Campos-Ortega J A. On the phenotype and development of mutants of early neurogenesis in D. melanogaster. Wilhelm Roux's Arch Dev Biol. 1983;192:62–74. doi: 10.1007/BF00848482. [DOI] [PubMed] [Google Scholar]
- 17.Matsunami N, Hamaguchi Y, Yamamoto Y, Kuze K, Kangawa K, Matsuo H, Kawaichi M, Honjo T. A protein binding to the J kappa recombination sequence of immunoglobulin genes contains a sequence related to the integrase motif. Nature. 1989;342:934–937. doi: 10.1038/342934a0. [DOI] [PubMed] [Google Scholar]
- 18.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagase T, Seki N, Ishikawa K, Tanaka A, Nomura N. Prediction of the coding sequences of unidentified human genes. V. The coding sequences of 40 new genes (KIAA0161–KIAA0200) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1996;3:17–24. doi: 10.1093/dnares/3.1.17. [DOI] [PubMed] [Google Scholar]
- 20.Newfeld S J, Smoller D A, Yedvobnick B. Interspecific comparison of the unusually repetitive Drosophila locus mastermind. J Mol Evol. 1991;32:415–420. doi: 10.1007/BF02101281. [DOI] [PubMed] [Google Scholar]
- 21.Nishimura M, Isaka F, Ishibashi M, Tomita K, Tsuda H, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter of mouse Hes2 gene, a homologue of Drosophila hairy and enhancer of split. Genomics. 1998;49:69–75. doi: 10.1006/geno.1998.5213. [DOI] [PubMed] [Google Scholar]
- 22.Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier L L, Gorman D M, Nolan G P, Miyajima A, Kitamura T. Applications of retrovirus-mediated expression cloning. Exp Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- 24.Petcherski A G, Kimble J. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature. 2000;405:364–368. doi: 10.1038/35012645. [DOI] [PubMed] [Google Scholar]
- 25.Petcherski A G, Kimble J. Mastermind is a putative activator for Notch. Curr Biol. 2000;10:R471–R473. doi: 10.1016/s0960-9822(00)00577-7. [DOI] [PubMed] [Google Scholar]
- 26.Ruvkun G, Hobert O. The taxonomy of developmental control in Caenorhabditis elegans. Science. 1998;282:2033–2041. doi: 10.1126/science.282.5396.2033. [DOI] [PubMed] [Google Scholar]
- 27.Sakai T, Furukawa T, Iwanari H, Oka C, Nakano T, Kawaichi M, Honjo T. Loss of immunostaining of the RBP-J kappa transcription factor upon F9 cell differentiation induced by retinoic acid. J Biochem (Tokyo) 1995;118:621–628. doi: 10.1093/oxfordjournals.jbchem.a124955. [DOI] [PubMed] [Google Scholar]
- 28.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 29.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 30.Smoller D, Friedel C, Schmid A, Bettler D, Lam L, Yedvobnick B. The Drosophila neurogenic locus mastermind encodes a nuclear protein unusually rich in amino acid homopolymers. Genes Dev. 1990;4:1688–1700. doi: 10.1101/gad.4.10.1688. [DOI] [PubMed] [Google Scholar]
- 31.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 32.Su W C, Kitagawa M, Xue N, Xie B, Garofalo S, Cho J, Deng C, Horton W A, Fu X Y. Activation of Stat1 by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature. 1997;386:288–292. doi: 10.1038/386288a0. [DOI] [PubMed] [Google Scholar]
- 33.Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- 34.Taniguchi Y, Furukawa T, Tun T, Han H, Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol Cell Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinmaster G. Notch signal transduction: a real rip and more. Curr Opin Genet Dev. 2000;10:363–369. doi: 10.1016/s0959-437x(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 36.Wharton K A, Johansen K M, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- 37.Wu L, Aster J C, Blacklow S C, Lake R, Artavanis-Tsakonas S, Griffin J D. MAML1, a human homologue of drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]