Abstract
Objective
To evaluate the application of RNAscope in the clinical diagnostic field compared to the current ‘gold standard’ methods employed for testing gene expression levels, including immunohistochemistry (IHC), quantitative real time PCR (qPCR), and quantitative reverse transcriptase PCR (qRT-PCR), and to detect genes, including DNA in situ hybridisation (DNA ISH).
Methods
This systematic review searched CINAHL, Medline, Embase and Web of Science databases for studies that were conducted after 2012 and that compared RNAscope with one or more of the ‘gold standard’ techniques in human samples. QUADAS-2 test was used for the evaluation of the articles’ risk of bias. The results were reviewed narratively and analysed qualitatively.
Results
A total of 27 articles (all retrospective studies) were obtained and reviewed. The 27 articles showed a range of low to middle risk of bias scores, as assessed by QUADAS-2 test. 26 articles studied RNAscope within cancer samples. RNAscope was compared to different techniques throughout the included studies (IHC, qPCR, qRT-PCR and DNA ISH). The results confirmed that RNAscope is a highly sensitive and specific method that has a high concordance rate (CR) with qPCR, qRT-PCR, and DNA ISH (81.8–100%). However, the CR with IHC was lower than expected (58.7–95.3%), which is mostly due to the different products that each technique measures (RNA vs. protein).
Discussion
This is the first systematic review to be conducted on the use of RNAscope in the clinical diagnostic field. RNAscope was found to be a reliable and robust method that could complement gold standard techniques currently used in clinical diagnostics to measure gene expression levels or for gene detection. However, there were not enough data to suggest that RNAscope could stand alone in the clinical diagnostic setting, indicating further prospective studies to validate diagnostic accuracy values, in keeping with relevant regulations, followed by cost evaluation are required.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40291-021-00570-2.
Key Points
| RNAscope is a novel technology that can be used to measure gene expression (RNA). |
| RNAscope could be used as a complementary technique alongside existing procedures to enhance the diagnosis of disease that occurs as a result of abnormal gene expression, for example to confirm any unclear results from gold standard methods. |
| For RNAscope to be used as a tool to diagnose disease, further research is required to fully validate the technique so that it complies with regulatory standards and to assess cost implications for the health service. |
Introduction
The Developmental History of RNAscope
Gene expression involves transcription of DNA into messenger RNA (mRNA) followed by translation of mRNA to protein. Other important RNA molecules, such as microRNAs and long non-coding RNAs, can also play a role in regulating gene expression and thus form a pivotal fingerprint in tracking cellular changes that occur in cancer and common syndromes, such as intestinal brush border lactase deficiency [1, 2]. Despite the abundance of RNA molecules within cells, and their importance as prognostic tools in cancer research, the development of methods to detect mRNA molecules has been relatively delayed compared to the other biomarkers, namely DNA and proteins. The reason for this delay is primarily due to the instability of RNA molecules, which means that they can be degraded rapidly before detection. This has greatly impacted the discovery and monitoring of the aforementioned diseases by RNA levels [3–5].
Several techniques such as Northern blotting, microarrays, quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), digital or quantitative real time PCR (qPCR), and traditional RNA in situ hybridisation (ISH) were developed over the last five decades to measure RNA molecule levels. However, these technologies have limitations. For example, Northern blotting and PCR-based techniques require RNA extraction— RNA molecules might be lost during this process. Furthermore, Northern blotting and PCR techniques are unable to determine the localisation of gene expression within cells/tissue. Traditional RNA ISH, which uses digoxigenin (DIG) or radioactive probes, was developed to detect RNA molecules internally based on branched DNA (bDNA) method and principle [3, 6]. However, a major limitation of traditional RNA ISH is that it cannot detect other than highly expressed genes, for example H19 (an imprinted maternally expressed transcript), because of the high degree of non-specific binding (lack of specificity) and resultant background noise (poor sensitivity) [3, 7]. Given these limitations, in 2012 RNAscope was introduced by Advanced Cell Diagnostics (ACD), Inc. as a novel improved technology of traditional RNA ISH [7, 8].
RNAscope Technique
Underlying Principle of the Technology
Similar to traditional RNA ISH, RNAscope is based on the basic principle that RNA probes can be designed to detect a particular RNA of interest by hybridising to its complementary sequence inside the cell (the cytoplasm in the case of mRNA molecules). In contrast to traditional ISH, where a single RNA sequence is conjugated with a label such as digoxigenin or a fluorophore, RNAscope uses a pair of ‘Z’ probes to detect the RNA of interest [9]. These ‘Z’ probes are comprised of three elements—the lower region that hybridises to RNA molecules, the spacer (linker) sequence that connects the lower region with the ‘Z’ probe tail, and the tail that binds to the pre-amplifier sequence (Fig. 1A) [3]. Once the bottom of the double ‘Z’ probes (RNA-specific sequence) bind to their target RNA sequence inside the cell, signal amplification is achieved through a series of sequential processes (Fig. 1B) [3]. Firstly, the pre-amplifiers attach to their binding sites at the top of each double ‘Z’ pair. Secondly, multiple amplifier sequences bind subsequently via complementary base pairing to the pre-amplifier sequence. Finally, labelled probes, which can be either chromogenic or fluorescent, conjugate to their specific sites on the amplifier molecules.
Fig. 1.
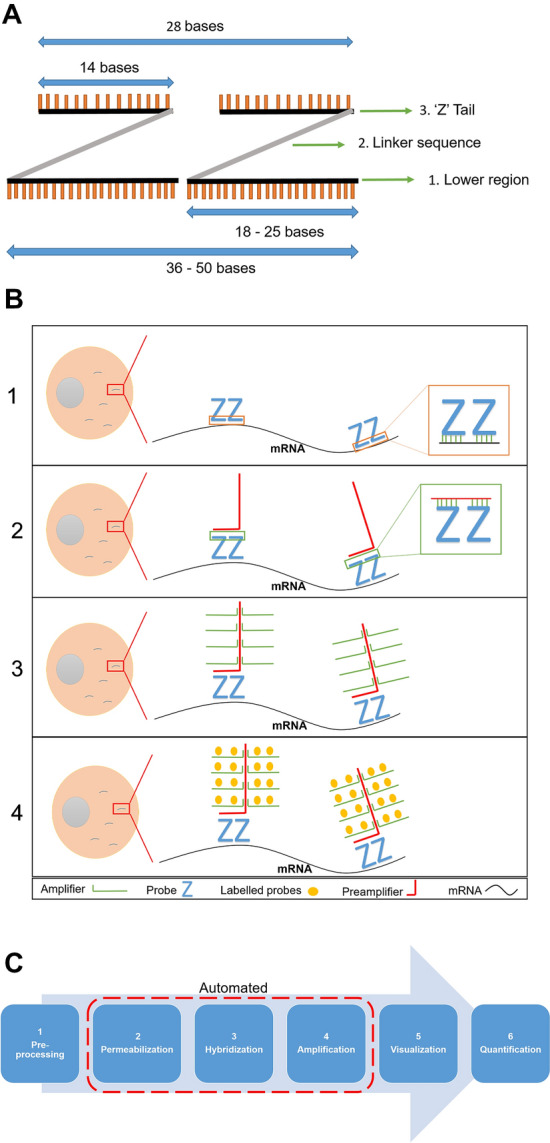
The elements of ‘Z’ probes. A The constituents of ‘Z’ probe dimers are: (1) the lower region that comprises 18–25 bases per each ‘Z’ probe; (2) linker sequence; (3) the tail that comprises 14 bases per each ‘Z’ probe. This figure panel was created with Powerpoint using data from Wang et al. [3]. B The sequential steps of RNAscope involve: (1) binding of double ‘Z’ probes to a complementary sequence; (2) attachment of pre-amplifier to double ‘Z’ pair tail; (3) binding of amplification molecules (amplifiers) to pre-amplifier; (4) attachment of the labelled probes to their specific sites on the amplifiers. This figure panel was created with Powerpoint using data from Erben and Buonanno [8] and Wang et al. [3]. C The presented flow-chart illustrates the RNAscope workflow process and highlights which parts can be automated (steps 2, 3 and 4). This figure panel was created with Powerpoint using data from [3, 10–12]
The unique design of the ‘Z’ probes constitutes the main reason for the high specificity of RNAscope, which can reach 100% [3, 10]. The assay requires ‘Z’ probes to form a dimer on the target RNA sequence so the pre-amplifier can bind, and the amplification cascade can start. The features of the ‘Z’ probe design are that it: (1) allows for single molecule detection, (2) facilitates recognising very short molecules and thus partially degraded molecules and samples, and (3) makes off-target binding very unlikely and thus suppresses background noise. The high sensitivity of RNAscope, which can also reach 100%, is due to the mechanism of the amplification process [3]. The unique process of signal amplification contributes significantly to the high sensitivity and specificity levels for RNAscope. Each RNA molecule should be hybridised to 20 ‘Z’ dimers (pre-amplifier). Each pre-amplifier in turn attaches to 20 amplifiers, which can subsequently be attached by 20 labelled probes per amplifier. This process results in up to 8,000 times signal amplification as 400 labelled probes will attach to each dimer.
Overall Workflow
The RNAscope workflow starts with slide preparation, which should be performed according to the type of tissue being used: formalin fixed paraffin embedded (FFPE) tissues (most commonly), tissue microarrays (TMA), fresh frozen tissues, or fixed cells [10, 11]. Prepared slides then proceed through three key steps where the main principle of RNAscope is applied: permeabilization, hybridization and signal amplification. Moreover, these three key steps can be performed automatically as part of an automated RNAscope workflow [10]. The workflow process ends with the visualisation of results using a bright-field or fluorescent microscope (depending on the probe type) and slides can be digitally scanned to facilitate quantification of the results, which can be performed either manually or by using a suitable computer software application [3, 12] (Fig. 1C).
RNAscope Controls
RNAscope quality is validated and assessed using positive and negative controls [10, 13]. The negative control probe utilises the bacterial gene dapB (dihydrodipicolinate B. subtilis reductase) to confirm the absence of background noise, as it is a gene that should not be present in any animal samples. On the other hand, a positive control is required to validate the detection of a signal resulting from expression of a gene that should be present in the tissue, such as a house-keeping gene. The positive control also acts as a measure of the tissue integrity—its failure to be detected would indicate degradation of RNA molecules. The most commonly used positive control is PPIB (peptidylprolyl isomerase B), which is employed for target genes that have moderate expression levels (10–30 copies per cell) [13, 14]. Polr2A (RNA polymerase II subunit A) is used for genes with a low level of expression (3–15 copies per cell). UBC (Ubiquitin C) is generally used for highly expressed genes (> 20 copies per cell), but can also be utilised for target genes with moderate expression [14].
Analysis of RNAscope Results
The analysis of RNAscope results involves quantification of the number of labelled dots within the tissue [8]. Each dot represents one RNA molecule, and thus the number of dots is indicative of the number of RNA molecules present—this is the critical factor to evaluate. However, it is noteworthy that sometimes, like in the case of highly expressed housekeeping genes, the dots can be found in clusters, which makes them difficult to distinguish separately. It is also important to highlight that the intensity and size of each dot reflects the number of double Z probes (as opposed to the number of transcripts) that are bound to the target molecule and thus will vary.
Scoring of RNAscope staining can be done either manually or by using computer software [8]. For manual scoring, standards are suggested by the manufacturer, where several regions on the slide should be quantitated in order to obtain a comprehensive result. Several computer software programs have been developed to read, analyse and quantify RNAscope results such as Halo, QuPath and Aperio software [15]. Using these programs requires scanning the whole slide comprehensively—images of the slide should be taken from at least three directions [8]. Halo is one of the gold standard programs that analyse ISH image results quantitatively. It is an adaptable platform with various advantages, including: scalability, powerful analytic capabilities and high processing speed, which are applicable for both TMA and FFPE slides [16, 17].
RNAscope Features
It is noteworthy that RNAscope can be used to assess heterogeneity between cells as it detects individual transcripts in a single cell. Furthermore, multiplex analysis can be performed to detect several genes within a single slide by using multiple probes with multiple channels—unique probes can be designed for each specific target, with each probe having a specific colour [18]. Additionally, RNAscope can be used in conjunction with immunohistochemistry (using either chromogenic or fluorescent detection) on the same tissue section as opposed to adjacent sections to allow the simultaneous detection of RNA and protein within the same tissue section [19]. Figure S1 (A–D) in the Electronic Supplementary Material (ESM) illustrates key features of RNAscope that are represented by results analysis.
Current Applications of RNAscope
Since its introduction in 2012, RNAscope has been used widely to study gene expression in the context of basic scientific research studies in diverse areas such as neuroscience, stem cells and developmental biology [12, 20, 21]. Furthermore, RNAscope has been applied in retrospective studies of clinical samples from non-infectious (e.g., cancer) and infectious (e.g., human papillomavirus (HPV) and, of great current interest, COVID-19) disease states [12, 20–23]. Interestingly, Neau et al. [24] have also highlighted the potential for integrating RNAscope to the biopharma field as a follow-up after gene expression analysis in the 3D culture process of organotypic cells, which is a vital tool in toxicology assessments and drug discovery. They also indicated that RNAscope has a promising future to be part of a comprehensive approach in tandem with omics data to assess histopathological samples. It is important to highlight that RNAscope can also be used in combination with other techniques such as microarray and immunohistochemistry (IHC) in a complementary way to confirm the results through producing data that cannot be achieved by IHC and microarray [10, 25].
Diagnosis of Disease Based on Analysis of Gene Expression
‘Gold standard’ Techniques and Their Limitations
Monitoring and analysis of gene expression is essential for the clinical diagnosis of a variety of diseases including cancers, infections (viruses, bacteria), cardiovascular, inflammatory, neurological and many more [2]. The currently used ‘gold standard’ methods for analysis of gene expression in clinical diagnostics have some challenges and drawbacks that need solutions and improvements. As mentioned earlier, traditional RNA ISH, which has been developed over the last 40 years, has several limitations and is a time-consuming method that requires complex procedures [3]. While conventional PCR and PCR-based methods provide robust information regarding absolute gene expression with a high degree of specificity and sensitivity, they do not provide spatial information of gene expression within cells or tissues [21].
IHC detects protein content via the use of specific antibodies that recognise a protein of interest [26, 27]. IHC is considered to be a cost-effective and robust method and is commonly used in the diagnostic setting, for example to detect E6/E7 proteins in HPV-driven cancers [28]. However, suitable antibodies for a protein of interest may not be commercially available and, thus, would have to be developed. Antibodies are available for only 25% of the human proteome, and new antibody development takes between 6 months to more than 1 year. This prolonged process impacts the detection of novel biomarkers or genetic signatures that are discovered as part of the clinical research process [26]. Another key limitation for IHC relates to antibody standardization. Promising antibodies that are used in the research field are not standardized, which can result in variability in the observed staining between studies [27]. Although antibodies that are used for IHC in the clinical diagnostic setting are standardized, the process to achieve standardization is lengthy, time consuming and expensive. A final limitation for IHC staining is that it lacks sensitivity for the target protein of interest in some cases, making it difficult to evaluate cases at the borderline of the limit of detection [29, 30].
Potential Advantages and Disadvantages of Using RNAscope in Clinical Diagnostic Testing
RNAscope has many advantages that indicate its potential to be utilised in clinical diagnostics [7, 21]: in principle, (1) it can detect the expression of any gene from any genome, which makes it suitable for diagnosis of infectious diseases [30]; (2) it can detect low levels of gene expression that exist inherently or due to tissue degradation as seen in clinical FFPE material [12]; (3) it has very high sensitivity and specificity [30]; (4) it provides both a quantitative level of gene expression and spatial information regarding where the gene is expressed within the tissue [7]; (5) RNAscope can be used in combination with IHC in the same tissue section to detect both RNAs and proteins that are implicated in disease diagnosis [19]; (6) the design and preparation of the required probes is relatively short at just 3 days to 2 weeks (Personal Communication from Andreas Rossbach, Advanced Cell Diagnostics (ACD)); (7) it can be performed in multiplex format to detect more than one gene (up to three genes) simultaneously within the same tissue [7], which in turn allows for different cell populations to be distinguished from each other [3]; and (8) RNAscope can be carried out in a high throughput manner and is suitable for automation [7]. Collectively, the incorporation of RNAscope into the clinical diagnostic field could have a significant impact on the diagnosis of many diseases.
The main drawbacks of the RNAscope technique, however, are the cost and workflow duration. There are large variations in costs for RNAscope among countries but using the UK as an example, the cost is estimated at around £65 per slide in comparison to £11 for IHC per slide (Personal Communication from CRUK Beatson Institute histology department). Also, according to University College of London (UCL), $48 is required for the analytical process per each RNAscope stained slide [31]. Additionally, using the Leica Bond Rx autostainer, the required time for a full run of RNAscope (30 slides) is around 9 h as compared to approximately 3 h for IHC. However, the manual protocol for RNAscope could be completed in a working day, taking approximately 6.5–7 h as compared to about 3 h for IHC (Personal Communication from CRUK Beatson Institute histology department).
Research Objective and Strategy
The primary objective of this systematic review is to assess RNAscope performance compared to the existing ‘gold standard’ technologies that are currently used for gene expression analysis (qPCR, qRT-PCR, RNA ISH and IHC) and gene detection (DNA ISH). To that end, this systematic review focuses on literature that has compared RNAscope to one or more of the existing techniques in human samples. The main criteria for comparison between RNAscope and the available technologies encompasses the concordance rate, sensitivity and specificity. In addition, this systematic review discusses some aspects and steps that are required to validate RNAscope for clinical diagnostic testing.
Methodology
Two reviewers (S.A. and S.S.) independently conducted the database searching, screening and data extraction from the identified articles in accordance with the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) guidelines [32].
Database Search
Initially, a population, intervention, comparable group, outcome and study design (PICOS) strategy was planned and followed to produce a robust research question and develop the required criteria for inclusion of eligible studies [33]. The population (P) in our study refers to human samples. The goal of this review was to measure multiple outcomes that related to diagnostic accuracy (with no condition to have all of them in the same article), thus the outcome was excluded. The included domains were the intervention group (I) indicated by the tested technique (RNAscope), the comparable group (C), which refers to the various techniques that are being used as the ‘gold standard’ techniques (immunohistochemistry (IHC), Northern blotting, microarray, qPCR, qRT-PCR and DNA ISH), and the study design (S), to include only articles that were primary scientific experimental research studies.
Literature searching for this systematic review was performed in four databases: CINAHL (Cumulative Index to Nursing and Allied Health) (EBSCOhost), Web of Science, Medline and Embase. The last conducted search was on 24 November 2020. The search was conducted in all the databases using the same key terms, Boolean operators and strategy; however, different wildcards, truncation, phrase searching and adjacency tools were used in accordance with guidelines for each database (Tables S1–S4, ESM). As RNAscope is a very recent method, it did not apply under any specific MeSH (Medical Subject Headings) term under RNAscope and RNA ISH terms.
Literature Screening
The first stage of assessing the articles available in databases was screening for the search terms in the title, abstract and keywords. The exclusion criteria were as follows: (1) studies published before 2012, (2) studies published in languages other than English, (3) the study objective was not to compare RNAscope to other technique/s, (4) the title and abstract do not include any of the included methods, (5) the intended meaning of RNA ISH was traditional RNA ISH rather than RNAscope, and (6) the samples used were non-human samples. Inclusion criteria were to retain any study that was conducted after 2012 that had full-text access that compared RNAscope to one or more of the ‘gold standard’ techniques, and the study aim was to evaluate the comparison of the methods.
Data Extraction and Quality Assessment
The results were extracted from the selected articles and tabulated under five main sections in accordance with the Cochrane Collaboration template [34]—general information, comparable techniques, methods, results and conclusions. The extracted data were collated and re-tabulated into three main tables: concordance rate and measures of diagnostic accuracy (sensitivity and specificity) of RNAscope (Table 1); the types of cancer tissues and genes that were studied (Table S6, ESM); and general information and details relating to the RNAscope methodology employed (Table S7, ESM).
Table 1.
Concordance rate and accuracy values of RNAscope
| Study no. | Ref. no. | Author and publication year | Study type | Sample size | Concordance rate (CR) | Sensitivity | Specificity | PPV | NPV | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IHC | PCR | FISH | Others | |||||||||
| 1 | [36] | Bakheet et al., 2020 | Retrospective | 194 | 86.4% | 93.2% | ||||||
| 2 | [27] | Bingham et al., 2015 | Retrospective | 127 | ||||||||
| 3 | [37] | Cioca et al., 2017 | Retrospective | 20 | ||||||||
| 4 | [38] | Coppock et al., 2018 | Retrospective | 112 | 59% | |||||||
| 5 | [26] | Craig et al., 2020 | Retrospective | 221 | 95% | 89% | 100% | |||||
| 6 | [46] | Daneshpajouhnejad et al., 2020 | Retrospective | 65 | 92.30% | 67.7% | 80% | |||||
| 7 | [47] | Dreyer et al., 2013 | Retrospective | 64 | 95.30% | 94.40% (of +ve) | 88.90% (of +ve) | |||||
| 8 | [39] | Drumheller et al., 2019 | Retrospective | 27 | 88.90% | |||||||
| 9 | [40] | Gafeer et al., 2018 | Retrospective | 20 | 60% | 75% | 92% | 27% | ||||
| 10 | [41] | Humphries et al., 2018 | Retrospective | 813 | ||||||||
| 11 | [48] | Kim et al., 2013 | Retrospective | 211 | 85.70% | 89% | 95.20% |
90.50% (SISH*) |
||||
| 12 | [42] | Kim et al., 2017 | Retrospective | 82 | 84% | |||||||
| 13 | [43] | Kim et al., 2018 | Retrospective | 101 | 66.30% | 48.4% | 97.30% | |||||
| 14 | [49] | Kulkarni et al., 2019 | Retrospective | 109 | 100% | 100% | ||||||
| 15 | [50] | Mendez-Pena et al., 2017 | Retrospective | 57 | 93% | 100% | 87% | 87% | 100% | |||
| 16 | [51] | Mills et al., 2017 | Retrospective | 127 | 97% | |||||||
| 17 | [24] | Mirghani et al., 2015 | Retrospective | 50 | 90% | 93% | 93% | 94% | 96% | |||
| 18 | [52] | Nakajima et al., 2017 | Retrospective | 555 |
91% 82% |
81.8% | 100% | 100% | ||||
| 19 | [53] | Randen-Brady et al., 2019 | Retrospective | 357 | Highly concordant | 93.40% | 92.4% | 95.5% | 89% | |||
| 20 | [54] | Roe et al., 2017 | Retrospective | 55 | 66.70% | 90.3% (traditional ISH) | EBV*: 86.4% | EBV: 100% | ||||
| CMV*: 100% | CMV: 50% | |||||||||||
| 21 | [55] | Rooper et al., 2016 | Retrospective | 82 | 100% | 100% | ||||||
| 22 | [56] | Rooper et al., 2020 | Retrospective | 320 | 92% | 89% | ||||||
| 23 | [57] | Schache et al., 2013 | Retrospective | 79 |
87.20% (qRT-PCR) |
97% | 93% | 91% | 98% | |||
| 24 | [44] | Shi et al., 2015 | Retrospective | 380 | 95% | 85% | ||||||
| 25 | [25] | Tretiakova et al., 2018 | Retrospective | 112 | 58.70% | |||||||
| 26 | [28] | Wang et al., 2013 | Retrospective | 163 | 88.90% | 97.30% | 100% | 100% | 95.20% | |||
| 27 | [45] | Yu et al., 2017 | Retrospective | 72 | 91.70% | |||||||
The study numbers (Study no.) in this table are used throughout the article to refer to these papers
FISH fluorescent in situ hybridization, IHC immunohistochemistry, NPV negative predictive value, PCR polymerase chain reaction, PPV positive predictive value, SISH silver-enhanced in situ hybridization, CMV cytomegalovirus, EBV Epstein-Barr virus
The aim of this systematic review was to assess a new diagnostic technique (RNAscope). Therefore, QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies) was selected to assess the quality and applicability of the included studies [35]. This method appraises the quality of four key criteria in the studies—sample selection, index test, reference standard, and flow and timing—by using several guiding questions in each domain. A modified version of the standard QUADAS-2 template (ESM 1) was used in this systematic review to evaluate each study. Specific cut off points were defined to assess the total risk of bias as follows: (1) the total risk was considered ‘low’ when three or four out of four domains were low risk, (2) a rating of ‘some concerns’ was given when two of the domains have a high or unclear risk of bias, and (3) overall risk was considered ‘high’ when three or four out of four domains had a high or unclear risk of bias.
Data Synthesis
Data were synthesized and the results were reviewed and analysed in a qualitative and narrative manner to answer the main research question. Meta-analysis could not be performed because of: (1) the heterogeneity of samples, comparable groups, obtained outcomes and (2) insufficient data being reported in included articles with respect to important parameters required for meta-analysis, including sensitivity, specificity, false positives, false negatives and concordance rate.
Results
Article Selection (Flow-Chart Results)
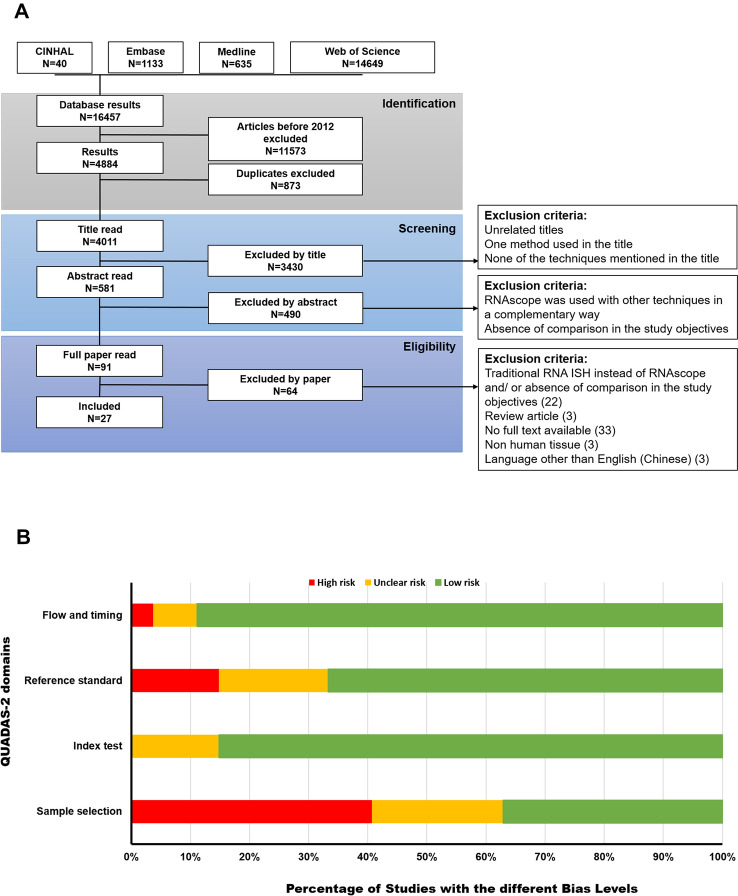
Using four different databases, a group of 16,457 articles was identified by the literature searching strategy. A total of 4,884 articles remained after the exclusion of published articles before 2012. After duplicates were removed, 4,011 titles were screened to end up with 581 articles. Based on the eligibility criteria, a further 490 articles were excluded after abstracts were screened. Finally, full-text screening led to the selection of 27 articles to be included in this systematic review (Fig. 2A).
Fig. 2.
PRISMA flow diagram and assessment of risk of bias. A The presented flow-chart outlines and summarizes the main research steps that were taken in the sequential selection of the articles included in the systematic review, including an explanation of the exclusion criteria for each step.
Adapted from PRISMA [32]. B The presented bar-chart illustrates the percentage of studies for each RoB level within each domain for the included studies as determined using QUADAS-2 tool. Green represents a low risk, yellow represents an unclear risk, and red represents a high risk
Quality Assessment—Risk of Bias (RoB)
The quality of each of the included studies was assessed using QUADAS-2 tool. Ten out of the 27 articles (37%) were classified to have some concerns in the risk of bias (RoB) assessment (Table S5). The majority of high and unclear risk of bias results were concentrated in the sample selection criteria and reference standard domains, whereas the index test and flow and timing domains had the lowest RoB assessment (Fig. 2B).
Study Characteristics
Techniques
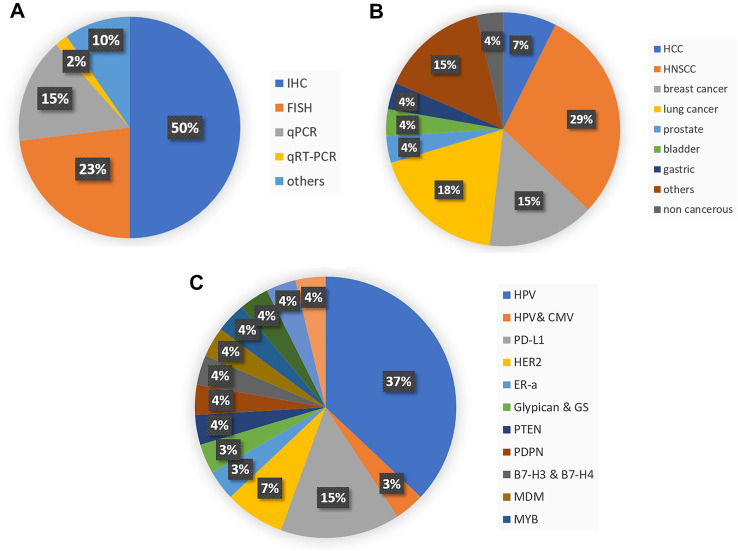
Table 1 and Fig. 3 represent the main data extracted from the selected articles. The publication dates extended from 2013 to 2020. All the selected articles compared RNAscope technique to one or more of the current gold standard techniques. The main ‘gold standard’ method compared to RNAScope was IHC, as evident by 11 articles (40.7%) that compared it to IHC only, and 15 articles (#1, 3, 4, 8–10, 12, 13, 24, 25, 27) (55.6%) that compared it to IHC and other techniques simultaneously (#2, 5–7, 11, 14–22, 26).
Fig. 3.
Evaluation of study characteristics from the 27 articles. The presented pie charts illustrate: A the percentage of studies using specified current gold standard techniques that were compared to RNAscope; B the percentages of studies using samples from specified types of cancer in the included articles; C the percentages of studies using specified markers within the included articles. AdCC adenoid cystic carcinoma, BC breast cancer, CMV cytomegalovirus, EBV Epstein-Barr virus, ERα estrogen receptor α, HNSCC head and neck squamous cell carcinoma, HCC hepatocellular carcinoma, HER-2 human epidermal growth factor receptor 2, NSCLC non-small-cell lung carcinoma, PDPN podoplanin, PTEN phosphatase and tensin homolog, PT phyllodes tumours, PD-L1 programmed death-ligand 1, SCC squamous cell carcinoma, SPARC secreted protein acidic and rich in cysteine, TTF1 Thyroid Transcription Factor 1
In addition to RNAscope, two gold standard techniques were used in seven studies, where five studies compared RNAscope to both IHC and fluorescent ISH (FISH) or DNA ISH (#6, 14, 18, 20, 22), and the other two studies compared RNAscope to both IHC and qPCR (#2, 17). Five studies compared RNAscope to three techniques; IHC, FISH and qPCR (#5, 7, 15, 16, 19), whereas one study compared RNAscope to four techniques, including IHC, qPCR, Chromogenic ISH (CISH) and dual ISH (#26). The remaining article compared RNAscope with duplicated techniques other than IHC, namely real-time DNA and qRT-PCR (#23) (Fig. 3A).
Tissues
26 out of 27 of the included studies (96.3%) used cancer tissue samples, out of which head and neck squamous cell carcinoma (HNSCC) was the most studied cancer type (in eight articles) (29.6%) (#5–8, 17, 19, 21, 23). Lung cancer was the second most common cancer type studied (in five articles) (18.5%) (#4, 9, 10, 18, 24) (Fig. 3B, C). Only one article studied CMV and EBV viruses obtained from inflammatory cases (#20). Although RNAscope is used to measure all types of RNA molecules, only mRNA was measured in all of the included articles.
Genes Studied and Biomarkers
The genes that were included in the selected articles were next scrutinised. Interestingly, ten articles (37%) focused on E6/E7 transcripts of HPV in HPV driven cancers (HNSCC, squamous cell carcinoma (SCC), and anogenital neoplasia) (#5–8, 15–17, 19, 21, 23). Several markers related to immune checkpoints, including programmed cell death ligand (PD-L1) (#4, 9, 10, 25) and B7-H3 and B7-H4 (#13), were also assessed. Another three papers investigated prognostic receptors in breast cancer, including epidermal growth factor receptor, HER-2 (#11, 26) and the nuclear hormone receptor, Erα (Estrogen receptor α) (#27). Two studies evaluated glycoproteins with prognostic values, like Podoplanin (PDPN) (#3) and glypican3 (GPC3) and glutamine synthetase (GS) (#1). Other genes studies included: the tumour suppressor genes, PTEN (phosphatase and tensin homolog) (#2) and SPARC (secreted protein acidic and rich in cysteine) (#12). The remaining four articles evaluated MDM2, anaplastic lymphoma kinase (ALK), MYB, Napsin A (the aspartic protease) and TTF1 (Thyroid Transcription Factor 1) genes, respectively (#14, 18, 22, 24) (Fig. 3C, Table S6).
Methods of Staining and Quantifications
Automated RNAscope was used in 11 studies (40.7%) (#4, 8–10, 13, 14, 16, 20, 22, 23, 26). RNAscope was conducted in a mixed way (manual and automated) in one article (3.7%) (#6). Full manual RNAscope was used in the remaining 15 articles (55.6%) (#1–3, 5, 7, 11, 12, 15, 17–19, 21, 24, 25, 27). The method for quantification was automated in four studies (#2, 10, 21, 26), mixed manual and automated in one study (#25), and manual in the remaining 21 studies. The method of quantification was unclear in one study (#7). Although only five studies used the automated scoring system, none of them used the same software. SpotStudio from ACD was the software of choice for Bingham et al. (#2); QuPath was used by Humphries et al. (#10); Rooper et al. (#21) carried the analysis out using ViewRNA program; Tretiakova et al. (#25) used web-based Spectrum Plus digital slide manager; and custom software was used by Wang et al. (#26).
RNAscope Controls
In terms of positive controls that were employed, four studies (14.8%) used UBC (#2, 16, 17, 23), while 14 studies (51.9%) used PPIB (#1, 5, 8, 10, 12, 13, 15, 18–20, 22, 24, 27). One study (3.7%) used Polr2A in tandem with UBC as the positive controls (#26). Three out of the eight remaining articles (11.1%) used appropriate controls without mentioning the probe that was used (#6, 9, 21). However, the last five articles (#3, 4, 11, 14, 25) (18.5%) did not mention the use of any positive control in their studies. With regard to negative controls, dapB was used in 16 (#1, 2, 5, 8, 10, 12, 13, 15–17, 19, 20, 23, 24, 26, 27) (59.3%) of the included articles. Appropriate controls were used in three (#6, 9, 21) (11.1%) of the 11 remaining articles, but without the mention of the specific probes. The last eight articles (#3, 4, 7, 11, 14, 18, 22, 25) (29.6%) did not mention the use of negative control in their studies. Chromogenic probes were used in 25 articles (92.6%) (#1–25), while two articles (7.4%) used the florescent probes (#26, 27) (Table S7, ESM).
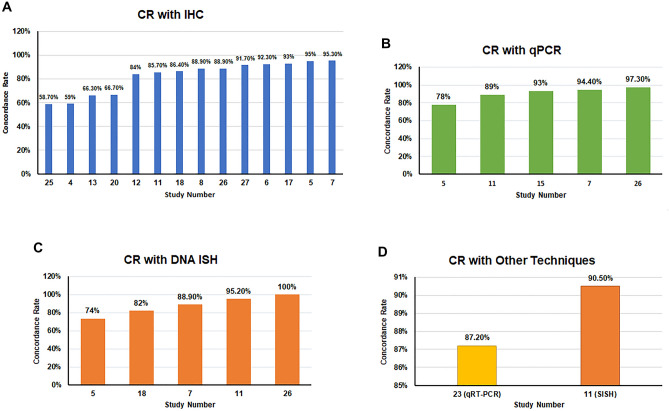
Concordance Rate (CR)
The CR was stated and extracted from 16 of the included articles (59.3%) (#4–8, 11–13, 15, 17, 18, 20, 23, 25–27). Fourteen papers (#4–8, 11–13, 17, 18, 20, 25–27) estimated CR of RNAscope with IHC, out of which IHC was co-compared to another technique in six (#7, 11, 17, 18, 20, 26) of the 14 studies. Five articles (#7, 11, 15, 17, 26) calculated CR against PCR. Although one study evaluated several techniques compared to RNAscope, the CR of RNAscope was reported against PCR method only (#15). Four of the 16 studies (#7, 11, 18, 26) calculated CR of RNAscope against DNA ISH or FISH among other techniques used in these studies. A high level of variability in CR (58.7–95.3%) was reported in the studies that compared RNAscope to IHC (Fig. 4A). However, studies that compared RNAscope to qPCR and DNA ISH demonstrated relatively close CR, within a range of 89–97.3% and 82–100%, respectively (Fig. 4B and C). The CR between RNAscope and qRT-PCR was reported only in one study as 78% (#23). Silver-enhanced in situ hybridization (SISH) technique also showed high concordance (90.5%) with RNAscope (#11) (Fig. 4D). One study reported low CR between RNAscope and IHC, but no numerical data was provided (#2). Similarly, two articles reported high CR between RNAscope and IHC without providing percentages (#2, 19).
Fig. 4.
Evaluation of the concordance rate (CR) between the results of RNAscope and IHC, qPCR, DNA ISH. The presented bar charts illustrate the CR results from: A 14 studies that compared RNAscope to IHC; B five studies that compared RNAscope to qPCR; C four studies that compared RNAscope to DNA ISH; and D two studies that compared RNAscope to other studies like qRT-PCR and SISH
The authors of the included studies where the CR between RNAscope and IHC was relatively and unexpectedly low (#4, 13, 20, 25) provided several possible reasons to explain these results. For example, Bingham et al. [29] referred to the existence of different mechanisms of gene regulation at both the transcriptional (mRNA) and posttranscriptional (protein) levels. Similarly, Kim et al. [43] referred to the possibility of inadequate translation of the required gene into protein, which might be due to impaired post-transcriptional processing of the mRNA transcript or the repression of translation initiation. In contrast, Tretiakova et al. [27] criticised the IHC technique, arguing that antibodies directed against PD-L1 require more standardization and validation.
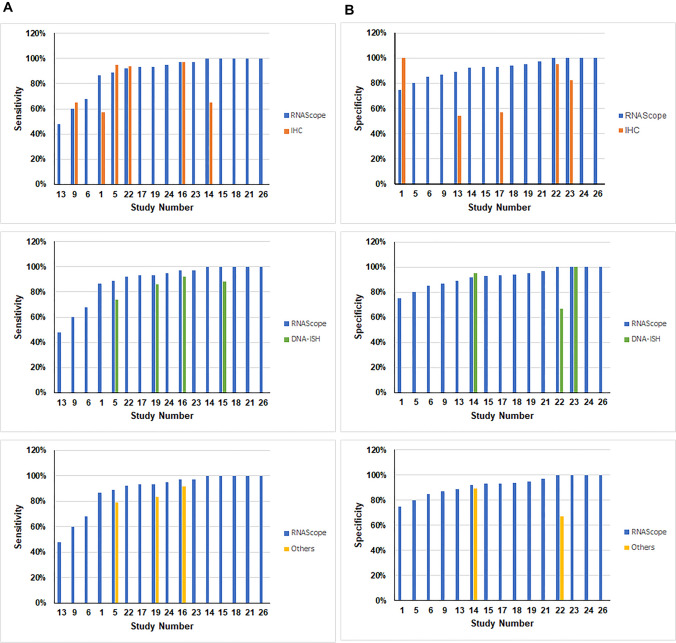
Accuracy of RNAscope (Sensitivity and Specificity)
Although determining the sensitivity and specificity of RNAscope was not considered as a primary aim within the included studies, 15 studies (55.6%) estimated both sensitivity and specificity ratios (#1, 5, 6, 9, 13–15, 17–19, 21–24, 26), and one study (3.7%) estimated only the sensitivity value of RNAscope (#16). Overall, the reported sensitivity and specificity results were relatively high in all of these studies. The sensitivity values ranged between 48 and 100%, with a median value of 94.3%, whereas the specificity ranged between 75 and 100%, with a median value of 93% (Fig. 5).
Fig. 5.
The sensitivity and specificity ratios of RNAscope versus other techniques. The presented bar charts illustrate: A sensitivity ratios for RNAscope (13 studies) and other techniques whose values were co-reported in the same studies. Top graph—IHC (co-reported in six studies); middle graph—DNA ISH (co-reported in four studies); lower graph—DNA PCR (co-reported in three studies) and B specificity ratios for RNAscope (11 studies) and other techniques whose values were co-reported in the same studies. Top graph—IHC (co-reported in five studies); middle graph—DNA ISH (co-reported in two studies); lower graph—DNA PCR (co-reported in two studies)
Eight out of these 15 articles also provided estimates of the sensitivity and specificity of the other included techniques (IHC, DNA ISH, etc.) (Table S8, ESM). The sensitivity of the RNAscope was reportedly similar or superior to the other techniques in six articles (#IHC: 1, 14, 16; DNA ISH: 5, 15, 16, 19; others: 5, 16, 19). However, IHC sensitivity outperformed RNAscope in three articles (#5, 9, 22) (Fig. 5A). The specificity of RNAscope exceeds the specificity of the other techniques in five articles (#IHC: 1, 5, 14, 22; DNA ISH: 5; others: 5, 19). On the other hand, the specificity ratios for IHC and DNA ISH were higher than RNAscope in two articles – (#9) and (#19), respectively (Fig. 5B).
Results and Clinical Outcomes/Diagnosis
The clinical utility of RNAscope was highlighted in some of the selected articles for its potential in providing accurate diagnosis and prognosis for certain conditions such as cancer and infections, particularly those that are caused by viruses. Nine articles (33.3%) recorded the relationship between RNAscope results and clinical outcomes (#1, 4–6, 9, 12, 13, 23, 25). Bakheet et al. [36] (#1) suggested that using RNAscope will improve the pathological and differential diagnoses of hepatocellular carcinoma at early stages. Coppock et al. [38] (#4) did not report any significant difference between the ability of RNAscope and IHC to predict patient survival rate (IHC predicted an average of 5.3 months; RNAscope predicted an average of 5.2 months). Craig et al. [28] (#5) demonstrated that the use of RNAscope in detecting HPV is superior to IHC as it is predicted to reduce the false positive/negative cases by almost half; although there was no difference between RNAscope and DNA ISH with respect to ability to detect the virus. In contrast, Daneshpajouhnejad et al. [46] (#6) nominated RNAscope over DNA ISH for the diagnostic process as it provides an interface with easier features for interpretation. Gafeer et al. [40] (#9) recommended using RNAscope in conjunction with IHC in the diagnostic process as it provides more accurate information to assist in determining the patients’ eligibility to receive immunotherapy. Kim et al. [43] and Kim et al. [42] (#12, 13) did not report a significant difference between RNAscope and IHC in predicting the disease recurrence rate as this was found to be almost the same for positive cases using both techniques. Similarly, Schache et al. [57] (#23) found qRT-PCR and RNAscope to be equally good with regards to predicting and discriminating patient survival rate as both techniques predicted the same survival rate for the patients. In contrast, the ability of RNAscope to predict patient survival rate in comparison to IHC was found to be unclear in the study by Tretiakova et al. [27] (#25).
RNAscope Advantages and Disadvantages
The included articles contained consideration of advantages and disadvantages for using RNAscope compared to the other gold standard techniques, and these are summarised in Table 2. The main advantage was that RNAscope was considered as an innovative technique demonstrating a high degree of accuracy and ability to detect any gene in a short time frame. However, the major disadvantage was cost because RNAscope is an expensive technique compared to IHC.
Table 2.
Advantages and disadvantages of RNAscope technique
| Factor | Study no.a |
|---|---|
| Advantages | |
| Identify gene expression at a single-cell level within a morphological context | 13, 18 |
| Does not depend on antibodies | 13 |
| Allows the detection of mRNA as a single gene copy | 12, 20 |
| High analytical accuracy, sensitivity and specificity | 1, 4, 7–9, 11,15, 17, 19–24 |
| More reliable than IHC | 3 |
| Suppress background noise and produce better resolution than IHC | 8, 15, 17, 20, 27 |
| Reduce the risk of false-positive results | 17 |
| Its results are easy to interpret | 5, 6, 8, 15, 17, 21 |
| It is a robust and quantitative technique | 11, 16, 27 |
| It can detect tissue heterogeneity and partially degraded RNA | 2, 27 |
| Quick to perform | 9, 11, 18 |
| It can be performed automatically and manually and saves time | 1, 14 |
| Disadvantages | |
| It is not suitable to discriminate between viral RNA transcripts and viral DNA | 7 |
| The stain will not take place well if the samples are with poor fixation quality and the cost is much higher compared to IHC | 11 |
| In cervical intraepithelial neoplasia (CIN) cases, the negativity of RNAscope does not guarantee the absence of HR-HPV | 16 |
| RNAscope was less specific differentiating AdCC from high grade basaloid sinonasal tumors | 22 |
| In the automated system, some areas in the slides need manual selection during the scoring process | 26 |
aThe study numbers (Study no.) in this table are used throughout the article to refer to the papers. See Table 1 for references
Discussion
According to the authors’ knowledge, this is the first systematic review to be conducted on the RNAscope technique comparing its use against gold-standard diagnostic methods. RNAscope is a relatively new technique, and while there are more than 500 papers that cited RNAscope [20], only 27 articles were identified that focused on evaluating RNAscope in comparison to current gold standard techniques that measure gene expression levels or detect genes.
Quality Assessment
The quality assessment findings demonstrated that all the included articles had an overall score of low or some concerns for RoB, demonstrating that this systematic review provides high quality data. However, it should be noted that personal bias can still be introduced [58].
The most affected domains with respect to RoB were sample selection and reference standard. The high scores in these domains were attributed to ambiguity related to sample selection (random vs. consecutive) because most of the samples were retrieved from biobanks or storage. Biobank samples might not be selected randomly or consecutively in the first place, which allows for potential bias. Articles also scored high or unclear RoB for the reference standard domain because RNAscope is still considered as a recent technique, for which a reference standard is yet to be developed. Each of the included articles used a different gold standard technique, but with variations in the procedures (reagents, conditions and scoring methods) that were employed. For this technique to be adopted into the clinical diagnostic field, a reliable and accurate reference standard for RNAscope should be designed so that the test accuracy can be normalised. Furthermore, producing a reliable and accurate reference standard requires careful consideration of the staining and quantification of elements of RNAscope, a process that involves two separate procedures that should be evaluated separately.
Although the possibility of bias can be reduced when automated systems are employed in either the staining or the scoring stages of RNAscope assays [59], less than half of the included studies in this systematic review utilized automated systems. This might be because the goal of the included studies was to validate RNAscope, for which a manual method was used in order to fully assess all stages of the technique. However, during the full-text screening process for this systematic review, many of the recently conducted articles that have used RNAscope as an experimental method in scientific research were noted to use either partial or fully automated systems. In the manual scoring method, no special training is required to produce reliable results [7]. However, manual scoring is a time-consuming method and at least two pathologists are needed validate the results [13, 25]. Automatic scoring methods are recommended over manual scoring, although the produced results will be robust either way if the proper guidelines are followed.
The quality of RNAscope was assessed through most of the included articles using positive and negative controls. Throughout this review, 14 studies used PPIB as a positive control, indicating that target mRNAs being studied were products of low and moderately expressed genes. Being able to accurately detect and monitor small changes in gene expression, including genes that are low in abundance, is critical with respect to the diagnosis and management of cancer and certain infectious diseases [2]. The high degree of specificity and sensitivity that is offered by RNAscope is highly advantageous in this regard. Furthermore, the relatively high CR for RNAscope with IHC for genes that are expressed at low levels including HER-2 gene emphasises the effectiveness of RNAscope for detecting such genes.
Study Characteristics
Cancer development is caused by changes in gene expression that lead to uncontrolled and inappropriate cell growth [60]. It is not surprising, therefore, that almost all of the included articles focused on cancer [61]. A variety of cancer types were covered in the included studies—the majority of articles concentrated on HNSCC, followed by the most common cancers worldwide according to the World Health Organization (WHO) (2020 record), namely lung and breast cancers [62].
The E6/E7 transcripts of HPV have a critical role in the development of cancer as they can transform cells and they have the ability to deregulate important tumour suppressor genes, including p53, Rb and others, which leads to uncontrolled cell proliferation and induction of oncogenesis [63]. E6/E7 was identified as the most investigated target in the selected articles in this systematic review, as evident in ten articles (#5–8, 15–17, 19, 21, 23) (37%). Although HPV is established to contribute to the development of cervical cancer (in 90% of cases) [64], through this systematic review, HPV was tested in the context of cervical cancer in only one of the included articles versus nine that were in the context of HNSCC. In light of this observation, it is noteworthy that there is an increasing body of evidence for HPV infection as a risk factor for HNSCC [65].
One of the main hallmarks of cancer is the evasion of the immune system [66]. Cancer can bypass the immune system by modulating key immune markers that are vital in suppressing the host anti-tumour response, in which effector T cells play a vital role [67]. Programmed cell death protein (PD-1) and B7-H3 are key examples of such markers, and it is of great interest that they were studied in some of the included articles (#4, 9, 10, 13, 25). The presence of PD-1 and B7-H3 are co-inhibitory to effector T-cell function—upon the interaction of effector T cells with tumour cells, the presence of PD-1 and B7-H3 on the surface of tumour cells results in inactivation, tolerance and anergy of the effector T cells, leading to uncontrolled cell growth in the cancer. The identification of such markers has allowed for the development of targeted therapies against these checkpoints, and immune checkpoint inhibition treatment has proven to be successful in various cancers [61]. However, treatment success is highly dependent upon investigation of the expression of these markers in patients to determine those who are most likely to respond to immune checkpoint inhibition. RNAscope could be a highly valuable tool in this diagnostic process.
Concordance Rate (CR)
The CRs between RNAscope and IHC varied considerably across the included articles. The main reason for lack of concordance was attributed to the difference between RNA and protein content (#4, 13, 20, 25). Furthermore, Yu et al. [45] highlighted that protein content, but not RNA, might change due to gene mutations. In their study of 62 genes in eight cancer types, Jia and Zhongming [68] drew attention to post-translational processes such as phosphorylation and glycosylation, which can affect protein, but not RNA expression. However, the fact that proteins are produced from raw RNA molecules (with coding and non-coding sequences) that could be translated differently into several proteins [69] explains the relatively low CR between RNAscope and IHC.
Interestingly, the CR between RNAscope and qPCR or qRT-PCR (presented in six studies) was relatively high (89–97.3%) compared to IHC (calculated in 14 studies). This is likely because RNAscope, qPCR, and qRT-PCR measure the same molecule, RNA. Further studies are required to fully evaluate RNAscope CR with IHC, qPCR and qRT-PCR, but interestingly, it was proposed by Bingham et al. [29] that combining RNAscope with IHC methods might produce more robust results than using either technique alone, resulting in greater accuracy. This notion is supported by Kang et al. [70]. DNA ISH has also demonstrated a strong CR (> 82%) with RNAscope, which is not unexpected as RNA is directly transcribed from DNA and only a small amount of data might be lost [69].
The Accuracy of RNAscope (Sensitivity and specificity)
Regardless of the sample size, most sensitivity and specificity ratios reported in this systematic review were relatively high, which is in keeping with other reports in the literature [3, 10] and suggests that RNAscope is a robust technique that would be suitable for the diagnostic field. Three articles that compared RNAscope to IHC reported relatively low sensitivity levels, but only Kim et al. [43] indicated the reasons for the observed low sensitivity of RNAscope (48.4%) compared to IHC (51.6%) might be due to: (1) the increased translation rate of mRNA to protein molecules, (2) a decrease in the elimination of activator proteins in the tissue, which function to increase gene transcription [71], and (3) small sample size introducing bias in the results.
The studies that reported a large difference in sensitivity of RNAscope compared to IHC (#1, 14) highlight the importance of using techniques that measure the same molecule (RNA) as a reference standard to validate RNAscope as opposed to techniques that measure a different molecule, for instance IHC, which measures protein. Since 2009, several techniques have been developed to measure RNA molecules. Single-cell RNA sequencing (sscRNA-seq) is the most notable of these—it has many of the advantages of RNAscope and is approved for clinical diagnostics [72]. However, none of the studies identified in the research process of this systematic review included sscRNA-seq as a comparative technique to RNAscope. Also, a gene expression profiling (GEP) method that utilises either microarray or sequencing technologies is used to show the pattern of the expressed genes by measuring mRNA levels [73, 74]. In 2013, Handorf et al. [75] compared the accuracy of IHC to GEP method, and it was around 71% compared to 91%, respectively.
Although the reported sensitivity and specificity values for RNAscope are high in included articles in this systematic review, there is insufficient data in the included studies to fully evaluate the suitability of RNAscope as an independent test—further appraisal of accuracy values (sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV)) for RNAscope as part of prospective studies is clearly needed before it can be adopted as a stand-alone test in the clinical diagnostic field. An important consideration relates to the level of expression of the gene being detected, for example HPV E6/E7 are highly expressed genes and so it is not surprising that RNAscope recorded high specificity results for their detection [76]. In order to fully evaluate whether RNAscope has superior specificity compared to the gold standard techniques, more studies on RNAscope focusing on genes with low expression levels should be conducted. Furthermore, it would be important to evaluate RNAscope effectiveness within low-risk populations as all the included articles in the systematic review relate to high-risk populations and sub-populations within these.
Comparing techniques that measure the same variable is recommended as part of the validation process for adoption of new techniques into the clinical diagnostic setting [77]. Furthermore, it is important to consider other variables in addition to CR and accuracy parameters as part of the validation process of a new technique, including benefit to the patient and test management.
Results and Clinical Outcomes
Within the included articles, ten studies reported the correlation between RNAscope results and clinical outcomes. Five studies (#4, 12, 13, 18, 23) out of the ten did not report any difference on the recurrence or survival rates. Two of the remaining three articles (#5, 9) recommend using RNAscope with IHC for better diagnosis and prediction of patients’ eligibility for treatment with immunotherapy. One study (#25) indicated that there was no clear correlation between using RNAscope and the survival rate. The last two studies (#6, 26) nominated a preference for using RNAscope over the other techniques for better diagnosis and evaluation of prognosis. This was indicated by Wang and his colleagues [78] in 2014 where they have measured HPV E6/E7 gene expression levels using RNAscope to predict the status of oropharyngeal SCC. They found that RNAscope was a good predictive method. On the basis of data and evidence in the articles included in this systematic review, the effectiveness of RNAscope as a prognostic tool remains to be fully determined and further studies are required to confirm this relationship.
Regulations
When introducing a new technique to the clinical diagnostic system, a long process of several steps should be followed to validate the technique analytically and clinically in addition to consideration of the ethical, legal and social implications of the test [77]. As part of the validation process, it should be considered as a quantitative test, and thus its trueness and robustness should be evaluated. RNAscope trueness should be measured correctly to evaluate the predicted bias either systematically or proportionally, and trueness should be adjusted using the appropriate correction factors. The robustness, which is represented by the precision of the quantitative test, should also be validated within either the run, the laboratory itself, or among several laboratories [79]. Adhering to these steps is critical to validate RNAscope and thus intercalate it into the clinical diagnostic field.
Costs
Cost-effectiveness is one of the most important aspects to consider with regards to implementing a new technique for the diagnostic system. Given the high cost of RNAscope, an incremental cost approach should be adopted as part of RNAscope cost-effectiveness analysis (CEA), which considers how cost relates to the potential outcome [80]. The incremental approach would consider how the direct costs (reagents (probes, staining kits, etc.) and equipment (e.g., specific machines and required software programs to analyse and quantify the data)) and other related costs (staff (including their training and recruitment) and building costs) would change if RNAscope were to be adopted. In the case of adopting RNAscope as a new technique, the outcome of any CEA study would recommend whether the benefits of implementing RNAscope would outweigh the costs of its introduction.
Limitations of this Systematic Review
This systematic review only included 27 studies that met the critera. The included studies did not all have the same objective. For example, some studies compared RNAscope to IHC alone, while some compared it to more than one different technique. Also, some articles involved more than one research question, which introduces bias during the data extraction process. Not all the included articles reported important parameters relating to test accuracy (sensitivity, specificity, PPV, NPV, concordance), which prevented meta-analysis and statistics from being performed and so it is difficult to draw firm conclusions. Ideally, the grey literature should be searched to avoid any source of bias in the results synthesis process. Furthermore, the bibliographies of relevant reviews should be searched manually. Due to time constraints, neither type of these searches was performed.
Another limitation of the current systematic review relates to publication bias, which might have been introduced throughout the search strategy, data extraction, quality assessment or data analysis procedures [81]. With respect to the selection criteria, included studies were restricted to full text articles that were available in the English language, which might have excluded articles in languages other than English as well as unpublished data in relevant topics. However, several procedures were followed throughout the multiple steps that were performed in generating this systematic review to avoid bias. For instance, the literature search was conducted on a large scale by using four different databases. Additionally, the screeding and selection criteria were comprehensive as they included all possible groups. Furthermore, the quality of the included articles was assessed using the QUADAS-2 tool to ensure the high quality and accuracy of the included articles. One limitation relating to the quality assessment tool (QUADAS-2) that might have introduced some bias is that RNAscope is still a new technique, and thus, no reference standard was available at the time this systematic review was conducted. For this reason, a new criterion was produced in the QUADAS-2 tool to avoid having many articles with a high RoB in the second domain (index test).
Conclusion
In conclusion, this systematic review is considered the first conducted review in this field. All the included articles focused on validating RNAscope against (an)other technique(s). This review demonstrated that RNAscope is a promising and accurate technique, as exemplified by high degrees of sensitivity and specificity. The authors of this study would recommend the adoption of RNAscope as a complementary method in the clinical field to confirm unclear results from other techniques, as it offers advantages and solutions for the current challenges of gold standard techniques. However, further studies comparing RNAscope to scRNA-seq and qRT-PCR techniques are needed to fully assess the effectiveness of RNAscope so it can be incorporated as a fully independent method to diagnose gene expression disorders. To validate the high specificity of RNAscope, more studies concentrating on low expressed genes should be conducted. Furthermore, diagnostic accuracy values of the technique should be evaluated in prospective studies to obtain firmer conclusions regarding sensitivity and specificity ratios. Finally, many validation steps (analytically, clinically and logistically) are needed to assess RNAscope more broadly and profoundly, and thus to apply it to the clinical diagnostic field.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The team of authors acknowledge Dr. Shameq Sayeed and Dr Tomoko Iwata for their helpful discussions and for commenting on the manuscript.
Declarations
Funding
No funding was received for the research or for producing the article.
Conflict of interest
The authors declare no conflicts of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material (data transparency)
All the data are available from the corresponding author upon request.
Code availability
Not applicable.
Authors' contributions
Sameeha Atout conceived and designed the analysis, collected the data, contributed data and analysis tools, performed the analysis, wrote the paper. Shaymaa Shuraab collected the data, contributed data and analysis tools, performed the analysis, reviewed and edited the manuscript. Carolyn Loveridge,the senior author who supervised the whole research project, participated in developing the idea, and reviewed and edited the manuscript.
References
- 1.Enguita FJ. New promising circulating RNA biomarkers for early diagnosis of lung adenocarcinoma. Ann Transl Med. 2019;3:S130. doi: 10.21037/atm.2019.05.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandvik AK, Alsberg BK, Nørsett KG, Yadetie F, Waldum HL, Lægreid A. Gene expression analysis and clinical diagnosis. Clin Chim Acta. 2006;363(1):157–164. doi: 10.1016/j.cccn.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 3.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14(1):22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baena-Del Valle JA, Zheng Q, Hicks JL, Fedor H, Trock BJ, Morrissey C, et al. Rapid loss of RNA detection by in situ hybridization in stored tissue blocks and preservation by cold storage of unstained slides. Am J Clin Pathol. 2017;148(5):398–415. doi: 10.1093/ajcp/aqx094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136(4):763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 6.Hittu M, Steenwinkel T, Dion W, Ostlund N, Raja K, Werner T. RNA in situ hybridization for detecting gene expression patterns in the abdomens and wings of drosophila species. Methods Protocols. 2021;4:1. doi: 10.3390/mps4010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaspar I, Ephrussi A. Strength in numbers: quantitative single-molecule RNA detection assays. Wiley Interdiscip Rev Dev Biol. 2015;4(2):135–150. doi: 10.1002/wdev.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erben L, Buonanno A. Detection and quantification of multiple RNA sequences using emerging ultrasensitive fluorescent in situ hybridization techniques. Curr Protoc Neurosci. 2019;87(1):e63. doi: 10.1002/cpns.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baumgart E, Schad A, Grabenbauer M (2001) In situ hybridization: general principles and application of digoxigenin-labeled cRNA for the detection of mRNAs. In: Beesley JE (eds) Immunocytochemistry and in situ hybridization in the biomedical sciences. Birkhäuser, Boston, MA. 10.1007/978-1-4612-0139-7_6
- 10.Chan S, Filéza-de-L'Etang A, Rangell L, Caplazi P, Lowe JB, Romeo V. A method for manual and automated multiplex RNAscope in situ hybridization and immunocytochemistry on cytospin samples. PLoS ONE. 2018;13(11):e0207619. doi: 10.1371/journal.pone.0207619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canene-Adams K. Preparation of formalin-fixed paraffin-embedded tissue for immunohistochemistry. Methods Enzymol. 2013;533:225–233. doi: 10.1016/b978-0-12-420067-8.00015-5. [DOI] [PubMed] [Google Scholar]
- 12.Anderson CM, Zhang B, Miller M, Butko E, Wu X, Laver T, et al. Fully automated rnascope in situ hybridization assays for formalin-fixed paraffin-embedded cells and tissues. J Cell Biochem. 2016;117(10):2201–2208. doi: 10.1002/jcb.25606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hongwei W. Multiplex fluorescent RNA in situ hybridization via RNAscope. In: Nan S, Hauptmann G, editors. In situ hybridization methods, neuromethods. New York: Humana Press; 2015. pp. 405–414. [Google Scholar]
- 14.Diagnostics AC. Control slides and control probes to assess technique, sample quality and RNA quality. 2021. https://acdbio.com/control-slides-and-control-probes-rnascope. Accessed 5 Jun 2021.
- 15.Acs B, Pelekanou V, Bai Y, Martinez-Morilla S, Toki M, Leung SCY, et al. Ki67 reproducibility using digital image analysis: an inter-platform and inter-operator study. Lab Invest. 2019;99(1):107–117. doi: 10.1038/s41374-018-0123-7. [DOI] [PubMed] [Google Scholar]
- 16.Holzer TR, Hanson JC, Wray EM, Bailey JA, Kennedy KR, Finnegan PR, et al. Cross-platform comparison of computer-assisted image analysis quantification of in situ mRNA hybridization in investigative pathology. Appl Immunohistochem Mol Morphol. 2019;27(1):15–26. doi: 10.1097/pai.0000000000000542. [DOI] [PubMed] [Google Scholar]
- 17.Halo: Halo (Image analysis platform). 2021. https://indicalab.com/?page_id=2637%20. Accessed 5 Jun 2021.
- 18.Diagnostics AC: RNAscope™ fluorescent multiplex assay. 2021. https://acdbio.com/rnascope-fluorescent-multiplex-assay. Accessed 20 Jun 2021.
- 19.Dikshit A, Zong H, Anderson C, Zhang B, Ma XJ. Simultaneous visualization of rna and protein expression in tissue using a combined RNAscope™ in situ hybridization and immunofluorescence protocol. Methods Mol Biol. 2020;2148:301–312. doi: 10.1007/978-1-0716-0623-0_19. [DOI] [PubMed] [Google Scholar]
- 20.Kersigo J, Pan N, Lederman JD, Chatterjee S, Abel T, Pavlinkova G, et al. A RNAscope whole mount approach that can be combined with immunofluorescence to quantify differential distribution of mRNA. Cell Tissue Res. 2018;374(2):251–262. doi: 10.1007/s00441-018-2864-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazzei M, Vascellari M, Zanardello C, Melchiotti E, Vannini S, Forzan M, et al. Quantitative real time polymerase chain reaction (qRT-PCR) and RNAscope in situ hybridization (RNA-ISH) as effective tools to diagnose feline herpesvirus-1-associated dermatitis. Vet Dermatol. 2019;30(6):491–e147. doi: 10.1111/vde.12787. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Babka AM, Kearney BJ, Radoshitzky SR, Kuhn JH, Zeng X. Molecular detection of SARS-CoV-2 in formalin fixed paraffin embedded specimens. bioRxiv. 2020 doi: 10.1101/2020.04.21.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravindranathan A, Cimini B, Diolaiti ME, Stohr BA. Preliminary development of an assay for detection of TERT expression, telomere length, and telomere elongation in single cells. PLoS ONE. 2018;13(12):e0206525. doi: 10.1371/journal.pone.0206525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neau L, Lorin C, Frentzel S, Hoeng J, Iskandar A, Leroy P, et al. Optimization of a novel in situ hybridization technology on 3D organotypic cell cultures. Appl In Vitro Toxicol. 2019;5(2):75–85. doi: 10.1089/aivt.2018.0021. [DOI] [Google Scholar]
- 25.Evans MF, Vacek PM, Sprague BL, Stein GS, Stein JL, Weaver DL. Microarray and RNA in situ hybridization assay for recurrence risk markers of breast carcinoma and ductal carcinoma in situ: Evidence supporting the use of diverse pathways panels. J Cell Biochem. 2020;121(2):1736–1746. doi: 10.1002/jcb.29409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirghani H, Casiraghi O, Amen F, He M, Ma XJ, Saulnier P, et al. Diagnosis of HPV-driven head and neck cancer with a single test in routine clinical practice. Mod Pathol. 2015;28(12):1518–1527. doi: 10.1038/modpathol.2015.113. [DOI] [PubMed] [Google Scholar]
- 27.Tretiakova M, Fulton R, Kocherginsky M, Long T, Ussakli C, Antic T, et al. Concordance study of PD-L1 expression in primary and metastatic bladder carcinomas: comparison of four commonly used antibodies and RNA expression. Mod Pathol. 2018;31(4):623–632. doi: 10.1038/modpathol.2017.188. [DOI] [PubMed] [Google Scholar]
- 28.Craig SG, Anderson LA, Moran M, Graham L, Currie K, Rooney K, et al. Comparison of molecular assays for HPV testing in oropharyngeal squamous cell carcinomas: a population-based study in Northern Ireland. Cancer Epidemiol Biomarkers Prev. 2020;29(1):31–38. doi: 10.1158/1055-9965.epi-19-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bingham V, Ong CW, James J, Maxwell P, Waugh D, Salto-Tellez M, et al. PTEN mRNA detection by chromogenic, RNA in situ technologies: a reliable alternative to PTEN immunohistochemistry. Hum Pathol. 2016;47(1):95–103. doi: 10.1016/j.humpath.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Portier BP, Gruver AM, Bui S, Wang H, Su N, et al. Automated quantitative RNA in situ hybridization for resolution of equivocal and heterogeneous ERBB2 (HER2) status in invasive breast carcinoma. J Mol Diagn. 2013;15(2):210–219. doi: 10.1016/j.jmoldx.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 31.(UCL) UCoL: Price list. 2021. https://www.ucl.ac.uk/ion/ucl-iqpath/price-list. Accessed 5 Jun 2021.
- 32.Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4(1):1. doi: 10.1186/2046-4053-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luijendijk HJ. How to create PICO questions about diagnostic tests. BMJ Evid-Based Med. 2021;26(4):155. doi: 10.1136/bmjebm-2021-111676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Care CEPaOo: EPOC resources for review authors. 2021. https://epoc.cochrane.org/resources/epoc-resources-review-authors. Accessed 5 Jun 2021.
- 35.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 36.Bakheet AMH, Zhao C, Chen JN, Zhang JY, Huang JT, Du Y, et al. Improving pathological early diagnosis and differential biomarker value for hepatocellular carcinoma via RNAscope technology. Hepatol Int. 2020;14(1):96–104. doi: 10.1007/s12072-019-10006-z. [DOI] [PubMed] [Google Scholar]
- 37.Cioca A, Cimpean AM, Ceausu RA, Tarlui V, Toma A, Marin I, et al. Evaluation of podoplanin expression in hepatocellular carcinoma using RNAscope and immunohistochemistry—a preliminary report. Cancer Genomics Proteom. 2017;14(5):383–387. doi: 10.21873/cgp.20048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coppock JD, Volaric AK, Mills AM, Gru AA. Concordance levels of PD-L1 expression by immunohistochemistry, mRNA in situ hybridization, and outcome in lung carcinomas. Hum Pathol. 2018;82:282–288. doi: 10.1016/j.humpath.2018.07.025. [DOI] [PubMed] [Google Scholar]
- 39.Drumheller B, Cohen C, Lawson D, Siddiqui MT. Automated RNA in situ hybridization for 18 high risk human papilloma viruses in squamous cell carcinoma of the head and neck: comparison with p16 immunohistochemistry. Appl Immunohistochem Mol Morphol. 2019;27(2):160–164. doi: 10.1097/pai.0000000000000550. [DOI] [PubMed] [Google Scholar]
- 40.Gafeer MM, Hosny Mohammed K, Ormenisan-Gherasim C, Choudhary F, Siddiqui MT, Cohen C. Diagnostic utility of PD-L1 expression in lung adenocarcinoma: immunohistochemistry and RNA in situ hybridization. Appl Immunohistochem Mol Morphol. 2018;26(8):e86–e90. doi: 10.1097/pai.0000000000000595. [DOI] [PubMed] [Google Scholar]
- 41.Humphries MP, McQuaid S, Craig SG, Bingham V, Maxwell P, Maurya M, et al. Critical Appraisal of programmed death ligand 1 reflex diagnostic testing: current standards and future opportunities. J Thorac Oncol. 2019;14(1):45–53. doi: 10.1016/j.jtho.2018.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim NI, Kim GE, Lee JS, Park MH. In phyllodes tumors of the breast expression of SPARC (osteonectin/BM40) mRNA by in situ hybridization correlates with protein expression by immunohistochemistry and is associated with tumor progression. Virchows Arch. 2017;470(1):91–98. doi: 10.1007/s00428-016-2048-0. [DOI] [PubMed] [Google Scholar]
- 43.Kim GE, Kim NI, Park MH, Lee JS. B7–H3 and B7–H4 expression in phyllodes tumors of the breast detected by RNA in situ hybridization and immunohistochemistry: association with clinicopathological features and T-cell infiltration. Tumour Biol. 2018;40(11):1010428318815032. doi: 10.1177/1010428318815032. [DOI] [PubMed] [Google Scholar]
- 44.Shi J, Liu H, Ma XJ, Chen Z, He MX, Luo Y, et al. Ribonucleic acid in situ hybridization is a more sensitive method than immunohistochemistry in detection of thyroid transcription factor 1 and napsin A expression in lung adenocarcinomas. Arch Pathol Lab Med. 2016;140(4):332–340. doi: 10.5858/arpa.2014-0644-OA. [DOI] [PubMed] [Google Scholar]
- 45.Yu X, Guo S, Song W, Xiang T, Yang C, Tao K, et al. Estrogen receptor α (ERα) status evaluation using RNAscope in situ hybridization: a reliable and complementary method for IHC in breast cancer tissues. Hum Pathol. 2017;61:121–129. doi: 10.1016/j.humpath.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Daneshpajouhnejad P, Miller JA, Maleki Z. Diagnostic utility of high-risk human papillomavirus mRNA in situ hybridisation in squamous cell carcinoma of unknown primary in the head and neck and implementing American Society of Clinical Oncology guideline recommendations. Cytopathology. 2020;31(6):547–554. doi: 10.1111/cyt.12896. [DOI] [PubMed] [Google Scholar]
- 47.Dreyer JH, Hauck F, Oliveira-Silva M, Barros MH, Niedobitek G. Detection of HPV infection in head and neck squamous cell carcinoma: a practical proposal. Virchows Arch. 2013;462(4):381–389. doi: 10.1007/s00428-013-1393-5. [DOI] [PubMed] [Google Scholar]
- 48.Kim MA, Jung JE, Lee HE, Yang H-K, Kim WH. In situ analysis of HER2 mRNA in gastric carcinoma: comparison with fluorescence in situ hybridization, dual-color silver in situ hybridization, and immunohistochemistry. Hum Pathol. 2013;44(4):487–494. doi: 10.1016/j.humpath.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 49.Kulkarni AS, Wojcik JB, Chougule A, Arora K, Chittampalli Y, Kurzawa P, et al. MDM2 RNA in situ hybridization for the diagnosis of atypical lipomatous tumor: a study evaluating DNA, RNA, and protein expression. Am J Surg Pathol. 2019;43(4):446–454. doi: 10.1097/pas.0000000000001199. [DOI] [PubMed] [Google Scholar]
- 50.Mendez-Pena JE, Sadow PM, Nose V, Hoang MP. RNA chromogenic in situ hybridization assay with clinical automated platform is a sensitive method in detecting high-risk human papillomavirus in squamous cell carcinoma. Hum Pathol. 2017;63:184–189. doi: 10.1016/j.humpath.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 51.Mills AM, Dirks DC, Poulter MD, Mills SE, Stoler MH. HR-HPV E6/E7 mRNA in situ hybridization: validation against PCR, DNA In situ hybridization, and p16 immunohistochemistry in 102 samples of cervical, vulvar, anal, and head and neck neoplasia. Am J Surg Pathol. 2017;41(5):607–615. doi: 10.1097/pas.0000000000000800. [DOI] [PubMed] [Google Scholar]
- 52.Nakajima N, Yoshizawa A, Kondo K, Rokutan-Kurata M, Hirata M, Furuhata A, et al. Evaluating the effectiveness of RNA in-situ hybridization for detecting lung adenocarcinoma with anaplastic lymphoma kinase rearrangement. Histopathology. 2017;71(1):143–149. doi: 10.1111/his.13198. [DOI] [PubMed] [Google Scholar]
- 53.Randén-Brady R, Carpén T, Jouhi L, Syrjänen S, Haglund C, Tarkkanen J, et al. In situ hybridization for high-risk HPV E6/E7 mRNA is a superior method for detecting transcriptionally active HPV in oropharyngeal cancer. Hum Pathol. 2019;90:97–105. doi: 10.1016/j.humpath.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 54.Roe CJ, Siddiqui MT, Lawson D, Cohen C. RNA in situ hybridization for epstein-barr virus and cytomegalovirus: comparison with in situ hybridization and immunohistochemistry. Appl Immunohistochem Mol Morphol. 2019;27(2):155–159. doi: 10.1097/pai.0000000000000568. [DOI] [PubMed] [Google Scholar]
- 55.Rooper LM, Gandhi M, Bishop JA, Westra WH. RNA in-situ hybridization is a practical and effective method for determining HPV status of oropharyngeal squamous cell carcinoma including discordant cases that are p16 positive by immunohistochemistry, but HPV negative by DNA in-situ hybridization. Oral Oncol. 2016;55:11–16. doi: 10.1016/j.oraloncology.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 56.Rooper LM, Lombardo KA, Oliai BR, Ha PK, Bishop JA. MYB RNA in situ hybridization facilitates sensitive and specific diagnosis of adenoid cystic carcinoma regardless of translocation status. Am J Surg Pathol. 2021;45(4):488–497. doi: 10.1097/pas.0000000000001616. [DOI] [PubMed] [Google Scholar]
- 57.Schache AG, Liloglou T, Risk JM, Jones TM, Ma XJ, Wang H, et al. Validation of a novel diagnostic standard in HPV-positive oropharyngeal squamous cell carcinoma. Br J Cancer. 2013;108(6):1332–1339. doi: 10.1038/bjc.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simundić AM. Bias in research. Biochem Med (Zagreb) 2013;23(1):12–15. doi: 10.11613/bm.2013.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maynard KR, Tippani M, Takahashi Y, Phan BN, Hyde TM, Jaffe AE, et al. dotdotdot: an automated approach to quantify multiplex single molecule fluorescent in situ hybridization (smFISH) images in complex tissues. Nucleic Acids Res. 2020;48(11):e66. doi: 10.1093/nar/gkaa312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Graham TA, Sottoriva A. Measuring cancer evolution from the genome. J Pathol. 2017;241(2):183–191. doi: 10.1002/path.4821. [DOI] [PubMed] [Google Scholar]
- 61.Goossens N, Nakagawa S, Sun X, Hoshida Y. Cancer biomarker discovery and validation. Transl Cancer Res. 2015;4(3):256–269. doi: 10.3978/j.issn.2218-676X.2015.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.(WHO) WHO: Cancer. 2021. https://www.who.int/news-room/fact-sheets/detail/cancer. Accessed 3 Mar 2021.
- 63.Pal A, Kundu R. Human papillomavirus E6 and E7: the cervical cancer hallmarks and targets for therapy. Front Microbiol. 2020;2020:10. doi: 10.3389/fmicb.2019.03116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arbyn M, Weiderpass E, Bruni L, de Sanjosé S, Saraiya M, Ferlay J, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health. 2020;8(2):e191–e203. doi: 10.1016/s2214-109x(19)30482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aggarwal N, Yadav J, Thakur K, Bibban R, Chhokar A, Tripathi T, et al. Human papillomavirus infection in head and neck squamous cell carcinomas: transcriptional triggers and changed disease patterns. Front Cell Infect Microbiol. 2020;2020:10. doi: 10.3389/fcimb.2020.537650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21(3):309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 67.Rodríguez P, Zea A, Ochoa A. Mechanisms of tumor evasion from the immune response. Cancer Chemother Biol Response Modif. 2003;21:351–364. doi: 10.1016/S0921-4410(03)21018-8. [DOI] [PubMed] [Google Scholar]
- 68.Jia P, Zhao Z. Impacts of somatic mutations on gene expression: an association perspective. Brief Bioinform. 2017;18(3):413–425. doi: 10.1093/bib/bbw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoerter JE, Ellis SR. Biochemistry, protein synthesis. StatPearls. Treasure Island (FL): StatPearls Publishing. Copyright © 2021, StatPearls Publishing LLC.; 2021.
- 70.Kang L, Liu A, Tian L. Linear combination methods to improve diagnostic/prognostic accuracy on future observations. Stat Methods Med Res. 2016;25(4):1359–1380. doi: 10.1177/0962280213481053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma J. Transcriptional activators and activation mechanisms. Protein Cell. 2011;2(11):879–888. doi: 10.1007/s13238-011-1101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hedlund E, Deng Q. Single-cell RNA sequencing: Technical advancements and biological applications. Mol Aspects Med. 2018;59:36–46. doi: 10.1016/j.mam.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 73.Metsis A, Andersson U, Baurén G, Ernfors P, Lönnerberg P, Montelius A, et al. Whole-genome expression profiling through fragment display and combinatorial gene identification. Nucleic Acids Res. 2004;32(16):e127. doi: 10.1093/nar/gnh126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fielden MR, Zacharewski TR. Challenges and limitations of gene expression profiling in mechanistic and predictive toxicology. Toxicol Sci. 2001;60(1):6–10. doi: 10.1093/toxsci/60.1.6. [DOI] [PubMed] [Google Scholar]
- 75.Handorf CR, Kulkarni A, Grenert JP, Weiss LM, Rogers WM, Kim OS, et al. A multicenter study directly comparing the diagnostic accuracy of gene expression profiling and immunohistochemistry for primary site identification in metastatic tumors. Am J Surg Pathol. 2013;37(7):1067–1075. doi: 10.1097/PAS.0b013e31828309c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suresh K, Shah PV, Coates S, Alexiev BA, Samant S. In situ hybridization for high risk HPV E6/E7 mRNA in oropharyngeal squamous cell carcinoma. Am J Otolaryngol. 2021;42(1):102782. doi: 10.1016/j.amjoto.2020.102782. [DOI] [PubMed] [Google Scholar]
- 77.Mattocks CJ, Morris MA, Matthijs G, Swinnen E, Corveleyn A, Dequeker E, et al. A standardized framework for the validation and verification of clinical molecular genetic tests. Eur J Hum Genet. 2010;18(12):1276–1288. doi: 10.1038/ejhg.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang H, Lessard L, Su N, Luo Y, Hoon D, Ma X-J. Abstract 4665: Quantitative in situ biomarker analysis via ultrasensitive RNA in situ hybridization and automated image analysis. Can Res. 2014;74(19 Supplement):4665. doi: 10.1158/1538-7445.AM2014-4665. [DOI] [Google Scholar]
- 79.Economou M, Schöni L, Hammer C, Galván JA, Mueller DE, Zlobec I. Proper paraffin slide storage is crucial for translational research projects involving immunohistochemistry stains. Clin Transl Med. 2014;12014:4. doi: 10.1186/2001-1326-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Payne K, Gavan SP, Wright SJ, Thompson AJ. Cost-effectiveness analyses of genetic and genomic diagnostic tests. Nat Rev Genet. 2018;19(4):235–246. doi: 10.1038/nrg.2017.108. [DOI] [PubMed] [Google Scholar]
- 81.Montori VM, Smieja M, Guyatt GH. Publication bias: a brief review for clinicians. Mayo Clin Proc. 2000;75(12):1284–1288. doi: 10.4065/75.12.1284. [DOI] [PubMed] [Google Scholar]
- 82.(ADC) ACD. A Guide for RNAscope Data Analysis. California, US. 2017. https://www.indicalab.com/wp-content/uploads/2018/04/MK_51_103_RNAScope_data_analysis_guide_RevB.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.