Abstract
Starvation for amino acids induces Gcn4p, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. In an effort to identify all genes regulated by Gcn4p during amino acid starvation, we performed cDNA microarray analysis. Data from 21 pairs of hybridization experiments using two different strains derived from S288c revealed that more than 1,000 genes were induced, and a similar number were repressed, by a factor of 2 or more in response to histidine starvation imposed by 3-aminotriazole (3AT). Profiling of a gcn4Δ strain and a constitutively induced mutant showed that Gcn4p is required for the full induction by 3AT of at least 539 genes, termed Gcn4p targets. Genes in every amino acid biosynthetic pathway except cysteine and genes encoding amino acid precursors, vitamin biosynthetic enzymes, peroxisomal components, mitochondrial carrier proteins, and autophagy proteins were all identified as Gcn4p targets. Unexpectedly, genes involved in amino acid biosynthesis represent only a quarter of the Gcn4p target genes. Gcn4p also activates genes involved in glycogen homeostasis, and mutant analysis showed that Gcn4p suppresses glycogen levels in amino acid-starved cells. Numerous genes encoding protein kinases and transcription factors were identified as targets, suggesting that Gcn4p is a master regulator of gene expression. Interestingly, expression profiles for 3AT and the alkylating agent methyl methanesulfonate (MMS) overlapped extensively, and MMS induced GCN4 translation. Thus, the broad transcriptional response evoked by Gcn4p is produced by diverse stress conditions. Finally, profiling of a gcn4Δ mutant uncovered an alternative induction pathway operating at many Gcn4p target genes in histidine-starved cells.
In response to environmental perturbations, Saccharomyces cerevisiae cells elicit rapid transcriptional reprogramming involving both activation and repression of gene expression. Transcriptional activator proteins function by binding to specific promoter elements, called upstream activating sequences (UASs) in yeast cells, and recruiting the transcriptional machinery. Thus, transcriptional stimulation requires the expression and function of an activator and the appropriate UAS element in the promoters of its target genes. A plethora of mechanisms are known to regulate the activity or expression of transcriptional activators in response to specific signals. For example, in cells grown on glucose, Gal80p inhibits the ability of Gal4p to activate transcription of genes encoding galactose-metabolizing enzymes, whereas Gal3p alleviates this inhibition on galactose medium (83). The transcriptional activators Pho4p, Swi5p, and Yap1p are regulated by the coupling of their nuclear localization to the levels of inorganic phosphate, cell cycle and mother-daughter status, or oxidative stress, respectively (reviewed in reference 52). Starvation for amino acids, purines, and glucose limitation induces the synthesis of Gcn4p, a bZIP transcriptional activator of amino acid biosynthetic genes in multiple pathways (35, 88, 108). This cross-pathway response is known as general amino acid control. Gcn2p, a translation initiation factor 2α (eIF2α) kinase, mediates the derepression of GCN4 mRNA translation in nutrient-starved cells. The activity of Gcn2p is induced by high levels of uncharged tRNA, implying that uncharged tRNA is a critical upstream signal for derepression of GCN4 translation under starvation conditions (37).
Previous studies showed that transcription of at least 35 genes encoding amino acid biosynthetic enzymes is induced by Gcn4p (36). Given that starvation for diverse nutrients induces GCN4 translation, it seemed plausible that Gcn4p would have many other targets besides amino acid biosynthetic genes. Indeed, computational searches of the yeast genome revealed that the Gcn4p binding site, or UASGCRE, is present at the promoters of numerous genes not directly connected with amino acid biosynthesis (data not shown). Additionally, it was shown previously that several adenine biosynthetic genes (51, 72, 88), ATR1 (11), and LPD1 (109) are induced by Gcn4p in amino acid-starved cells. These observations led us to investigate whether the transcriptional activation function of Gcn4p greatly transcends the amino acid biosynthetic genes.
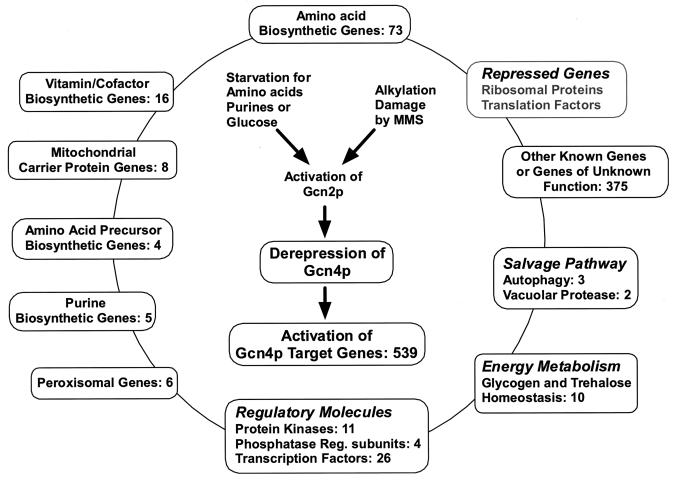
We used whole-genome expression profiling (62, 93) to identify the complete set of genes regulated by Gcn4p. Our results showed that more than 1,000 genes were induced in wild-type (WT) cells in response to starvation for histidine by treatment with 3-aminotriazole (3AT), a competitive inhibitor of His3p. To evaluate the contribution of Gcn4p to this massive regulatory response, we compared the expression profiles of isogenic WT and gcn4Δ strains treated with 3AT. We also compared the expression profiles under nonstarvation conditions of a WT strain and a GCN4c mutant that expresses high levels of Gcn4p constitutively (GCN4c/GCN4 experiment). These comparative profiling experiments indicated that at least 60% of the genes induced by 3AT are dependent on Gcn4p for high-level activation. As expected, these Gcn4p-dependent genes, called Gcn4p targets and numbering 539, encompass a larger number of amino acid biosynthetic genes than previously identified. They also include genes involved in cofactor biosyntheses, organelle biogenesis, mitochondrial transport, autophagy, and glycogen homeostasis. Furthermore, Gcn4p induced genes encoding 11 protein kinases and 26 transcription factors. Interestingly, treatment with the alkylating agent methyl methanesulfonate (MMS) induced GCN4 translation and a large proportion of Gcn4p target genes. Thus, it appears that Gcn4p is a master regulator of gene expression that evokes a broad range of transcriptional and signaling responses under conditions of nutrient limitation and other forms of cell stress.
MATERIALS AND METHODS
Supplementary data.
The complete data set for all of the experiments analyzed in Fig. 1 is available at http://www.rii.com/tech/pubs/mcb2001.htm. Copies of figures, including Fig. 1 in alternate colors, can be obtained at the above website.
FIG. 1.
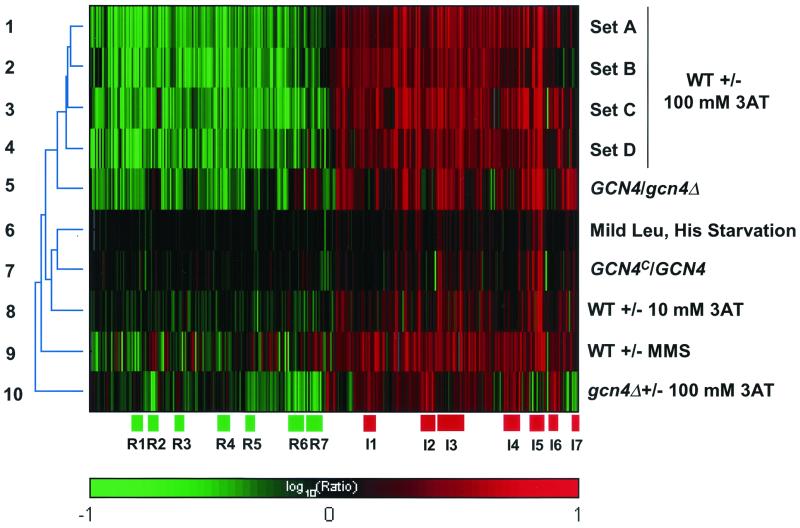
Hierarchical two-dimensional clustering analysis of the results from 10 microarray experiments involving 60 hybridizations. The color scale at the bottom represents the log10 ratio of hybridization signals obtained with the experimental samples to signals from control RNA samples, ranging from −1.0 (brightest green, 10-fold repressed) to 1.0 (brightest red, 10-fold induced). The log10 ratios for 2,528 genes with P values of ≤0.05 and at least twofold changes in expression were plotted along the x axis for the different experiments listed along the y axis. Details of experiments 1 to 3, 5, 7, and 10 are given in Table 2. Experiment 4 was conducted independently of experiments 1 and 2 with strain R491, and the data shown are the average expression ratios from one pair of hybridizations. The data from experiments 8 (WT ± 10 mM 3AT) (66) and 9 (WT with or without MMS) (48) were described previously. The data for experiment 6 were obtained by comparing the expression profiles of auxotrophic strain R491 grown with limiting amino acids and the same strain grown with abundant amino acids. The dendrogram on the left depicts the relatedness of expression profiles for the different experiments. Blocks R1, R3, and R4 shown below row 10 depict clusters of genes that were repressed by 3AT in WT but not in a gcn4Δ mutant and showed strong Gcn4p dependence in the GCN4/gcn4Δ experiment. Gene clusters R2, R5, R6, and R7 were repressed by 3AT in both WT and gcn4Δ strains and showed no Gcn4p dependence in the GCN4/gcn4Δ experiment. Clusters I1, I2, I4, and I6 depict genes that were induced in both WT and gcn4Δ strains and showed no Gcn4p dependence in the GCN4/gcn4Δ experiment. Clusters I3 and I5 represent canonical Gcn4p target genes that were induced by 3AT in WT but showed little or no induction in the gcn4Δ strain and displayed Gcn4p dependence in the GCN4/gcn4Δ and GCN4c/GCN4 experiments. Cluster I7 was repressed in the gcn4Δ strain and showed little or no induction in WT treated with 100 mM 3AT but was induced by 10 mM 3AT and displayed strong Gcn4p dependence in the GCN4/gcn4Δ experiment; hence, it was judged to contain Gcn4p target genes. A different version of this figure using alternative colors can be obtained at http://www.rii.com/tech/pubs/mcb2001.htm.
Strains and media.
All strains were derived from S288c and are listed in Table 1. Strain R176 was transformed with p238 (74) containing the constitutive GCN4 allele or vector YCp50 to construct strains R4760 and R6257, respectively. For all microarray experiments, cells were cultured in the synthetic complete (SC) medium whose composition is as follows: 1.6 g of yeast nitrogen base without ammonium sulfate and amino acids, 5 g of ammonium sulfate, 11 g of succinic acid, 6.9 g of sodium hydroxide, 1.4 g of a mixture of amino acids (called “C” powder; described below), and 20 g of dextrose per liter. The pH of the medium was adjusted to 5.8 with sodium hydroxide. C powder contains 1 g each of adenine, histidine, methionine, uracil, and arginine; 2.5 g of phenylalanine; 3 g each of lysine and tyrosine; 4 g each of tryptophan, leucine, and isoleucine; 5 g each of glutamic acid and aspartic acid; 7.5 g of valine; 10 g of threonine; and 20 g of serine. Complete (YPD) and minimal (SD) media were described previously (96).
TABLE 1.
Strains used in this study
| Name | Genotype |
|---|---|
| R491 | MATa/α trp1-63/trp1-63 ura3Δ0/ura3Δ0 |
| R176 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 met15Δ0/MET15 lys2Δ0/LYS2 |
| R6257 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 met15Δ0/MET15 lys2Δ0/LYS2 [YCp50 (CEN4 URA3)] |
| R4760 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ met15Δ0/MET15 lys2Δ0/LYS2 [p238 (GCN4-ΔuORF CEN4 URA3)] |
| KNY124 | MATα leu2-3,112 ura3-52 ino1 trp1::hisG gcn4Δ-103 |
| KNY164 | MATα leu2-3,112 ura3-52 ino1 trp1::hisG |
| KNY201 | MATα leu2 ura3-52 trp1 prb1-1122 pep4-3 gal2 |
| KNY202 | MATα leu2 ura3-52 trp1 prb1-1122 pep4-3 gal2 gcn4-103 |
| H187 | MATaura3-52 ino1 <GCN4-lacZ URA3> |
| H1895 | MATaleu2-3,112 ura3-52 trp1-63 gcn2Δ <GCN4-lacZ TRP1> |
| F113 | MATaura3-52 ino1 CAN1R |
| H2079 | MATaura3-52 ino1 CAN1Rgcn1Δ |
| H2512 | MATaura3-52 ino1 CAN1Rgcn20Δ::hisG |
Growth conditions.
Treatment of cells with 100 mM 3AT for 1 h was conducted as described previously (66). For the GCN4c/GCN4 experiment, strains R4760 and R6257 were grown in SC medium lacking uracil to a density of 107 cells/ml, harvested, and lysed as described previously for 3AT treatment (66). To impose a mild amino acid starvation, strain R176, auxotrophic for leucine and histidine, was grown overnight in SC medium to an optical density at 600 nm (OD600) of 0.5 and used to inoculate SC medium containing 0.65 g of C powder (0.5× amino acids) or 1.4 g of C powder (1× amino acids) per liter. Cells were grown to a density of 107 cells/ml, harvested, and lysed as described previously (66).
Generation and analysis of microarray data.
Poly(A)+ RNA preparation, cDNA labeling, cDNA microarray production, hybridization, washing, scanning, and image analysis were conducted as described previously (43, 66). Each individual microarray data set was generated by a fluor-reversed set of hybridizations as described previously (66), herein referred to as pairs of hybridizations. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer was performed as described previously (42). Use of an error model to determine P values for the data was described previously (43). The MMS data from the work of Jelinsky and Samson (48) were imported into the Rosetta Resolver system, and data were analyzed using the methods described in reference 44 for handling single-channel data.
GCN4-lacZ induction by MMS.
Strain H187 was grown in duplicate in YPD medium to an OD600 of 0.9, MMS (Aldrich) was added to one culture to 0.07% (vol/vol), and both cultures were incubated with shaking at 30°C for 1 h. The MMS treatment reduced cell viability to 31% of that of untreated cultures, as judged by colony formation on YPD medium. Cells were collected by centrifugation and washed with sterile water, and the β-galactosidase activity was assayed in cell extracts as described previously (71). Strain H1895, bearing GCN4-lacZ integrated in the chromosome, was transformed to uracil prototrophy with p585 (GCN2 CEN4 ARS1), p2201 (gcn2-m2 CEN6 ARSH4), p614 (gcn2-psk CEN4 ARS1), or a vector (URA3 2μm). As expected, p585 complemented the 3AT-sensitive phenotype of H1895 while the other plasmids did not. Strains F113 (WT), H2079 (gcn1Δ), and H2512 (gcn20Δ) were transformed with p180 (GCN4-lacZ CEN4 ARS1). The transformants of H1895, F113, H2079, and H2512 were cultured in SC-Ura medium with or without MMS (0.07%) and assayed for β-galactosidase activity as described above.
RESULTS
Gcn4p stimulates transcription of a large fraction of the yeast genome.
We used cDNA microarray technology to compare the genome-wide expression profiles of a WT strain (KNY164) grown in SC medium and the same strain grown in SC medium containing 100 mM 3AT (WT ± 3AT experiment). Gcn4p is expressed at low levels on SC medium and rapidly induced in response to histidine starvation imposed by 3AT (1, 36). The results showed that, of the 3,940 genes which were measured with a statistical significance (P value) of 0.05 or less, 949 genes (24% of the total) were induced by 3AT by a factor of 2 or more (Table 2, set C, rows 1 to 4). Interestingly, almost an equal fraction (28%) of genes were repressed by 3AT in the same experiment (Table 2, set C, rows 5 to 7). Multiple experiments were carried out under identical growth conditions using a second, nonisogenic GCN4 strain (R491). Whereas 322 genes were highly induced (≥4-fold) by 3AT in KNY164, a somewhat smaller number of genes were induced to this extent in the second strain (Table 2, sets A and B versus C). This may be attributable to the greater sensitivity of the ink-jet platform used for set C compared with that of cDNA spotted arrays (see reference 42 for a comparison of the two technologies). Nevertheless, large fractions of the genome were induced or repressed by 3AT in both GCN4 strains. A hierarchical two-dimensional clustering analysis of genes showing ≥2-fold changes in gene expression and with a P value of ≤0.05 was conducted to determine whether the same group of genes was induced or repressed by 3AT in all experiments described above. As shown in Fig. 1, the vast majority of genes that were found to be induced (shown in red) or repressed (shown in green) by 3AT using data set C behaved similarly using data sets A, B, and D (compare rows 1 to 4).
TABLE 2.
Summary of expression profiles in different experiments
| Type of expression | Log10 ratio (fold) | No. of responding genes
|
|||||
|---|---|---|---|---|---|---|---|
| WT ± 3ATa
|
GCN4/gcn4Δ ratiob | GCN4/GCN4c ratioc | gcn4Δ ± 3ATd | ||||
| Set A | Set B | Set C | |||||
| Induced | ≥10 | 23 | 44 | 99 | 63 | 7 | 18 |
| ≥4 | 164 | 177 | 322 | 210 | 54 | 80 | |
| ≥2 | 735 (18%)e | 771 (16%) | 949 (24%) | 612 (20%) | 158 (26%) | 568 (18%) | |
| <2 | 2,481 | 2,991 | 2,991 | 1,836 | 452 | 2,186 | |
| Repressed | ≥10 | 9 | 37 | 52 | 14 | 3 | 5 |
| ≥4 | 225 | 336 | 347 | 101 | 8 | 72 | |
| ≥2 | 886 (22%) | 955 (20%) | 1,092 (28%) | 636 (21%) | 65 (11%) | 496 (15%) | |
| Total ORFs consideredf: | 4,102 | 4,717 | 3,940 | 3,087 | 610 | 3,250 | |
Expression in WT strain treated with 100 mM 3AT relative to expression in untreated strain. Set A, average expression ratio from 11 independent pairs of hybridizations; set B; average expression ratio from 8 independent pairs of hybridizations; set C; average expression ratio from 2 independent pairs of hybridizations. Sets A and B and GCN4/GCN4c ink-jet data sets were derived from cDNA arrays, and sets C and gcn4Δ ± 3AT data sets were derived from the use of FlexJet oligonucleotide arrays. Data sets A and B were derived from two separate RNA preparations from strain R491. Data set C was obtained from strain KNY 164. All strains are in an S288c background.
Expression ratio of a WT GCN4 strain (KNY164) to an isogenic gcn4Δ strain (KNY124), both treated with 100 mM 3AT.
Expression ratio of strain R4760 overexpressing GCN4 constitutively (GCN4c) to strain R6257 bearing vector alone (GCN4).
Expression ratio of gcn4Δ strain (KNY124) treated with 100 mM 3AT to the untreated strain.
Values in parentheses are percentages of total ORFs considered.
Total number of ORFs that are above the P value cutoff (P = 0.05).
An additional experiment was carried out using a lower concentration of 3AT (10 mM) to impose less severe histidine starvation on GCN4 strain R491. Additionally, we determined expression profiles of strain R491, auxotrophic for leucine and histidine, in medium containing limiting amounts of the amino acids (0.5×) versus medium replete with amino acids (1×). The two-dimensional clustering analysis in Fig. 1 shows that the majority of genes that were induced or repressed by the mild amino acid limitation imposed in these two experiments (rows 6 and 8) also were induced or repressed, respectively, in the multiple experiments using 100 mM 3AT (rows 1 to 4). As might be expected, the magnitude of induction or repression for most genes was diminished in the last two experiments compared to the 100 mM 3AT experiments (Fig. 1).
To evaluate whether the majority of genes induced by 3AT were dependent on Gcn4p for this response, we compared the expression profiles of GCN4 strain KNY164 and isogenic gcn4Δ strain KNY124 when both were treated with 100 mM 3AT (GCN4/gcn4Δ experiment). The results indicated that 612 genes were expressed at levels ≥2-fold higher in GCN4 than in gcn4Δ cells (Table 2, GCN4/gcn4Δ). The two-dimensional clustering analysis (Fig. 1) revealed that most genes showing higher expression in GCN4 than in gcn4Δ cells treated with 3AT (red bars) also were induced by 3AT in the GCN4 cells (compare row 5 with rows 1 to 4). The same correlation holds when comparing the genes with lower expression in GCN4 than in gcn4Δ cells (green bars) and those that were repressed by 3AT in GCN4 cells (Fig. 1).
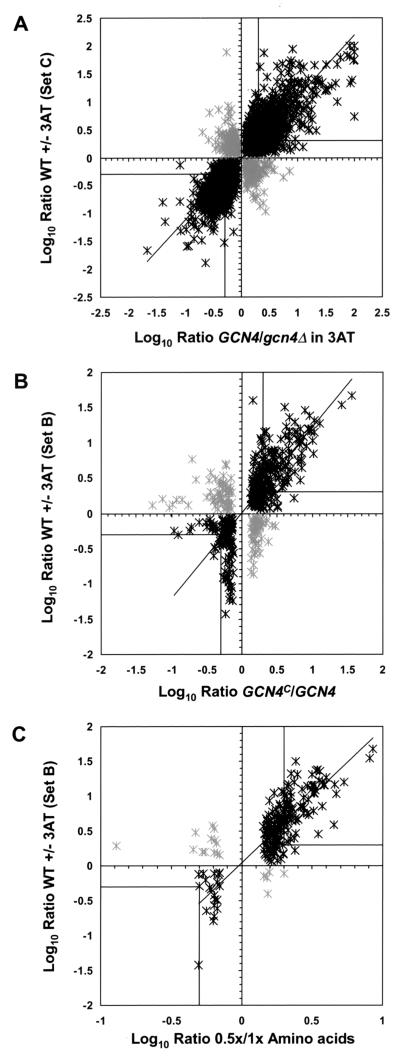
To compare the data obtained in the WT ± 3AT (set C) and GCN4/gcn4Δ experiments in greater detail, the log-ratio scatter plot shown in Fig. 2A was constructed for the 2,372 genes for which statistically significant data (P value of ≤0.05) were obtained in both experiments. Overall, the expression ratios in the two experiments were highly correlated, with a correlation coefficient of 0.81. Closer inspection of the plot revealed that 64% of the 635 genes induced ≥2-fold by 3AT in GCN4 cells also showed ≥2-fold-higher expression in GCN4 than in gcn4Δ cells. The 408 genes with this behavior are enclosed in the box in the upper right quadrant of the plot. Additionally, 64% of the 781 genes repressed by a factor of ≥2 by 3AT in GCN4 cells also showed reduced expression in GCN4 cells compared with that in gcn4Δ cells (Fig. 2A, black stars enclosed in the box in the lower left quadrant). A small fraction of genes that were induced (54 genes) or repressed (47 genes) in the WT ± 3AT experiment were negatively correlated in the GCN4/gcn4Δ experiment (gray stars in Fig. 2A). We conclude that a large fraction of genes induced by 100 mM 3AT are dependent on Gcn4p for maximal expression under these starvation conditions. Furthermore, Gcn4p contributes to the repression of most genes whose expression is reduced by severe histidine limitation.
FIG. 2.
Log10 ratio scatter plots comparing expression profiles from different experiments. The log10 ratios of expression for all genes with P values of ≤0.05 in two experiments being compared were plotted against one another. Black stars depict genes whose expression profiles were correlated, while gray stars depict genes with negatively correlated expression profiles. A trend line was generated for the genes depicted by black stars. Genes induced (log10 ratio ≥ +0.30) or repressed (log10 ratio ≤ −0.30) twofold or more are enclosed in a box in the upper right or the lower left quadrants, respectively. (A) Comparison of data set C (WT ± 100 mM 3AT) with the GCN4/gcn4Δ data set. (B) Comparison of data set B (WT ± 100 mM 3AT) with the GCN4c/GCN4 data set. (C) Comparison of data set B (WT ± 100 mM 3AT) with the 0.5×/1× amino acid data set.
We used an independent means of identifying Gcn4p-inducible genes by comparing the expression profiles in WT and GCN4c mutant cells. This mutant expresses induced levels of WT Gcn4p under nonstarvation conditions (74). The cluster analysis in Fig. 1 shows that the profile of gene induction associated with constitutive expression of Gcn4p (row 7) most closely resembles that elicited by moderate amino acid starvation (rows 6 and 8). The log-ratio scatter plot in Fig. 2B for the 346 genes represented in both the GCN4c/GCN4 and WT ± 3AT (set B) experiments shows that 68% of the genes induced ≥2-fold by 3AT in WT cells also showed ≥2-fold-higher expression in GCN4c than in GCN4 cells (black stars enclosed in the box in the upper right quadrant in Fig. 2B). This correlation provides additional evidence that the majority of genes induced by 3AT are under Gcn4p control.
Interestingly, the scatter plot in Fig. 2B showed a poor correlation between genes that were repressed by 100 mM 3AT in WT cells (stars below zero in the y axis) and those repressed by constitutive expression of Gcn4p under nonstarvation conditions (stars to the left of zero in the x axis). Expression of only 12% of the genes was reduced in both experiments (Fig. 2B, black stars enclosed in the box in the lower left quadrant). Moreover, the genes that were most highly repressed by 3AT in WT cells (stars with the most negative y coordinates) showed little or no repression in the GCN4c/GCN4 strain. Thus, high-level expression of Gcn4p under nonstarvation conditions in the GCN4c mutant was insufficient to evoke the extensive repression of genes that occurred in WT cells under severe histidine starvation conditions. Similarly, the scatter plot in Fig. 2C shows that relatively few genes were repressed by moderate Leu-His starvation. Hence, the widespread repression of genes observed in the WT ± 3AT experiments seems to require a combination of high-level Gcn4p and severe amino acid limitation.
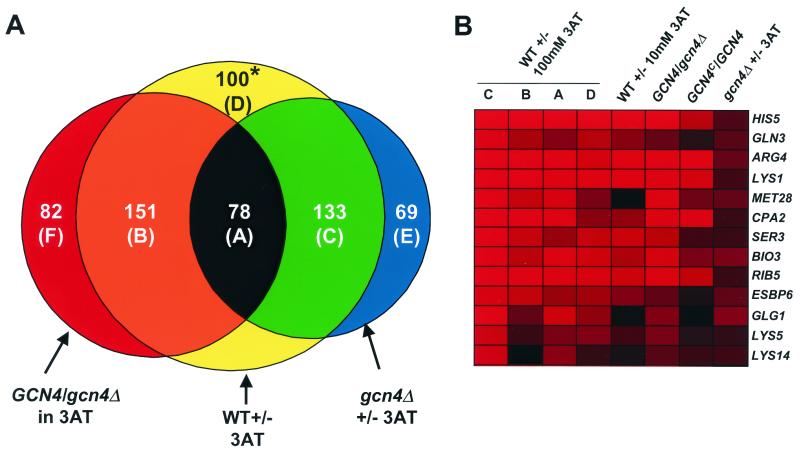
Finally, we examined a gcn4Δ mutant in the presence or absence of 100 mM 3AT to determine which genes can be induced or repressed by severe histidine starvation in the absence of Gcn4p (gcn4Δ ± 3AT experiment). The cluster analysis in Fig. 1 shows that many genes that were induced by 3AT in GCN4 cells also were induced in the gcn4Δ mutant (rows 1 to 4 versus row 10). In addition, many such genes were dependent on Gcn4p for maximal induction by 3AT in the WT, showing higher expression in GCN4 than in gcn4Δ cells (compare rows 5 and 10). The overlap between these different gene sets is depicted graphically in Fig. 3A for 613 genes that produced significant data (P ≤ 0.05) in the WT ± 100 mM 3AT (data set C), GCN4/gcn4Δ, and gcn4Δ ± 100 mM 3AT experiments and had an induction ratio of 2 or more in one of these experiments. There were 229 genes induced in both the WT ± 100 mM 3AT and the GCN4/gcn4Δ experiments, indicating a dependence on Gcn4p for maximal induction (Fig. 3A, sectors A and B). Interestingly, 78 of these genes also were induced by 3AT in the gcn4Δ mutant (sector A in Fig. 3A), including canonical Gcn4p target genes encoding amino acid biosynthetic enzymes, such as HIS5 and ARG4. As shown in Fig. 3B, the magnitude of 3AT induction of this latter class of genes was reduced in the gcn4Δ mutant. Hence, many genes displayed a strong, but incomplete, dependence on Gcn4p for induction by 100 mM 3AT.
FIG. 3.
A fraction of Gcn4p target genes is induced by 3AT in gcn4Δ cells. (A) Venn diagram depicting the overlap among 613 genes for which statistically significant data were obtained (P ≤ 0.05) and that showed an induction ratio of ≥2 in the WT ± 3AT (set C), GCN4/gcn4Δ, or gcn4Δ ± 3AT experiments. There are 229 Gcn4p target genes (sectors A and B) found at the intersection between 462 genes induced by 3AT in the WT (sectors A, B, C, and D) and 311 genes showing Gcn4p dependence for induction in the GCN4/gcn4Δ experiment (sectors F, B, and A). There were 280 genes induced by 3AT in the gcn4Δ ± 3AT experiment (sectors A, C, and E). The 151 genes in sector B are Gcn4p targets that were not induced by 3AT in the gcn4Δ strain and thus are completely dependent on Gcn4p for induction. Sector A contains the subset of Gcn4p target genes that were also induced by 3AT in the gcn4Δ mutant. These 78 genes are subject to dual regulation and require Gcn4p only for maximal induction by 3AT. Sector C is comprised of the 133 genes induced by 3AT in both gcn4Δ and WT cells which showed little or no dependence on Gcn4p in the GCN4/gcn4Δ experiment (induction ratio of <2-fold). These genes are induced by 3AT independently of Gcn4p. Thus, ∼50% of the genes induced by 3AT in the WT are designated Gcn4p targets because of their dependence on Gcn4p for maximal induction ([A + B]/[A + B + C + D]), and 34% of these target genes (A/[A + B]) can be induced by 3AT even in the absence of Gcn4p. The genes in sector F showed greater expression in WT cells than in gcn4Δ cells in the presence of 3AT but were not induced by 3AT in the WT. This class includes additional Gcn4p targets that depend on Gcn4p primarily to prevent repression in histidine-starved cells (see text). The genes in sector E were induced only in gcn4Δ cells and thus belong to the class of genes induced by 3AT independently of Gcn4p. The results obtained for the 100 genes in sector D marked with an asterisk are paradoxical in that any genes induced by 3AT in the WT should be either dependent on (sector A or B) or independent of (sector C) Gcn4p for their induction. Close examination of this group revealed that 28 genes were induced only in the single WT ± 3AT experiment used to construct this diagram (data set C) but not in other experiments of the same type (data set A, B, or D); hence, these 28 genes probably are not 3AT inducible. Fifty of the remaining genes in sector D showed induction ratios between 1.5 and 2.0 in the GCN4/gcn4Δ experiment and most likely belong to sector A or B, comprising the Gcn4p targets. Fifteen of the remaining genes had induction ratios between 1.5 and 2.0 in the gcn4Δ ± 3AT experiment and probably belong to sector C or E, comprising the Gcn4p-independent 3AT-inducible genes. Among the 133 genes assigned to sector C, 53 had induction ratios between 1.5- and 2-fold in the GCN4/gcn4Δ experiment, suggesting that they belong to sector A: Gcn4p targets that are also inducible by a Gcn4p-independent mechanism. (B) Color display plot of the expression ratios of selected Gcn4p target genes that were induced ≥1.9-fold by 3AT in the gcn4Δ strain. The log10 ratios of expression are depicted using the color code described for Fig. 1, with the brightest red depicting log10 ratios of ≥1.0 and black signifying no significant change in expression (P > 0.05). Data were taken from the different experiments listed across the top (as defined in Fig. 1 and Table 2) for the genes listed on the right.
The data in Fig. 1 also uncovered a sizable group of genes that were highly induced by 3AT in the gcn4Δ mutant and showed little or no dependence on Gcn4p for maximal induction in the WT (Fig. 1, clusters I1, I2, I4, and I6 in row 10). The lack of Gcn4p dependence for these genes can be seen from the GCN4/gcn4 results shown in Fig. 1 (dull red, black, or green bars in row 5). Genes in this category fall into sector C of the Venn diagram shown in Fig. 3A, representing roughly half of the genes that are induced by 3AT in gcn4Δ cells. Either Gcn4p plays no role in their induction, or they can be induced equally well by a Gcn4p-independent mechanism in cells lacking Gcn4p. A number of genes in this group are known to be induced by hydrogen peroxide, including CTA1, CTT1, GTT2, HSP26, HSP42, and HSP78, consistent with the fact that 3AT inhibits catalase activity (30). Other highly induced Gcn4p-independent genes include BTN2, GAL7, IME2, INO1, NRG1, RPN4, and SSA4.
The results of the gcn4Δ ± 3AT experiment also confirmed that most of the genes that were repressed by 3AT treatment of WT cells were dependent on Gcn4p for maximal repression. Such genes showed a repression ratio of ≥2 in both the WT ± 3AT and the GCN4/gcn4Δ experiments but experienced little or no repression in the gcn4Δ ± 3AT experiment (Fig. 1, clusters R1, R3, and R4 in rows 1 to 5 and 10). However, a sizable group of genes were also strongly repressed in the gcn4 mutant (Fig. 1, clusters R2, R5, R6, and R7 in row 10) and displayed minimal Gcn4p dependence for this response in the GCN4/gcn4Δ experiment (black or dull red bars, row 5). Included in this category are ALD6, ADH1, ACS2, RPE1, SAM2, and ALG7. It should be noted that treatment of a gcn4Δ mutant with 100 mM 3AT severely impedes growth because high-level induction of histidine biosynthetic enzymes cannot occur in the absence of Gcn4p. Hence, this represents a more extreme starvation condition than that when WT cells were treated with 100 mM 3AT.
Summarizing the results described thus far, expression of 539 genes was induced ≥2-fold in at least one of the WT ± 3AT experiments and also displayed significant Gcn4p dependence for this response, showing an induction ratio of ≥2.0 in the GCN4/gcn4Δ or GCN4c/GCN4 experiments. Henceforth, we refer to this large group of genes as the Gcn4p targets. As noted above, many genes induced by 3AT in the WT were induced equally well, or more strongly, in the gcn4Δ mutant (sector C in Fig. 3A). It is conceivable that some of these genes are Gcn4p targets that can be induced to high levels by histidine starvation through an alternative mechanism in cells lacking Gcn4p. Hence, the number of Gcn4p target genes may have been underestimated by demanding dependence on Gcn4p for induction by 3AT.
Interestingly, a small subset of 29 Gcn4p target genes were not strongly induced in WT cells by 100 mM 3AT but required Gcn4p to maintain high-level expression in starved cells; hence, these genes were repressed by 100 mM 3AT in the gcn4Δ strain (Fig. 1, cluster I7, rows 1 to 4 versus row 10). Among the genes exhibiting this behavior are ILV1, ILV2, LEU1, and BAT1 (see below in Fig. 6C). One way to interpret this behavior is to propose that the promoters of these genes contain regulatory elements that mediate reduced transcription in response to severe histidine starvation and that Gcn4p counteracts this repression. Consistent with this explanation, these genes were induced most effectively under the less extreme starvation conditions of 10 mM 3AT and also by the GCN4c allele in nonstarved cells (Fig. 1, cluster I7).
FIG. 6.
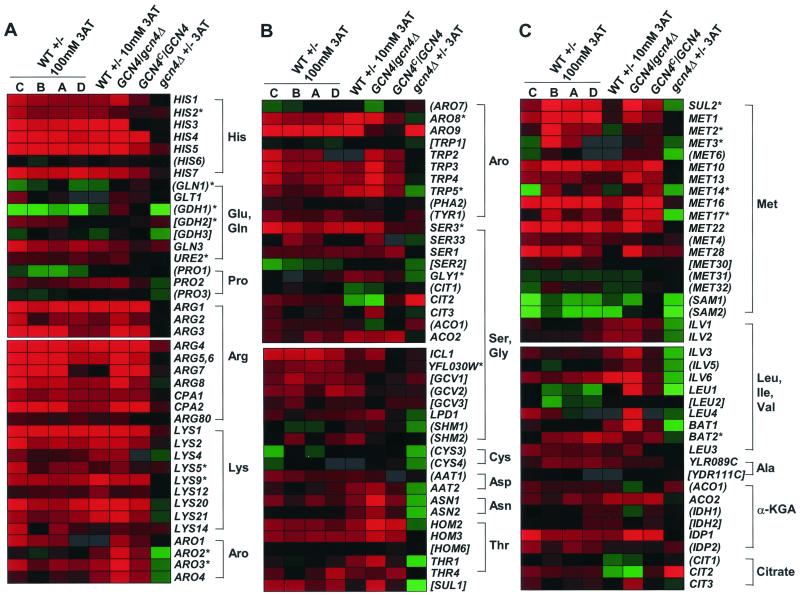
Color display plots of the expression ratios of genes involved in amino acid biosynthesis. Genes that are known or predicted to participate in the biosynthesis of amino acids, or amino acid precursors, are indicated on the right, and the log10 ratios of expression for the experiments listed along the top are displayed using the color code described for Fig. 1. (Gray indicates that no data were obtained for that gene.) Genes that were not judged to be Gcn4p targets are shown in parentheses, and those for which insufficient data exist to assess Gcn4p dependence are enclosed in brackets. (Expression of TRP1 and LEU2 could not be assessed because most of the strains that we analyzed have trp1 and leu2 mutations. Although strains R6257 and R4760 used in the GCN4c/GCN4 experiment carry TRP1, statistically significant data were not obtained for TRP1 in that experiment.) Asterisks indicate Gcn4p target genes lacking a recognizable Gcn4p binding site (TGASTCW, TGACTGA, or TGATTCA) in the −20 to −600 region of the promoter. (A) Results for genes in the His, Glu, Gln, Pro, Arg, Lys, or aromatic (Aro) amino acid biosynthetic pathways. (B) Results for genes in the aromatic, Ser, Gly, Cys, Asp, Asn, and Thr biosynthetic pathways. (C) Results for genes in the Met, Leu, Ile, Val, Ala, α-ketoglutarate (α-KGA), and citrate biosynthetic pathways.
The microarray results revealed that transcription of GCN4 was not induced by 3AT treatment for 1 h. However a twofold increase in GCN4 mRNA was previously observed at 2 h of 3AT induction (1). Our microarray data also showed that expression of the Gcn4p translational activators, GCN1, GCN3, and GCN20, was not substantially altered, whereas GCN2 expression was increased twofold by 3AT treatment, consistent with an earlier observation (92). Thus, stimulation of GCN4 mRNA translation, via the upstream open reading frames (ORFs) and activation of Gcn2p by uncharged tRNA (reviewed in references 38 and 39), seems to be the predominant mechanism for inducing Gcn4p during the first hour of histidine starvation.
Relationship between Gcn4p-dependent gene expression and occurrence of Gcn4p binding sites in the promoter.
If the Gcn4p-induced genes identified above are regulated directly by Gcn4p, they should contain one or more copies of the UASGCRE in their promoters. Previous in vitro studies showed that Gcn4p binds to the TGA(C/G)TCA sequence, with the critical central C · G base pair flanked by TGA half-sites (34, 77). Gcn4p can also bind to naturally occurring variants of this sequence (TGATTCA, TGACTCT, TGACTGA, and TGACTAT) found in the ILV2 and HIS4 promoters (2, 34), and ATGACTCT was found to be a functional UASGCRE in the HIS3 promoter in vivo (34). A computer algorithm used to scan the promoters of several amino acid biosynthetic genes, including known genes under Gcn4p control, predicted a consensus Gcn4p site, RRRWGASTCA (with R = purine, W = T or A, and S = G or C), that closely matched the previous findings (41). Hence, we used a motif search program called CoSMoS (28) to calculate what fraction of the genes with the greatest dependence on Gcn4p for induction by 3AT contained a copy of the sequence TGASTCW or one of the known functional variants in the 5′ noncoding DNA. Of the 210 genes showing an induction ratio of ≥4-fold in the GCN4/gcn4Δ experiment, 52% contained one or more copies of the UASGCRE located between 20 and 600 nucleotides upstream from the translation start site. It is possible that Gcn4p-dependent genes which lack a Gcn4p binding site in this interval would contain a functional UASGCRE in the coding region or 3′ noncoding sequences. Alternatively, these genes may be induced indirectly by Gcn4p, as numerous transcriptional activators are among the Gcn4p targets (see below).
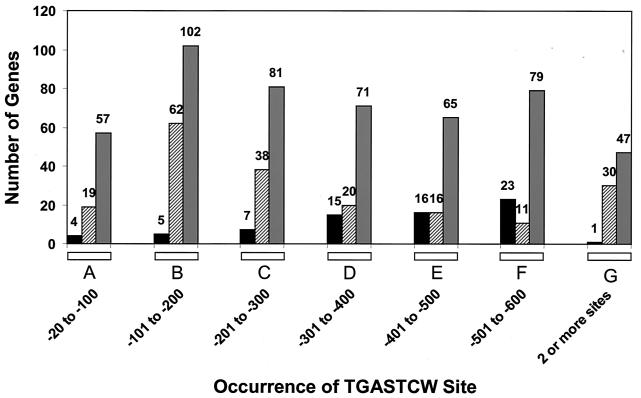
In a complementary approach, we scanned the complete genome sequence for those genes containing UASGCRE in the 5′ noncoding DNA. We found that 7 genes harbored three copies, 78 contained two copies, and 822 had a single UASGCRE in the −600 to −20 interval. Of the genes containing two or more copies of UASGCRE (for which we also obtained sufficient data to analyze their expression and Gcn4p dependence), 64% showed Gcn4p-dependent induction by 3AT in the GCN4/gcn4 experiment, whereas only a single gene in this group showed Gcn4p-dependent repression by 3AT (Fig. 4, column G). For the genes containing only a single UASGCRE located between −20 and −300, ∼50% showed Gcn4p-dependent induction while only 6% displayed Gcn4p-dependent repression (Fig. 4, columns A to C). For the remaining genes containing only a single UASGCRE upstream of −300, there was a nearly equal probability of ∼22 to 25% that the gene was induced or repressed by 3AT in a Gcn4p-dependent manner (Fig. 4, columns D to F). Thus, genes containing a UASGCRE within 300 nucleotides upstream of the gene are much more likely to be induced than to be repressed by Gcn4p in 3AT-treated cells. We interpret this strong bias to indicate that induced genes that fit these criteria (numbering 149) are activated by direct binding of Gcn4p to the UASGCRE in the promoter. Furthermore, the role of Gcn4p in gene repression is probably indirect (see below).
FIG. 4.
Correlation between Gcn4p-dependent induction by 3AT and the presence of Gcn4p binding sites in the 5′ noncoding DNA. The location of the Gcn4p binding site TGASTCW was determined for all genes identified in the GCN4/gcn4Δ experiment. Genes that harbor two or more binding sites between −20 and −600 were assigned to a single group (column G), and those with a single site were divided into separate groups (columns A to F) depending on the location of the site. The gray bars represent the total numbers of genes in each category; the striped bars depict the numbers of genes with induction ratios of ≥2-fold, and the black bars represent the numbers with repression ratios of ≥2-fold, in the GCN4/gcn4Δ experiment.
Ribosomal proteins (RP) are strongly repressed when Gcn4p is highly induced in amino acid-starved cells.
About 1,000 genes were repressed by a factor of 2 or more in 3AT-treated cells, and our analysis showed that 66% of these genes showed an unmistakable dependence on Gcn4p for strong repression by 100 mM 3AT. The 90 RPL and RPS genes encoding the RP formed the largest group of genes with a common function that was repressed by 3AT in a Gcn4p-dependent manner. This group also included numerous genes encoding general translation initiation factors. This behavior can be rationalized as a mechanism for coordinating a decrease in ribosome production and protein synthesis with the induction of amino acid biosynthetic capacity under conditions of amino acid limitation. In this sense, it is analogous to the stringent response of Escherichia coli (8). Repression of the RP genes occurs in response to various conditions of starvation or stress (7, 10, 19, 48). What seems unique here is that Gcn4p contributes to the magnitude of the repression under amino acid starvation conditions. Most of the RPL and RPS genes lack the UASGCRE in the promoter; thus, Gcn4p probably contributes indirectly to their repression in histidine-starved cells.
As noted above, high-level Gcn4p expression in the GCN4c mutant did not elicit strong gene repression under nonstarvation conditions, suggesting that amino acid limitation is additionally required. To explain this dual requirement for repression, we propose that promoters of RP genes are down-regulated under amino acid starvation conditions by a mechanism that involves the activator Rap1p (71) and signal transduction by protein kinase A (PKA) (54). Induction of Gcn4p in amino acid-starved cells would intensify the response by sequestering one or more transcription factors required at RP promoters (squelching) (84). Squelching alone by overexpressing Gcn4p in nonstarvation conditions (GCN4c mutant) would be insufficient for strong repression. Moreover, when strain R491 was grown with limiting amounts of the required amino acids, either the induction of Gcn4p was not extensive enough or the starvation was not severe enough to elicit strong repression of RP genes.
Overlap between the induction profiles of MMS and 3AT.
It was shown recently that MMS treatment induced the transcription of about 40 genes involved in amino acid metabolism among a total of 1,324 genes induced ≥2-fold (48). We compared the expression profiles during 3AT and MMS treatment and found that, of the 409 genes induced by 3AT, 309 (90%) were also induced by MMS, whereas only 28% of the MMS-induced genes were induced by 3AT (Fig. 1, row 9 versus rows 1 to 4). Based on this comparison, it seemed likely that Gcn4p was responsible for activating a substantial fraction (∼28%) of the MMS-induced genes. As the steady-state level of GCN4 mRNA was unchanged by MMS treatment (48), we predicted that MMS would induce GCN4 at the translational level.
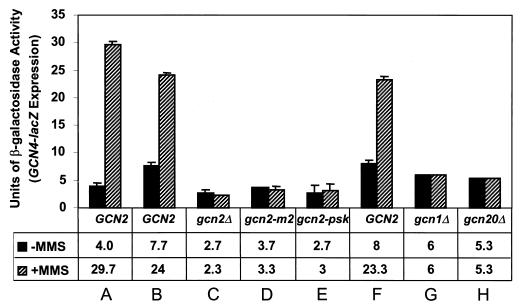
In agreement with this prediction, treatment of WT strain H187 on nutrient-rich medium with 0.07% (vol/vol) MMS induced a GCN4-lacZ reporter 7.4-fold (Fig. 5, GCN2, column A). Somewhat lower induction ratios were observed for two other WT strains (GCN2, columns B and F). Because strain H187 is prototrophic for all amino acids, we conclude that MMS does not induce GCN4-lacZ expression indirectly by interfering with amino acid uptake in an auxotroph. Importantly, GCN4-lacZ induction by MMS was completely impaired in the gcn2Δ strain (Fig. 5, columns B and C), lacking the eIF2α kinase Gcn2p required for translational induction of GCN4. Uncharged tRNA activates Gcn2p in amino acid-starved cells by binding to a domain related to histidyl-tRNA synthetase (HisRS) located adjacent to the kinase domain (107). As shown in Fig. 5, the MMS induction of GCN4-lacZ was defective in a gcn2-m2 mutant with point mutations in the HisRS-like domain that impair tRNA binding and kinase activity (107, 110). MMS induction of GCN4-lacZ also was absent in the gcn2-psk mutant, bearing a two-codon substitution in a degenerate kinase domain located N-terminal to the authentic kinase domain (106), and in gcn1Δ and gcn20Δ strains lacking positive effectors required for activation of Gcn2p in amino acid-starved cells (26, 65, 103). These results indicate that the same regulatory elements are required for induction of GCN4 translation in response to MMS or 3AT treatment. The fact that MMS induction of GCN4-lacZ required the tRNA-binding activity of Gcn2p could indicate that MMS interferes with aminoacylation of one or more tRNAs and thereby generates the same activating ligand as does amino acid starvation. Alternatively, binding of uncharged tRNA may be an unconditional prerequisite for Gcn2p activation, and methylation of Gcn2p (or an unknown negative regulator of Gcn2p) by MMS could lower the threshold of uncharged tRNA required to activate the kinase.
FIG. 5.
MMS induces GCN4-lacZ expression dependent on translational activators of GCN4. The β-galactosidase activity expressed from a GCN4-lacZ fusion was assayed in extracts from untreated (black bars) or MMS-treated (striped bars) cultures of prototrophic strain H187 grown in YPD medium (column A). The same fusion was assayed in extracts from gcn2Δ strains harboring plasmids bearing GCN2 (column B), no allele (column C), the gcn2-m2 allele (column D), or the gcn2-psk allele (column E) cultured in SC medium with or without MMS. Finally, GCN4-lacZ was assayed in isogenic WT (column F), gcn1Δ (column G), or gcn20Δ (column H) strains grown in SC medium with or without MMS. Error bars depict the standard errors of the means of activities measured from at least three independent cultures or transformants.
The checkpoint proteins Mec1p, Rad53p, and Dun1p are required for a response to DNA damage (reviewed in reference 104). We found that null mutations in these genes did not impair MMS induction of GCN4-lacZ expression (data not shown). Thus, it appears that Gcn2p is activated in MMS-treated cells independently of the major signal transduction system for responding to DNA damage. Interestingly, certain genes induced by Gcn4p are involved in the repair of DNA damage, including RAD5, RAD14, RAD26, RAD55, ECM32, NTG1, and APN1. Thus, transcriptional stimulation of these genes by Gcn4p might be important for efficient repair of MMS-induced DNA damage. However, we found that gcn4Δ and gcn2Δ mutants were not more sensitive to the toxic effects of MMS than were isogenic WT strains (data not shown). Hence, the induction of Gcn4p target genes is not a critical aspect of the cellular response to DNA damage by MMS under laboratory growth conditions.
Amino acid biosynthetic genes induced by Gcn4p.
We used the MIPS functional categories (69) in an effort to assign the 539 Gcn4p target genes to different pathways. There are 119 genes in the MIPS amino acid biosynthesis category, and 78 of them were induced by 3AT in a WT strain. In accordance with previous work (36), 73 were identified here as Gcn4p target genes. All amino acid biosynthetic pathways contain multiple genes induced by Gcn4p in our studies, except for the Cys pathway, which has none. And even for the Cys pathway, genes involved in the biosynthesis of the precursors serine and homocysteine were induced by Gcn4p. As discussed below, there were other instances where genes involved in producing precursors for amino acid biosynthetic pathways were induced by Gcn4p. Taken together, all of these biosynthetic genes account for 77 of the 539 Gcn4p target genes that we identified.
(i) Histidine pathway.
The microarray data showed that six of the seven histidine biosynthetic genes were Gcn4p targets (Fig. 6A, His), as expected from previous work (36, 57). HIS2 showed the weakest induction by 3AT and the least dependence on Gcn4p (Fig. 6A), consistent with the absence of a consensus Gcn4p site in the −20 to −600 region. HIS6 was judged not to be a Gcn4p target, based primarily on data from the WT ± 3AT (set C) and GCN4/gcn4Δ experiments (Fig. 6A), despite the presence of a Gcn4p binding site. The magnitude of HIS gene induction by 3AT was not significantly greater than that of many other amino acid biosynthetic pathways (e.g., Arg, Lys, and Met [Fig. 6A and B]), consistent with the absence of histidine-specific transcriptional repression in yeast. Histidine, tryptophan, and adenine biosyntheses require phosphoribosyl pyrophosphate (PRPP) (51), and numerous TRP and ADE genes also are Gcn4p target genes (Fig. 6B and data not shown). However, four of the five PRS genes encoding PRPP synthetase (33) were repressed more than twofold by 3AT, whereas PRS2 expression was not substantially affected (data not shown).
(ii) Glutamate family: Glu/Gln, Arg, Pro, and Lys.
Glutamate and glutamine are the key amino group donors in the biosynthesis of amino acids, nucleotides, and other nitrogen-containing compounds (51). Glutamine is produced from glutamate and ammonia in a reaction catalyzed by Gln1p, an enzyme shown previously to be induced by Gcn4p (70). Glutamate can be synthesized from α-ketoglutarate and ammonia by the isozymes encoded by GDH1 and GDH3 or from α-ketoglutarate and glutamine by glutamate synthase (Glt1p) (64). In our experiments, GLT1 was weakly induced by Gcn4p, whereas GDH1 and GLN1 were repressed, and GDH3 showed little response to 3AT (Fig. 6A, Glu and Gln).
Gln3p is a transcriptional activator of GLN1 (70), GLT1 (102), and many other genes involved in utilization of poor nitrogen sources (64). Interestingly, GLN3 was found to be a Gcn4p target (Fig. 6A), and it contains a consensus UASGCRE in its promoter. Perhaps GLN1 was not induced by 3AT because Gln3p was impaired by its negative regulator Ure2p, owing to the presence of ammonia as a nitrogen source (64). In fact, URE2 was induced by Gcn4p in response to 3AT treatment (Fig. 6A, Glu and Gln). In this event, the modest induction of GLT1 that we observed likely involved Gln3p-independent activation by Gcn4p. Consistently, GLT1 contains a UASGCRE in its promoter. Presumably, induction of GLN3 by Gcn4p in cells starved for glutamine would induce GLN1 and GLT1 because Ure2p would be inactive (64).
Glutamate biosynthesis requires α-ketoglutarate as the precursor. Citrate is converted to α-ketoglutarate by the sequential action of the tricarboxylic acid (TCA) cycle enzymes aconitase and NAD-dependent isocitrate dehydrogenase, encoded by ACO1 and IDH1/IDH2, respectively. Expression of ACO1 was moderately induced by 3AT, but independently of Gcn4p, and IDH1 was not judged to be a Gcn4p target (Fig. 6C, α-KGA). Although IDH2 was induced about twofold by 3AT in the WT, its Gcn4p dependence could not be ascertained. However, the NADP-dependent isocitrate dehydrogenases encoded by IDP1 and IDP2 can functionally substitute for the IDH1 and IDH2 products (32), and IDP1 was found to be a Gcn4p target. Similarly, ACO2/YJL200C, encoding an isozyme of aconitase, is a Gcn4p target (Fig. 6C, α-KGA). Two of the three genes encoding citrate synthase, CIT2 and CIT3 (encoding the peroxisomal and mitochondrial isozymes, respectively), were judged to be Gcn4p targets. Although CIT2 was strongly induced by 3AT in both the WT and gcn4Δ strains, its expression was induced ∼2-fold in the GCN4c/GCN4 experiment (Fig. 6C, Citrate). IDP1, ACO2, CIT2, and CIT3 contain UASGCRE elements in their 5′ noncoding sequences, consistent with direct activation by Gcn4p. We conclude that Gcn4p-mediated induction of IDP1, ACO2, CIT2, and CIT3 may play an important role in stimulating α-ketoglutarate synthesis for glutamate production during amino acid starvation on glucose medium.
Arginine and proline are synthesized directly from glutamate (51). Of the three genes involved in proline biosynthesis, only PRO2 was judged to be a Gcn4p target. Surprisingly, PRO1 was repressed by 3AT (Fig. 6A, Pro), consistent with a previous report that PRO1 is not induced by histidine starvation (59). As expected (36), we found that nine of the arginine biosynthetic genes (ARG1, -2, -3, -4, -5, 6, -7, and -8; CPA1; and CPA2) are Gcn4p targets and that most were strongly induced by 3AT (Fig. 6A, Arg).
Lysine is synthesized from α-ketoglutarate by a pathway of eight enzymes (51). In agreement with previous findings (36), all eight LYS genes were found to be Gcn4p targets (Fig. 6A, Lys). LYS14, encoding a pathway-specific activator (24), was found to be a Gcn4p target (Fig. 6A, Lys), suggesting that derepression of LYS genes by Gcn4p is mediated partly by induction of Lys14p (at least under lysine starvation conditions). It is probable that Gcn4p also activates LYS genes directly (86) and that consensus (LYS1, -12, and -2) or functional (LYS4, -9, -20, and -21) variants of UASGCRE occur in the promoters of all LYS genes except LYS5 and LYS9.
(iii) Aromatic family: Trp, Phe, and Tyr.
The aromatic amino acids tryptophan, tyrosine, and phenylalanine are synthesized from chorismate (51). As shown previously (21, 36, 50), all four genes encoding enzymes required for chorismate synthesis (ARO1, ARO2, ARO3, and ARO4) and three of the four genes encoding enzymes that convert chorismate to tryptophan (TRP2, TRP3, TRP4, and TRP5) were found to be Gcn4p targets. ARO2 belongs to the class of Gcn4p targets that were not strongly induced by 3AT but required Gcn4p to prevent its repression in severely starved cells (Fig. 6A and B, Aro). As noted previously (94), ARO7 was not induced by Gcn4p. Although expression of TRP1 could not be ascertained (see legend to Fig. 6), there is previous evidence that its expression is not induced by Gcn4p (5).
TYR1 and PHA2, whose products carry out the second steps in tyrosine and phenylalanine biosynthesis, respectively, were not judged to be Gcn4p targets, although TYR1 was induced by 3AT independently of Gcn4p (Fig. 6B, Aro). ARO8, encoding aromatic aminotransferase I, which functions in the last step of Tyr and Phe biosynthesis (100), was strongly regulated by Gcn4p, in accordance with previous findings (46).
ARO9 encodes aromatic aminotransferase II, which functions in the first step of tryptophan degradation (46). The ARO10 product may catalyze the second step (decarboxylation) of this pathway, and both ARO9 and ARO10 were induced by tryptophan on medium containing a poor nitrogen source (45). Both genes displayed significant dependence on Gcn4p for their induction by 3AT and also were highly induced in the gcn4Δ strain (Fig. 6B, Aro, and data not shown).
(iv) Serine family: Ser, Gly, and Cys.
In addition to their roles in protein synthesis, serine and glycine serve as precursors for the one-carbon units carried by tetrahydrofolate (THF) derivatives. The latter participate as coenzymes in single-carbon transfer reactions in purine and pyrimidine biosynthesis, amino acid metabolism, and methyl group biogenesis. During growth on fermentable carbon sources, the majority of serine is derived from 3-phosphoglycerate, a glycolytic intermediate, by sequential action of SER3/SER33, SER1, and SER2. Our data show that SER1 is a Gcn4p target, in agreement with previous results (68), as are SER3 and -33, whereas SER2 was repressed by 3AT (Fig. 6B, Ser). As SER2 contains two consensus UASGCREs in its promoter, it is possible that Gcn4p can induce this gene in serine-deprived cells but is prevented from doing so in histidine-starved cells by a serine-specific transcriptional repression. Glycine can be produced from serine by serine hydroxymethyltransferase (SHMT); however, the major synthetic route is catalyzed by threonine aldolase encoded by GLY1 (61). GLY1 belongs to the category of Gcn4p-dependent genes that are not highly induced by 3AT but show reduced expression in starved gcn4Δ cells (Fig. 6B, Gly).
An alternative pathway for serine and glycine biosynthesis operates during growth on nonfermentable carbon sources, proceeding from citrate to glycine via glyoxylate in reactions catalyzed by aconitase (encoded by ACO1/2), isocitrate lyase (encoded by ICL1), and alanine-glyoxylate aminotransferase (probably encoded by YFL030W). A portion of the glycine is decarboxylated by glycine decarboxylase (encoded by GCV1, GCV2, GCV3, and LPD1), forming 5,10-methylene THF, which serves as the one-carbon donor for production of serine from glycine by SHMT (51). As indicated above, SHMT can also utilize serine to generate glycine and 5,10-methylene-THF. ACO2, ICL1, YFL030W, GCV1, GCV2, LPD1, and SHM2 (encoding the cytoplasmic SHMT) were all induced by 3AT, and of these genes, ACO2, ICL1, YFL030W, and LPD1 were judged to be Gcn4p targets (Fig. 6B, Ser/Gly). The presence of a Gcn4p site(s) upstream of GCV1, GCV2, GCV3, SHM2, ACO2, ICL1, and LPD1 is consistent with direct activation of these genes by Gcn4p.
It is thought that cysteine is synthesized in yeast exclusively from serine and homocysteine, an intermediate of methionine biosynthesis, by the products of CYS3 and CYS4 (78). Surprisingly, CYS3 and CYS4 were either repressed or unaffected by 3AT (Fig. 6B, Cys). Nevertheless, Gcn4p might stimulate cysteine biosynthesis by induction of the serine and methionine biosynthetic pathways, leading to increased production of the precursors of cysteine.
(v) Aspartate family: Asp, Asn, Thr, and Met.
Aspartate is synthesized by transamination of oxaloacetate via glutamate, in a reaction catalyzed by aspartate aminotransferase, encoded by AAT1 (mitochondrial) and AAT2 (cytoplasmic and peroxisomal). The regulation of AAT1 by Gcn4p was equivocal, whereas AAT2 was judged to be a Gcn4p target (Fig. 6B, Asp). Both genes contain consensus Gcn4p sites in their promoters. Asparagine is synthesized from aspartate by asparagine synthetase, encoded by ASN1 and ASN2. In accordance with previous findings (15), both genes were found to be Gcn4p targets (Fig. 6B, Asn) and to contain Gcn4p sites. Threonine is synthesized from aspartate by the products of HOM3, HOM2, HOM6, THR1, and THR4. In keeping with published findings (36), HOM3, HOM2, THR1, and THR4 were found to be Gcn4p targets (Fig. 6B, Thr), and they contain several UASGCREs in their promoters. AAT2, ASN1, ASN2, and THR1 are examples of genes that were not induced by 100 mM 3AT but were dependent on Gcn4p to prevent repression under these starvation conditions (Fig. 6B).
The methionine biosynthetic genes have been well characterized in yeast (for a review, see reference 98). Previous studies have shown that Gcn4p regulates MET16 and MET17 (75), encoding two enzymes in the pathway, and MET4 (73), encoding a transcriptional activator of the MET genes. There were also previous indications that Gcn4p induces MET3, MET14, and MET6, although only in methionine-starved cells (36). Our findings confirmed that MET16, MET17, MET3, and MET14 are Gcn4p targets (Fig. 6C, Met). MET6 belongs to the class of genes dependent on Gcn4p to prevent its repression in 100 mM 3AT. Additionally, we found that MET10, MET1, MET13, MET22, and MET2, encoding other Met pathway enzymes, and MET28, encoding a transcriptional activator of the pathway, are Gcn4p targets, as are SUL1 and SUL2, encoding high-affinity sulfate transporters. The transcriptional activators MET31 and MET32 do not appear to be regulated by Gcn4p; while MET4 was strongly induced by 3AT, it showed little dependence on Gcn4p in this response (Fig. 6C, Met). Although Gcn4p was thought to have a limited role in MET gene expression under methionine-limiting conditions (98), data from our studies and others (75) indicate strong Gcn4p-dependent induction of MET genes in cells starved for histidine or tryptophan.
Because Gcn4p induces Met4 and Met28 expression, it may indirectly activate MET genes by stimulating these pathway-specific activators. Additionally, Gcn4p can activate MET16 and MET17/25 independently of Met4p, suggesting direct activation of these genes by Gcn4p (75). The cellular level of S-adenosylmethionine (AdoMet) is the regulatory signal for methionine abundance, and at high levels of AdoMet, the SCFMet30 complex targets Met4p for degradation and thereby represses MET gene transcription (91). Interestingly, SAM1 and SAM2, encoding AdoMet synthetase, were repressed two- to fivefold by 3AT (Fig. 6C, Met). It is possible, therefore, that repression of AdoMet synthetase in 3AT medium decreases the AdoMet pool and activates MET gene transcription by reducing SCFMet30-mediated degradation of Met4p.
(vi) Pyruvate family: Ile, Val, Leu, and Ala.
Isoleucine, valine, and leucine are synthesized from threonine and pyruvate by the sequential action of ILV1, ILV2, ILV6, ILV5, ILV3, BAT1, BAT2, LEU4, LEU1, and LEU2. Our data show that all of these genes, excluding ILV5, are Gcn4p targets (Leu, Ile, and Val in Fig. 6C). These results confirm previous findings (36), except for ILV3 and ILV6, which had not been analyzed in this regard. LEU2 expression was not induced by 3AT, and its Gcn4p dependence could not be established in our strains (see the legend to Fig. 6). However, there is previous evidence that LEU2 is not under general control (40). The expression pattern of ILV5 resembles that of those genes that require Gcn4p only to prevent their repression by 100 mM 3AT. In fact, all of the genes in these pathways exhibit strong Gcn4p dependence but relatively low 3AT induction ratios (Fig. 6C). Hence, they may have promoter elements in common that mediate reduced expression in response to severe amino acid starvation.
Leu3p is a transcriptional activator of all three LEU genes and probably also ILV2 and ILV5. As LEU3 is induced by Gcn4p (109a) (Fig. 6C), the activation of LEU4, ILV2, and ILV5 by Gcn4p could be indirect. LEU4 transcription is also activated directly by Gcn4p (36). Additionally, all of the genes involved in Leu, Ile, or Val biosynthesis, except for BAT2, ILV2, and LEU2, contain a consensus Gcn4p binding site, suggesting a direct role for Gcn4p in their induction. Since the biosynthesis of alanine and threonine, precursors of this pathway, seems to be induced by Gcn4p, this may provide an additional stimulatory effect of Gcn4p on the biosynthesis of Ile, Val, and Leu.
Biosynthesis of alanine is thought to occur by transamination of pyruvate (51). YLR089c and YDR111c have sequence similarity to bacterial alanine aminotransferases and likely encode isozymes of the corresponding yeast enzyme. Whereas YLR089c was judged to be a Gcn4p target gene, data for YDR111c were below the P value threshold, and its dependence on Gcn4p could not be ascertained (Fig. 6C, Ala).
(vii) Aminoacyl-tRNA synthetases.
It was shown previously that KRS1/GCD5, ILS1, and MES1 genes, encoding aminoacyl-tRNA synthetases for Lys, Ile, and Met, respectively, were induced by Gcn4p in amino acid-starved cells (36, 58). We found that KRS1 and the genes encoding AspRS (DPS1), ArgRS (YDR341C), and a protein related to ThrRS (YGL219C) are all Gcn4p targets. Presumably, the induced levels of these enzymes lead to higher levels of the corresponding aminoacylated tRNAs under conditions of histidine limitation. By contrast, transcription of the genes encoding GlnRS (GLN4), PheRS (FRS1 and FRS2), and SerRS (SES1) was repressed by 3AT. Thus, regulation of these enzymes is akin to that of other components of the translational machinery. Expression of MES1, ILS1, DED81 (encoding AsnRS), TYS1 (encoding TyrRS), and YHR020W (encoding a protein related to ProRS) was not substantially altered by 3AT, although DED81 probably depends on Gcn4p to prevent its repression in 100 mM 3AT. Thus, the different genes encoding aminoacyl-tRNA synthetases exhibit diverse responses to 3AT and a varying dependence on Gcn4p, which is not currently understood.
Purine-pyrimidine biosynthetic enzymes.
The purine biosynthetic genes ADE1, ADE2, ADE3, ADE4, ADE8, ADE12, and ADE17 were induced by 3AT, and Gcn4p contributed to this response at ADE1, ADE3, ADE8, ADE12, and ADE17. Previously, it was reported that ADE4 transcription was induced in a mutant strain containing high constitutive levels of Gcn4p (72) and that ADE1, -2, -4, -5, -7, and -8 were moderately induced by Gcn4p upon 3AT treatment (88). As the purine ring of ATP is partially consumed in histidine biosynthesis, an increase in adenine nucleotide biosynthesis could be viewed as a strategy to support increased histidine biosynthesis. On the other hand, the pathway to AMP consumes PRPP, glycine, aspartate, glutamine, and THF derivatives, and its induction could be viewed as counterproductive under amino acid starvation conditions. It was shown previously that adenine limitation in medium replete with amino acids induces GCN4 mRNA translation and that mutations in GCN4 or its translational activator GCN1 or GCN2 impair cell growth under adenine starvation conditions (88). Thus, the contribution of Gcn4p to ADE gene expression in adenine-starved cells, demonstrable for ADE8 in particular, seems to be required for adequate adenine nucleotide biosynthesis under adenine starvation conditions and may have little to do with amino acid biosynthesis.
The ADE genes have one or more TGACTC elements in their promoters, consistent with a direct role for Gcn4p in activating these genes. However, Bas1p is an activator of multiple ADE genes (except ADE3 [17]) and it also binds to TGACTC elements (14, 99). We identified BAS1 as a Gcn4p target gene, and it contains TGACTG (a weak Gcn4p binding site) at −272 and a consensus Gcn4p site at −1038. Accordingly, the induction of Bas1p by Gcn4p in response to histidine or purine limitation may contribute to the activation of ADE genes under these starvation conditions. As Bas1p additionally activates HIS4 (3, 99), HIS7 (97), and SHM2 and MTD1 transcription (17), Gcn4p-dependent activation of one or more of these genes in 3AT medium could involve a contribution from the induced levels of Bas1p. Since ADE3 expression is independent of Bas1p (14), Gcn4p presumably activates this gene directly in histidine-starved cells.
The genes URA1 through -8, encoding the pyrimidine biosynthetic enzymes, were repressed by 3AT treatment, along with PRP1, encoding a transcriptional activator of the URA genes. This could be viewed as a means of limiting consumption of aspartate, glutamine, and PRPP, precursors of the pyrimidine pathway, under amino acid starvation conditions. Paradoxically, URA10 was highly induced by 3AT in a Gcn4p-dependent manner and contains a single Gcn4p binding site. URA10 contributes about 20% of the orotate phosphoribosyltransferase activity, with the remainder coming from URA5 (16).
Vitamin-cofactor biosynthetic pathways.
An unanticipated finding of this study is that numerous vitamin biosynthetic genes are induced during amino acid starvation in a Gcn4p-dependent manner. These genes are required for biosynthesis of biotin, NAD, THF, riboflavin, pyridoxal phosphate, and coenzyme A. Because vitamins function as cofactors for various enzymes of intermediary metabolism, we propose that vitamin biosynthesis is induced by Gcn4p to support increased amino acid production.
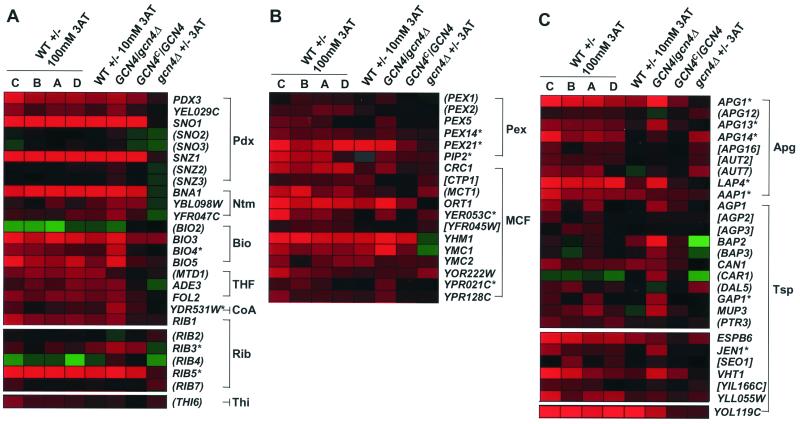
Pyridoxal phosphate is synthesized from pyridoxine by the sequential action of pyridoxine kinase and pyridoxine (pyridoxamine) phosphate oxidase (67). PDX3, encoding the latter enzyme, and YEL029C, whose product has ∼38% identity to human pyridoxine kinase, were both identified as Gcn4p targets (Fig. 7A, Pdx). It was shown recently that fungal proteins highly related to yeast Snz1p (and perhaps Sno1p) are involved in pyridoxine (vitamin B6) biosynthesis (22, 79), although an enzymatic activity has not been ascribed to them. 3AT treatment led to 20- to 50-fold induction of the SNZ1-SNO1 pair, which was completely Gcn4p dependent (Fig. 7A, Pdx). These two genes are divergently transcribed from a common promoter, and their transcription is induced in late stationary phase (80). Although two other highly related gene pairs occur in yeast, SNZ2-SNO2 and SNZ3-SNO3, only SNZ1-SNO1 transcription was induced by Gcn4p (Fig. 7A, Pdx), and consistently, only the SNZ1-SNO1 promoter has consensus Gcn4p sites.
FIG. 7.
Color display plots of the expression ratios for genes involved in selected pathways that may contribute indirectly to amino acid biosynthesis or accumulation. The log10 ratios of expression for the genes indicated on the right in the experiments listed across the top are displayed using the color code defined for Fig. 1. (A) Results for genes known or suspected to be involved in biosynthesis of pyridoxal phosphate (Pdx), nicotinamide (Ntm), biotin (Bio), THF, riboflavin (Rib), and thiamine (Thi). (B) Results for genes involved in peroxisome (Pex) biogenesis or belonging to the MCF. (C) Results for genes involved in autophagy (Apg) or belonging to the transporter (Tsp) family.
Nicotinamide, derived from tryptophan, is a component of NAD and NADP, two coenzymes involved in dehydrogenase reactions. BNA1/HAD1 (encoding 3-hydroxyanthranilate 3,4-dioxygenase) and YBL098W and YFR047C, the two other predicted genes in this pathway (56), were all identified as Gcn4p targets in our experiments (Fig. 7A, Ntm). BIO3, BIO4, and BIO5, whose products are involved in biotin synthesis, also were found to be Gcn4p targets, although BIO2 was not (Fig. 7A, Bio), despite the presence of a Gcn4p site in its promoter. Biotin is the carrier of carboxyl groups in enzymatic carboxylation reactions. It was reported previously that ornithine transcarbomylase, encoded by ARG3, requires biotin (20). It is unclear which of the many ATP-dependent carboxylation reactions in amino acid biosynthetic pathways require biotin as a carrier.
Interestingly, MTD1 and ADE3, encoding the enzymes catalyzing formation of 5,10-methenyl-THF and 10-formyl-THF (a precursor of adenine biosynthesis) from 5,10-methylene-THF, both were induced by 3AT. ADE3 is clearly a Gcn4p target, whereas the Gcn4p dependence of MTD1 is less certain (Fig. 7A, THF). FOL2, an enzyme involved in folic acid synthesis, also was found to be a Gcn4p target, and it contains a consensus Gcn4p binding site in its promoter. Thus, several enzymes involved in formation of THF derivatives, besides SHMT and glycine decarboxylase, mentioned above, are regulated by Gcn4p. Gcn4p as mentioned above, may induce SHMZ and MTD1 indirectly via induction of Bas1p.
Pantothenate, a component of the acyl group carrier coenzyme A, is synthesized from pantoate. YDR531W, encoding pantothenate kinase, the first committed step in coenzyme A biosynthesis (6), is a Gcn4p target gene (Fig. 7A, CoA). Riboflavin (vitamin B2) is the precursor for flavin mononucleotide and flavin adenine dinucleotide, both of which function as coenzymes in various reactions of intermediary metabolism. Riboflavin is synthesized from GTP by the sequential action of the products of RIB1, RIB7, RIB2, RIB3, RIB4, and RIB5 genes. Our results showed that transcription of RIB1, -3, and -5 is Gcn4p dependent (Fig. 7A, Rib). Although THI6, encoding an enzyme in thiamine biosynthesis, was induced by 3AT, it was only weakly Gcn4p dependent (Fig. 7A, Thi).
Organellar involvement in amino acid biosynthesis. (i) Peroxisomal genes.
Another unexpected finding was that Gcn4p activates peroxisomal genes. The yeast peroxisome is the sole site of β-oxidation for catabolism of fatty acids. Recent evidence indicates that lysine biosynthetic enzymes Lys1p and Lys4p are localized in peroxisomes and that pex8 and pex15 mutants are leaky lysine auxotrophs (28). Thus, lysine biosynthesis occurs, at least partly, in peroxisomes and is dependent on the functions of Pex8p and Pex15p. We found that PEX1, PEX2, PEX5, PEX11, PEX14, PEX21, and PXA2 were induced in 3AT medium and that all but PEX1 and PEX2 were dependent on Gcn4p for this response (Fig. 7B, Pex). The products of these genes function in peroxisome biogenesis or are located in the peroxisomal membrane, and Pxa2p is required for fatty acid transport across the peroxisomal membrane. Interestingly, PIP2, encoding a transcriptional activator of oleate-induced genes (including PEX genes) and peroxisome proliferation (53, 90), was identified as a Gcn4p target. We speculate that Gcn4p induces peroxisome proliferation in amino acid-starved cells as a means of stimulating lysine biosynthesis.
(ii) Mitochondrial carrier proteins.
Portions of certain amino acid biosynthetic pathways, including Arg, Lys, Ile, Val, and Leu, take place in the mitochondria; thus, precursors and intermediates in these pathways must be shuttled between the two compartments (51). The yeast genome encodes about 35 members of the mitochondrial carrier family (MCF) involved in small molecule transport between cytosol and mitochondria (82). We found that Gcn4p induces transcription of 10 members of this family: ARG11/ORT1, YHM1, OAC1, YMC1, YMC2, CRC1, YER053C, YOR222W, YPR021C, and YPR128C (Fig. 7B, MCF; also data not shown). Transcription of two others, CTP1 and YFR045W, was induced by 3AT, but their Gcn4p dependence is unclear. The fact that a mutation in ARG11/ORT1 produced a leaky arginine auxotrophy (13) established the importance of this MCF protein in arginine biosynthesis. OAC1 encodes the mitochondrial transporter for oxaloacetate and sulfate (81). Given that oxaloacetate is the immediate precursor of aspartate, export of oxaloacetate (a TCA cycle intermediate) from mitochondria by Oac1p may be important for efficient aspartate synthesis by Aat2p in the cytosol. If enzymes of the Met pathway responsible for sulfate assimilation are mitochondrial, then transport of sulfate into this organelle by Oac1p could also facilitate Met biosynthesis.
Autophagy genes.
In response to starvation for nitrogen, carbon, sulfate, phosphate, or amino acids, cytosolic proteins are targeted to the vacuole for bulk degradation in membrane-bound autophagosomes, in a process known as autophagy (55, 76). Our analysis revealed that APG1, APG13, and APG14 are Gcn4p targets; however, APG14 also was induced by 3AT in the gcn4Δ strain (Fig. 7C, Apg). Thus, APG14 shows Gcn4p-dependent and -independent induction in response to histidine starvation. APG16 was induced by 3AT independently of Gcn4p. A consensus Gcn4p site is present upstream of APG13, a degenerate site (TGACTT) occurs at APG1, and no Gcn4p site is evident at APG14.
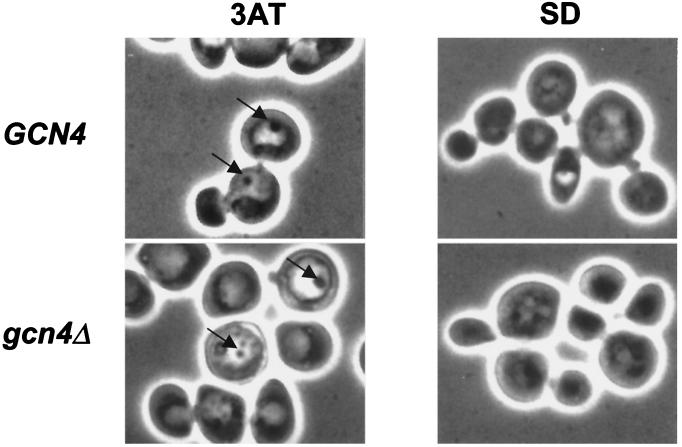
Consistent with transcriptional induction of APG genes by Gcn4p, 3AT treatment induced dense bodies exhibiting Brownian motion inside the vacuoles, known as autophagic vesicles (Fig. 8, GCN4, SD versus 3AT). However, gcn4Δ cells also produced autophagic vesicles (Fig. 8), suggesting that Gcn4p is not critically required for the response. We found that apg1Δ, apg13Δ, and apg14Δ strains were not impaired for growth in the presence of 3AT or sulfometuron methyl (SM), an inhibitor of branched-chain amino acid biosynthesis (data not shown), indicating that autophagy is not required for growth of GCN4 cells under amino acid starvation conditions where cell division is still occurring. Presumably, the salvage of amino acids from proteins by autophagy would lessen the impact of more severe amino acid starvation conditions than were imposed here. LAP4, encoding a vacuolar aminopeptidase, and AAP1, encoding alanine/arginine aminopeptidase, were found to be Gcn4p targets. The induction of these vacuolar proteases may accelerate the degradation of proteins transported to the vacuole by the autophagy pathway.
FIG. 8.
Autophagy is induced by amino acid starvation. Strains KNY201 (GCN4) and KNY202 (gcn4Δ) were cultured in YPD medium to an OD600 of 1.0, harvested, washed with minimal (SD) medium, resuspended in SD medium or in SD medium containing 40 mM 3AT (3AT), and incubated with shaking for 4.5 h. Aliquots of cells were removed and visualized by phase-contrast microscopy. The arrows indicate dense bodies inside the vacuoles that exhibited Brownian motion, judged to be autophagic vesicles.
Amino acid transporters.
Yeast cells transport external amino acids through general and specific amino acid permeases. It might be expected that expression of these genes would be induced by Gcn4p to increase amino acid uptake under starvation conditions. Indeed, the genes encoding the general amino acid permease (GAP1), a basic amino acid permease (CAN1), leucine permease (BAP2), and the broad substrate permease (AGP1) were strongly induced by 3AT in a Gcn4p-dependent manner (Fig. 7C, Tsp). Additionally, MUP3, encoding a low-affinity Met permease, and LYP1 (data not shown), encoding a lysine permease, were identified as Gcn4p targets. Expression of several amino acid-specific permeases was repressed or remained unchanged in response to 3AT. Presumably, Gcn4p-mediated activation of the GAP1 and AGP1 general permeases would stimulate increased transport of most amino acids under starvation conditions. VHT1, a member of the allantoate permease family involved in biotin uptake, and YGL186C (data not shown), a member of the purine/cytosine permease family, also showed Gcn4p-dependent induction. Thus, uptake of vitamins, purines, and pyrimidines may all be induced in response to amino acid limitation.
Activation of genes encoding transcription factors.
We observed induction and repression of numerous transcription factors when cells were cultured in 3AT medium. Of the 159 known or predicted DNA-binding transcription factors in the MIPS functional catalog (69), 26 were identified as Gcn4p targets. These include regulators of amino acid and purine biosynthesis (ARG80, LEU3, LYS14, MET4, MET28, BAS1, and GLN3), TCA cycle intermediates (RTG3), and peroxisome biogenesis (PIP2), most of which are discussed above. Additional Gcn4p targets included transcriptional activators involved in utilization of poor nitrogen sources (GAT1 and UGA3), catabolism of maltose (MAL13), heat shock response (HSF1), copper homeostasis (CUP9), meiosis (RIM101), and Ty transcription (TEA1). For most of the latter, known target genes were not induced in a Gcn4p-dependent manner. Perhaps the influence of Gcn4p would be observed only under the physiological conditions that normally stimulate these activators and their target genes, e.g., by heat shocking amino acid-starved cells.
Regulation of protein kinases and phosphatases by Gcn4p.
Eleven genes in the MIPS protein kinase group were found to be Gcn4p targets, including APG1 (discussed above), DBF20, STE11, and NPR1. In nutrient-poor medium, Npr1p is active and promotes Gap1p stabilization and degradation of the tryptophan permease Tat2p (95). Perhaps induction of Npr1p by Gcn4p can augment the induction of GAP1 transcription by Gcn4p. Consistently, TAT2 expression was repressed by 3AT. Two of the three genes encoding PKA catalytic subunits, TPK1/SRA3 and TPK2, were also identified as Gcn4p targets. It was shown recently that a tpk1 mutant has reduced expression of the Gcn4p target genes BAT1 and ILV5 in YPD medium (87). Additionally, several other genes that are dependent on TPK1 or TPK2 for high-level expression (87) were identified as Gcn4p targets. Thus, Gcn4p may increase PKA expression, which in turn can promote increased expression of a subset of the genes induced in a Gcn4p-dependent manner.
Several regulatory subunits of protein phosphatases were identified as Gcn4p targets, including GIP1, PTP1, and SAP4. The essential type 1 phosphatase Glc7p has multiple roles, including stimulation of glycogen accumulation, and it was implicated previously as a negative regulator of general amino acid control (105). As Gip1p interacts with Glc7p (85), the induction of GIP1 transcription by Gcn4p could influence the negative effect of Glc7p on the general control response.
Role for Gcn4p in glycogen metabolism.
Forty-five genes encoding enzymes or regulatory proteins involved in energy generation (69) were identified as Gcn4p targets. These included GLG1, GSY1, GSY2, and GLC3, involved in the biosynthesis of the polysaccharide glycogen. Glycogen accumulates under various nutrient starvation conditions or with decreasing growth rates (25, 60), leading to the notion that it serves as a storage carbohydrate and is used as an energy source when cells resume rapid growth. Interestingly, GPH1 and GDB1/YPR184W (encoding glycogen phosphorylase and glycogen debranching enzyme, respectively), involved in the breakdown of glycogen, also were induced by Gcn4p in 3AT medium. Thus, Gcn4p seems to induce the enzymatic capacity to maintain glycogen homeostasis.
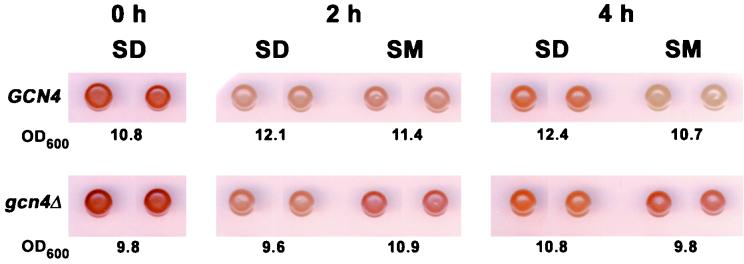
We used a semiquantitative assay based on iodine staining of intact cells to monitor glycogen levels (9) under amino acid starvation conditions. WT or gcn4Δ strains were grown to saturation in minimal (SD) medium and resuspended at the original cell density in fresh SD medium or in SD medium containing SM to impose branched-chain amino acid starvation. After 2 or 4 h of incubation, in which less than one cell doubling occurred, aliquots of cells were removed and assayed for glycogen by iodine staining. As expected, the stationary-phase cells prior to resuspension in fresh medium stained intense brown, indicating high glycogen content (Fig. 9, 0 h). Following incubation of the GCN4 cells in fresh SD medium, the glycogen levels decreased in the presence or absence of SM; however, glycogen levels were consistently higher in the gcn4Δ strain treated with SM (Fig. 9, 4 h). These results suggest that Gcn4p prevents accumulation of glycogen under amino acid starvation conditions. Presumably, inducing the catabolic enzymes encoded by GPH1 and GDB1/YPR184W overrides the simultaneous induction of glycogen anabolic enzymes by Gcn4p. We speculate that the glucose released by glycogen breakdown during amino acid starvation would stimulate amino acid precursor biosynthesis via glycolysis and early steps of the TCA cycle. In contrast to our results obtained with amino acid-starved cells, it was reported previously that glucose deprivation leads to glycogen accumulation dependent on Gcn2p (108).
FIG. 9.
Evidence that Gcn4p promotes reduced glycogen levels during amino acid starvation. Strains KNY164 (GCN4) and KNY124 (gcn4Δ) were cultured in SD medium for 40 h, harvested, washed with SD medium, resuspended in either SD medium or SD medium containing SM, and incubated with shaking at 30°C. Aliquots of cells corresponding to 3.0 OD600 units were harvested at 0, 2, and 4 h and assayed for glycogen by iodine staining for 11 min (0 h) or 14 min (2 h and 4 h). The OD600 values of the cultures at different time points are indicated.
DISCUSSION
Gcn4p is a master regulator of gene expression.
We used whole-genome expression profiling to investigate the cellular response to amino acid starvation, focusing on the role of the transcriptional activator Gcn4p. Combining data from multiple experiments, we found that histidine starvation by treatment with 100 mM 3AT led to a global alteration in gene expression, with 23% of the genome surveyed (1,400 out of 6,038 genes) being induced and 23% of the genes being repressed by a factor of 2 or more. By comparing the expression profiles among isogenic GCN4, GCN4c, and gcn4Δ strains, we determined that 3AT induction of about 60% of the 897 genes that yielded significant data in multiple experiments was dependent on Gcn4p for maximal induction. These 539 genes were designated Gcn4p targets.
We found that Gcn4p activates 78 genes encoding amino acid and purine biosynthetic enzymes (Fig. 10). There are Gcn4p targets in every amino acid biosynthetic pathway except for cysteine. We also found that Gcn4p induced four genes encoding precursors for glutamate, glutamine, or cysteine biosynthesis. Not every gene in a particular biosynthetic pathway is under Gcn4p regulation; however, enzymes that are not induced transcriptionally may still be activated allosterically in starved cells. For example, Aro7p and Trp2p catalyze the first steps in the biosynthesis of Phe/Tyr and Trp, respectively, at the branch point after the common steps in the Aro pathway (51). Gcn4p does not induce ARO7 transcription, but it does induce TRP2 (5). Interestingly, Aro7p is allosterically stimulated by tryptophan (5). Thus, Aro7p activity should be elevated as a consequence of increased Trp biosynthesis engendered by Gcn4p activation of TRP2 expression.
FIG. 10.
Schematic representation of functional categories of Gcn4p target genes. Starvation for any of several nutrients or treatment with MMS leads to activation of protein kinase Gcn2p with attendant derepression of GCN4 translation. The induced level of Gcn4p elicits transcriptional activation of at least 539 genes, designated Gcn4p targets. These genes (depicted in black) were induced ≥2-fold in at least one of the four WT ± 3AT experiments and had an induction ratio of ≥2.0 in the GCN4/gcn4Δ or GCN4c/GCN4 experiments. The numbers of Gcn4p targets in different functional categories also are indicated. A large number of genes were repressed by a factor of ≥2 in response to 3AT treatment and were dependent on Gcn4p for maximal repression under these conditions. Prominent among these repressed genes were those encoding RP and translation initiation factors (shown in gray). Gene names and functions were obtained from the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/) and the Yeast Proteome Database (12), and genes were assigned to groups based on the MIPS functional categories (69).
Sixteen vitamin or cofactor biosynthetic genes were identified as Gcn4p targets, presumably reflecting the requirement for these compounds by amino acid or purine biosynthetic enzymes. Six genes required for peroxisome proliferation and eight mitochondrial carrier proteins were identified as Gcn4p targets, most likely because of their respective roles in lysine biosynthesis and in shuttling of pathway intermediates between the mitochondria and cytoplasm (Fig. 10). Three genes involved in autophagy and two genes encoding vacuolar proteases were induced by Gcn4p, presumably to mobilize amino acid salvage pathways. The observed induction of six genes encoding amino acid transporters could provide another nonbiosynthetic means of increasing amino acid pools. We confirmed that autophagy is triggered under amino acid starvation conditions that induce Gcn4p; however, because Gcn4p was not required for the appearance of autophagic vesicles, it may only enhance the autophagy response. Ten genes encoding enzymes involved in biosynthesis or breakdown of the storage carbohydrate glycogen were identified as Gcn4p targets (Fig. 10). Interestingly, gcn4Δ cells accumulated higher levels of glycogen than did WT cells under amino acid starvation conditions, suggesting that Gcn4p induction produces a net breakdown of glycogen.
A large number of genes encoding regulatory proteins were identified as Gcn4p targets, including genes encoding transcription factors (26 genes), protein kinases (11 genes), and protein phosphatase regulatory subunits (4 genes) (Fig. 10). Thus, induction of Gcn4p may elicit a cascade of regulatory events leading to gene activation or repression and the mobilization of various signal transduction pathways. It is possible that these secondary regulatory responses require an additional environmental stimulus, in which case Gcn4p would serve to intensify (or dampen) the response when there is also amino acid limitation.
Genes whose transcription is regulated directly by Gcn4p should possess one or more Gcn4p binding sites, UASGCREs. We found that 235 of the 539 Gcn4p target genes contain a predicted Gcn4p binding site between positions −20 and −600 relative to the start codon. The remaining Gcn4p targets may contain Gcn4p sites in the coding region or 3′ untranslated region, or they may be regulated indirectly through induction of other transcription factors by Gcn4p. The latter explanation is consistent with the fact that a much higher proportion of the canonical Gcn4p targets encoding amino acid biosynthetic enzymes contain a Gcn4p binding site (∼84%), compared to the occurrence of the UASGCRE among Gcn4p target genes at large (∼44%).
Genes that were induced by Gcn4p were much more likely to contain a binding site in the 5′-proximal 300 nucleotides than were those whose maximal repression is Gcn4p dependent. Hence, Gcn4p probably does not bind to the promoters of genes repressed by 3AT. Rather, we predict that such genes contain promoter elements that function ineffectively in amino acid-starved cells. The high level of Gcn4p may squelch a transcription factor and exacerbate reduced promoter activity at these genes in starved cells.
It is remarkable that Gcn4p function is required, directly or indirectly, for activation of at least 539 genes, since microarray studies of the transcriptional response to the activators Gal4p, Yap1p, Pdr1p/Pdr3p, Mac1p, and Ace1p (10, 18, 19, 29, 63, 89) yielded only a handful of target genes. Ndt80p was implicated in activation of about 200 genes, and mutant analysis showed that most of these genes were only partially dependent on Ndt80p (10); moreover, Zap1p was implicated in the activation of 111 genes (63), still far below the number of Gcn4p target genes identified here.
Induction of Gcn4p target genes by other treatments.
While this work was in progress, Jia et al. determined the expression profile of a subset of the yeast genome (1,529 genes) in cells treated with SM, an inhibitor of branched-chain amino acid biosynthesis (49). Treatment of yeast cells with SM, like treatment with 3AT, induces GCN4 translation (36). In accordance with our findings, SM induced multiple amino acid and vitamin biosynthetic genes (49). In contrast to 3AT, however, SM repressed genes encoding enzymes of methionine and purine biosynthesis and sulfur assimilation pathways. Additionally, SM altered the expression of fewer genes in a gcn4Δ mutant (49) than did 3AT treatment of the gcn4Δ strain reported here. Additional work is required to understand the nature of the Gcn4p-independent response to 3AT treatment.
Stimulated by the previous finding that many amino acid biosynthetic genes were induced by MMS (48), we discovered that MMS induces GCN4 translation dependent on Gcn2p. Interestingly, γ-irradiation, which causes DNA damage, did not induce the Gcn4p target genes (47). Moreover, we found that induction of Gcn4p by MMS did not require the DNA damage checkpoint proteins Dun1p, Mec1p, and Rad53p. Thus, protein methylation rather than DNA damage may be the stimulus that triggers increased GCN4 translation in MMS-treated cells. On the other hand, UV irradiation stimulates the transcriptional activation function of Gcn4p, and possibly GCN4 translation, and gcn4 mutants show a modest increase in UV sensitivity (23). Hence, certain forms of DNA damage seem to activate Gcn4p at different levels, and MMS may stimulate both the activation function of Gcn4p and its synthesis.
A sizable number of Gcn4p target genes were induced by the drug rapamycin (31). Rapamycin inhibits the TOR1 and TOR2 proteins, which negatively regulate Gln3p function. Thus, rapamycin induces Gln3p target genes involved in the utilization of poor nitrogen sources (7, 31). Of the 228 genes induced twofold or more by rapamycin (31), we found that 71 (31%) are Gcn4p targets. These include certain arginine biosynthetic genes, the putative pyridoxine biosynthetic gene SNO1, and the general amino acid permease GAP1. However, none of the lysine biosynthetic genes, all prominent Gcn4p targets, were induced by rapamycin, indicating that only a fraction of Gcn4p targets are induced by rapamycin. A recent study indicates that Gcn4p is required for full induction of a subset of the genes induced by rapamycin in cells growing on rich medium, including DAL5, DAL1, and HIS3. Induction of DAL5 and DAL1 by rapamycin is also dependent on Gln3p. Interestingly, rapamycin induced GCN4 expression on rich medium to only ∼1/10 the level achieved by 3AT on minimal medium (101). The small amount of Gcn4p induced by rapamycin may be inadequate for induction of most Gcn4p target genes, but it may contribute substantially to activation of promoters containing the UASGCRE that are bound simultaneously by Gln3p.
Brown and colleagues described a set of about 868 genes that were regulated in a stereotypical manner in response to diverse stress conditions, designated the environmental stress response (ESR) (27). About 586 genes were repressed and 282 genes were induced in the ESR. Ninety-four of the genes induced in the ESR were identified here as Gcn4p targets, including ARO9, ARO10, HSP12, HSP78, CTT1, GTT1, GSY2, APG1, NPR1, and PKA1. It is possible that Gcn4p contributes to the induction of these 94 genes under stress conditions besides amino acid starvation that induce the ESR. This would be akin to the contribution of Gcn4p to induction of Gcn4p targets by rapamycin discussed above.
Gcn4p-independent response to amino acid starvation.
A sizable number of the genes induced by 3AT in the WT were not dependent on Gcn4p for this induction, showing expression ratios close to unity in the GCN4/gcn4Δ comparison and high-level induction by 3AT in a gcn4Δ mutant (Fig. 3A, sectors C and E). Thus, these genes are induced in histidine-starved cells by a transcriptional mechanism independent of Gcn4p. Interestingly, a number of Gcn4p targets also were induced by 3AT in the gcn4Δ mutant. These genes are dependent on Gcn4p for maximal induction by 3AT, showing lower induction ratios in the gcn4Δ mutant than in WT cells. Several canonical Gcn4p targets involved in amino acid biosynthesis belong to this group of genes that respond to an alternative induction pathway in histidine-starved cells, including HIS5, GLN3, ARG4, MET28, and BIO3 (Fig. 3B). ARO9, ARO10, YOR391C, and YNR069C are other Gcn4p target genes with particularly high 3AT induction ratios in the gcn4Δ mutant. It is possible that the transcriptional machinery itself is altered to permit increased transcription from certain promoters under starvation conditions independently of Gcn4p. For example, an altered form of the SAGA complex containing histone acetyltransferase Gcn5p accumulates in histidine-starved cells (4). Identification of the factor(s) mediating this Gcn4p-independent transcriptional response would shed new light on the regulatory networks that govern transcriptional control in amino acid-starved cells.
ACKNOWLEDGMENTS
We thank Steve Elledge, Orna Cohen-Fix, Ted Weinert, and Yoshinori Ohsumi for yeast strains. We thank Allan Jones and Chris Armour for coordination of the microarray experiments, including the GCN4 constitutive experiment; Tom Dever, Ronda Rolfes, Orna Cohen-Fix, Minerva Garcia, and members of the Hinnebusch laboratory for helpful discussions; and Tom Dever and Klaus Nielsen for critical reading of the manuscript. We also thank Zhixiong Xue for communicating results prior to publication. We thank the Center for Information Technology at NIH for assistance with Excel programming, and we acknowledge the Saccharomyces Genome Database and Yeast Protein Database for access to gene annotations.
REFERENCES
- 1.Albrecht G, Mosch H U, Hoffmann B, Reusser U, Braus G H. Monitoring the Gcn4 protein-mediated response in the yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:12696–12702. doi: 10.1074/jbc.273.21.12696. [DOI] [PubMed] [Google Scholar]
- 2.Arndt K, Fink G R. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5′ TGACTC 3′ sequences. Proc Natl Acad Sci USA. 1986;83:8516–8520. doi: 10.1073/pnas.83.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arndt K T, Styles C, Fink G R. Multiple global regulators control HIS4 transcription in yeast. Science. 1987;237:874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- 4.Belotserkovskaya R, Sterner D E, Deng M, Sayre M H, Lieberman P M, Berger S L. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braus G H. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol Rev. 1991;55:349–370. doi: 10.1128/mr.55.3.349-370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calder R B, Williams R S, Ramaswamy G, Rock C O, Campbell E, Unkles S E, Kinghorn J R, Jackowski S. Cloning and characterization of a eukaryotic pantothenate kinase gene (panK) from Aspergillus nidulans. J Biol Chem. 1999;274:2014–2020. doi: 10.1074/jbc.274.4.2014. [DOI] [PubMed] [Google Scholar]
- 7.Cardenas M E, Cutler N S, Lorenz M C, Di Como C J, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cashel M, Rudd K E. The stringent response. In: Neidhardt F C, Ingraham J L, Magasanik B, Low K B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1410–1438. [Google Scholar]
- 9.Chester V E. Heritable glycogen-storage deficiency in yeast and its induction by ultra-violet light. J Gen Microbiol. 1968;51:49–56. doi: 10.1099/00221287-51-1-49. [DOI] [PubMed] [Google Scholar]
- 10.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 11.Coleman S T, Tseng E, Moye-Rowley W S. Saccharomyces cerevisiae basic region-leucine zipper protein regulatory networks converge at the ATR1 structural gene. J Biol Chem. 1997;272:23224–23230. doi: 10.1074/jbc.272.37.23224. [DOI] [PubMed] [Google Scholar]
- 12.Costanzo M C, Hogan J D, Cusick M E, Davis B P, Fancher A M, Hodges P E, Kondu P, Lengieza C, Lew-Smith J E, Lingner C, Roberg-Perez K J, Tillberg M, Brooks J E, Garrels J I. The Yeast Proteome Database (YPD) and Caenorhabditis elegans Proteome Database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crabeel M, Soetens O, De Rijcke M, Pratiwi R, Pankiewicz R. The ARG11 gene of Saccharomyces cerevisiae encodes a mitochondrial integral membrane protein required for arginine biosynthesis. J Biol Chem. 1996;271:25011–25018. doi: 10.1074/jbc.271.40.25011. [DOI] [PubMed] [Google Scholar]
- 14.Daignan-Fornier B, Fink G R. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci USA. 1992;89:6746–6750. doi: 10.1073/pnas.89.15.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang V D, Valens M, Bolotin-Fukuhara M, Daignan-Fornier B. Cloning of the ASN1 and ASN2 genes encoding asparagine synthetases in Saccharomyces cerevisiae: differential regulation by the CCAAT-box-binding factor. Mol Microbiol. 1996;22:681–692. doi: 10.1046/j.1365-2958.1996.d01-1715.x. [DOI] [PubMed] [Google Scholar]
- 16.de Montigny J, Kern L, Hubert J C, Lacroute F. Cloning and sequencing of URA10, a second gene encoding orotate phosphoribosyl transferase in Saccharomyces cerevisiae. Curr Genet. 1990;17:105–111. doi: 10.1007/BF00312853. [DOI] [PubMed] [Google Scholar]
- 17.Denis V, Daignan-Fornier B. Synthesis of glutamine, glycine and 10-formyl tetrahydrofolate is coregulated with purine biosynthesis in Saccharomyces cerevisiae. Mol Gen Genet. 1998;259:246–255. doi: 10.1007/s004380050810. [DOI] [PubMed] [Google Scholar]
- 18.DeRisi J, van den Hazel B, Marc P, Balzi E, Brown P, Jacq C, Goffeau A. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 2000;470:156–160. doi: 10.1016/s0014-5793(00)01294-1. [DOI] [PubMed] [Google Scholar]
- 19.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 20.Dixon B, Rose A H. A specific requirement for biotin in the synthesis of ornithine carbamoyltransferase by yeast. Biochem J. 1966;99:513–520. doi: 10.1042/bj0990513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncan K, Edwards R M, Coggins J R. The Saccharomyces cerevisiae ARO1 gene. An example of the co-ordinate regulation of five enzymes on a single biosynthetic pathway. FEBS Lett. 1988;241:83–88. doi: 10.1016/0014-5793(88)81036-6. [DOI] [PubMed] [Google Scholar]
- 22.Ehrenshaft M, Bilski P, Li M Y, Chignell C F, Daub M E. A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc Natl Acad Sci USA. 1999;96:9374–9378. doi: 10.1073/pnas.96.16.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelberg D, Klein C, Martinetto H, Struhl K, Karin M. The UV response involving the ras signaling pathway and AP-1 transcription factors is conserved between yeast and mammals. Cell. 1994;77:381–390. doi: 10.1016/0092-8674(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 24.Feller A, Ramos F, Pierard A, Dubois E. In Saccharomyces cerevisiae, feedback inhibition of homocitrate synthase isoenzymes by lysine modulates the activation of LYS gene expression by Lys14p. Eur J Biochem. 1999;261:163–170. doi: 10.1046/j.1432-1327.1999.00262.x. [DOI] [PubMed] [Google Scholar]
- 25.Francois J, Parrou J L. Reserve carbohydrate metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:125–145. doi: 10.1111/j.1574-6976.2001.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Barrio M, Dong J, Ufano S, Hinnebusch A G. Association of GCN1/GCN20 regulatory complex with the conserved N-terminal domain of eIF2α kinase GCN2 is required for GCN2 activation in vivo. EMBO J. 2000;19:1887–1899. doi: 10.1093/emboj/19.8.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasch A P, Spellman P T, Kao C M, Carmel-Harel O, Eisen M B, Storz G, Botstein D, Brown P O. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geraghty M T, Bassett D, Morrell J C, Gatto G J, Jr, Bai J, Geisbrecht B V, Hieter P, Gould S J. Detecting patterns of protein distribution and gene expression in silico. Proc Natl Acad Sci USA. 1999;96:2937–2942. doi: 10.1073/pnas.96.6.2937. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Gross C, Kelleher M, Iyer V R, Brown P O, Winge D R. Identification of the copper regulon in Saccharomyces cerevisiae by DNA microarrays. J Biol Chem. 2000;275:32310–32316. doi: 10.1074/jbc.M005946200. [DOI] [PubMed] [Google Scholar]
- 30.Halliwell B, Gutteridge J M C. Free radicals in biology and medicine. 2nd ed. Oxford, United Kingdom: Clarendon Press; 1989. [Google Scholar]
- 31.Hardwick J S, Kuruvilla F G, Tong J K, Shamji A F, Schreiber S L. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haselbeck R J, McAlister-Henn L. Function and expression of yeast mitochondrial NAD- and NADP-specific isocitrate dehydrogenases. J Biol Chem. 1993;268:12116–12122. [PubMed] [Google Scholar]
- 33.Hernando Y, Carter A T, Parr A, Hove-Jensen B, Schweizer M. Genetic analysis and enzyme activity suggest the existence of more than one minimal functional unit capable of synthesizing phosphoribosyl pyrophosphate in Saccharomyces cerevisiae. J Biol Chem. 1999;274:12480–12487. doi: 10.1074/jbc.274.18.12480. [DOI] [PubMed] [Google Scholar]
- 34.Hill D E, Hope I A, Macke J P, Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986;234:451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- 35.Hinnebusch A G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci USA. 1984;81:6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinnebusch A G. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae. In: Broach J R, Jones E W, Pringle J R, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 319–414. [Google Scholar]
- 37.Hinnebusch A G. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 199–244. [Google Scholar]
- 38.Hinnebusch A G. Translational regulation of yeast GCN4: a window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 39.Hinnebusch A G. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 185–243. [Google Scholar]
- 40.Hsu Y P, Kohlhaw G B, Niederberger P. Evidence that α-isopropylmalate synthase of Saccharomyces cerevisiae is under the “general” control of amino acid biosynthesis. J Bacteriol. 1982;150:969–972. doi: 10.1128/jb.150.2.969-972.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes J D, Estep P W, Tavazoie S, Church G M. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- 42.Hughes T R, Mao M, Jones A R, Buchard J, Marton M J, Shannon K W, Lefkowitz S M, Ziman M, Schelter J M, Meyer M R, Kobayashi S, Dai H, He Y D, Stephaniants S B, Cavet G, Walker W L, West A, Coffey E, Shoemaker D D, Stoughton R, Blanchard A P, Friend S H, Linsley P S. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 43.Hughes T R, Marton M J, Jones A R, Roberts C J, Stoughton R, Armour C D, Bennett H A, Coffey E, Dai H, He Y D, Kidd M J, King A M, Meyer M R, Slade D, Lum P Y, Stepaniants S B, Shoemaker D D, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend S H. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 44.Hughes T R, Roberts C J, Dai H, Jones A R, Meyer M R, Slade D, Burchard J, Dow S, Ward T R, Kidd M J, Friend S H, Marton M J. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 45.Iraqui I, Vissers S, Andre B, Urrestarazu A. Transcriptional induction by aromatic amino acids in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:3360–3371. doi: 10.1128/mcb.19.5.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iraqui I, Vissers S, Cartiaux M, Urrestarazu A. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol Gen Genet. 1998;257:238–248. doi: 10.1007/s004380050644. [DOI] [PubMed] [Google Scholar]
- 47.Jelinsky S A, Estep P, Church G M, Samson L D. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol Cell Biol. 2000;20:8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jelinsky S A, Samson L D. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia M H, Larossa R A, Lee J M, Rafalski A, Derose E, Gonye G, Xue Z. Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol Genomics. 2000;3:83–92. doi: 10.1152/physiolgenomics.2000.3.2.83. [DOI] [PubMed] [Google Scholar]
- 50.Jones D G, Reusser U, Braus G H. Molecular cloning, characterization and analysis of the regulation of the ARO2 gene, encoding chorismate synthase, of Saccharomyces cerevisiae. Mol Microbiol. 1991;5:2143–2152. doi: 10.1111/j.1365-2958.1991.tb02144.x. [DOI] [PubMed] [Google Scholar]
- 51.Jones E W, Fink G R. Regulation of amino acid and nucleotide biosynthesis in yeast. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 181–300. [Google Scholar]
- 52.Kaffman A, O'Shea E K. Regulation of nuclear localization: a key to a door. Annu Rev Cell Dev Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- 53.Karpichev I V, Luo Y, Marians R C, Small G M. A complex containing two transcription factors regulates peroxisome proliferation and the coordinate induction of β-oxidation enzymes in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:69–80. doi: 10.1128/mcb.17.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein C, Struhl K. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol Cell Biol. 1994;14:1920–1928. doi: 10.1128/mcb.14.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klionsky D J, Emr S D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kucharczyk R, Zagulski M, Rytka J, Herbert C J. The yeast gene YJR025c encodes a 3-hydroxyanthranilic acid dioxygenase and is involved in nicotinic acid biosynthesis. FEBS Lett. 1998;424:127–130. doi: 10.1016/s0014-5793(98)00153-7. [DOI] [PubMed] [Google Scholar]
- 57.Kuenzler M, Balmelli T, Egli C M, Paravicini G, Braus G H. Cloning, primary structure, and regulation of the HIS7 gene encoding a bifunctional glutamine amidotransferase:cyclase from Saccharomyces cerevisiae. J Bacteriol. 1993;175:5548–5558. doi: 10.1128/jb.175.17.5548-5558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lanker S, Bushman J L, Hinnebusch A G, Trachsel H, Mueller P P. Autoregulation of the yeast lysyl-tRNA synthetase gene GCD5/KRS1 by translational and transcriptional control mechanisms. Cell. 1992;70:647–657. doi: 10.1016/0092-8674(92)90433-d. [DOI] [PubMed] [Google Scholar]
- 59.Li W, Brandriss M C. Proline biosynthesis in Saccharomyces cerevisiae: molecular analysis of the PRO1 gene, which encodes γ-glutamyl kinase. J Bacteriol. 1992;174:4148–4156. doi: 10.1128/jb.174.12.4148-4156.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lillie S H, Pringle J R. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980;143:1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J Q, Nagata S, Dairi T, Misono H, Shimizu S, Yamada H. The GLY1 gene of Saccharomyces cerevisiae encodes a low-specific L-threonine aldolase that catalyzes cleavage of L-allo-threonine and L-threonine to glycine—expression of the gene in Escherichia coli and purification and characterization of the enzyme. Eur J Biochem. 1997;245:289–293. doi: 10.1111/j.1432-1033.1997.00289.x. [DOI] [PubMed] [Google Scholar]
- 62.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 63.Lyons T J, Gasch A P, Gaither L A, Botstein D, Brown P O, Eide D J. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc Natl Acad Sci USA. 2000;97:7957–7962. doi: 10.1073/pnas.97.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Magasanik B. Regulation of nitrogen utilization. In: Broach J R, Jones E W, Pringle J R, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 283–317. [Google Scholar]
- 65.Marton M J, Crouch D, Hinnebusch A G. GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol Cell Biol. 1993;13:3541–3556. doi: 10.1128/mcb.13.6.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marton M J, DeRisi J L, Bennett H A, Iyer V, Meyer M R, Roberts C J, Stoughton R, Burchard J, Slade D, Dai H, Bassett D E, Hartwell L H, Brown P O, Friend S H. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 67.McCormick D B, Chen H. Update on interconversions of vitamin B-6 with its coenzyme. J Nutr. 1999;129:325–327. doi: 10.1093/jn/129.2.325. [DOI] [PubMed] [Google Scholar]
- 68.Melcher K, Rose M, Kunzler M, Braus G H, Entian K D. Molecular analysis of the yeast SER1 gene encoding 3-phosphoserine aminotransferase: regulation by general control and serine repression. Curr Genet. 1995;27:501–508. doi: 10.1007/BF00314439. [DOI] [PubMed] [Google Scholar]
- 69.Mewes H W, Hani J, Pfeiffer F, Frishman D. MIPS: a database for protein sequences and complete genomes. Nucleic Acids Res. 1998;26:33–37. doi: 10.1093/nar/26.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mitchell A P, Magasanik B. Three regulatory systems control production of glutamine synthetase in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2767–2773. doi: 10.1128/mcb.4.12.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moehle C M, Hinnebusch A G. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mosch H U, Scheier B, Lahti R, Mantsala P, Braus G H. Transcriptional activation of yeast nucleotide biosynthetic gene ADE4 by GCN4. J Biol Chem. 1991;266:20453–20456. [PubMed] [Google Scholar]
- 73.Mountain H A, Bystrom A S, Korch C. The general amino acid control regulates MET4, which encodes a methionine-pathway-specific transcriptional activator of Saccharomyces cerevisiae. Mol Microbiol. 1993;9:221–223. doi: 10.1111/j.1365-2958.1993.tb01684.x. [DOI] [PubMed] [Google Scholar]
- 74.Mueller P P, Hinnebusch A G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986;45:201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- 75.O'Connell K F, Surdin-Kerjan Y, Baker R E. Role of the Saccharomyces cerevisiae general regulatory factor CP1 in methionine biosynthetic gene transcription. Mol Cell Biol. 1995;15:1879–1888. doi: 10.1128/mcb.15.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ohsumi Y. Molecular mechanism of autophagy in yeast, Saccharomyces cerevisiae. Philos Trans R Soc Lond B Biol Sci. 1999;354:1577–1580. doi: 10.1098/rstb.1999.0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oliphant A R, Brandl C J, Struhl K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Mol Cell Biol. 1989;9:2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ono B I, Hazu T, Yoshida S, Kawato T, Shinoda S, Brzvwczy J, Paszewski A. Cysteine biosynthesis in Saccharomyces cerevisiae: a new outlook on pathway and regulation. Yeast. 1999;15:1365–1375. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1365::AID-YEA468>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 79.Osmani A H, May G S, Osmani S A. The extremely conserved pyroA gene of Aspergillus nidulans is required for pyridoxine synthesis and is required indirectly for resistance to photosensitizers. J Biol Chem. 1999;274:23565–23569. doi: 10.1074/jbc.274.33.23565. [DOI] [PubMed] [Google Scholar]
- 80.Padilla P A, Fuge E K, Crawford M E, Errett A, Werner-Washburne M. The highly conserved, coregulated SNO and SNZ gene families in Saccharomyces cerevisiae respond to nutrient limitation. J Bacteriol. 1998;180:5718–5726. doi: 10.1128/jb.180.21.5718-5726.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Palmieri L, Vozza A, Agrimi G, De Marco V, Runswick M J, Palmieri F, Walker J E. Identification of the yeast mitochondrial transporter for oxaloacetate and sulfate. J Biol Chem. 1999;274:22184–22190. doi: 10.1074/jbc.274.32.22184. [DOI] [PubMed] [Google Scholar]
- 82.Paulsen I T, Sliwinski M K, Nelissen B, Goffeau A, Saier M H., Jr Unified inventory of established and putative transporters encoded within the complete genome of Saccharomyces cerevisiae. FEBS Lett. 1998;430:116–125. doi: 10.1016/s0014-5793(98)00629-2. [DOI] [PubMed] [Google Scholar]
- 83.Platt A, Reece R J. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J. 1998;17:4086–4091. doi: 10.1093/emboj/17.14.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ptashne M, Gann Alexander A F. Activators and targets. Nature. 1990;346:329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- 85.Ramaswamy N T, Li L, Khalil M, Cannon J F. Regulation of yeast glycogen metabolism and sporulation by Glc7p protein phosphatase. Genetics. 1998;149:57–72. doi: 10.1093/genetics/149.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramos F, Dubois E, Pierard A. Control of enzyme synthesis in the lysine biosynthetic pathway of Saccharomyces cerevisiae. Eur J Biochem. 1988;171:171–176. doi: 10.1111/j.1432-1033.1988.tb13773.x. [DOI] [PubMed] [Google Scholar]
- 87.Robertson L S, Causton H C, Young R A, Fink G R. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc Natl Acad Sci USA. 2000;97:5984–5988. doi: 10.1073/pnas.100113397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rolfes R J, Hinnebusch A G. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol Cell Biol. 1993;13:5099–5111. doi: 10.1128/mcb.13.8.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roth F P, Hughes J D, Estep P W, Church G M. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- 90.Rottensteiner H, Kal A J, Filipits M, Binder M, Hamilton B, Tabak H F, Ruis H. Pip2p: a transcriptional regulator of peroxisome proliferation in the yeast Saccharomyces cerevisiae. EMBO J. 1996;15:2924–2934. [PMC free article] [PubMed] [Google Scholar]
- 91.Rouillon A, Barbey R, Patton E E, Tyers M, Thomas D. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30) complex. EMBO J. 2000;19:282–294. doi: 10.1093/emboj/19.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roussou I, Thireos G, Hauge B M. Transcriptional-translational regulatory circuit in Saccharomyces cerevisiae which involves the GCN4 transcriptional activator and the GCN2 protein kinase. Mol Cell Biol. 1988;8:2132–2139. doi: 10.1128/mcb.8.5.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schena M, Shalon D, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 94.Schmidheini T, Mosch H U, Graf R, Braus G H. A GCN4 protein recognition element is not sufficient for GCN4-dependent regulation of transcription in the ARO7 promoter of Saccharomyces cerevisiae. Mol Gen Genet. 1990;224:57–64. doi: 10.1007/BF00259451. [DOI] [PubMed] [Google Scholar]
- 95.Schmidt A, Beck T, Koller A, Kunz J, Hall M N. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sherman F, Fink G R, Lawrence C W. Methods of yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1974. [Google Scholar]
- 97.Springer C, Kunzler M, Balmelli T, Braus G H. Amino acid and adenine cross-pathway regulation act through the same 5′-TGACTC-3′ motif in the yeast HIS7 promoter. J Biol Chem. 1996;271:29637–29643. doi: 10.1074/jbc.271.47.29637. [DOI] [PubMed] [Google Scholar]
- 98.Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tice-Baldwin K, Fink G R, Arndt K T. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science. 1989;246:931–935. doi: 10.1126/science.2683089. [DOI] [PubMed] [Google Scholar]
- 100.Urrestarazu A, Vissers S, Iraqui I, Grenson M. Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination. Mol Gen Genet. 1998;257:230–237. doi: 10.1007/s004380050643. [DOI] [PubMed] [Google Scholar]
- 101.Valenzuela L, Aranda C, Gonzalez A. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J Bacteriol. 2001;183:2331–2334. doi: 10.1128/JB.183.7.2331-2334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Valenzuela L, Ballario P, Aranda C, Filetici P, Gonzalez A. Regulation of expression of GLT1, the gene encoding glutamate synthase in Saccharomyces cerevisiae. J Bacteriol. 1998;180:3533–3540. doi: 10.1128/jb.180.14.3533-3540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vazquez de Aldana C R, Marton M J, Hinnebusch A G. GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2α kinase GCN2 in amino acid-starved cells. EMBO J. 1995;14:3184–3199. doi: 10.1002/j.1460-2075.1995.tb07321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weinert T. DNA damage checkpoints update: getting molecular. Curr Opin Genet Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- 105.Wek R C, Cannon J F, Dever T E, Hinnebusch A G. Truncated protein phosphatase GLC7 restores translational activation of GCN4 expression in yeast mutants defective for the eIF-2α kinase GCN2. Mol Cell Biol. 1992;12:5700–5710. doi: 10.1128/mcb.12.12.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wek R C, Ramirez M, Jackson B M, Hinnebusch A G. Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol Cell Biol. 1990;10:2820–2831. doi: 10.1128/mcb.10.6.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wek S A, Zhu S, Wek R C. The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15:4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang R, Wek S A, Wek R C. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol Cell Biol. 2000;20:2706–2717. doi: 10.1128/mcb.20.8.2706-2717.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zaman Z, Bowman S B, Kornfeld G D, Brown A J, Dawes I W. Transcription factor GCN4 for control of amino acid biosynthesis also regulates the expression of the gene for lipoamide dehydrogenase. Biochem J. 1999;340:855–862. [PMC free article] [PubMed] [Google Scholar]
- 109a.Zhou K, Brisco P R G, Hinkkanen A E, Kohlhaw G B. Structure of yeast regulatory gene LEU3 and evidence that LEU3 itself is under general amino acid control. Nucleic Acids Res. 1987;15:5261–5273. doi: 10.1093/nar/15.13.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhu S, Sobolev A Y, Wek R C. Histidyl-tRNA synthetase-related sequences in GCN2 protein kinase regulate in vitro phosphorylation of eIF-2. J Biol Chem. 1996;271:24989–24994. doi: 10.1074/jbc.271.40.24989. [DOI] [PubMed] [Google Scholar]