Summary
Recent studies have demonstrated the potential to recover ancient human mitochondrial DNA and nuclear DNA from cave sediments. However, the source of such sedimentary ancient DNA is still under discussion. Here we report the case of a Bronze Age human skeleton, found in a limestone cave, which was covered with layers of calcite stone deposits. By analyzing samples representing bones and stone deposits from this cave, we were able to: i) reconstruct the full human mitochondrial genome from the bones and the stones (same haplotype); ii) determine the sex of the individual; iii) reconstruct six ancient bacterial and archaeal genomes; and finally iv) demonstrate better ancient DNA preservation in the stones than in the bones. Thereby, we demonstrate the direct diffusion of human DNA from bones into the surrounding environment and show the potential to reconstruct ancient microbial genomes from such cave deposits, which represent an additional paleoarcheological archive resource.
Subject areas: Biological sciences, Evolutionary biology, Paleobiology, Paleogenetics, Anthropology, Archeology
Graphical abstract

Highlights
-
•
Bronze Age human skeletal remains were found unburned in Wimsener cave in Germany
-
•
The bones were covered with layers of calcite stone deposits
-
•
The ancient human and microbial DNA diffused from the bones to the stones
-
•
The mitochondrial haplogroup and sex were revealed by analyzing the stones only
Biological sciences; Evolutionary biology; Paleobiology; Paleogenetics; Anthropology; Archeology
Introduction
Ancient DNA (aDNA) has become a powerful tool to study the ancestral history, not only of hominins, but also other animals, plants, and even microbes (Capo et al., 2021; Epp et al., 2015). In addition to the skeletal and mummified remains also sedimentary materials were recently identified as a promising source of aDNA, that still containing Pleistocene Neanderthal and Denisovan mitochondrial DNA and nuclear DNA (Gelabert et al., 2021; Slon et al., 2017; Vernot et al., 2021). There were different postulations on the source of such sedimentary ancient DNA (sedaDNA), e.g., macrofossils, small bone fragments, excreta, and decayed soft tissues (Haile et al., 2007; Slon et al., 2017; Willerslev et al., 2003). However, we still miss a case where we see a direct link between the human sedaDNA and its source.
Here we report the finding of human skeletal remains (1306-1017 calBCE) dating back to the Urnfield culture of the late Bronze Age within the underwater river cave named either Wimsener Höhle (Straub, 2006; Straub and Lehmkuhl, 2009) or Friedrichshöhle near Hayingen (Swabian Alb, Baden-Württemberg, Germany, Figure 1). The discovery of an unburned skeleton itself is remarkable because the predominant tradition of Urnfield Cultures is cremation of the deceased. The finding of contemporary pottery and other human bone fragments in the entrance lake of the cave suggest the possibility of a cultic site (Straub and Lehmkuhl, 2009). Similar finds from other caves in the Swabian Alb point to a religious phenomenon of the Late Bronze Age, namely burials or ritual acts in caves (Rebay-Salisbury, 2010).
Figure 1.
Skeleton finding site
(A) Location of the Wimsener caves in Baden-Württemberg, Germany.
(B) Longitudinal section of the first 200 meters of the Wimsener caves; the scale bar refers to the cave length.
(C) Cross section of the “Schatzkammer”; the red star refers to the location of the tibia.
(D) Photograph showing the difficult accessibility of the finding site; the red arrow indicates the protruding part of the sampled tibia.
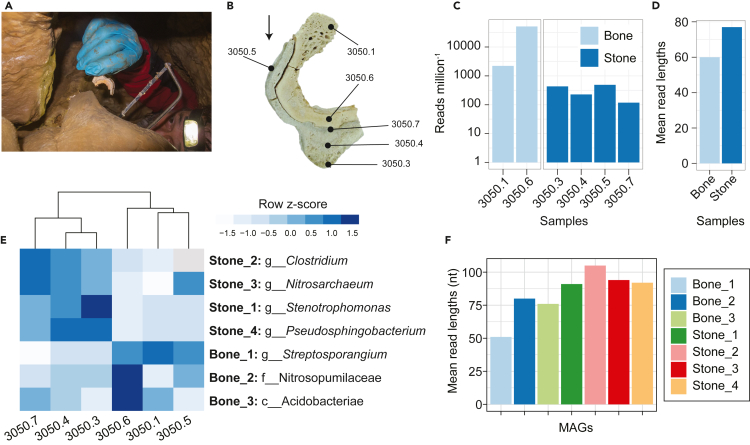
The commingled skeletal remains were found covered with heavy layers of calcite deposits, as a result of continuous water dripping from the cave ceiling, forming bulges toward the direction of the gravity (Figures 2A and 2B). Because most of the skeleton was heavily covered by calcite deposits, only a sample of the more accessible tibia was taken (Figure 1D). To further investigate this archeological finding molecularly, we used a slice of the tibia sample with the surrounding calcite layer (Figure 2B). Different parts of the bone and stone were subsampled and subjected to metagenomic sequencing (Table S1, Figure 2, and STAR Methods). On these remains we could demonstrate the diffusion of ancient human and microbial DNA from the skeleton to the surrounding environment.
Figure 2.
Metagenomic analysis of human and microbial DNA of cave human bone and its surrounding stone deposits
(A) On-site sampling of human tibia covered with a layer of calcite deposits.
(B) Close-up cross-section of sampled tibia slice, indicating the sampling locations for DNA analysis; the black arrow refers to the direction of gravity.
(C) Number of metagenomic reads of each sample mapped to the human autosomal DNA (Ref genome hg19).
(D) Mean read lengths of the mapped reads against the human genome (hg19).
(E) Heatmap showing the abundance of different metagenome-assembled genomes (MAGs) in different samples (genome copies per million reads). The color intensity indicates row z-scores.
(F) Mean read lengths of the mapped reads the MAGs. For the (E and F) the names of MAGs refer to the source of the MAGs (stone or bone) e.g., “Stone_1” means that this genome was assembled from the stone sample.
Results
Bone and stone contain ancient human DNA of the same individual
Analysis of the human DNA (Table S2), by comparing the DNA against the human reference genome hg19 (Rosenbloom et al., 2015), revealed the presence of human DNA reads, not only in the bone samples, but also in all stone samples. The human DNA of the stones accounted for ∼1% of the human DNA found in the bone samples (Figure 2C and Table S2). In general, the human DNA fragment lengths of the stone samples were higher than the bone samples (Figure 2D), with comparable DNA damage levels, i.e., percentage of terminal cytosine deamination to thymine (Figure S1). In addition, the ancient human DNA in the stone and the bone displayed the same molecular sex (male, XY), which would not be possible with classical anthropological investigations. To further confirm that the human DNA in stone belongs to the skeleton, we enriched it for mitochondrial DNA using a hybridization capture assay (STAR Methods). This enabled us to reconstruct the mitochondrial genome from the stone samples, with > 7x coverage (Tables S2 and S3), revealing the identical mitochondrial haplotype of the bone samples (i.e., J1C1). Further and to gain a glimpse into the origin of the individual, we performed principal component analysis (PCA) against selected modern Eurasian individuals and other Bronze Age individuals (Mittnik et al., 2019). The PCA analysis showed our individual falls within the European diversity similar to other Bronze Age individuals from the same region (Figure S2). Overall, we could confidently demonstrate that the human DNA diffused from the bone into the stone and preserved in the calcite for millennia.
Ancient microbial genomes reconstructed from the bone and stone
Based on the human DNA results, we assumed that such calcite deposits could represent a time capsule and still contain ancient molecular information other than human DNA. Therefore, we extended our analysis to the microbial communities of both samples, by performing general microbial profiling and de novo assembly of the short metagenomic reads into longer contigs to reconstruct metagenome-assembled genomes (MAGs) (See STAR Methods and Table S4). In general, the microbial profiles of the bone and stone were similar with a high richness of Archaea and Actinobacteria which is typical for cave environments (Figure S3). The bone and stone samples were then pooled independently for de novo assembly, which resulted in three high-quality and four medium-quality prokaryotic MAGs, from both samples (three genomes from the bone and four from the stone, please refer to Table S4 and Figure 2E). All reconstructed MAGs displayed DNA damage, except for one MAG from the bone samples (Figure S4). Similar to what was observed with human DNA, the presence of the MAGs in both the bone and stone indicate microbial DNA diffusion. It was also observed that the read lengths were longer in general in the stone compared to bone (Figures 2F and S4). These genomes represent microbes that may have been involved in initial postmortem decomposition and initiation of secondary stalactite formation. For instance, members of Clostridium and Streptosporangium are reported among the most abundant postmortem bone degraders (Eriksen et al., 2020; Philips et al., 2017). Moreover, Streptosporangium has also been implicated in bone tunneling in waterlogged environments (Eriksen et al., 2020; Kim et al., 2020; Turner-Walker, 2009). Although other microbes, like Stentrophomonas and Rhodococcus, have been shown to crystalize calcium carbonate in cave environments (Enyedi et al., 2020), which may suggest a potential role in stalactite formation (Pacton et al., 2013).
Discussion
In this case study, we took bone and calcite stone deposits from a Bronze Age human skeleton and subjected them to DNA analysis. We recovered well-preserved ancient human DNA and microbial DNA from the stone and bone, enough to determine the sex of the individual, and to reconstruct human mitochondrial- and microbial genomes.
We identified the same human DNA in both bone and stone samples and thereby demonstrated the direct diffusion of human DNA from bones into the surrounding environment. This presents an additional explanation model for the reported presence of ancient human DNA in cave sediments (Slon et al., 2017; Vernot et al., 2021). Earlier, in 2003, Willerslev and colleagues reported the presence of Euryapteryx curtus DNA in sand samples collected from the interior of a moa bone in New Zealand (Willerslev et al., 2003). Later in 2007, Haile et al. also demonstrated the potential post-depositional vertical migration of ancient DNA across sedimentary strata (Haile et al., 2007). However, it is yet-to-be determined how the stalactite deposits can better preserve aDNA fragments from the past. In general, DNA can adsorb to different mineralogical elements because of its negative charge (Freeman and Sand, 2020). In addition, cave environments maintain a constant temperature and humidity as well as aphotic zones which allow less exposure to conventional DNA damages (Stahlschmidt et al., 2019; Zepeda Mendoza et al., 2016). It may also be possible that the leached DNA gets less exposed to the postmortem microbial decomposition, because of the fact that the bones are continuously degraded because of their content of organic substrates (e.g., proteins).
In this report we aim to bring attention to such valuable mineralogical deposits that are found attached to archaeological findings in cave environments. We showed that they are not only representing mineralogical deposits, but rather an extension to the archeological findings that retain and preserve its historical information in the form of aDNA. Currently these deposits are mainly used as dating proxies for archeological findings that are beyond the limits of the radiocarbon dating (14C), e.g., using Uranium–Thorium dating (U-Th). Considering our finding, these deposits could help to avoid future destructive sampling of similar archeological remains, offering an additional paleogenetic archive that can be used to reconstruct ancient human genomes and ancient microbial communities. It remains to be explored how far such deposits are capable of preserving, in addition, other ancient biomolecules, e.g., proteins or lipids.
Finally, our message to our community of archeologists and anthropologists is to consider innovative sampling resources (e.g., sediments, mineralogical deposits, or water) during excavations, especially for molecular analyses that often involve destructive procedures.
Limitations of the study
The main limitation of the study is that it is based on a single individual from a single site, and it is also rare to find ancient skeletal samples covered with such calcite stone deposits. In addition, the total amount of human DNA reads obtained from the calcite stone deposits is two order of magnitude less than the bone. This might suggest accompanying the DNA library preparation from similar samples a subsequent application of a human nuclear/mitochondrial DNA capture approach.
STAR★Methods
Key resources table
Resource availability
Lead contact
-
•
Further information on materials, datasets, and protocols should be directed to and will be fulfilled by the Lead Contact, Frank Maixner (frank.maixner@eurac.edu).
Material availability
-
•
This study did not generate new unique reagents.
Experimental model and subject details
In this study, we metagenomically analyzed samples of human skeletal remains (1306-1017 calBCE) dating back to the Urnfield culture of the late Bronze Age within the underwater river cave named either Wimsener Höhle or Friedrichshöhle near Hayingen (Swabian Alb, Baden-Württemberg, Germany). The bone samples were found covered with layers of calcite deposits, which was also metagenomically analyzed.
Method details
DNA extraction and library preparation
Amounts of 20-100 mg of bone/stone were sampled at the locations indicated in Figure 2B (Please refer to Table S1 for further details). EDTA/Proteinase K mixtures were added to the samples and incubated for 24 h at 40°C, followed by purification and recovery modified protocol from Tang et al. (Tang et al., 2008). DNA extracts were quantified using QUANTUS (Promega, USA), then 20 μl of each sample were converted into double-indexed pair-end DNA libraries following a special protocol for highly degraded ancient DNA (Meyer and Kircher, 2010). All previous steps were carried out in the ancient DNA laboratory of the institute of mummy studies at EURAC research in Bolzano, Italy. Further, selected indexed samples were pooled and enriched for human mitochondrial genome (myBaits®, https://arborbiosci.com), following the manufacturer’s instructions (Selected samples are indicated in Table S2).
DNA sequencing and post-sequencing processing
The indexed libraries were pooled and subjected to next generation DNA sequencing using HiSeqX (2 × 150 pair end). After demultiplexing, we used the tool fastp (Chen et al., 2018) to trim the adapters and low quality sequences, and to merge pair-end reads with at least 10 nucleotides overlap. The overall workflow is described in the Figure S5.
Human DNA analysis
First, for each sample we used the tool SeqKit (Shen et al., 2016) to deduplicate the merged sequences, and to remove sequences shorter than 25 nucleotides. Next, we mapped these reads against the human reference genome (build hg19) (Rosenbloom et al., 2015) and the human reference mtDNA genome (rCRS) (Andrews et al., 1999) using Burrows-Wheeler Aligner (BWA) (Li and Durbin, 2010). SAMtools was used to filter for minimum mapping quality of 30. We used QualiMap to generate basic mapping statistics (Okonechnikov et al., 2016). To check the authenticity of the mapped reads being of ancient origin and not modern contamination we used the tool mapDamage 2.0 (Jónsson et al., 2013) to quantify the percentages of C to T and G to A substitution. In the case of mitochondrial DNA, we additionally used option “--rescale” to rescale the quality scores of the damaged mis-incorporated sites, and the tool Schmutzi to estimate the level of contamination based on deamination patterns (Renaud et al., 2015). For haplogroup assignment, we first converted the rescaled bam files into variant calling format (VCF) and then employed HaploGrep 2.0 (Weissensteiner et al., 2016).
To compare our sample to other individuals, we used SAMtools mpileup and pileupCaller (https://github.com/stschiff/sequenceTools) to call the 1240K targeted SNPs (David Reich lab, https://reich.hms.harvard.edu/datasets), with pseudodiploid method. Then, we used the EIGENSOFT tools “mergeit” to merge the data and “smartpca” to perform the principal component analysis (PCA). Finally, our ancient sample as well as selected Bronze Age individuals from Mittnik et al. (Mittnik et al., 2019) were projected to the modern Eurasian and Middle Eastern individuals.
To determine the sex of the individual, we used the mapped human DNA reads to compute the karyotype frequency of X and Y chromosomes, using a Maximum likelihood method (Skoglund et al., 2013).
Microbial DNA analysis
For general microbiome profiling, we applied MetaPhlAn 3.0 on the merged reads, with options of “--min_mapq_val 30” and “--read_min_len 25” (Beghini et al., 2021).
Additionally, we used the quality-filtered unmerged reads to perform de novo sequence assembly, following a co-assembly approach by combining samples into two groups, bone (n = 2) and stone (n = 4), and using the MetaWRAP pipeline (Uritskiy et al., 2018). In detail, we used the metagenomic assembler MEGAHIT (Li et al., 2015) and SPAdes with “--meta” option (Nurk et al., 2017), to assemble the pair-end reads into longer contigs. All contigs shorter than 1000 nt were excluded from downstream analyses. We used three different metagenomic binning tools, MetaBAT2 (Kang et al., 2019), MaxBin2 (Wu and Singer, 2021), and CONCOCT (Alneberg et al., 2014), to cluster the contigs into bins based on abundance and kmer frequency. Then, we checked the completeness and contamination of the resulting bins using CheckM (Parks et al., 2015), and kept only the high- (completeness > 90% and contamination < 5%) and medium (completeness 50-90% and contamination < 10%) quality bins for further analysis. Further, we used the “reassemble_bins” module to remap the raw reads against each of the bins, then reassembled the mapped reads using SPAdes assembler. This step was proved to increase the completeness and reduce the contamination of the bins. In order to assess the abundance of each bin in each of the samples, we implemented the MetaWRAP module “quant_bins”, which quantifies the abundance of each bin and expresses it as a unit of “genome copies per million reads”.
To taxonomically classify each bin, we used GTDB-Tk v1.5.0 and the module “classify_wf” (Chaumeil et al., 2019; Zhou et al., 2020), which identifies 120 marker genes in bacteria and 122 marker genes in archaea, performs a multiple sequence alignment of the markers, and finally assigns taxonomy based on the phylogenetic placement with known reference genomes. Average nucleotide identity (ANI)/relative evolutionary divergence between reference genomes and metagenomic bins (henceforth referred to as metagenomes-assembled genomes, MAGs) was also performed using GTDB-Tk.
Finally, to differentiate the MAGs of ancient origin from those modern ones, we used Bowtie2 (Langmead and Salzberg, 2012) to map the quality filtered raw reads against these MAGs, then we sorted and indexed the resulted bam files using SAMtools (Li et al., 2009), and implemented DamageProfiler (Neukamm et al., 2021) to check for ancient DNA damage patterns, i.e. C-to-T and G-to-A substitutions, resulting from cytosine deamination.
Quantification and statistical analysis
The abundance of MAGs in each of the samples were shown as row z-scores calculated and visualized, in R-Studio (https://www.rstudio.com/) using the pheatmap package.
Acknowledgments
Support was provided by the European Regional Development Fund 2014–2020_CALL-FESR 2017 Research and Innovation_Autonomous Province of Bolzano - South Tyrol_Project: FESR1078-MummyLabs. The Sampling campaign was broadcasted in the ZDF documentary “Terra X -Geheimnisse aus der Tiefe” (https://www.zdf.de/dokumentation/terra-x/geheimnisse-aus-der-tiefe-mit-florian-huber-100.html). The authors thank the Department of Innovation, Research and University of the Autonomous Province of Bozen/Bolzano, Italy for covering the Open Access publication costs.
Author contributions
Conceptualization, M.S.S., M.F., and F.M.; Methodology, M.S.S. and F.M.; Formal Analysis, M.S.S.; Investigation, M.S.S.; Resources, A.L., R.S., G.W., M.F., A.Z., and F.M.; Writing – Original Draft, M.S.S.; Writing – Review & Editing, M.S.S., G.W., M.F., A.Z. A.T., and F.M.; Visualization, M.S.S.; Supervision, F.M.; Project Administration, M.F., A.Z., and F.M.; Funding Acquisition, M.F., A.Z., and F.M.
Declaration of interests
The authors declare no competing interests.
Published: January 1, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103397.
Contributor Information
Mohamed S. Sarhan, Email: mohamed.sarhan@eurac.edu.
Frank Maixner, Email: frank.maixner@eurac.edu.
Supplemental information
Data and code availability
-
•
The metagenomic shotgun sequencing data have been deposited at ENA: PRJEB47715 and are publicly available as of the date of publication.
-
•
All codes used in this study and other previously published genomic data is available at the sources referenced in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alneberg J., Bjarnason B.S., de Bruijn I., Schirmer M., Quick J., Ijaz U.Z., Lahti L., Loman N.J., Andersson A.F., Quince C. Binning metagenomic contigs by coverage and composition. Nat. Methods. 2014;11:1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Beghini F., McIver L.J., Blanco-Míguez A., Dubois L., Asnicar F., Maharjan S., Mailyan A., Manghi P., Scholz M., Thomas A.M. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife. 2021;10:e65088. doi: 10.7554/eLife.65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo E., Giguet-Covex C., Rouillard A., Nota K., Heintzman P.D., Vuillemin A., Ariztegui D., Arnaud F., Belle S., Bertilsson S. Lake sedimentary DNA research on past terrestrial and aquatic biodiversity: Overview and recommendations. Quaternary. 2021;4:6. [Google Scholar]
- Chaumeil P.A., Mussig A.J., Hugenholtz P., Parks D.H. GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedi N.T., Makk J., Kótai L., Berényi B., Klébert S., Sebestyén Z., Molnár Z., Borsodi A.K., Leél-Őssy S., Demény A. Cave bacteria-induced amorphous calcium carbonate formation. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-65667-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp L.S., Gussarova G., Boessenkool S., Olsen J., Haile J., Schrøder-Nielsen A., Ludikova A., Hassel K., Stenøien H.K., Funder S. Lake sediment multi-taxon DNA from North Greenland records early post-glacial appearance of vascular plants and accurately tracks environmental changes. Quat. Sci. Rev. 2015;117:152–163. [Google Scholar]
- Eriksen A.M.H., Nielsen T.K., Matthiesen H., Carøe C., Hansen L.H., Gregory D.J., Turner-Walker G., Collins M.J., Gilbert M.T.P. Bone biodeterioration—the effect of marine and terrestrial depositional environments on early diagenesis and bone bacterial community. PLoS One. 2020;15:e0240512. doi: 10.1371/journal.pone.0240512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman C., Sand K. Survival of environmental DNA in natural environments: Surface charge and topography of minerals as driver for DNA storage. bioRxiv. 2020 doi: 10.1101/2020.01.28.922997. 2020.2001.2028.922997. [DOI] [Google Scholar]
- Gelabert P., Sawyer S., Bergström A., Margaryan A., Collin T.C., Meshveliani T., Belfer-Cohen A., Lordkipanidze D., Jakeli N., Matskevich Z., et al. Genome-scale sequencing and analysis of human, wolf, and bison DNA from 25,000-year-old sediment. Curr. Biol. 2021 doi: 10.1016/j.cub.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haile J., Holdaway R., Oliver K., Bunce M., Gilbert M.T.P., Nielsen R., Munch K., Ho S.Y., Shapiro B., Willerslev E. Ancient DNA chronology within sediment deposits: Are paleobiological reconstructions possible and is DNA leaching a factor? Mol. Biol. Evol. 2007;24:982–989. doi: 10.1093/molbev/msm016. [DOI] [PubMed] [Google Scholar]
- Jónsson H., Ginolhac A., Schubert M., Johnson P., Orlando L. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D.D., Li F., Kirton E., Thomas A., Egan R., An H., Wang Z. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7:e7359. doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Cho Y., Lee J., Kim H.S., Jung J.Y., Kim E.S. Metagenomic analysis of postmortem-bone using next-generation sequencing and forensic microbiological application §. Microbiol. Soc. Korea. 2020;56:10–18. [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357. doi: 10.1038/nmeth.1923. https://www.nature.com/articles/nmeth.1923#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5448. pdb prot5448. [DOI] [PubMed] [Google Scholar]
- Mittnik A., Massy K., Knipper C., Wittenborn F., Friedrich R., Pfrengle S., Burri M., Carlichi-Witjes N., Deeg H., Furtwangler A., et al. Kinship-based social inequality in Bronze Age Europe. Science. 2019;366:731–734. doi: 10.1126/science.aax6219. [DOI] [PubMed] [Google Scholar]
- Neukamm J., Peltzer A., Nieselt K. DamageProfiler: Fast damage pattern calculation for ancient DNA. Bioinformatics. 2021 doi: 10.1093/bioinformatics/btab190. [DOI] [PubMed] [Google Scholar]
- Nurk S., Meleshko D., Korobeynikov A., Pevzner P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K., Conesa A., Garcia-Alcalde F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacton M., Breitenbach S.F., Lechleitner F.A., Vaks A., Rollion-Bard C., Gutareva O., Osintcev A., Vasconcelos C. The role of microorganisms in the formation of a stalactite in Botovskaya Cave, Siberia–paleoenvironmental implications. Biogeosciences. 2013;10:6115–6130. [Google Scholar]
- Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips A., Stolarek I., Kuczkowska B., Juras A., Handschuh L., Piontek J., Kozlowski P., Figlerowicz M. Comprehensive analysis of microorganisms accompanying human archaeological remains. GigaScience. 2017;6:gix044. doi: 10.1093/gigascience/gix044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay-Salisbury K. In: Body Parts and Bodies Whole: Changing Relations and Meanings. Rebay-Salisbury K., Sørensen M.L.S., Hughes J., editors. Oxbow; Oxford: 2010. Cremations: Fragmented Bodies in the Bronze and Iron Ages; pp. 64–71. [Google Scholar]
- Renaud G., Slon V., Duggan A.T., Kelso J. Schmutzi: Estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome Biol. 2015;16:224. doi: 10.1186/s13059-015-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. The UCSC genome browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Le S., Li Y., Hu F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One. 2016;11:e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P., Storå J., Götherström A., Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 2013;40:4477–4482. [Google Scholar]
- Slon V., Hopfe C., Weiss C.L., Mafessoni F., de la Rasilla M., Lalueza-Fox C., Rosas A., Soressi M., Knul M.V., Miller R., et al. Neandertal and denisovan DNA from pleistocene sediments. Science. 2017;356:605–608. doi: 10.1126/science.aam9695. [DOI] [PubMed] [Google Scholar]
- Stahlschmidt M.C., Collin T., Fernandes D., Bar-Oz G., Belfer-Cohen A., Gao Z., Jakeli N., Matskevich Z., Meshveliani T., Pritchard J. Ancient mammalian and plant DNA from late quaternary stalagmite layers at Solkota Cave, Georgia. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-43147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub R. Archäologische Funde aus der Wimsener Höhle (Kat. Nr.-7722/01), Schwäbische Alb–Frühgeschichtliche Nutzung oder Quellenkult vor 3.200 Jahren. Mitt. Verb. dt. Höhlen-u. Karstforscher. 2006;52:45–51. [Google Scholar]
- Straub R., Lehmkuhl A. Spätbronzezeitlicher Skelettfund aus der Wimsener Höhle, Schwäbische Alb. Baden-Württemberg. Mitteilungen des Verbands der deutschen Höhlen- und Karstforscher. 2009;553:86–90. [Google Scholar]
- Tang J.-N., Zeng Z.-G., Wang H.-N., Yang T., Zhang P.-J., Li Y.-L., Zhang A.-Y., Fan W.-Q., Zhang Y., Yang X. An effective method for isolation of DNA from pig faeces and comparison of five different methods. J. Microbiol. Methods. 2008;75:432–436. doi: 10.1016/j.mimet.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Turner-Walker G. Nederlandse Archeologische Rapporten (NAR); 2009. Degradation Pathways and Conservation Strategies for Ancient Bone from Wet, Anoxic Sites; pp. 659–675. [Google Scholar]
- Uritskiy G.V., DiRuggiero J., Taylor J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6:158. doi: 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B., Zavala E.I., Gómez-Olivencia A., Jacobs Z., Slon V., Mafessoni F., Romagné F., Pearson A., Petr M., Sala N. Unearthing neanderthal population history using nuclear and mitochondrial DNA from cave sediments. Science. 2021;372:eabf1667. doi: 10.1126/science.abf1667. [DOI] [PubMed] [Google Scholar]
- Weissensteiner H., Pacher D., Kloss-Brandstatter A., Forer L., Specht G., Bandelt H.J., Kronenberg F., Salas A., Schonherr S. HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44:W58–W63. doi: 10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willerslev E., Hansen A.J., Binladen J., Brand T.B., Gilbert M.T., Shapiro B., Bunce M., Wiuf C., Gilichinsky D.A., Cooper A. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science. 2003;300:791–795. doi: 10.1126/science.1084114. [DOI] [PubMed] [Google Scholar]
- Wu Y.W., Singer S.W. Recovering individual genomes from metagenomes using MaxBin 2.0. Curr. Protoc. 2021;1:e128. doi: 10.1002/cpz1.128. [DOI] [PubMed] [Google Scholar]
- Zepeda Mendoza M.L., Lundberg J., Ivarsson M., Campos P., Nylander J.A., Sallstedt T., Dalen L. Metagenomic analysis from the interior of a speleothem in Tjuv-Ante's cave, northern Sweden. PLoS One. 2016;11:e0151577. doi: 10.1371/journal.pone.0151577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Xu Q., He S., Ye W., Cao R., Wang P., Ling Y., Yan X., Wang Q., Zhang G. GTDB: An integrated resource for glycosyltransferase sequences and annotations. Database (Oxford) 2020;2020 doi: 10.1093/database/baaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The metagenomic shotgun sequencing data have been deposited at ENA: PRJEB47715 and are publicly available as of the date of publication.
-
•
All codes used in this study and other previously published genomic data is available at the sources referenced in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.