Abstract
SARS-CoV-2, an RNA virus, has been prone to high mutations since its first emergence in Wuhan, China, and throughout its spread. Its genome has been sequenced continuously by many countries, including Pakistan, but the results vary. Understanding its genomic patterns and connecting them with phenotypic features will help in devising therapeutic strategies. Thus, in this study, we explored the mutation landscape of 250 Pakistani isolates of SARS-CoV-2 genomes to check the genome diversity and examine the impact of these mutations on protein stability and viral pathogenesis in comparison with a reference sequence (Wuhan NC 045512.2). Our results revealed that structural proteins mainly exhibit more mutations than others in the Pakistani isolates; in particular, the nucleocapsid protein is highly mutated. In comparison, the spike protein is the most mutated protein globally. Furthermore, nsp12 was found to be the most mutated NSP in the Pakistani isolates and worldwide. Regarding accessory proteins, ORF3A is the most mutated in the Pakistani isolates, whereas ORF8 is highly mutated in world isolates. These mutations decrease the structural stability of their proteins and alter different biological pathways. Molecular docking, the dissociation constant (KD), and MM/GBSA analysis showed that mutations in the S protein alter its binding with ACE2. The spike protein mutations D614G-S943T-V622F (−75.17 kcal/mol), D614G-Q677H (−75.78 kcal/mol), and N74K-D614G (−73.84 kcal/mol) exhibit stronger binding energy than the wild type (−66.34 kcal/mol), thus increasing infectivity. Furthermore, the simulation results strongly corroborated the predicted protein servers. Our analysis findings also showed that E, M, ORF6, ORF7A, ORF7B, and ORF10 are the most stable coding genes; they may be suitable targets for vaccine and drug development.
Keywords: SARS-CoV-2, COVID-19, ACE2, Mutation, Phosphorylation serine/threonine kinases
1. Introduction
In December 2019, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus appeared in Wuhan, China, instigated a global coronavirus pandemic (COVID-19) [1,2]. Till October 2021, 241,199,505 cases have been reported with about 4,910,487 deaths recorded (WHO, 2020, (https://covid19.who.int). The transmission of SARS-CoV-2 is very high, with common symptoms attributed to this disease includes, high fever, breathing complexities, dry cough, and acute respiratory distress [[3], [4], [5], [6], [7], [8]].
SARS-CoV-2 is a single-stranded, positive-sense RNA virus with a genome size of ∼29.8 kb which encodes 16 non-structural proteins (nsp1–16) to form the replicase machinery of the virus [5,9]. It also codes for four structural proteins, namely Spike (S), envelope (E), membrane (M), and nucleocapsid (N). Besides structural and non-structural protein, SARS_CoV-2 codes for seven accessory proteins (ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, and ORF10), which modulate host responses against viral infections to assist effective infection and pathogenesis [[10], [11], [12], [13], [14]].
Genome sequencing insights have shown nucleotide substitution rate as ∼1 × 10−3 per year for SARS-CoV-2 [15,16]. Variations in different proteins of SARS-CoV-2, particularly the spike glycoproteinlead to a drift in the antigenicity of vaccines and other therapeutics [17]. The mutation rate of RNA viruses, including, SARS-CoV-2 is exceptionally high, which in turn, increase the virulence and transmission of these viruses [16,[18], [19], [20]]. These mutations occur in almost all the proteins of SARS-CoV-2, thereby rendering this virus to escape the host immune system more efficiently and consequently increases the pathogenicity and transmissibility [[21], [22], [23], [24], [25], [26], [27], [28]]. These amino acid substitutions also alter the protein structures and consequently viral replication. The depletion of 9 amino acids in Orf6 caused the shifting of transmembrane localization that results in IFN resistance during antiviral therapy [29]. Mutations in the proteins affect the structure, function, and binding with the other proteins necessary for disease progression. Other mutations result in a new RNA polymerase variant that is reliant on RNA [30]. Some of the reported mutations also expedite the transmission of the virus i.e., D614G mutation in the spike protein has been reported to be associated with unusual increased in the transmission fitness of SARS-CoV-2 [23,[31], [32], [33], [34]]. The genome of the virus is unceasingly mutating since its outbreak in late 2019 and many variants are reported until now. The B.1.617 lineage was identified in India in October 2020 [24,35]. It then became a dominant strain in India and spread to other countries of the world as well. Furthermore, the B1.617.1, B.1.617.2, and B.1.617.3 subtypes of this lineage also harbours different mutations in the N-terminal domain (NTD) and the receptor-binding domain (RBD) of the spike protein [27,36].
Since the virus mutates often, it is hard to develop effective therapeutics which can hinder all the variants. To come up with a successful treatment approach or vaccination, it is necessary to have a greater understanding of both the virus and host genome [37]. An idea of the efficacy of the vaccines being researched may be extrapolated from sequencing the entire genome sequences of isolates collected from diverse locations across the world [38]. The race for a final magic drug still continues and according to the updated data provided by the COVID-19 vaccine and therapeutics tracker, 408 clinical trials of 138 vaccine candidates are under consideration and are in different clinical phases (https://covid19.trackvaccines.org/). Globally 6.65 billion doses of COVID-19 vaccines are administered among which 2.83 billion are fully vaccinated which counts for only 36% of the whole population. Understanding the mutational landscape of SARS-CoV-2 would provide a better view of the therapeutic interventions. For instance, Abbas et al., used computational approaches to study the impact of different mutations on the binding and infectivity [18,20,39,40]. Moreover, using these approaches many small molecules drugs are tested against different proteins of SARS-CoV-2 [[41], [42], [43], [44], [45], [46]]. Therefore, herein we used computational approaches to study the reported 250 genomes of SARS-CoV-2 isolated from Pakistan. In Pakistan, the first case of COVID-19 was reported on February 26, 2020. As of October 2021, about 1.27 million cases were reported, with total mortality of 28,280 were recorded in Pakistan. Hence, we used GSAID to extract the mutational spectrum in different proteins and their impact on the structure and function was estimated. Furthermore, protein-protein docking and molecular dynamics simulation approaches were used to grasp the impact of the most prevalent mutations on the binding and stability of the spike protein particulalry. Consequently, the current study provides distinguishable features for the discovery of novel therapeutics and Pakistan-specific anti-COVID-19 strategies.
2. Materials and methods
2.1. Collection of genome data for SARS-CoV-2
The genome sequence of 250 Pakistani isolates OF SARS-CoV-2 was downloaded from a global initiative on sharing all influenza data (GISAID) (https://www.gisaid.org/) [47] and NCBI [48]. Only coding sequences were analyzed for mutational analysis at the protein level in the present study.
2.2. Multiple sequence alignment
DNASTAR, Lasergene, and Clustal W tools align the 250 Pakistani SARS-CoV-2 isolates [49,50]. The previously described isolate of SARS-CoV-2 (Wuhan NC 045512.2) was utilized as a reference sequence for mutational analysis at the amino acid level.
2.3. Sequence-based mutational effects prediction
The I-mutant online tool was utilized to assess the structural stability change [51]. Mutpred2 was also utilized to examine the molecular consequences and functional impacts of mutations found [52].
2.4. Wild type structure retrieval and variants modelling
The wild-type structure of SARS-CoV-2 spike protein (6VSB) was obtained from the Protein Data Bank, and Chimera v 1.15 software was used for SARS-CoV-2 spike protein variant modeling [53,54]. The wild type structure 6VSB was used as a template that shares 97% identity with the sequence of the variant. The.FASTA format was used as input and BLAST was carried out to select the template based on sequence identity and coverage.
2.5. Protein-protein docking
HDOCK (http://hdock.phys.hust.edu.cn/) online server algorithm was used for protein-protein interaction of both wild type-ACE2 and variants-ACE2, to check the respective binding efficiencies [55]. HDOCK uses a hybrid algorithm that combines both template-based and free approaches to estimate the docking conformation and affinity.
2.6. Dissociation constant (KD) determination
Estimation of the strength of biomolecular association is an important parameter in biological processes. Herein, to estimate the strength of the wild type and mutants RBD with ACE2 PRODIGY (PROtein binDIng enerGY prediction)(https://wenmr.science.uu.nl/prodigy/) webserver was used [56].
2.7. Molecular dynamic simulation
MD simulation of the wild type and mutant complexes was completed by using the AMBER20 simulation package [57,58]. The default parameters used by Abbas et al., were employed to complete the 200ns simulation for each complex [59,60]. Post-simulation analyses such as RMSD and RMSF were calculated using CPPTRAJ and PTRAJ modules [61].
2.8. Binding free energy calculations
The binding energy between WT and mutant spike protein with the human receptor ACE2 was calculated using the MM/GBSA approach [62]. The spike mutants include N74K, D614G, V622F, Q677H, S943T, D115G with the human receptor ACE2 using the HAWKDOCK server [63].
The energy is calculated with the following equation:
Each component of the total free energy was estimated using the following equation:
3. Results and discussion
3.1. Retrieved genome of SARS-CoV-2
A total of 250 SARS-CoV-2 isolates from Pakistan were obtained via the Global Initiative on GISAIDand the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/). Only coding sequences were examined for mutational analysis at the protein level (Table 1 ).
Table 1.
Retrieved genomes of SARS-CoV-2 from Pakistan and their mutational landscape.
| S. No | Isolates | ORF1AB | ORF1A | Spike | N | M | ORF3A | ORF8 | ORF10 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MW447643.1 | Q63L, L3293F, P4075, SA4489V, P4716L | Q63L P4075S | D614G | Q57H | ||||
| 2 | MW447622.1 | G82S, L2564F, P4716L, A4921V | G82S L2564F | D614G | |||||
| 3 | MW447639.1 | G82S, P4716L | G82S | D614G | Q57H | ||||
| 4 | MT240479.1 | R207C, V378I, P2965L, L3606F | R207C V378I P2965L L3606F | ||||||
| 5 | MW447634.1 | K261 N, P4716L, T5451I | K261 N | D614G | Q57H | ||||
| 6 | MW447614.1 | F275L, V2133A, P4716L | D614G | S194L | Q57H | ||||
| 7 | MW242667.1 | K292E, V627F, T2018K, L3606F | K292E V627F T2016K L3606F | P13L | V30L | ||||
| 8 | MT500122.1 | D448 N, N449, deletion | D448 N N449delete | ||||||
| 9 | MW447612.1 | G934C, P4716L | D614G | R209I | Q57H | ||||
| 10 | MW031799.1 | S944L, T1246I, K1305 N, G3278S, P4716L | S944LT1246I K1305 N G3278S | D614G | R203K | ||||
| 11 | MW031800.1 | S944L, T1246I, K1305 N, P4716L | S944L T1246I K1305 N G3278S |
D614G | R203K | ||||
| 12 | MW031801.1 | S944L, T1246I, K1305 N, P4716L | S944L T1246I G3278S | D614G | R203K | ||||
| 13 | MW031802.1 | S944L, T1246I, K1305 N, P4716L | S944L T1246I K1305 N G3278S |
N74K D614G | R203K | ||||
| 14 | MW447609.1 | L1175I, P4716L | D614G | R203K | Q57H | ||||
| 15 | MW447644.1 | T1754I, T2846I, P4716L | T1754I T2846I | D614G | K21 N | ||||
| 16 | MW447645.1 | T2408I, P4716L, L6082F | V2133A T2408I | D614G | S194LT379I | Q57H | |||
| 17 | MW447642.1 | A2618V, P4716L | A2618V | D614G | R209I | Q57H T223I | W45L | ||
| 18 | MW447611.1 | Q2702H, K3353R P4716L, V4979L | Q2702H K3353R | D614G | S327L | Q57H | |||
| 19 | MT879619.1 | Q2702H, P4716L, A5561T | Q2702H L3606F | D614G | S327L | Q57H | V30L | ||
| 20 | MT262993.1 | (T3153Y, WFF3157, deleted) (L6418, YLDAY6424 N deleted) | (T3153YWFF3157 deleted) | ||||||
| 21 | MW447636.1 | P3447S, P4716L | P3447S | D614G | R209I | Q57H | |||
| 22 | MW031803.1 | P4716L | D614G | ||||||
| 23 | MW400961.1 | P4716L | D614G V622F S943T |
Q57H | |||||
| 24 | MW447638.1 | P4716L, V6600F | D614G, Q677H | S194L | Q57H | ||||
| 25 | MW447640.1 | P4716L, V6600F | A2956V | D614G | S194L | H25Y | |||
| 26 | MW447641.1 | P4716L | D614G | R209I | Q57H | ||||
| 27 | MW447644.1 | P4716L | R209I | ||||||
| 28 | MW403500.1 | D614G | S194L | Q57H | |||||
| 29 | MW411960.1 | D614G | R209I | ||||||
| 30 | MW447610.1 | D614G | Q57H | ||||||
| 31 | MW447619.1 | D614G | S194L | Q57H | |||||
| 32 | MW447620.1 | D614G | R209I | Q57H | |||||
| 34 | MW447618.1 | S202 N | L84S | ||||||
| 35 | MW447621.1 | D614G D1153G |
R209I | Q57H |
3.2. Worldwide mutational distribution of SARS-CoV-2 proteins
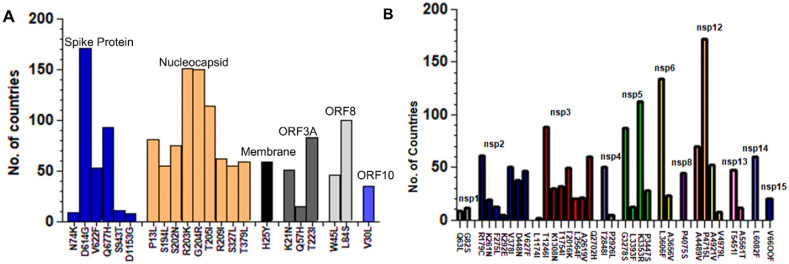
SARS-CoV-2 protein mutations were examined globally as part of a global initiative program for sharing all influenza data (GISAID). We observed mutations in most of the proteins of the SARS-CoV-2 virus. However, some proteins like envelop, ORF6, ORF7A, and ORF7B showed resistance to mutations. Additionally, specific proteins were more prone to mutations than others. Among the Structural proteins, spike protein was mutated the most, followed by nucleocapsid. Within spike, D614G was found to be mutated in 171 countries. Mutation R203K in nucleocapsid protein was observed in 151 countries (Fig. 1 A). Among the NSPs, the mutation P4715L in NSP12 was observed in 172 countries globally, followed by L3606F in NSP3, which occurred in 134 countries. The L1174I mutation in NSP3 was observed only in 2 countries (Fig. 1B). Among the accessory proteins, ORF8 was found to be the most mutated protein. Several mutations have been reported in the ORF8 protein, but L84S is a highly recurred mutation observed in 100 countries (Fig. 1A).
Fig. 1.
Worldwide mutational distribution of SARS-CoV-2 proteins. (A) Among the Structural proteins, D614G mutation in spike protein was found the most reoccurring mutation in 171Countries while L84S in ORF8 was observed in 100 countries. (B) Among the Non-Structural proteins, the mutation P4715L in NSP12 was observed in 172 countries of the world, followed by L3606F in NSP3 which occurred in 134 countries while the L1174I mutation in NSP3 was observed only in 2 countries.
3.3. Mutational spectrum of the structural proteins of Pakistani SARS-CoV-2 isolates
Mutational analysis of the structural proteins of 250 Pakistani isolates of SARS-COV2 harbors six different amino acid substitutions in spike protein, one in membrane protein, nine amino acids substitutions, and a large deletion in nucleocapsid phosphoprotein. At the same time, no mutation was observed in envelope protein. The spike protein of 29 out 50 samples of SARS-COV2 Pakistani isolates harbors a signature D614G mutation indicative of the widespread mutation in Pakistani isolates. The D614G mutation in spike protein has been reported to strengthen the folding stability of the spike protein. This mutation was first reported in China and is the second-largest mutation in the United States, and its ratio is increasing as time goes on. The D614G mutation of the spike protein where Aspartic acid (D) with a polar negative side charged amino acid is substituted with Glycine (G) with a nonpolar side chain amino acid. This D614G mutation of spike protein has reported a critical mutation that makes the SARS-CoV-2 more contagious and enhances its infectivity [64]. The other less common mutation that co-occurred with D614G was N74K, V622F, Q677H, S943T, and D1153G (Table 1). Surprisingly, in the spike protein, the D614G substitution was associated with two consecutive series of two substitutions R203K, and G204R of the nucleocapsid phosphoprotein in five isolates (MW031799.1, MW031800.1, MW031801.1, MW0831802.1, and MW031803.1). These two mutations in nucleocapsid phosphoprotein were previously reported in the United States [64]. The amino acid arginine (R) and Lysine (L) are both positively charged, and its substitution may not affect the function of the nucleocapsid phosphoprotein. In contrast, the Glycine (a nonpolar) substitution by arginine (R) may affect the hydrophilicity of the nucleocapsid phosphoprotein of SARS-CoV2. Furthermore, a more significant deletion was observed in the nucleocapsid phosphoprotein of isolate MT240479.1 (Table 1). While the envelop protein was 100% identical in all isolates compared with the reference sequence (Wuhan NC_045512.2).
3.4. Mutational spectrum of the accessories proteins of Pakistani SARS-CoV-2 isolates
Among accessories protein of SARS-CoV-2 of Pakistani isolates, three proteins (ORF3A, ORF8, and ORF10) showed different amino acid substitution while no deletion was observed in these proteins of the selected isolates (Table 1). ORF3a protein (274 amino acids) of SARS-CoV-2 is expressed abundantly in infected cells and plays an essential role in pathogenesis. In these SARS-COV2 isolates, a signature Q57H substitution in ORF3 protein was observed in 23 out of 50 samples which is the second frequent mutation in these Pakistani isolates. This substitution of Q57H was first reported in Singapore and occurred in 70% of US Covid-19 cases [64]. The prevalence of Q57H substitution in ORF3 protein was reported relatively high (62.7%) in South Africa [65]. The Q57H substitution in ORF3 co-occurs with two other amino acids mutations K21 N and T223I, in isolates MW447644.1, MW447642.1 respectively. The SARS-COV-2, ORF8 has been reported to reduce the translocation of IRF3 to the nucleus and consequently reduce expression of the interferon production and to down-regulate MHC-1 in cells [66,67]. Interestingly, in Pakistani SARS-COV-2 isolates, a new mutation in ORF8, W45L was observed in the present study, while 35 out of 50 isolates have the ORF8-L genotype. The substitution of Tryptophan (W) by lysine (L) may affect the hydrophilicity of the ORF8 of SARS-CoV-2 protein. In ORF10, single substitution V30L has been observed in two isolates MT 879619.1 and MW242667.1. While the other accessories protein ORF6, ORF7A, and ORF7B were found 100% identical to the reference sequence Wuhan NC_045512.2).
3.5. Mutational spectrum of non-structural proteins of Pakistani SARS-CoV-2 isolates
Mutational analysis of non-structural proteins of 50 Pakistani SARS-CoV-2 isolates harbored 75 substitution mutations and deletions compared to the reference sequence (NC_045512.2). Among substitution, two in nsp1 (Q63L, G82S), seven in nsp2 (R197C, K261 N, F275L, K292E, V378I, D448 N, V627F) and one deletion at N449, Nine amino acids substitutions in nsp3 (L1174I, T1246I, K1308 N, T1754I, T2016K, T2408F, L2564F, A2619V, Q2702H), two substitutions (T2848I, P2926L and five amino acid deletion 3153–3157) in nsp4, four in nsp5 (G3278S, L3393F, K3353R, P3447S), Two in nsp6 (L3606F, A3656V), one in nsp8 (P4075S), four in nsp12 (A4489V, P4715L, A4921V, V4979L), two in nsp13 (T5451I, A5561T), one substitution (L6082F), seven amino acid deletion (6418–6424) in nsp14, one in nsp15 (V6600F) while no mutation was observed in nsp7, nsp9, nsp10, and nsp16. Among the substation, two substations of nsp3 T1246I and K1308 N co-occurred with substitution G3278S and P4715L of nsp5 and nsp12, respectively in isolates MW031799.1, MW031800.1, MW031801.1, MW031802.1. In isolate MT500122 the substation D448 N has preceded a deletion of N449 amino acid in nsp2. Furthermore, two consecutive deletions of five amino acids FYWFF (3153–3157) and seven amino acids LYLDAYN (6418–6424) in nsp4 have been observed in the isolate MT262993 as compared to the reference sequence (Table 1).
3.6. Identification of hyper-variable genomic hotspot in SARS-CoV-2 genomes of Pakistani isolates
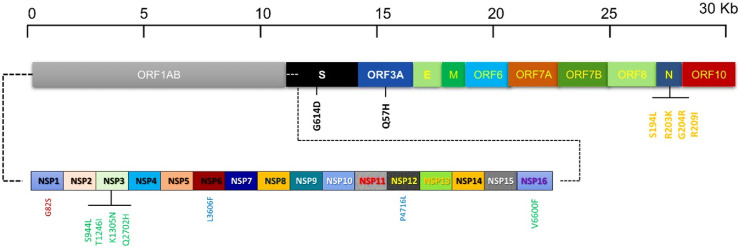
Interestingly, among all recurrent mutations in structural protein, the spike protein has one D614G (82%), nucleocapsid had three S194L (20%), R203K (14%), G204R (14%) R209I (25%) mutation hotspot (Fig. 2 ). The accessory protein the ORF3A has one mutation hotspot Q57H (63%). Among the non-structural proteins, nsp3 have four mutation hotspots S944L (11%), T1246I (11%), K1305 N (8%), NSP12 have one P4715L (57%), NSP15 have one V6600F (5%) mutation hotspots in Pakistani isolates as compared with the reference (Wuhan NC_045512.2) (Fig. 2).
Fig. 2.
Schematic representation illustrating the distribution of recurrent mutations in structural (S, E, M, N), Accessories (ORF3A, ORF6, ORF7A, ORF7B, ORF8, ORF10), and nonstructural (nsp1 to nsp 16) proteins along with the SARS-CoV-2 genome. Recurrent mutations are represented by vertical lines.
3.7. Mutational effects on structure stability
It has been determined that 27 mutations decrease structural stability, whereas six mutations enhance structural stability due to I-mutant server analysis of 96 missense mutations. These proteins' structural stability was expected to be lowered by most mutations in surface glycoprotein and nucleocapsid phosphoprotein, whereas specific changes in ORF1AB and ORF1A were projected to improve it (Table 2 ).
Table 2.
Prediction of the mutational effects on the structural stability. A negative value indicates destabilizing effect.
| Index | Protein | Amino Acid Changes | SVM2 Prediction Effect | DDG Value (kcal/mol) |
|---|---|---|---|---|
| 1 | Surface Glycoprotein | N74K | Decrease | −0.49 |
| 2 | Surface Glycoprotein | D614G | Decrease | −0.93 |
| 3 | Surface Glycoprotein | V622F | Decrease | −1.01 |
| 4 | Surface Glycoprotein | Q677H | Decrease | −0.67 |
| 5 | Surface Glycoprotein | S943T | Decrease | −0.24 |
| 6 | Surface Glycoprotein | D1153 | Decrease | −1.08 |
| 7 | Nucleocapsid phosphoprotein | P13L | Decrease | −0.48 |
| 8 | Nucleocapsid phosphoprotein | R203K | Decrease | −0.93 |
| 9 | Nucleocapsid phosphoprotein | G204R | Decrease | −0.52 |
| 10 | Nucleocapsid phosphoprotein | T205I | Decrease | −0.53 |
| 11 | ORF1AB Polyprotein | R207C | Decrease | −0.60 |
| 12 | ORF1AB Polyprotein | K292E | Decrease | −0.42 |
| 13 | ORF1AB Polyprotein | V378I | Decrease | −0.23 |
| 14 | ORF1AB Polyprotein | S944L | Decrease | 0.63 |
| 15 | ORF1AB Polyprotein | T1246I | Increase | 0.14 |
| 16 | ORF1AB Polyprotein | K1305 N | Increase | −0.10 |
| 17 | ORF1AB Polyprotein | Q2702H | Decrease | −0.68 |
| 18 | ORF1AB Polyprotein | G3278S | Decrease | −0.89 |
| 19 | ORF1AB Polyprotein | L3606F | Decrease | −1.00 |
| 20 | ORF1AB Polyprotein | A4489L | Decrease | −0.67 |
| 21 | ORF1AB Polyprotein | P4715L | Decrease | −0.83 |
| 22 | ORF1A Polyprotein | R207C | Decrease | −0.60 |
| 23 | ORF1A Polyprotein | K292E | Decrease | −0.42 |
| 24 | ORF1A Polyprotein | S318I | Increase | 0.36 |
| 25 | ORF1A Polyprotein | D448 N | Decrease | −0.98 |
| 26 | ORF1A Polyprotein | V627F | Decrease | −1.19 |
| 27 | ORF1A Polyprotein | S944L | Increase | 0.63 |
| 28 | ORF1A Polyprotein | T1246I | Increase | 0.14 |
| 29 | ORF1A Polyprotein | K1305 N | Increase | −0.10 |
| 30 | ORF1A Polyprotein | P2965L | Decrease | −0.67 |
| 31 | ORF1A Polyprotein | G3278S | Decrease | −0.89 |
| 32 | ORF1A Polyprotein | L3606F | Decrease | −1.00 |
| 33 | 0RF3A Polyprotein | Q57H | Decrease | −0.90 |
3.8. Molecular consequences of reported mutations in different pathways
Additionally, three mutations occurring in ORFIA were predicted to alter the molecular consequences, including Loss of Disulfide linkage, Loss of Catalytic site, Loss of Allosteric site, Gain of Proteolytic cleavage, and Gain of Sulfation. Similarly, one mutation each in Membrane Glycoprotein and ORF8 were predicted to alter transmembrane protein,altered DNA binding, as well as altered disordered interface respectively Table 3 .
Table 3.
Prediction of effects of the mutation on different biological pathways.
| Protein Name | Mutation | Effects |
|---|---|---|
| Membrane Glycoprotein | H125Y | Altered Transmembrane protein |
| Altered Ordered interface | ||
| ORF8 | W45L | Altered Ordered interface |
| Altered Disordered interface | ||
| Altered DNA binding | ||
| Altered Transmembrane protein | ||
| ORF1A | T2408I | Altered Transmembrane protein |
| Gain of strand | ||
| Loss of Disulfide linkage at C2404 | ||
| Gain of N-linked glycosylation at N2405 | ||
| Loss of Catalytic site at C2404 | ||
| Loss of GPI-anchor amidation at N2405 | ||
| P2965L | Altered Transmembrane protein | |
| Altered Ordered interface | ||
| Loss of Relative solvent accessibility | ||
| Altered Metal binding | ||
| P3447S | Altered Transmembrane protein | |
| Altered Ordered interface | ||
| Loss of Allosteric Site at Y3445 | ||
| Gain of Strand | ||
| Loss of Relative solvent accessibility | ||
| Gain of Proteolytic cleavage at D3450 | ||
| Loss of Pyrrolidone carboxylic acid at Q3452 | ||
| Gain of Sulfation at Y3445 | ||
| ORF1AB | L3606 | Loss of Helix |
| Gain of Loop | ||
| Altered Ordered interface | ||
| Gain of Sulfation at Y3607 | ||
| Gain of Strand |
3.9. Modelling of the wild type and spike mutant structures
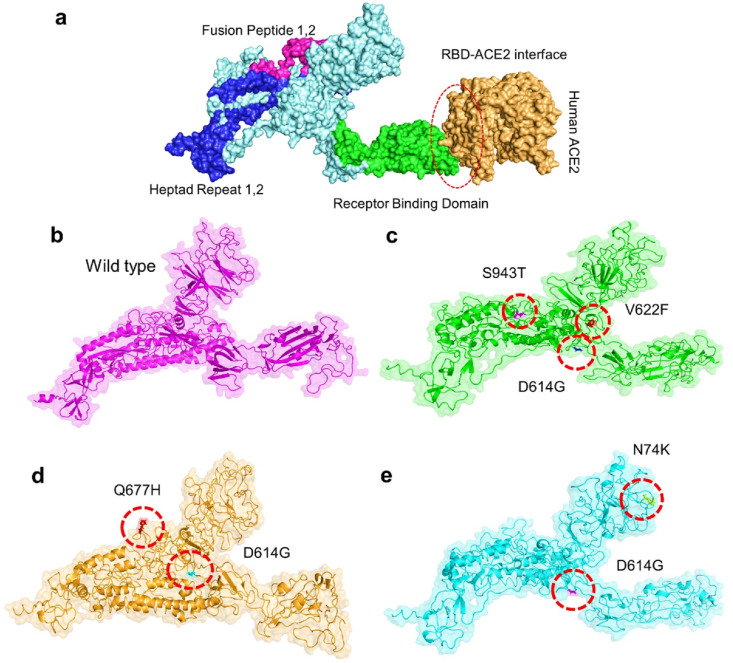
Continued mutations in SARS-CoV-2 Genome cause potential differences in its transmission, virulence, and pathogenesis compared to their wild counterparts. Recently reported mutations in the spike proteins of SARS-CoV-2 from South Africa (Lys417Asn, Glu484Lys, Asn501Tyr) (501Y.V2Variant) and Brazil (Lys417Asn, Glu484Lys, Asn501Tyr) had led these strains to evade the vaccines successfully [18]. Therefore, it was necessary to explore the mutations in the spike protein of Pakistani isolates. SARS-CoV-2 virus Spike proteins have an RBD domain, which aids in binding to ACE2 receptors on the host cell. This binding sets off a chain reaction of events that lead to fusing the host cell and viral membranes for cell penetration. SARS-spike CoV-2's RBD was submitted to comparative binding and biophysical study upon interaction with ACE2 because of its relevance in virulence and pathogenesis. RCSB was used to get the structure of the Spike and ACE2 proteins (PDB ID:6M0J). Docking the spike protein with the human ACE2 receptor, HDOCK online server was used. To verify whether the binding affinity of the RBD domain for the human ACE2 can be affected by the six mutations present in the spike protein of Pakistani isolates, we made a double mutant, which consists of the N74K-D614G, N74K-Q677H, D614G-V622F, D614G-S943F, D614G-V622F–S943F, and D614G-Q677H. Structural organization, domain separation, and binding interface of spike protein are shown in Fig. 3 a. The mutants were generated by Chimera and are given in Fig. 3b–e.
Fig. 3.
Representation of spike glycoprotein structure (PDB ID:6M0J) and SARS-CoV-2 binding domain (a) Different domains of spike protein with different colors. The green color shows the RBD domain while the orange color shows the ACE2 receptor. The right-side figure shows the binding interface along with its stick representation of the key hydrogen interactions of ACE2 and the spike RBD. (b) wild type spike protein, (c) D614G, V622F, S943T (d) D614G, Q677H (e) N74K, D614G. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.10. Docking of the wild type and mutant spike structures
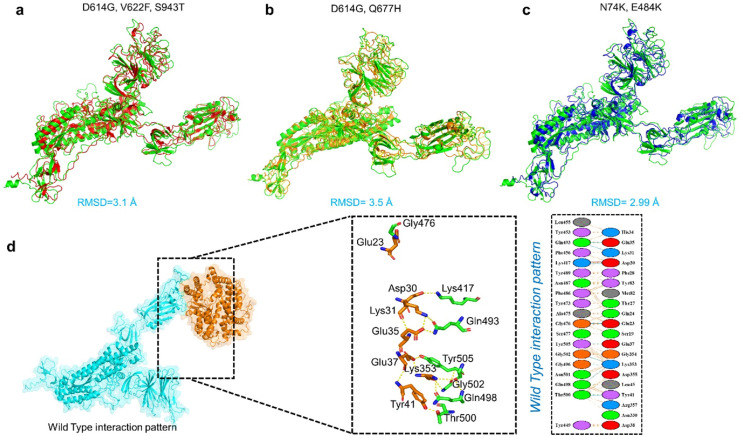
Keeping in mind the involvement of spike protein in a particular process, including attachment and pathogenesis, we investigated the critical analysis of both wild and mutant versions of the spike protein (Spike RBD) with ACE2 receptor. Structural study of protein interactions and their binding energy is an important initial step in biological process knowledge. Binding affinity, which defines whether a complex is formed under specific conditions, is essential to regulate molecular interactions, create new treatments (i.e. directing rational drug design) and anticipate the impact variation on the protein interfaces [68]. Prior to docking of Spike and ACE2 superimposition of the mutants was performed to check the structural variations caused by the mutations. The generated mutants were superimposed on wild-type spike protein, and the RMSD values were recorded (Fig. 4 a, b, c).
Fig. 4.
(a, b, c) Superimposed structure of wild-type spike protein (green) with D614G-V622F–S943F (red), and D614G-Q677H (orange) and. N74K-D614 (blue). The RMSD of each superimposition was reported to be 3.1 Å, 3.5 Å, and 2.99 Å. (d) Docking complex of wild-type spike protein with ACE2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
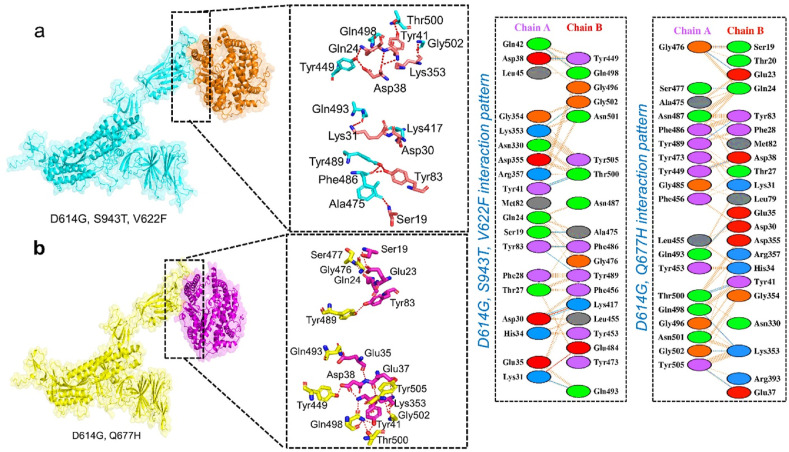
Therefore, we used HDOCK to perform the docking of ACE2 with the wild-type and double mutant spike protein (D614G-V622F–S943F D614G-Q677H and N74K-D614G) to explain how these mutations led to the higher infectivity of SARS-CoV-2 variants. HDOCK resulted in a docking score of −12.40 kcal/mol for the ACE2-wild type spike complex. PDBsum interaction study revealed that the interface is made up of 37 residues. 18 residues are supplied by ACE2 and 15 by spike-RBD, which are responsible for interface formation. One salt bridge, 11 hydrogen bonds, and 125 non-bonded interactions were found at the interface. The hydrogen bonds formed by the ACE2-spike (RBD) wild include Gln493-Glu35, Gln493-Lys31, Lys417-Asp30, Gly476-Glu23, Tyr505-Glu37, Gly502-Lys353, Gly496- Lys353, Gln498- Lys353, and double hydrogen bond between Thr500-Tyr41(Fig. 4d). One salt bridge was reported between Asp30 (ACE2) and Lys417 (spike RBD).
The HDOCK docking score for D614G-V622F–S943F (ACE2-spike RBD) was reported as −13.70 kcal/mol. This distinction between wild-type and mutant was investigated further by examining the molecular interactions. The interaction analysis showed two salt bridges and an increased number of hydrogen bonds (12 hydrogen bonds) in mutant form. The enhanced interaction is due to substituted residue D614G, V622F, S943F in the spike protein complex (Fig. 5 a). The key residues Lys31-Glu484 and Asp30-Lys417 established the salt bridges. Among the hydrogen bonds Thr747-Asp609, Lys417-His195, Glu406-His195, Thr415-Arg195, Gln755-His493, Asn501-Gln86, Gln498-Gln86, Tyr489-Ser77, Tyr489-Lys74, and Arg457-Ser109 residues are involved. As previously stated, the D614G-V622F–S943F mutation is responsible for enhancing the dissemination and infectivity of the SARS-CoV-2 variant and increasing the binding affinity and infectivity. The HDOCK docking score for D614G, Q677H (ACE2-spike RBD), was −13.42 kcal/mol. The difference between the HDOCK score of wild-type and mutant is explained in terms of molecular interactions. Among the hydrogen bonds Gly476-Ser19, Gly476-Glu23, Ser477-Gln24, Tyr489-Tyr83, Tyr449-Asp38, Gln493-Glu35, Thr500-Tyr41, Gln489-Lys353, Gly502-Lys353, and Tyr505-Glu37 residues are involved (Fig. 5b). As a result, the D614G, Q677H double mutant enhanced binding efficiency, and this mutant, as well as other interface residues, are important hotspots for therapeutic development against SARS-CoV-2 variants.
Fig. 5.
Docking complexes of D614G-V622F–S943F spike protein with ACE2. (b) Docking complexes of D614G-Q677H spike protein with ACE2.
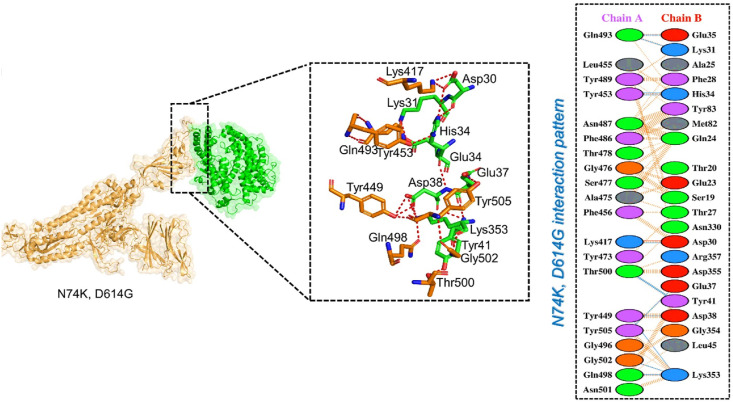
The predicted score of HDOCK for the ACE2-N74K, D614G mutant spike protein was −12.50 which is comparable with the wild-type complex. According to a PDBsum study of the complex, the interface is formed by 37 residues, 18 of which are supplied by ACE2 and 15 by the spike-RBD. According to the interaction study, both structures create 1 salt bridge, 11 hydrogen bonds, and 125 non-bonded interactions. The hydrogen bonds formed by the ACE2- N74K, D614G spike (RBD) include Gln493-Glu35, Gln493-Lys31, Tyr453-His34, Lys417-Asp30, Thr500-Tyr41, Tyr449-Asp38, Tyr505-Glu37, Gly502-Lys353, Tyr449- Lys353, Gln498-Lys353, and double hydrogen bond between Thr500-Tyr41(Fig. 6 ). In this complex, most of the hydrogen binding pattern is similar to the wild-type complex.
Fig. 6.
Docking complexes of ACE2-N74K, D614G spike protein with ACE2. The right panel shows the 2D interaction pattern generated with PDBsum.
3.11. Estimation of KD
Estimation of the KD revealed that D614G-S943T-V622F (2.0 E−10) variant binds stronger than all. Moreover, the strength of the wild type and mutants was ranked as D614G-Q677H variant (1.0 E−09), wild type (1.9 E−09) while for the N74K-D614G variant the predicted KD value was 2.2 E−09. Conclusively this shows that the two mutants possess stronger affinity than the wild type which might be responsible for the increased cases in Pakistan. The docking score and the predicted KD values are given in Table 4 .
Table 4.
The predicted docking scores and KD values for each complex.
| Complex | ΔG (kcal mol−1) | KD (M) at 38.0 °C |
|---|---|---|
| Wild type | −12.40 | 1.9E−09 |
| D614G-S943T-V622F Variant | −13.42 | 2.0E−10 |
| D614G-Q677H Variant | −13.70 | 1.0E−09 |
| N74K-D614G Variant | −12.50 | 2.2E−09 |
3.12. Dynamic stability and flexibility analysis
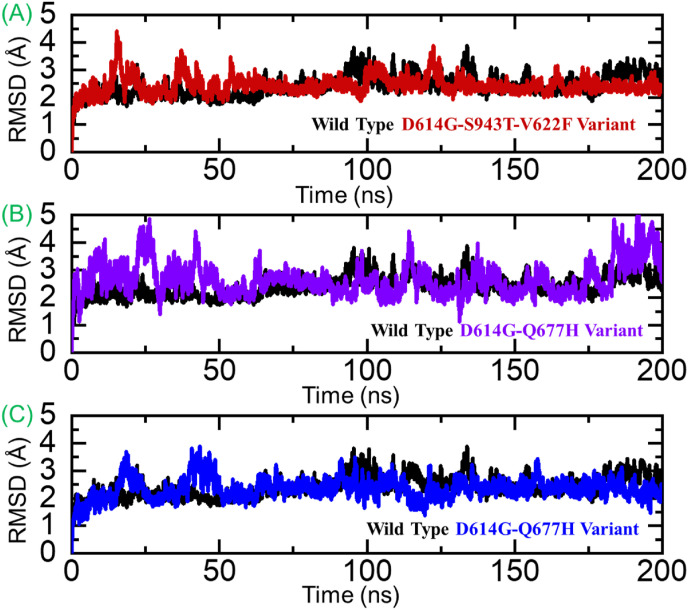
To understand the dynamic behavior of the wild type and these variants, MD simulation of each complex was performed. The stability of each complex was assessed as root mean square deviation (RMSD) and residual flexibility as root mean square fluctuation. The RMSD graphs shown in Fig. 7 A-C demonstrate that the wild type exhibit more stable dynamics than the variants. From Fig. 7A, it can be seen that the wild type complex attained the equilibrium at 25ns and reached stability at 2.0 Å. On the other hand, the D614G-S943T-V622F variant complex demonstrated unstable dynamics at different times interval thus corroborated with the servers’ predicted results. Unlike the D614G-S943T-V622F variant, the other two variants i.e., D614G-Q677H and N74K-D614G reported a significantly destabilizing effect throughout the simulation. From Fig. 7B and C, it can be seen that these two complexes demonstrated significant structural perturbation and hence reports uniform results with the aforementioned results predicted by different servers.
Fig. 7.
Dynamic stability analysis of the wild type and mutant complexes. (A) RMSD of the wild type. (B) RMSD of D614G-S943T-V622F, (C) RMSD of D614G-Q677H and (D) RMSD N74K-D614G variant complex.
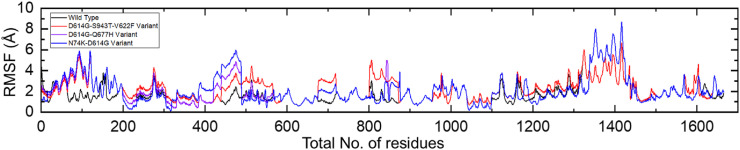
On the other hand, the RMSF results demonstrated a differential flexibility index for each complex thus showing the impact of these mutations on the structural-dynamic features. The RMSF results of each complex are shown in Fig. 8 .
Fig. 8.
Residual flexibility of the wild type, D614G-S943T-V622F, D614G-Q677H, and RMSD N74K-D614G variant complexes.
3.13. Binding free energy calculations
To connect protein conformational changes with the binding, induced by mutations in the proteins, we used the MM/GBSA technique to calculate the binding energy of spike RBD to hACE2. The binding comparison revealed significant variation in the spike RBD binding between the wild type, D614G-V622F–S943F, and D614G-Q677H double mutants. As given in Table 5 , the total binding energy for the D614G-Q677H variant was −75.78 kcal/mol followed by the D614G-S943T-V622F variant −75.17 kcal/mol and −73.84 kcal/mol for the N74K-D614G variant. The total binding energy for the wild type was −66.34 kcal/mol. The vdW results reported a comparable result however the electrostatic energies are reported to be higher in the case of the variants which strongly corroborate with the previous papers which reported an increment in the electrostatic energy by variants. The notion of electrostatic energy has been a key factor in the enhanced binding [20,59,60,69]. In sum, the polar solvation energy supplies a non-favorable contribution by comparing with non-polar which reported a good contribution. The dynamic's environment's charge density seems to engage with itself infrequently, explaining the non-significant effect in binding. In brief, the solvation energy of variants adhering to ACE2 is unfavorable, whilst gas-phase energy is critical for the variants' strong binding to ACE2. Decisively this shows that the re-ranking of the complex using MM/GBSA validated that the mutants possess a stronger affinity towards ACE2 than the wild type thus enhancing the transmission and infectivity. The MM/GBSA results including vdW (van der Waal), electrostatic (ELE), GB (generalized Born), SA (Surface Area), and the total binding energy are given in Table 5.
Table 5.
MM/GBSA analysis of the wild type and mutant complexes. All the results are calculated in kcal/mol.
| Complex | VDW | ELE | GB | SA | TOTAL |
|---|---|---|---|---|---|
| Wild Type | −108.39 | −659.94 | 716.45 | −14.46 | −66.34 |
| D614G-S943T-V622F Variant | −114.64 | −697.17 | 751.1 | −14.46 | −75.17 |
| D614G-Q677H Variant | −109.08 | −700.02 | 747.87 | −14.55 | −75.78 |
| N74K-D614G Variant | −109.81 | −687.13 | 737.56 | −14.46 | −73.84 |
4. Conclusions
In summary, this study used genome-based analysis and molecular modeling approaches to determine a basis for the increased infection and transmission of SARS-CoV-2 in Pakistan. The data revealed that the D614G mutation particularly contributes to the enhanced infectivity. A significant bonding reprogramming was identified, and the structural dynamic features strongly corroborated the predicted results. However, such polymorphic sites should be further studied to explore the evolutionary paradigm and any emerging variants. This work provides a strong impetus for the development of novel drugs against the strains circulating in Pakistan and around the world.
Data availability
All the data is available on RCSB, UniProt, and any simulation data would be provided on reasonable demand. The accession numbers to access this data are given in the manuscript.
Ethical approval
Not Applicable.
Declaration of competing interest
Declared None.
Acknowledgments
Dong-Qing Wei is supported by grants from the Key Research Area Grant 2016YFA0501703 of the Ministry of Science and Technology of China, the National Science Foundation of China (Grant No. 32070662, 61832019, 32030063), the Science and Technology Commission of Shanghai Municipality (Grant No.: 19430750600), as well as SJTU JiRLMDS Joint Research Fund and Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2021ZD02). The computations were partially performed at the PengCheng Lab. and the Center for High-Performance Computing, Shanghai Jiao Tong University.
References
- 1.Benvenuto D., Giovanetti M., Ciccozzi A., Spoto S., Angeletti S., Ciccozzi M. The 2019-new coronavirus epidemic: evidence for virus evolution. J. Med. Virol. 2020;92:455–459. doi: 10.1002/jmv.25688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stadnytskyi V., Bax C.E., Bax A., Anfinrud P. The airborne lifetime of small speech droplets and their potential importance in SARS-CoV-2 transmission. Proc. Natl. Acad. Sci. U.S.A. 2020;117:11875–11877. doi: 10.1073/pnas.2006874117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klompas M., Baker M.A., Rhee C. Airborne transmission of SARS-CoV-2: theoretical considerations and available evidence. JAMA. 2020;324:441–442. doi: 10.1001/jama.2020.12458. [DOI] [PubMed] [Google Scholar]
- 5.Hilal El Idrissi H. COVID-19: what you need to know. Gene. Rep. 2020;20:100756. doi: 10.1016/j.genrep.2020.100756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahman H.S., Aziz M.S., Hussein R.H., Othman H.H., Salih Omer S.H., Khalid E.S., Abdulrahman N.A., Amin K., Abdullah R. The transmission modes and sources of COVID-19: a systematic review. Intern. J. Surg. Open. 2020;26:125–136. doi: 10.1016/j.ijso.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang D., Comish P., Kang R. The hallmarks of COVID-19 disease. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haque S.M., Ashwaq O., Sarief A., Azad John Mohamed A.K. A comprehensive review about SARS-CoV-2. Future Virol. 2020;15:625–648. doi: 10.2217/fvl-2020-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day T., Gandon S., Lion S., Otto S.P. On the evolutionary epidemiology of SARS-CoV-2. Curr. Biol. 2020;30:R849–R857. doi: 10.1016/j.cub.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinesh D.C., Chalupska D., Silhan J., Veverka V., Boura E. bioRxiv; 2020. Structural Basis of RNA Recognition by the SARS-CoV-2 Nucleocapsid Phosphoprotein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duchene S., Featherstone L., Haritopoulou-Sinanidou M., Rambaut A., Lemey P., Baele G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus evol. 2020;6:veaa061. doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussain I., Pervaiz N., Khan A., Saleem S., Shireen H., Wei D.-Q., Labrie V., Bao Y., Abbasi A.A. Evolutionary and structural analysis of SARS-CoV-2 specific evasion of host immunity. Gene Immun. 2020:1–11. doi: 10.1038/s41435-020-00120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., Swann O.C., Kugathasan R., Penn R., Brown J.C., Sanchez-David R.Y. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021:1–11. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 18.Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.-Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021 doi: 10.1002/jcp.30367. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan A., Khan M., Saleem S., Babar Z., Ali A., Khan A.A., Sardar Z., Hamayun F., Ali S.S., Wei D.-Q. Phylogenetic analysis and structural perspectives of RNA-dependent RNA-polymerase inhibition from SARs-CoV-2 with natural products. Interdiscipl. Sci. Comput. Life Sci. 2020;12:335–348. doi: 10.1007/s12539-020-00381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A. Khan, D.-Q. Wei, K. Kousar, J. Abubaker, S. Ahmad, J. Ali, F. Al-Mulla, S.S. Ali, N. Nizam-Uddin, A.M. Sayaf, Preliminary Structural Data Revealed that the SARS-CoV-2 B. 1.617 Variant's RBD Binds to ACE2 Receptor Stronger than the Wild Type to Enhance the Infectivity, ChemBioChem. [DOI] [PMC free article] [PubMed]
- 21.Garcia-Beltran W.F., Lam E.C., Denis K.S., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., Sigal A., Schmidt A.G., Iafrate A.J., Naranbhai V., Balazs A.B. medRxiv; 2021. Multiple SARS-CoV-2 Variants Escape Neutralization by Vaccine-Induced Humoral Immunity. 2021.2002.2014.21251704. [Google Scholar]
- 22.Nagano K., Tani-Sassa C., Iwasaki Y., Takatsuki Y., Yuasa S., Takahashi Y., Nakajima J., Sonobe K., Ichimura N., Nukui Y. medRxiv; 2021. SARS-CoV-2 R. 1 Lineage Variants Prevailed in Tokyo in March 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou B., Thao T.T.N., Hoffmann D., Taddeo A., Ebert N., Labroussaa F., Pohlmann A., King J., Portmann J., Halwe N.J. bioRxiv; 2020. SARS-CoV-2 Spike D614G Variant Confers Enhanced Replication and Transmissibility. [Google Scholar]
- 24.Hoffmann M., Hofmann-Winkler H., Krüger N., Kempf A., Nehlmeier I., Graichen L., Sidarovich A., Moldenhauer A.-S., Winkler M.S., Schulz S., Jäck H.-M., Stankov M.V., Behrens G.M.N., Pöhlmann S. bioRxiv; 2021. SARS-CoV-2 Variant B.1.617 Is Resistant to Bamlanivimab and Evades Antibodies Induced by Infection and Vaccination. 2021.2005.2004.442663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A. SARS-CoV-2 variants B. 1.351 and P. 1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393. doi: 10.1016/j.cell.2021.03.036. e2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh J., Rahman S.A., Ehtesham N.Z., Hira S., Hasnain S.E. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 2021:1–3. doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- 27.Focosi D., Tuccori M., Baj A., Maggi F. Multidisciplinary Digital Publishing Institute; 2021. SARS-CoV-2 Variants: A Synopsis of in Vitro Efficacy Data of Convalescent Plasma, Currently Marketed Vaccines, and Monoclonal Antibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98:495. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy Á., Pongor S., Győrffy B. Different mutations in SARS-CoV-2 associate with severe and mild outcome. Int. J. Antimicrob. Agents. 2021;57:106272. doi: 10.1016/j.ijantimicag.2020.106272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu S., Tian C., Liu P., Guo D., Zheng W., Huang X., Zhang Y., Liu L. Effects of SARS‐CoV‐2 mutations on protein structures and intraviral protein–protein interactions. J. Med. Virol. 2021;93:2132–2140. doi: 10.1002/jmv.26597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T., Farzan M., Choe H. BioRxiv; 2020. The D614G Mutation in the SARS-CoV-2 Spike Protein Reduces S1 Shedding and Increases Infectivity. [Google Scholar]
- 32.Groves D.C., Rowland-Jones S.L., Angyal A. The D614G mutations in the SARS-CoV-2 spike protein: implications for viral infectivity, disease severity and vaccine design. Biochem. Biophys. Res. Commun. 2021;538:104–107. doi: 10.1016/j.bbrc.2020.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohammad A., Alshawaf E., Marafie S.K., Abu-Farha M., Abubaker J., Al-Mulla F. Higher binding affinity of furin for SARS-CoV-2 spike (S) protein D614G mutant could be associated with higher SARS-CoV-2 infectivity. Int. J. Infect. Dis. 2021;103:611–616. doi: 10.1016/j.ijid.2020.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751. doi: 10.1016/j.cell.2020.09.032. e738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernal J.L., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., Myers R., Campbell C., Amirthalingam G., Edmunds M., Zambon M., Brown K., Hopkins S., Chand M., Ramsay M. medRxiv; 2021. Effectiveness of COVID-19 Vaccines against the B.1.617.2 Variant; p. 2021. 2005.2022.21257658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Mulligan M.J., Landau N.R. BioRxiv; 2021. The Spike Proteins of SARS-CoV-2 B. 1.617 and B. 1.618 Variants Identified in India Provide Partial Resistance to Vaccine-Elicited and Therapeutic Monoclonal Antibodies. [Google Scholar]
- 37.Ruan Y., Wei C.L., Ling A.E., Vega V.B., Thoreau H., Thoe S.Y.S., Chia J.-M., Ng P., Chiu K.P., Lim L. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ali A., Vijayan R. Dynamics of the ACE2–SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-71188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan M.T., Khan A., Rehman A.U., Wang Y., Akhtar K., Malik S.I., Wei D.-Q. Structural and free energy landscape of novel mutations in ribosomal protein S1 (rpsA) associated with pyrazinamide resistance. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-44013-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khan A., Khan M.T., Saleem S., Junaid M., Ali A., Ali S.S., Khan M., Wei D.-Q. Structural Insights into the mechanism of RNA recognition by the N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein. Comput. Struct. Biotechnol. J. 2020 doi: 10.1016/j.csbj.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhardwaj V.K., Singh R., Das P., Purohit R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Comput. Biol. Med. 2021;128:104117. doi: 10.1016/j.compbiomed.2020.104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh R., Bhardwaj V.K., Sharma J., Purohit R., Kumar S. In-silico evaluation of bioactive compounds from tea as potential SARS-CoV-2 nonstructural protein 16 inhibitors. J. tradition. complemen. med. 2021 doi: 10.1016/j.jtcme.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma J., Kumar Bhardwaj V., Singh R., Rajendran V., Purohit R., Kumar S. An in-silico evaluation of different bioactive molecules of tea for their inhibition potency against non structural protein-15 of SARS-CoV-2. Food Chem. 2021;346:128933. doi: 10.1016/j.foodchem.2020.128933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh R., Bhardwaj V.K., Sharma J., Kumar D., Purohit R. Identification of potential plant bioactive as SARS-CoV-2 Spike protein and human ACE2 fusion inhibitors. Comput. Biol. Med. 2021;136:104631. doi: 10.1016/j.compbiomed.2021.104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miryala S.K., Basu S., Naha A., Debroy R., Ramaiah S., Anbarasu A., Natarajan S. Identification of bioactive natural compounds as efficient inhibitors against Mycobacterium tuberculosis protein-targets: a molecular docking and molecular dynamics simulation study. J. Mol. Liq. 2021;341:117340. [Google Scholar]
- 46.Basu S., Veeraraghavan B., Ramaiah S., Anbarasu A. Novel cyclohexanone compound as a potential ligand against SARS-CoV-2 main-protease. Microb. Pathog. 2020;149:104546. doi: 10.1016/j.micpath.2020.104546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data–from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li M., Yu T.-f. The application of DNAStar soft in research of animal virus. J. Sci. Teach. Coll. Univ. 2010;30:61–63. [Google Scholar]
- 50.Thompson J.D., Gibson T.J., Higgins D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. protoc. bioinform. 2003:2.3. 1–2.3. 22. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 51.Wang L., Zheng J., Luo Y., Xu T., Zhang Q., Zhang L., Xu M., Wan J., Wang M.B., Zhang C. Construction of a genomewide RNA i mutant library in rice. Plant biotechnol. j. 2013;11:997–1005. doi: 10.1111/pbi.12093. [DOI] [PubMed] [Google Scholar]
- 52.Pejaver V., Urresti J., Lugo-Martinez J., Pagel K.A., Lin G.N., Nam H.-J., Mort M., Cooper D.N., Sebat J., Iakoucheva L.M. BioRxiv; 2017. MutPred2: Inferring the Molecular and Phenotypic Impact of Amino Acid Variants; p. 134981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rose P.W., Prlić A., Altunkaya A., Bi C., Bradley A.R., Christie C.H., Costanzo L.D., Duarte J.M., Dutta S., Feng Z. Nucleic acids research; 2016. The RCSB Protein Data Bank: Integrative View of Protein, Gene and 3D Structural Information; p. gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 55.Yan Y., Tao H., He J., Huang S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020;15:1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 56.Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A.M., Vangone A. PRODIGY: a web server for predicting the binding affinity of protein–protein complexes. Bioinformatics. 2016;32:3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 57.Case D.A., Cheatham T.E., III, Darden T., Gohlke H., Luo R., Merz K.M., Jr., Onufriev A., Simmerling C., Wang B., Woods R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pearlman D.A., Case D.A., Caldwell J.W., Ross W.S., Cheatham T.E., III, DeBolt S., Ferguson D., Seibel G., Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 59.Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021;236:7045–7057. doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khan A., Gui J., Ahmad W., Haq I., Shahid M., Khan A.A., Shah A., Khan A., Ali L., Anwar Z. The SARS-CoV-2 B. 1.618 variant slightly alters the spike RBD–ACE2 binding affinity and is an antibody escaping variant: a computational structural perspective. RSC Adv. 2021;11:30132–30147. doi: 10.1039/d1ra04694b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 62.Sun H., Li Y., Tian S., Xu L., Hou T. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys. Chem. Chem. Phys. 2014;16:16719–16729. doi: 10.1039/c4cp01388c. [DOI] [PubMed] [Google Scholar]
- 63.Weng G., Wang E., Wang Z., Liu H., Zhu F., Li D., Hou T. HawkDock: a web server to predict and analyze the protein–protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. 2019;47:W322–W330. doi: 10.1093/nar/gkz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang R., Chen J., Gao K., Hozumi Y., Yin C., Wei G.-W. Analysis of SARS-CoV-2 mutations in the United States suggests presence of four substrains and novel variants. Commun. bio. 2021;4:1–14. doi: 10.1038/s42003-021-01754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laamarti M., Alouane T., Kartti S., Chemao-Elfihri M., Hakmi M., Essabbar A., Laamarti M., Hlali H., Bendani H., Boumajdi N. Large scale genomic analysis of 3067 SARS-CoV-2 genomes reveals a clonal geo-distribution and a rich genetic variations of hotspots mutations. PLoS One. 2020;15 doi: 10.1371/journal.pone.0240345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rashid F., Dzakah E.E., Wang H., Tang S. The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon beta. Virus Res. 2021;296:198350. doi: 10.1016/j.virusres.2021.198350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y., Zhang J., Chen Y., Luo B., Yuan Y., Huang F., Yang T., Yu F., Liu J., Liu B. BioRxiv; 2020. The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion through Potently Downregulating MHC-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith G.R., Sternberg M.J. Prediction of protein-protein interactions by docking methods. Curr. Opin. Struct. Biol. 2002;12:28–35. doi: 10.1016/s0959-440x(02)00285-3. [DOI] [PubMed] [Google Scholar]
- 69.Khan A., Khan T., Ali S., Aftab S., Wang Y., Qiankun W., Khan M., Suleman M., Ali S., Heng W. Biomedicine & Pharmacotherapy; 2021. SARS-CoV-2 New Variants: Characteristic Features and Impact on the Efficacy of Different Vaccines; p. 112176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data is available on RCSB, UniProt, and any simulation data would be provided on reasonable demand. The accession numbers to access this data are given in the manuscript.