Abstract
The development of genetic tools allowed for the validation of the pro-aging and pro-disease functions of senescent cells in vivo. These discoveries prompted the development of senotherapies—pharmaceutical interventions aimed at interfering with the detrimental effect of senescent cells—that are now entering the clinical stage. However, unequivocal identification and examination of cellular senescence remains highly difficult because of the lack of universal and specific markers. Here, to overcome the limitation of measuring individual markers, we describe a detailed two-phase algorithmic assessment to quantify various senescence-associated parameters in the same specimen. In the first phase, we combine the measurement of lysosomal and proliferative features with the expression of general senescence-associated genes to validate the presence of senescent cells. In the second phase we measure the levels of pro-inflammatory markers for specification of the type of senescence. The protocol can help graduate-level basic scientists to improve the characterization of senescence-associated phenotypes and the identification of specific senescent subtypes. Moreover, it can serve as an important tool for the clinical validation of the role of senescent cells and the effectiveness of anti-senescence therapies.
Introduction
In response to certain types of stress, cells can enter a state of stable cell cycle arrest known as cellular senescence1,2. Examples of stresses leading to senescence are telomere dysfunction, which can originate from repetitive cell divisions3, hyperactive oncogene signalling4, direct DNA damage—such as from γ-irradiation—and mitochondrial dysfunctions5. Many of these stresses activate a persistent DNA damage response (DDR) and most induce expression of cell cycle inhibitors, most frequently p53/p21WAF1/Cip1 and p16INK4a/RB1. In addition to being irreversibly arrested, senescent cells develop a heterogeneous secretory program termed the senescence-associated secretory phenotype (SASP)6.
Depending on the context, senescent cells and the SASP can positively or negatively influence biological processes in vivo. Transient activation of senescence suppresses tumorigenesis7 and promotes wound repair in skin8 and limb patterning in developing embryos9,10. In contrast, persistent senescent cells are associated with tissue dysfunction and chronic inflammation11, and can promote the onset and progression of diverse age-associated disorders including atherosclerosis12, neurodegeneration13,14, osteoarthritis15, type 2 diabetes16,17 and cancer18. Importantly, the SASP of pathological senescent cells is often enriched in pro-inflammatory factors6. Targeted elimination of senescent cells (senolysis) or neutralization of their undesirable properties (senomorphics) can alleviate some of these pathologies and improve tissue function13,15,19. Owing to these exciting results, the feasibility of using senotherapeutics in humans is currently being tested in clinical trials20.
Ideally, a biomarker should unequivocally detect senescent cells in tissues to determine the burden of senescent cells and efficacy of senotherapeutic interventions. However, virtually all senescence-associated markers are neither specific nor universal for all cell types, making identification, specification and quantification of senescent cells a challenging task.
Development of the protocol
The first documented biomarker of senescence is enhanced activity of the acidic lysosomal β-galactosidase, termed senescence-associated beta-galactosidase or SA-β-Gal21. SA-β-Gal is active in most senescent cells and is absent in the majority of proliferating and quiescent cells. However, it can also be detected in macrophages22, as well as in hair follicles and sebaceous glands21. Moreover, additional stress conditions such as long-term confluence and prolonged serum starvation can trigger SA-β-Gal activity23. Senescent cells are generally characterized by increased lysosomal content occurring as a consequence of defective lysosomal turnover24 or of multi-faceted changes in autophagy25. Senescent lysosomes accumulate high levels of lipofuscin (LF) aggregates, auto-fluorescent lipopigments formed by lipids, metals and misfolded proteins26. LF can be a biomarker of senescence, even in archival formalin fixed and paraffin embedded (FFPE) material27,28. Of note, LF accumulation in aged post-mitotic tissues occurs most probably in the context of post-mitotic cellular senescence (PoMiCS), which is suspected to play a significant role in driving tissue aging29,30, but can be present in non-senescent post-mitotic tissues and thus is not strictly senescence-specific31.
One commonly used marker of senescent cells is the CDK4/6 inhibitor p16INK4a. p16INK4a was discovered in 1993 as an inhibitor of progression from G1 to S phase during the cell cycle32, and subsequently shown to have a role in senescence maintenance33,34. p16INK4a is frequently expressed late after senescence induction, while expression of p21WAF1/Cip1, a CDK4/6 and CDK2 inhibitor, occurs earlier34. Thus, p16INK4a might not be a suitable senescence marker in samples where early senescent cells are present. p21WAF1/Cip1 is also involved in DNA damage signaling34,35, and thus can be expressed by non-senescent cells upon DNA damage. Expression of p21WAF1/Cip1 and p16INK4a can also depend on the type of senescence program. For example, developmental senescent cells express p21WAF1/Cip1 but not p16INK4a9,10, while the reverse is observed in melanocytic nevi36,37. Accordingly, by comparing whole-transcriptome datasets obtained from different types of senescent cells, we and others have recently shown that neither p21WAF1/Cip1 nor p16INK4a are consistently upregulated. However, these analyses allowed the identification of a ‘core’ senescence signature—a set of genes that are commonly differentially expressed in all the senescent cells analyzed5,38. Since none of these genes appear to be uniquely expressed by senescent cells, the levels of multiple transcripts should be measured at the same time and in the same sample to avoid false positives. Importantly, the analysis and comparison of whole-transcriptome datasets allowed the definition of gene sets that associate specifically with individual senescent subtypes5,38.
The absence of proliferation, often measured by lack of DNA synthesis or of Ki-67 expression (see ‘Experimental design’), is unable to discriminate senescence from other states of growth arrest, such as quiescence or terminal differentiation24. DDR proteins such as γ-H2AX and 53BP1 are often present in senescent cells, but these markers also appear in non-senescent cells with transient DNA damage124. Moreover, induction of senescence can occur in the absence of DNA damage, for example as a consequence of mitochondrial dysfunctions39. The SASP is highly complex and heterogeneous, with a composition that is influenced by many intrinsic and extrinsic factors5,38. Recently, we have used mass spectrometry in cell culture and human plasma and identified a core set of SASP proteins that are commonly produced and secreted by different types of senescent cells38. Since none of the individual SASP proteins are senescence-specific, it remains essential to use a panel of core SASP proteins for validation of the senescent phenotype. Despite the existence of certain core proteins, the expression and secretion of most SASP factors remain variable and context-dependent. Pathological SASP—the collection of factors expressed and secreted by senescent cells in disease conditions—is enriched in pro-inflammatory factors8. Thus, the measurement of certain cytokines and chemokines can help to predict detrimental senescence-associated functions.
In summary, considering the lack of specificity and universality of senescence-associated features, the use of multiple markers remains the sole option for accurate validation of senescence in cultured cells and in vivo. Moreover, considering the phenotypical differences among the different senescence programs, the use of pro-inflammatory markers can be an essential step to predict the pathological function of senescent cells. Here, we aim to achieve senescence validation and specification by combining the measurement of multiple markers in the same specimen using a two-phase algorithmic protocol (Fig. 1). We provide details to analyze human specimens, but with minor adjustments the protocol can be adapted for use in other species. The protocol, while being more time-consuming than the ones currently in use40-43, can minimize the risk of under- or overestimating the number of senescent cells, and also provide preliminary information on potential senescence-associated pathological functions.
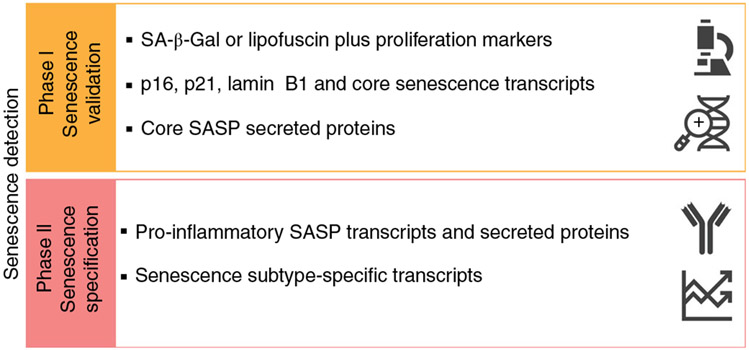
Fig. 1 ∣. Schematic representation of the protocol.
Phase 1 aims to validate the presence of senescence, phase 2 to specify the senescence subtype.
Experimental design
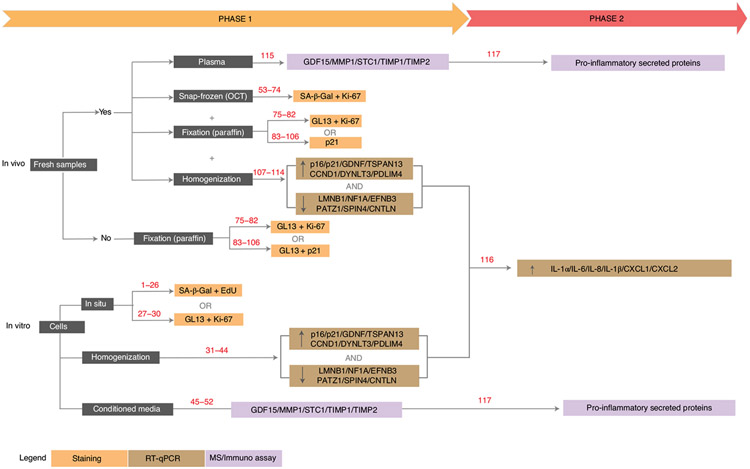
A detailed graphical summary of the protocol is depicted in Fig. 2. Phase 1 of our protocol aims to validate the presence of senescent cells by staining for SA-β-Gal and/or LF along with markers of proliferation, and by measuring the transcription and secretion of the most common senescence-associated proteins. Phase 2 aims to specify the type of senescent cells by measuring the transcription and secretion of pro-inflammatory factors. The first procedure in our protocol is sample preparation, and this depends upon whether the specimens were derived from cultured cells or in vivo–derived tissues, and whether the in vivo samples can be processed immediately after harvesting.
Fig. 2 ∣. Step-by-step protocol.
A summary of the procedures described in the two-phase protocol, highlighting the distinction between in vitro and in vivo samples. Phase 1 focuses on analyzing the expression of general markers of senescence. Phase 2 measures markers that are specific for senescence subtypes. MS, mass spectrometry. Procedure step numbers are shown in red.
In vivo sample preparation of solid tissues
If fresh biopsies (of any tissue) are available, each specimen is split into three parts. The first part is snap frozen in optimal cutting temperature compound (OCT) for SA-β-Gal, LF and indirect immunohistochemical (p21WAF1/Cip1, p16INK4a and others) or proliferative marker staining. The second part is treated with appropriate fixatives for LF, p21WAF1/Cip1 staining, markers of proliferation and/or additional histological analysis. The third part is homogenized and cells are lysed for RNA extraction. RT-PCR is carried out to determine the levels of p16INK4a, p21WAF1/Cip1 and core senescence mRNAs and also to elucidate the type of senescent cells that are present. If only fixed samples in paraffin are available, then co-staining for LF and other indirect markers for senescence (p21WAF1/Cip1, p16INK4a and others) or proliferation is the only option.
Plasma collection
For the collection of plasma to measure protein biomarkers, we recommend the guidelines developed by the clinical proteomic tumor analysis consortium (CPTAC). CPTAC has developed highly standardized and rigorously validated standard operating procedures for the collection, processing and storage of plasma, which help remove bias and improve reproducibility during the collection of patient samples. These procedures are available online from the National Cancer Institute (NIH/NCI) at https://brd.nci.nih.gov/brd/sop/show/341 with the protocol/SOP termed ‘Prospective Biospecimen Collection Protocol, Blood Collection and Processing for Plasma and Whole Cell Components’. Briefly, it is recommended that plasma is collected by venipuncture into EDTA-containing vacutainer tubes followed by inverting the tubes 8–10 times. Tubes must be kept at 4 °C after collection and during subsequent sample processing steps. First, samples are centrifuged for 15 min at 1,500g within 30 min after collection, transferred to 15 mL tubes and centrifuged at 2,000g for 15 min at 4 °C to remove cell debris. Plasma is then transferred to 2 mL cryovials and immediately snap-frozen and stored in liquid nitrogen. This entire procedure should be completed within 90 min of blood collection.
Cell culture sample preparation and conditioned media collection
Proper cell culture maintenance is required to ensure in vitro results generated in our protocol are valid. Fibroblasts cultured in our laboratory and used for previous validation of core senescence genes were grown in DMEM. However, different cell types might require different media for optimal growth. Routine mycoplasma testing should also be regularly carried out as mycoplasma are known to influence inflammatory signaling in mammalian cells. Contamination will invalidate experiments that analyze the SASP through either mRNA or proteomic analysis. Medium changes should occur at least twice a week. For cell passaging, cells are typically washed once in PBS, then detached using trypsin. Cells are then resuspended with medium and counted using trypan blue to ensure dead cells are excluded.
For SA-β-Gal and/or LF staining, and then co-staining for markers of proliferation, we recommend plating cells on coverslips in a 24-well plate. This procedure ensures enough cells can be used for statistical quantification and limits the amount of reagents required. For SASP analysis using either mRNA or conditioned medium, we recommend plating cells in 6-well plates, which should allow sufficient amounts of mRNA or protein to be the collected. For mRNA extraction, cells are washed once with PBS, trypsinized and resuspended with media. The cells are centrifuged and the pellet is resuspended in RNA lysis buffer. For conditioned medium collection, cells are required to be cultured for 24 h in serum-free, phenol red-free medium before collection. This is important as the presence of serum can compromise the detection of SASP proteins secreted from cells. Phenol red can also interfere with protein quantification during the procedure. The conditioned medium is then concentrated using centrifugal spin filters. The cells should also be counted as described, since this data will be required for quantification purposes.
Double staining for SA-β-Gal and proliferation markers
The SA-β-gal assay is used to detect lysosomal β-galactosidase activity at pH 6. Senescent cells display high lysosomal activity, allowing detection of the enzyme’s activity at this suboptimal pH21. As the optimal pH of lysosomal β-galactosidase is 4, the pH of the β-gal staining solution must be carefully monitored, as use of the assay at a pH < 6 can lead to positivity in non-senescent cells21. This procedure can be carried out using cultured cells or in vivo tissues. Samples are briefly fixed and incubated overnight in staining solution at pH 6. One component of this solution is 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-gal). X-gal is cleaved by β-galactosidase to yield an insoluble blue product44, which can be visualized the next day using light microscopy and quantified.
5-ethynyl-2′-deoxyuridine (EdU) is a thymidine analog that, when administered to cells, can incorporate into DNA during replication. A fluorescent azide is then added that binds the EdU and allows visualization of replicating or newly replicated DNA by microscopy or other means of fluorescence detection45. The number of EdU-positive cells is then quantified using fluorescence microscopy. Ki-67 is a protein expressed during all phases of the cell-cycle and is commonly used as a marker for cell proliferation46.
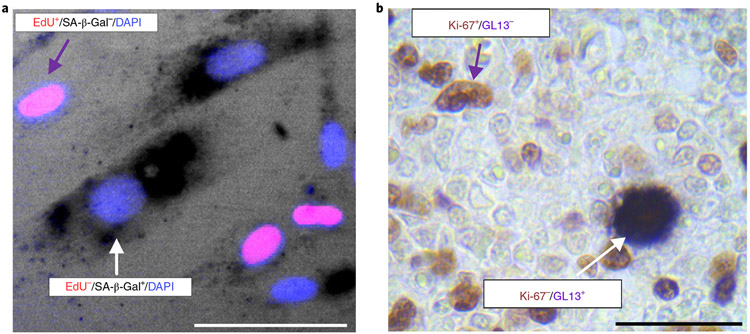
For cultured cells, SA-β-gal can be co-stained with either EdU or Ki-67. For SA-β-gal / EdU co-staining (Fig. 3a), EdU is normally added for 24 h before the cells are harvested for SA-β-gal staining. The incubation time for EdU should be adapted to the doubling of the cell population in question, and reduced or increased accordingly. Proliferating cell populations display greater numbers of EdU-positive cells than senescent populations. However, it should be noted that some residual EdU incorporation can occur from DNA repair activity in senescent cells. For SA-β-gal / Ki-67 co-staining, cells are first stained with SA-β-gal, re-fixed after SA-β-gal staining is completed and representative pictures have been taken, then processed for Ki-67 immunostaining. For both types of co-staining, we recommend the use of two controls. The first, a non-senescent and non-confluent proliferating sample, should be included as a positive control for EdU/Ki-67 and negative control for SA-β-gal staining. Confluent cell populations should be avoided as they are unable to replicate and are often positive for SA-β-gal activity21. The second, a cell population exposed to γ-irradiation or genotoxic drugs, should be used as a positive control for SA-β-gal and a negative control for EdU/Ki-67 staining5,18.
Fig. 3 ∣. Double staining SA-β-Gal or LF and proliferation markers.
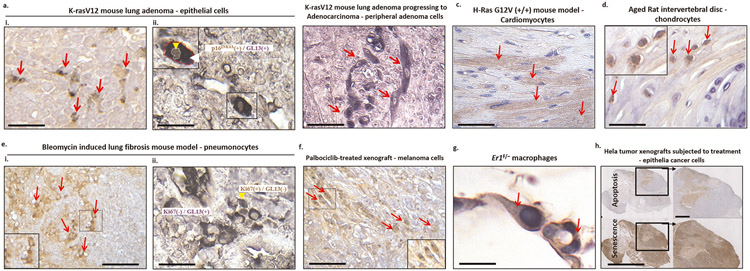
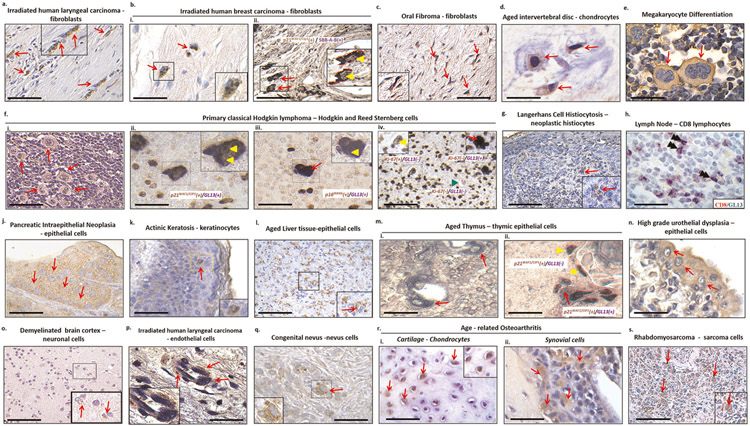
a, Human BJ fibroblasts were treated with 4 μM vorinostat (SAHA) for 8 d. Drug was refreshed daily. Eight days post-treatment, mutually exclusive staining between SA-β-Gal and EdU was observed. The purple arrow denotes an EdU+/SA-β-Gal− cell and the white arrow depicts an EdU−/SA-β-Gal+ (senescent) cell. Scale bar, 150 μm. b, Double immunohistochemical/hybrid histo-immunohistochemical staining in human primary classical Hodgkin lymphomas (cHLS). A mutually exclusive staining pattern between GL13 and Ki-67 in neoplastic Hodgkin and Reed-Sternberg cells was observed. The purple arrow denotes a Ki-67+/GL13− cell and the white arrow depicts a Ki-67−/GL13+ (senescent) cell. Alkaline phosphatase (AP) chromogenic reaction: dark blue cytoplasmic product; diaminobenzidine (DAB) reaction: nuclear brown signal; counterstain: nuclear fast red. Scale bar, 50 μm.
For in vivo tissues, as already mentioned, SA-β-gal staining can be used exclusively on flash-frozen tissues previously embedded in OCT. Moreover, it is critical that the SA-β-gal staining assay is done as soon as possible after the frozen tissue has been sectioned, as the activity of the enzyme is quickly lost upon exposure to higher temperatures. EdU incorporation in vivo can be done in actively proliferating organs (e.g., intestine), but the combination of EdU and SA-β-gal staining for tissues remains unreliable and will not be covered in this protocol. An alternative, Ki-67 / SA-β-gal double staining, should be the preferred combination for senescence detection in tissues. However, because co-staining procedures are difficult and produce a significant amount of artifactual false positives and negatives, we recommend a sequential staining (i.e., independent staining on sequential and adjacent sections) for frozen in vivo samples without pre-fixation.
Double staining for LF and proliferation markers
Another readout of enhanced lysosomal activity in senescent cells is staining for LF, which has the advantage of remaining detectable in paraffin-embedded material and is thus applicable in archival (FFPE) material. The staining can utilize the Sudan Black B (SBB) dye or a superior biotinylated SBB analog (GL13), which we use here. Caution should be taken when preparing the staining solution. The presence of crystals indicates improper dilution of the reagent in ethanol or saturation of the solution due to ethanol evaporation. Both can affect the staining properties and the effectiveness of the dye. During incubation with GL13, it is crucial to avoid evaporation of ethanol, as this can lead to crystal formation and nonspecific background staining. Subsequently, the GL13-LF interaction is visualized by detecting the biotin moiety attached to GL13, following principles of immunocyto (histo)chemistry or immunofluorescence17. As a result, irrespective of the approach—chromogenic or fluorescent—small and randomly distributed cytoplasmic granules or larger and more complex occasionally paranuclear structures are evident under the microscope17. Positive controls (samples with established senescence), negative controls (samples with absence of senescence) and technical negative controls (samples in which key reagents have been omitted during the staining procedure) should always be utilized17.
For cultured cells, LF can be easily co-stained with EdU or Ki-67, and the procedure is comparable to that described in the section covering SA-β-gal staining. For in vivo samples, LF staining has the advantage of being compatible with double co-staining approaches. Tissue samples can first undergo Ki-67 immunocytochemistry and the LF staining assay can then be applied to the same section (Fig. 3b). It is possible to co-stain LF with other senescence-associated markers such as p21WAF1/Cip1.
RT-qPCR
RT-qPCR comprises both phases of the proposed algorithm. RNA is first extracted from cell or tissue lysates, which can be carried out using any suitable commercial kit. cDNA is then synthesized from extracted RNA and again, can be carried out using a commercial kit, before genes of interest are amplified. We use the UPL probe system from Roche for RT-PCR and provide primer sequences and probe sequences designed using the associated website (https://lifescience.roche.com/en_nl/articles/Universal-ProbeLibrary-System-Assay-Design.html). SYBR green assays can also be used, but this would require investigators to design their own primers.
Phase 1 includes the measurement of the mRNA levels of p16INK4a, p21WAF1/Cip1, lamin B1 and a selected number of genes as part of a core senescence signature to validate the presence of senescence. In phase 2, mRNA levels of genes associated with specific subtypes of senescent cells are evaluated for senescence specification. Because of the lack of assays for the absolute quantification of most of the mRNAs measured, it is important that control samples (e.g., non-senescent cells, healthy tissue) are included in the analysis to determine relative fold changes. It is also critical that suitable ‘house-keeping’ genes are used for normalization47. In line with the MIQE (Minimum Information for Publication of Quantitative Real-Time PCR Experiments), we recommend using at least two housekeeping genes48,49. Here, we used the geometric mean of α-tubulin and β-actin, but the choice of housekeeping genes should be adapted to the experimental system. A list of housekeeping genes to be used in common senescence cellular models has been previously published by our group47.
SASP analysis in plasma and conditioned media
Emerging proteomic approaches are promising areas for the development of clinically relevant senescence biomarkers in human plasma50. Using unbiased and comprehensive mass spectrometry, a recently described ‘Atlas of the Senescence-Associated Secretory Phenotype’ was shown to have significant overlap with biomarkers of aging in human plasma, leading to a selective and evolving biomarker panel for measuring senescence-derived proteins in human plasma38.
Our panel of SASP proteins can feasibly be measured by multiple proteomic approaches, ranging from comprehensive mass spectrometry–based approaches to targeted antibody-based approaches available to most labs, such as antibody arrays, western blots and ELISAs. Measurement of SASP proteins is included in both phases of the protocol. In phase 1, we propose to use a selection of ‘core’ SASP factors for senescence validation. In phase 2, we propose to measure a panel of pro-inflammatory factors—also evaluated at the mRNA level—in order to specify the subtypes of senescent cells. The secretion of other factors can be added to the analysis to discriminate additional senescence subtypes38, but will not be covered in this protocol because of lack of definitive validations.
Applications of the methods
Our two-phase algorithm is designed to systematically define cellular senescence induction in cultured cells and in vivo. In both preclinical and clinical settings, it is common to define a senescent cell on the basis of individual markers which can under- or overestimate the number of senescent cells. Our detailed protocol is designed to guide researchers to more thoroughly define cellular senescence in their studies. Our protocol will be important for basic research in many fields, including aging and oncology, and will help evaluate the clinical relevance of senescence in a time when senotherapies are being tested in patients.
In phase 1 of our in vivo protocol, we provide several options, depending on the availability of sample preparation. If fresh samples are accessible, both snap-frozen samples in OCT without fixation and fixed samples in paraffin can be obtained. In that case, researchers have the option to use the combination of SA-β-gal or GL13 with other senescence and proliferation markers. If only FFPE samples are available, our protocol suggests the combination of GL13 with other senescence or proliferative markers. In phase 2, the type of senescence in the samples can also be identified. We provide here the specifics to identify cells with or without a pro-inflammatory SASP, as the secretion of certain cytokines and chemokines can directly relate to pathological senescence. Other subtypes of senescent cells could be identified by adding additional senescence-associated signatures. However, these additional signatures remain poorly characterized and will not be covered in the protocol.
Comparisons with other methods
Our protocol is a combination of established methods to detect cellular senescence, and aims to minimize the number of false positives and negatives and identify pro-inflammatory features. Wherever possible, we propose to perform double staining for markers in the same cell or tissue to validate the senescence status in situ (SA-β-gal or GL13 plus other markers for senescence or proliferation). Importantly, and in contrast to other multi-marker approaches, the tools necessary for our algorithmic approach are easy to obtain and the methods do not require special equipment, thus remaining highly accessible to most laboratories. Previous multi-marker approaches have combined flow cytometry and high-content image analysis to identify senescent cells in both cells and tissue, such as combinations of SA-β-gal with either Ki67 or γ-H2AX40. However this methodology requires expensive equipment and trained personal.
Limitations
There are still several limitations to this protocol. First, combining all the markers in the same sample is not yet possible without incurring potential false positives or negatives. Second, tissue analysis for SA-β-gal is limited unless the sample is quickly frozen and analyzed to maintain enzymatic activity, which can limit further histological analysis due to poor tissue structure preservation. Third, GL13 is diluted in 70% (vol/vol) ethanol, rendering this assay inconsistent with analysis of living cells17. Fourth, antigen retrieval is necessary for many antibodies targeting other senescence markers (e.g., p21WAF1/Cip1), which is more easily performed in paraffin-embedded samples, making it difficult to combine SA-β-gal with additional markers. Fifth, the identification of senescence subtypes remains limited, mainly because it is currently impossible to isolate and purify individual senescent cells from human tissues. Sixth, our two-phase protocol is more complicated than many others, especially for studies not focused on cellular senescence, which might be a barrier for wide application.
Materials
Reagents
Biological materials
Cell lines of human or mouse origin, e.g., BJ cells (ATCC, cat. no. CRL-2522; RRID: CVCL_3653)
Primary cells from human or mouse tissues, e.g., mouse lung fibroblasts generated in the lab from tissue from in-house animal experimental studies ! CAUTION Cells used in cell culture should be tested on a regular basis for mycoplasma infection
Human or mouse tissues snap frozen and/or embedded in paraffin, e.g., mouse kidneys from in-house animal experimental studies ! CAUTION Any experiments involving live animals must conform to relevant Institutional and National regulations. ! CAUTION All experiments on human tissues must have ethics approval in accordance with governmental and institutional regulations. Informed consent must be obtained for all human samples ▲CRITICAL SA-β-Gal staining in tissues can be performed on fresh slides from snap-frozen tissues only
Common reagents
PBS (Gibco, cat. no. 10010-023)
Triton X-100 (Acros organics, cat. no. 215682500) ! CAUTION Triton X-100 is harmful if swallowed and an irritant in direct contact with eyes. Wear protective clothing and avoid exposure.
16% (wt/vol) formaldehyde (Thermo Fisher Scientific, cat. no. 28908) ! CAUTION Formaldehyde is harmful if swallowed, inhaled or in direct contact with skin. Ensure ventilation is adequate and wear protective clothing to prevent exposure.
Tris base (Roche, cat. no. 11814273001)
DAPI (Sigma-Aldrich, cat. no. D9542)
ProLong gold antifade mountant (Thermo Fisher Scientific, cat. no. P36934)
Cell culture
DMEM + GlutaMAX (Gibco, cat. no. 31966)
FBS (Sigma-Aldrich, cat. no. F7524)
0.5% (wt/vol) trypsin-EDTA solution (10×; Sigma-Aldrich, cat. no. T4174)
SA-β-Gal staining
25% (wt/vol) glutaraldehyde (Thermo Fisher Scientific, cat. no. MERC8.20603.1000) ! CAUTION Glutaraldehyde is harmful if swallowed or inhaled and an irritant to skin and eyes. Ensure ventilation is adequate and avoid exposure.
X-gal (Thermo Fisher Scientific, cat. no. 10113253)
Dimethylformamide (DMF; Merck Millipore, cat. no. 1.10983.1000) ! CAUTION DMF is flammable and harmful if inhaled or in direct contact with skin and eyes. Ensure ventilation is adequate, wear protective clothing avoid exposure. Prevent contact with flames.
Citric acid monohydrate (Sigma-Aldrich, cat. no. C1909) ! CAUTION Citric acid monohydrate is harmful and an irritant to eyes. Wear protective clothing to avoid exposure.
Sodium dibasic phosphate (Thermo Fisher Scientific, cat. no. 424380010)
Potassium ferricyanide (Thermo Fisher Scientific, cat. no. 10414583)
Potassium ferrocyanide (Thermo Fisher Scientific, cat. no. 10216460)
Hydrogen chloride (Merck, cat. no. 1.00317.1000)
Sodium hydroxide (Fisher, cat. no. S/4920/60)
Sodium chloride (Merck Millipore, cat. no. 1.06404.1000)
Magnesium chloride (Fisher Chemicals, cat. no. M/0550/53)
OCT (Sakura, cat. no. 4583)
EdU staining
EdU (Lumiprobe, cat. no. 10540)
Copper(II) sulfate pentahydrate (Sigma-Aldrich, cat. no. 209198) ! CAUTION Harmful if swallowed and an irritant if in direct contact with skin and eyes. Wear protective clothing and avoid exposure. Very toxic to aquatic life and should be disposed of in appropriate containers to a waste disposal plant.
Sulfo-cyanine-3-azide (Lumiprobe, cat. no. D1330)
Sodium ascorbate (Sigma-Aldrich, cat. no. A4034)
Immunohistochemistry
Xylene (VWR, cat. no. 28975) ! CAUTION Xylene causes skin, eye and respiratory irritation. Wear protective clothing and avoid exposure. Ensure ventilation is adequate.
Ethanol, absolute (VWR, cat. no. 83813) ! CAUTION Ethanol is flammable and an irritant if in direct contact with eyes. Wear protective clothing and avoid exposure. Prevent contact with flames.
Tween-20 (Sigma-Aldrich, cat. no. P1379)
Methanol (VWR, cat. no. 20903) ! CAUTION Methanol is flammable and harmful if swallowed, inhaled or in direct contact with skin. Wear protective clothing and avoid exposure. Ensure ventilation is adequate. Prevent contact with flames.
Acetone (VWR, cat. no. E646) ! CAUTION Acetone is flammable and an irritant in direct contact with eyes. Wear protective clothing and prevent contact with flames.
Normal goat serum (Sanquin, cat. no. M1530)
Hydrogen peroxide (Thermo Fisher Scientific, cat. no. H325) ! CAUTION Hydrogen peroxide is harmful if swallowed or in direct contact with eyes. Wear protective clothing and avoid exposure. Hydrogen peroxide is very toxic to aquatic life and should be disposed of in appropriate containers to a waste disposal plant.
Ki-67 antibody (rabbit polyclonal; Abcam, cat. no. ab16667; RRID: AB_302459)
p21WAF1/Cip1 antibody (rabbit, clone M-19; SantaCruz Biotechnology, cat. no. SC 471; RRID: AB_632123)
p16INK4a antibody (mouse monoclonal F-4; SantaCruz Biotechnology, cat. no. SC 74401)
Citric acid monohydrate (Sigma-Aldrich, cat. no. C1909)
Tri-sodium citrate dihydrate (Merck, cat. no. 1064480500)
Secondary goat-anti-rabbit antibody biotin conjugated (Vector, cat. no. BA-1000; RRID: AB_2313606)
ABC/PO (avidin-biotin peroxidase) complex (Vector, cat. no. PK-4000; RRID: AB_2336818)
DAB substrate (Sigma-Aldrich, cat. no. D5637)
30% (wt/wt) hydrogen peroxide solution (Sigma-Aldrich, cat. no. 216763)
Hematoxylin (Sigma-Aldrich, cat. no. 51275)
LF staining
TBS (Tris Buffered Saline; 20×): 2.7 M NaCl, 0.16 M Tris–HCl ultrapure, pH = 7.4)
GL13 reagent (SenTraGor, Lab Supplies Scientific–P. Galanis & Co., cat. nos. AR8850020, AR8850040 and AR8850080)
Ultravision Quanto Detection System HRP DAB kit (Thermo Fisher Scientific, cat. no. TL-125-QHD)
Primary anti-biotin antibody [Hyb-8] (Abcam, cat. no. ab201341; RRID: AB_2861249)
Glycergel (DakoCytomation, cat. no. C056330-2)
Secondary goat anti-mouse antibody AP conjugated (Invitrogen, cat. no. G21060; RRID: AB_2536528)
Secondary goat anti-mouse antibody AF-568 (ThermoFisher Scientific, cat. no. A-11004; RRID: AB_2534072)
NBT/BCIP ready-to-use tablets (Roche, cat. no. 11697471001)
Levamisole hydrochloride (Sigma-Aldrich (Merck), cat. no. 31742)
RT-PCR
Isolate II RNA Mini Kit (Bioline, cat. no. BIO-52073)
High-Capacity cDNA Reverse Transcriptase Kit (Thermo Fisher Scientific, cat. no. 4368813)
Sensifast Probe Lo-ROX kit (Bioline, cat. no. BIO-84020)
UPL Probe Library (Roche, cat. no. 04683633001)
RNAse Away (Thermo Fisher Scientific, cat. no. 10666421)
Table 1 ∣.
Primer and probe sequences for senescence and housekeeping genes
| Primer sequence (5′–3′) | Probe sequence (5′–3′) | |
|---|---|---|
| Gene (up) | ||
| p16INK4a | F: GAGCAGCATGGAGCCTTC R: CGTAACTATTCGGTGCGTTG |
Sense: CTCCAGCA Antisense: TGCTGGAG |
| p21WAF1/Cip1 | F: TCACTGTCTTGTACCCTTGTGC R: GGCGTTTGGAGTGGTAGAAA |
Sense: GGGAGCAG Antisense: CTGCTCCC |
| GDNF | F: ATGTCCAACCTAGGGTCTGC R: CATCCCATAACTTCATCTTAAAGTCC |
Sense: CCGCCGCC Antisense: GGCGGCGG |
| TSPAN13 | F: TCAACCTGCTTTACACCTTGG R: AATCAGCCCGAAGCCAAT |
Sense: CTCCAGCA Antisense: TGCTGGAG |
| CCND1 | F: GCTGTGCATCTACACCGACA R: TTGAGCTTGTTCACCAGGAG |
Sense: AGGAGCTC Antisense: GAGCTCCT |
| DYNLT3 | F: GTGCTCTACCGGCGTGTC R: CAGCATTGAAGCCAACCTC |
Sense: CTCCTCCA Antisense: TGGAGGAG |
| PDLIM4 | F: GGATCCACATCGATCCTGAG R:GCTTGGTCTGCCATCTTCTG |
Sense: GCCTGCTG Antisense: CAGCAGGC |
| Gene (down) | ||
| LMNB1 | F: GTGCTGCGAGCAGGAGAC R: CCATTAAGATCAGATTCCTTCTTAGC |
Sense: CCCAGCAG Antisense: CTGCTGGG |
| NF1A | F: CCAGATTAGGGGGATGGTATG R: CCCCTTATCTTTCTAAGCATTTCC |
Sense: GGGAGCAG Antisense: CTGCTCCC |
| EFNB3 | F: TGGAACTCGGCGAATAAGAG R: CGATCTGAGGGTACAGCACA |
Sense: CTCTGCCT Antisense: AGGCAGAG |
| PATZ1 | F: GCAACTTCTGCAGTATCTGTAACC R: ACCGTGGTGGGTTTTAACAT |
Sense: CTCTGCCT Antisense: AGGCAGAG |
| SPIN4 | F: ACTATTTCCCTACAGCAGAACAGG R: GCTTGCCCACGAGACTGT |
Sense: CCTGGAGA Antisense: TCTCCAGG |
| CNTLN | F: AAGGCAGCAATACAAGAATTGAA R: TTGACCTCATCATCTTCACCAG |
Sense: TGGCAGAG Antisense: CTCTGCCA |
| Gene (reference) | ||
| α-tubulin | F: CTTCGTCTCCGCCATCAG R: CGTGTTCCAGGCAGTAGAGC |
Sense: GCCTGCTG Antisense: CAGCAGGC |
| β-actin | F: CCAACCGCGAGAAGATGA R: CCAGAGGCGTACAGGGATAG |
Sense: CAGCCTGG Antisense: CCAGGCTG |
Table 2 ∣.
Core SASP panel proteins
| UniprotID | Gene | Protein name |
|---|---|---|
| Q99988 | GDF15 | Growth/differentiation factor 15 (GDF-15) (Macrophage inhibitory cytokine 1) (MIC-1) (NSAID-activated gene 1 protein) (NAG-1) (NSAID-regulated gene 1 protein) (NRG-1) (Placental TGF-beta) |
| P03956 | MMP1 | Interstitial collagenase (EC 3.4.24.7) (Fibroblast collagenase) (Matrix metalloproteinase-1) (MMP-1) |
| P52823 | STC1 | Stanniocalcin-1 (STC-1) |
| P01033 | TIMP1 | Metalloproteinase inhibitor 1 (Erythroid-potentiating activity) (EPA) (Fibroblast collagenase inhibitor) (Collagenase inhibitor) (Tissue inhibitor of metalloproteinases 1) (TIMP-1) |
| P16035 | TIMP2 | Metalloproteinase inhibitor 2 (CSC-21K) (Tissue inhibitor of metalloproteinases 2) (TIMP-2) |
SASP protein measurements
Serum-free, phenol red-free medium (Gibco, cat. no. 21063-029)
Calcium- and magnesium-free PBS (Gibco, cat. no. 10010-023)
BCA protein assay kit (Pierce, cat. no. 23225)
Equipment
CO2 incubator (or Tri-Gas Incubator (N2/O2/CO2), e.g., for primary cells that grow at lower oxygen concentration (Panasonic, cat. no. MCO-19M)
Tissue culture coverslips (Sarstedt, cat. no. 83.1840)
24-well tissue culture plate (Sarstedt, cat. no. 83.3922)
HeraTherm IGS60 incubator (no CO2; Thermo Fisher Scientific, cat. no. 51028130)
pH meter (VWR, cat. no. Inolab pH7110)
Brightfield and phase-contrast microscope (ThermoFisher Scientific, cat. no. AMEX1000)
Automated cell counter (ThermoFisher Scientific, cat. no. Countess II)
Amicon Ultra-15 centrifugal filter unit (MerckMillipore, cat. no. UFC900308)
Insulin disposable syringe 1 ml (Insumed 30G, PIC Solutions) carrying a 13 mm (membrane 0.22 μm) filter (Thermo Scientific Nalgene Syringe Filter 0.2 μm Pes, cat. no.720-1320)
Fluorescent microscope (Leica, cat. no. DMI6000)
Coplin jars (VWR, cat. no. 720-0707)
Hydrophobic barrier pen (Sigma-Aldrich, cat. no. Z672548)
Nanodrop spectrophotometer (Thermo Fisher Scientific, cat. no. ND-2000)
LightCycler 480 Multiwell Plate 384, white (Roche, cat. no. 4729749001)
Lightcycler 480 sealing foil (Roche, cat. no. 4729757001)
Lightcycler 480 Instrument II (Roche, cat. no. 05015243001)
Microtome (ThermoFisher Scientific, cat. no. HM 340 E)
Cryostat (Leica, cat. no. CM3050)
Mortar and pestle (Carl Roth, cat. no. CYH1.1)
Waterbath (Leica, cat. no. HI1210)
Microwave (Sharp, cat. no. R-239(IN)-A)
Reagent setup
▲CRITICAL Apart from the reagents that need to be made fresh, the remaining reagents should be stable for at least several months if stored properly. Reagents are stored at room temperature (RT: 20–25 °C) unless stated otherwise.
2% (wt/vol) formaldehyde + 0.2% (wt/vol) glutaraldehyde fixative solution
Prepare a 4% (wt/vol) formaldehyde solution by diluting the 16% (wt/vol) formaldehyde stock solution 4× in PBS. Add 800 μL of 25% (wt/vol) glutaraldehyde stock solution to 50 mL of 4% (wt/vol) formaldehyde solution, then add 49.2 mL PBS to a total volume of 100 mL.
5% (wt/vol) paraformaldehyde/PBS solution, pH = 7.4 with or without 0.2% (wt/vol) glutaraldehyde
Dissolve 5 g of paraformaldehyde (PFH) in 100 mL of PBS in a glass beaker. Heat and stir the mixture until it becomes transparent. Let the solution cool and adjust pH to 7.4 using HCl and NaOH. To achieve 0.2% glutaraldehyde concentration when required, add 1.6 mL of 25% (wt/vol) glutaraldehyde stock solution to a total volume of 100 mL final solution.
X-gal stock solution, 20 mg/mL
Prepare fresh. Weigh 20 mg of X-gal and dissolve in 1 mL of DMF. ▲CRITICAL This solution is light-sensitive.
Citric acid stock solution, 0.1 M
Dissolve 4.2 g in 200 mL ddH2O.
Sodium phosphate (dibasic) stock solution, 0.2 M
Dissolve 10.7 g in 200 mL ddH2O.
Citric acid/sodium phosphate buffer, 0.2 M
Add 36.85 mL citric acid stock solution (0.1 M) to 63.15 mL sodium phosphate (dibasic) stock solution (0.2 M). Adjust pH to 6.0 using HCl and NaOH.
Potassium ferricyanide stock solution, 100 mM
Dissolve 3.3 g in 100 mL ddH2O. ▲CRITICAL Store at 4 °C. This solution is light-sensitive.
Potassium ferrocyanide stock solution, 100 mM
Dissolve 4.2 g in 100 mL ddH2O. ▲CRITICAL Store at 4 °C. This solution is light-sensitive.
Sodium hydroxide stock solution, 10 M
Dissolve 20 g in 50 ml ddH2O.
Sodium chloride stock solution, 5 M
Dissolve 58.4 g in 200 mL ddH2O.
Magnesium chloride stock solution, 1 M
Dissolve 19.0 g in 200 mL ddH2O.
Potassium chloride stock solution, 1 M
Dissolve 7.5 g in 100 mL ddH2O.
EdU stock solution, 5 mM
Dissolve 12.5 mg in 10 mL ddH2O. ▲CRITICAL Store aliquots at −20 °C
Sulfo-Cy3-azide stock solution, 4 mM
Dissolve 2.9 mg in 1 mL ddH2O. ▲CRITICAL Store aliquots at 4 °C. This solution is light-sensitive.
Cu(II)sulfate (pentahydrate) stock solution, 200 mM
Dissolve 500 mg in 10 mL ddH2O. ▲CRITICAL Store aliquots at 4 °C.
TBS (Tris-Buffered Saline, 20×): 2.7 M NaCl, 0.16 M Tris-HCl ultrapure, pH = 7.4.)
For 1,000 mL: dissolve 20 g Tris-HCl ultrapure and 162 g NaCl in 900 mL ddH2O. Adjust pH to 7.4 using HCl 6 N and add ddH2O to a final volume of 1,000 mL. For a 1× solution, mix 50 mL. of the 20× solution with 950 mL ddH2O and adjust pH to 7.4.
TRIS-HCl stock solution (pH = 7.6), 500 mM
Dissolve 6.0 g in 70 mL ddH2O. Set pH at 7.6 using 37% (vol/vol) HCl, then add ddH2O to a total volume of 100 mL. Dilute the stock solution 5× in ddH2O to get 100 mM Tris (pH = 7.6).
TRIS-HCl stock solution (pH = 7.8), 500 mM
Dissolve 6.0 g in 70 mL ddH2O. Set pH at 7.8 using 37% (vol/vol) HCl, then add ddH2O to a total volume of 100 mL. Dilute the stock solution 10× in ddH2O to get 0.05 M Tris (pH = 7.8).
10% (vol/vol) Triton X-100 in PBS stock solution
Add 1 mL of Triton X-100 to 9 mL PBS to get a total volume of 10 mL. Dilute the stock solution 100× in PBS to get 0.1% (vol/vol) Triton X-100. Dilute the stock solution 20× in PBS to get 0.5% (vol/vol) Triton X-100.
DAPI stock solution, 2 mg/mL
Dissolve 10 mg of DAPI in 5 mL ddH2O. Dilute the DAPI stock solution 1000× in ddH2O to get 2 μg/ mL DAPI. ▲CRITICAL Store aliquots at 4 °C. This solution is light-sensitive.
10% (vol/vol) Triton X-100 in TBS
Add 1 mL of Triton X-100 to 9 mL TBS to get a total volume of 10 mL. Dilute the stock solution 20× in TBS to get 0.5% (vol/vol) Triton X-100.
DAB stock solution, 70 mM
Dissolve 500 mg of DAB in 100 mL ddH2O. ▲CRITICAL Store aliquots at −20 °C.
DAB substrate
In a tube combine 5 mL DAB (70 mM), 45 mL Tris-HCl (0.05 M; pH = 7.8) and 5 μL 30% (vol/vol) H2O2. ▲CRITICAL Prepare this solution fresh.
Citrate buffer 1× (pH = 6.0), 10 mM
Dissolve 12.6 g citric acid monohydrate and 11.8 g tri-sodium citrate dihydrate in 800 mL of ddH2O and adjust the pH to 6.0 using HCL and NaOH. Add ddH2O to a total volume of 1 L to get a 0.1 M citrate buffer (10×). For 10 mM citrate buffer (1×), mix 100 mL of the 0.1 M citrate buffer with 900 mL ddH2O.
Levamisole hydrochloride, 100 mM
Dissolve 2.4 g levamisole hydrochloride in 100 mL ddH2O. ▲ CRITICAL Prepare solution fresh.
KTBT buffer stock solution, 0.05 M Tris-HCl (pH = 7.5), 0.15 M NaCl, 0.01 M KCl
To 900 mL of ddH2O, add 8.7 g of NaCl, 7.9 g of TRIS-HCl and 0.75 g of KCl (1 M) 860 mL. Mix and adjust volume to 1,000 mL. Do not adjust the pH.
Procedure
Phase 1: validating senescence in in vitro samples
▲CRITICAL The first phase in our algorithm is to validate senescence in samples. For cell culture samples (Steps 1–49), this can be done by co-staining for SA-β-Gal and EdU (Steps 1–26), or for LF and p21WAF1/Cip1, p16INK4a or Ki-67 (Steps 27–30A–D). This co-staining is followed by analyzing mRNA levels of p16INK4a, p21WAF1/Cip1, lamin B1 and a subset of genes that are part of the core senescence signature we identified previously on the basis of by fold change in expression compared to non-senescent tissues or cells and p-values > 0.05)5 (Steps 31–44). Lastly, we perform proteomic analysis of core SASP proteins in the conditioned media (Steps 45–52).
SA-β-Gal and EdU staining ● Timing 28–42 h
Put sterile coverslips in the wells of a 24-well plate.
Upon senescence induction (see Box 1 for the most common methods to induce senescence in cells), re-plate 2 × 104 induced cells per well (with coverslip) of a 24-well plate with 500 μL culture medium.
Incubate the cells overnight at 37 °C with 5% CO2 and 5% O2.
Replace the culture medium with new medium containing 10 μM EdU.
-
Incubate the cells for 12–24 h, or more depending on the doubling time of the culture, at 37 °C with 5% CO2 and 5% O2.
? TROUBLESHOOTING
Wash cells twice with 500 μL PBS.
Aspirate the PBS and fix the cells for 3–5 min at RT using 500 μL/well of 2% (wt/vol) formaldehyde + 0.2% (wt/vol) glutaraldehyde in PBS.
Wash cells twice with 500 μL PBS.
-
Prepare SA-β-gal staining solution fresh according to the table below, and adjust the amount depending on the total number of samples:
Component Volume per well (μL) Final Concentration X-gal in dimethylformamide (20 mg/mL) 25 1 mg/mL Citric acid/sodium phosphate buffer, pH = 6.0 (0.2 M) 100 40 mM Potassium ferrocyanide (100 mM) 25 5 mM Potassium ferricyanide (100 mM) 25 5 mM Sodium chloride (5 M) 15 150 mM Magnesium chloride (1 M) 1 2 mM Water 310 Total 500 ▲CRITICAL STEP Verify that the pH of the SA-β-gal staining solution is pH 6.0.
? TROUBLESHOOTING
-
Aspirate the PBS and add 500 μL staining solution to each well and seal the plate with parafilm to avoid evaporation.
▲CRITICAL STEP Evaporation leads to the formation of crystals.
-
Incubate the plate at 37° C in a dry incubator without CO2 for 12–16 h.
▲CRITICAL STEP CO2 affects the pH and will artifactually modify the results.
Wash twice with 500 μL PBS.
Check the results under a light microscope. Positive cells present a blue perinuclear staining. Compare results of senescent cells versus their appropriate controls for the treatment used.
Upon successful staining of SA-β-gal, wash twice with 500 μL PBS.
-
Fix the cells for 10 min with 500 μL of 4% (wt/vol) formaldehyde in PBS.
■PAUSE POINT Fixed cells can be stored at 4 °C for up to 1 week.
Incubate the cells in 100 mM Tris (pH = 7.6) in PBS for 10 min.
Permeabilize the cells in 500 μL of 0.1% (vol/vol) Triton X-100 in PBS for 10 min.
Wash twice with 500 μL PBS.
- Prepare EdU staining solution fresh, according to the table below, adjusting the volume depending on the total number of samples:
Component Volume (μL) Final Concentration Cu(II)SO4 (200 mM) 0.5 2 mM Sulfo-Cy3-azide (4 mM) 0.05 4 μM Sodium ascorbate (200 mg/mL) 5 20 mg/mL PBS 44.45 Total 50 -
Add 50 μL of the EdU staining solution to a piece of parafilm after the SA-β-Gal staining has concluded. Lift the coverslip and put it with the cells facing downwards on top of the EdU staining solution. Incubate for 30 min in the dark at RT.
▲CRITICAL STEP Ensure that there are no bubbles.
Put the coverslips back in the original wells (with the cells facing up) and wash twice with 500 μL PBS.
Incubate the cells with 2 μg/mL DAPI for 20 min in the dark at RT.
Wash twice with 500 μL PBS.
Mount the coverslips with mounting medium onto glass slides and let them dry overnight (protected from light).
Visualize SA-β-Gal staining and EdU incorporation using a fluorescence microscope equipped with bright field. Use a filter appropriate for Cy3 (excitation/emission: 552/570 nm).
Quantify senescence using the following formula (at least 300 cells from three independent experiments need to be counted): EdU positive cells (%) = [EdU positive cell count (Cy3) / total cell count (DAPI)] × 100 SA-β-Gal cells (%) = [SA-β-Gal positive cell count (bright field) / total cell count (DAPI)] × 100. Double-positive cells might be present in certain conditions. Double-negative cells might be present in quiescent cells.
Box 1 ∣. Common methods to induce senescence in cells in vitro.
γ-irradiation: expose cells to one dose of 10 Gγ γ-irradiation using a Cesium137 source irradiator. Incubate for a further 7–10 d.
Doxorubicin: incubate cells with 250 nM doxorubicin for 24 h. Wash cells once with fresh medium and incubate cells for a further 7–10 d.
Oncogenic RAS-induced senescence: infect cells with viral particles expressing oncogenic HRASV12. Incubate cells for at least another 14 d.
-
Replicative senescence: serially passage cells until growth arrested. At each passage monitor population doubling level (PDL) using the equation:
PDL = x + 3.32(log y – log I)
x = previous population doubling level
y = number of cells counted at subculture
I = initial number of cells plated at previous PDL
LF staining
-
27
Plate and treat cells as described in Steps 1–2. When cells are ready, remove the coverslip from the well with thin edged forceps and wash three times in a clean well of new plate filled each time with fresh PBS.
-
28
Fix cells mounted on coverslips for 3–5 min at RT using 500 μL/well of 1–5% (wt/vol) formaldehyde/PBS solution. The concentration should be optimized to get the best staining results and can also depend on the cell type and whether preservation of detailed morphological features of the cells is needed. Alternatively, use 100% methanol, 100% ethanol or 2% (wt/vol) formaldehyde + 0.2% (wt/vol) glutaraldehyde in PBS, for 3–5 min at RT.
■PAUSE POINT Fixed cells can be stored at 4 °C for up to 1 week.
-
29
Permeabilize fixed cells with 0.5% (vol/vol) Triton X-100 for 3–5 min at RT.
-
30
Carry out LF staining as directed in Options A–D. Options A and C are more suitable for screening for senescence using a light or fluorescence microscope, respectively. Options B and D can be applied to verify the expression of a marker/factor within senescent cells, using a light or fluorescence microscope, respectively.
? TROUBLESHOOTING- Hybrid cytochemistry/immunocytochemistry ● Timing 5–7 h
- Wash cells twice with PBS and then once with TBS.
-
Place the coverslips carrying cells (hereafter referred to as primary coverslips) on top of a histological glass slide, so that cells are facing upwards. This allows accurate handling of the material. Beyond this step, primary coverslips are always positioned on the slide and cells are constantly covered with drops of different solutions applied in the next steps.▲CRITICAL STEP Incubations should be performed by placing the slide in a humidified chamber commonly used for immunohistochemistry that is either commercially available or made by covering the inner walls of a tightly sealed chamber with moistened paper.
- Block endogenous hydrogen peroxidase. We use the UltraVision Hydrogen Peroxide Block included in the Ultravision Quanto Detection System HRP DAB kit in the dark for 10 min at RT.
- Wash twice with TBS for 30 s and once for 5 min at RT.
- Rinse primary coverslips with 50% (vol/vol) ethanol for 5 min at RT.
- Rinse primary coverslips with 70% (vol/vol) ethanol for 5 min at RT.
-
Prepare the GL13 staining solution as follows: dissolve 40 mg of the compound (SenTraGor) in 7.4 mL 100% ethanol in a glass container and seal it with parafilm. Incubate the container at 56 °C in a water bath for 120 min until it dissolves completely. The dye can be stored for a maximum of 2–3 months at RT in the dark.▲CRITICAL STEP Evaporation of ethanol leads to the formation of crystals. The latter indicates the reagent is not completely dissolved.
-
Put a drop of GL13 solution on each of the primary coverslips with the help of a syringe carrying a 13 mm (membrane 0.22 μm) filter. Cover each primary coverslip with a clean second coverslip devoid of cells (from now on secondary coverslip), using thin-edged forceps.▲CRITICAL STEP This step is crucial to avoid evaporation of ethanol.
- Monitor the staining reaction under the microscope until detection of the signal (variably sized blue-black or brown cytoplasmic granules after an average time of 5–10 min).
- Remove the secondary coverslip and wash twice with 50% (vol/vol) ethanol for 5 min at RT.
-
Repeat washing twice with fresh 50% (vol/vol) ethanol for 5 min at RT.▲CRITICAL STEP The second and third washes are essential for removal of nonspecific staining.
- Wash twice with TBS for 30 s and once for 5 min at RT.
- Incubate with 0.5% (vol/vol) Triton X/TBS for 3 min at RT.
- Wash with TBS for 5 min at RT.
-
Add the anti-biotin antibody to the primary coverslips. We use the [Hyb-8] ab201341 primary antibody from Abcam, diluted 1/300 in TBS for 60 min at 37 °C.▲CRITICAL STEP Omit the anti-biotin antibody as a negative control. If nonspecific background staining occurs, the following options are available: (i) increase antibody dilution, (ii) decrease duration of incubation and (iii) an antigen blocking step can precede incubation with the antibody; for instance, incubation with the Ultra Protein Block included in the Ultravision Quanto Detection System HRP DAB kit for 2–5 min at RT can be performed before adding the primary antibody.
- Wash three times in TBS for 5 min at RT.
- Signal development (first step): for primary antibody amplification use Quanto included in the Ultravision Quanto Detection System HRP DAB kit for 10 min at RT.
- Wash three times in TBS for 5 min at RT.
- Signal development (second step): incubate with the HRP Polymer Quanto included in the Ultravision Quanto Detection System HRP DAB kit for 10 min at RT.
- Wash twice in TBS for 30 s and once for 5 min at RT.
-
Signal development (third step): apply DAB Plus Chromogen diluted 1/300 in DAB Plus Substrate (Ultravision Quanto Detection System HRP DAB kit).▲CRITICAL STEP Beyond Step 30A(xvii), alternative signal detection systems can be used.
- Monitor the staining reaction under the microscope at RT until brown signal develops (~30–60 s).
- Wash in tap water for 5 min at RT.
- Counterstain with Hematoxylin diluted 1/4 in deionized water.
- Wash in tap water for 5 min at RT.
- Apply permanent mounting media glycergel.
-
Observe under the light microscope.? TROUBLESHOOTING
- Co-staining (double): immunocytochemistry and hybrid cytochemistry/immunocytochemistry ● Timing 9–12h
- Perform immunocytochemistry of choice (Ki-67, p21WAF1/Cip1, p16INK4a and others), using DAB as chromogen. Specifically, incubate the cells in TBS for 5 min at RT and block endogenous hydrogen peroxidase, using the UltraVision Hydrogen Peroxide Block included in the Ultravision Quanto Detection System HRP DAB kit for 10 min at RT and in dark conditions.
- Then wash twice in TBS for 30 s and once more for 5 min at RT.
- Incubate with the Ultra Protein Block included in the Ultravision Quanto Detection System HRP DAB kit for 7 min at RT.
- Wash three times in TBS for 5 min at RT.
- Incubate with the primary antibody of choice: anti-p21WAF1/Cip1 mouse (F-5)(sc-6246) Santa-Cruz, dilution: 1/200; anti-p16INK4a mouse [(sc-74401) Santa-Cruz, dilution: 1/100]; anti-Ki67 rabbit polyclonal antibody (ab16667, Abcam, dilution 1/500), for 60 min at 37 °C.
- After washing twice in TBS for 1 min each at RT, incubate with the primary antibody amplifier Quanto included in the Ultravision Quanto Detection System HRP DAB kit, for 10 min at RT.
- Subsequently, wash three times in TBS for 5 min at RT and incubate with the HRP Polymer Quanto included in the Ultravision Quanto Detection System HRP DAB kit, for 10 min at RT. Following two washes in TBS for 30 s and once for 5 min at RT, apply DAB Plus Chromogen diluted 1/100 in DAB Plus Substrate (Ultravision Quanto Detection System HRP DAB kit).
- Monitor the staining reaction under the light microscope until detection of the dark brown signal (average time 30–60 s) at RT. Once the brown immunocytochemical signal develops, continue with Hybrid Cytochemistry/Immunocytochemistry as described in Steps 30B (ix–xxvi), applying an alkaline phosphatase (AP)-mediated detection assay.
- Wash in tap water for 5 min at RT.
- Rinse the coverslips once in 50% (vol/vol) ethanol for 5 min at RT.
- Rinse once in 70% (vol/vol) ethanol for 5 min at RT.
- Incubate with GL13 (see Steps 30A(vii–viii) at RT.
- Monitor the staining reaction under the light microscope.
- Rinse three times in 50% (vol/vol) ethanol.
- Repeat washing step three times in fresh 50% (vol/vol) ethanol.
- Rinse twice in TBS for 30 s and once for 5 min at RT.
- Incubate with the anti-biotin antibody (see Step 30A(xv)).
- Rinse three times in TBS for 5 min at RT.
-
Incubate with the secondary goat anti-mouse antibody conjugated with AP diluted 1/800 in TBS, for 60 min at RT.▲CRITICAL STEP Determination of optimal dilution and incubation time of the secondary antibody to avoid nonspecific staining is crucial.
- Rinse three times in TBS for 5 min at RT.
- Rinse twice in TBS for 30 s and once for 5 min at RT.
- Apply the NBT/BCIP substrate (1 tablet in 10 mL ddH2O) plus 20 μL of 100 mM levamisole. Monitor the staining reaction under the light microscope until detection of the dark blue/purple signal (5–7 min) at RT.
- Rinse twice in KTBT buffer for 5 min at RT.
- Rinse twice in tap water for 5 min each at RT.
- Apply the permanent mounting media glycergel.
-
Observe under the light microscope.? TROUBLESHOOTING
- Hybrid cytochemistry/immunofluorescence ● Timing 4–6 h
- Follow Steps 30A(i and ii), but omit Step 30A(iii) (blocking of endogenous hydrogen peroxidase).
- Continue with Steps 30A(iv–xvi).
-
Incubate with goat anti-mouse secondary antibody; we use 568 Alexa Fluor conjugated, diluted 1/200 in TBS for 60 min at RT.▲CRITICAL STEP Determination of optimal dilution and incubation time of the secondary antibody to avoid nonspecific staining is crucial. Beyond this step, avoid light exposure.
- Wash five times in TBS for 5 min at RT.
- Incubate with DAPI diluted 1/1,000 in TBS for 5 min at RT.
- Wash twice in TBS for 5 min at RT.
- Apply permanent mounting medium glycergel.
-
Observe under the fluorescence microscope.? TROUBLESHOOTING
- Co-staining (double) immnofluorescence and hybrid cytochemistry/immunofluorescence ● Timing 5–6 h
- Perform Steps 30A(i–xiv), but omit Step 30A(iii), then proceed to the following steps.
-
Incubate the coverslips with a mixture of the anti-biotin [Hyb-8] antibody and the antibody against the antigen of choice (anti-Ki67 rabbit polyclonal antibody (ab16667, Abcam, dilution 1/500)) in TBS for 60 min at 37 °C.▲CRITICAL STEP Omit the anti-biotin antibody as a negative control. If nonspecific background staining occurs, the following options are available: (i) increase antibody dilution, (ii) decrease duration of incubation and (iii) a blocking step of nonspecific binding can precede incubation with the primary antibody; for instance incubation with the Ultra Protein Block included in the Ultravision Quanto Detection System HRP DAB kit for 2–5 min at RT. Primary antibodies should be raised in different species.
- Wash three times in TBS for 5 min at RT.
-
Incubate cells with a mixture of two secondary antibodies (goat anti-mouse secondary antibody 568 Alexa Fluor conjugated and the other secondary antibody to detect the antigen of choice), diluted 1/200 in TBS, for 60 min at RT.▲CRITICAL STEP Use secondary antibodies conjugated with two different fluorochromes. Determine the optimal dilution and incubation time of the secondary antibody to avoid nonspecific staining. Additional blocking steps are optional. Beyond this step, avoid light exposure.
- Wash five times in TBS for 5 min at RT.
- Incubate with DAPI diluted 1/1,000 in TBS for 5 min at RT.
- Wash twice in TBS for 5 min at RT.
- Apply permanent mounting medium glycergel.
-
Observe under the fluorescence microscope.? TROUBLESHOOTING
RT-qPCR ● Timing 3 h
-
31
Remove medium from cells and wash twice each with 1 mL 1× PBS solution.
-
32
Add 500 μL trypsin per well to the cells and incubate at 37 °C for 5 min.
-
33
Resuspend the cells in 1 mL medium and place the mixture in a 1.5 mL eppendorf tube. Centrifuge at 300g for 5 min at RT.
-
34
Aspirate the supernatant and resuspend the pellet in lysis buffer used for RNA extraction.
-
35
Isolate RNA with a commercial kit. We use the ISOLATE II RNA Mini Kit (Bioline).
▲CRITICAL STEP Ensure that RNA is always kept on ice to prevent degradation and equipment is kept RNAse free by cleaning with RNase AWAY.
? TROUBLESHOOTING
-
36
Elute RNA in 20 μL of RNase-free water and measure RNA concentration on a Nanodrop spectrophotometer.
■PAUSE POINT RNA can be stored at −80 °C for up to 1 year.
-
37
Synthesize cDNA from equal amounts of RNA using a commercial kit. We use the High-capacity cDNA Reverse Transcription Kit from Thermo Fisher Scientific.
-
38
Add 80 μL of RNase-free water to cDNA samples to bring the final volume to 100 μL.
■PAUSE POINT cDNA can be stored at −20 °C for up to 1 year.
-
39
Measure mRNA expression of via RT-PCR. Primers and probes for p16INK4a, p21WAF1/Cip1, lamin B1 and selected genes from the core senescence signature, as well as β-actin and α-tubulin, are listed in Table 1.
▲CRITICAL STEP In line with MIQE guidelines, we recommend using at least two housekeeping genes for reference. The geometric mean of β-actin and α-tubulin can be used as a general combination, but the choice of the right reference will depend upon the experimental system and requires direct testing by the investigator. Examples of reference genes to be used for different types of senescent cells can be found in this study30.
-
40We use the Universal Probe Library (UPL) from Roche and SensiFAST Probe Lo-Rox master mix from Bioline for cDNA amplification. For each sample, add reagents into a white 384-well plate, as described below. All samples should be measured in duplicate.
Reagent Volume (μL) SensiFAST Probe Lo-Rox 5 Forward primer (100 μM) 0.05 Reverse primer (100 μM) 0.05 UPL probe 0.1 RNase free water 2.3 cDNA 2.5 Total 10 -
41
Seal the plate and ensure that the film evenly covers all the wells on the 384-well plate.
-
42
Spin the plate at 2,000g for 1 min at RT.
-
43Place the plate in a Lightcycler 480 Instrument II (Roche) and amplify cDNA using the following parameters:
Temperature (°C) Time (s) Cycles 95 420 1 95 5 40 60 30 40 37 60 1 ▲CRITICAL STEP For each gene, always include a sample containing Rnase-free water instead of cDNA to ensure amplified products are not primer dimers or from contamination.
? TROUBLESHOOTING
-
44
Calculate the ΔCt value relative to the reference genes and use the appropriate control to calculate the ΔΔCt value for the specific senescence-inducing treatment.
Collection of cell culture conditioned medium for proteomic analysis ● Timing 25 h
▲CRITICAL The composition of the SASP can vary dramatically depending on the length of time after senescence induction. Prior to starting the protocol, it is therefore important to decide the length of time after senescence induction for which the SASP should be collected and measured. For primary human lung fibroblasts (IMR90), we typically culture the cells for 10 d following γ-irradiation before starting the collection protocol below. Note: other cell types can be used, and other senescence inducers can similarly be applied (with potentially adjusted induction periods).
▲CRITICAL Prepare both senescent and non-senescent (control) secreted protein samples for each senescence condition tested in multiple biological replicates (at least three). SASP protein components are identified based on significantly changed abundance in the conditioned medium of senescent cells versus non-senescent control cells.
-
45
Establish cultures of senescent and non-senescent (control) cells (106 cells per replicate) in T75 flasks. For primary human lung fibroblasts, we typically continue to the next step 10 d following induction of senescence with γ-irradiation (10 Gy γ-ray).
-
46
Aspirate and discard conditioned medium from cells.
-
47
Wash cells twice with 5 mL warm PBS.
▲CRITICAL STEP Wash cells thoroughly but gently in order to remove residual proteins derived from serum and other media supplements.
-
48
Culture cells for 24 h in serum-free, phenol red-free medium containing no protein supplements.
▲CRITICAL STEP Culture medium used during this step must be free of serum and as many other protein components/contaminants as possible. Abundant exogenous protein contamination can severely limit the identification and quantification of true secreted proteins in the proteomic analyses (due to potential dynamic range problems and ionization suppression). If the culture medium must contain protein components, these proteins must be excluded from later proteomic data analysis.
▲CRITICAL STEP Phenol red in the medium at this step can interfere with quantification of protein content by BCA assay in subsequent steps.
-
49
Collect conditioned medium containing secreted proteins from each T75 flask.
-
50
Perform cell counts for every flask and record for later use.
▲CRITICAL STEP Recording cell counts for every flask is essential for accurate downstream protein quantification. During proteomic analysis, secreted protein levels are normalized to cell counts.
-
51
Concentrate and buffer-exchange conditioned medium samples by ultrafiltration with Amicon Ultra centrifugal filters with a 3 kDa molecular weight cutoff according to the manufacturer’s protocol. The exchange buffer should be calcium- and magnesium-free PBS. The final volume of each sample from one T75 flask, see Step 46) should be less than 0.5 mL. Measure protein concentration using a BCA assay following the manufacturer’s instructions. The expected final concentration is 200–700 μg/mL of protein.
■PAUSE POINT Samples can be stored long-term at −80 °C.
? TROUBLESHOOTING
-
52
These protein samples can now be used for downstream proteomic analysis of core SASP factors summarized in Table 2.
? TROUBLESHOOTING
Phase 1: validating senescence in tissue samples
▲CRITICAL For in vivo samples (Steps 53–115), senescence validation entails double-staining for SA-β-Gal and Ki-67 in sequential sections, co-staining of LF and Ki-67 and sole staining of p21WAF1/Cip1, RT-PCR analyses for p16INK4a and p21WAF1/Cip1 mRNAs, as well as measurements of core SASP secreted proteins in human plasma.
SA-β-Gal staining ● Timing 13–17 h
▲CRITICAL The fresh tissues are immediately embedded in OCT and flash frozen in liquid nitrogen, after which they can be stored at −80 °C for up to 1 year.
▲CRITICAL The staining should be done immediately after sectioning; within 1 h is recommended.
▲CRITICAL The sectioned slides should also be kept at −20 °C until all sectioning is done.
-
53
Wash the slides in the glass coplin staining jar with prechilled PBS on ice (2 × 5 min).
? TROUBLESHOOTING
-
54
Fixation and staining conditions might vary, depending on the tissue. For example, for mouse kidney, fix with 2% (wt/vol) formaldehyde + 0.2% (wt/vol) glutaraldehyde in PBS for ~10 min on ice.
-
55
Wash the slides briefly with pre-chilled milliQ water.
-
56Prepare SA-β-Gal staining solution fresh, according to the table below and the total number of samples.
Component Volume per sample Final Concentration X-gal in dimethylformamide (20 mg/mL) 1 mL 1 mg/mL Citric acid/sodium phosphate buffer, pH = 6.0 (0.2 M) 4 mL 40 mM Potassium ferrocyanide (100 mM) 1 mL 5 mM Potassium ferricyanide (100 mM) 1 mL 5 mM Sodium chloride (5 M) 0.6 mL 150 mM Magnesium chloride (1 M) 40 μL 2 mM Water 12.4 mL Total 20 mL -
57
Pre-warm the staining solution to 37 °C and add it to the staining jar.
-
58
Incubate in a 37 °C non-CO2 incubator overnight.
▲CRITICAL STEP Incubation time needs to be optimized so that staining is still negative in the control group, but positive in treated groups.
? TROUBLESHOOTING
-
59
Check the results under a normal light microscope. Positive cells present a blue perinuclear staining. Compare results of tissues versus their appropriate control for the treatment used.
-
60
Upon successful staining, wash twice with 500 μL PBS.
-
61
Mount the slides with coverslips and mounting media and image using a bright field microscope.
■PAUSE POINT Alternatively, the samples can be stored at 4 °C for 1 month.
Ki-67 immunofluorescence ● Timing 24 h
▲CRITICAL The staining should be done using slides adjacent to the slides used for SA-β-galactosidase staining.
-
62
Fix the samples with pre-chilled fixative (methanol or acetone) on ice for 10 min.
-
63
Wash the slides in the glass staining jar with pre-chilled PBS on ice (2 × 5 min)
-
64
Draw a circle around the samples with a hydrophobic barrier pen.
-
65
Wash the slides twice with 1% (wt/vol) normal goat serum in PBS-T (PBS with 0.4% (vol/vol) Triton X-100) for 10 min each.
▲CRITICAL STEP The species of the animal serum should be the same as the species of the secondary antibody.
-
66
If endogenous peroxidase activity is suspected, quench the samples with 3% (vol/vol) H2O2 in PBS for 15 min.
-
67
Block nonspecific binding by incubating the samples with 10% (wt/vol) normal goat serum in PBS-T for 30 min at RT.
-
68
Add primary antibody diluted in 5% (wt/vol) normal goat serum in PBS-T and incubate the samples overnight at 4 °C in a humidified chamber.
▲CRITICAL STEP Use the recommended dilution of the antibody specified on the datasheet. If not specified, the typical starting dilution can be 2–5 μg/mL.
-
69
Wash the slides twice with 1% (wt/vol) normal goat serum in PBS-T for 10 min each.
-
70
Add a secondary antibody diluted in 5% (wt/vol) normal goat serum in PBS-T and incubate at RT for 1–2 h.
-
71
Wash the slides twice with 1% (wt/vol) normal goat serum in PBS-T for 10 min each.
-
72
Incubate the samples with a DNA binding dye such as DAPI.
-
73
After dye incubation, wash the slides once for 5 min with PBS.
-
74
Mount the slides with anti-fade mounting media and imaged using a fluorescence microscope.
■PAUSE POINT The samples can be stored at 4 °C for 2 weeks.
LF staining
-
75
Preparation of the material: fix tissues for 12–24 h (depending on the size) at RT. Among fixatives we prefer: (i) 1–5% (wt/vol) paraformaldehyde/PBS solution, pH = 7.4 with or without 0.2% (wt/vol) glutaraldehyde or (ii) 10% (wt/vol) buffered formalin solution, depending on the tissue.
▲CRITICAL STEP Avoid overfixation (limit 24 h), which can influence the staining results.
-
76
Embed the tissue in paraffin and cut sections (4–5 μm thick) using a microtome.
-
77Deparaffinize paraffin-embedded sections as follows:
- Incubate in an incubator at 60 °C for 30 min.
- Rinse in xylene for 15 min at RT.
-
78Rehydrate the sections gradually by immersing sequentially in:
- 100% ethanol for 15 min at RT
- 96% (vol/vol) ethanol for 10 min at RT
- 80% (vol/vol) ethanol for 5 min at RT
- 70% (vol/vol) ethanol for 3 min at RT
-
50% (vol/vol) ethanol for 3 min at RT▲CRITICAL STEP These rehydration steps take place in staining coplin jars. Beyond this step, OCT-embedded cryosections with or without fixation can also be used.
-
79
Block endogenous hydrogen peroxidase. We use the UltraVision Hydrogen Peroxide Block included in the Ultravision Quanto Detection System HRP DAB kit in the dark for 10 min at RT.
▲CRITICAL STEP Incubations should be performed in a chamber to avoid drying of the tissue.
-
80
Permeabilize with 0.5% (vol/vol) Triton X-100 for 3–5 min at RT.
▲CRITICAL STEP An antigen retrieval step (optional) using citrate buffer pH = 6.0 for 10 min in a microwave can improve the overall quality of the staining, depending on the tissue.
-
81
Wash twice with TBS for 30 s and once for 5 min at RT.
-
82Carry out LF staining as directed in Step 30A (screening for sensescence using a light microscope) or B (to verify the expression of a marker/factor within senescent cells, using a light microscope).
- Hybrid histochemistry/immunohistochemistry ● Timing 8–10 h
- Rinse with 50% (vol/vol) ethanol for 5 min at RT.
- Rinse with 70% (vol/vol) ethanol for 5 min at RT.
- Prepare the GL13 staining solution as described in Step 30A(vii).
-
Incubate with the GL13 solution at RT. A drop of freshly prepared reagent is placed on the section using a syringe that carries a 13 mm (membrane 0.22 μm) filter covered with a coverslip.▲CRITICAL STEP Covering with the coverslip is crucial to avoid evaporation of ethanol and should be performed in a chamber.
-
Monitor the staining reaction under a light microscope until detection of the signal (variably sized blue-black or brown cytoplasmic granules), which should take ~5–10 min, depending on the tissue.▲CRITICAL STEP Upon weak signal, repetition of the staining procedure with the GL13 solution can result in enhancement of the reaction especially when overfixation of the tissue has taken place.
- Wash twice with 50% (vol/vol) ethanol for 5 min at RT.
-
Wash twice more with fresh 50% (vol/vol) ethanol for 5 min at RT.▲CRITICAL STEP The additional washes are required for removal of nonspecific staining.
- Incubate with 0.5% (vol/vol) Triton X/TBS for 3 min at RT.
- Wash in TBS for 5 min at RT.
- Incubate the slides with anti-biotin antibody. We use the [Hyb-8] ab201341 primary antibody from Abcam, diluted 1/300 in TBS, for 60 min at 37 °C or overnight at 4 °C.
-
Wash three times in TBS for 5 min at RT.▲CRITICAL STEP The following steps describe our preferred method of signal development; other signal detection systems (kits) that are available in the market can be used instead of Steps 82A(xii–xvi).
- Signal development: first step: for primary antibody amplification use Quanto included in the Ultravision Quanto Detection System HRP DAB kit for 10 min at RT.
- Wash three times in TBS for 5 min at RT.
- Signal development: second step: incubate with the HRP Polymer Quanto included in the Ultravision Quanto Detection System HRP DAB kit for 10 min at RT.
- Wash twice in TBS for 30 s and once for 5 min at RT.
- Signal development: third step: apply DAB Plus Chromogen diluted 1/300 in DAB Plus Substrate (Ultravision Quanto Detection System HRP DAB kit).
- Monitor the staining reaction under a light microscope until brown (~30–60 s) at RT.
- Wash the slides in tap water for 5 min at RT.
- Counterstain with hematoxylin diluted 1/4 in deionized water.
- Wash in tap water for 5 min at RT.
- Apply permanent mounting media Glycergel.
-
Observe under the light microscope.? TROUBLESHOOTING
- Co-staining (double): immunohistochemistry—hybrid histochemistry/immunohistochemistry ● Timing 26–28 h
- Incubate with the antibody of choice [anti-p21WAF1/Cip1 mouse (F-5)(sc-6246) Santa-Cruz, dilution: 1/200; anti-p16INK4a mouse [(sc-74401) Santa-Cruz, dilution: 1/100; anti-Ki67 rabbit polyclonal antibody (ab16667, Abcam, dilution 1/500)] overnight at 4 °C.
- Wash twice in TBS for 5 min at RT. Then incubate with the primary antibody amplifier Quanto included in the Ultravision Quanto Detection System HRP DAB kit, for 10 min at RT.
- Wash three times in TBS for 5 min at RT and incubate with the HRP Polymer Quanto included in the Ultravision Quanto Detection System HRP DAB kit, for 10 min at RT.
- Following three washes in TBS for a total of 6 min at RT, apply DAB Plus Chromogen diluted 1/300 in DAB Plus Substrate (Ultravision Quanto Detection System HRP DAB kit).
- Subsequently, monitor the staining reaction under the light microscope until detection of the dark brown signal (average time 30–60 s) at RT. Finally, wash in tap water for 5 min at RT.
- Rinse sections once in 50% (vol/vol) ethanol for 5 min at RT.
- Rinse once in 70% (vol/vol) ethanol for 5 min at RT.
- Incubate with GL13 as specified in Step 30A(viii) at RT.
- Monitor the staining reaction under the light microscope.
- Rinse three times in 50% (vol/vol) ethanol.
- Repeat washing three times in fresh 50% (vol/vol) ethanol.
- Rinse twice in TBS for 30 s and once for 5 min at RT.
- Incubate with the anti-biotin antibody as described in Step 30A(xv).
- Rinse three times in TBS for 5 min at RT.
-
Incubate with the secondary goat anti-mouse antibody AP conjugated diluted 1/800 in TBS, for 60 min at RT.▲CRITICAL STEP It is crucial to determine the optimal dilution and incubation time of the secondary antibody to avoid nonspecific staining.
- Rinse three times in TBS for 5 min at RT.
- Rinse twice in TBS for 30 s and once for 5 min at RT.
- Apply the NBT/BCIP substrate (dilution of 1 tablet in 10 mL ddH2O (Roche kit, cat no: REF 001,11,471,697)) with the addition of 20 μL of 100 mM Levamisole. Monitor the staining reaction under the light microscope until detection of the dark blue/purple signal (~5–7 min) at RT.
- Rinse twice in KTBT buffer for 5 min at RT.
- Rinse in tap water for 5 min at RT.
- Rinse in tap water for 5 min at RT.
- Apply the permanent mounting medium glycergel.
-
Observe under the light microscope.? TROUBLESHOOTING
p21WAF1/Cip1 immunohistochemistry ● Timing 28 h
-
83
Dry the slides (not previously stained) in a 55° C incubator for 4–6 h.
-
84Deparaffinize and rehydrate the slides as follows:
- Xylene for 10 min (×2)
- 100% ethanol for 5 min
- 90% (vol/vol) ethanol for 5 min
- 70% (vol/vol) ethanol for 5 min
- PBS for 10 min.
-
85
Incubate the slides with 10 mM citrate buffer (pH = 6.0) in the microwave (5 min at high energy followed by 10 min at low energy).
? TROUBLESHOOTING
-
86
Cool down the slides for 20 min at 4 °C.
-
87
Transfer the slides to a coplin staining jar containing PBS.
-
88
Rinse the slides with PBS for 5 min and repeat three times.
-
89
Draw a circle around the samples with a hydrophobic barrier pen.
-
90
Incubate the slides at RT in freshly prepared 3% (vol/vol) H2O2 in methanol for 10 min to block endogenous peroxidase activity.
-
91
Rinse slides with PBS/Tween-20 for 5 min and repeat three more times.
-
92
Incubate the samples with 10% (wt/vol) normal goat serum in PBS for 10 min.
-
93
Incubate the samples with the primary antibody rabbit anti p21WAF1/Cip1 (1/100 in PBS with 5% (wt/vol) normal goat serum in PBS) in a humid chamber overnight at 4 °C.
-
94
Rinse the slides with PBS/Tween-20 for 5 min and repeat 3 more times.
-
95
Incubate the samples with the secondary antibody goat anti rabbit (1/200 in PBS with 5% (wt/vol) normal goat serum in PBS) in a humid chamber for 30 min at RT.
-
96
Prepare the ABC/PO-complex solution freshly 60 min before use.
-
97
Rinse the slides with PBS/Tween-20 for 5 min and repeat three more times.
-
98
Incubate the samples with ABC/PO-complex 1/25 in PBS for 30 min.
-
99
Rinse the slides with PBS for 5 min and repeat three more times.
-
100
Incubate the samples with freshly prepared DAB substrate for 10 min.
-
101
Rinse the slides after incubation with the DAB solution with distilled water once in the flow cabinet.
-
102
Wash the slides in running distilled water for 5 min.
-
103
Counterstain the samples with hematoxylin for 30 s.
-
104
Wash the slides in running distilled water for 10 min.
-
105Dehydrate the slides as follows:
- 70% (vol/vol) ethanol for 5 min
- 90% (vol/vol) ethanol for 5 min
- 100% ethanol for 5 min
- Xylene for 10 min (×2)
-
106
Mount the slides with mounting medium and image with a light microscope.
■PAUSE POINT Mounted slides can be stored at RT for at least 1 month.
RT-qPCR ● Timing 2 h
-
107
Clean a mortar and pestle with RNAse decontamination solution such as RNase AWAY.
-
108
Remove the tissue from the −80° C freezer and immediately place it on dry ice.
▲CRITICAL STEP Ensure the tissue is kept on dry ice and is not allowed to thaw, to prevent endogenous RNAse activation.
-
109
Fill the mortar with liquid nitrogen and place the tissue in the liquid nitrogen.
! CAUTION Protective clothing, gloves and goggles should be worn to prevent liquid nitrogen burns.
-
110
Grind the tissue using the pestle until a fine powder is obtained.
▲CRITICAL STEP Ensure the liquid nitrogen does not evaporate while the tissue is being ground to prevent the sample from thawing. Add more liquid nitrogen if needed and continue grinding.
-
111
Once a fine powder is obtsained, transfer the powder to a new liquid nitrogen-cooled tube and keep on dry ice. Re-clean the mortar and pestle with RNase AWAY for each sample.
■PAUSE POINT Ground tissue powder can be stored at −80 °C for up to 1 year.
-
112
Transfer up to 10 mg of ground tissue into a 1.5 mL Eppendorf tube and keep on dry ice.
-
113
Resuspend ground tissue in 150 μL lysis buffer used for RNA extraction.
-
114
Carry out RT-PCR against p16INK4a and p21WAF1/Cip1 as described in Steps 31–44.
Measurement of core SASP proteins in plasma ● Timing 48–72 h
-
115Using comprehensive and unbiased mass spectrometry analysis, we have identified core SASP proteins that are commonly secreted by senescent cells38, and a selection of these proteins (Fig. 2) should be used to validate the presence of senescence. These senescence protein markers can be probed in collected plasma using the following:
- Mass spectrometric targeted assays or discovery proteomics
- Immunoassays (ELISA, immunohistochemistry, western blot, antibody arrays)
- Aptamer-based assays
Phase 2: specifying the pro-inflammatory phenotype of senescent cells
▲CRITICAL Phase 2 of our protocol is to specify the pro-inflammatory phenotype of senescent cells. This step consists of the evaluation of the cellular mRNA and extracellular protein levels of six pro-inflammatory SASP factors. These SASP factors are part of the IL-1-dependent inflammatory arm57, and are normally expressed by DNA damage-induced senescent cells. Cells without a persistent DDR58, such as cells induced to senescence via mitochondrial dysfunction or enforced upregulation of the CDK4/6 inhibitors p16 and p2139,58, do not express these pro-inflammatory SASP factors. These factors are commonly described as the mediator of the pro-disease functions of senescent cells in vivo59.
Pro-inflammatory SASP transcript measurements ● Timing 2 h
-
116
Using cDNA generated in Step 37 or 114, measure mRNA levels of SASP genes by RT-PCR as described in Steps 38–44. Table 3 lists the primer and probe sequences.
Table 3 ∣.
Primer and probe sequences of SASP genes
| Gene | Primer sequence (5′–3′) | Probe sequence (5′–3′) |
|---|---|---|
| IL-1A | F: GGTTGAGTTTAAGCCAATCCA R: TGCTGACCTAGGCTTGATGA |
Sense: CAGAGGAA Antisense: TTCCTCTG |
| IL-6 | F: CAGGAGCCCAGCTATGAACT R: GAAGGCAGCAGGCAACAC |
Sense: CTGGGGCT Antisense: AGCCCCAG |
| IL-8 | F: GAGCACTCCATAAGGCACAAA R: ATGGTTCCTTCCGGTGGT |
Sense: GCCAGGAA Antisense: TTCCTGGC |
| IL-1B | F: TACCTGTCCTGCGTGTTGAA R: TCTTTGGGTAATTTTTGGGATCT |
Sense: AGCTGGAG Antisense: CTCCAGCT |
| CXCL1 | F: CATCGAAAAGATGCTGAACAGT R: ATAAGGGCAGGGCCTCCT |
Sense: GGTGGCTG Antisense: CAGCCACC |
| CXCL2 | F: CCCATGGTTAAGAAAATCATCG R: CTTCAGGAACAGCCACCAAT |
Sense: CTTCCTCC Antisense: GGAGGAAG |
Pro-inflammatory SASP protein measurements ● Timing 48–72 h
-
117Measure pro-inflammatory SASP protein levels (IL1A/IL6/IL8/IL1B/CXCL1/CXCL2) following either Option A (cultured cells) or B (plasma).
- Cell culture-conditioned medium
- Collect cell culture-conditioned medium and measure SASP protein levels as described in Steps 45–52.
- Plasma
- Measure pro-inflammatory SASP protein levels in plasma as described in Step 116.
Troubleshooting
Troubleshooting advice can be found in Table 4.
Table 4 ∣.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 5 | Low percentage of EdU+ cells in control group | Cells have low proliferative capacity | Prolong the incubation time with EdU |
| 9 | False negative staining | Insufficient X-gal | Ensure you are using the right concentration of X-gal |
| Insufficient time of incubation | Titrate the time for X-gal solution incubation | ||
| pH of X-gal solution is >6.0 | Add appropriate amounts of citric acid or sodium phosphate buffer to adjust the final pH | ||
| False-positive staining | pH of X-gal solution is <6.0 | Add appropriate amounts of citric acid or sodium phosphate buffer to adjust the final pH | |
| Improper confluence of cells | Make sure cells are 50-70% confluent | ||
| 30A(xxvii),30B(xxvi),30C(viii),30D(ix), 82A(xxii),82B(xxiii) | Absent or weak signal | Low duration of GL13 reagent incubation | Increase incubation time of GL13 reagent in a step-wise manner |
| Incubate GL13 reagent at 37 °C. Increasing the temperature from RT to 37 °C might result in enhancement of the GL13-LF interaction and eventually to a stronger signal | |||
| Insufficient penetration of GL13 reagent into the cells | Use a permeabilization step before the addition of the GL13 reagent. Incubate with 0.5% (vol/vol) Triton X-100/TBS for 5 min at 4 °C. Then continue with next steps | ||
| Reduced anti-biotin antibody reaction | Increase incubation time with anti-biotin antibody | ||
| Increase anti-biotin antibody concentration. (Decrease the dilution of the anti-biotin solution) | |||
| High nonspecific background staining | Insufficient washing | Thoroughly wipe away excess GL13 reagent immediately after the incubation step is complete | |
| Increase washing time in ethanol and TBS solutions. Add several wash steps either to remove excess of GL13 (ethanol solution) or to avoid non specific reaction of the anti-biotin antibody and the detection system (TBS solution) | |||
| Duration of GL13 reagent incubation is too long | Reduce GL13 reagent incubation time in a stepwise manner | ||
| High concentration of the anti-biotin antibody | Decrease the anti-biotin antibody concentration. (Increase the dilution of the anti-biotin solution) | ||
| Decrease the anti-biotin antibody incubation time | |||
| High concentration of the secondary antibody | Decrease the concentration of the secondary antibody. (Increase the dilution of the secondary antibody) | ||
| Increased DAB exposure | Reduce DAB exposure time | ||
| 35,43 | No amplification detected | Poor RNA quality or improper qPCR settings | Make sure that the RNA is not degraded and the protocol for cDNA amplication is correct |
| 51 | BCA shows high background and poor standard curve | Phenol red still present in the cell culture medium | Repeat collection of conditioned medium in phenol red-free medium, or thoroughly buffer exchange the sample |
| 52 | Albumin is one of the major proteins detected by mass spectrometry | Serum proteins (from FBS) are still present in the cell culture medium | More thoroughly wash cells with PBS prior to moving cells into serum-free medium (for collection of secreted proteins) |
| Many proteins are more abundant in control samples | Non-senescent (control) cell counts are significantly greater than senescent cell counts | Normalize the sample loading or protein quantification to cell counts | |
| 53 | False-negative staining | The enzyme activity of SA-β-Gal is lost | Make sure that the tissues are fresh and stain them immediately done after sectioning |
| 58 | False-positive staining | Overstaining | Make sure that the staining time is not too long, otherwise false positive signals can be detected in the control |
| 85 | False-negative staining | Antigen retrieval is not done properly | Optimize the antigen retrieval |
Timing
Phase 1: validating senescence in in vitro samples
Steps 6–10, SA-β-Gal staining: ~40 min–1 h
Steps 14–26, EdU staining: ~2 h
Steps 27–30, LF staining:~ 7–15 d, depending on the type of senescence (see Box 1)
Steps 30A(vii and viii), 30B(xii), 30C(ii), 30D(i), Preparation and application of the GL13 staining solution: ~120 min
Steps 30A(xv), 30B(xvii), 30C(ii), 30D(ii), Incubation with the primary anti-biotin antibody: ~60 min
Steps 30A(xvii–xxi), 30B(xii–xxiv), Signal development: ~30 min
Steps 30B(xix), 30C(iii), 30D(iv), Incubation with the secondary antibody: ~60 min
Steps 31–44, RT-qPCR: 3 h
Steps 45–52, Collection of cell culture-conditioned medium and proteomic analysis
Quantification by antibody-based approaches such as ELISA or western blot: ~48 h
Quantification by mass spectrometry–based approaches: ~72 h
Phase 1: validating senescence in tissue samples
Steps 53–61, SA-β-Gal staining: 13–17 h
Steps 62–74, Ki-67 immunofluorescence: 24 h
Steps 75–82, LF staining: 22–52 h
Steps 75–81, Material preparation (tissue fixation, embedding, deparaffinization, rehydration, blocking of endogenous hydrogen peroxidase and permeabilization): 14–26 h
Step 82A(i–xxii), Hybrid histochemistry/immunohistochemistry: 8–10 h
Step 82B(i–xxiii), Immunohistochemistry—hybrid histochemistry/immunohistochemistry: 26– 28 h
Steps 83–106, p21WAF1/Cip1 immunohistochemistry: 28 h
Steps 107–114, RT-qPCR: 2 h
Step 115, Measurement of core SASP proteins in plasma
Quantification by antibody-based approaches such as ELISA or western blot: ~48 h
Quantification by mass spectrometry–based approaches: ~72 h
Phase 2: specifying the pro-inflammatory SASP phenotype
Step 116, Measure mRNA levels of pro-inflammatory SASP genes by RT-PCR: ~2 h
Step 117, Pro-inflammatory SASP protein measurements
Measure pro-inflammatory SASP protein levels in conditioned medium: ~48–72 h
Measure pro-inflammatory SASP protein levels in plasma: ~48–72 h
Anticipated results
We combine the staining of EdU and SA-β-Gal (Steps 1–26) to initially identify senescent cells. The percentage of EdU−SA-β-Gal+ cells varies based on cell type and senescence inducers. In BJ foreskin fibroblasts treated with doxorubicin, we can observe >90% EdU−SA-β-Gal+ (senescent) cells. In vehicle-treated or untreated fibroblasts, we can observe >90% EdU+SA-β-Gal−cells. However, in vehicle-treated or untreated melanocytes, we can only detect 30% EdU+SA-β-Gal−cells since the melanocytes proliferate more slowly than fibroblasts. Moreover, in the kidneys of doxorubicin-treated mice, we can observe that ~15% of the cells are positive for SA-β-Gal. In the kidneys of cisplatin-treated mice, a maximum of 4% of the cells are positive for p21 staining.
We also stain cells and tissues with GL13 alone or concurrently with other indirect markers (p16INK4A, p21WAF1/Cip1) or Ki67 to detect senescent cells in a variety of senescence settings comprising cellular systems, animal models and clinical samples. In senescent cells in culture, 50–65% of the population is positive for GL13 staining, with 35–55% simultaneously positive for nuclear p16INK4A or p21WAF1/Cip1 and GL13 (Extended Data Fig. 1). A mutually exclusive staining pattern between nuclear Ki67 positivity and GL13 staining is also evident in these cells (Extended Data Fig. 1). Lower percentages of senescent cells are expected in tissues from animal models (range of senescent cells: 8–26% of total cells – see figure legend of Extended Data Fig. 2 for details) and human primary (non-treated) classical Hodgkin lymphomas (range of senescent cells: 1–34% of total cells) (Extended Data Fig. 3).
With a panel of markers (GDF15, MMP1, STC1, TIMP1, TIMP2), we measure the SASP in conditioned medium from human cells or in human plasma samples. We anticipate the levels of SASP proteins to be higher in conditions or individuals with higher levels of senescent cells. For example, SASP proteins will be elevated in older individuals60 compared with younger individuals.
Extended Data
Extended Data Fig. 1∣. Sensitive detection of senescence in a variety of cell types using GL13 in cellular systems.
Positive GL13 staining is depicted in normal (a and b) and cancerous (f and l) cells of epithelial origin, normal (c, d, j, o), premalignant (e) and malignant mesenchymal (g, h, m, n) cells as well as in differentiating megakaryocytes (k)61 and peripheral blood mononuclear cells (PBMCs) from patients suffering from a chronic aged-related disease (p). Chromogenic assays: Diaminobenzidine (DAB)-brown cytoplasmic signal (a, b, ci-ii, a, d-gi, h, l, mi, n-p) and Alkaline phosphatase (BCIP/NBT)-blue purple cytoplasmic signal (white arrowheads in ciib, gii, k); Fluorescent assay: granular red cytoplasmic signal (j, mii -white arrowheads). Double staining experiments showing nuclear p16INK4A or p21WAF1/Cip1 expression (yellow arrowheads) in senescent cells that are concurrently positive with GL13 (white arrowheads) (ciib, gii and miii). Black arrowheads depict double negative cells. giii: Mutually exclusive staining pattern between nuclear Ki67 positivity (yellow arrowhead) and GL13 staining (white arrowhead). Images adopted from: a, c, g, h, m and n28; b62; j63. Counterstain: Hematoxylin (chromogenic assay) and DAPI (fluorescent assay). Scale bars: 5 μm (l), 10μm (a-c, f-k-m-p,), 15μm (e) and 20 μm (d).
Extended Data Fig. 2 ∣. Sensitive detection of senescence using GL13 in a variety of tissues obtained from different animal models.
Identification of senescent cells in the following animal models with established senescence: a. K-rasV12 induced lung adenoma mouse model (images ai and aii adopted from28; for material depicted in panel b see64), c. H-Ras G12V knock-in mouse model (65,66), d. Aged Rat (age: 22 months), e. Bleomycin induced lung fibrosis mouse model (images adopted from28), f. Palbociclib induced tumor (melanoma) mouse model (image adopted from28), g. Floxed Ercc1 allele in Lys2-Ercc1F/– mouse model (image adopted from67) h. Hela tumor xenografts subjected to G-quadruplex ligand 20A treatment (image adopted from68). Red arrows denote GL13 positive cells. Double staining assays revealing nuclear p16INK4A expression (yellow arrowheads) in concurrently positive with GL13 senescent cells (white arrowheads) (aii, adopted from28), as well as inverse staining between Ki67 (yellow arrowhead) and GL13 (white arrowhead) eii, adopted from28). Chromogenic assays: DAB-brown cytoplasmic signal (ai, c, d, ei, f-h) and Alkaline phosphatase (BCIP/NBT)-blue purple cytoplasmic signal (aii, b and eii). Counterstain (when applied): Hematoxylin for DAB and Nuclear Fast Red for BCIP/NBT. Scale bars: 25 μm (g), 50μm (aii, b-d, eii), 100μm (ai, ei, f,) and 200 μm (h).
Extended Data Fig. 3 ∣. Sensitive detection of senescence using GL13 in a variety of tissues from human clinical samples.
Senescent cells of different cell origin (mesenchymal: a-d, p, r, s; megakaryocytes: e; lymphoid/histiocytic: f-h; epithelial: j-n; neuronal:o, nevoid:q) identified via GL13 staining in various clinical settings (red arrows and double black arrowheads in image h). Chromogenic assays: DAB-brown cytoplasmic signal (a, bi, c-g, j, k, l, n-s) and Alkaline phosphatase (BCIP/NBT)-blue purple cytoplasmic signal (bii, fii-iv, h, m); Double staining experiments showing nuclear p16INK4A or p21WAF1/Cip1 expression (yellow arrowheads) in senescent cells that are simultaneously positive with GL13 (red arrows) (bi, fii-iii, mii). Mutually exclusive staining pattern between nuclear Ki67 positivity (yellow arrowhead) and GL13 reactivity (red arrows) is evident (fiv). Green arrowhead depicts a double negative cell (fiv). Images adopted from: b, q28; f, g, j69; h70; k71; m72; o73; s74. Counterstain: Hematoxylin for DAB and Nuclear Fast Red for BCIP/NBT. Scale bars: 25 μm (e, p), 50μm (bii, d, f, h, m, n, q, r), 100μm (a, bi, g, j, k, o) and 200 μm (c, l, s).
Acknowledgements
We thank M. Koster for continuous help with immunostaining, D. Kletsas for providing material and all the members of the Demaria lab for fruitful discussions. This work was supported by Dutch Cancer Foundation (KWF) grant no. 10989 (MD); Horizon 2020 Marie Sklodowska-Curie individual fellowship (JK); Horizon 2020 Marie Sklodowska-Curie training program no. 722729 (SYNTRAIN; KE and VG); the Welfare Foundation for Social & Cultural Sciences, Athens, Greece (KIKPE; KE and VG); H. Pappas donation; Hellenic Foundation for Research and Innovation (HFRI) grants no. 775 and 3782 (KE and VG); National Public Investment Program of the Ministry of Development and Investment of Greece/ General Secretariat for Research and Technology, in the framework of the Flagship Initiative to address SARS-CoV-2 (GSRT No 2020ΣE01300001) and NKUA-SARG grant 70/3/8916 (KE and VG); National Institutes of Health under award numbers U01 AG060906 and U01 AG060906-02S1 (BS), P01 AG017242 and R01 AG051729 (JC) and K99 AG065484 (NB).
Footnotes
Competing interests
M.D. is co-founder and advisor of Cleara Biotech. J.C. is co-founder and shareholder of Unity Biotechnology.
Extended data is available for this paper at https://doi.org/10.1038/s41596-021-00505-5.
Related links
Key references using this protocol
Evangelou, K. et al. Aging Cell 16, 192–197 (2017): https://doi.org/10.1111/acel.12545
Hernandez-Segura, A. et al. Curr. Biol. 27, 2652–2660 (2017): https://doi.org/10.1016/j.cub.2017.07.033
Data availability
The data that support the anticipated results are available from the corresponding author upon reasonable request. The source of figures and corresponding data is mentioned in each legend.
References
- 1.Calcinotto A et al. Cellular senescence: aging, cancer, and injury. Physiol. Rev 99, 1047–1078 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Gorgoulis V et al. Cellular senescence: defining a path forward. Cell v179, 813–827 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Shay JW Role of telomeres and telomerase in aging and cancer. Cancer Discov. 6, 584–593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serrano M, Lin AW, McCurrach ME, Beach D & Lowe SW Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16(INK4a). Cell 88, 593–602 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Hernandez-Segura A et al. Unmasking transcriptional heterogeneity in senescent cells. Curr. Biol 27, 2652–2660.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coppé J-P et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6, e301 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharpless NE et al. Loss of p16Ink4a with retention of p19 predisposes mice to tumorigenesis. Nature 413, 86–91 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Demaria M et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell 31, 722–733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muñoz-Espín D et al. Programmed cell senescence during mammalian embryonic development. Cell 155, 1104 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Storer M et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell 155, 1119 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Baker DJ et al. Naturally occurring p16 Ink4a-positive cells shorten healthy lifespan. Nature 530, 184–189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Childs BG et al. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science 354, 472–477 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bussian TJ et al. Clearance of senescent glial cells prevents tau-dependent pathology and cognitive decline. Nature 562, 578–582 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang P et al. Senolytic therapy alleviates Aβ-associated oligodendrocyte progenitor cell senescence and cognitive deficits in an Alzheimer’s disease model. Nat. Neurosci 22, 719–728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeon OH et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat. Med 23, 775–781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguayo-Mazzucato C et al. Acceleration of β Cell Aging Determines Diabetes and Senolysis Improves Disease Outcomes. Cell Metab. 30, 129–142.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson PJ et al. Targeted Elimination of Senescent Beta Cells Prevents Type 1 Diabetes. Cell Metab. 29, 1045–1060.e10 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Demaria M et al. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov 7, 165–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paez-Ribes M, González-Gualda E, Doherty GJ & Muñoz-Espín D Targeting senescent cells in translational medicine. EMBO Mol. Med 11, e10234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borghesan M, Hoogaars WMH, Varela-Eirin M, Talma N & Demaria M A senescence-centric view of aging: implications for longevity and disease. Trends Cell Biol. 30, 777–791 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Dimri GP et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall BM et al. p16(Ink4a) and senescence-associated β-galactosidase can be induced in macrophages as part of a reversible response to physiological stimuli. Aging 9, 1867–1884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang N & Hu M The limitations and validities of senescence associated-β-galactosidase activity as an aging marker for human foreskin fibroblast Hs68 cells. Exp. Gerontol 40, 813–819 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Hernandez-Segura A, Nehme J & Demaria M Hallmarks of cellular senescence. Trends Cell Biol. 28, 436–453 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Kang C & Elledge SJ How autophagy both activates and inhibits cellular senescence. Autophagy 12, 898–899 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terman A & Brunk UT Lipofuscin: mechanisms of formation and increase with age. APMIS 106, 265–276 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Georgakopoulou EA et al. Specific lipofuscin staining as a novel biomarker to detect replicative and stress-induced senescence. A method applicable in cryo-preserved and archival tissues. Aging 5, 37–50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evangelou K et al. Robust, universal biomarker assay to detect senescent cells in biological specimens. Aging Cell 16, 192–197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Zglinicki T, Wan T & Miwa S Senescence in post-mitotic cells: a driver of aging? Antioxid. Redox Signal. 10.1089/ars.2020.8048 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sapieha P & Mallette FA Cellular senescence in postmitotic cells: beyond growth arrest. Trends Cell Biol. 28, 595–607 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Brunk UT & Terman A Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Radic. Biol. Med 33, 611–619 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Serrano M, Hannon GJ & Beach D A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366, 704–707 (1993). [DOI] [PubMed] [Google Scholar]
- 33.Hara E et al. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol. Cell. Biol 16, 859–867 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcorta DA et al. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc. Natl Acad. Sci. USA 93, 13742–13747 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karimian A, Ahmadi Y & Yousefi B Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair 42, 63–71 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Michaloglou C et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436, 720–724 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Gray-Schopfer VC et al. Cellular senescence in naevi and immortalisation in melanoma: a role for p16? Br. J. Cancer 95, 496–505 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basisty N et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 18, e3000599 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiley CD et al. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 23, 303–314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biran A et al. Quantitative identification of senescent cells in aging and disease. Aging Cell 16, 661–671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noren Hooten N & Evans MK Techniques to induce and quantify cellular senescence. J. Vis. Exp 2017, 55533 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hernandez-Segura A, Brandenburg S & Demaria M Induction and validation of cellular senescence in primary human cells. J. Vis. Exp 2018, e57782 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Debacq-Chainiaux F, Erusalimsky JD, Campisi J & Toussaint O Protocols to detect senescence-associated beta-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc 4, 1798–1806 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Juers DH, Matthews BW & Huber RE LacZ β-galactosidase: Structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 21, 1792–1807 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng C et al. Evaluation of 5-ethynyl-2′-deoxyuridine staining as a sensitive and reliable method for studying cell proliferation in the adult nervous system. Brain Res. 1319, 21–32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scholzen T & Gerdes J The Ki-67 protein: from the known and the unknown. J. Cell. Physiol 182, 311–322 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Hernandez-Segura A, Rubingh R & Demaria M Identification of stable senescence-associated reference genes. Aging Cell 18, e12911 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bustin SA et al. The need for transparency and good practices in the qPCR literature. Nat. Methods 10, 1063–1067 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Bustin SA et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem 55, 611–622 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Basisty N, Kale A, Patel S, Campisi J & Schilling B The power of proteomics to monitor senescence-associated secretory phenotypes and beyond: toward clinical applications. Expert Rev. Proteomics 17, 297–308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schilling B, Gibson BW & Hunter CL Generation of high-quality SWATH® acquisition data for label-free quantitative proteomics studies using tripleTOF® mass spectrometers. in Proteomics 223–233 (Humana Press, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keshishian H et al. Quantitative, multiplexed workflow for deep analysis of human blood plasma and biomarker discovery by mass spectrometry. Nat. Protoc 12, 1683–1701 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohl TO & Ascoli CA Immunometric antibody sandwich enzyme-linked immunosorbent assay. Cold Spring Harb. Protoc 2017, 450–457 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Hofman FM & Taylor CR Immunohistochemistry. Curr. Protoc. Immunol 103, 21.4.1–21.4.26 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Mahmood T & Yang PC Western blot: technique, theory, and trouble shooting. N. Am. J. Med. Sci 4, 429–434 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gold L et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 5, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodier F et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol 11, 973–979 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coppé JP et al. Tumor suppressor and aging biomarker p16 INK4a induces cellular senescence without the associated inflammatory secretory phenotype. J. Biol. Chem 286, 36396–36403 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laberge RM et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat. Cell Biol 17, 1049–1061 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka T et al. Plasma proteomic signature of age in healthy humans. Aging Cell 17, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muñoz-Espín D & Serrano M Cellular senescence: From physiology to pathology. Nat. Rev. Mol. Cell Biol 15, 482–496 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Sasaki M, Sato Y & Nakanuma Y An impaired biliary bicarbonate umbrella may be involved in dysregulated autophagy in primary biliary cholangitis. Lab. Investig 98, 745–754 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Gnani D et al. An early-senescence state in aged mesenchymal stromal cells contributes to hematopoietic stem and progenitor cell clonogenic impairment through the activation of a pro-inflammatory program. Aging Cell 18, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vitos-Faleato J et al. Requirement for epithelial p38α in KRAS-driven lung tumor progression. Proc. Natl Acad. Sci. USA 117, 2588–2596 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schuhmacher AJ et al. A mouse model for Costello syndrome reveals an Ang II-mediated hypertensive condition. J. Clin. Invest 118, 2169–2179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen X et al. Endogenous expression of HrasG12V induces developmental defects and neoplasms with copy number imbalances of the oncogene. Proc. Natl Acad. Sci. USA 106, 7979–7984 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goulielmaki E et al. Tissue-infiltrating macrophages mediate an exosome-based metabolic reprogramming upon DNA damage. Nat. Commun 11, 42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beauvarlet J et al. Modulation of the ATM/autophagy pathway by a G-quadruplex ligand tips the balance between senescence and apoptosis in cancer cells. Nucleic Acids Res. 47, 2739–2756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Myrianthopoulos V et al. Senescence and senotherapeutics: a new field in cancer therapy. Pharmacol. Ther 193, 31–49 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Papalampros A et al. Unique spatial immune profiling in pancreatic ductal adenocarcinoma with enrichment of exhausted and senescent T cells and diffused CD47-SIRPα expression. Cancers 12, 1825 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Azazmeh N et al. Chronic expression of p16INK4a in the epidermis induces Wnt-mediated hyperplasia and promotes tumor initiation. Nat. Commun 11, 2711 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barbouti A et al. In situ evidence of cellular senescence in thymic epithelial cells (TECs) during human thymic involution. Mech. Ageing Dev 177, 88–90 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Kritsilis M et al. Ageing, cellular senescence and neurodegenerative disease. Int. J. Mol. Sci 19, 2937 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Giatromanolaki A, Kouroupi M, Balaska K & Koukourakis MI Immunohistochemical detection of senescence markers in human sarcomas. Pathol. Res. Pract 216, 152800 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the anticipated results are available from the corresponding author upon reasonable request. The source of figures and corresponding data is mentioned in each legend.