Abstract
PURPOSE
Family cancer history is an important component of genetic testing guidelines that estimate which patients with breast cancer are most likely to carry a germline pathogenic variant (PV). However, we do not know whether more extensive family history is differentially associated with PVs in specific genes.
METHODS
All women diagnosed with breast cancer in 2013-2017 and reported to statewide SEER registries of Georgia and California were linked to clinical genetic testing results and family history from two laboratories. Family history was defined as strong (suggestive of PVs in high-penetrance genes such as BRCA1/2 or TP53, including male breast, ovarian, pancreatic, sarcoma, or multiple female breast cancers), moderate (any other cancer history), or none. Among established breast cancer susceptibility genes (ATM, BARD1, BRCA1, BRCA2, CDH1, CHEK2, NF1, PALB2, PTEN, RAD51C, RAD51D, and TP53), we evaluated PV prevalence according to family history extent and breast cancer subtype. We used a multivariable model to test for interaction between affected gene and family history extent for ATM, BRCA1/2, CHEK2, and PALB2.
RESULTS
A total of 34,865 women linked to genetic results. Higher PV prevalence with increasing family history extent (P < .001) was observed only with BRCA1 (3.04% with none, 3.22% with moderate, and 4.06% with strong history) and in triple-negative breast cancer with PALB2 (0.75% with none, 2.23% with moderate, and 2.63% with strong history). In a multivariable model adjusted for age and subtype, there was no interaction between family history extent and PV prevalence for any gene except PALB2 (P = .037).
CONCLUSION
Extent of family cancer history is not differentially associated with PVs across established breast cancer susceptibility genes and cannot be used to personalize genes selected for testing.
INTRODUCTION
Germline genetic testing is common after a breast cancer diagnosis,1 and may increase further as germline-targeted therapies emerge.2,3 Recent studies have defined the prevalence and penetrance of pathogenic variants (PVs) in several cancer susceptibility genes among population-based breast cancer patients.4,5 However, testing multiple genes substantially increases the yield of uncertain (variant of uncertain significance [VUS]) results, particularly among groups that have had limited testing access such as racial/ethnic minorities.3,6 VUS results may contribute to anxiety and suboptimal treatment recommendations.7,8 Thus, there is rationale for careful consideration of which genes to test.
CONTEXT
Key Objective
Can we use family cancer history to select genes for germline testing in women with breast cancer?
Knowledge Generated
Among 34,865 female patients with breast cancer who underwent clinical germline genetic testing, there was no substantial difference between the established breast cancer genes (ATM, BARD1, BRCA1, BRCA2, CDH1, CHEK2, NF1, PALB2, PTEN, RAD51C, RAD51D, and TP53) in association of the extent or type of family cancer history with carrying a pathogenic variant.
Relevance
Family cancer history cannot be used to select specific genes for germline testing in women with breast cancer.
Family cancer history is an important component of genetic testing guidelines that aim to identify which patients are most likely to carry a PV.9 More extensive family history (eg, more relatives diagnosed with cancer or at younger ages) has been associated with higher PV prevalence in high-penetrance genes such as BRCA1 and BRCA2 (BRCA1/2).10 Yet, we do not know whether family cancer history is differentially associated with specific PVs: for example, whether more extensive family cancer history predicts a PV in BRCA1 but not the lower-penetrance ATM. A better understanding of the relationship between family cancer history and the prevalence of PVs in different genes might inform selection of a smaller, more personalized testing panel for each patient. We examined the association between family cancer history and PV prevalence by gene among a population-based cohort of women diagnosed with breast cancer from 2013 to 2017. Our hypothesis was that PVs in moderate-penetrance breast cancer genes such as ATM, CHEK2, and PALB2 would be less associated with the extent of family cancer history than PVs in high-penetrance genes such as BRCA1/2. If confirmed, such a finding would suggest that patients who have a family cancer history that is most consistent with a high-penetrance gene PV might be spared testing of moderate-penetrance genes—and the higher probability of a VUS result that comes with testing more genes.3,6
METHODS
All women diagnosed with breast cancer from January 1, 2013, to December 31, 2017, in Georgia or California and reported to SEER cancer registries in Georgia (the Georgia Cancer Registry) and in California (the Los Angeles Cancer Surveillance Program, the Greater Bay Area Cancer Registry, and the Cancer Registry of Greater California) were linked to clinical germline genetic testing results from four laboratories (Ambry Genetics, Aliso Viejo, CA; GeneDx, Gaithersburg, MD; Invitae, San Francisco, CA; and Myriad Genetics, Salt Lake City, UT) that performed the substantial majority of testing according to clinician and patient surveys.1,11 Two of these laboratories (Ambry Genetics and Myriad Genetics), comprising 75% of tested patients, had previously abstracted the family cancer history reported on testing request forms by ordering clinicians for research use,12,13 and only patients who linked to a test from one of these two laboratories were included for analysis. For a subset of women who participated in the earlier iCanCare study,14 patient self-reported family cancer history was compared with that provided by laboratories. All research was approved by institutional review boards associated with the SEER registries.
As previously described,1,3,8,11 the analytic data set combined genetic results with demographic and clinical variables from SEER registries. The results were reported by gene with the interpretation provided to the ordering clinician, as follows: PV or likely PV; VUS; and benign or likely benign. We focused on genes associated with breast cancer risk in the CARRIERS and Breast Cancer Association Consortium studies4,5: BRCA1, BRCA2, ATM, BARD1, CDH1, CHEK2, NF1, PALB2, PTEN, RAD51C, RAD51D, and TP53.
Family cancer history was provided in terms of degree (first-degree relative [FDR] or second-degree relative), number and sex of affected relative/s, and their cancer diagnoses. On the basis of testing guidelines and our prior work,9,14 we categorized the extent of family cancer history as strong (male FDR/second-degree relative with breast cancer; FDR with sarcoma, pancreatic, or ovarian cancer; two or more female FDRs with breast cancer; three or more FDRs with any cancer), moderate (any family cancer history not categorized as strong), or none. This definition of strong family cancer history was based on features that are associated with PVs in high-penetrance genes (BRCA1/2 for male breast, pancreatic, and ovarian cancer; TP53 for sarcoma) and that are recognized by practice guidelines as indications for genetic testing.9 As a sensitivity analysis, we evaluated the effect of categorizing family cancer history in a manner more focused on breast cancer, given that all tested patients had a breast cancer diagnosis. In this alternative categorization, family cancer history was defined as follows: any relative with breast cancer, but no relatives with any other cancer (breast cancer only); any relative with a nonbreast cancer, but none with breast cancer (nonbreast cancer only); relatives with breast cancer and with nonbreast cancers (in different individuals); and no family cancer history.
We evaluated PV prevalence by gene according to family cancer history extent and breast cancer subtype (hormone receptor–negative and human epidermal growth factor receptor 2–negative [triple-negative] v non–triple-negative), given differential PV prevalence by subtype.4,5 We used a multivariable model of PV prevalence, controlling for diagnosis age and subtype, to test for interaction between five genes in which PVs are relatively common (BRCA1/2, ATM, CHEK2, and PALB2) and extent of family cancer history.
RESULTS
A total of 34,865 women linked to genetic results. Among these, 1,016 (2.9%) had previously reported their family cancer history as a component of their participation in the earlier iCanCare study,14 and this prior report enabled us to compare self-reported to laboratory-reported family cancer history. For a reported family history of any relative affected by cancer (v no relatives affected), the concordance between self-report and laboratory report was 95% (Fleiss kappa = 0.903).
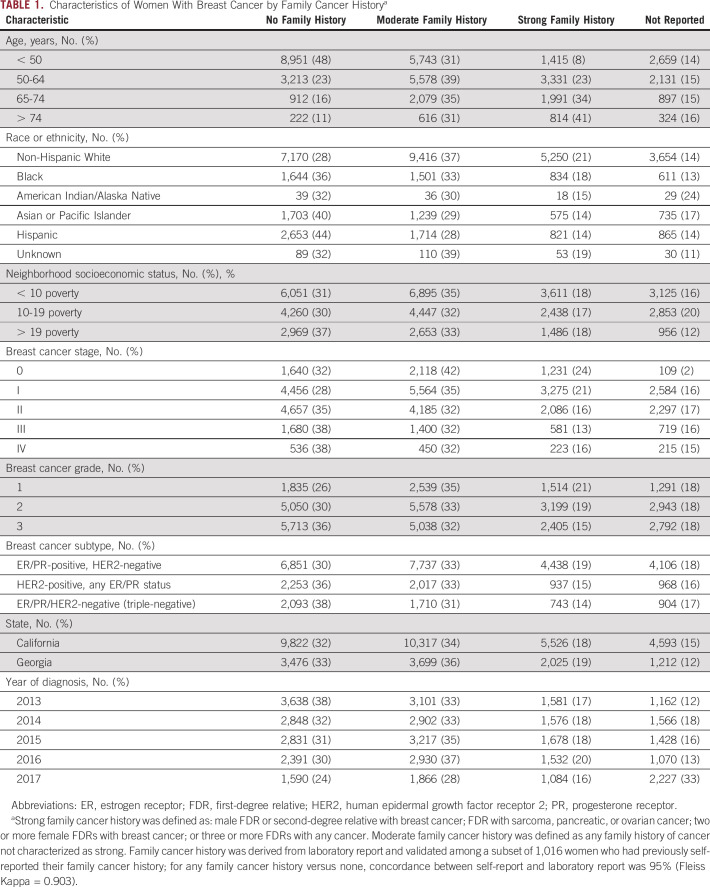
Table 1 shows patient characteristics; family cancer history was missing for approximately 15%-20%, with no pattern except a higher rate in 2017 (33%) than in earlier years (12%-18%). A report of no family cancer history was more common among Black (36%), Asian or Pacific Islander (40%), and Hispanic (44%) patients than among non-Hispanic White (28%) patients.
TABLE 1.
Characteristics of Women With Breast Cancer by Family Cancer Historya
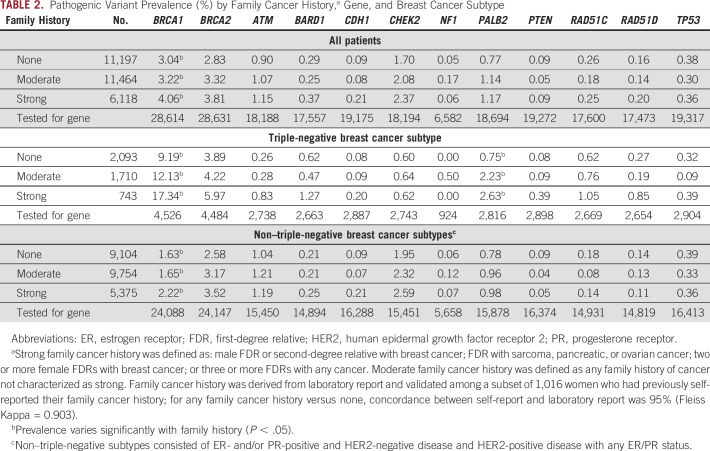
Table 2 shows PV prevalence by family cancer history, gene, and subtype. Higher prevalence with increasing extent of family cancer history (P < .001) was observed with BRCA1 (among all patients: 3.04% with none, 3.22% with moderate, and 4.06% with strong family cancer history) and PALB2 (among patients with triple-negative breast cancer: 0.75% with none, 2.23% with moderate, and 2.63% with strong family cancer history).
TABLE 2.
Pathogenic Variant Prevalence (%) by Family Cancer History,a Gene, and Breast Cancer Subtype
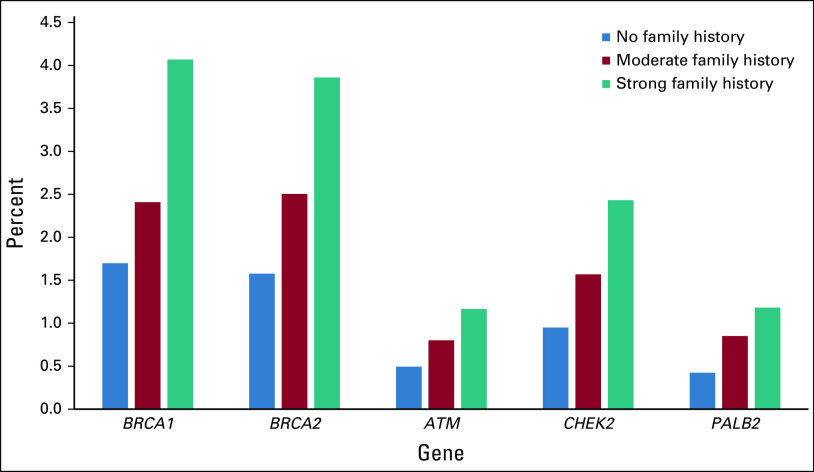
Figure 1 shows adjusted PV prevalence by gene and family cancer history. In a model of PV prevalence by gene, adjusted for diagnosis age and breast cancer subtype, there was no interaction between extent of family cancer history and PV prevalence for any gene except PALB2 with moderate family cancer history (P = .037).
FIG 1.
Adjusted pathogenic variant prevalence by affected gene and family cancer history. Marginal distributions from multivariable logistic model, including family cancer history, subtype, and age as covariates. A statistically significant interaction (P < .05) between family cancer history and gene was seen only for PALB2.
In our sensitivity analysis of using a breast cancer–focused categorization of family cancer history, the distribution of patients with a family cancer history of both breast and nonbreast cancers (Data Supplement) was similar to the distribution of patients with a family cancer history that we categorized as strong (Table 1). In the Data Supplement, higher PV prevalence with increasing extent of family cancer history (P < .05) was observed for BRCA1, BRCA2, PALB2, and RAD51C. However, in a multivariable model of PV prevalence by gene, adjusted for diagnosis age and breast cancer subtype (Data Supplement), there was no significant interaction between extent of family cancer history and PV prevalence for any gene except PALB2 (P = .012). These modeling results were consistent with those presented in Figure 1, in which we used the strong/moderate categorization of family cancer history.
DISCUSSION
Among 34,865 patients with breast cancer, we found minimal evidence that the extent of family cancer history is differentially associated with PVs across breast cancer susceptibility genes. Although we used a definition of strong family cancer history (eg, male breast cancer and ovarian cancer) on the basis of testing guidelines that were developed primarily for BRCA1/2, PV distribution by family cancer history was fairly uniform in other genes. In a sensitivity analysis that categorized family cancer history differently, as breast cancer versus nonbreast cancer, we obtained very similar results. We conclude that the extent of family cancer history cannot be used to exclude any established breast cancer susceptibility gene (BRCA1, BRCA2, ATM, BARD1, CDH1, CHEK2, NF1, PALB2, PTEN, RAD51C, RAD51D, and TP53) from testing in a woman with breast cancer. This finding is robust to differences in how family cancer history is categorized.
Notably, racial/ethnic minority patients were more likely than non-Hispanic White patients to have a report of no family cancer history. Potential explanations for this difference might include deficits in family history-taking by clinicians, which would be consistent with known racial/ethnic disparities in hereditary risk assessment referrals and genetic testing receipt,11,15 and/or higher competing mortality risks related to social determinants of health. Prior work has shown that limited family structure (eg, less information about cancer patterns in a family because of small family size or early deaths from other causes) can lead clinicians to underestimate the likelihood of BRCA1/2 PV carriage,16 and this may warrant consideration when evaluating diverse patients and families for cancer genetic testing.
Study limitations include family cancer history collection from genetic test request forms rather than patient self-report; however, there was high concordance with self-reported family cancer history in a subset of patients for whom self-report was available,14 consistent with prior work.12 Patients were tested clinically, and thus may not represent untested patients; they resided in two states, which may not represent the entire United States. These limitations are balanced by notable strengths, including a diverse, contemporary sample from population-based SEER registries and detailed genetic results from testing laboratories.
Questions remain about the clinical utility of detecting PVs in lower-penetrance genes such as ATM and CHEK2, with uncertainty about optimal breast screening regimens and no gene-specific evidence as yet to support risk-reducing surgery or targeted therapies.9,17-19 Thus, we recommend pretest counseling about the advantages and disadvantages of including lower-penetrance genes. However, the extent of family cancer history should not be used to estimate that a patient will test positive for a PV in one breast cancer susceptibility gene versus another.
ACKNOWLEDGMENT
The authors thank Lynne S. Penberthy, MD, and Valentina I. Petkov, MD, at the National Cancer Institute; Nicola Schussler at Information Management Services; Delores Bowman, RN, and Rachel Klein, MS, CGC, at Bioreference/GeneDx; Edward Esplin, MD, PhD, and Rebecca Truty, PhD, at Invitae; Holly LaDuca, MS, Virginia Speare, PhD, and Carolyn Horton, MS, at Ambry Genetics; and Jonathan Nelson, BS, and Larry Hardebeck, BS, at Myriad Genetics for their collaboration on the genetic test data linkage to SEER data. Written permission was obtained to include the names of all acknowledged individuals.
Research funding to her institution for an unrelated project was provided to Allison Kurian, MD, MSc, by Myriad Genetics. Lily Hoang, BS, Amal Yussuf, BS, and Jill Dolinsky, MS, report employment by Ambry Genetics. Krystal Brown, PhD, and Thomas Slavin, MD, report employment by Myriad Genetics.
Allison W. Kurian
Research Funding: Myriad Genetics (Inst)
Other Relationship: Ambry Genetics, Color Genomics, GeneDx/BioReference, InVitae, Genentech
Jonathan S. Berek
Leadership: Oncoquest
Research Funding: Tesaro (Inst), Karyopharm Therapeutics (Inst), Immunogen (Inst)
Lily Hoang
Employment: Ambry Genetics
Amal Yussuf
Employment: Ambry Genetics
Jill Dolinsky
Employment: Ambry Genetics
Krystal Brown
Employment: Myriad Genetics, Adela
Thomas Slavin
Employment: Myriad Genetics
Leadership: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
No other potential conflicts of interest were reported.
DISCLAIMER
The ideas and opinions expressed herein are those of the author(s) and do not necessarily reflect the opinions of the State of California, Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their contractors and subcontractors.
SUPPORT
Supported by the National Cancer Institute (NCI) of the National Institutes of Health under award numbers P01 CA163233 to the University of Michigan and R01 CA225697 to Stanford University. The collection of cancer incidence data in Georgia was supported by contract HHSN261201800003I, Task Order HHSN26100001 from the NCI and cooperative agreement 5NU58DP006352-03-00 from the Centers for Disease Control and Prevention (CDC). The collection of cancer incidence data used in this study was supported by the California Department of Public Health pursuant to California Health and Safety Code Section 103885; CDC's National Program of Cancer Registries, under cooperative agreement 5NU58DP006344; the National Cancer Institute's SEER Program under contract HHSN261201800032I awarded to the University of California, San Francisco, contract HHSN261201800015I awarded to the University of Southern California, and contract HHSN261201800009I awarded to the Public Health Institute, Cancer Registry of Greater California.
AUTHOR CONTRIBUTIONS
Conception and design: Allison W. Kurian, Paul Abrahamse, Kevin C. Ward, Dennis Deapen, Jonathan S. Berek, Timothy P. Hofer, Steven J. Katz
Financial support: Allison W. Kurian, Steven J. Katz
Administrative support: Steven J. Katz
Provision of study materials or patients: Ann S. Hamilton, Dennis Deapen
Collection and assembly of data: Allison W. Kurian, Paul Abrahamse, Kevin C. Ward, Ann S. Hamilton, Dennis Deapen, Lily Hoang, Amal Yussuf, Jill Dolinsky, Timothy P. Hofer, Steven J. Katz
Data analysis and interpretation: Allison W. Kurian, Paul Abrahamse, Kevin C. Ward, Dennis Deapen, Jonathan S. Berek, Jill Dolinsky, Krystal Brown, Thomas Slavin, Timothy P. Hofer, Steven J. Katz
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Allison W. Kurian
Research Funding: Myriad Genetics (Inst)
Other Relationship: Ambry Genetics, Color Genomics, GeneDx/BioReference, InVitae, Genentech
Jonathan S. Berek
Leadership: Oncoquest
Research Funding: Tesaro (Inst), Karyopharm Therapeutics (Inst), Immunogen (Inst)
Lily Hoang
Employment: Ambry Genetics
Amal Yussuf
Employment: Ambry Genetics
Jill Dolinsky
Employment: Ambry Genetics
Krystal Brown
Employment: Myriad Genetics, Adela
Thomas Slavin
Employment: Myriad Genetics
Leadership: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
No other potential conflicts of interest were reported.
REFERENCES
- 1.Kurian AW, Ward KC, Hamilton AS, et al. : Uptake, results, and outcomes of germline multiple-gene sequencing after diagnosis of breast cancer.JAMA Oncol 4:1066–10722018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tutt ANJ, Garber JE, Kaufman B, et al. : Adjuvant olaparib for patients with BRCA1- or BRCA2-mutated breast cancer.N Engl J Med 384:2394–24052021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurian AW, Ward KC, Abrahamse P, et al. : Time trends in receipt of germline genetic testing and results for women diagnosed with breast cancer or ovarian cancer, 2012-2019.J Clin Oncol 39:1631–16402021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breast Cancer Association Consortium. Dorling L, Carvalho S, et al. : Breast cancer risk genes—Association analysis in more than 113,000 women.N Engl J Med 384:428–4392021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu C, Hart SN, Gnanaolivu R, et al. : A population-based study of genes previously implicated in breast cancer.N Engl J Med 384:440–4512021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caswell-Jin JL, Gupta T, Hall E, et al. : Racial/ethnic differences in multiple-gene sequencing results for hereditary cancer risk.Genet Med 20:234–2392018 [DOI] [PubMed] [Google Scholar]
- 7.Domchek S, Brower J, Symecko S, et al. : Uptake of oophorectomy in women with findings on multigene panel testing: Results from the Prospective Registry of Multiplex Testing (PROMPT).J Clin Oncol 382020(suppl; abstr 1508) [Google Scholar]
- 8.Kurian AW, Li Y, Hamilton AS, et al. : Gaps in incorporating germline genetic testing into treatment decision-making for early-stage breast cancer.J Clin Oncol 35:2232–22392017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daly MB, Pal T, Berry MP, et al. : Genetic/familial high-risk assessment: Breast, ovarian, and pancreatic, version 2.2021, NCCN clinical practice guidelines in oncology.J Natl Compr Canc Netw 19:77–1022021 [DOI] [PubMed] [Google Scholar]
- 10.Frank TS, Deffenbaugh AM, Reid JE, et al. : Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: Analysis of 10,000 individuals.J Clin Oncol 20:1480–14902002 [DOI] [PubMed] [Google Scholar]
- 11.Kurian AW, Ward KC, Howlader N, et al. : Genetic testing and results in a population-based cohort of breast cancer patients and ovarian cancer patients.J Clin Oncol 37:1305–13152019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaDuca H, McFarland R, Gutierrez S, et al. : Quality of clinician-reported cancer history when ordering genetic testing.JCO Clin Cancer Inform 2:1–112018 [DOI] [PubMed] [Google Scholar]
- 13.Kurian AW, Hughes E, Handorf E, et al. : Breast and ovarian cancer penetrance estimates derived from germline multiple-gene sequencing results in women.JCO Precis Oncol 10.1200/PO.16.00066 [DOI] [PubMed] [Google Scholar]
- 14.Kurian AW, Griffith KA, Hamilton AS, et al. : Genetic testing and counseling among patients with newly diagnosed breast cancer.JAMA 317:531–5342017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson JM, Pepin A, Thomas R, et al. : Racial disparities in breast cancer hereditary risk assessment referrals.J Genet Couns 29:587–5932020 [DOI] [PubMed] [Google Scholar]
- 16.Weitzel JN, Lagos VI, Cullinane CA, et al. : Limited family structure and BRCA gene mutation status in single cases of breast cancer.JAMA 297:2587–25952007 [DOI] [PubMed] [Google Scholar]
- 17.Tung N, Domchek SM, Stadler Z, et al. : Counselling framework for moderate-penetrance cancer-susceptibility mutations.Nat Rev Clin Oncol 13:581–5882016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tung NM, Robson ME, Ventz S, et al. : TBCRC 048: Phase II study of olaparib for metastatic breast cancer and mutations in homologous recombination-related genes.J Clin Oncol 38:4274–42822020 [DOI] [PubMed] [Google Scholar]
- 19.Robson M: Management of women with breast cancer and pathogenic variants in genes other than BRCA1 or BRCA2.J Clin Oncol 39:2528–25342021 [DOI] [PMC free article] [PubMed] [Google Scholar]