Abstract
Estrogen-dependent recruitment of coactivators by estrogen receptor alpha (ERα) represents a crucial step in the transcriptional activation of target genes. However, studies of the function of individual coactivators has been hindered by the presence of endogenous coactivators, many of which are potentially recruited in the presence of agonist via a common mechanism. To circumvent this problem, we have generated second-site suppressor mutations in the nuclear receptor interaction domain of p160 coactivators which rescue their binding to a transcriptionally defective ERα that is refractory to wild-type coactivators. Analysis of these altered-specificity receptor-coactivator combinations, in the absence of interference from endogenous coregulators, indicated that estrogen-dependent transcription from reporter genes is critically dependent on direct recruitment of a p160 coactivator in mammalian cells and that the three p160 family members serve functionally redundant roles. Furthermore, our results suggest that such a change-of-specificity mutation may act as a transposable protein-protein interaction module which provides a novel tool with which to dissect the functional roles of other nuclear receptor coregulators at the cellular level.
Estrogen receptor alpha (ERα) is a ligand-inducible transcription factor which belongs to the nuclear receptor superfamily (10, 25). Upon binding to its natural ligand, 17β-estradiol, activated ERα has been proposed to recruit a number of putative coactivators which lead to transcriptional activation through physical or enzymatic modification of local chromatin structure and recruitment of the basal transcription machinery at target gene promoters (13, 28). Recruitment of coactivators is mediated by two distinct transcriptional activation domains (ADs): ligand-independent AF1 at the N terminus and ligand-dependent AF2 at the C terminus, which is encompassed by the ligand binding domain (LBD) (8, 37). A large number of putative coactivators which are capable of binding nuclear receptors in a ligand-dependent manner have been isolated through a variety of genetic and biochemical methods. Among them are the p160 family of coactivators, SRC1, TIF2/GRIP1, and RAC3/AIB1/ACTR/p/CIP (14, 27). Together with CBP/p300 and P/CAF, they form a subgroup of nuclear receptor coregulators which possess histone acetyltransferase activity. Several other functionally distinct nuclear receptor coregulators include the TRAP/DRIP complexes (24), TIF1α, PGC-1, SRA (14, 27), and ASC-2/RAP250/NRC1 (4, 19, 22).
A common feature of most, if not all, putative nuclear receptor coactivators is the presence of one or more copies of the LXXLL motif (where L stands for leucine and X is any amino acid), a signature sequence which confers agonist-dependent binding to nuclear receptors (15, 18, 38). From crystallographical studies, the LXXLL motif was shown to be encompassed in a two-turn, amphipathic α-helical structure which docks to a hydrophobic groove on the surface of agonist-bound nuclear receptor LBDs (9, 29, 34). Notably, the coactivator docking sites, which formally define AF2 of ERα, PPARγ, and TRβ, appear to share striking similarity and this conservation is likely to extend to other members of the nuclear receptor superfamily, as predicted by sequence and structural comparisons (41, 43). Although a number of features at the receptor-coactivator interface had been noted which may confer binding specificity to isolated LXXLL-containing α-helices (9, 11, 23, 26), preferential binding of a given coactivator to a single nuclear receptor is rarely observed in the context of full-length protein. Given the common mechanism of receptor-coregulator interaction, it has been difficult to assign specific functional roles to a designated coregulator in nuclear receptor transactivation in mammalian cell culture systems.
We are particularly interested in determining the relative importance of putative coactivators in ERα transactivation. It has been reported that exogenous expression of p160 coactivators, CBP/p300, ASC-2/RAP250/NRC1, or PGC-1 potentiates the ability of ERα to stimulate transcription from reporter genes (6, 17, 19, 36, 40). On the other hand, there is evidence that the TRAP/DRIP complex is also involved in mediating nuclear receptor transactivation (12, 32). Notably, the TRAP220/DRIP205 component, which possesses two LXXLL motifs, is thought to anchor the complex to agonist-bound nuclear receptors, including ERα (3, 31, 47, 48).
Our overall goal was to examine the ability of specific p160 family members to mediate transcription by ERα in the absence of interference from endogenous coactivators. In mammalian cells, endogenous coactivators are usually sufficient to support estrogen-dependent transcriptional activation of reporter genes. As a result, it is not feasible to determine whether exogenously expressed coactivators potentiate ERα transactivation by direct interaction or in combination with endogenous coregulators which are already in direct contact with the receptor. Through genetic selection in yeast, we isolated a mutant SRC1 which is capable of interacting with mERα V380H, a transcriptionally defective receptor refractory to wild-type coactivators. By using this altered-specificity receptor-coactivator pair, we demonstrated that ERα transactivation is dependent upon direct recruitment of SRC1 and its subsequent interaction with CBP/p300 in mammalian cells. Furthermore, we obtained evidence that all p160 coactivator family members serve redundant functions by examining mutant versions of TIF2 and RAC3 which carry the same altered-specificity mutation.
MATERIALS AND METHODS
Plasmids. (i) mERα.
The point mutation V380H in the mouse ERα (mERα) LBD was introduced by recombinant PCR using PfuTurbo DNA polymerase (Stratagene). A PCR fragment was introduced into plasmid pSP6MORK (8) digested with NdeI and BglII. The full-length mutant receptor was subsequently subcloned into pSG5 as an EcoRI fragment, designated pSG5 MORK V380H, for transient transfection. The LBD of mERα V380H was fused to the Gal4 DNA binding domain (DBD) by subcloning an XbaI restriction fragment from pSG5 MORK V380H into pSG-Gal MORK (23). The LBD of the wild-type receptor and the V380H mutant receptor was fused to the Gal4 AD by cloning PCR fragments encompassing Ser313 and Ile599 of mERα into pGAD424 (Clontech) digested with EcoRI and BamHI.
(ii) hRARα.
To generate the construct pSG-Gal hRARα LBD, a PCR fragment encompassing Ser154 to Pro462 of human retinoic acid receptor alpha (hRARα) was cloned into pSG-Gal digested with EcoRI and BglII. The point mutation I258H was introduced by recombinant PCR using PfuTurbo DNA polymerase (Stratagene). A PCR fragment containing the mutation was inserted into pSG-Gal hRARα LBD which had been digested with SacI and SmaI.
(iii) SRC1.
The construct pSG5 SRC1e m13, in which the first and third LXXLL motifs had been mutated to LXXAA, was described previously (17). An XhoI site was introduced at nucleotide position 2040 by recombinant PCR in order to generate the construct pSG5 SRCX1e m13 (where X denotes the new XhoI site). A double-FLAG epitope tag was introduced into the N terminus of SRC1e by transferring an SmaI-CelII fragment from pSG5 FLAG SRC1e (E. Kalkhoven, unpublished data) into pSG5 SRCX1e m13 to give pSG5 FLAG SRCX1e m13. In order to generate pGBDU-SRCX1 m13 RID, in which the SRC1 m13 receptor interaction domain encompassing Pro570 and Asp782 was fused to the Gal4 DBD, a PCR fragment was subcloned into pGBDU-C1 (a gift from P. James; 16) which had been digested with EcoRI and SalI. The VHC mutation was transferred from the yeast library construct into full-length SRC1e by subcloning an XhoI-EcoRV fragment into pSG5 FLAG SRCX1e m13 to generate pSG5 FLAG SRCX1e VHC. The point mutation 1689A was introduced into pSG5 FLAG SRCX1e VHC by subcloning an XhoI-EcoRV recombinant PCR fragment. The construct pSG5 FLAG SRCX1e VHC ΔAD1 was generated by subcloning a BamHI-MscI restriction fragment from pSG5 SRC1e ΔAD1 which lacked residues 900 to 950 (1).
(iv) TIF2.
The construct pSG5 TIF2 m123, in which all three LXXLL motifs had been mutated to LXXAA, was a gift from H. Gronemeyer. Full-length TIF2 m123 was subcloned into pSG-FLAG (a vector based on pSG5; B. Belandia, unpublished data) where an N-terminal FLAG epitope tag was placed in frame with the TIF2 open reading frame to give pSG FLAG TIF2 m123. The second LXXLL motif was reverted back to the wild type to give pSG FLAG TIF2 m13 by subcloning a PstI-digested recombinant PCR fragment. The VHC mutation was introduced into pSG FLAG TIF2 m13 by subcloning a PstI-digested recombinant PCR fragment in order to generate the construct pSG FLAG TIF2 VHC.
(v) RAC3.
The construct pCMX-F.RAC3 was a gift from J. D. Chen. Full-length RAC3 was subcloned into pSG-FLAG, where an N-terminal FLAG epitope tag was placed in frame with the RAC3 open reading frame to give pSG FLAG RAC3. Mutations of the LXXLL motifs to LXXAA, either individually or in all possible combinations, were generated by recombinant PCR and subcloned into pSG FLAG RAC3 as either HindIII-SpeI or XhoI-SpeI fragments. The new XhoI site at nucleotide position 1881 was introduced during mutagenesis of LXXLL motif 1. The VHC mutation was introduced into pSG FLAG RAC3 m13 by subcloning an XhoI-SpeI recombinant PCR fragment to produce pSG FLAG RAC3 VHC.
Library construction and yeast two-hybrid screening.
A library was constructed based on plasmid pGBDU-SRC1 m13 RID, in which the codons for Leu690 and Leu694 were randomized by using the QuickChange Site-Directed Mutagenesis kit (Stratagene) with complementary primers 5′-cagaacggcataaaattnnscaccggctcnnscaggagggtagcccctcag-3′. Escherichia coli strain DH5α was transformed by electroporation with mutated plasmids, and 2,800 independent colonies were harvested from which the library DNA was prepared. Sequencing of randomly selected clones indicated that approximately 80% of the library contained the targeted mutations.
The yeast two-hybrid screening was performed by using strain PJ69-4A (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ) (16) transformed with pGAD mERα V380H and the mutant library. We selected 59,000 transformants on plates with synthetic medium lacking uracil, leucine, and histidine and containing 100 nM 17β-estradiol and 5 mM 3-aminotriazole in order to suppress any spontaneous activation of the HIS3 reporter gene by the Gal4 DBD fusion proteins of the library. One colony was recovered, and the ligand-dependent interaction between the putative clone and mERα V380H was verified by growth on plates with synthetic medium lacking uracil, leucine, and adenine, either in the absence or in the presence of 100 nM 17β-estradiol.
Cell culture and transient-transfection experiments.
HeLa, COS-1, and 293-T cells were routinely maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS; Gibco BRL). For transient-transfection reporter assays, HeLa and COS-1 cells were plated in 96-well microtiter plates in phenol red-free DMEM containing 5% charcoal-dextran-stripped FBS. Cells were transfected by calcium phosphate coprecipitation as described earlier (8). For each individual well, the transfected DNA included a pRL-CMV control plasmid (0.5 ng; Promega); a p2×ERE-TATA-GL3, p2×ERE-pS2-GL3, or p5Gal-E1B-GL3 reporter (10 ng); and a pSG5-based expression plasmid encoding either full-length mERα or a Gal4 fusion of mERα (2 ng) plus or minus designated coactivators (15 ng). A constant amount of DNA was maintained in each well with an appropriate amount of the pSG5-based expression vector. After 16 h, the cells were washed and then maintained in medium containing 5% charcoal-dextran-stripped FBS and phenol red-free DMEM in the presence or absence of ligand for 24 h. Subsequently, cells were harvested and extracts were assayed for luciferase activity with the LucLite luciferase reporter assay kit (Packard) and for Renilla luciferase activity by using 250 ng of Coelenterazine (dissolved in dimethyl sulfoxide and diluted in 0.5 M HEPES [pH 7.8]–40 mM EDTA) (Calbiochem) per well as the reaction substrate. The Renilla luciferase activity was used to correct for differences in transfection efficiency.
For transient transfection of 293-T cells, a calcium phosphate coprecipitation method (Profection; Promega) was used in accordance with the manufacturer's protocol. For each 10-cm-diameter dish, 20 μg of a pSG5-based expression plasmid was transfected (12 μg for wild-type or mutant mERα and 8 μg for designated coactivators).
Immunoprecipitation and Western blot analysis.
After 24 h of incubation, 293-T cells transfected with combinations of wild-type or mutant mERα and p160 coactivators were lysed by using buffer A (20 mM Tris-HCl [pH 8], 75 mM NaCl, 5 mM EDTA [pH 8], 1% Nonidet P-40) which contained a cocktail of protease inhibitors (Roche). For each 10-cm-diameter culture dish, 1 ml of buffer A was used. The crude lysate was centrifuged at 10,000 × g for 20 min at 4°C, and the supernatant was precleared by incubation with protein G-Sepharose (Pharmacia) for 30 min at 4°C. The cleared lysate was divided into 400-μl aliquots and subjected to immunoprecipitation with the addition of 25 μg of Anti-FLAG M2 agarose (Kodak) plus or minus 17β-estradiol (1 μM final concentration) in a total volume of 1 ml. After incubation at 4°C for 5 h, agarose beads were washed four times with buffer A and once with phosphate-buffered saline. The immunoprecipitated complexes were eluted by boiling in sodium dodecyl sulfate gel loading buffer.
The protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane. For detection of mERα, monoclonal antibody H222 (a gift of Geoff Greene) was used at a 1:2,000 dilution. For detection of the FLAG epitope, an anti-FLAG M2 monoclonal antibody (Kodak) was used at a 1:1,000 dilution. This was followed by a horseradish peroxidase-conjugated secondary antibody (DAKO) at a 1:3,000 dilution. Bound antibodies were visualized with ECL reagent (Amersham).
RESULTS
Experimental design.
To investigate the functional consequence of a direct interaction between p160 coactivators and ERα in mammalian cells, we first generated mutant receptors which were transcriptionally defective by disrupting the coactivator interaction surface. This was followed by directed genetic selection for altered-specificity SRC1 mutants that were capable of binding the mutant receptors. The ERα mutants were unable to activate reporter genes presumably because they could not interact with endogenous coactivators (Fig. 1A). We then wished to determine whether transcriptional activation would be conditional upon exogenous expression of altered-specificity SRC1. Rescue of transcriptional activity would suggest that direct recruitment of SRC1 is required for ERα transactivation, whereas failure to do so would imply either that a direct interaction between ERα and SRC1 is not crucial or that transcription must be dependent on the recruitment of other coactivators to the AF2 surface of ERα.
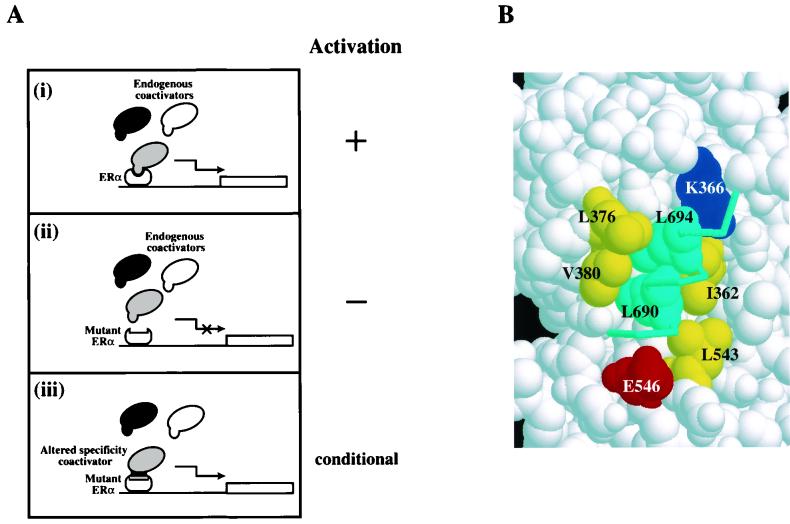
FIG. 1.
Estrogen-dependent gene activation through ERα-coactivator interaction. (A) Model for gene activation by wild-type and mutant ERα. (i) In mammalian cells, wild-type ERα interacts with endogenous coactivators in a ligand-dependent manner to activate the transcription of a reporter gene. Different shadings represent distinct species of coactivators which are capable of interacting with agonist-bound ERα and whose relative functional importance is unresolved. (ii) Disruption of the coactivator interaction surface of ERα prevents its binding to any endogenous coactivators. The mutant ERα is therefore unable to activate transcription. (iii) Conditional gene activation may be achieved by coexpression of the mutant ERα with its altered-specificity coactivator partner on the assumption that the coactivator is directly recruited by ERα under physiological conditions. (B) A close-up view of the agonist-bound hERα-GRIP1 NR box II peptide cocrystal structure showing the receptor-peptide interface. The residues that form the coactivator-interacting surface in the receptor moiety are yellow (hydrophobic), red (acidic), and blue (basic) and are numbered as in mERα. The peptide is in cyan. The two leucine residues, in close contact with V380 of mERα, are shown in space-filled mode to highlight the interaction. The model was generated by RasMol and was based on the coordinates under Protein Data Bank entry code 3ERD.
Mutant mERα impaired for interaction with transcriptional coactivators.
The molecular determinants of the mERα-coactivator interface have been established in biochemical and crystallographic studies, and selected residues which mediate the protein-protein interactions are highlighted in Fig. 1B (2, 23, 34). We have previously analyzed the role of V380, a conserved residue on the surface of mERα LBD, in coactivator binding. While the V380D mutant receptor failed to bind SRC1e, binding by the V380A mutant receptor was unaffected. In contrast, replacement of L543 with alanine was sufficient to abolish coactivator interaction (23). This prompted us to conclude that V380 is not essential at the mERα coactivator-interacting surface and mutations at V380 which abolish coactivator binding might be more amenable to complementation.
We generated one additional mutant receptor, V380H, which satisfied the criteria for potential complementation by altered-specificity SRC1e. V380H was unable to interact with SRC1e in vivo and in vitro and displayed greatly reduced transcriptional activity when transiently transfected into mammalian cells (see below). The structural integrity of the mutant receptor was demonstrated by (i) normal binding affinity for 17β-estradiol (Kd = 0.87 nM for V380H and 0.33 nM for the wild-type receptor) and (ii) binding to a consensus estrogen response element from the vitellogenin A2 promoter (data not shown). Furthermore, V380H was expressed at a similar level in 293-T cells compared with the wild-type control (see below). Taken together, the results show that the tertiary structure of the V380H mutant remained intact and its reduced transcriptional activity could be attributed to an impairment in coactivator recruitment.
Screen for an altered-specificity mutant of SRC1e capable of interacting with mERα mutants.
The crystal structure of the agonist-bound human ERα LBD complexed with the GRIP1 nuclear receptor box II peptide indicates that V376 of human ERα (which corresponds to V380 of mERα) interdigitates with L690 and L694 of GRIP1 (34). We assumed that similar van der Waals contacts exist between V380 of mERα and L690 and L694 of SRC1e since the residues which constitute the receptor-coactivator interface are highly conserved (Fig. 1B). Furthermore, the failure of both V380D and V380H mutant receptors to bind wild-type SRC1e was most likely due to disruption of these contacts. In order to isolate altered-specificity SRC1e mutants capable of interacting with mERα V380D or V380H, we randomly mutated SRC1e at L690 and L694, which form part of the second LXXLL motif. The second LXXLL motif was chosen because it was shown to preferentially interact with mERα (17, 23). The mutant library was based on a construct encompassing the entire receptor-interacting domain of SRC1e, where the first and third LXXLL motifs were rendered nonfunctional by mutation to LXXAA (SRC1 m13) (Fig. 2A). This ensured that interaction with the mutant receptors would be restored solely by mutations based on the second LXXLL motif and not by cooperation with a wild-type motif. It also justified the use of SRC1 m13 as a wild-type reference for receptor-coactivator interaction and function in subsequent experiments.
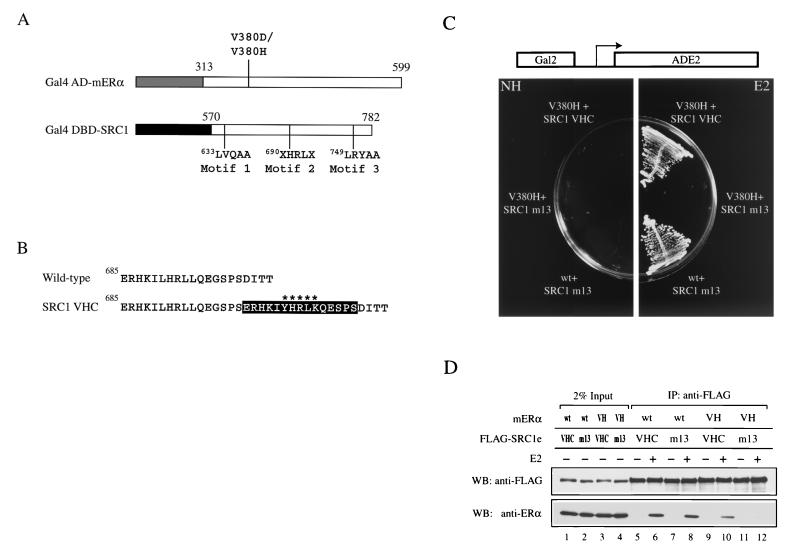
FIG. 2.
Altered-specificity SRC1e. (A) Schematic representation of constructs used in the yeast two-hybrid screen for SRC1 mutants which suppress mutations in V380 of mERα. The numbers indicate amino acid positions in the full-length protein. The letter X represents any amino acids and signifies the two randomized positions. In addition, LXXLL motifs 1 and 3 of SRC1 were rendered nonfunctional by mutation to LXXAA and the construct was denoted by the suffix m13. The lightly shaded box represents the AD of Gal4 (amino acids 768 to 881), and the darkly shaded box represents the DBD of Gal4 (amino acids 1 to 147). (B) Sequence comparison of wild-type SRC1 and SRC1 VHC. The black box encompasses the 15-amino-acid insertion found immediately C terminal to the wild-type LXXLL motif. The variant motif YXXLK is marked with asterisks. (C) Ligand-dependent interaction of mERα V380H with SRC1 VHC in vivo. Expression of Gal4 AD-V380H and Gal4 DBD-SRC1 VHC two-hybrid proteins conferred E2 (100 nM)-dependent growth of yeast strain PJ69-4A on synthetic medium lacking adenine by activating the ADE2 gene, which was under the control of the Gal2 promoter. PJ69-4A transformed with plasmids encoding Gal4 AD-wild type (wt) mERα and Gal4 DBD-SRC1 m13 acted as a positive control. Plates were incubated at 30°C for 2 days. NH, no hormone. (D) Ligand-dependent interaction of mERα V380H with SRC1e VHC in vitro. Full-length mERα and FLAG epitope-tagged wild-type or mutant SRC1e was transiently expressed in 293-T cells, and the whole-cell lysate was subjected to immunoprecipitation (IP) with an anti-FLAG antibody immobilized on agarose beads in the absence or presence of 1 μM E2. SRC1e was detected by Western blotting using an anti-FLAG antibody. The coimmunoprecipitated mERα was detected by using anti-ERα antibody H222. VH, V380H; WB, Western blot.
A yeast two-hybrid screen was performed in which a library of SRC1e mutants, representing 2,800 independent clones, were selected for binding to Gal4 activation domain fusions of mERα LBD containing either the V380D or V380H mutation. We were unable to recover any suppressor allele for the V380D mutant receptor. However, one suppressor allele, designated SRC1 VHC for V380H complement, was recovered among 59,000 transformants for the V380H mutation. To gain insight into the molecular basis of complementation, the DNA sequence of the mutant SRC1 allele was determined. To our surprise, the mutant allele consisted of a wild-type LXXLL motif, followed immediately by a C-terminal insertion of 15 amino acids containing a variant motif, YXXLK (Fig. 2B). It was plausible that two mutagenic primers were incorporated in tandem during the library construction. This was supported by the observation that codons which encoded L690 and L694 in the mutant allele differed from those of the wild-type. The mutant allele represented a rare species in the library and provided an explanation for the recovery of a single allele from an apparent saturation screen.
Next, we verified the binding properties of the suppressor mutant. Ligand-dependent interaction between SRC1 VHC and mERα V380H in yeast was confirmed by the ability of the two-hybrid proteins to activate a Gal2-ADE2 reporter and thereby conferred growth on Ade− medium (Fig. 2C). Full-length SRC1e VHC coimmunoprecipitated with mERα V380H in a ligand-dependent manner in vitro (Fig. 2D). The strength of interaction was approximately 50% compared with that of the wild-type receptor-coactivator pair (Fig. 2D, compare lanes 8 and 10). In addition, ligand-dependent interaction between SRC1e VHC and wild-type mERα was also detected because of the presence of an intact LXXLL motif (Fig. 2D, lanes 5 and 6). Taken together, our results have identified an altered-specifity mutant SRC1e through directed genetic selection in yeast which is capable of interacting with mERα V380H in vivo and in vitro.
Functional rescue of mERα V380H by altered specificity SRC1e.
Having established that SRC1e VHC interacts with mERα V380H, we next asked whether it could restore the transcriptional activity of the mutant receptor. The full-length wild-type or V380H mutant receptor was transiently transfected into HeLa cells and tested for the ability to activate a 2×ERE-TATA-luciferase reporter. The V380H mutant receptor had markedly reduced transcriptional activity, indicating that it was severely compromised in its interaction with endogenous coactivators (Fig 3A). In the presence of exogenously expressed SRC1e m13 and SRC1e VHC, the transcriptional activity of the wild-type receptor was potentiated by approximately four- to sixfold (Fig. 3A). Exogenous expression of SRC1e VHC led to a 14-fold induction of V380H agonist-dependent activity. The level of reporter gene activation was comparable to that achieved by the wild-type receptor-coactivator pair and represented a complete functional rescue of V380H. A similar profile of transcriptional activation was obtained when the full-length wild-type or V380H mutant receptor was tested on a 2×ERE-pS2-luciferase reporter in HeLa cells (Fig. 3B). Taken together, our results demonstrated the functional rescue of V380H by SRC1e VHC in the context of two different promoters, which implied that direct recruitment of SRC1e by mERα might be sufficient to elicit transcriptional activation in mammalian cells.
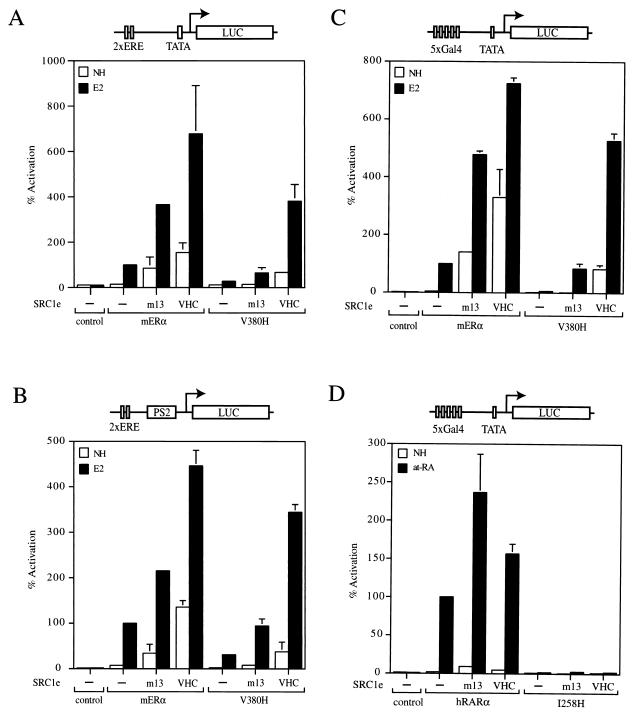
FIG. 3.
Specific functional rescue of mERα V380H by SRC1e VHC. (A) Wild-type and mutant full-length receptors were transiently transfected into HeLa cells together with the p2×ERE-TATA-GL3 reporter in the absence (−) or presence of full-length SRC1e m13 or SRC1e VHC. The pRL-CMV plasmid, which encoded the Renilla luciferase gene driven by a cytomegalovirus promoter, was cotransfected as an internal control. After transfection, cells were treated with the ethanol vehicle alone (no hormone [NH]) or 17β-estradiol (E2) at 10 nM for 24 h. Subsequently, cells were assayed for firefly luciferase (LUC) and Renilla luciferase activities. Normalized values are expressed as percentages of the activity of wild-type mERα alone in the presence of E2 (100%). The results shown are averages of at least two independent experiments assayed in quadruplicate plus the standard errors. (B) Full-length wild-type or mutant mERα was transiently transfected into HeLa cells together with the p2×ERE-pS2-GL3 reporter. Experimental procedures and data presentation are as described for panel A. (C) Wild-type or mutant chimeric receptors consisting of the mERα LBD fused to the Gal4 DBD were transiently transfected into HeLa cells together with the p5Gal-E1B-GL3 reporter. Experimental procedures and data presentation are as described for panel A. (D) Wild-type and mutant chimeric receptors consisting of the hRARα LBD fused to the Gal4 DBD were transiently transfected into HeLa cells together with the p5Gal-E1B-GL3 reporter. After transfection, cells were treated with the ethanol vehicle alone (NH) or all-trans retinoic acid (at-RA) at 100 nM for 24 h. Presentation of data is as described for panel A, except that normalized values are expressed as percentages of the activity of wild-type hRARα alone in the presence of at-RA (100%).
We next investigated whether SRC1e VHC could rescue V380H AF2 activity in the absence of AF1, which is located at the N terminus of the receptor, by testing the ability of Gal4 DBD-ER LBD chimeric receptors to activate a Gal4 reporter gene in HeLa cells. Exogenously expressed SRC1e m13 potentiated the transcriptional activity of Gal4-ERα by fivefold (Fig. 3C). A sevenfold potentiation of the wild-type chimeric receptor activity by SRC1e VHC was also observed (Fig. 3C). The Gal4-V380H mutant had negligible transcriptional activity and was partially rescued by exogenously expressed SRC1e m13 (Fig. 3C). Remarkably, coexpression of SRC1e VHC potentiated the activity of the mutant chimeric receptor by more than 80-fold and the level of reporter gene activation was comparable to that observed with the wild-type receptor-coactivator pair. Similar results were obtained with COS-1 cells (see Fig. 4D) and 293-T cells (data not shown). Taken together, our results clearly established that mERα V380H could be functionally rescued by SRC1e, which we attribute to the restoration of AF2 activity in the mutant receptor.
FIG. 4.
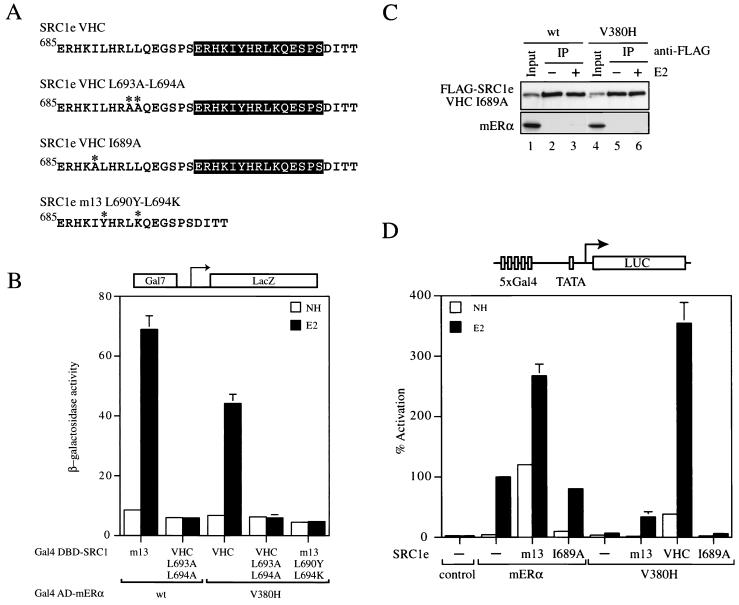
Molecular determinants of the mERα V380H-SRC1e VHC interaction. (A) Sequence comparison of SRC1e mutants. Mutated residues are marked with asterisks. (B) Yeast two-hybrid interaction assay using the Gal7-lacZ reporter in strain PJ69-4A. Transformants with the indicated constructs were grown overnight in selective medium in the absence (no hormone [NH]) or presence of 1 mM 17β-estradiol (E2). β-Galactosidase activity was measured by using o-nitrophenyl-β-d-galactopyranoside as the substrate and is expressed in Miller units. The results shown represent the average activity of two independent transformants. wt, wild type. (C) The I689A mutation in SRC1e VHC abolished its in vitro binding to wild-type and V380H mutant mERα. Coimmunoprecipitation was carried out as described for Fig. 2D. The input control represents 2% of the whole-cell extract employed in the immunoprecipitation (IP) reaction. (D) Wild-type and mutant chimeric receptors consisting of the mERα LBD fused to the Gal4 DBD were transiently transfected into COS-1 cells together with the p5Gal-E1B-GL3 reporter in the absence (−) or presence (+) of full-length SRC1e m13, SRC1e VHC, or SRC1e VHC 1689A as indicated. The pRL-CMV plasmid was cotransfected as an internal control. Data are presented as described for Fig. 3A.
Next, we asked whether SRC1e VHC was able to rescue the transcriptional activity of other nuclear receptors that bear mutations analogous to V380H in mERα. As predicted by sequence analysis and by inspection of the hRARαLBD crystal structure, I258 in hRARα occupies a position in helix 5 of the LBD similar to that of V380 in mERα. When I258 was replaced with histidine (I258H), a chimeric receptor containing the LBD of the mutant receptor fused to the Gal4 DBD was unable to activate a reporter gene when transiently transfected into HeLa cells (Fig. 3D). In addition, coexpression of SRC1e VHC had no effect on the transcriptional activity of the I258H mutant. Hence, the functional rescue of mERα V380H by SRC1e VHC appears to be highly specific and is most likely due to recognition of features of the mERα LBD that are not present in other nuclear receptors.
Molecular determinants of the mERα V380H-SRC1e VHC interaction.
Given the composite nature of the second-site suppressor mutation, we attempted to determine the relative contribution of the wild-type LXXLL motif and the variant YXXLK motif present in SRC1e VHC in mutant receptor-coactivator interaction. By using a yeast two-hybrid interaction assay, we found that SRC1e VHC L693A-L694A in which the wild-type LXXLL motif had been mutated to LXXAA was unable to bind either the wild-type or the V380H mutant receptor (Fig. 4A and B). Furthermore, an SRC1e mutant which contained a single copy of the variant motif but was devoid of any wild-type motif failed to rescue the interaction with V380H (Fig. 4A and B). It was shown that I689 in SRC1e made extensive van der Waals contacts with the ERα coactivator docking surface (34) and that this −1 position relative to the LXXLL motif is frequently occupied by hydrophobic residues, indicating functional importance (15). When I689 of SRC1e VHC was replaced with alanine, the mutant coactivator was unable to bind both the wild-type and V380H mutant receptors in vitro (Fig. 4A and C). In keeping with the loss of interaction, the I689A mutant neither potentiated the transcriptional activity of wild-type mERα nor functionally rescued V380H in transiently transfected COS-1 cells (Fig. 4D). In conclusion, the variant motif YXXLK is not sufficient to mediate mutant receptor-coactivator interaction. These results suggest that the LXXLL motif and its flanking residues are likely to function in conjunction with the 15-amino-acid insertion in SRC1e VHC as an integral module and are indispensible for its interaction with mERα V380H.
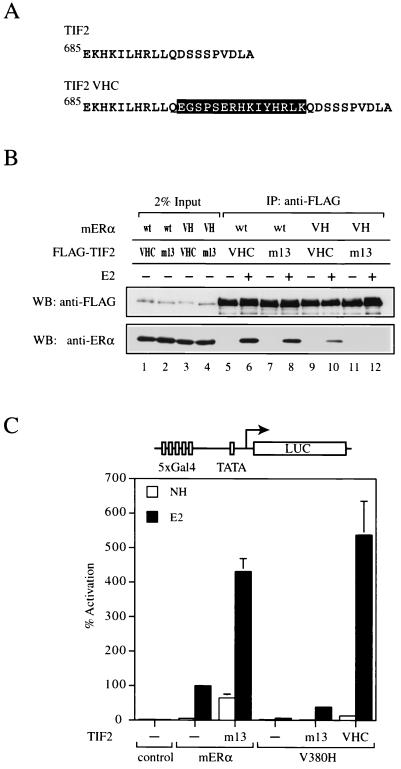
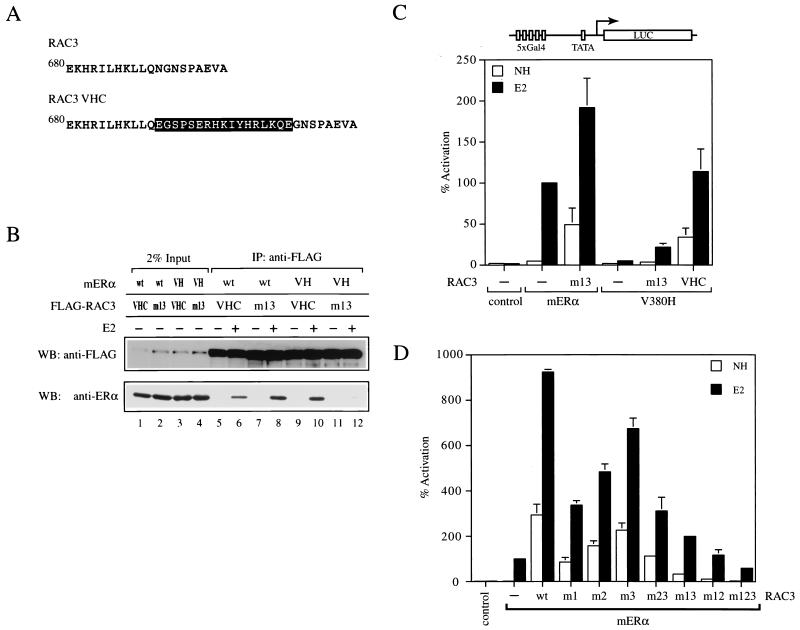
Functional rescue of mERα V380H by TIF2 and RAC3 altered-specificity mutants.
To explore the possibility that the suppressor mutation in SRC1e VHC functions as a protein-protein interaction module, we introduced analogous mutations into other p160 coactivator family members. The sequence conservation among the three p160 coactivators in the vicinity of the second LXXLL motif allowed us to place the 15-amino-acid insertion found in SRC1e VHC at a similar position C terminal to the LXXLL motif in TIF2 and RAC3 (Fig. 5A and 6A). The mutants, designated TIF2 VHC and RAC3 VHC, were then tested for the ability to interact with mERα V380H. Both TIF2 VHC and RAC3 VHC coimmunoprecipitated with mERα V380H in a ligand-dependent manner in vitro (Fig. 5B, lanes 9 and 10, and 6B, lanes 9 and 10). As we found for SRC1e VHC, they also bound to wild-type mERα (Fig. 5B, lanes 5 and 6, and 6B, lanes 5 and 6). These data suggest that the suppressor mutation originally recovered in SRC1e can function in other p160 coactivators when placed in a similar context and confers the ability to interact with mERα V380H.
FIG. 5.
Analysis of altered-specificity TIF2. (A) Sequence comparison of wild-type TIF2 and TIF2 VHC. The 15-amino-acid insertion found in SRC1e VHC (encompassed by the black box) was placed immediately C terminal to TIF2 wild-type LXXLL motif 2 as indicated. (B) Ligand-dependent interaction of mERα V380H with TIF2 VHC in vitro. Coimmunoprecipitation (IP) was carried out as described for Fig. 2D. VH, V380H; wt, wild type; WB, western blot. (C) Wild-type and mutant chimeric receptors consisting of the mERα LBD fused to the Gal4 DBD were transiently transfected into HeLa cells together with the p5Gal-E1B-GL3 reporter in the absence (−) or presence of full-length TIF2 m13 or TIF2 VHC as indicated. The pRL-CMV plasmid was cotransfected as an internal control. Data are presented as described for Fig. 3A. NH, no hormone.
FIG. 6.
Analysis of altered-specificity RAC3. (A) Sequence comparison of wild-type RAC3 and RAC3 VHC. The 15-amino-acid insertion found in SRC1e VHC (encompassed by the black box) was placed immediately C terminal to RAC3 wild-type LXXLL motif 2 as indicated. (B) Ligand-dependent interaction of mERα V380H with RAC3 VHC in vitro. Coimmunoprecipitation (IP) was carried out as described for Fig. 2D. VH, V380H; wt, wild type; WB, western blot. (C) Wild-type and mutant chimeric receptors consisting of the mERα LBD fused to the Gal4 DBD were transiently transfected into HeLa cells together with the p5Gal-E1B-GL3 reporter in the absence (−) or presence of full-length RAC3 m13 or RAC3 VHC as indicated. The pRL-CMV plasmid was cotransfected as an internal control. Data are presented as described for Fig. 3A. NH, no hormone. (D) Potentiation of mERα transcriptional activity by RAC3 mutants. Wild-type and mutant chimeric receptors consisting of the mERα LBD fused to the Gal4 DBD were transiently transfected into HeLa cells together with the p5Gal-E1B-GL3 reporter in the absence (−) or presence of of full-length RAC3 mutants as indicated. m1 denotes nonfunctional LXXLL motif 1, and the same nomenclature scheme applies to all of the other mutants. The pRL-CMV plasmid was cotransfected as an internal control. Data are presented as described for Fig. 3A.
Next, we tested whether TIF2 VHC, and RAC3 VHC could rescue the ability of mERα V380H to stimulate transcription from reporter genes. In HeLa cells, coexpression of TIF2 VHC led to a 90-fold induction of V380H transcriptional activity on a Gal4 reporter gene (Fig. 5C). The level of gene activation achieved was comparable to that of the wild-type receptor-coactivator pair, indicating complete functional rescue. Similarly, RAC3 VHC was able to induce the transcriptional activity of mERα V380H by 22-fold (Fig. 6C), approximately 60% of that achieved by the wild-type counterparts. We therefore concluded that TIF2 VHC and RAC3 VHC are capable of rescuing the transcriptional activity of mERα V380H, albeit to various degrees. Furthermore, our results imply that recruitment of SRC1e, TIF2, or RAC3 is sufficient to mediate the AF2 activity of mERα.
One possibility for the incomplete functional rescue of mERα V380H by RAC3 VHC was that the second LXXLL motif was not preferentially used for ERα-RAC3 interaction. As a result, the suppressor mutation may not be presented in an optimal conformation, which might, as a consequence, hinder the rescue. In SRC1e and TIF2, the second LXXLL motif was clearly preferred for interaction with ERα and retention of this motif alone allowed SRC1e and TIF2 to function as efficiently as their wild-type counterparts (17, 40). To gain insight into the preference of LXXLL motifs in RAC3 by mERα, a complete series of RAC3 mutants were generated in which the LXXLL motifs were rendered nonfunctional by mutation to LXXAA either individually or in all possible combinations. We then tested the abilities of these mutants to potentiate the transcriptional activity of Gal4-ERα in HeLa cells. Unlike SRC1e and TIF2, mutation of a single LXXLL motif impaired the ability of RAC3 to function as a coactivator, with the effects most pronounced when motif 1 was mutated (Fig. 6D). When only one LXXLL motif was retained, none of the mutants were able to recapitulate the full activity of wild-type RAC3. Mutation of all three motifs eliminate the ability of RAC3 to potentiate ERα activity. Our functional data correlate well with other studies in which ERα-RAC3 interaction was examined (6, 20) and led us to postulate that cooperation of multiple LXXLL motifs might be necessary to foster ERα-RAC3 interaction. Hence, the incomplete functional rescue of mERα V380H by RAC3 VHC could be attributed to the absence of cooperating motifs for the functional motif in RAC3 VHC, which resulted in suboptimal interaction with the receptor.
By inserting the altered-specificity mutation from SRC1e VHC into TIF2 and RAC3, we showed that this mutation functions as a protein-protein interaction module which confers the ability to suppress the V380H mutation in mERα. More importantly, our results demonstrate that SRC1e, TIF2, and RAC3 are functionally redundant and that direct recruitment of a single species of p160 coactivator by the ERα LBD is sufficient to instigate agonist-dependent transcriptional activation.
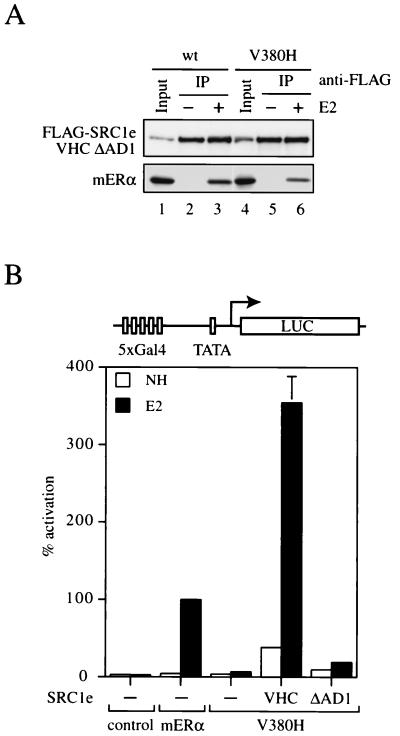
Role of CBP/p300 in functional rescue of mERα V380H by SRC1e VHC.
AD1 of SRC1e and other p160 coactivator family members was shown to physically interact with CBP and p300 (5, 17, 40). To directly address whether AD1, and thereby recruitment of CBP/p300 or other coactivator proteins, is central to the function of p160 coactivators, we utilized a SRC1e VHC construct which lacks AD1 and tested its ability to mediate transactivation by the V380H mutant receptor. The deletion mutant was expressed at a level comparable to that of the wild-type control (data not shown), and the deletion did not grossly affect the structure of the coactivator, as exemplified by its binding to both wild-type and V380H mutant receptors in vitro (Fig. 7A). However, expression of SRC1e VHC ΔAD1 failed to rescue the transcriptional activity of a chimeric receptor consisting of the mERα V380H LBD fused to the Gal4 DBD in transiently transfected COS-1 cells (Fig. 7B). Similar results were also obtained with HeLa cells (data not shown). This demonstrates that recruitment of CBP/p300 and/or other coactivator proteins through the AD1 region of SRC1e VHC is essential for functional rescue of the transcriptionally defective V380H mutant. Our data further suggest that p160 coactivators serve as ligand-dependent adapter proteins whose primary function is to recruit other coactivators, such as CBP/p300, to the promoter where ERα is bound.
FIG. 7.
Functional analysis of SRC1e VHC ΔAD1. (A) Ligand-dependent interaction of mERα V380H with SRC1e VHC ΔAD1 in vitro. Coimmunoprecipitation was carried out as described for Fig. 2D. The input control represents 2% of the whole-cell extract employed in the immunoprecipitation (IP) reaction. wt, wild type. (B) Wild-type and mutant chimeric receptors consisting of the mERα LBD fused to the Gal4 DBD were transiently transfected into HeLa cells together with the p5Gal-E1B-GL3 reporter in the absence (−) or presence of full-length SRC1e VHC or SRC1e VHC ΔAD1 as indicated. The pRL-CMV plasmid was cotransfected as an internal control. Data are presented as described for Fig. 3A. NH, no hormone.
DISCUSSION
Genetic selection for second-site suppressor mutations has been used to study the significance of specific protein-protein interaction in both prokaryotic and eukaryotic systems and was employed here to probe the functional roles of p160 coactivators in ERα action (7, 21, 35, 42). Numerous proteins have been postulated to function as coregulators of the agonist-dependent transcriptional activity of ERα; however, attempts to decipher their function and biological relevance in cells have been hindered by their common mode of interaction with the ERα AF2 surface. In this study, we focused on the p160 family of coactivators and circumvented this problem by selecting a mutant version of SRC1 that interacts with mERα V380H, a mutant receptor incapable of interacting with endogenous p160 coactivators. Our strategy for the identification of SRC1 altered-specificity mutants relied on the clear indication from the available cocrystal structures that V380 of mERα is most likely to interdigitate with L690 and L694 of SRC1 (9, 29, 34). As a result, targeted random mutations were made at these two residues in SRC1. Our genetic selection in yeast yielded a single suppressor allele, SRC1 VHC, which specifically restored binding to mERα V380H. Interestingly, this suppressor allele contains an insertional mutation, which indicates that mutations at L690 and L694 of SRC1 alone are not sufficient to reconstitute a functional interface with mERα V380H (see below).
Transcriptional activation by ERα through direct recruitment of p160 coactivator.
The coexpression of SRC1 VHC fully restored the transcriptional activity of mERα V380H. Although a large number of proteins have been reported to interact with agonist-bound ERα via the LXXLL motifs, our results suggest that ERα transactivation through AF2 is primarily dependent on direct recruitment of p160 coactivators. We further demonstrated, by using a version of SRC1 VHC which lacks its CBP/p300 binding domain (ΔAD1), that the recruitment of CBP/p300 and/or other coactivator proteins via the AD1 region is an obligatory second step in SRC1-mediated gene activation. Recent reports concerning activation of the estrogen-responsive pS2 gene support our hypothesis that the p160 coactivators are primary mediators of ERα AF2 activity. In chromatin immunoprecipitation experiments, ACTR/RAC3 was found to associate with the pS2 promoter upon hormone treatment and, importantly, the cessation of hormone-induced gene activation was accompanied by the dissociation of ACTR/RAC3 and CBP from the promoter (6).
In addition to p160 coactivators and CBP/p300, it has been proposed that the TRAP/DRIP complex serves an important role as a mediator for a number of transcription factors, including nuclear receptors (24). The TRAP/DRIP complex appears to be recruited to the nuclear receptor AF2 surface via LXXLL motifs in TRAP220 (3, 32, 47). This led to the hypothesis that either there is competition between the TRAP/DRIP complex and p160 coactivators for the AF2 surface (39) or, alternatively, there may be sequential recruitment, first of p160 coactivators and then of the TRAP/DRIP complex (12, 46). More recently, it was demonstrated that p160 coactivators and the TRAP/DRIP complex could be recruited concomitantly to the same estrogen-responsive promoter (33). Nevertheless, there appeared to be a strict requirement for a p160 coactivator in ERα-mediated activation of the endogenous cathepsin D gene which cannot be replaced by directed recruitment of TRAP220 (33). This is in agreement with our observation that restoration of SRC1 binding to a defective ERα AF2 surface is sufficient to fully rescue its activity, which points to a critical role of p160 coactivators in ERα-regulated transcription.
A model for mERα V380H-SRC1 VHC interaction.
Several lines of evidence suggest that both the wild-type LXXLL motif and the 15-amino-acid insertion found in SRC1 VHC are necessary for mutant receptor-coactivator interaction. First, disruption of the wild-type LXXLL motif in SRC1 VHC abolished its ability to bind mERα V380H. Second, mutant SRC1 that was devoid of the wild-type LXXLL motif but contained a single copy of the variant motif YXXLK found in SRC1 VHC was unable to interact with mERα V380H. Finally, SRC1 VHC was unable to rescue the transcriptional activity of mERα V380H in the presence of antiestrogens such as tamoxifen and ICI 182780, implying that SRC1 VHC could not interact with an antagonist-bound receptor (data not shown). Tamoxifen binding forces helix 12 to adopt a position which occludes the docking site for the wild-type LXXLL motif, with minimal effects on the rest of the ERα LBD structure (34). The last observation, therefore, suggests that mERα V380H-SRC1 VHC interaction employs a variant interface which is likely to be based on the one utilized by their wild-type counterparts. Although the interaction between the wild-type LXXLL motif with the remodeled coactivator docking surface in V380H is severely impaired, it is tempting to speculate that it remains as a recognition or anchoring module for the mutant mERα-SRC1 interaction. Nevertheless, stable equilibrium binding requires the sequence insertion in the mutant SRC1 VHC allele which might interact directly with the histidine or arginine side chain where V380 is normally found. Alternatively, the sequence insertion may contact a second site on the receptor surface which is only available in the presence of ligand. We favor the latter model based on our observations that SRC1 VHC is a more potent coactivator of the wild-type receptor (Fig. 3), which might be attributed to enhanced intereaction with SRC1 VHC. One candidate for the second contact site is helix 1 of the receptor LBD, which has recently been shown to undergo subtle conformational change upon ligand binding (30).
It is important to note that the sequence insertion in SRC1 VHC is unlikely to alter the structural integrity of the protein. This is because the SRC1 moiety (residues 623 to 710) in the holo-PPARγ-SRC1 complex is unstructured except for the short helices which contain the LXXLL motifs (29). Therefore, the 15-amino-acid insertion is likely to be accommodated in the random coil region without major disruption to the tertiary structure. Setting aside the question of the precise nature of the mutant receptor-coactivator interaction, it is clear that the altered-specificity mutation in SRC1 VHC does not constitute a promiscuous protein binding motif. This is supported by the observation that SRC1 VHC was unable to rescue an hRARα mutant which bears a mutation analogous to V380H in mERα.
Introduction of the SRC1 VHC suppressor mutation into other p160 coactivators allowed us to generate mutant versions of TIF2 and RAC3 which could interact with mERα V380H. This suggests that the suppressor mutation may function as a transposable protein-protein interaction module, enabling heterologous proteins to interact with mERα V380H. It may be possible to use this module to study other nuclear receptor interacting proteins which contain the LXXLL motif, such as TRAP220, and enable us to probe the functional consequence of direct recruitment of TRAP220 by ERα in the future.
Functional redundancy of p160 coactivators.
In mammalian cells, the agonist-dependent transcriptional activity of mERα V380H could be rescued by mutant versions of SRC1, TIF2, or RAC3. Hence, the recruitment of any one of the p160 coactivators appears to be sufficient to instigate ERα transactivation. This clearly suggests that the three p160 proteins are functionally redundant and that expression of one family member could potentially compensate for the absence of others. Therefore, our data are in line with the relatively mild phenotype of SRC1-null mice, which has been attributed to the upregulation of TIF2 gene expression in selected tissues (45). In contrast, our results do not support an earlier proposal that p/CIP might be functionally distinct from other p160 family members, based on cell microinjection experiments using immunoglobulins against SRC1 and p/CIP (38).
The observations that mice lacking SRC3/RAC3 do not display phenotypes similar to that of SRC1-null mice led to the suggestion that they may have distinct roles in vivo (44, 45). However, it is important to note that our proposal about functional redundancy of the p160 coactivators addresses their role in ERα transactivation at the cellular level and is compatible with these animal models, where distinct phenotypes have been attributed to differential gene expression (44). It is apparent that phenotypes of the SRC1- and SRC3/RAC3-null mice may be complicated by the existence of both cell-autonomous and cell-nonautonomous effects. For example, SRC3/RAC3-null mice have a lower level of systemic estrogen, which predictably affects multiple aspects of the sexual maturation and reproductive function of female mice (44). We therefore propose that the use of altered-specificity mutants may complement existing animal models in the study of the cell-autonomous function of individual receptor-coactivator pairs in the complex network of nuclear receptor-coregulator interactions.
ACKNOWLEDGMENTS
We thank Hinrich Gronemeyer, Don Chen, Philip James, Borja Belandia, and David Heery for plasmid and yeast strain gifts; Geoff Greene for the H222 antibody; I. Goldsmith and staff for oligonucleotides; and G. Clark and staff for DNA sequencing. We also thank Caroline Hill, Jesper Svejstrup, Roger White, and members of the Molecular Endocrinology Laboratory for discussions and comments on the manuscript.
This work was supported by the Imperial Cancer Research Fund.
REFERENCES
- 1.Bevan C L, Hoare S, Claessens F, Heery D M, Parker M G. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol Cell Biol. 1999;20:8383–8392. doi: 10.1128/mcb.19.12.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J-A, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 3.Burakov D, Wong C W, Rachez C, Cheskis B J, Freedman L P. Functional interactions between the estrogen receptor and DRIP205, a subunit of the heteromeric DRIP coactivator complex. J Biol Chem. 2000;275:20928–20934. doi: 10.1074/jbc.M002013200. [DOI] [PubMed] [Google Scholar]
- 4.Caira F, Antonson P, Pelto-Huikko M, Treuter E, Gustafsson J A. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J Biol Chem. 2000;275:5308–5317. doi: 10.1074/jbc.275.8.5308. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 7.Crispino J D, Lodish M B, MacKay J P, Orkin S H. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol Cell. 1999;3:219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- 8.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Croce L, Okret S, Kersten S, Gustafsson J A, Parker M, Wahli W, Beato M. Steroid and nuclear receptors. Villefranche-sur-Mer, France, May 25–27, 1999. EMBO J. 1999;18:6201–6210. doi: 10.1093/emboj/18.22.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P J, Stallcup M R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 12.Fondell J D, Guermah M, Malik S, Roeder R G. Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc Natl Acad Sci USA. 1999;96:1959–1964. doi: 10.1073/pnas.96.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freedman L P. Increasing the complexity of coactivation in nuclear receptor signaling. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 14.Glass C K, Rosenfeld M G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 15.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 16.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor coactivator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Douarin B, Nielsen A L, Garnier J-M, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S K, Anzick S L, Choi J E, Bubendorf L, Guan X Y, Jung Y K, Kallioniemi O P, Kononen J, Trent J M, Azorsa D, Jhun B H, Cheong J H, Lee Y C, Meltzer P S, Lee J W. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J Biol Chem. 1999;274:34283–34293. doi: 10.1074/jbc.274.48.34283. [DOI] [PubMed] [Google Scholar]
- 20.Leo C, Li H, Chen J D. Differential mechanisms of nuclear receptor regulation by receptor-associated coactivator 3. J Biol Chem. 2000;275:5976–5982. doi: 10.1074/jbc.275.8.5976. [DOI] [PubMed] [Google Scholar]
- 21.Li M, Moyle H, Susskind M M. Target of the transcriptional activation function of phage lambda cI protein. Science. 1994;263:75–77. doi: 10.1126/science.8272867. [DOI] [PubMed] [Google Scholar]
- 22.Mahajan M A, Samuels H H. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol Cell Biol. 2000;20:5048–5063. doi: 10.1128/mcb.20.14.5048-5063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mak H Y, Hoare S, Henttu P M A, Parker M G. Molecular determinants of the estrogen receptor-coactivator interface. Mol Cell Biol. 1999;19:3895–3903. doi: 10.1128/mcb.19.5.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malik S, Roeder R G. Transcriptional regulation through mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 25.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R E. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna N J, Lanz R B, O'Malley B W. Nuclear receptor coregulators: cellular and molecular biology. Endocrinol Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 28.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 29.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 30.Pissios P, Tzameli I, Kushner P, Moore D D. Dynamic stabilization of nuclear receptor ligand binding domains by hormone or corepressor binding. Mol Cell. 2000;6:245–253. doi: 10.1016/s1097-2765(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 31.Rachez C, Gamble M, Chang C-P B, Atkins G B, Lazar M A, Freedman L P. The DRIP complex and SRC-1/p160 coactivators share similar nuclear receptor binding determinants but constitute functionally distinct complexes. Mol Cell Biol. 2000;20:2718–2726. doi: 10.1128/mcb.20.8.2718-2726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 33.Shang Y, Hu X, DiRenzo J, Lazar M A, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 34.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 35.Tansey W P, Herr W. Selective use of TBP and TFIIB revealed by a TATA-TBP-TFIIB array with altered specificity. Science. 1997;275:829–831. doi: 10.1126/science.275.5301.829. [DOI] [PubMed] [Google Scholar]
- 36.Tcherepanova I, Puigserver P, Norris J D, Spiegelman B M, McDonnell D P. Modulation of estrogen receptor-alpha transcriptional activity by the coactivator PGC-1. J Biol Chem. 2000;275:16302–16308. doi: 10.1074/jbc.M001364200. [DOI] [PubMed] [Google Scholar]
- 37.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent non-acidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 38.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 39.Treuter E, Johansson L, Thomsen J S, Warnmark A, Leers J, Pelto-Huikko M, Sjoberg M, Wright A P, Spyrou G, Gustafsson J A. Competition between thyroid hormone receptor-associated protein (TRAP) 220 and transcriptional intermediary factor (TIF) 2 for binding to nuclear receptors. Implications for the recruitment of TRAP and p160 coactivator complexes. J Biol Chem. 1999;274:6667–6677. doi: 10.1074/jbc.274.10.6667. [DOI] [PubMed] [Google Scholar]
- 40.Voegel J J, Heine M J, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weatherman R V, Fletterick R J, Scanlan T S. Nuclear-receptor ligands and ligand-binding domains. Annu Rev Biochem. 1999;68:559–581. doi: 10.1146/annurev.biochem.68.1.559. [DOI] [PubMed] [Google Scholar]
- 42.Winnier A R, Meir J Y, Ross J M, Tavernarakis N, Driscoll M, Ishihara T, Katsura I, Miller D M., 3rd UNC-4/UNC-37-dependent repression of motor neuron-specific genes controls synaptic choice in Caenorhabditis elegans. Genes Dev. 1999;13:2774–2786. doi: 10.1101/gad.13.21.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wurtz J-M, Bourguet W, Renaud J-P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 44.Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O'Malley B W. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc Natl Acad Sci USA. 2000;97:6379–6384. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J, Qiu Y, DeMayo F, Tsai S, Tsai M-J, O'Malley B W. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) Gene Sci. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 46.Yang W, Rachez C, Freedman L P. Discrete roles for peroxisome proliferator-activated receptor γ and retinoid X receptor in recruiting nuclear receptor coactivators. Mol Cell Biol. 2000;20:8008–8017. doi: 10.1128/mcb.20.21.8008-8017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan C-X, Ito M, Fondell J D, Fu Z-Y, Roeder R G. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Y, Qi C, Jain S, Rao M S, Reddy J K. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]