Fig. 3.
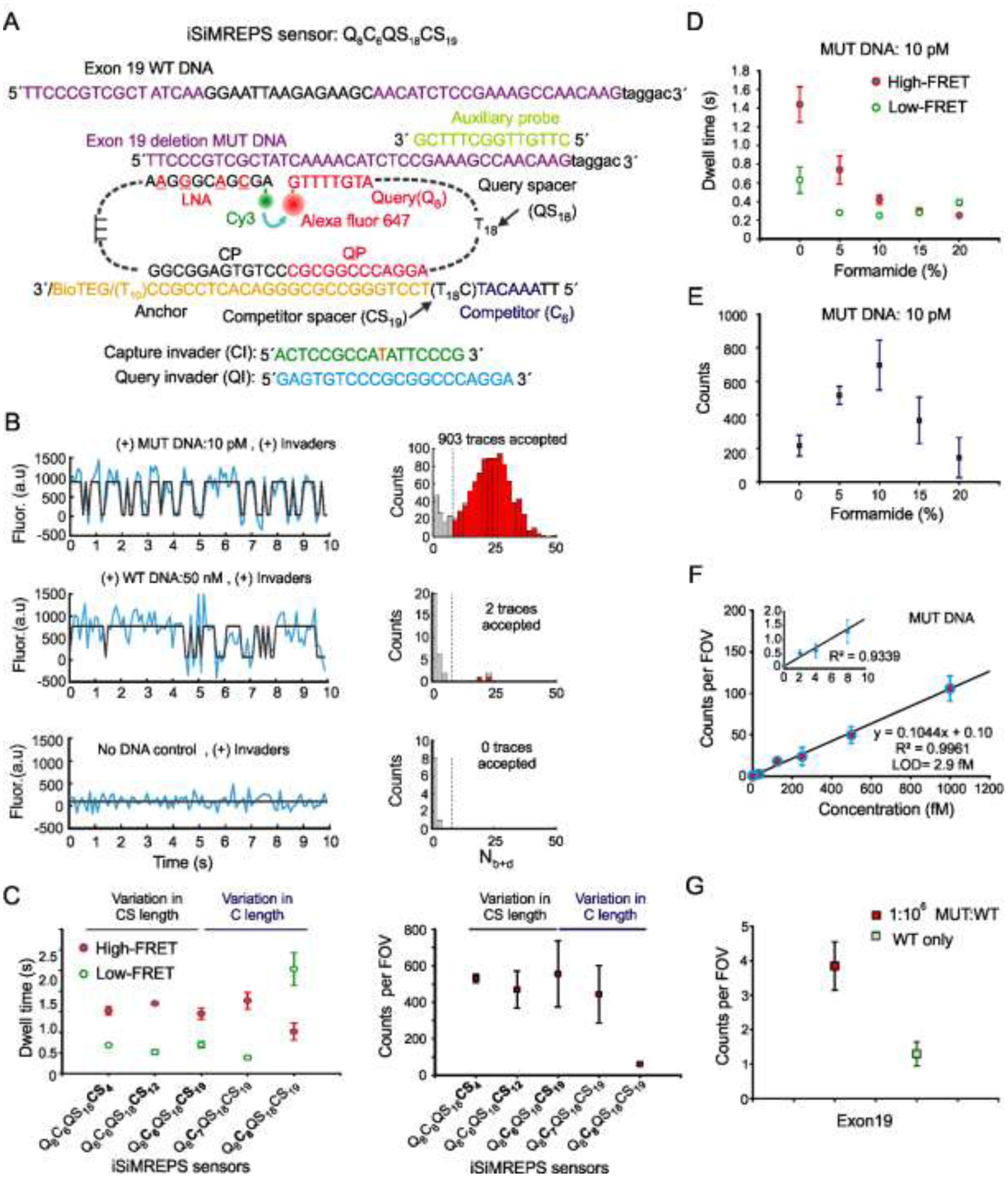
Development of smFRET-based iSiMREPS for detection of EGFR exon 19 deletion mutant DNA. (A) Schematic of the sensor design of exon 19 deletion mutant and WT DNA detection. (B) Representative single-molecule kinetic fingerprints and histograms of the number of candidate molecules per field-of-view (FOV) showing a given number of binding and dissociation events (Nb+d) after applying thresholds for FRET intensity, signal-to-noise, and dwell times of target bound and non-target-bound states in presence of 10 pM exon 19 deletion mutant DNA (top), 50 nM WT DNA (middle), and no DNA control (bottom). (C) The average dwell times spent in the high-FRET and low-FRET states for different iSiMREPS sensors designs and accepted counts per FOV at a standard acquisition of 10 s. Lifetime averages were calculated using an exponential decay fitting model. (D, E) The average dwell times of high- and low-FRET states and the number of candidate mutant DNA bound molecules per FOV for each formamide v/v% condition with a standard data acquisition of 10 s after applying kinetic thresholds. (F) Standard curve for exon 19 deletion mutant DNA showing an LOD of ~2.9 fM. Linear fits were constrained to a y-intercept of accepted counts at 0 fM, yielding R2 values = 0.9961. (G) Comparison of counts from low MUT allelic fraction and WT only conditions for determining specificity. The specificity obtained was 99.9999% over the MUT fraction of 0.0001%. All data are presented as the mean ± s.d of n = ≥ 3 independent measurements. For each independent measurement, 10 FOVs were collected. Panels A, D, E and G were adapted with permission from reference 13.
