Abstract
Objective
Alström syndrome is an autosomal recessive genetic disease caused by a mutation in the ALMS1 gene. Alström syndrome is clinically characterized by multisystem involvement, including sensorineural deafness, cone-rod dystrophy, nystagmus, obesity, insulin resistance, type 2 diabetes and hypogonadism. The diagnosis is thus challenging for patients without this characteristic set of clinical symptoms. We explored the effectiveness of whole-exome sequencing in the diagnosis of Alström syndrome.
Methods
A girl with symptoms of Alström syndrome was tested and diagnosed with the disease by whole-exome sequencing.
Results
Whole-exome sequencing revealed two novel variants, c.6160_6161insAT: p.Lys2054Asnfs*21(exon 8) and c.10823_10824 delAG:p.Glu 3608Alafs*9 (exon16) in the ALMS1 gene, leading to premature termination codons and the domain of ALMS1 protein. Blood sample testing of her asymptomatic parents revealed them to be heterozygous carriers of the same mutations. Assembly showed that the mutations on both alleles were located in conserved sequences. A review of the ALMS1 gene nonsense mutation status was performed.
Conclusion
We herein report two novel variants of the ALMS1 gene discovered in a Chinese Alström syndrome patient that expand the mutational spectrum of ALMS1 and provided new insight into the molecular mechanism underlying Alström syndrome. Our findings add to the current knowledge concerning the diagnosis and treatment of Alström syndrome.
Keywords: ALMS1, mutations, Alström syndrome, nonsense mutation
Introduction
Alström syndrome (AS) is an autosomal recessive hereditary disease with an estimated prevalence rate of one to nine per million. The major clinical feature of AS is its multiple organ involvement with the development of cone-rod dystrophy, progressive hearing impairment, trunk obesity, hyperinsulinemia, type 2 diabetes mellitus (T2DM), hypertriglyceridemia, cardiomyopathy and progressive lung, liver and kidney damage (1). Mutations in the ALMS1 gene encoding the ALMS1 protein have been shown to be the main cause of AS (2). While AS patients can exhibit their first clinical symptoms as soon as early infancy, the actual age of the onset and the severity of the symptoms can vary significantly, even among family members who share the same ALMS1 variants (3,4). The rapid progress in molecular analysis technology has made an early diagnosis possible.
We herein report a patient with mutations in the ALMS1 gene that led to poor vision, sensorineural deafness, short of stature, obese, nystagmus, photophobia, seizures, hypertriglyceridemia, cardiomyopathy and liver damage. She was diagnosed using whole-exome sequencing combined with a urine analysis.
Materials and Methods
Whole-exome sequencing
The genomic DNA was sheared using a Covaris Ultra Sonicator (Covaris, Woburn, USA). The DNA library was constructed using an Illumina TruSeq DNA Sample Preparation Kit (Illumina, San Diego, USA) following the manufacturer's protocol and amplified using pre-capture Ligation-mediated polymerase chain reaction (LM-PCR). An Agilent DNA1,000 Chip (Agilent Technologies, Palo Alto, USA) was used for the library quality assessment. The amplified library was then hybridized for exon regions using a SeqCap EZ Human Exome Kit v2.0 (Roche, Basel, Switzerland) according to the User's Guide (Joy Orient, Beijing, China). The library was then washed and amplified using post-capture LM-PCR. An Agilent DNA 1,000 Chip was used to assess the quality, concentration and size distribution of the captured library, and Quantitative Real-time (qPCR) was used to measure the enrichment and quality. The exon-enriched DNA library was sequenced on the Illumina HiSeq 2,500 platform (Illumina) according to the manufacturer's protocol for 150-bp paired-end reads. The raw image file was processed using bcl2fastq (Illumina) for base calls, which generated the raw data. Only genotypes with quality scores ≥20 were included for further data analyses.
Data analyses
Paired-end reads were aligned to the NCBI human reference genome (GRCh37/hg19) using the Burrows-Wheeler alignment tool (BWA, version 0.7.15; https://sourceforge.net/projects/bio-bwa/) software program. The sequence reads were subjected to Samtools (version 1.6) (http://samtools.sourceforge.net/) and Pindel analyses (http://gmt.genome.wustl.edu/packages/pindel/) to analyze the single-nucleotide polymorphisms (SNPs) and indels relative to the reference sequence. The identified variants were further annotated and filtered by an Ingenuity Variant Analysis (https://variants.ingenuity.com). Common variants were filtered based on their frequencies (MAF <0.05) in the databases of the Exome Aggregation Consortium (http://exac.broadinstitute.org; ExAC), the Exome Sequencing Project (https://esp.gs.washington.edu; ESP) or the 1000 Genomes project (http://www.1000genomes.org; 1,000 G).
Conserved sequence analyses
The UCSC Genome Browser on Human Dec. 2013 (GRCh38/hg38) Assembly was used to analyze whether or not the locations of mutations on both alleles were in conserved sequences.
Protein hydrophobicity analyses
We used the ProtScale (https://web.expasy.org/protscale/) and ProtParam (https://web.expasy.org/protparam/) software programs to predict protein hydrophobicity and stability. We used the Hphob./Kyte & Doolittle scale to estimate the hydrophilicity index of each amino acid and the grand average of hydropathicity (GRAVY) value to indicate the hydrophilicity of the protein. A GRAVY value <0 indicated hydrophilicity, while a GRAVY value >0 indicated hydrophobicity. We used the instability index (0-100) to determine the protein stability, with a higher instability index indicating greater instability.
Protein structure prediction on the iterative threading assembly refinement (I-TASSER) server for Lys 2054 Asnfs*21
The Lys 2054 Asnfs*21 mutant structure is predicted by the I-TASSER with default parameters (5).
Results
Clinical features
A five-year-old girl with a history of learning difficulties was referred to the neurology department of the Children's Hospital of Shanghai (Shanghai City, China) for an evaluation. The patient has suffered from poor vision and sensorineural deafness since birth.
A physical examination revealed a short stature and obesity (height 113 cm, weight 30 kg). Her blood pressure was 90/50 mmHg, and her heart rate was regular with a pulse of 107 beats per minute. Nystagmus and photophobia were exhibited during infancy. Vision loss progressed rapidly during early childhood, resulting in a best-corrected visual acuity of 20/40. An examination of the fundus revealed a shallow optic disc with greyish pigmentation and attenuation of retinal vessels. Electrocardiography (EKG) readings showed low blunt T wave at I, V5, and V6 leads. Electroencephalogram (EEG) findings were normal. A blood test revealed a plasma triglycerides (TG) level of 0.98 mmol/L (normal <1.7 μmol/L) and a high-density lipoprotein cholesterol level of 1.22 mmol/L (normal 0.9-1.55 μmol/L). There were minor changes in the liver function, as the level of alanine aminotransferase was 73 U/L (normal 5-40 U/L), and the level of aspartate aminotransferase was 50 U/L (normal 8-40 U/L). The patient's glycosylated hemoglobin (HbA1c) was 16.20 (normal 4-6), glycosylated albumin was 37.84% (normal 11%-16%), and fasting blood glucose was 12.6 (normal 3.9-6.1 mmol/L). Bilateral sensorineural hearing loss was confirmed by audiometry. Both of her parents were healthy.
Identification of novel disease-causing ALMS1 mutations
The sequences were analyzed using the DNASTAR software program (http://www.dnastar.com/) to confirm the mutations discovered in all samples. The findings showed that the ALMS1 mutation c.6160_6161insAT:p.Lys2054Asnfs*21 was derived from the patient's mother, while the patient's father had another mutation: c.10823_10824 delAG:p.Glu 3608Alafs*9 (see Fig. 1 and 2 for details). The two gene mutation sites distribution in ALMS1 gene, c.6160_6161insAT:p.Lys2054Asnfs*21 located in exon 8 and c.10823_10824 delAG:p.Glu 3608Alafs*9 located in exon 16 (see Fig. 3).
Figure 1.
Sanger sequencing results concerning the position of c.6160_6161insAT: p.Lys2054Asnfs*21on the ALMS1 gene. Electropherograms showing the DNA sequence at position c.6160_6161insAT: p.Lys2054Asnfs*21 of the ALMS1 gene in this patient (top). Her mother carried an insertion of AT in this position (bottom), whereas her father had normal findings (middle).
Figure 2.
Sanger sequencing results at the position of c.10823_10824 delAG: p.Glu3608Alafs*9 on the ALMS1 gene. Electropherograms showing the DNA sequence at position c.10823_10824 delAG: p.Glu3608Alafs*9 of the ALMS1 gene in this patient (top). Her father carried a deletion of AG in this position (middle), whereas her mother had normal findings (bottom).
Figure 3.
Sequence features of ALMS1. a: Represents the position of the exon of the ALMS1 gene and the mutation at the two sites in the exon. b: The N-terminal polyglutamate (PolyE) tract was polymorphic, CC: predicted coiled-coil domain, LZ: leucine zipper motif, pNLS: potential nuclear localization signal
Distribution of the nonsense mutation in the ALMS1 gene
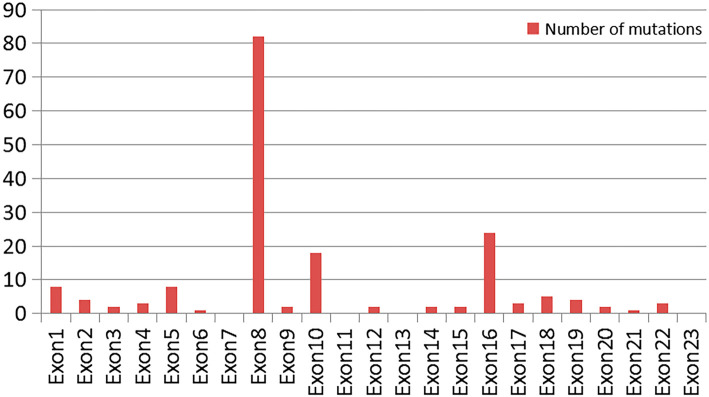
These nonsense mutations were mainly concentrated in exons 8, 10 and 16 on the ALMS1 gene (see Fig. 4).
Figure 4.
A comparison of the nonsense mutations on each exon of the ALMS1 gene.
Conserved sequence analyses
After the genetic diagnosis, an analysis using the UCSC Genome Browser on Human Dec. 2013 (GRCh38/hg38) Assembly showed that the mutations on both alleles were located in conserved sequences (see Fig. 5).
Figure 5.
Conserved amino acid sequences of ALMS1 (amino acids 2,054 and 3,608).
Protein hydrophobicity
The hydropathic index for each amino acid on ALMS1 protein was estimated by the Hphob./Kyte & Doolittle Method using the ProtScale Software program. The grand average hydropathicity of the wild-type ALMS1 protein was predicted by the ProtParam software program to be -0.720. The grand average hydropathicity of the predicted protein after the p.Lys2054Asnfs*21 mutation was -0.681. The grand average hydropathicity of the predicted protein after the p.Glu3608Alafs*9 mutation was -0.637. Both sites were shown to be capable of inducing a decrease in protein hydrophilicity after the mutation, and the instability of the structure was increased.
Protein structure prediction on the I-TASSER server for Lys 2054 Asnfs*21
The I-TASSER server suggested that the ALMS1 protein is composed of many regions, which consists of many α-helix (helix region), random coil region (coil region), and β chain region (strand region). The helix and coil regions are relatively unstable, while the strand region is relatively stable. The Lys 2054 Asnfs*21 mutant structure was predicted using the I-TASSER server with default parameters. According to the protein sequence feature and the predicted structure, in the region between 625 and 2,200 amino acids, there were 32 fragment repeats, most of them likely predicted α-helix region, which were relatively unstable. (see Fig. 6).
Figure 6.
The Lys 2054 Asnfs*21 mutant structure was predicted by the I-TASSER server with default parameters. According to the protein sequence feature and the predicted structure, in the region between 625 and 2,200 amino acids, there were 32 fragment repeats, were most likely predicted as α-helix region.
Discussion
AS is a rare autosomal recessive disease (OMIM 203800) characterized by cone-rod dystrophy, obesity, progressive bilateral sensorineural hearing impairment, acute infantile-onset cardiomyopathy and/or adolescent or adult-onset restrictive cardiomyopathy, insulin resistance/T2DM, non-alcoholic fatty liver disease (NAFLD) and chronic progressive kidney disease. Cone-rod dystrophy presents as progressive visual impairment, photophobia and nystagmus, usually starting between birth and 15 months of age (6). The original phenotype was first reported in 1959 with an estimated prevalence rate of 1 to 9 cases per 1,000,000 individuals (7). Multiple organ involvement is the main clinical characteristic of AS, leading to a reduction in life expectancy to less than 50 years old. Dilated cardiomyopathy and renal failure are the major causes of death. Cardiac problems in AS include systolic dysfunction, myocardial fibrosis, dilated cardiomyopathy and reduced myocardial mass (8). The symptom onset can vary among patients with AS, even amongst family members with the same mutations.
Mutations in the ALMS1 gene were first identified as the cause of AS in 1997 (9). The ALMS1 (OMIM 606844) gene is located at the short arm of chromosome 2 (2p13.2), with 224,161 bases forming a total of 23 exons. The encoded ALMS1 protein is composed of 4,169 amino acids and functions in microtubule organization, particularly in the formation and maintenance of cilia (10). ALMS1 protein is a large protein that lacks known catalytic domains. It has several sequence features with unknown functions, including a large tandem repeat domain (TRD), three short predicted coiled-coil (CC) domains and a C-terminus dubbed the ALMS1 motif. The N-terminal polyglutamate (PolyE) tract is polymorphic and is followed by seven alanine residues. The CC domain, the leucine zipper(LZ) motif domain, and the potential nuclear localization signal(pNLS)domain also play an important role in ALMS1 protein (11).
Collin et al. first discovered in 2002 that a mutation in the ALMS1 gene can cause AS (12). Currently, there are 1,318 mutations in ALMS1 gene entries reported in the ClinVar Database, with most related to AS. Different types of mutations occur at the ALMS1 locus, including insertions or deletions and single-nucleotide substitutions, which may lead to premature stop codons or missense mutations at conserved residues. A number of ALMS1 gene mutations have been detected in patients with AS, with almost all being nonsense or frameshift mutations leading to a premature stop codon (4,13). We reviewed the literature concerning ALMS1 gene mutations, including 553 missense mutations and 177 nonsense mutations. These nonsense mutations included 42 homozygous nonsense mutations and 134 nonsense mutations after frameshift. Marshall, Shenje and Das Bhowmik et al. (14-16) reported c.1054C>T (p.Arg352Ter*), c.1900C>T (p.Gln634Ter*), c.2822T>A (p.Leu941Ter*) on the ALMS1 gene. These site mutations were shown to be associated with homozygous nonsense mutations. Shenje, Nicola and Hearn et al. (17,18) reported c.4292_4293CA[2] (p.His1432fs), c.1735delA (p.Arg579Glyfs), c.2141_2142delCT (p.Ser714Tyrfs) on the ALMS1 gene. According to the ClinVar Database, about 98% of nonsense mutations are pathogenic or likely pathogenic, and about 91% are related to AS. These nonsense mutations are mainly concentrated at exons 8, 10 and 16.
The ALMS1 gene is expressed in most tissues of the body and most regions of the brain (http://www.brain-map.org/). In mice, knockdown of the ALMS1 gene by short interfering RNA has been shown to stunt the growth of cilia on kidney epithelial cells, preventing them from increasing the calcium influx in response to mechanical stimuli, a potential mechanism underlying AS's extensive clinical symptoms (19). Nesmith et al. studied a genomic AS model by targeting the zebrafish ALMS1 gene using CRISPR/Cas9. The resulting heritable mutation ablated protein production and resulted in systemic defects that recapitulated the human syndrome, including defects in neurosensory, cardiac and renal systems (20).
In infants and young children, visual dysfunction is usually the earliest symptoms of AS. Nystagmus and photophobia appear within a few weeks after birth. In the retina, the rod cell count may be preserved initially but decline with age. In addition, progressive bilateral sensorineural hearing loss develops in 89% of AS patients (21,22). In the present patient, nystagmus was the first clinical symptom, followed by progressive visual impairment beginning at one year old. Progressive sensorineural hearing impairment began at three years old and was exacerbated whenever the patient had a cold.
Although severe cognitive impairment is not a common feature of AS, some delays in the development of learning skills occur in approximately 45% of affected children. Thus far, nearly 700 cases have been described worldwide. In addition, AS patients may experience absence seizures and general sleep disturbance (6,23,24), as was observed in our patient, who has had episodes of epilepsy seizures at two years old and had been experiencing learning difficulties in school.
Nonsense mutations can generate premature translation termination codons (PTCs), resulting in truncated proteins. Due to the production of truncated proteins, PTCs usually inactivate the gene function. Degradation of the nonsense-mediated mRNA decay (NMD) pathway usually results in a significant decrease in the amount of mRNA, causing defects in the encoded protein or amount and thereby leading to disease phenotypes (25). A molecular study of our AS patient revealed two novel ALMS1 mutations: p.K2054Nfs*21 at exon 8 and p.Q3608Qfs*7 at exon 16, both of which are capable of causing a premature termination codon in the ALMS1 protein. An examination of the molecular status of her parents revealed that her phenotypically normal parents also carried heterozygous mutations in the ALMS1 gene. The compound heterozygous mutations of c.6160_6161insAT p. Lys2054Asnfs*21in ALMS1 resulted in a premature stop codon and the loss of 2095 amino acids.
The Lys 2054 Asnfs*21 mutant structure was predicted by the I-TASSER server with default parameters. According to the protein sequence feature and the predicted structure, in the region between 625 and 2,200 amino acids, there were 32 fragment repeats, and most of them likely predicted α-helix region, which were relatively unstable. The introduction of the Lys 2054 Asnfs*21 mutation led to the loss of the last two fragment repeats, resulting in the deformation of the beta sheet structure due to the frameshift, which can cause structural instability. The compound heterozygous mutations c.10823_10824 delAG:p.Glu3608Alafs*9 in ALMS1 resulted in a premature stop codon and the loss of 552 amino acids.
We used hydrophobicity analysis to predict protein stability after gene mutation. The grand average hydropathicity of the wild-type ALMS1 protein was predicted by the ProtParam software program to be -0.720. The grand average hydropathicity of the predicted protein after the p.Lys2054Asnfs*21 mutation was -0.681. The grand average hydropathicity of the predicted protein after the p.Glu3608Alafs*9 mutation was -0.637. Both sites were thus capable of inducing a decrease in protein hydrophilicity after mutation and destabilized the structure.
There are no specific treatments for AS, and current management consists of only symptomatic therapies. Protein-bound iodine-4050 (PBI-4050) is a new molecular entity that has demonstrated anti-inflammatory and anti-fibrotic activities in preclinical models, including animal models of human diseases characterized by progressive fibrosis in the kidney, heart, liver and lungs. Studies in T2DM with metabolic syndrome and idiopathic pulmonary fibrosis further support the anti-inflammatory and anti-fibrotic activities of PBI-4050, suggesting that PBI-4050 has potential utility for treating the pathological inflammatory and fibrotic features of ALMS1 (26). The development of therapeutic approaches for diseases caused by nonsense mutations has focused on small-molecule read-through agents. The goal of this type of therapy is to induce the translation machinery to recode a PTCs into a sense codon so that translation continues in the correct reading frame in order to complete the synthesis of a full-length, potentially functional protein (27,28). Since part of AS is caused by nonsense mutations, we may be able to treat AS caused by nonsense mutations with drugs, although this merits further investigation.
Conclusion
We analyzed the mutational characteristics of the ALMS1 gene associated with AS reported thus far. We also studied the clinical phenotype and genotype of a Chinese girl diagnosed with AS and assessed its pathogenesis. We found that the patient had a complex heterozygous mutation of the ALMS1 gene: p.Lys2054Asnfs*21 (exon 8) and p.Glu3608Alafs*9 (exon16). Mutations at both sites were deemed capable of causing nonsense mutations. The Lys 2054 Asnfs*21 mutant structure was predicted by the I-TASSER server with default parameters. According to the protein sequence feature and the predicted structure, in the region between 625 and 2,200 amino acids, there were 32 fragment repeats, and most of them likely predicted as α-helix, which were relatively unstable. The introduction of the Lys 2054 Asnfs*21 mutation led to the loss of the last two fragment repeats, resulting in the deformation of the beta sheet structure due to the frameshift, which can cause structural instability. We predicted this structure for the first time.
Through our hydrophobic analysis, the affinity of the two truncated proteins was found to be lower than that of the normal protein, and the stability was decreased, which affected the protein transport function. These findings expanded the mutational spectrum of ALMS1 and provided new insight into the molecular mechanism underlying AS, which may aid in the diagnosis and treatment of AS in the future.
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
The authors state that they have no Conflict of Interest (COI).
Financial Support
This work was supported by grants from Shanghai Science and Technology Fund (No. 16410723400), Key Subject Program from Shanghai Municipal Commission of Health and Family Planning (No. 2016ZB0102), Key disciplines of top priority in Shanghai (2017ZZ02019), Research project of Shanghai children's hospital (2016YMS005) and Shanghai Hospital Development Center (SHDC12015113).
Chunmei Wang and Xiaona Luo contributed equally to this work.
Acknowledgement
We would like to express our gratitude to the patient and her parents for their support.
References
- 1.Alvarez-Satta M, Castro-Sanchez S, Valverde D. Alström syndrome: current perspectives. Appl Clin Genet 8: 171-179, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kılınç S, Yücel-Yılmaz D, Ardagil A, et al. Five novel ALMS1 gene mutations in six patients with Alström syndrome. Pediatr Endocrinol Metab 6: 681-687, 2018. [DOI] [PubMed] [Google Scholar]
- 3.Marshall JD, Maffei P, Collin GB, et al. Alström syndrome: genetics and clinical overview. Curr Genomics 3: 225-235, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marshall JD, Muller J, Collin GB, et al. Alström syndrome: Mutation Spectrum of ALMS1. Hum Mutat 36: 660-668, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jianyi Y, Yang Z. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Research 43: W174-W181, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paisey RB, Steeds R, Barrett T, et al. GeneReviewsⓇ [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2019. 2003 Feb 7 [updated 2019 Jun 13].
- 7.Alstrom CH, Hallgren B, Nilsson LB, et al. Retinal degeneration combined with obesity, diabetes mellitus and neurogenous deafness: a specific syndrome (not hitherto described) distinct from the Laurence-Moon-Bardet-Biedl syndrome: a clinical, endocrinological and genetic examination based on a large pedigree. Acta Psychiatr Neurol Scand Suppl 129: 1-35, 1959. [PubMed] [Google Scholar]
- 8.Brofferio A, Sachdev V, Hannoush H, et al. Characteristics of cardiomyopathy in Alström syndrome: prospective single-center data on 38 patients. Mol Genet Metab 121: 336-343, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collin GB, Marshall JD, Cardon LR, et al. Homozygosity mapping at Alström syndrome to chromosome 2p. Hum Mol Genet 2: 213-219, 1997. [DOI] [PubMed] [Google Scholar]
- 10.Kılınç S, Yücel-Yılmaz D, Ardagil A, et al. Five novel ALMS1 gene mutations in six patients with Alström syndrome. Pediatr Endocrinol Metab 6: 681-687, 2018. [DOI] [PubMed] [Google Scholar]
- 11.Hearn T. ALMS1 and Alström syndrome: a recessive form of metabolic, neurosensory and cardiac deficits. Mol Med (Berl) 1: 1-17, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collin GB, Marshall JD, Ikeda A, et al. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alström syndrome. Nat Genet 1: 74-78, 2002. [DOI] [PubMed] [Google Scholar]
- 13.Tsai MC, Yu HW, Liu T, et al. Rare compound heterozygous frameshift mutations in ALMS1 gene identified through exome sequencing in a Taiwanese patient with Alström syndrome. Front Genet 18: 110, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marshall JD, Muller J, Collin GB, et al. Alström Syndrome: Mutation Spectrum of ALMS1. Hum Mutat 36: 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shenje LT, Andersen P, Halushka MK, et al. Mutations in Alström protein impair terminal differentiation of cardiomyocytes. Nat Commun 5: 3416, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das Bhowmik A, Gupta N, Dalal A, et al. Whole exome sequencing identifies a homozygous nonsense variation in ALMS1 gene in a patient with syndromic obesity. Obes Res Clin Pract 2: 241-246, 2017. [DOI] [PubMed] [Google Scholar]
- 17.Edwards NC, Moody WE, Yuan M, et al. Diffuse left ventricular interstitial fibrosis is associated with sub-clinical myocardial dysfunction in Alström Syndrome: an observational study. Orphanet J Rare Dis 10: 83, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hearn T, Renforth GL, Spalluto C, et al. Mutation of ALMS1, a large gene with a tandem repeat encoding 47 amino acids, causes Alström syndrome. Nat Genet 31: 79-83, 2002. [DOI] [PubMed] [Google Scholar]
- 19.Li G, Vega R, Nelms K, et al. A role for Alström syndrome, alms1, in kidney ciliogenesis and cellular quiescence. PLoS Genet 3: e8, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nesmith JE, Hostelley TL, Leitch CC, et al. Genomic knockout of alms1 in zebrafish recapitulates Alström syndrome and provides insight into metabolic phenotypes. Hum Mol Genet 28: 2212-2223, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L, Dong B, Chen X, et al. Identification of a novel ALMS1 mutation in a Chinese family with Alström syndrome. Eye (Lond) 23: 1210-1212, 2009. [DOI] [PubMed] [Google Scholar]
- 22.Ozantürk A, Marshall JD, Collin GB, et al. The phenotypic and molecular genetic spectrum of Alström syndrome in 44 Turkish kindreds and a literature review of Alström syndrome in Turkey. J Hum Genet 60: 51, 2015. [DOI] [PubMed] [Google Scholar]
- 23.Piñeiro-Gallego T, Cortón M, Ayuso C, Baiget M, Valverde D. Molecular approach in the study of Alström syndrome: analysis of ten Spanish families. Mol Vis 18: 1794-1802, 2012. [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss S, Cohen L, Ben-Yosef T, Ehrenberg M, Goldenberg-Cohen N. Late diagnosis of Alström syndrome in a Yemenite-Jewish child. Ophthalmic Genet 40: 7-11, 2019. [DOI] [PubMed] [Google Scholar]
- 25.Ottens F, Gehring NH. Physiological and pathophysiological role of nonsense-mediated mRNA decay. Pflugers Arch 468: 1013-1028, 2016. [DOI] [PubMed] [Google Scholar]
- 26.Baig S, Veeranna V, Bolton S, et al. Treatment with PBI-4050 in patients with Alström syndrome: study protocol for a phase 2, single-Centre, single-arm, open-label trial. BMC Endocr Disord 18: 88, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagel-Wolfrum K, Moller F, Penner I, et al. Targeting nonsense mutations in diseases with translational read-through-inducing drugs (TRIDs). BioDrugs 30: 49-74, 2016. [DOI] [PubMed] [Google Scholar]
- 28.Linde L, Boelz S, Nissim-Rafinia M, et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. Clin Invest 117: 683-689, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]