Abstract
Background:
Several epigenome-wide association studies (EWAS) of ambient particulate matter with aero-dynamic diameter ≤ 2.5 µm (PM2.5) have been reported. However, EWAS of PM2.5 elements (PEs), reflecting different emission sources, are very limited.
Objectives:
We performed EWAS of short- and intermediate-term exposure to PM2.5 and 13 PEs. We hypothesized that significant changes in DNAm may vary by PM2.5 mass and its elements.
Methods:
We repeatedly collected blood samples in the Normative Aging Study and measured leukocyte DNA methylation (DNAm) with the Illumina HumanMethylation450K BeadChip. We collected daily PM2.5 and 13 Pes at a fixed central site. To estimate the associations between each PE and DNAm at individual cytosine-phosphate-guanine (CpG) sites, we incorporated a distributed-lag (0–27 d) term in the setting of median regression with subject-specific intercept and examined cumulative lag associations. We also accounted for selection bias due to loss to follow-up and mortality prior to enrollment. Significantly differentially methylated probes (DMPs) were identified using Bonferroni correction for multiple testing. We further conducted regional and pathway analyses to identify significantly differentially methylated regions (DMRs) and pathways.
Results:
We included 695 men with 1,266 visits between 1999 and 2013. The subjects had a mean age of 75 years. The significant DMPs, DMRs, and pathways varied by to PM2.5 total mass and PEs. For example, PM2.5 total mass was associated with 2,717 DMPs and 10,470 DMRs whereas Pb was associated with 3,173 DMPs and 637 DMRs. The identified pathways by PM2.5 mass were mostly involved in mood disorders, neuroplasticity, immunity, and inflammation, whereas the pathways associated with motor vehicles (BC, Cu, Pb, and Zn) were related with cardiovascular disease and cancer (e.g., “PPARs signaling”).
Conclusions:
PM2.5 and PE were associated with methylation changes at multiple probes and along multiple pathways, in ways that varied by particle components.
Keywords: PM2.5, PM2.5 elements, DNA methylation, Epigenome-wide association study, Distributed-lag, Pathway analyses
1. Introduction
Exposure to ambient fine particulate matter (an aerodynamic diameter ≤2.5 µm; PM2.5), have been linked with death (Kloog et al. 2013) and multi-systemic diseases (Brook et al. 2010; Gu et al. 2020; Hunt et al. 2003; Nassan et al. 2021; Turner et al. 2011), in short-, intermediate-, and long-term time windows. Moreover, PM2.5 is a complex mixture of many particulate elements (PEs) that differ in their physicochemical, toxicological properties (Council 2004), reflecting specific emission sources. Emerging epidemiological studies have found that PM2.5-related health effects vary by different PEs (Dai et al. 2016; Franklin et al. 2008; Wang et al. 2020b; White et al. 2019). However, the underlying molecular alterations caused by PM2.5 and its PEs have not been adequately investigated.
DNA methylation (DNAm), a chemical modification of DNA with a methyl group addition predominantly at a cytosine-phosphate-guanine (CpG) site (Robertson and Jones 2000), has been reported to be associated with human health, such as inflammation (Gonzalez-Jaramillo et al. 2019), oxidative stress (Menezo et al. 2016), aging (Gonzalo 2010), cardiovascular disease (Pepin et al. 2019), and cancer (Wang and Lei 2018). Meanwhile, DNAm has been linked with PM2.5 across different time windows (Eze et al. 2020; Gondalia et al. 2019; Gruzieva et al. 2019; Jiang et al. 2014; Li et al. 2018; Panni et al. 2016; Plusquin et al. 2017). For instance, Gondalia et al. assessed epigenome-wide association studies (EWAS) of monthly mean PM2.5 before health examinations across twelve elderly cohorts and found three suggestive significant CpGs by random-effects meta-analysis (Gondalia et al. 2019). Panni et al. studied EWAS of PM2.5 in both short- and intermediate-term time windows across three elderly cohorts. The random-effects meta-analysis observed 12 significant CpGs (Panni et al. 2016).
Since PM2.5 is a combination of different PEs with different characteristics, these PEs may be associated with DNAm at different sites. To date, only five studies have assessed the associations between PEs and DNAm, among which three (Baccarelli et al. 2009; Chen et al. 2015; Hou et al. 2014) focusing on short-term and one (Madrigano et al. 2011) focusing on intermediate-term effects of PEs on specific loci or repetitive elements. And only one (Dai et al. 2017) examined the long-term effects of PEs on DNAm in an epigenome-wide scope, which was conducted by our group previously. We investigated EWAS of one-year moving averages of PEs and observed 20 significant CpGs for iron, 8 for nickel, and 1 for vanadium (Dai et al. 2017). Pathway analysis also suggested specific effects on DNAm for each PE. For example, iron and nickel were associated with type II diabetes mellitus and insulin signaling pathways. However, we did not examine the associations of PM2.5 total mass and DNAm. And linear mixed-effect models that we used are not optimal for DNAm data, which is usually non-normally distributed. Additionally, studies examining whether the associations between PM2.5/PEs and DNAm vary by different time windows remain quite limited. To date, no EWAS of short- and intermediate-term exposure to PEs has been performed.
The present study, therefore, sought to investigate the associations between short- and intermediate-term exposure to PM2.5/PEs and DNAm by conducting EWAS and quantile regression analyses, using whole-blood samples in the same population. We hypothesized that the changes in DNAm varied by PM2.5 and the PEs.
2. Methods
2.1. Study population
The NAS is a closed and ongoing cohort established in 1963 by the U.S. Veterans Administration in the Greater Boston Area (Bell et al. 1966). The participants were aged 21–82 years and were free of any known chronic diseases at enrollment. They have undergone health examinations in a clinical center, including blood collection, every 3–5 years.
In the present study, we included subjects who lived in the Greater Boston Area during the study period and had visits with DNA samples collected starting in 1999 (Wang et al. 2020b). We dropped non-white participants (~3%) to diminish study heterogeneity that may be introduced by diverse genetic ancestry (Wang et al. 2020b). All study participants provided written informed consent before enrollment and sample collection. This study was approved by the Harvard T.H. Chan School of Public Health and the Institutional Review Boards of the Department of Veterans Affairs.
2.2. DNAm measures
DNA samples were extracted using the IQAamp DNA Blood Kit (Qiagen, CA, U.S.) from the buffy coat of the whole blood collected between 1999 and 2013. We measured DNAm by the Illumina Infinium Human Methylation450K BeadChip (450 K; Illumina Inc., San Diego, CA, U.S.), which provides information on ~ 485,000 CpG sites. To minimize batch effects, we randomized the samples across 450 K BeadChip and 96-well plates based on a two-stage age-stratified algorithm so that age distributed similarly across plates (Dai et al. 2017).
We preprocessed DNAm data via the ewastools package in Github (Heiss and Just 2018; 2019). At the sample level, we finally included 1,559 high-quality samples (Heiss and Just 2018). We then corrected dye-bias using a regression on the logarithm of internal control probes described in Xu et al. (Xu et al. 2017). At the probe level, we first dropped the probes with a detection p-value > 0.01 based on an estimation of the background distribution using non-specific fluorescence (Heiss and Just 2019). We then excluded probes with outliers or low quality (Xu et al. 2016). We also dropped sex-chromosome probes, non-CpG probes, probes with a single nucleotide polymorphism (SNPs) at the CpG site (minor allele frequency (MAF) ≥ 5%), probes with SNPs at single-base extension (MAF ≥ 5%), and probes containing an SNP (MAF ≥ 5%) (Cardenas et al. 2019). In addition, we excluded probes with any annotated SNPs within ten base pairs of the CpG site as provided in the Illumina annotation regardless of MAF (The 1000 Genomes Projects Consortium 2012), cross-reactive probes (Chen et al. 2013), and probes that were non-unimodal in the dip test (Hartigan and Hartigan 1985). As a result, 360,272 high-quality probes were included in this study. The steps for probe cleaning are shown in more detail in Table S1.
We normalized DNAm data by controlling for the normalization factors in the outcome regression instead of using other commonly used approaches, such as beta-mixture quantile normalization (Teschendorff et al. 2013). This normalization approach ensures a better adjustment for batch effects as their impact often varies across probes, and we have applied it previously (Wang et al. 2020a; Wang et al. 2020b). Specifically, we used an elastic-net regularized generalized linear model. We chose the mixing parameter to be 0.5 and did the grid search over lambda because it was generally a good choice and led to good results. We then extracted the top five important experimental covariates (i.e., Non-polymorphic Red, Specificity I Red, Bisulfite Conversion I Red, Bisulfite Conversion II, Extension Red) which has the fewest number of coefficients equal to 0 (BeadArray controls report 2015).
DNAm level was expressed as the ratio of methylated cytosines over the sum of the methylated and unmethylated cytosines at the position and then multiplied by 100 (mean %5-methylcytosine, i.e., %5-mC). Thus, the DNAm level ranged from 0- to 100 %5-mC.
2.3. PM2.5 and its elements (PEs) measures
We collected daily ambient PM2.5 and 13 PEs at a stationary monitoring site located at the roof of the Harvard University Countway Library that is less than 1 km from the health examination clinical center. We measured PM2.5 total mass using a Harvard Impactor Sampler (Koutrakis et al. 1993). PEs included black carbon (BC), iron (Fe), lead (Pb), zinc (Zn), copper (Cu), aluminum (Al), calcium (Ca), silicon (Si), potassium (K), nickel (Ni), vanadium (V), sodium (Na), and sulfur (S). We focused on these PEs because their concentrations were mostly above detection limits and representative of different sources of emission (Dai et al. 2017; Wang et al. 2020b) – S is a regional pollutant primarily from power plants and regional pollution; K is from wood burning; BC, Cu, Pb, and Zn are components from motor vehicles (Pb is no longer in gasoline, but can be emitted from brake wear and ground down tire weights, and is present in re-suspended dust from motor vehicles); Ca and Fe are tracers of road dust; Al and Si are mostly from soil; Ni and V originate from oil combustion; Na is a major component of sea salt. We measured BC using an Aethalometer (Magee Scientific Inc., Berkeley CA, U.S.) (Lepeule et al. 2014) and the other 12 PEs using the Energy Dispersive X-ray Fluorescence Spectrometers (Epsilon 5, PAN-alytical, Almelo, Netherlands) (Wang et al. 2020b).
To estimate the short- and intermediate-term effects of PM2.5 and its 13 PEs on DNAm, we considered a single time window from day 0 (the current day of the event) to day 27 (27 days prior to the event) a priori based on previous studies (Gondalia et al. 2019; Mehta et al. 2015; Panni et al. 2016).
2.4. Covariates assessment
The covariates were selected a priori based on the relevant literature (Dai et al. 2016; Wang et al. 2020b): 1) sociodemographic factors including age (years), years of education; 2) lifestyle factors including smoking status (ever/never), cigarette pack-years, alcohol consumption (<2 or ≥ 2 drinks/day); body mass index (BMI, kg/m2); the metabolic equivalent of task (hours/week); 3) biological factors: the estimated cell-type compositions (CD4 + T lymphocytes, CD8 + T lymphocytes, natural killer cells, B cells, and monocytes) by the Houseman method (Houseman et al. 2012); 4) technical factors such as batch effects and five normalization factors; and 5) weather variables including season (March-May; June-August; September-November; December-February), and ambient temperature and relative humidity measured at the Boston Logan Airport weather station, which is 8 km away from the clinical center (Dai et al. 2016). For the covariates mentioned above, only years of education had four missing. The bootstrapping-based algorithm was used to impute the missing values (Honaker et al. 2011).
2.5. Statistical analyses
We used three different approaches of EWAS to analyze the accumulative effects of short- and intermediate-term exposure to PM2.5 and its 13 PEs on DNAm: site-by-site, regional, and pathway analyses.
2.5.1. Site-by-site analyses
To identify statistically differentially methylated probes (DMPs) due to PM2.5 and PEs, we performed site-by-site analyses. Since most of the DNAm levels were not normally distributed, statistical inferences based on classical least squares regression may be inappropriate. Therefore, we used median regression, which is influenced neither by outliers nor skewness in the distribution of the dependent variable (Koenker and Hallock 2001), to analyze the associations between particles and DNAm. In the median regression, we included in the models constrained polynomial distributed lag terms, which used a 3-degree polynomial structure to constrain how the size of the particle effects vary with the lags between exposure and DNAm measurement from the same day up to lags of 27 days (Schwartz 2000). Only the overall effect across the 28-d time window was reported. The distributed lag terms account for the latency of the effects of particles and the overall effects minimize the unstable estimate due to the high degree of collinearity between lags at neighboring days (Schwartz 2000).
We fitted median regression for longitudinal data by the Koenker et al. method (Koenker 2004) because approximately 80% of the participants had repeated DNAm measures. Briefly, this method enables one to fit fixed-effects and correlated random-effects within the same subject and uses Bootstrap inference. In addition, we adjusted for two selection biases: 1) healthier men were more likely to return for subsequent exams after 1999 and 2) mortality that occurred prior to the first DNAm visit (1999). We applied inverse probability weighting (Hogan and Lancaster 2004; Robins et al. 1995) via logistic regression to calculate the two probabilities given chronological age, education, BMI, blood pressure, smoking status, cigarette pack-years, alcohol consumption, C-reactive protein, asthma, chronic bronchitis, and emphysema at previous visit (Wang et al. 2020a, 2020b; McCracken et al. 2010). We then multiplied the two inverse probability weights. Therefore, the visits in this study remained representative of the original population.
We fitted the following models to assess the cumulative effects of ambient PM2.5 total mass only without considering its elements (as shown in formula (1)) and the cumulative effects of each of the 13 PEs one at a time, adjusting for PM2.5 total mass in the model (as shown in formula (2)).
| (1) |
| (2) |
where Yij is the DNAm level of subject i at visit j. βl in formula (1), and φ2l in formula (2) are the lag-specific coefficients of PM2.5 total mass, with l ∈ [0, 27] lag days starting from the day of the visit to the previous 27 days. Σβl is the cumulative effect of PM2.5 total mass on DNAm over 28 days (lags 0–27). Similarly, φ1l in formula (2) are the lag-specific co-efficients of each of the PEs, with l ∈ [0, 27] lag days. Σφ1l is the cumulative effect of each of the PE’s exposure on DNAm over 28 days (lags 0–27). Xij are the covariates we mentioned above (Covariates Assessment) for subject i at visit j. δi in formula (1) and ωi in formula (2) are the random intercepts for participant i. ∊ij in formula (1) and εij in formula (2) are the residuals. Median regressions with distributed lags, correlated random-effects, and inverse probability weighting were performed using the rqpd package in R (Bind et al. 2016).
We reported our results as the median difference in DNAm (%5-mC) per one interquartile range (IQR) increase in PM2.5 and its elements after 28 days (0–27) exposure. The results from individual CpGs were adjusted for multiple testing by Bonferroni threshold p-value < 4.96 × 10−9 (0.05/(360,272 × 28) for PM2.5 total mass and < 3.81 × 10−10 (0.05/(360,272 × 28 × 13) for each of the 13 PEs.
2.5.2. Regional and pathway analyses
In addition to examining the associations of PM2.5/PEs and DNAm at the individual probe level, we also investigated statistically differentially methylated regions (DMRs) and pathways in relation to PM2.5/PEs. To identify the DMRs, we applied the combp function from the ENmix package in R Bioconductor (Xu et al. 2020b) because the comb-p tool has the best sensitivity and highest control of false-positive rate compared to other DMR tools (Mallik et al. 2019). The input contained individual CpG sites with their p-values from site-by-site analyses as well as information on chromosomal locations (Xu et al. 2020b). We defined a significant DMR as one with three or more probes and its Sidak p-value < 0.05 (Li et al. 2020).
Ingenuity Pathway Analysis (IPA) database (QIAGEN Inc.) was used to identify canonical pathways significantly enriched with genes located around the corresponding top100 CpGs associated with PM2.5 total mass and its elements. We calculated permutation p-values using the results of 10,000 random shuffles of association p-values for the CpGs on the 450 K array (Xu et al. 2020a). We defined significant pathways if p-value < 0.05 and gene set contains ≥ 3genes.
2.5.3. Sensitivity analyses
We conducted two sensitivity analyses in the site-by-site analyses to check the robustness of our results. First, instead of adjusting the top five important experimental covariates in the median regression models, we additionally adjusted the following two experimental covariates (i.e., Bisulfite Conversion I Green and Hybridization High Medium). Second, we only considered the selection bias due to healthier men being more likely to return for subsequent exams after 1999 when DNA samples began being collected. We compared the effect sizes and p-values of the top one probe for PM2.5 total mass and each PE from the main analyses with the ones from the two sensitivity analyses.
3. Results
3.1. Population description
We included 695 men with 1,266 visits. The characteristics of the study population are presented in Table 1. Eighty-two percent of the participants had more than one visit. The mean age [standard deviation (SD)] at the first and the last visits were 73 (7) and 76 (5) years, respectively. The men were generally highly educated, with a mean of 15 years (SD = 3) of education. Thirty-one percent of subjects never smoked, and 80% had fewer than two drinks per day across all visits.
Table 1.
Characteristics of elderly white men from the Normative Aging Study, 1999–2013.
| Variables | First visit (N = 695) |
Second visit (N = 432) |
Third visit (N = 136) |
Fourth visit (N = 3) |
All visits (N = 1,266) |
|---|---|---|---|---|---|
| Age, Mean ± SD | 73 ± 7 | 76 ± 7 | 80 ± 6 | 76 ± 5 | 75 ± 7 |
| BMI (kg/m2), Mean ± SD | 28.14 ± 4.15 | 27.90 ± 4.14 | 27.81 ± 4.33 | 30.02 ± 7.45 | 28.03 ± 4.17 |
| Years of education, Mean ± SD | 15.03 ± 3.00 | 15.27 ± 3.09 | 15.64 ± 3.07 | 16.67 ± 5.51 | 15.18 ± 3.05 |
| Smoking status, n (%) | |||||
| Never | 220 (31.65%) | 133 (30.79%) | 48 (35.29%) | 1 (33.33%) | 402 (31.75%) |
| Ever | 475 (68.35%) | 299 (69.21%) | 88 (64.71%) | 2 (66.67%) | 864 (68.25%) |
| Pack-years, Mean ± SD | 20.71 ± 24.62 | 20.64 ± 24.17 | 17.65 ± 22.11 | 5.00 ± 8.66 | 20.32 ± 24.19 |
| Alcohol consumption (drinks/day), n (%) | |||||
| <2 | 564 (81.15%) | 354 (81.94%) | 110 (80.88%) | 2 (66.67%) | 1,030 (81.30%) |
| ≥ 2 | 131 (18.85%) | 78 (18.06%) | 26 (19.12%) | 1 (33.33%) | 236 (18.70%) |
| Season, n (%) | |||||
| Spring (March-May) | 169 (24.31%) | 108 (25.00%) | 34 (25.00%) | 0 (0%) | 311 (23.85%) |
| Summer (June-August) | 174 (25.04%) | 115 (26.62%) | 31 (22.79%) | 0 (0%) | 320 (25.28%) |
| Fall (September-November) | 238 (34.15%) | 137 (31.71%) | 41 (30.15%) | 1 (33.33%) | 417 (32.88%) |
| Winter (December-February) | 114 (16.50%) | 72 (16.67%) | 30 (22.06%) | 2 (66.67%) | 218 (17.27%) |
| Estimated cell type (%), Mean ± SD | |||||
| Granulocytes | 57.20 ± 9.46 | 58.58 ± 9.63 | 62.78 ± 9.63 | 59.27 ± 4.39 | 58.27 ± 9.46 |
| Monocytes | 10.79 ± 2.77 | 10.19 ± 3.00 | 10.13 ± 2.65 | 9.88 ± 2.87 | 10.52 ± 2.85 |
| B cells | 1.56 ± 2.61 | 1.17 ± 2.02 | 1.31 ± 3.50 | 0.83 ± 0.73 | 1.40 ± 2.54 |
| CD4 + T lymphocytes | 8.53 ± 4.37 | 8.44 ± 4.63 | 7.10 ± 3.99 | 9.17 ± 5.56 | 8.35 ± 4.44 |
| CD8 + T lymphocytes | 8.69 ± 3.08 | 8.62 ± 3.19 | 7.16 ± 2.79 | 7.51 ± 0.77 | 8.50 ± 3.12 |
| Natural killer cells | 7.21 ± 3.80 | 7.54 ± 4.40 | 7.06 ± 3.92 | 7.94 ± 3.27 | 7.31 ± 4.02 |
Abbreviations: BMI, body mass index; SD, standard deviation.
3.2. Concentrations of PM2.5 and 13 PEs concentrations
Table 2 describes of daily concentrations of PM2.5 and its elements during the study period (1999–2013). The mean (SD) concentration of daily PM2.5 was 10.3 (6.6) µg/m3, with an IQR of 7.3 µg/m3. Among the examined PEs, S (power plants and regional pollution) accounted for the largest proportion of PM2.5 total mass (10.4%). The major components of motor vehicles (BC, Pb, Zn, Cu) accounted for 8.0%, although other motor vehicle components not measured would increase the total from this source. The PEs from sea salt (Na) and soil (Al, Si) were responsible for 1.9% and 1.3%, respectively. Tracers of road dust (Ca, Fe), wood-burning (K), and oil combustion (Ni and V) took up 1.1%, 0.35%, and 0.1% of PM2.5 total mass, respectively.
Table 2.
Distributions of the daily concentrations of PM2.5, its 13 PEs, and meteorological data during the study period from the Normative Aging Study, 1999–2013.
| Particles | Mean (SD) | Min | Median (25%, 75%) | Max | IQR |
|---|---|---|---|---|---|
| PM2.5 (µg/m3) | 10.32 (6.56) | 1.07 | 8.45 (5.60, 12.91) | 52.72 | 7.31 |
| S (ng/m3) | 1070.95 (883.31) | 0.79 | 776.24 (485.44, 1398.32) | 6663.98 | 912.88 |
| BC (ng/m3) | 801.68 (413.05) | 187.38 | 707.91 (481.64, 1040.87) | 2426.88 | 559.23 |
| Pb (ng/m3) | 5.81 (6.04) | 0.01 | 5.15 (3.22, 7.53) | 94.59 | 4.31 |
| Zn (ng/m3) | 12.69 (10.04) | 0.01 | 10.07 (6.49, 15.60) | 87.6 | 9.11 |
| Cu (ng/m3) | 3.55 (3.04) | 0.01 | 3.33 (1.82, 4.86) | 53.08 | 3.04 |
| Na (ng/m3) | 192.52 (139.77) | 0.01 | 161.26 (97.20, 255.54) | 1270.69 | 158.34 |
| Al (ng/m3) | 51.86 (39.91) | 0.01 | 44.26 (30.73, 63.31) | 601.71 | 32.58 |
| Si (ng/m3) | 78.39 (67.65) | 1.10 | 64.59 (41.87, 94.48) | 914.02 | 52.6 |
| Ca (ng/m3) | 34.34 (22.03) | 0.37 | 30.76 (21.91, 39.69) | 269.33 | 17.78 |
| Fe (ng/m3) | 74.97 (56.51) | 5.36 | 65.90 (48.10, 88.74) | 699.72 | 40.64 |
| K (ng/m3) | 36.02 (21.90) | 0.42 | 30.95 (22.05, 44.88) | 215.31 | 22.82 |
| Ni (ng/m3) | 3.03 (3.16) | 0.01 | 2.06 (1.05, 4.07) | 26.32 | 3.02 |
| V (ng/m3) | 3.50 (3.08) | 0.01 | 2.61 (1.26, 4.73) | 23.14 | 3.47 |
| Temperature (℃) | 12.33 (8.85) | −18.89 | 12.78 (5.60, 19.40) | 31.10 | 13.80 |
| Relative humidity (%) | 68.13 (15.94) | 19.92 | 67.83 (56.38, 81.17) | 99.33 | 24.79 |
Abbreviations: PM2.5, particulate matter with an aerodynamic diameter ≤ 2.5 µm; S, sulfur; BC, black carbon; Pb, lead; Zn, zinc; Cu, copper; Na, sodium; Al, aluminum; Si, silicon; Ca, calcium; Fe, iron; K, potassium; Ni, nickel; V, vanadium; IQR, interquartile range; SD, standard deviation.
3.3. EWAS of PM2.5 and 13 PEs
3.3.1. Significantly differentially methylated probes
In the site-by-site analyses, we observed multiple significant DMPs for PM2.5 total mass and its 13 PEs across 28 days of exposure (see Table 3). For example, 3,173 significant DMPs were observed due to Pb exposure; 164 significant DMPs were identified by Fe (see Table 3). We presented the significant DMPs ranked by p-value with their annotated genes in Tables S2–15. We also compared the significant DMPs by PM2.5 total mass and the PEs. We found that among the 2,717 significant DMPs by PM2.5 total mass, there were 53 that were also associated with at least one of the 13 PEs (see Fig. 1). Among the 53 DMPs, 45 were associated with PEs from motor vehicles, and three (i.e., cg26201011, cg15415507, and cg07776285) were associated with PEs from more than one emission source.
Table 3.
The number of differentially methylated probes (DMPs) of cumulative effects (lag 0–27d) from exposure to PM2.5 and its elements in the site-by-site analyses.
| Particles | Number of DMPs | |
|---|---|---|
| Sources | PM2.5 mass | 2,717 |
| Motor vehicles | BC | 394 |
| Cu | 615 | |
| Pb | 3,173 | |
| Zn | 281 | |
| Oil combustion | Ni | 24 |
| V | 20 | |
| Road dust | Ca | 2 |
| Fe | 164 | |
| Soil | Al | 31 |
| Si | 19 | |
| Wood burning | K | 343 |
| Power plants | S | 270 |
| Sea salt | Na | 1,707 |
Abbreviations: PM2.5, particulate matter with an aerodynamic diameter ≤ 2.5 µm; BC, black carbon; Cu, copper; Pb, lead; Zn, zinc; Ni, nickel; V, vanadium; Ca, calcium; Fe, iron; Al, aluminum; Si, silicon; K, potassium; S, sulfur; Na, sodium; DMP, differentially methylated probes.
Fig. 1.
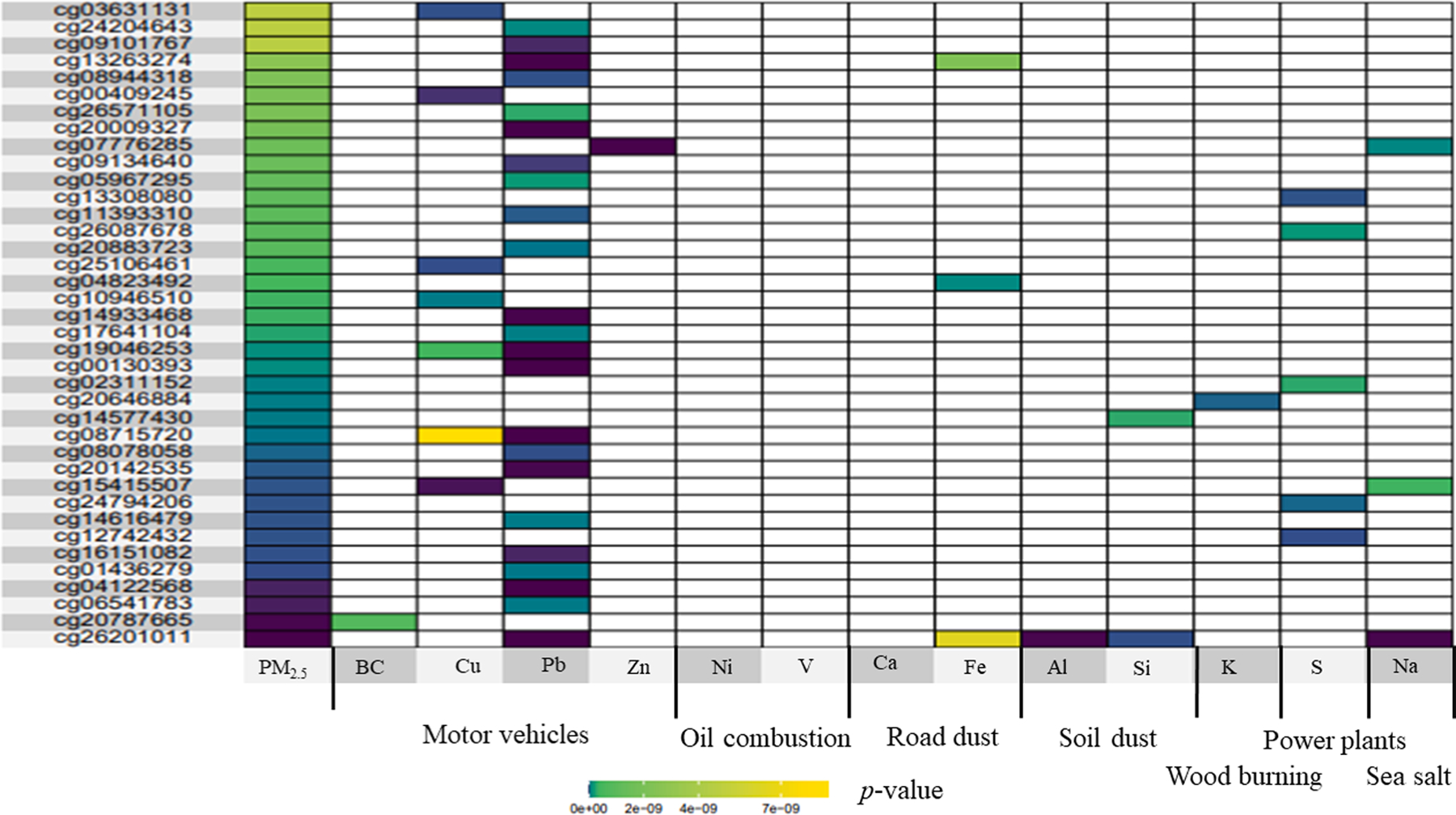
The heatmap of 53 differentially methylated probes that were associated with both PM2.5 mass and at least one of its 13 PEs. Abbreviations: PM2.5, particulate matter with an aerodynamic diameter ≤ 2.5 µm; BC, black carbon; Cu, copper; Pb, lead; Zn, zinc; Ni, nickel; V, vanadium; Ca, calcium; Fe, iron; Al, aluminum; Si, silicon; K, potassium; S, sulfur; Na, sodium.
We then extracted the top three DMPs associated with PM2.5 total mass and PEs (N = 42; i.e., 3 × 14). We found that among the 42 DMPs, 6 were located in the genes’ promoter, with all related with PEs, not PM2.5 total mass (see Table 4). For example, 28-day cumulative exposure to BC was associated with an increase in DNAm at cg24899205 (0.47 %5-mC, p = 0.00 × 10−12), which was annotated to the TXNRD2 gene’s promoter; whereas 28-day cumulative to S was associated with a decrease in DNAm at cg22056044 (−2.32 %5-mC, p = 0.00 × 10−12), which was mapped in the TM9SF3 gene’s promoter. For these 6 probes, we compared the effect sizes for their related PEs in Table 4 with the effect size for age and found that the absolute values of the effects sizes per IQR increase for all the PEs were greater than those for a one-year increase of age (Table S16).
Table 4.
The top three DMPs associated with one IQR increase in PM2.5 and its PEs if they are located in the genes’ promoter in the site-by-site analyses.
| Sources | Particles | CpG | UCSC gene a | Chr a | Position | p-value | Difference in DNAm b |
|---|---|---|---|---|---|---|---|
| Motor vehicles | BC | cg24899205 | TXNRD2 | 22 | 19,929,184 | 0.00 × 10−16 | 0.47 |
| Cu | cg12298562 | RBP4 | 10 | 95,361,132 | 0.00 × 10−16 | −1.68 | |
| Cu | cg24912904 | ACTN3 | 11 | 66,314,217 | 0.00 × 10−16 | −0.97 | |
| Zn | cg01910869 | EDNRB | 13 | 78,493,182 | 0.00 × 10−16 | 0.75 | |
| Oil combustion | Ni | cg19821536 | OR6B3 | 2 | 24,098,376 | 1.33 × 10−12 | −1.71 |
| Power plants | S | cg22056044 | TM9SF3 | 10 | 98,347,055 | 0.00 × 10−16 | −2.32 |
Note: BC, black carbon; Cu, copper; Zn, zinc; Ni, nickel; S, sulfur.
The University of California Santa Cruz UCSC database (GRCh37/hg19)
Difference in DNAm level (% 5-mC) per one IQR increase in particles
3.3.2. Significant regions and pathways
We identified multiple DMRs for PM2.5 mass and its PEs across 28 days of exposure (see Table S17–30). For example, 10,470 DMRs were observed due to PM2.5 total mass; 11,293 DMRs were found by Na. We compared the top 10 DMRs and their annotated genes from PM2.5 total mass and PEs and found no common DMRs or annotated genes.
In the pathway analyses, we also identified multiple significant pathways in relation to PM2.5 total mass and PEs: 17 for PM2.5, 45 for BC, 1 for Cu, 1 for Pb, 3 for Zn, 0 for Ni, 1 for V, 6 for Ca, 5 for Fe, 1 for Al, 1 for S, 4 for Si, 1 for K, and 0 for Na. We presented the significant pathways ranked by p-value in Tables S31–42. We also compared the enriched pathways by PM2.5 total mass and its PEs and found there were 12 common pathways identified by PM2.5 and some of its elements, for example, “Gg alpha signaling” and “RhoGDI signaling” (see Table 5).
Table 5.
Common enriched pathways by PM2.5 and its elements.
| Common pathways | Particles | |
|---|---|---|
| Calcium Signaling | PM2.5 | Zn |
| Dopamine-DARPP32 Feedback in cAMP Signaling | PM2.5 | Ca |
| Glucocorticoid Receptor Signaling | PM2.5 | Al |
| Gq alpha Signaling | PM2.5 | BC |
| Aryl Hydrocarbon Receptor Signaling | BC | Al |
| Thrombin Signaling | BC | Al |
| Xenobiotic Metabolism Signaling | BC | Ca |
| Production of Nitric Oxide and Reactive Oxygen Species in Macrophages | BC | Ca |
| Human Embryonic Stem Cell Pluripotency | BC | Si |
| P2Y Purigenic Receptor Signaling Pathway | BC | Si |
| Wnt/β-catenin Signaling | Zn | Fe |
| RhoGDI Signaling | Al | Si |
Note: PM2.5, particulate matter with an aerodynamic diameter ≤ 2.5 µm; Al, aluminum; Ca, calcium; Si, silicon; BC, black carbon; Fe, iron; Zn, zinc.
3.4. Sensitivity analyses
We extracted the top one probes from PM2.5 and its 13 PEs in the main analyses and compared their effect sizes and p-values with that from two sensitivity analyses. It showed no substantive changes (see Table S43). The effect sizes in the main analyses were close to the ones in sensitivity analyses #1. And the effect sizes were almost the same in the main analyses as in the sensitivity analyses #2.
4. Discussion
To our knowledge, this is the first EWAS of short- and intermediate-term exposure to PM2.5 and PEs. We identified multiple DMPs, DMRs, and pathways cumulatively associated with these particles across lag 0–27 days. Moreover, the identified DMPs, DMRs, and pathways by PM2.5 were different from those by its elements. Specifically, the significant pathways suggest that PM2.5 total mass was related to DNAm involved in mood disorders, neuroplasticity, immunity, and inflammation, whereas the pathways associated with motor vehicle emissions (BC, Cu, Pb, and Zn) were mostly annotated to cardiovascular disease and cancer pathways.
We start our discussion from the significant pathways associated with PM2.5 total mass. Pathways of “dopamine-DARPP32 feedback in cAMP signaling”, “synaptic long-term depression”, and “synaptic long-term potentiation” were involved in mood disorders (Amare et al. 2017; Bliss and Cooke 2011) and neuroplasticity (Morimoto et al. 2014). Other pathways such as “CD28 signaling in T helper cells”, “role of NFAT in regulation of the immune response”, and “Phospholipase C signaling” were associated with the immunity and inflammatory system (Dong and Flavell 2000; Zhu et al. 2018). Pathways associated with PEs were different from those associated with PM2.5 total mass. More specifically, elements from motor vehicle emissions (BC, Cu, Pb, Zn) were associated with pathways related to cardiovascular disease, such as “Cardiac β-adrenergic signaling”, “relaxin signaling”, “PPARs signaling”, “endothelin-1 signaling”, “leptin signaling in obesity”, “blood coagulation”, “eNOS signaling” (Johnson and Faraci 2011; Orekhov et al. 2016; Sandoval et al. 2014; St-Louis and Massicotte 1985; Yang and Barouch 2007), and cancer, for instance, “small cell lung cancer signaling”, “ceramide signaling”, “p53 signaling”, “aryl hydrocarbon receptor signaling”, “glioma signaling”, and “P2Y purinergic receptor signaling pathway”, and “Wnt/β-catenin signaling” (Feng et al. 2013; MacDonald et al. 2009; Schulien et al. 2020; Sheridan and Ogretmen 2021). The pathways associated with road dust (Ca and Fe) and soil (Al and Si) were involved in cancer, for example, “HIPPO signaling” (Zygulska et al. 2017) and “RhoGDI signaling” (Harding and Theodorescu 2010). These findings help us understand that specific PEs may affect health via different pathways compared with the PM2.5 total mass and suggest the necessity to examine the associations between DNAm and different PM2.5 elements. We note that our PEs do not include organic carbon, nitrate, or ammonium particles, which may explain why we see PM2.5 total mass associated with pathways (and DMR and DMP) not associated with the particle elements we had.
Many toxicological studies have linked particles and the pathways that we identified in this study. For example, Jiang et al. have examined exposure to PM2.5 in Nrf2−/− mice and found a reduction in PPARα (Jiang et al. 2020) in the liver. Zheng et al. have suggested that exposure to PM2.5 reduces the expression of PPARγ in both liver tissues and hepatic stellate cells (Zheng et al. 2013; Zheng et al. 2015). PPARs are ligand-inducible transcription factors and consist of three nuclear receptor isoforms, i.e., PPARγ, PPARα, and PPARδ. All three isoforms have a wide spectrum of beneficial effects on the prevention and treatment of cardiovascular disease (Orekhov et al. 2016). Pan et al. have assessed Pb-induced toxicity mechanisms via the skin permeability in mice and shown the overexpression of Rho GDP-dissociation inhibitor 2 (RhoGDI2) (Pan et al. 2010). Although we did not find the association between Pb and RhoGDI2 in the present study, we identified “RhoGDI signaling” by Al and Si. This pathway has been reported to mediate several processes during tumorigenesis and cancer progression, such as pancreatic and bladder cancer (Harding and Theodorescu 2010). Therefore, the intervention of this pathway has been proposed as a novel cancer therapeutic strategy (Harding and Theodorescu 2010).
It is worth mentioning that inflammation is involved in most of the pathways by PM2.5. We also find the pathways of inflammation-related diseases such as arthritis (due to BC exposure) in this study. These associations have been reported in both mechanistic and epidemiological studies. For example, in an experimental study, Liu et al. have found that PM2.5 might increase the risk of osteoarthritis via the production of IL-6 synthesis (Liu et al. 2021). Adami et al. have shown a positive association between air pollution and rheumatoid arthritis severity in a case-crossover study (Park et al. 2021). We note that these pathways are not necessarily associated with poor health endpoints. But our findings do suggest that particles may influence health by triggering different biological changes, and elements from motor vehicles are related with DNAm in more pathways compared with elements from other emission sources.
To date, only a limited number of research studies investigated associations between PM2.5 elements and DNAm and most of them focused on DNAm at specific loci or repeated elements. Our previous study was the only one examining PEs and DNAm in an epigenome-wide but focusing on a one-year moving average time window (Dai et al. 2017). The number of the identified DMPs and pathways was much less in our previous work compared with this present study (We did not perform the regional analyses in our previous study). Our previous work identified a total of 29 DMPs (20 for Fe, 8 for Ni, and 1 for V) and 9 significant pathways for Fe, Ni, and V. We did not find any common DMPs, but a few overlapping pathways in our two studies, such as pathways in cancer by Fe. The different findings (especially the significant DMPs) between the two studies indicate that the exposure windows are crucial when we examine the particles ~ DNAm relationships.
This study has some limitations: 1) The concentrations of ambient PM2.5 total mass and PEs were estimated from monitors at a stationary site because there were not enough monitors in the study area and AOD from MISR could only help with S and organic carbon prediction. The misclassification from a single ambient site occurs because of geographic differences in ambient exposure. In addition, the subjects with more outdoor activity would have higher exposure to ambient particles. All these may lead to the inevitable exposure misclassification. Our group is working on models predicting short-term metals’ exposures, which will be used in future. 2) We do not have data on gene expression, and thus we are not able to determine the regulation directions between DNAm and the coded protein. 3) This study only included elderly white men. The generalizability of our findings to other age groups, sex, and races are uncertain although studies that assess the modification effects of age, sex, and races in the association between particles and DNAm are limited (Ding et al. 2017; Ladd-Acosta et al. 2019; Wu et al. 2021). 4) IPA database is built based on multiple tissues; the role of the identified pathways in leukocytes may not be relevant for all diseases. 5) As in other cohort studies, this study is subject to selection bias such as index event bias and depletion of susceptible participants (Dudbridge et al. 2019; Fireman et al. 2020; Stovitz et al. 2018). However, we addressed this issue by using the inverse probability weighting to account for two main selection biases, i.e., we controlled for key predictors of mortality and drop-out. 6) It is quite challenging to determine potential confounders since this is an agnostic study (without pre-specifying any outcomes within the DNAm sites covered by the analytical platform we used). In our follow-up studies with specific health outcomes or pathways, we will be able to deal with these confounders in our models.
The notable strengths of this study include: 1) It was a relatively large EWAS to assess short- and intermediate-term effects of PM2.5 total mass and its elements. 2) Statistical inference using median regression does not require the normally distributed assumption for the residuals. 3) Repeated measurements of methylation give us a variation of the exposure and outcome within-subject. Hence, it increases statistical power. Also, since the visits are approximately 4 years apart, there is a large amount of variation in exposure between-subjects. This reduces the dependence of the analysis on between-subject variation in exposure.
5. Conclusions
In summary, this EWAS of short- and intermediate-term exposure to PM2.5 and its 13 PEs indicate that PM2.5 mass alone is not a sufficient metric when understanding the health effects of particles. PM2.5 total mass was associated with pathways in mood disorders, neuroplasticity, immunity, and inflammation whereas elements from motor vehicles (BC, Cu, Pb, and Zn) were mostly involved in cardiovascular disease and cancer. More EWAS of PEs with diverse study populations from different areas, especially from low-income, high-pollution regions are needed, to enrich present findings.
Supplementary Material
Acknowledgments
We acknowledge Dr. Zongli Xu for conducting pathway analyses using Ingenuity Pathway Analysis (IPA) database and all members of the Normative Aging Study (NAS) team.
Funding
This work was supported by the National Institute of Environmental Health Sciences (R01ES027747, R01ES025225, P30ES009089, R01ES031259), HSPH-NIEHS Center for Environmental Health (ES000002), and the U.S. EPA (RD-835872). Its contents are solely the responsibility of the grantee and do not necessarily represent the official views of the National Institute of Environmental Health Sciences or the U.S. EPA. Further, Neither the National Institute of Environmental Health Sciences nor the U.S. EPA endorses the purchase of any commercial products or services mentioned in the publication.
Footnotes
CRediT authorship contribution statement
Cuicui Wang: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Andres Cardenas: Software, Methodology, Funding acquisition, Writing – review & editing. John N. Hutchinson: Software, Resources, Writing – review & editing. Allan Just: Data curation, Resources, Writing – review & editing. Jonathan Heiss: Data curation, Resources, Writing – review & editing. Lifang Hou: Methodology, Writing – review & editing. Yinan Zheng: Methodology, Writing – review & editing. Brent A. Coull: Methodology, Writing – review & editing. Anna Kosheleva: Data curation, Writing – review & editing. Petros Koutrakis: Funding acquisition, Writing – review & editing. Andrea A. Baccarelli: Funding acquisition, Writing – review & editing. Joel D. Schwartz: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2021.106955.
Referencess
- BeadArray controls reporter, 2015. San Diego, CA: Illumina; October1000000004009v00. [Google Scholar]
- Amare AT, Schubert KO, Klingler-Hoffmann M, Cohen-Woods S, Baune BT, 2017. The genetic overlap between mood disorders and cardiometabolic diseases: a systematic review of genome wide and candidate gene studies. e1007 e1007 Transl. Psychiatry 7 (1). 10.1038/tp.2016.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J, 2009. Rapid DNA methylation changes after exposure to traffic particles. Am. J. Respir. Crit. Care Med 179 (7), 572–578. 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell B, Rose CL, Damon A, 1966. The Veterans Administration longitudinal study of healthy aging. Gerontologist 6 (4), 179–184. 10.1093/geront/6.4.179. [DOI] [PubMed] [Google Scholar]
- Bind M-A, Coull BA, Baccarelli A, Tarantini L, Cantone L, Vokonas P, Schwartz J, 2016. Distributional changes in gene-specific methylation associated with temperature. Environ. Res 150, 38–46. 10.1016/j.envres.2016.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss TV, Cooke SF, 2011. Long-term potentiation and long-term depression: a clinical perspective. Clinics (Sao Paulo) 66 (Suppl 1), 3–17. 10.1590/s1807-59322011001300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook RD, Rajagopalan S, Pope CA, Brook JR, Bhatnagar A, Diez-Roux AV, Holguin F, Hong Y, Luepker RV, Mittleman MA, Peters A, Siscovick D, Smith SC, Whitsel L, Kaufman JD, 2010. Particulate matter air pollution and cardiovascular disease: An update to the scientific statement from the American Heart Association. Circulation 121 (21), 2331–2378. 10.1161/CIR.0b013e3181dbece1. [DOI] [PubMed] [Google Scholar]
- Cardenas A, Sordillo JE, Rifas-Shiman SL, Chung W, Liang L, Coull BA, Hivert M-F, Lai PS, Forno E, Celedón JC, Litonjua AA, Brennan KJ, DeMeo DL, Baccarelli AA, Oken E, Gold DR, 2019. The nasal methylome as a biomarker of asthma and airway inflammation in children. Nat. Commun 10 (1) 10.1038/s41467-019-11058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Qiao L, Li H, Zhao Y, Zhang Y, Xu W, Wang C, Wang H, Zhao Z, Xu X, Hu H, Kan H, 2015. Fine particulate matter constituents, nitric oxide synthase DNA methylation and exhaled nitric oxide. Environ. Sci. Technol 49 (19), 11859–11865. 10.1021/acs.est.5b02527. [DOI] [PubMed] [Google Scholar]
- Chen Y. a., Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R, 2013. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8 (2), 203–209. 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Council N, 2004. Research priorities for airborne particulate matter: IV Continuing Research Progress, Washington, D.C. [Google Scholar]
- Dai L, Koutrakis P, Coull BA, Sparrow D, Vokonas PS, Schwartz JD, 2016. Use of the adaptive LASSO method to identify PM2.5 components associated with blood pressure in elderly Men: the Veterans Affairs Normative Aging Study. Environ. Health Perspect 124 (1), 120–125. 10.1289/ehp.1409021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Mehta A, Mordukhovich I, Just AC, Shen J, Hou L, Koutrakis P, Sparrow D, Vokonas PS, Baccarelli AA, Schwartz JD, 2017. Differential DNA methylation and PM2.5 species in a 450K epigenome-wide association study. Epigenetics 12 (2), 139–148. 10.1080/15592294.2016.1271853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, Jin Y, Liu X, Ye H, Zhu Z, Zhang Y, Wang T, Xu Y, 2017. Dose- and time-effect responses of DNA methylation and histone H3K9 acetylation changes induced by traffic-related air pollution. Sci. Rep 7 (1) 10.1038/srep43737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Flavell RA, 2000. Cell fate decision: T-helper 1 and 2 subsets in immune responses. Arthritis Res 2, 179–188. 10.1186/ar85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F, Allen RJ, Sheehan NA, Schmidt AF, Lee JC, Jenkins RG, Wain LV, Hingorani AD, Patel RS, 2019. Adjustment for index event bias in genome-wide association studies of subsequent events. Nat. Commun 10 (1) 10.1038/s41467-019-09381-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eze IC, Jeong A, Schaffner E, Rezwan FI, Ghantous A, Foraster M, Vienneau D, Kronenberg F, Herceg Z, Vineis P, Brink M, Wunderli J-M, Schindler C, Cajochen C, Röösli M, Holloway JW, Imboden M, Probst-Hensch N, 2020. Genome-wide DNA methylation in peripheral blood and long-term exposure to source-specific transportation noise and air pollution: the SAPALDIA study. Environ. Health Perspect 128 (6), 067003. 10.1289/EHP6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cao Z, Wang X, 2013. Role of aryl hydrocarbon receptor in cancer. BBA 1836 (2), 197–210. 10.1016/j.bbcan.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Fireman B, Gruber S, Zhang Z, et al. , 2020. Consequences of depletion of susceptibles for hazard ratio estimators based on propensity scores. Epidemiology 31, 806–814. 10.1097/EDE.0000000000001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin M, Koutrakis P, Schwartz P, 2008. The role of particle composition on the association between PM2.5 and mortality. Epidemiology 19, 680–689. 10.1097/ede.0b013e3181812bb7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondalia R, Baldassari A, Holliday KM, et al. , 2019. Methylome-wide association study provides evidence of particulate matter air pollution-associated DNA methylation. Environ. Int 132, 104723 10.1097/ede.0b013e3181812bb7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Jaramillo V, Portilla-Fernandez E, Glisic M, Voortman T, Ghanbari M, Bramer W, Chowdhury R, Nijsten T, Dehghan A, Franco OH, Nano J, 2019. Epigenetics and inflammatory markers: a systematic review of the current evidence. Int J Inflam 2019, 1–14. 10.1155/2019/6273680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, 2010. Epigenetic alterations in aging. J. Appl. Physiol. (1985) 109:586–597. 10.1152/japplphysiol.00238.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruzieva O, Xu C-J, Yousefi P, Relton C, Merid SK, Breton CV, Gao L.u., Volk HE, Feinberg JI, Ladd-Acosta C, Bakulski K, Auffray C, Lemonnier N, Plusquin M, Ghantous A, Herceg Z, Nawrot TS, Pizzi C, Richiardi L, Rusconi F, Vineis P, Kogevinas M, Felix JF, Duijts L, den Dekker HT, Jaddoe VWV, Ruiz JL, Bustamante M, Antó JM, Sunyer J, Vrijheid M, Gutzkow KB, Grazuleviciene R, Hernandez-Ferrer C, Annesi-Maesano I, Lepeule J, Bousquet J, Bergström A, Kull I, Söderhäll C, Kere J, Gehring U, Brunekreef B, Just AC, Wright RJ, Peng C, Gold DR, Kloog I, DeMeo DL, Pershagen G, Koppelman GH, London SJ, Baccarelli AA, Melén E, 2019. Prenatal particulate air pollution and DNA methylation in newborns: an epigenomewide meta-analysis. Environ. Health Perspect 127 (5), 057012. 10.1289/EHP4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Shi Y, Zhu Y, Chen N, Wang H, Zhang Z, Chen T, Guo Y, 2020. Ambient air pollution and cause-specific risk of hospital admission in China: A nationwide time-series study. PLoS Med 17 (8), e1003188. 10.1371/journal.pmed.1003188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding MA, Theodorescu D, 2010. RhoGDI signaling provides targets for cancer therapy. Eur. J. Cancer 46 (7), 1252–1259. 10.1016/j.ejca.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartigan JA, Hartigan PM, 1985. The dip test of unimodality. The annals of Statistics 70–84.
- Heiss JA, Just AC, 2018. Identifying mislabeled and contaminated DNA methylation microarray data: an extended quality control toolset with examples from GEO. Clin Epigenetics 10, 73. 10.1186/s13148-018-0504-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss JA, Just AC, 2019. Improved filtering of DNA methylation microarray data by detection p values and its impact on downstream analyses. Clin Epigenetics 11, 15. 10.1186/s13148-019-0615-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan JW, Lancaster T, 2004. Instrumental variables and inverse probability weighting for causal inference from longitudinal observational studies. Stat. Methods Med. Res 13 (1), 17–48. 10.1191/0962280204sm351ra. [DOI] [PubMed] [Google Scholar]
- Honaker J, King G, Blackwell M, 2011. Amelia II: A program for missing data. J. Stat. Softw 45, 1–47. 10.18637/jss.v045.i07. [DOI] [Google Scholar]
- Hou L, Zhang X, Zheng Y, et al. , 2014. Altered methylation in tandem repeat element and elemental component levels in inhalable air particles. Environ. Mol. Mutagen 55, 256–265. 10.1002/em.21829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT, 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf 13 (1) 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A, Abraham JL, Judson B, Berry CL, 2003. Toxicologic and epidemiologic clues from the characterization of the 1952 London smog fine particulate matter in archival autopsy lung tissues. Environ. Health Perspect 111 (9), 1209–1214. 10.1289/ehp.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Li D, Piao J, Li J, Sun H, Chen L, Chen S, Pi J, Zhang R, Chen R, Leng S, Chen W, Zheng Y, 2020. Real-ambient exposure to air pollution exaggerates excessive growth of adipose tissue modulated by Nrf2 signal. Sci. Total Environ 730, 138652. 10.1016/j.scitotenv.2020.138652. [DOI] [PubMed] [Google Scholar]
- Jiang R, Jones MJ, Sava F, Kobor MS, Carlsten C, 2014. Short-term diesel exhaust inhalation in a controlled human crossover study is associated with changes in DNA methylation of circulating mononuclear cells in asthmatics. Part. Fibre Toxicol 11 (1) 10.1186/s12989-014-0071-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AW, Faraci FM, 2011. Trans-forming endothelial nitric oxide synthase in hypertension: more than meets the eye. Hypertension 58 (3), 359–360. 10.1161/HYPERTENSIONAHA.111.177139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloog I, Ridgway B, Koutrakis P, Coull BA, Schwartz JD, 2013. Long- and short-term exposure to PM2.5 and mortality: using novel exposure models. Epidemiology 24 (4), 555–561. 10.1097/EDE.0b013e318294beaa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenker R, 2004. Quantile regression for longitudinal data. J Multivar Anal 91 (1), 74–89. 10.1016/j.jmva.2004.05.006. [DOI] [Google Scholar]
- Koenker R, Hallock KF, 2001. Quantile regression. J Economic Perspect 15 (4), 143–156. 10.1257/jep.15.4.143. [DOI] [Google Scholar]
- Koutrakis P, Sioutas C, Ferguson ST, Wolfson JM, Mulik JD, Burton RM, 1993. Development and evaluation of a glass honeycomb denuder/filter pack system to collect atmospheric gases and particles. Environ. Sci. Technol 27 (12), 2497–2501. [Google Scholar]
- Ladd-Acosta C, Feinberg JI, Brown SC, Lurmann FW, Croen LA, Hertz-Picciotto I, Newschaffer CJ, Feinberg AP, Fallin MD, Volk HE, 2019. Epigenetic marks of prenatal air pollution exposure found in multiple tissues relevant for child health. Environ. Int 126, 363–376. 10.1016/j.envint.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepeule J, Bind M-A, Baccarelli AA, Koutrakis P, Tarantini L, Litonjua A, Sparrow D, Vokonas P, Schwartz JD, 2014. Epigenetic influences on associations between air pollutants and lung function in elderly men: the normative aging study. Environ. Health Perspect 122 (6), 566–572. 10.1289/ehp.1206458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chen R, Cai J, Cui X, Huang N, Kan H, 2018. Short-term exposure to fine particulate air pollution and genome-wide DNA methylation: A randomized, double-blind, crossover trial. Environ. Int 120, 130–136. 10.1016/j.envint.2018.07.041. [DOI] [PubMed] [Google Scholar]
- Li QS, Sun Y, Wang T, 2020. Epigenome-wide association study of Alzheimer’s disease replicates 22 differentially methylated positions and 30 differentially methylated regions. Clin Epigenetics 12, 149. 10.1186/s13148-020-00944-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-F, Chi M-C, Lin C-Y, Lee C-W, Chang T-M, Han C-K, Huang Y-L, Fong Y-C, Chen H-T, Tang C-H, 2021. PM2.5 facilitates IL-6 production in human osteoarthritis synovial fibroblasts via ASK1 activation. J. Cell. Physiol 236 (3), 2205–2213. 10.1002/jcp.v236.310.1002/jcp.30009. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X, 2009. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26. 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigano J, Baccarelli A, Mittleman MA, Wright RO, Sparrow D, Vokonas PS, Tarantini L, Schwartz J, 2011. Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ. Health Perspect 119 (7), 977–982. 10.1289/ehp.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallik S, Odom GJ, Gao Z, et al. , 2019. An evaluation of supervised methods for identifying differentially methylated regions in Illumina methylation arrays. Brief Bioinform 20, 2224–2235. 10.1093/bib/bby085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken J, Baccarelli A, Hoxha M, Dioni L, Melly S, Coull B, Suh H, Vokonas P, Schwartz J, 2010. Annual ambient black carbon associated with shorter telomeres in elderly men: Veterans Affairs Normative Aging Study. Environ. Health Perspect 118 (11), 1564–1570. 10.1289/ehp.0901831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AJ, Kubzansky LD, Coull BA, Kloog I, Koutrakis P, Sparrow D, Spiro A, Vokonas P, Schwartz J, 2015. Associations between air pollution and perceived stress: the Veterans Administration Normative Aging Study. Environ Health 14 (1) 10.1186/1476-069X-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezo YJR, Silvestris E, Dale B, Elder K, 2016. Oxidative stress and alterations in DNA methylation: two sides of the same coin in reproduction. Reprod Biomed Online 33 (6), 668–683. 10.1016/j.rbmo.2016.09.006. [DOI] [PubMed] [Google Scholar]
- Morimoto SS, Wexler BE, Liu J, Hu W, Seirup J, Alexopoulos GS, 2014. Neuroplasticity-based computerized cognitive remediation for treatment-resistant geriatric depression. Clinical Trial 5 (1) 10.1038/ncomms5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassan FL, Wang C, Kelly RS, Lasky-Su JA, Vokonas PS, Koutrakis P, Schwartz JD, 2021. Ambient PM2.5 species and ultrafine particle exposure and their differential metabolomic signatures. Environ. Int 151, 106447. 10.1016/j.envint.2021.106447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orekhov AN, Mukhamedova N, Ivanova EA, Rizzo M, 2016. PPAR in Cardiovascular Disorders. PPAR Res 2016, 1–2. 10.1155/2016/6293629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T-L, Wang P-W, Al-Suwayeh SA, Chen C-C, Fang J-Y, 2010. Skin toxicology of lead species evaluated by their permeability and proteomic profiles: a comparison of organic and inorganic lead. Toxicol. Lett 197 (1), 19–28. 10.1016/j.toxlet.2010.04.019. [DOI] [PubMed] [Google Scholar]
- Panni T, Mehta AJ, Schwartz JD, Baccarelli AA, Just AC, Wolf K, Wahl S, Cyrys J, Kunze S, Strauch K, Waldenberger M, Peters A, 2016. Genome-wide analysis of DNA methylation and fine particulate matter air pollution in three study populations: KORA F3, KORA F4, and the Normative Aging Study. Environ. Health Perspect 124 (7), 983–990. 10.1289/ehp.1509966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Choi S, Kim K, et al. 2021. Association of Particulate Matter with Autoimmune Rheumatic Diseases among Adults in South Korea. Rheumatology (Oxford) Keab127. 10.1093/rheumatology/keab127. [DOI] [PMC free article] [PubMed]
- Pepin ME, Ha C-M, Crossman DK, Litovsky SH, Varambally S, Barchue JP, Pamboukian SV, Diakos NA, Drakos SG, Pogwizd SM, Wende AR, 2019. Genome-wide DNA methylation encodes cardiac transcriptional reprogramming in human ischemic heart failure. Lab. Invest 99 (3), 371–386. 10.1038/s41374-018-0104-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusquin M, Guida F, Polidoro S, Vermeulen R, Raaschou-Nielsen O, Campanella G, Hoek G, Kyrtopoulos SA, Georgiadis P, Naccarati A, Sacerdote C, Krogh V, Bas Bueno-de-Mesquita H, Monique Verschuren WM, Sayols-Baixeras S, Panni T, Peters A, Hebels DGAJ, Kleinjans J, Vineis P, Chadeau-Hyam M, 2017. DNA methylation and exposure to ambient air pollution in two prospective cohorts. Environ. Int 108, 127–136. 10.1016/j.envint.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD, Jones PA, 2000. DNA methylation: past, present and future directions. Carcinogenesis 21, 461–467. 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- Robins JM, Rotnitzky A, Zhao LP, 1995. Analysis of semiparametric regression models for repeated outcomes in the presence of missing data 90:106–121. [Google Scholar]
- Sandoval YH, Atef ME, Levesque LO, et al. , 2014. Endothelin-1 signaling in vascular physiology and pathophysiology. Curr. Vasc. Pharmacol 12, 02–214. 10.2174/1570161112666140226122054. [DOI] [PubMed] [Google Scholar]
- Schulien I, Hockenjos B, van Marck V, Ayata CK, Follo M, Thimme R, Hasselblatt P, 2020. Extracellular ATP and Purinergic P2Y(2) Receptor Signaling Promote Liver Tumorigenesis in Mice by Exacerbating DNA Damage. Cancer Res 80 (4), 699–708. 10.1158/0008-5472.CAN-19-1909. [DOI] [PubMed] [Google Scholar]
- Schwartz J, 2000. The distributed lag between air pollution and daily deaths. Epidemiology 11 (3), 320–326. 10.1097/00001648-200005000-00016. [DOI] [PubMed] [Google Scholar]
- Sheridan M, Ogretmen B, 2021. The role of ceramide metabolism and signaling in the regulation of mitophagy and cancer therapy. Cancers (Basel) 13 (10), 2475. 10.3390/cancers13102475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Louis J, Massicotte G, 1985. Chronic decrease of blood pressure by rat relaxin in spontaneously hypertensive rats. Life Sci 37 (14), 1351–1357. 10.1016/0024-3205(85)90251-6. [DOI] [PubMed] [Google Scholar]
- Stovitz SD, Banack HR, Kaufman JS, 2018. ‘Depletion of the susceptibles’ taught through a story, a table and basic arithmetic. BMJ Evid Based Med 23 (5), 199. 10.1136/bmjebm-2018-110972. [DOI] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, et al. , 2013. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29, 189–196. 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Projects Consortium, 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65. 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MC, Krewski D, Pope CA, Chen Y, Gapstur SM, Thun MJ, 2011. Long-term ambient fine particulate matter air pollution and lung cancer in a large cohort of never-smokers. Am. J. Respir. Care Med 184 (12), 1374–1381. 10.1164/rccm.201106-1011OC. [DOI] [PubMed] [Google Scholar]
- Wang C, Just A, Heiss J, Coull BA, Hou L, Zheng Y, Sparrow D, Vokonas PS, Baccarelli A, Schwartz J, 2020a. Biomarkers of aging and lung function in the normative aging study. Aging (Albany NY) 12 (12), 11942–11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Koutrakis P, Gao X.u., Baccarelli A, Schwartz J, 2020b. Associations of annual ambient PM2.5 components with DNAm PhenoAge acceleration in elderly men: The Normative Aging Study. Environ. Pollut 258, 113690. 10.1016/j.envpol.2019.113690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Ni W, Yao Y, Just A, Heiss J, Wei Y, Gao X.u., Coull BA, Kosheleva A, Baccarelli AA, Peters A, Schwartz JD, 2021. DNA methylation-based biomarkers of age acceleration and all-cause death, myocardial infarction, stroke, and cancer in two cohorts: The NAS, and KORA F4. EBioMedicine 63, 103151. 10.1016/j.ebiom.2020.103151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-P, Lei Q-Y, 2018. Metabolic recoding of epigenetics in cancer. Cancer Commun. (Lond) 38 (1), 25. 10.1186/s40880-018-0302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AJ, Keller JP, Zhao S, Carroll R, Kaufman JD, Sandler DP, 2019. Air pollution, clustering of particulate matter components, and breast cancer in the Sister Study: a U.S.-wide cohort. Environ. Health Perspect 127 (10), 107002. 10.1289/EHP5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Qie R, Cheng M, Zeng Y, Huang S, Guo C, Zhou Q, Li Q, Tian G, Han M, Zhang Y, Wu X, Li Y, Zhao Y, Yang X, Feng Y, Liu D, Qin P, Hu D, Hu F, Xu L, Zhang M, 2021. Air pollution and DNA methylation in adults: A systematic review and meta-analysis of observational studies. Environ. Pollut 284, 117152. 10.1016/j.envpol.2021.117152. [DOI] [PubMed] [Google Scholar]
- Xu Z, Langie SAS, De Boever P, Taylor JA, Niu L, 2017. RELIC: a novel dye-bias correction method for Illumina Methylation BeadChip. BMC Genomics 18 (1). 10.1186/s12864-016-3426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Niu L, Li L, Taylor JA, 2016. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. e20 e20 Nucleic Acids Res 44 (3). 10.1093/nar/gkv907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Sandler DP, Taylor JA, 2020a. Blood DNA Methylation and Breast Cancer: A Prospective Case-Cohort Analysis in the Sister Study. J. Natl Cancer Inst 112, 87–94. 10.1093/jnci/djz065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Xie C, Taylor JA, et al. , 2020b. ipDMR: Identification of differentially methylated regions with interval p-values. Bioinformatics 37, 711–713. 10.1093/jnci/djz065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Barouch LA, 2007. Leptin signaling and obesity: cardiovascular consequences. Circ. Res 101 (6), 545–559. 10.1161/CIRCRESAHA.107.156596. [DOI] [PubMed] [Google Scholar]
- Zheng Z.e., Xu X, Zhang X, Wang A, Zhang C, Hüttemann M, Grossman LI, Chen LC, Rajagopalan S, Sun Q, Zhang K, 2013. Exposure to ambient particulate matter induces a NASH-like phenotype and impairs hepatic glucose metabolism in an animal model. J. Hepatol 58 (1), 148–154. 10.1016/j.jhep.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z.e., Zhang X, Wang J, Dandekar A, Kim H, Qiu Y, Xu X, Cui Y, Wang A, Chen LC, Rajagopalan S, Sun Q, Zhang K, 2015. Exposure to fine airborne particulate matters induces hepatic fibrosis in murine models. J. Hepatol 63 (6), 1397–1404. 10.1016/j.jhep.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Jones C, Zhang G, 2018. The Role of Phospholipase C Signaling in Macrophage-Mediated Inflammatory Response. J Immunol Res 2018, 1–9. 10.1155/2018/5201759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zygulska AL, Krzemieniecki K, Pierzchalski P, 2017. Hippo pathway - brief overview of its relevance in cancer. J. Physiol. Pharmacol 68, 311–335. 10.1155/2018/5201759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


