Abstract
The general transcription factor IIB (TFIIB) is required for transcription of class II genes by RNA polymerase II. Previous studies demonstrated that mutations in the Saccharomyces cerevisiae SUA7 gene, which encodes TFIIB, can alter transcription initiation patterns in vivo. To further delineate the functional domain and residues of TFIIB involved in transcription start site utilization, a genetic selection was used to isolate S. cerevisiae TFIIB mutants exhibiting downstream shifts in transcription initiation in vivo. Both dominant and recessive mutations conferring downstream shifts were identified at multiple positions within a highly conserved homology block in the N-terminal region of the protein. The TFIIB mutations conferred downstream shifts in transcription initiation at the ADH1 and CYC1 promoters, whereas no significant shifts were observed at the HIS3 promoter. Analysis of a series of ADH1-HIS3 hybrid promoters and variant ADH1 and HIS3 promoters containing insertions, deletions, or site-directed base substitutions revealed that the feature that renders a promoter sensitive to TFIIB mutations is the sequence in the immediate vicinity of the normal start sites. We discuss these results in light of possible models for the mechanism of start site utilization by S. cerevisiae RNA polymerase II and the role played by TFIIB.
Accurate and efficient transcription of eukaryotic protein-coding (class II) genes involves the concerted action of RNA polymerase II (RNAPII) and a host of accessory proteins. A subset of these proteins are known as the general transcription factors (GTFs) and include TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (reviewed in reference 34). The GTFs are being intensively studied with the objective of determining their respective functions during the different stages of RNAPII transcription, which include (i) formation of a preinitiation complex (PIC) on the promoter, (ii) melting of the promoter DNA, (iii) transcription initiation, (iv) clearance of RNAPII from the promoter, (v) elongation of the nascent transcript, and (vi) transcription termination.
Most promoters of class II genes contain both upstream regulatory elements and TATA elements. TATA elements, containing the consensus sequence TATAa/tAa/t, are located upstream of the mRNA start sites and are specific binding sites for the TATA-binding protein (TBP) subunit of TFIID (32, 36). For most class II promoters, formation of an active PIC is thought to occur by the initial binding of TFIID to the TATA element, in some cases accompanied by TFIIA. It is proposed that PIC formation then proceeds by either an ordered stepwise association of the remaining factors and RNAPII or by the direct recruitment of RNAPII holoenzyme (reviewed in reference 34). Upon PIC formation, the promoter DNA can be melted in an energy-dependent step, facilitating the initiation of mRNA synthesis and clearance of RNAPII from the promoter. In higher eukaryotes, transcription initiation usually occurs at a discrete start site located about 25 to 30 bp downstream of the TATA element. In contrast, transcription initiation in the yeast Saccharomyces cerevisiae frequently occurs at multiple sites within a window of 45 to 120 bp downstream of the TATA element (reviewed in references 17 and 45).
TFIIB plays an essential role in RNAPII transcription. The TFIIB polypeptide comprises a protease-sensitive N-terminal region that is highly conserved, followed by a protease-resistant C-terminal core domain that contains two imperfect direct repeats (2, 4, 31, 33). The N-terminal region contains a putative zinc-ribbon motif (W. Zhu, Q. Zeng, C. M. Colangelo, M. Lewis, M. F. Summers, and R. A. Scott, Letter, Nat. Struct. Biol. 3:122–124, 1996) and is required for interaction with TFIIF and RNAPII (3, 9, 12, 18, 35). The C-terminal core domain binds TBP (8, 18, 30) and the TBP-associated factor TAF40 (16) and interacts with DNA both immediately upstream and downstream of the TATA element (25, 26). In light of these multiple sets of interactions, TFIIB is often viewed as a bridging factor between promoter-bound TFIID and the remainder of the general transcription machinery. TFIIB may also play a role in the response to transcriptional activator proteins, as mutations in both the N-terminal and the C-terminal domains that reportedly impair activation have been identified (40, 42, 43, 47) and many transcriptional activator proteins directly bind TFIIB (10, 11, 13, 19, 28, 29, 41, 48).
In S. cerevisiae, TFIIB is encoded by the SUA7 gene. SUA7 was initially identified and characterized by Hampsey and coworkers in a suppressor analysis of respiration-deficient strains that contained an aberrant ATG translational initiation codon in the leader region of the CYC1 gene (cyc1-5000) (37). In those studies, two sua7 mutations that mapped to the N-terminal region of the protein and suppressed the respiration-deficient phenotype by conferring a downstream shift in transcription initiation at cyc1 were identified (38). Subsequent structure-function studies of yeast TFIIB have identified additional mutations in the N-terminal region of yeast TFIIB that alter start site utilization (3, 35).
To further investigate the domain of S. cerevisiae TFIIB involved in transcription start site utilization, we combined the genetic approach of Hampsey and coworkers with error-prone PCR and plasmid-shuffling to directly select for TFIIB mutants that confer downstream shifts in transcription initiation in vivo. The mutants isolated in this selection, combined with previously identified mutants altering start site utilization, define a highly conserved discrete domain within the N-terminal region of TFIIB involved in transcription start site recognition. To gain insight into the mechanism by which TFIIB affects start site utilization, a series of variant ADH1 and HIS3 promoters were constructed and analyzed in order to determine which feature of a promoter renders it sensitive to TFIIB mutations that confer downstream shifts in initiation. The results reported here demonstrate that the sensitivity of a promoter to TFIIB mutations is determined by neither the upstream regulatory elements, TATA elements, general promoter architecture, nor intrinsic strength of the start sites but rather is dictated by the identity of bases at or in the immediate vicinity of the normal start sites.
MATERIALS AND METHODS
S. cerevisiae strains and media.
All S. cerevisiae strains used in this study are derivatives of S288C. Plasmid-shuffle strain FP227 (MATα ura3-52 trp1Δ63 sua7Δ1 cyc1-5000 + [p316/yIIB(URA3)]) was constructed by two-step gene replacement of the CYC1 gene in strain FP153 (MATα ura3-52 trp1Δ63 sua7Δ1 + [p316/yIIB(URA3)]) using the cyc1-5000-integrating plasmid pSF13. For the analysis of the effects of TFIIB mutations on ADH1-HIS3 hybrid promoters and variant ADH1 and HIS3 promoters containing insertions, deletions, or site-directed base substitutions, the relevant ADH1 or HIS3 reporter plasmids (all containing URA3 as a selectable marker) were transformed into the following strains: FP251 (MATα ura3-52 trp1Δ63 his3Δ200 lys2 sua7Δ1 + [p314/yIIB(TRP1)]); FP252 (MATα ura3-52 trp1Δ63 his3Δ200 lys2 sua7Δ1 + [p314/V79L(TRP1)]); FP253 (MATα ura3-52 trp1Δ63 his3Δ200 lys2 sua7Δ1 + [p314/R64G(TRP1)]); and FP254 (MATα ura3-52 trp1Δ63 his3Δ200 lys2 sua7Δ1 + [p314/E62G(TRP1)]). Complete lineages of strains are available upon request.
Rich media (YPD plates and liquid) and 5-FOA (synthetic complete medium containing 5-fluoroorotic acid) plates were prepared as described previously (44). Casamino Acids medium (CAA) contained 0.6% Casamino Acids, 0.68% yeast nitrogen base without amino acids, 25 μg of adenine per ml, 25 μg of uracil per ml, 80 μg of tryptophan per ml, either 2% glucose or 2% lactate, and 2% agar for solid medium.
Plasmids.
Plasmid p314/yIIB-Mlu is a derivative of p314/yIIB, which contains the entire SUA7 promoter and coding region in the yeast-Escherichia coli shuttle vector pRS314 (CEN6 TRP1) and has been described previously (3). To generate p314/yIIB-Mlu, the megaprimer method of PCR site-directed mutagenesis was used to introduce a unique MluI restriction site into the coding region at the position corresponding to amino acids 94 and 95 with no resulting change in protein sequence (1). Integrating plasmid pSF13, containing the S. cerevisiae cyc1-5000 gene, was constructed by replacing the 385-bp XmaI-EcoRI CYC1 promoter fragment in plasmid pM142 with the corresponding fragment isolated from plasmid pM330 (5).
The construction of all ADH1 and HIS3 promoter derivatives was based on plasmids p316/A and p316/H. Plasmid p316/A contains the ADH1 promoter (fragment −500 to +12) fused to the HIS3 coding region (fragment +1 to +900) in vector pRS316 (CEN6 URA3). Plasmid p316/H contains the HIS3 gene (fragment −440 to +900) cloned into the pRS316 vector. The two AT-rich elements in the ADH1 promoter in p316/A (TATAAATA, positions −128 to −121, and TATTAAT, positions −110 to −104) were converted to XbaI sites by site-directed mutagenesis to generate plasmids p316/D1 (−128) and p316/D2 (−110). To generate ADH1 promoter derivatives with variable distance between the TATA element and the transcription start sites, MluI restriction sites were introduced at positions −52, −64, −80, or −92 of the ADH1 promoter. The digestion and subsequent cloning of the different ADH1 promoter fragments generated a battery of pRS316-based plasmids containing 12-, 28-, and 40-bp deletions (p316/A-12, p316/A-28, p316/A-40, respectively) or insertions (p316/A+12, p316/A+28, p316/A+40, respectively) in the ADH1 promoter. The construction of the ADH1-HIS3 promoter hybrids was based on the introduction of MluI restriction sites in both the ADH1 and HIS3 promoters 14 bp upstream from the first major transcription start site (position −52 in ADH1 and position −37 in HIS3). The promoter fragment swap generated plasmids p316/AH (ADH1 promoter upstream of the HIS3 transcription initiation region) and p316/HA (HIS3 promoter upstream of the ADH1 initiation region). Plasmids p316/AH and p316/HA were also used as templates in the PCR-based site-directed mutagenesis of the sequences adjacent to the transcription start sites.
Construction of mutant sua7 libraries.
The entire SUA7 insert in plasmid p314/yIIB-Mlu was amplified by PCR under error-prone conditions. Reaction mixtures (100 μl) contained 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 0.01% gelatin, 7 mM MgCl2, 1 mM MnCl2, 1 mM dCTP, 1 mM dTTP, 200 μM dATP, 200 μM dGTP, 200 ng of p314/yIIB-Mlu template, 100 pmol each of the M13 forward and reverse primers, and 5 U of Taq polymerase (Perkin-Elmer). Reactions were carried out for 30 cycles of 92°C for 1 min, 55°C for 1 min, and 72°C for 2 min. For the construction of the N-terminal mutant library, the 1.9-kb PCR product was digested with NdeI and MluI, and the approximately 300-bp fragment (encoding amino acids 1 to 94) was gel purified and cloned into the NdeI and MluI sites of p314/yIIB-Mlu. For the construction of the C-terminal mutant library, the 1.9-kb PCR product was digested with MluI and PstI, and the approximately 900-bp fragment (encoding amino acids 95 to 345) was gel purified and cloned into the MluI and PstI sites of p314/yIIB-Mlu. Approximately 25,000 primary E. coli transformants were obtained for each library. Nucleotide sequencing of 10 randomly selected clones revealed the mutation frequency to be on average one mutation per 200 bp.
Phenotypic analyses of mutant TFIIB strains.
Mutant sua7 plasmids identified in the genetic selection were reintroduced into strain FP227 by transformation, and the cells were plated on CAA medium lacking uracil and tryptophan. The plates were incubated for 3 days at 30°C to select for cells containing both the mutant plasmid (TRP1-containing) and the endogenous plasmid containing the URA3 and SUA7 genes. The Ura+ Trp+ transformants were then purified once on CAA lacking Trp and uracil. To test for dominant growth phenotypes on lactate-containing medium, the strains were streaked on CAA-lactate lacking uracil and Trp, and the plates were incubated at 30°C for 9 days. To analyze the growth properties of strains containing the mutant sua7 plasmid as the sole source of TFIIB, the Ura+ Trp+ strains were streaked on 5-FOA medium and the plates were incubated at room temperature for 4 days. 5-FOA-resistant colonies were purified once by streaking on plates of CAA without Trp and incubating the plates at room temperature for 3 to 4 days. The mutant strains were tested for their relative growth rates on CAA-lactate medium without Trp (30°C for 7 to 10 days) and on rich medium at high temperature (37°C for 3 days) and low temperature (16°C for 4 days).
Mapping of in vivo mRNA start sites.
Plasmids containing known TFIIB substitution mutants were introduced into strain FP153 by transformation, and the endogenous plasmid containing wild-type TFIIB was removed by plasmid shuffling on 5-FOA medium. To analyze CYC1 transcripts, cells were grown in liquid CAA-lactate medium lacking Trp and total RNA was prepared as described previously (39). To analyze ADH1 and HIS3 transcripts, total RNA was prepared from cells grown in liquid CAA-glucose medium lacking Trp. Primer extension analysis was carried out using 30 μg of total RNA under conditions previously described (39). The oligonucleotide primer for the analysis of CYC1 mRNA contained the sequence 5′-dGTCTTGAAAAGTGTAGCACC-3′, corresponding to positions +53 to +34 (where +1 is the initiating ATG). The oligonucleotide primer for the analysis of ADH1 mRNA contained the sequence 5′-dGTATTCCAACTTACCGTGGGATTCG-3′, corresponding to positions +63 to +39 (where +1 is the initiating ATG). To analyze transcripts from the ADH1-HIS3 hybrid promoters and variant ADH1 and HIS3 promoters containing insertions, deletions, or site-directed base substitutions (all fused to the HIS3 coding region), strains FP251 to FP254 containing the relevant reporter plasmids were grown in liquid CAA medium without uracil and total RNA was prepared and analyzed by primer extension. The oligonucleotide primer contained the sequence 5′-dGGTTTCATTTGTAATACGC-3′, corresponding to HIS3 positions +45 to +26 (where +1 is the initiating ATG).
RESULTS
Isolation of S. cerevisiae TFIIB mutants conferring downstream shifts in transcription initiation.
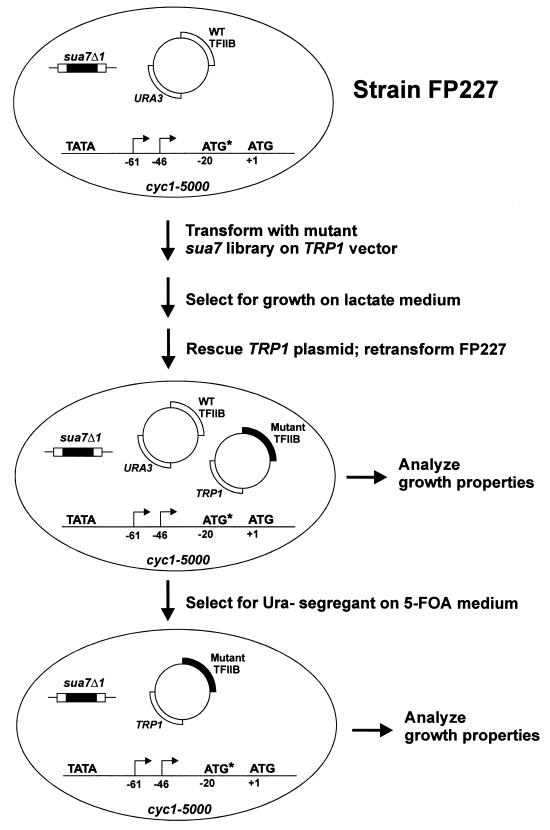
To isolate TFIIB mutants conferring alterations in transcription initiation, two mutant sua7 libraries that contained substitutions in either the highly conserved N-terminal region (residues 1 to 94) or the C-terminal core domain (residues 95 to 345) were constructed. The libraries were generated utilizing PCR amplification under error-prone conditions to randomly introduce mutations into the SUA7 gene on a plasmid containing the TRP1 gene as a selectable marker (see Materials and Methods). The sua7 libraries were independently introduced into strain FP227, which contains a chromosomal deletion of the essential SUA7 gene, a plasmid containing the URA3 and SUA7 genes to maintain cell viability, and a chromosomal cyc1-5000 gene (Fig. 1). The cyc1-5000 mutation, characterized by Hampsey and coworkers, contains an aberrant ATG translational initiation codon positioned upstream and out of frame with the normal CYC1 initiation codon (21). The reduced amount of iso-1-cytochrome c in this strain results in an inability to grow on media containing nonfermentable carbon sources such as lactate. Importantly, downstream shifts in transcription initiation can generate transcripts that initiate between the aberrant and normal ATG codons, thereby partially restoring iso-1-cytochrome c protein levels and conferring the ability to grow on medium containing lactate as the sole carbon source (37). For each sua7 library, approximately 7 × 105 Trp+ transformants of strain FP227 were plated on medium lacking tryptophan and containing lactate as the sole carbon source. After incubation at 30°C for 10 days, approximately 70 colonies were observed with the N-terminal library and approximately half that number for the C-terminal library. To determine whether growth on lactate medium was conferred by a mutant sua7 plasmid, plasmids containing the TRP1 gene and a candidate sua7 gene were rescued (53 colonies from the N-terminal library and 33 colonies from the C-terminal library) and reintroduced into strain FP227, and the transformants were again tested for growth on CAA/lactate-Trp. For the N-terminal library, 32 of the 53 rescued plasmids conferred the ability to grow on CAA-lactate medium without Trp when retested, whereas 0 of 33 rescued plasmids from the C-terminal library conferred the ability to grow on CAA-lactate without Trp. These results suggest that the C-terminal core domain of TFIIB does not play a significant role in the mechanism of start site utilization.
FIG. 1.
Scheme used for the genetic selection of TFIIB mutants that confer downstream shifts in transcription initiation in vivo.
To begin a characterization of the TFIIB mutants conferring growth on CAA-lactate medium lacking Trp, the 32 isolated sua7 plasmids were sequenced. The results revealed the presence of 24 different mutant sua7 genes among these plasmids (Table 1). The mutant sua7 genes contained single or multiple amino acid substitutions clustered to residues Glu-62, Arg-64, Arg-78, and Val-79, with Arg-78 being the most frequently altered residue. Among the 15 mutants with multiple amino acid substitutions, 5 contained a mutation of Tyr-30, which included Y30N, Y30D or Y30F substitutions. Interestingly, these substitutions were always found in combination with a substitution at residue Arg-78 or Val-79.
TABLE 1.
Growth properties of sua7 mutant strains
| TFIIB | Growth ratea on YPD at 30°C | Temperature sensitivityb | Phenotype for growth on lactate |
|---|---|---|---|
| Wild type | ++++ | ||
| E62Q | ++++ | Cs | Recessive |
| E62V | +++ | Cs | Recessive |
| E62V/I7L | +++ | Cs | Recessive |
| E62G | +++ | Cs | Recessive |
| E62G/S39R | +++ | Cs | Recessive |
| R64G | +++ | Cs | Recessive |
| R64G/E26G | +++ | Cs | Recessive |
| R78C | +++ | Cs | Dominant |
| R78C/D58A | +++ | Cs | Dominant |
| R78C/D75V | +++ | Cs | Dominant |
| R78C/R10G/V29A | +++ | Cs | Dominant |
| R78H/E40V | ++ | Cs, ts | Dominant |
| R78L | ++ | Cs, ts | Dominant |
| R78L/Y30D | +++ | Cs, ts | Dominant |
| R78L/Y30F | + | Cs, ts | Dominant |
| R78L/Y30N | +++ | Cs, ts | Dominant |
| R78L/N72I | + | Cs, ts | Dominant |
| R78S | +++ | Cs | Dominant |
| R78S/I7R | +++ | Cs | Dominant |
| V79L | ++ | Cs, ts | Dominant |
| V79D | ++ | Cs, ts | Dominant |
| V79D/T23A | ++ | Cs, ts | Dominant |
| V79G/R64G/Y30F | ++ | Cs, ts | Dominant |
| V79A/R64G/Y30N/G73D | ++ | Cs | Dominant |
Plus signs represent growth rates relative to that of the wild type (++++).
Cs, cold sensitive (impaired growth at 16°C); ts, temperature sensitive (impaired growth at 37°C).
To further characterize the TFIIB mutants, the growth properties of strains containing the mutant plasmids were determined in either the presence or the absence of wild-type TFIIB. Strain FP227 was transformed with the 24 mutant plasmids, and the transformants were plated on CAA medium lacking uracil and tryptophan to select for the presence of both the mutant plasmid (containing TRP1) and the plasmid containing SUA7 and URA3. The Ura+ Trp+ transformants were initially tested for their ability to grow on CAA medium lacking uracil and tryptophan and containing lactate as the sole carbon source. Seventeen of the 24 mutants exhibited a dominant phenotype for growth on lactate-containing medium in the presence of wild-type SUA7 (Table 1). These 17 mutants all contained substitutions at residue Arg-78 or Val-79. To analyze the growth properties of strains containing only the mutant sua7 plasmid, the Ura+ Trp+ strains were plated on 5-FOA medium to select for cells that had lost the URA3-containing plasmid with wild-type SUA7 (7). All 24 of the mutant strains gave rise to 5-FOA-resistant colonies, demonstrating that all of the mutants could support at least minimal cell growth in the absence of wild-type TFIIB. The mutant strains were then tested for their growth properties on CAA-lactate medium lacking Trp as well as on rich medium at high (37°C) and low (16°C) temperatures. As expected, all of the mutant strains grew on CAA-lactate medium lacking Trp, confirming that the seven mutants with substitutions at residue Glu-62 or Arg-64 exhibited recessive phenotypes with respect to growth on lactate medium (Table 1). All of the mutants exhibited a cold-sensitive phenotype on rich medium, consistent with the phenotype of previously isolated mutants exhibiting downstream shifts in transcription initiation, while 10 mutants also exhibited a temperature-sensitive phenotype. In addition, many of the mutants that displayed a dominant phenotype for growth on lactate medium (substitutions at Arg-78 and Val-79) grew slower than the recessive mutants (Table 1). In this regard, it was noteworthy that mutant R78L grew slowly on YPD at 30°C, whereas the R78L/Y30D and R78L/Y30N mutants grew significantly better. These results suggest a possible functional relationship between residues Arg-78 and Tyr-30.
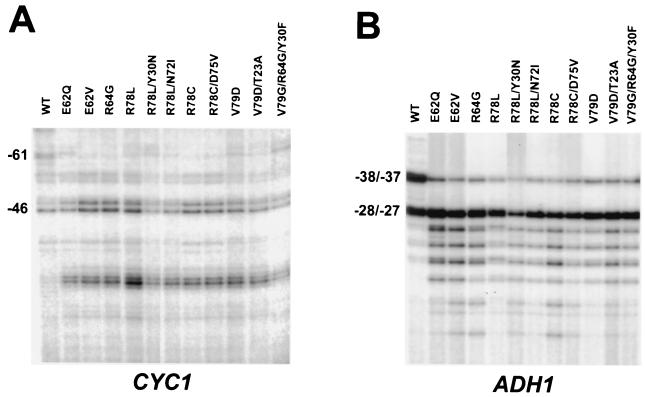
To directly analyze the effects of the TFIIB mutants on transcription initiation, primer extension analysis was used to map the transcription start sites at the CYC1 and ADH1 promoters in the mutant strains. Eleven representative strains, each containing a mutant sua7 plasmid as the sole source of TFIIB, were grown in either liquid CAA-lactate medium lacking Trp (for analysis of CYC1) or CAA-glucose medium lacking Trp (for analysis of ADH1). Total RNA was prepared and analyzed by primer extension using a CYC1-specific primer (Fig. 2A) or an ADH1-specific primer (Fig. 2B). For both promoters, the results revealed a decreased utilization of a major transcriptional start site proximal to the TATA element, which was accompanied by more pronounced utilization of minor start sites further downstream from the TATA element. These results confirm that the TFIIB mutants confer downstream shifts in transcription initiation at the CYC1 and ADH1 promoters in vivo. In contrast, no significant downstream shifts were detected at the HIS3 promoter (Fig. 3B and data not shown), consistent with previous observations (3, 5).
FIG. 2.
TFIIB mutations confer downstream shifts in transcription initiation at the CYC1 and ADH1 promoters. Total RNA (30 μg) from yeast strains containing the indicated TFIIB mutations were analyzed by primer extension utilizing a CYC1-specific (A) or ADH1-specific (B) oligonucleotide primer. The numbers to the left of each panel indicate the major transcription start sites and the distance from the translation-initiating ATG (+1).
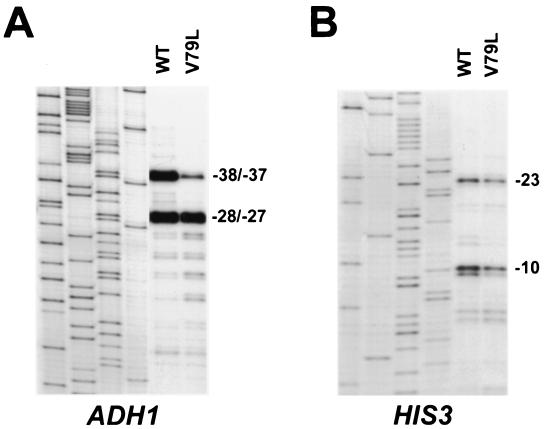
FIG. 3.
Differential effect of the V79L TFIIB mutation on transcription initiation at the ADH1 and HIS3 promoters. Primer extension analyses were performed using an ADH1-specific (A) or HIS3-specific (B) primer as described in the legend to Fig. 2. The first four lanes in each panel contain sequencing ladders of each promoter region (left to right: G, A, T, C) obtained with the same primers used in the primer extension analyses.
The ADH1 promoter contains a single functional TATA element and both TFIIB-sensitive and TFIIB-insensitive major transcription start sites.
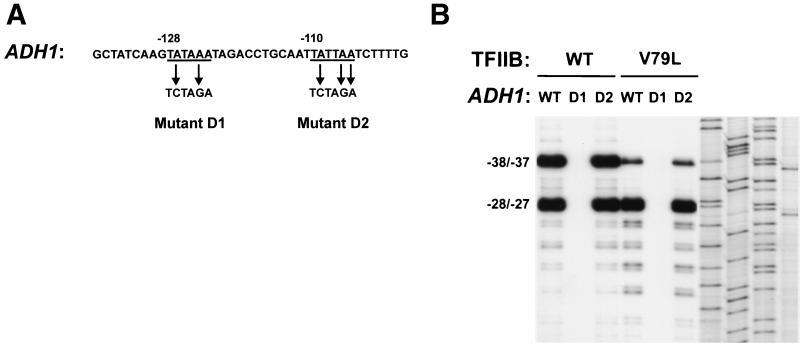
The ADH1 promoter contains three major transcription start sites. In our strains these sites map to positions −37, −28, and −27 (where +1 corresponds to the translation-initiating ATG) and an additional minor start site maps to position −38. Although the TFIIB mutants clearly confer decreased utilization of the −38 and −37 sites along with more pronounced utilization of minor start sites further downstream, the utilization of the −28 and −27 sites appears relatively constant in both wild-type and mutant TFIIB strains (Fig. 2B and 3A). One possible explanation for this result is that the ADH1 promoter contains two functional TATA elements, one that directs initiation to the −38 and −37 sites and a more downstream TATA that directs initiation to the −28 and −27 sites. The relatively constant amount of the −28 and −27 transcripts could be explained by TFIIB mutations causing downstream shifts for initiation directed by both the upstream and downstream TATA elements. In this view, a downstream shift in initiation directed by the more downstream TATA element would give rise to the observed far-downstream transcripts as well as a reduction in the amount of the −28 and −27 transcripts. However, a downstream shift for initiation directed by the more upstream TATA element would give rise to a reduction in the amount of the −38 and −37 transcripts along with an increase in the amount of the more downstream −28 and −27 transcripts, thereby restoring essentially wild-type levels of the −28 and −27 transcripts. Alternatively, the ADH1 promoter could contain a single functional TATA element and the constant level of the −28 and −27 transcripts might be due to these sites being insensitive to the effects of the TFIIB mutations. To distinguish between these possibilities, we sought to identify the functional TATA element(s) in the ADH1 promoter. Examination of the sequence upstream of the −38 start site revealed two potential TATA elements located between positions −128 and −123 (TATAAA) and between positions −110 and − 105 (TATTAA) (Fig. 4A). Site-directed mutagenesis was utilized to disrupt each of these motifs with the substitution of an XbaI restriction site (Fig. 4A). To facilitate primer extension analysis of the variant ADH1 promoters in strains containing a wild-type chromosomal ADH1 gene, plasmids that contained the wild-type or mutant ADH1 promoters fused to the HIS3 coding region were constructed. The plasmids were then introduced into wild-type or mutant TFIIB strains that contained a deletion of the chromosomal HIS3 gene. Primer extension analysis of total RNA using a HIS3-specific primer revealed that substitutions at positions −127 and −124 (D1) abolished all detectable ADH1 transcription whereas substitutions at positions −109, −107, and −106 (D2) had no detectable effect (Fig. 4B). These results demonstrate that the ADH1 promoter contains a single functional TATA element located between positions −128 and −123 and suggest that the −38 and −37 start sites and the −28 and −27 start sites differ in their intrinsic sensitivity to the effects of TFIIB mutations.
FIG. 4.
The ADH1 promoter contains a single functional TATA element. (A) Sequence of the ADH1 promoter region from positions −137 to −98 (where +1 corresponds to the initiating ATG) and specific mutations introduced to disrupt the two potential TATA elements (D1, D2). (B) Primer extension analyses were performed using an ADH1-specific primer and 30 μg of total RNA isolated from yeast strains containing the indicated ADH1 promoter construct and either wild-type or mutant (V79L) TFIIB. The last four lanes contain the sequencing ladder of the ADH1 promoter obtained with the same primer used in the primer extension analysis (left to right; G, A, T, C).
Analysis of ADH1-HIS3 hybrid promoters and ADH1 promoters with altered distance between the TATA element and the start sites.
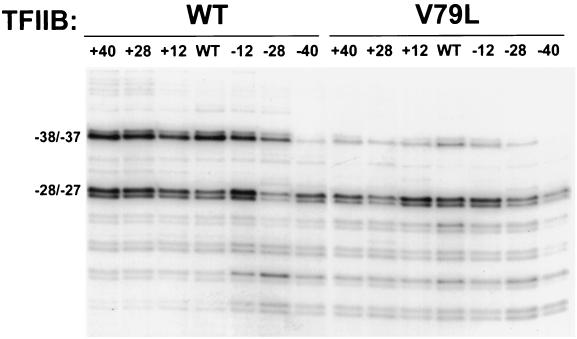
Mutations in TFIIB can cause downstream shifts in transcription initiation at the ADH1 promoter, whereas no significant shifts are detected at the HIS3 promoter (Fig. 3B; data not shown) (3, 5). To gain insight into the mechanism by which TFIIB mutants alter transcription initiation, we sought to determine which features of the ADH1 and HIS3 promoters render them susceptible and insensitive, respectively, to downstream shifts. We initially tested whether alterations in the spacing between the ADH1 TATA element and the start sites result in altered sensitivity to TFIIB mutations. A series of variant ADH1 promoters containing insertions or deletions of 12, 28, or 40 bp between the TATA element and the start sites were constructed and introduced into both wild-type and V79L mutant TFIIB strains (Fig. 5). Primer extension analysis demonstrated that deletion of 12 or 28 bp or insertion of 12, 28, or 40 bp between the TATA element and the initiation region had no significant effect on the initiation pattern in the wild-type TFIIB strain or on the downstream shifts observed for the mutant strain (Fig. 5). Deletion of 40 bp resulted in a significant reduction in the amount of the upstream −38 and −37 transcripts in the wild-type TFIIB strain, but downstream transcripts were still observed in the V79L strain (Fig. 5). The reduction in the amount of the −38 and −37 transcripts in the 40-bp deletion construct was not unexpected since the −38 and −37 sites in this construct are positioned 50 bp downstream of the TATA element, close to the minimal distance required between a yeast TATA element and an initiation site. Nonetheless, these results suggest that the spacing between the ADH1 TATA element and the initiation region does not play a significant role in determining the sensitivity of the start sites to TFIIB mutations.
FIG. 5.
Analysis of ADH1 promoters with altered distances between the TATA element and the start sites. Primer extension analyses were performed with variants of the ADH1 promoter containing insertions or deletions of 12, 28, or 40 bp. The −40 and +40 constructs contain a deletion and duplication, respectively, of the region between positions −92 and −52. The −28 and +28 constructs contain a deletion and duplication, respectively, of the region between positions −92 and −64. The −12 and +12 constructs contain a deletion and duplication, respectively, of the region between positions −92 and −80.
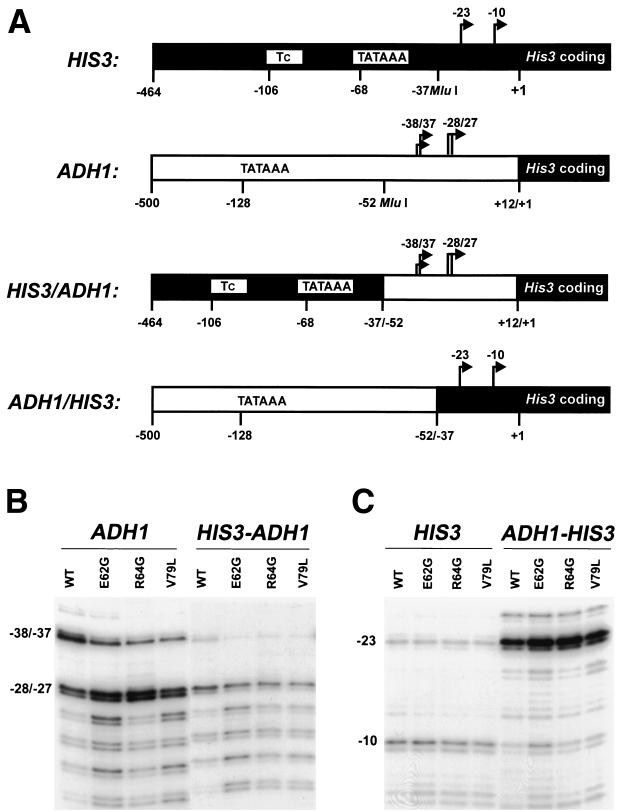
To further investigate which features of the ADH1 and HIS3 promoters determine their susceptibility to downstream shifts, reciprocal ADH1-HIS3 hybrid promoters were constructed and analyzed. The HIS3 promoter contains two major transcription start sites that in our yeast strains map to positions −23 and −10, where +1 corresponds to the translation-initiating ATG (Fig. 3B). The −10 start site is governed by a traditional TATA element located at position −68, whereas the −23 start site is governed by the TC element, which appears to contain several weak TATA elements located between positions −106 and −77 (24). Through the introduction of an MluI restriction site 14 bp upstream of the ADH1 and HIS3 initiation regions, reciprocal HIS3-ADH1 and ADH1-HIS3 hybrid promoters that contain the start sites from one promoter substituted at the position of the normal start sites of the other promoter were constructed (Fig. 6A). Reporter plasmids containing the ADH1, HIS3-ADH1, HIS3, or ADH1-HIS3 promoters were introduced into wild-type and TFIIB mutant strains, and the transcripts were analyzed by primer extension. Compared to the ADH1 promoter in the wild-type TFIIB strain, fusion of the HIS3 sequences to the ADH1 initiation region resulted in a significantly lower ratio of the amount of the −38 and −37 transcripts to the amount of the −28 and −27 transcripts (Fig. 6B, lanes 1 and 5). As noted above for the ADH1 40-bp deletion construct, the reduction in the amount of the −38 and −37 transcripts in the HIS3-ADH1 promoter was expected since these sites are positioned only 45 bp downstream of the HIS3 TATA element. The HIS3-ADH1 promoter also yielded lower overall levels of transcripts in all strains tested, consistent with the lower intrinsic activity of the HIS3 promoter. Significantly, introduction of the ADH1 or HIS3-ADH1 promoters into the TFIIB mutant strains resulted in a further reduction in the level of the −38 and −37 transcripts, which was accompanied by an increase in the amount of more-downstream transcripts (Fig. 6B). These results indicate that downstream shifts in transcription initiation can occur when the ADH1 start sites reside at their normal positions in the ADH1 promoter or when they are substituted at the position of their counterparts in the HIS3 promoter.
FIG. 6.
The differential effect of TFIIB mutations on the ADH1 and HIS3 promoters is determined by the promoter regions containing the respective transcription start sites. (A) Schematic representation of the ADH1 and HIS3 promoter regions and the reciprocal HIS3-ADH1 and ADH1-HIS3 hybrid promoters. The numbers indicate the distances from the initiating ATG, and the arrows indicate the positions of the major transcription start sites. (B) Primer extension analyses were performed using a HIS3-specific primer and 30 μg of total RNA isolated from yeast strains containing the ADH1 or HIS3-ADH1 promoter constructs and either wild-type TFIIB or the indicated TFIIB mutant. (C) Primer extension analyses were performed as described in the legend to panel B using yeast strains containing the HIS3 or ADH1-HIS3 promoter constructs and either wild-type TFIIB or the indicated TFIIB mutant.
In contrast to the nearly equal levels of the −23 and −10 HIS3 transcripts observed with the normal HIS3 promoter, fusion of the ADH1 sequences to the HIS3 initiation region resulted in almost exclusive utilization of the −23 start site, along with an expected increase in the overall transcript levels (Fig. 6C, lanes 1 and 5). Moreover, the positioning of the ADH1 TATA element 90 bp upstream of the HIS3 −23 site resulted in an increased amount of a minor transcript that maps to HIS3 position −27 (Fig. 6C, lane 5). Importantly, no significant downstream shifts in initiation were observed when either the HIS3 or the ADH1-HIS3 promoters were introduced into mutant TFIIB strains (Fig. 6C). Taken together, the results from the hybrid promoter analyses suggest that the sensitivity of the ADH1 and HIS3 promoters to the effects of TFIIB mutations is determined by the sequences surrounding their respective transcription start sites.
Base substitutions in the vicinity of transcription start sites can alter their sensitivity to TFIIB mutations.
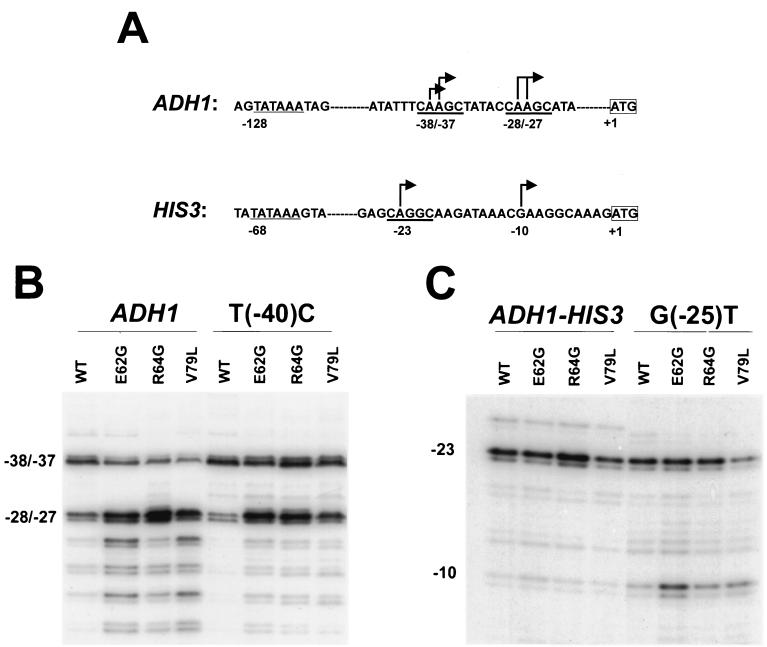
The preceding results strongly suggest that the DNA sequence surrounding a transcription start site determines whether that site is utilized to a lesser extent in mutant TFIIB strains with a concomitant increase in the usage of more-downstream sites. In light of these results, we addressed whether base substitutions in the vicinity of a start site could alter the sensitivity of that site to downstream shifts in mutant TFIIB strains. As noted earlier, the ADH1 −38 and −37 start sites and the ADH1 −28 and −27 start sites differ in their sensitivity to TFIIB mutations, with the −38 and −37 sites being significantly less utilized than the −28 and −27 sites in mutant TFIIB strains. Comparison of the sequences encompassing the −38 and −37 start sites and the −28 and −27 start sites revealed that all four start sites reside within an identical 5-bp CAAGC motif (Fig. 7A). In contrast to the C that is immediately upstream of the motif containing the −28 and −27 sites (position −30), a T is present at the homologous position upstream of the motif containing the −38 and −37 sites (position −40) (Fig. 7A). To determine whether this base difference contributes to the differential sensitivity of these sites to TFIIB mutations, a mutant ADH1 promoter containing a T-to-C substitution at position −40 was constructed and analyzed. In a wild-type TFIIB strain, the T(−40)C mutant promoter yielded a slightly higher amount of the −38 and −37 transcripts than the normal ADH1 promoter and reduced levels of the −28 and −27 and further downstream minor transcripts (Fig. 7B, lanes 1 and 5). In mutant TFIIB strains, the normal ADH1 promoter displayed the classical downstream initiation pattern, yielding significantly reduced levels of the −38 and −37 transcripts relative to the −28 and −27 transcripts accompanied by increased levels of the more downstream transcripts (Fig. 7B, lanes 2 to 4). In contrast, the T(−40)C promoter yielded nearly equal levels of the −38 and −37 transcripts and the −28 and −27 transcripts and significantly reduced levels of the more-downstream transcripts (Fig. 7B, lanes 6 to 8). These results suggest that the T(−40)C substitution increases utilization of the −38 and −37 sites in a wild-type TFIIB strain and renders these sites less sensitive to downstream shifts in mutant TFIIB strains.
FIG. 7.
Base substitutions in the vicinity of transcription start sites can alter their sensitivity to TFIIB mutations. (A) Presented is the DNA sequence of the transcription initiation regions in the ADH1 and HIS3 promoters. Underlined in bold is the CAPuGC motif present at both of the ADH1 start sites and at the HIS3 −23 start site. (B) Primer extension analyses were performed as described in the legend to Fig. 6 using yeast strains containing the ADH1 or T(−40)C mutant ADH1 promoter constructs and either wild-type TFIIB or the indicated TFIIB mutant. (C) Primer extension analyses were performed using yeast strains containing the ADH1-HIS3 or G(−25)T mutant ADH1-HIS3 promoter constructs and either wild-type TFIIB or the indicated TFIIB mutant.
Having observed that the ADH1 T(−40)C substitution conferred decreased sensitivity to the effects of TFIIB mutations, we tested whether a base substitution in the vicinity of the HIS3 −23 start site could conversely confer increased sensitivity to TFIIB mutations. As shown earlier, no significant downstream shifts are observed with the HIS3 −23 and −10 start sites. Examination of the sequence encompassing the HIS3 −23 start site revealed that it resides within the sequence CAGGC, sharing homology with the CAAGC motif present at the ADH1 start sites (Fig. 7A). Immediately upstream of this motif, the HIS3 −23 site contains a G at position −25, in contrast to the T and C present immediately upstream of this motif at the ADH1 −38 and −37 sites and the ADH1 −28 and −27 sites, respectively. Since the ADH1 −38 and −37 start sites are sensitive to TFIIB mutations and contain a T immediately upstream of the 5-bp motif, we tested whether substituting a T at HIS3 position −25 could confer increased sensitivity to TFIIB mutations. A variant ADH1-HIS3 promoter containing the G(−25)T substitution was constructed, and the variant and normal ADH1-HIS3 promoter constructs were introduced into wild-type and mutant TFIIB strains. As seen earlier, the ADH1-HIS3 promoter exhibited almost exclusive utilization of the −23 site in the wild-type TFIIB strain and yielded an extremely low level of the −10 transcript when introduced into the mutant TFIIB strains (Fig. 7C, lanes 1 to 4). In contrast, the G(−25)T variant promoter yielded a slightly increased level of the −10 transcript in the wild-type TFIIB strain and exhibited significant downstream shifts in the mutant TFIIB strains, with approximately equal levels of the −23 and −10 transcripts being observed in the E62G or V79L strains (Fig. 7C, lanes 5 to 8). These results indicate that the G(−25)T substitution renders the HIS3 −23 start site more sensitive to downstream shifts in mutant TFIIB strains, and combined with the results from the variant ADH1 T(−40)C promoter, demonstrate that the sensitivity of transcription start sites to the effects of TFIIB mutations is influenced by the DNA sequence immediately upstream.
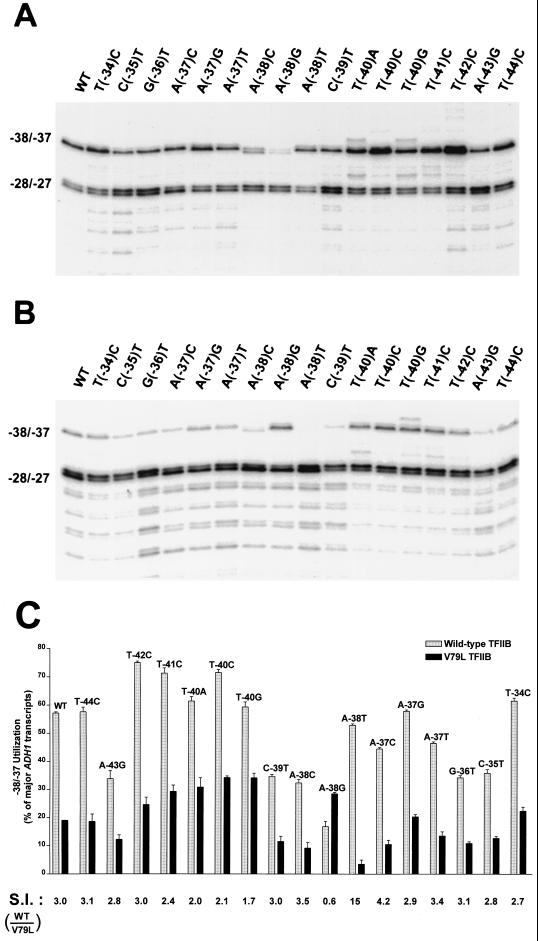
The observation that the ADH1 T(−40)C substitution increases the utilization of the −38 and −37 sites in a wild-type TFIIB strain and renders these sites less sensitive to downstream shifts in mutant TFIIB strains suggests that the sensitivity of start sites to the effects of TFIIB mutations may be correlated to the intrinsic strength of the start site. Alternatively, sensitivity may be conferred by the identity of bases at specific positions in the vicinity of the start site and not be strictly correlated to the strength of the start site. To determine whether the sensitivity of a start site to TFIIB mutations is dictated by overall strength or specific sequences, 16 additional variant ADH1 promoters that contained single-base substitutions upstream, downstream, or in the position of the −38 and −37 start sites (from positions −34 to −44) were constructed. Plasmids containing the normal or variant ADH1 promoters were introduced into a wild-type TFIIB strain or the V79L mutant strain, and the transcripts were analyzed by primer extension and quantitated by phosphorimaging. In the wild-type TFIIB strain, the −38 and −37 transcripts constituted 57% of the major transcripts yielded by the normal ADH1 promoter (Fig. 8A and C). Of the 17 substitutions analyzed, 3 significantly increased utilization of the −38 and −37 start sites in the wild-type TFIIB strain [T(−42)C (75%), T(−41)C (71%), and T(−40)C (72%)] and 8 decreased −38 and −37 utilization [A(−43)G (34%), C(−39)T (35%), A(−38)C (32%), A(−38)G (17%), A(−37)C (45%), A(−37)T (46%), G(−36)T (34%), and C(−35)T (35%)], while 6 had minimal effects [T(−44)C (57%), T(−40)A (61%), T(−40)G (59%), A(−38)T (53%), A(−37)G (58%), and T(−34)C (61%)] (Fig. 8A and C). In addition, the A(−38)C and A(−38)G substitutions conferred an altered initiation pattern at the −38 and −37 sites, abolishing utilization of the −38 start site and creating a new start site at position −36 (Fig. 8A).
FIG. 8.
The sensitivity of transcription start sites to TFIIB mutations is determined by the sequence in the immediate vicinity of the start sites and not by their intrinsic strength. Primer extension analyses were carried out as described in the legend to Fig. 6 using yeast strains containing the ADH1 or the indicated variant ADH1 promoter construct and either wild-type TFIIB (A) or the V79L TFIIB mutant (B). (C) Quantitation of −38 and −37 start site utilization. Phosphorimaging and Molecular Analyst software were used to quantitate ADH1 transcripts from three independent primer extension analyses. Utilization of the −38 and −37 sites was plotted as the percentage of major ADH1 transcripts (−38 plus −37 divided by the sum of −38, −37, −28, and −27). The sensitivity of the −38 and −37 sites to the effects of the V79L TFIIB mutation was defined by the sensitivity index (S.I. = utilization of the −38 and −37 sites in the wild-type TFIIB strain divided by that in the V79L strain).
In the V79L mutant TFIIB strain, the −38 and −37 transcripts constituted 19% of the major transcripts yielded by the normal ADH1 promoter. We defined the sensitivity index (S.I.) of the −38 and −37 sites in the normal ADH1 promoter as 3.0, corresponding to the ratio of their utilization in the wild-type strain (57%) to that in the V79L strain (19%) (Fig. 8B and C). Substitutions that significantly decreased the sensitivity of the −38 and −37 sites to the effects of the V79L TFIIB mutation included T(−40)G (S.I. = 1.7), T(−40)A (S.I. = 2.0), and T(−40)C (S.I. = 2.1) (Fig. 8B and C). Surprisingly, introduction of the A(−38)G construct into the V79L strain restored efficient −38 and −37 utilization, which had been abolished when this construct was in the wild-type TFIIB strain (S.I. = 0.6) (Fig. 8). In addition, two substitutions significantly increased the sensitivity of the start sites to the V79L TFIIB mutation [A(−38)T (S.I. = 15.0) and A(−37)C (S.I. = 4.2)] (Fig. 8B and C). Taken together, the results in Fig. 8 demonstrate that the sensitivity of the ADH1 −38 and −37 start sites to the effects of TFIIB mutations is not correlated to the overall strength of the start sites, but rather, is determined by the identity of bases at or in the immediate vicinity of the start sites.
DISCUSSION
In this study we investigated the domain and residues of S. cerevisiae TFIIB involved in the utilization of transcription start sites and the underlying reason for why TFIIB mutations cause downstream shifts in transcription initiation at some but not other promoters. To identify residues of TFIIB involved in the utilization of start sites, we modified the genetic approach of Hampsey and coworkers to directly select for TFIIB mutants conferring downstream shifts in transcription initiation in vivo (37). This approach resulted in the isolation of 24 sua7 alleles that conferred downstream shifts in transcription initiation at the CYC1 and ADH1 promoters in vivo. The mutants contained single or multiple amino acid substitutions in the highly conserved N-terminal region of the TFIIB protein and in all cases involved a substitution for Glu-62, Arg-64, Arg-78, or Val-79. Seventeen of the mutants, all with substitutions for Arg-78 or Val-79, conferred a dominant phenotype for the ability to grow on lactate in the presence of wild-type TFIIB. The remaining seven mutants, all containing substitutions for Glu-62 or Arg-64, displayed a recessive phenotype for growth on lactate medium. All of the mutants also exhibited a cold-sensitive phenotype.
It is noteworthy that among the 15 mutants with multiple amino acid substitutions, 5 of them contained substitutions of Tyr-30. Single-substitution mutants of Tyr-30 were not identified in the selection, but rather, these substitutions were always found in combination with a substitution of Arg-78 or Val-79. We think it unlikely that these Tyr-30 substitutions are fortuitous for several reasons. First, three different substitutions for this residue were identified. Second, the Tyr-30 substitutions were always in combination with an Arg-78 or Val-79 substitution. Third, in two cases the Tyr-30/Arg-78 double-mutant strain grew better than the corresponding Arg-78 mutant strain. Further work will be required to determine the role of Tyr-30 in the function of both wild-type TFIIB and the Arg-78 and Val-79 mutant proteins.
In previous studies utilizing site-directed mutagenesis, we also reported substitutions of residues Trp-63 and Phe-66 that conferred downstream shifts in transcription initiation. Thus, substitutions of residues Glu-62, Trp-63, Arg-64, Phe-66, Arg-78, or Val-79 have been identified that confer downstream shifts in transcription initiation, thereby defining a highly conserved discrete domain within the N-terminal region of TFIIB involved in the utilization of start sites. This domain is positioned adjacent and C terminal to a zinc-ribbon motif in TFIIB that is required for stable RNAPII-TFIIB binding (35). Continued biochemical analyses are needed to determine the primary biochemical alteration associated with the mutant TFIIB proteins.
As observed previously, TFIIB mutations conferred downstream shifts in transcription initiation at the ADH1 and CYC1 promoters but not at the HIS3 promoter (Fig. 3). This differential effect of TFIIB mutations on these promoters could be due to differences in (i) the upstream regulatory regions, (ii) the TATA elements, (iii) the spacing between the TATA elements and the start sites, (iv) specific sequences between the TATA elements and the start sites, and/or (v) specific sequences in the immediate vicinity of the start sites. To shed light on the mechanism by which TFIIB influences the utilization of start sites, we sought to exploit the promoter-specific effect of the TFIIB mutations and determine which of the above promoter features renders the ADH1 and HIS3 promoters sensitive and insensitive, respectively, to TFIIB mutations. We initially examined whether the distance between the TATA element and the start sites plays a role in establishing the sensitivity of those sites to TFIIB mutations. Analysis of a series of ADH1 promoters containing insertions or deletions between the TATA element and the initiation region demonstrated that alteration in the spacing of this region had no significant effect on the sensitivity of this promoter to TFIIB mutations (Fig. 5). A sequence-dependent basis for the differential sensitivity to TFIIB mutations was initially suggested by the observation that the ADH1 −38 and −37 start sites and the ADH1 −28 and −27 start sites, separated by only 10 bp, differ in their sensitivity to TFIIB mutations (Fig. 4). Results from the analysis of reciprocal HIS3-ADH1 and ADH1-HIS3 hybrid promoters supported this view, demonstrating that the differential sensitivity of the HIS3 and ADH1 promoters to TFIIB mutations was not due to differences in promoter architecture or sequences far upstream of the start sites, but rather was due to differences in the specific sequences in the immediate vicinity of the respective start sites (Fig. 6). Examination of the sequence of the ADH1 and HIS3 start sites revealed that the ADH1 −38 and −37 sites and the ADH1 −28 and −27 sites reside within a CAAGC motif while the HIS3 −23 site is contained within the sequence CAGGC, both sharing homology with a PyAA/TPu proposed consensus sequence for S. cerevisiae RNAPII start sites (14, 20, 22). The suggestion that the differential sensitivity of these start sites to TFIIB mutations is due to sequence differences in the immediate vicinity of the sites was further supported by the demonstration that a T(−40)C substitution conferred decreased sensitivity of the ADH1 −38 and −37 sites to TFIIB mutations, whereas a G(−25)T substitution rendered the HIS3 −23 start site more sensitive (Fig. 7).
The preceding results demonstrated that base substitutions in the immediate vicinity of start sites could alter their sensitivity to TFIIB mutations. However, since the ADH1 T(−40)C substitution rendered the −38 and −37 start sites less sensitive to downstream shifts in mutant TFIIB strains but also modestly increased their utilization in a wild-type TFIIB strain, it was unclear whether the sensitivity of start sites to TFIIB mutations was dictated by specific sequences or simply by the intrinsic strength of the start site. The analysis of 16 additional variant ADH1 promoters containing single-base substitutions upstream, downstream, or in the position of the −38 and −37 start sites demonstrated that sensitivity was not correlated with overall strength, but rather was dictated by the identity of bases at or in the immediate vicinity of the start sites (Fig. 8). This was evidenced by (i) the T(−42)C substitution, which conferred the highest utilization of the −38 and −37 sites in a wild-type TFIIB strain yet the same degree of sensitivity seen for the normal ADH1 promoter (S.I. = 3.0) in the V79L mutant TFIIB strain; (ii) the T(−40)A, T(−40)C, and T(− 40)G substitutions, which conferred little or no change in −38 and −37 utilization in the wild-type strain but decreased sensitivity (S.I. = 1.7 to 2.1) in the V79L strain; (iii) the A(−38)T substitution, which conferred no change in −38 and −37 usage in the wild-type strain yet dramatically increased sensitivity in the V79L strain (S.I. = 15); and (iv) the A(−38)G substitution, which abolished utilization of the −38 site in the wild-type strain yet surprisingly supported efficient utilization in the V79L strain. The last result also provides the first example of a start site sequence that is utilized preferentially in a mutant TFIIB strain, suggesting that TFIIB mutations may not only impair normal start site recognition by RNAPII but also confer altered specificity as well.
As noted earlier, transcription initiation in higher eukaryotes usually occurs at a discrete site located about 30 bp downstream of the TATA element, whereas transcription initiation in S. cerevisiae often occurs at multiple sites within a window of 45 to 120 bp downstream of the TATA element. The ability of S. cerevisiae RNAPII to initiate within an extended window downstream of the TATA element reflects a fundamental difference in the initiation mechanism compared to that in higher eukaryotic cells, and the basis for this difference is unknown. Thus, it remains of significant interest to determine both the mechanism by which S. cerevisiae RNAPII initiates transcription at such a distance from the TATA element and the manner in which wild-type and mutant forms of TFIIB affect the utilization of potential start sites. One possibility to account for the difference in the mammalian and yeast initiation patterns is that the positioning of yeast PICs on a promoter is fundamentally different from that of mammalian PICs. Yeast PICs could potentially adopt a much more elongated conformation than mammalian PICs and/or loop out intervening DNA in order to contact downstream start sites. Although this remains a possibility, it has been reported that promoter melting for the S. cerevisiae GAL1 and GAL10 genes in vivo is comparable to that seen in other eukaryotes, occurring approximately 20 bp downstream of the TATA element (15). Moreover, the extent of melting was reported to be independent of the distance between the TATA element and the transcription start sites. Thus, the positioning and promoter melting of yeast PICs appears to be similar to that of mammalian PICs. In light of these results, Giardina and Lis proposed that S. cerevisiae RNAPII may be released from the PIC and scan downstream DNA for preferred start sites (15).
Results from both genetic and biochemical studies strongly suggest that RNAPII and TFIIB are solely responsible for determining the start site of transcription in S. cerevisiae. Genetic studies have shown that mutations in the largest subunit of RNAPII (Rpb1) can confer downstream shifts in transcription initiation that are similar to those observed with TFIIB mutations (6). In contrast, deletion of the Rpb9 subunit of RNAPII, or mutation disrupting a C-terminal zinc-ribbon in Rpb9, confers an upstream shift in transcription initiation (23, 46). Biochemical studies by Kornberg and colleagues have shown that the pairwise exchange of S. cerevisiae TFIIB and RNAPII with their homologs from Schizosaccharomyces pombe is sufficient to convert the transcription initiation pattern to that of S. pombe (27). Thus, an attractive extension to the scanning model is that a TFIIB-RNAPII interaction governs the start site recognition properties of RNAPII and that TFIIB mutants confer downstream shifts in transcription initiation by impairing start site recognition. Initially proposed by Hampsey and colleagues, such impairment could cause RNAPII to bypass sites normally used for initiation and allow for initiation at sites further downstream (38). Our results here demonstrate that the sequence at or in the immediate vicinity of a start site determines the tendency of that site to be underutilized in mutant TFIIB strains accompanied by increased usage of further downstream start sites. These results are consistent with the view that a TFIIB-RNAPII functional interaction, either direct or through the action of another factor, affects the efficiency by which potential start sites are recognized and utilized by a scanning polymerase. The incorporation of TFIIB mutants into additional genetic, biochemical, and molecular studies should prove useful in determining the precise mechanism by which S. cerevisiae RNAPII recognizes and utilizes potential transcription start sites.
ACKNOWLEDGMENTS
We thank Michael Hampsey for CYC1 plasmids, Lynne Pajak and Tom Smith for technical assistance, and Michelle Ammerman, Tim Pardee, and Lynn Ziegler for helpful discussions.
This work was supported by a grant from National Science Foundation (MCB-9905418) and a Public Health Service grant from the National Institutes of Health (GM51124) to A.S.P.
REFERENCES
- 1.Aiyar A, Leis J. Modification of the megaprimer method of PCR mutagenesis: improved amplification of the final product. BioTechniques. 1993;14:366–367. [PubMed] [Google Scholar]
- 2.Bagby S, Kim S, Maldonado E, Tong K I, Reinberg D, Ikura M. Solution structure of the C-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA-binding protein. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- 3.Bangur C S, Pardee T S, Ponticelli A S. Mutational analysis of the D1/E1 core helices and the conserved N-terminal region of yeast transcription factor IIB (TFIIB): identification of an N-terminal mutant that stabilizes TATA-binding protein-TFIIB-DNA complexes. Mol Cell Biol. 1997;17:6784–6793. doi: 10.1128/mcb.17.12.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barberis A, Muller C W, Harrison S C, Ptashne M. Delineation of two functional regions of transcription factor TFIIB. Proc Natl Acad Sci USA. 1993;90:5628–5632. doi: 10.1073/pnas.90.12.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berroteran R W, Hampsey M. Genetic analysis of yeast iso-1-cytochrome c structural requirements: suppression of Gly6 replacements by an Asn52 Ile replacement. Arch Biochem Biophys. 1991;288:261–269. doi: 10.1016/0003-9861(91)90193-m. [DOI] [PubMed] [Google Scholar]
- 6.Berroteran R W, Ware D E, Hampsey M. The sua8 suppressors of Saccharomyces cerevisiae encode replacements of conserved residues within the largest subunit of RNA polymerase II and affect transcription start site selection similarly to sua7 (TFIIB) mutations. Mol Cell Biol. 1994;14:226–237. doi: 10.1128/mcb.14.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 8.Buratowski S, Zhou H. Functional domains of transcription factor TFIIB. Proc Natl Acad Sci USA. 1993;90:5633–5637. doi: 10.1073/pnas.90.12.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushnell D A, Bamdad C, Kornberg R D. A minimal set of RNA polymerase II transcription protein interactions. J Biol Chem. 1996;271:20170–20174. doi: 10.1074/jbc.271.33.20170. [DOI] [PubMed] [Google Scholar]
- 10.Chiang Y C, Komarnitsky P, Chase D, Denis C L. ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. J Biol Chem. 1996;271:32359–32365. doi: 10.1074/jbc.271.50.32359. [DOI] [PubMed] [Google Scholar]
- 11.Colgan J, Ashali H, Manley J L. A direct interaction between a glutamine-rich activator and the N terminus of TFIIB can mediate transcriptional activation in vivo. Mol Cell Biol. 1995;15:2311–2320. doi: 10.1128/mcb.15.4.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang S M, Burton Z F. RNA polymerase II-associated protein (RAP) 74 binds transcription factor (TF) IIB and blocks TFIIB-RAP30 binding. J Biol Chem. 1996;271:11703–11709. doi: 10.1074/jbc.271.20.11703. [DOI] [PubMed] [Google Scholar]
- 13.Franklin C C, McCulloch A V, Kraft A S. In vitro association between the Jun protein family and the general transcription factors, TBP and TFIIB. Biochem J. 1995;305:967–974. doi: 10.1042/bj3050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furter-Graves E M, Hall B D. DNA sequence elements required for transcription initiation of the Schizosaccharomyces pombe ADH gene in Saccharomyces cerevisiae. Mol Gen Genet. 1990;223:407–416. doi: 10.1007/BF00264447. [DOI] [PubMed] [Google Scholar]
- 15.Giardina C, Lis J T. DNA melting on yeast RNA polymerase II promoters. Science. 1993;261:759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- 16.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 17.Guarente L. Regulatory proteins in yeast. Annu Rev Genet. 1987;21:425–452. doi: 10.1146/annurev.ge.21.120187.002233. [DOI] [PubMed] [Google Scholar]
- 18.Ha I, Roberts S, Maldonado E, Sun X, Kim L U, Green M, Reinberg D. Multiple functional domains of human transcription factor IIB: distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev. 1993;7:1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]
- 19.Hadzic E, Desai-Yajnik V, Helmer E, Guo S, Wu S, Koudinova N, Casanova J, Raaka B M, Samuels H H. A 10-amino-acid sequence in the N-terminal A/B domain of thyroid hormone receptor alpha is essential for transcriptional activation and interaction with the general transcription factor TFIIB. Mol Cell Biol. 1995;15:4507–4517. doi: 10.1128/mcb.15.8.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn S, Hoar E T, Guarente L. Each of three “TATA elements” specifies a subset of transcription initiation sites at the CYC1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1985;82:8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hampsey M, Na J G, Pinto I, Ware D E, Berroteran R W. Extragenic suppressors of a translational initiation defect in the cyc1 gene of Saccharomyces cerevisiae. Biochimie. 1991;73:1445–1455. doi: 10.1016/0300-9084(91)90177-3. [DOI] [PubMed] [Google Scholar]
- 22.Healy A M, Helser T L, Zitomer R S. Sequences required for transcriptional initiation of the Saccharomyces cerevisiae CYC7 gene. Mol Cell Biol. 1987;7:3785–3791. doi: 10.1128/mcb.7.10.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hull M W, McKune K, Woychik N A. RNA polymerase II subunit RPB9 is required for accurate start site selection. Genes Dev. 1995;9:481–490. doi: 10.1101/gad.9.4.481. [DOI] [PubMed] [Google Scholar]
- 24.Iyer V, Struhl K. Mechanism of differential utilization of the HIS3 TR and TC TATA elements. Mol Cell Biol. 1995;15:7059–7066. doi: 10.1128/mcb.15.12.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagrange T, Kapanidis A N, Tang H, Reinberg D, Ebright R H. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S, Hahn S. Model for binding of transcription factor TFIIB to the TBP-DNA complex. Nature. 1995;376:609–612. doi: 10.1038/376609a0. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Flanagan P M, Tschochner H, Kornberg R D. RNA polymerase II initiation factor interactions and transcription start site selection. Science. 1994;263:805–807. doi: 10.1126/science.8303296. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y S, Ha I, Maldonado E, Reinberg D, Green M R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 29.MacDonald P N, Sherman D R, Dowd D R, Jefcoat S C, Jr, DeLisle R K. The vitamin D receptor interacts with general transcription factor IIB. J Biol Chem. 1995;270:4748–4752. doi: 10.1074/jbc.270.9.4748. [DOI] [PubMed] [Google Scholar]
- 30.Maldonado E, Ha I, Cortes P, Weis L, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II: role of transcription factors IIA, IID, and IIB during formation of a transcription-competent complex. Mol Cell Biol. 1990;10:6335–6347. doi: 10.1128/mcb.10.12.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malik S, Lee D K, Roeder R G. Potential RNA polymerase II-induced interactions of transcription factor TFIIB. Mol Cell Biol. 1993;13:6253–6259. doi: 10.1128/mcb.13.10.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakajima N, Horikoshi M, Roeder R G. Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol Cell Biol. 1988;8:4028–4040. doi: 10.1128/mcb.8.10.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 34.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 35.Pardee T S, Bangur C S, Ponticelli A S. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J Biol Chem. 1998;273:17859–17864. doi: 10.1074/jbc.273.28.17859. [DOI] [PubMed] [Google Scholar]
- 36.Parker C S, Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region specific DNA-binding activity. Cell. 1984;36:357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- 37.Pinto I, Ware D E, Hampsey M. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell. 1992;68:977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- 38.Pinto I, Wu W H, Na J G, Hampsey M. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J Biol Chem. 1994;269:30569–30573. [PubMed] [Google Scholar]
- 39.Ponticelli A S, Struhl K. Analysis of Saccharomyces cerevisiae his3 transcription in vitro: biochemical support for multiple mechanisms of transcription. Mol Cell Biol. 1990;10:2832–2839. doi: 10.1128/mcb.10.6.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts S G, Ha I, Maldonado E, Reinberg D, Green M R. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993;363:741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- 41.Sauer F, Fondell J D, Ohkuma Y, Roeder R G, Jackle H. Control of transcription by Kruppel through interactions with TFIIB and TFIIE beta. Nature. 1995;375:162–164. doi: 10.1038/375162a0. [DOI] [PubMed] [Google Scholar]
- 42.Shaw S P, Carson D J, Dorsey M J, Ma J. Mutational studies of yeast transcription factor IIB in vivo reveal a functional surface important for gene activation. Proc Natl Acad Sci USA. 1997;94:2427–2432. doi: 10.1073/pnas.94.6.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaw S P, Wingfield J, Dorsey M J, Ma J. Identifying a species-specific region of yeast TFIIB in vivo. Mol Cell Biol. 1996;16:3651–3657. doi: 10.1128/mcb.16.7.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherman F. Getting started with yeast. In: Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. Vol. 194. San Diego, Calif: Academic Press, Inc.; 1991. pp. 3–20. [Google Scholar]
- 45.Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987;49:295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- 46.Sun Z W, Tessmer A, Hampsey M. Functional interaction between TFIIB and the Rpb9 (Ssu73) subunit of RNA polymerase II in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:2560–2566. doi: 10.1093/nar/24.13.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu W H, Hamsey M. An activation-specific role for transcription factor TFIIB in vivo. Proc Natl Acad Sci USA. 1999;96:2764–2769. doi: 10.1073/pnas.96.6.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xing L, Gopal V K, Quinn P G. cAMP response element-binding protein (CREB) interacts with transcription factors IIB and IID. J Biol Chem. 1995;270:17488–17493. doi: 10.1074/jbc.270.29.17488. [DOI] [PubMed] [Google Scholar]