Abstract
The Crithidia fasciculata cycling sequence binding protein (CSBP) binds with high specificity to sequence elements in several mRNAs that accumulate periodically during the cell cycle. Mutations in these sequence elements abolish both cycling of the mRNA and binding of CSBP. Two genes, CSBPA and CSBPB, encoding putative subunits of CSBP have been cloned and were found to be present in tandem on the same DNA molecule and to be closely related. CSBPA and CSBPB are predicted to encode proteins with sizes of 35.6 and 42.0 kDa, respectively. Both CSBPA and CSBPB proteins have a predicted coiled-coil domain near the N terminus and a novel histidine and cysteine motif near the C terminus. The latter motif is conserved in other trypanosomatid species. Gel sieving chromatography and glycerol gradient sedimentation results indicate that CSBP has a molecular mass in excess of 200 kDa and an extended structure. Recombinant CSBPA and CSBPB also bind specifically to the cycling sequence and together can be reconstituted to give an RNA gel shift similar to that of purified CSBP. Proteins in cell extracts bind to an RNA probe containing six copies of the cycling sequence. The RNA-protein complexes contain both CSBPA and CSBPB, and the binding activity cycles in near synchrony with target mRNA levels. CSBPA and CSBPB mRNA and protein levels show little variation throughout the cell cycle, suggesting that additional factors are involved in the cyclic binding to the cycling sequence elements.
Gene organization and expression in trypanosomes differ significantly from those of higher eukaryotes. Trypanosome genes typically lack introns, although a single example of an intron-containing gene in Trypanosoma brucei and Trypanosoma cruzi has been reported recently (15). Also, protein-coding genes are generally organized into long polycistronic transcription units. Polycistronic transcripts are not observed, however, since cleavage, trans-splicing of a 39-nucleotide (nt) spliced leader sequence to the 5′ end of all mRNAs, and polyadenylation of 3′ ends occur rapidly to produce individual mRNAs. Consequently, most regulation of mRNA levels is exerted posttranscriptionally (23).
The most extensive studies of polycistronic transcription units are those of telomeric expression sites used by the variant surface glycoprotein (VSG) genes during the bloodstream phase of T. brucei. Expression site-associated gene (ESAG) mRNA and VSG mRNA levels differ by more than 100-fold, despite their being transcribed at the same level (13). For parasites such as T. brucei, which undergo morphological and physiological transformations during differentiation of mammalian bloodstream forms to that characteristic of the midgut of the insect vector, many genes show a high level of stage-specific expression. In the case of VSG mRNA, sequences within the 3′ untranslated region (UTR) of the mRNA were shown to influence gene expression (2, 12). Expression of the T. brucei procyclins, which are the major surface glycoproteins of the insect form of the parasite, was also shown to be strongly regulated at the posttranscriptional level due to sequences within the 3′ UTR (11). The levels of abundance of life cycle-specific mRNAs in other trypanosomatid species have also been shown to be dependent on sequences within their 3′ UTRs (1, 7, 20). Much less is known about the possible role of 5′ UTR sequences in gene expression in trypanosomatids. Recent studies of the amastin and tuzin gene cluster in T. cruzi have shown that a short open reading frame in the tuzin 5′ UTR acts in concert with the tuzin spliced leader acceptor site to decrease expression of a downstream reporter gene (22).
Replication of the mitochondrial DNA or kinetoplast DNA (kDNA) in the trypanosomatid Crithidia fasciculata occurs in approximate synchrony with nuclear DNA synthesis (6, 17). In most other eukaryotes, no cell cycle coordination of nuclear and mitochondrial DNA synthesis has been observed. Rather, mitochondrial DNA replication appears to take place throughout the cell cycle (3, 8, 24). To begin to understand the basis for this coordination in Crithidia, we have examined mRNA levels of several nuclear and kDNA replication genes. The mRNA levels of genes encoding the large and middle subunits of the nuclear single-strand DNA-binding protein (RPA1), dihydrofolate reductase-thymidylate synthetase (DHFR-TS), the kinetoplast type II DNA topoisomerase (TOP2), and histone H1-like protein (KAP3) all cycle in parallel as cells progress through the cell cycle (10, 17). The mRNA levels are maximal just prior to S phase and then decline sharply as DNA synthesis is completed. For the RPAI and TOP2 genes, the rate of synthesis of their protein products was shown to closely parallel their mRNA levels (5, 9). Mutational analysis of cloned versions of TOP2 and RPAI identified small sequence elements in the 5′ UTR that are required for the periodic accumulation of these mRNAs (5, 14). An octamer consensus sequence or cycling sequence present in the 5′ UTR of these mRNAs is required for cycling of the TOP2 and RPAI mRNA levels during the cell cycle. The central hexamer (AUAGAA) in this sequence is absolutely conserved in these mRNAs, and mutations within the hexamer abolish cyclic accumulation of the mRNAs. Insertion of six copies of the octamer into the 5′ UTR of a reporter gene has also been shown to confer cyclic accumulation on the mRNA. We have recently purified a protein, termed the cycling sequence binding protein (CSBP; previously referred to as CEBP), that specifically binds to mRNAs containing wild-type cycling sequences, but not to cycling sequences containing single-base substitutions. Two proteins were purified previously based on specific binding to the hexamer sequence (14). We have identified genes encoding each protein (CSBPA and CSBPB) and have initiated studies of the role of these proteins in regulating mRNA levels.
MATERIALS AND METHODS
Cloning of CSBPA and CSBPB genes.
The CSBP proteins were purified as described previously (14). Partial sequence analysis of tryptic peptides of each protein were obtained by the Harvard Microchemistry Facility and used to design degenerate primers for PCR amplification of chromosomal sequences. A 189-bp CSBPA PCR product was obtained by using forward primer E54 (TACCAGACSGARAARCC) and reverse primer E47 (CCSCCBGCGTTGTTRTTRTT). A 266-bp CSBPB PCR product was obtained by using forward primer E57 (TACGCSGAGGTSAARGAYCC) and reverse primer E53 (CGSGGSCCRTCGTTSACNGC). These PCR products were cloned and sequenced and then used as probes to screen a λGEM11 C. fasciculata genomic library (18). Five of six phage clones contained inserts that hybridized to both CSBPA and CSBPB probes. A 6.3-kb fragment was subcloned from one λ clone as a NotI-BglII fragment into pGEM11 plasmid (Promega) cut with NotI and BamHI to yield plasmid pJH37. An adjacent genomic fragment was cloned as a SacI-BglII fragment into pGEM11 cut with SacI and BamHI to yield plasmid pJH37.2. Inserts in both plasmids were sequenced with the New England Biolabs transposon-based Genome Priming System GPS-1. Products of PCR cycle sequencing reactions were sequenced by the UCLA DNA Sequencing Facility.
Overexpression and purification of recombinant CSBPA and CSBPB.
To produce recombinant His-tagged CSBP proteins, the genes encoding CSBPA and CSBPB were cloned in the pET22b(+) expression vector (Novagen) by PCR amplification. Plasmid pJH3748 was constructed by combining the inserts in plasmids pJH37 and pJH37.2 and thus contains the contiguous Crithidia genomic fragment shown in Fig. 1. pJH3748 was used as the template for PCR amplification of CSBPA with the primers F21 (GACCAATACACACACCATATGAAGGCGAAC) and F22 (AGCGAAGCGGGCTCGAGTGCCTTCACGCCC). The forward primer F21 contained an NdeI site upstream of the initiation codon. The reverse primer F23 contained an XhoI site engineered to remove the stop codon of CSBPA and to allow the synthesis of a six-histidine tag. Amplification with Vent polymerase (New England Biolabs) by using 0.1 μg of pJH3748 DNA and oligonucleotides F21 and F23 was carried out in a thermal cycler for 25 cycles (45 s at 94°C, 30 s at 55°C, and 1 min at 72°C). The resulting 1-kb PCR product was digested with NdeI and XhoI and ligated with gel-purified pET22b(+) DNA digested with NdeI and XhoI to generate the bacterial expression plasmid pBM1 encoding His-tagged CSBPA.
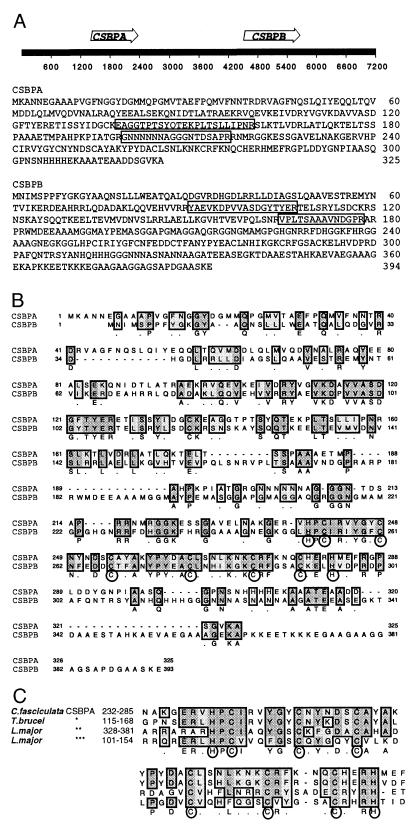
FIG. 1.
CSBP sequence analysis. (A) Genomic organization and predicted amino acid sequences of the CSBPA and CSBPB genes. CSBPA and CSBPB proteins have predicted molecular masses of 35,574 and 41,982 Da, respectively. The locations of coding sequences on 7.2 kb of genomic sequence are indicated by arrows. Sequences obtained by amino-terminal sequence analysis of tryptic peptides are enclosed in boxes. (B) Clustal alignment of the predicted amino acid sequences of CSBPA and CSBPB. Identical residues are indicated by dark shading, and similar residues are indicated by light shading. Conserved histidine and cysteine residues near the C terminus are circled. (C) Conservation of the cysteine- and histidine-rich domain of C. fasciculata CSBP in other trypanosomatid species. A consensus sequence is shown underneath, with the conserved histidine and cysteine residues enclosed in circles. Asterisks indicate translated sequences from the T. brucei genomic DNA fragment (∗; accession no. AQ942261) and the L. major predicted gene on chromosome 1 (∗∗ and ∗∗∗; accession no. AAC24631).
Cloning of CSBPB into the pET22b(+) vector was performed in a similar manner, with F24 (CAGACGGACCCACACATATGAACATCATGT) as the forward primer containing the NdeI restriction site and F26 (CACCACCGCATCTCGAGCTCCTTCGAGGCC) as the reverse primer with an XhoI-cut site. Amplification was carried out with Vent polymerase under the conditions described for CSBPA amplification. The 1.2-kb PCR product was digested with NdeI and XhoI and cloned into NdeI-XhoI-cut pET22b(+) vector as described earlier for pBM1 to generate the plasmid pBM3 for expression of His-tagged CSBPB.
For overexpression, plasmids pBM1 and pBM3 were individually transformed into Escherichia coli BL21(DE3), a strain that expresses the T7 polymerase gene under the control of a lacUV5 promoter. Expression in E. coli (100-ml culture) was induced at an optical density at 600 nm (OD600) of 0.7 by addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Both proteins were expressed at a high level, but were found largely in inclusion bodies. To purify inclusion bodies, cells harvested from 100-ml cultures were resuspended in 10 ml of buffer A (20 mM Tris [pH 7.5], 10 mM EDTA, 1% Triton X-100) and lysed by sonication. The inclusion bodies were harvested by centrifugation at 10,000 × g for 10 min at room temperature and further purified by repeated washing with buffer A. Inclusion bodies were solubilized in buffer B (50 mM 3-[cyclohexylamino]-1-propanesulfonic acid [CAPS; pH 11.0], 0.3% N-laurylsarcosine, 0.1 mM dithiothreitol [DTT]) at a final protein concentration of 2 to 5 mg/ml, incubated in buffer B for 15 min at room temperature, and centrifuged at 12,000 × g for 15 min. The supernatant containing the solubilized protein was extensively dialyzed against buffer C containing 20 mM Tris (pH 7.9), 50 mM NaCl, and 0.1 mM DTT.
The His-tagged proteins were purified by Ni-column chromatography with His-Bind resin (Novagen) according to the manufacturer's protocol. Renatured CSBPA or CSBPB proteins were loaded onto a 1-ml His-Bind column. After the column had been washed thoroughly with buffer D containing 20 mM Tris (pH 7.9), 500 mM NaCl, and 80 mM imidazole, the His-tagged proteins were finally eluted with buffer D containing 500 mM imidazole.
In vitro transcription and gel retardation assay.
Gel retardation assays were performed with 32P-labeled RNA probes containing six copies of either the wild-type consensus octamer cycling sequence recognized by CSBP or six copies with a single-base substitution mutant sequence to which CSBP does not bind. Wild-type and mutant 6X octamer RNA probes were synthesized by in vitro transcription reactions with NotI-linearized plasmids pRM16 and pRM20 as described previously (14). Gel retardation assays were performed with 32P-labeled RNA probes in the presence of 6.7 mg of heparin per ml as a nonspecific competitor and the RNase inhibitor RNAsin (10 U/reaction). Binding reactions were carried out at 28°C for 15 min, after which the RNA-protein complexes were resolved by electrophoresis at 4°C for 35 min (Fig. 2C) or 45 min (Fig. 2A and B) at 200 V on a pre-electrophoresed (200 V for 20 min, 0.5× Tris-borate-EDTA [TBE]buffer), 0.75-mm-thick, 6% (acrylamide/bisacrylamide ratio of 60:1) nondenaturing polyacrylamide minigel (Bio-Rad). Alternatively (Fig. 3B), complexes were resolved by electrophoresis at 4°C for 130 min at 150 V on a pre-electrophoresed (150 V for 45 min, 0.5× TBE buffer), 0.5-mm-thick, 4% (acrylamide/bisacrylamide ratio of 60:1) nondenaturing 15-cm polyacrylamide gel (Hoefer). After electrophoresis, the gels were fixed in 10% isopropanol–5% acetic acid solution, dried, and exposed to X-ray films at −70°C with intensifying screens.
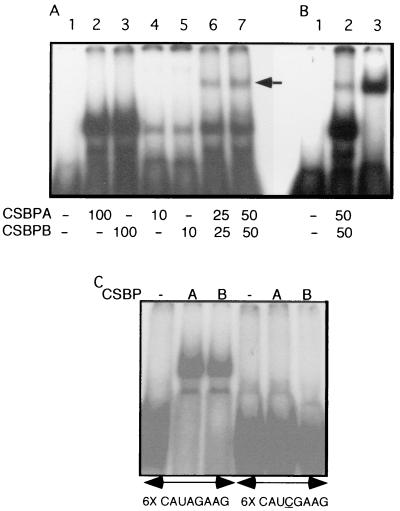
FIG. 2.
Recombinant CSBPA and CSBPB binding to the 6X octamer RNA. (A) 6X octamer RNA probe (lane 1) was incubated with various concentrations of recombinant CSBPA (lanes 2 and 4) and CSBPB (lanes 3 and 5) alone or together (lanes 6 and 7). The amount of each protein (in micrograms) is indicated underneath the figure. The arrow shows the slower-migrating RNA-protein complex following incubation with CSBPA and CSBPB together. (B) Comparison of the retarded complexes obtained following incubation of the 6X octameric probe (lane 1) with CSBPA and CSBPB together (lane 2) or with the purified CSBP from C. fasciculata (lane 3). (C) Binding specificity of recombinant CSBPA and CSBPB. 6X wild-type and mutant octameric RNAs were used as a probe in gel retardation assays. Purified recombinant CSBPA or CSBPB (0.1 μg per reaction) was used to investigate RNA binding specificity. Mutant octamer differs from the wild type in having an A-to-C mutation (underlined) in the fourth position of each of the six octamer sequences.
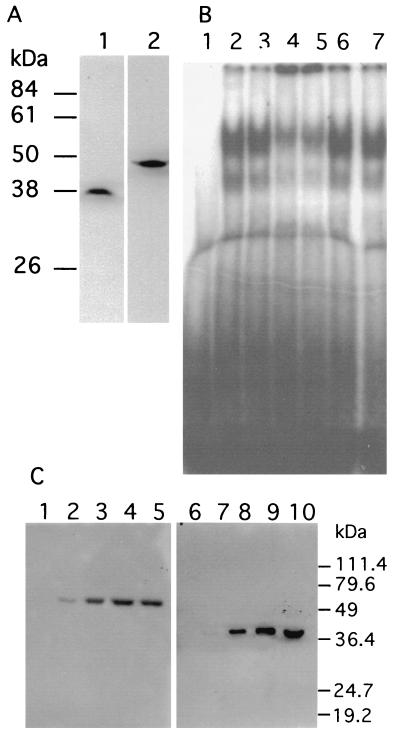
FIG. 3.
Immunological characterization of CSBP. (A) Specificity of antibodies against CSBPA and CSBPB. Immunoblots of Crithidia whole-cell extracts were probed with affinity-purified antibodies against CSBPA (lane 1) or CSBPB (lane 2). (B) Supershift of gel-retarded complexes by antibodies against CSBPA and CSBPB. Crithidia whole-cell extracts were preincubated for 15 min in the absence (lane 2) or presence of preimmune serum (lane 3), 0.5 μg of affinity-purified anti-CSBPA (lane 4), or 0.4 μg of affinity-purified anti-CSBPB (lane 5) or anti-SSE1 (lane 6) and anti-RPA1 (lane 7) antisera. Each reaction mixture contained approximately 0.5 μg of immunoglobulin G. The reaction mixtures were then further incubated for another 15 min at 30°C following the addition of 32P-labeled 6X octameric RNA probe and analyzed by electrophoresis on a 4% nondenaturing polyacrylamide gel. Lane 1 shows probe alone in absence of cell extract. (C) Immunoprecipitation of CSBPA and CSBPB. Crithidia nuclear extract (300 μg) was used in immunoprecipitation reactions either with rabbit preimmune serum (lanes 1 and 6) or with 2.5 (lanes 2 and 7), 10 (lanes 3 and 8), or 40 (lanes 4 and 9) μg of affinity-purified polyclonal anti-CSBPA antibody. Immunoprecipitation reactions were also performed in the presence of 20 μg of RNase A (lanes 5 and 10) by using 40 μg of the anti-CSBPA antibody. The immunoprecipitates were then Western blotted and probed with either horseradish peroxidase-conjugated anti-CSBPB peptide antibody (lanes 1 to 5) or with biotinylated anti-CSBPA peptide antibody (lanes 6 to 10). The biotinylated antibody was detected with streptavidin-peroxidase.
Gel sieving chromatography and glycerol gradient sedimentation analysis.
CSBP was purified as previously described (14) through a UNO Q anion-exchange column (Bio-Rad) purification step. The most active UNO Q fraction was stabilized by addition of bovine serum albumin (BSA), concentrated in a Centricon 30 concentrator, diluted to reduce the salt concentration, and then reconcentrated. The final buffer contained 50 mM Tris, 150 mM NaCl, 1 mM DTT, 20% glycerol, and 80 μg of BSA per ml. Fifty-microliter samples were chromatographed on a Superose 12 fast protein liquid chromatography column in the buffer described above at 0.2 ml/min at 23°C. Marker proteins were chromatographed in 50 μl of the same buffer under identical conditions. Also, 50-μl UNO Q samples were diluted threefold with buffer lacking glycerol, loaded onto a 4.9-ml 10 to 30% glycerol gradient, and centrifuged in an SW 50.1 rotor at 42,000 rpm at 23°C for 3.5 h. Fractions were collected by dripping from the bottom of the tube. Marker proteins were centrifuged and collected under the same conditions. All column and gradient fractions were assayed by RNA gel retardation (14). Finally, the peak fraction from Superose 12 was diluted threefold and then centrifuged on a glycerol gradient, as described above, and the peak fraction from a glycerol gradient was chromatographed on Superose 12, as described above.
Antibodies.
Rabbit polyclonal antibodies were raised against the purified recombinant CSBPA and CSBPB proteins. Purified CSBPA (460 μg) and CSBPB (340 μg) were used for raising the polyclonal antisera in rabbits (Bethyl Laboratories). The antibodies were further purified from the antisera by affinity column chromatography on a column containing 3 mg of purified CSBPA or CSBPB linked to CNBr-activated Sepharose CL4B (Pharmacia Biotech) as per the manufacturer's protocol. The antisera were incubated at 55°C for 20 min and then allowed to cool to room temperature. The antisera were then centrifuged in a microcentrifuge, and the supernatants were loaded onto affinity columns (1 ml) preequilibrated with Tris-buffered saline (TBS) buffer (25 mM Tris [pH 8.0], 137 mM NaCl, 27 mM KCl). After the columns had been washed with TBS buffer containing 300 mM NaCl, the columns were eluted with 6 ml of 0.1 M glycine (pH 2.5). Fractions with a 500-μl volume were collected into tubes containing 25 μl of 10-mg/ml BSA and 50 μl of 1 M Tris base for pH neutralization.
Rabbit anti-CSBPA peptide antibodies and anti-CSBPB peptide antibodies were prepared by Bethyl Laboratories, Inc., against the peptides CETMPAHKPIATGRGN and CGAAGAAGGAGSAPDGAASKE, respectively. Each peptide was conjugated to keyhole limpet hemocyanin (KLH) as a carrier, using maleimide chemistry, and injected into animals as an immunogen. Hyperimmune sera were affinity purified to capture antibodies specific for each peptide. A portion of the anti-CSBPA antibodies was biotinylated, and a portion of the anti-CSBPB antibodies was conjugated to horseradish peroxidase.
Quantitative immunoblotting.
To estimate the relative amounts of CSBPA and CSBPB per cell, a culture of C. fasciculata was harvested, suspended in sodium dodecyl sulfate (SDS) gel loading buffer, electrophoresed through a 12% polyacrylamide gel, and immunoblotted as described previously (4). Blots were probed with either affinity-purified anti-CSBPA or anti-CSBPB antibodies and subsequently probed with 125I-labeled protein A (ICN). Various amounts of purified recombinant CSBPA and CSBPB were electrophoresed on the same gel and transferred to the same blot to permit quantitation of the amount of each protein in cell lysates. Radioactive bands were quantitated with a Molecular Dynamics PhosphorImager.
RESULTS
CSBPA and CSBPB gene cloning.
Both CSBPA and CSBPB were cloned from a lambda genomic library by hybridizing plaque lifts with DNA probes made by PCR amplification of chromosomal DNA. Degenerate PCR primers were designed based on partial amino acid sequences of tryptic peptides derived from the purified proteins. Surprisingly, both the CSBPA and CSBPB probes hybridized to DNA from a single phage clone, indicating that both genes are contained on the same DNA. Sequence analysis of 7.2 kb of the DNA insert confirmed this conclusion and showed that the CSBPA and CSBPB genes are oriented in the same direction, with their coding sequences separated by 2.1 kb (Fig. 1A). CSBPA and CSBPB are predicted to encode proteins with sizes of 35.6 and 42.0 kDa, respectively, and correspond to the proteins previously estimated to have molecular masses of 38 kDa (CSBPA) and 48 kDa (CSBPB), based on their migration on SDS gels (14). The differences between the predicted molecular masses and those estimated by SDS gel electrophoresis possibly reflect effects of the overall amino acid composition of the proteins (16).
Inspection of these sequences revealed that the CSBPA and CSBPB genes are related, as shown in the Clustal alignment of the predicted amino acid sequences (Fig. 1B). Both the CSBPA and CSBPB proteins are predicted to have a coiled-coil structure in the amino-terminal region between residues 71 and 114 in CSBPA and the corresponding region in CSBPB (residues 52 to 95). In addition, a histidine and cysteine motif is conserved in the C-terminal portion of these proteins. Database searches revealed the presence of the latter motif in predicted protein sequences from Leishmania major and T. brucei as well (Fig. 1C). Interestingly, this motif is present in two copies in the L. major predicted protein.
Recombinant CSBPA and CSBPB.
Both CSBPA and CSBPB were subcloned into expression vectors and expressed as six-histidine-tagged proteins in an E. coli strain expressing the T7 RNA polymerase. The recombinant proteins were purified by metal chelate chromatography and used to characterize their RNA-binding properties. In previous studies (14), only CSBPA was detected in nuclear extracts analyzed by Northwestern blotting. However, in RNA gel shift experiments, both recombinant proteins were found to bind an RNA probe containing six copies of the consensus octamer cycling sequence (6X octamer RNA) (Fig. 2A). Each protein alone gave a gel-shifted species (Fig. 2A, lanes 2 to 5) that migrated faster than that observed for purified CSBP (Fig. 2B, lane 3). Incubation of recombinant CSBPA and CSBPB together prior to addition of the 6X octamer probe produced a small amount of an additional species that comigrated with that produced by CSBP purified from C. fasciculata (Fig. 2A, lanes 6 and 7; and B, lanes 2 and 3). These results suggest that an oligomeric form of CSBP can be assembled from the recombinant CSBPA and CSBPB proteins. This in vitro-assembled complex produces an RNA gel shift identical to that of purified CSBP (Fig. 2B, lanes 2 and 3). CSBPA and CSBPB are relatively abundant and are present in C. fasciculata in nearly equimolar amounts. Quantitative Western blots indicate that there are approximately 230,000 molecules of CSBPA and 170,000 copies of CSBPB per cell (data not shown).
Purified CSBP was shown previously to be highly specific in its binding to the octameric cycling sequence (14). Single-nucleotide substitutions in the first 5 nt of the conserved central hexamer abolished binding. Experiments performed with each of the recombinant proteins showed similar binding specificity. Figure 2C shows RNA gel shifts by recombinant CSBPA and CSBPB proteins obtained with either wild-type 6X octamer probe or a 6X octamer probe in which each copy of the octamer had an A-to-C mutation in the third nucleotide of the central hexamer. An RNA gel shift is only observed with the wild-type probe. These results indicate that both CSBPA and CSBPB are highly sequence specific in their binding to the cycling element sequence.
Immunological characterization of CSBPA and CSBPB.
Rabbit polyclonal antibodies prepared against recombinant CSBPA and CSBPB were affinity purified and used for immunological studies of the endogenous C. fasciculata proteins. The antibodies against CSBPA and CSBPB each specifically recognized single proteins in Crithidia cell extracts (Fig. 3A). Each of the proteins migrates at a position corresponding to that of the purified Crithidia protein.
To confirm the presence of both CSBPA and CSBPB in RNA gel-shifted complexes, we have used the CSBPA and CSBPB antibodies to attempt to supershift the complexes. As shown in Fig. 3B, antibodies against CSBPA and CSBPB shifted a large fraction of the complexes from whole-cell extracts into the well (lanes 4 and 5), whereas preimmune antisera (lane 3) or antisera against either SSE1 protein (lane 6) or RPA1 (lane 7) did not. A small fraction of the complexes could not be supershifted and may represent an additional comigrating complex that remains to be characterized. The presence of both CSBPA and CSBPB in the RNA gel-shifted complexes appears to be a consequence of their direct interaction with each other, since immunoprecipitation of CSBPA from nuclear extracts also coprecipitates CSBPB, even in the presence of RNase A (Fig. 3C). However, the possibility that the association of these proteins could involve a small RNA species that is protected from cleavage by RNase is not excluded.
Purified CSBP exists as an oligomeric complex.
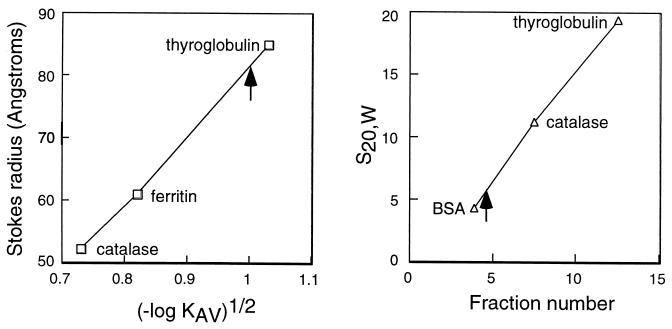
The possibility that the CSBPA and CSBPB proteins represent subunits of an oligomeric complex was investigated further by gel sieving chromatography and glycerol gradient sedimentation analysis of purified CSBP. The molecular mass of the complex can be calculated from the Stokes radius measured by gel sieving chromatography and the sedimentation coefficient measured by velocity gradient sedimentation (21). CSBP binding activity eluted from a Superose 12 column consistent with a Stokes radius of 80 Å and sedimented in a glycerol gradient at 6.3S (Fig. 4). In the data shown here, the CSBP sample analyzed by gel sieving chromatography had been purified by glycerol gradient sedimentation of the UNO Q fraction and the sample analyzed by glycerol gradient sedimentation had been purified by Superose 12 gel sieving chromatography of the UNO Q fraction. Essentially identical results were obtained upon direct analysis of the UNO Q fraction by each method. Calculation of the molecular mass of the CSBP complex based on these data indicates a mass of 209 kDa with a frictional ratio of 2.0. Quantitative Western blots of purified CSBP indicated that CSBPA and CSBPB were present in approximately equimolar amounts in the purified complex (data not shown). Within the error of these measurements, the mass of purified CSBP suggests that the complex could possibly be an A3B3 hexamer. The predicted molecular masses of CSBPA and CSBPB are 35.6 and 42.0, which would result in a molecular mass of 233 kDa for such a complex. The unusually high frictional ratio suggests a highly asymmetric structure for the oligomeric complex (21).
FIG. 4.
Superose 12 gel sieving chromatography and glycerol gradient sedimentation analysis of purified CSBP. (A) Superose 12 gel sieving chromatography of glycerol gradient fraction of CSBP performed as described in Materials and Methods. The marker proteins run under the same conditions were thyroglobulin (85 Å), ferritin (61 Å), and catalase (52.2 Å). CSBP binding activity eluted at the position shown by the arrow. (B) Glycerol gradient sedimentation of the Superose 12 fraction of CSBP. The marker proteins run in parallel gradients were thyroglobulin (19.4S), catalase (11.3S), and BSA (4.4S). In both experiments, fractions were assayed for CSBP binding activity by RNA gel retardation with the 6X octamer probe. The peak of CSBP binding activity is indicated by an arrow.
Cycling of CSBP binding activity.
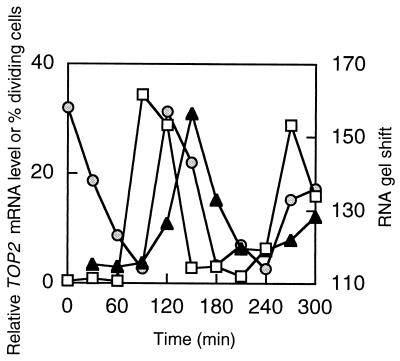
We have examined whole-cell extracts of a synchronous culture for binding to the 6X octamer probe. Samples taken at 30-min intervals were used for RNA gel shifts and quantitated by PhosphorImager analysis. Binding to the RNA probe is shown in Fig. 5, along with results of quantitative Northern analysis of a representative target mRNA (TOP2). The percentage of dividing cells is also shown as an indication of cell synchrony. Binding to the 6X octamer probe cycles and reaches a maximum following cell division and just prior to attainment of the maximum level of the target mRNA. The rapid decline in specific RNA-binding activity from 120 to 180 min is closely followed by a sharp decline in the target mRNA level. Binding to an RNA probe derived from the TOP2 5′ UTR was previously observed to cycle in the same manner (14). No binding of these extracts was observed when the 6X octamer probe contained a single nucleotide substitution in each copy of the octamer.
FIG. 5.
Cell cycle-dependent binding to 6X octamer RNA. A C. fasciculata culture was synchronized by hydroxyurea arrest, and nuclear extracts were prepared at 30-min intervals following release from arrest. Circles, RNA gel shift with 8 μg of nuclear extract by a 32P-labeled 6X octamer probe; triangles, TOP2 mRNA level determined by Northern blotting with a 32P-labeled probe for TOP2 coding sequence; squares, percentage of cells having two nuclei as a measure of cell synchronization. RNA gel shifts and Northern blots were quantitated by PhosphorImager analysis.
Expression of CSBP genes during the cell cycle.
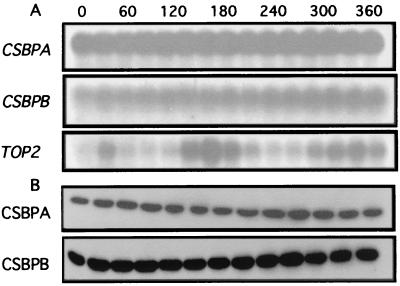
To investigate the basis for the cyclic binding of CSBP to RNA probes containing cycling sequence elements, we performed both Northern and Western blotting of cell extracts prepared at 30-min intervals following release from a hydroxyurea block. Figure 6A shows the results of Northern blots obtained with coding sequence probes for CSBPA, CSBPB, and a target gene (TOP2). Surprisingly, the CSBPA and CSBPB mRNA levels are observed to be relatively constant throughout the cell cycle, whereas the TOP2 mRNA cycles strongly, as reported previously (17). We note that neither CSBPA nor CSBPB 5′ or 3′ UTRs contain the cycling sequence elements identified in the TOP2, RPA1, DHFR-TS, and KAP3 mRNAs (5, 9, 10, 17).
FIG. 6.
Expression of CSBPA and CSBPB during the cell cycle. (A) Northern blots of mRNAs expressed from CSBPA, CSBPB, and TOP2 at 30-min intervals in a synchronous culture of C. fasciculata. Ten micrograms of total RNA was loaded in each lane of the gel. The resulting blot was probed three separate times with probes for the individual coding sequences. The estimated sizes of the CSBPA and CSBPB mRNAs are 2.7 and 2.3 kb, respectively. (B) Western blots of whole-cell extracts (120 μg per lane) probed with antisera against CSBPA and CSBPB.
Western blots of the cell extracts with antibodies against CSBPA and CSBPB indicate that the CSBPA and CSBPB protein levels are also relatively constant throughout the cell cycle, consistent with the mRNA levels (Fig. 6B). These results suggest that factors other than the levels of the CSBPA and CSBPB polypeptides may modulate the CSBP binding activity during the cell cycle. Possibilities include a minor subunit that may not have been detected or that may have been lost during purification, structural rearrangement of the oligomeric complex, or a posttranslational modification of one of the subunits. While phosphorylation of one or both subunits is an attractive hypothesis, we have been unable to detect phosphorylation of either CSBPA or CSBPB.
DISCUSSION
The recent purification of CSBP based on specific RNA binding to cycling sequence elements in target mRNAs has allowed the cloning of genes encoding the subunits of this oligomeric complex. The unexpected finding that the CSBPA and CSBPB genes are closely related and present in tandem in the genome suggests that this relationship resulted from gene duplication and subsequent divergence. Both proteins are predicted to have retained a two-stranded coiled-coil domain near the amino end of each protein and a novel histidine-cysteine motif near the carboxyl terminus. The latter motif may represent a metal ion-binding motif and is also present in proteins of T. brucei and L. major based on predicted amino acid sequences from genomic DNA sequences.
Analysis of CSBP by both glycerol gradient sedimentation and gel sieving chromatography indicates that CSBP exists as an oligomeric complex, possibly an A3B3 hexamer. Both CSBPA and CSBPB were observed to be present in nearly equimolar amounts in purified CSBP and were found to coimmunoprecipitate from cell extracts. From gradient sedimentation alone, CSBP was shown previously to sediment at a rate close to that of T7 RNA polymerase, a 98-kDa protein (14). However, gel sieving chromatography indicated a higher molecular mass for the complex, and the combined S value and Stokes radius yield a molecular mass of 209 kDa with a frictional ratio of 2.0. This unusually large frictional ratio is consistent with the cosedimentation of CSBP with a much smaller protein and its coelution in gel sieving chromatography with much larger proteins, and it underscores the need for both measurements in determining the molecular mass of highly asymmetric proteins (21).
Recombinant forms of CSBPA and CSBPB bind to the cycling sequence RNA probe with the same sequence specificity as the oligomeric CSBP complex and together can produce a gel-shifted RNA-protein complex that migrates at the same rate as that produced by purified CSBP. Only a small amount of this species is formed upon preincubation of recombinant CSBPA and CSBPB, indicating inefficient assembly of the oligomeric complex under the conditions used here. However, efficient in vivo assembly has been achieved recently by coexpression of CSBPA and CSBPB in E. coli (unpublished observations).
Two lines of evidence implicate CSBP binding to cycling element sequences in target mRNAs in the modulation of these mRNA levels during the cell cycle. First, CSBP binds to cycling element sequences with high specificity. Mutations in cycling elements present in the 5′ UTR of TOP2 and RPA1 gene constructs were shown previously to abolish cycling of these mRNAs, and the same mutations also abolish binding by CSBP to the cycling elements (14). Second, the CSBP binding activity in cell extracts cycles and peaks just prior to the attainment of peak levels of the target mRNAs. The presence of both CSBPA and CSBPB in the specific RNA binding activity in cell extracts is demonstrated by antibody supershift experiments. Together these results strongly support a role for CSBP in the cell cycle regulation of mRNAs containing cycling sequence elements. Additional proteins are likely involved as well, including proteins that might facilitate the periodic binding of CSBP to cycling sequences and ones that might initiate degradation of target mRNAs.
Earlier studies have shown that mutation of the cycling elements in reporter gene transcripts results in a high level of the mRNA throughout the cell cycle, similar to the maximum mRNA level attained with wild-type cycling elements (5, 14, 19). This result implies that the cycling elements are negative regulatory elements. When the elements are mutated, the down-regulation that normally occurs prior to and following DNA replication does not occur. Consequently, the mutant transcript is at a high level throughout the cell cycle. A possible role for CSBP in modulating target mRNAs might be to confer protection on specific wild-type mRNAs during S phase, when these mRNAs are observed at their maximum levels. The proposed protection could result either directly from interaction of CSBP with the mRNA or indirectly from the recruitment of protective factors by CSBP. Ongoing experiments aimed at manipulating CSBP levels by gene disruption or constitutive expression from an episome should provide additional insight into the proposed role of CSBP in mRNA regulation.
ACKNOWLEDGMENTS
We thank Nancy Sturm for her comments on the manuscript.
This work was supported by National Institutes of Health grant GM53254 to D.S.R.
REFERENCES
- 1.Aly R, Argaman M, Halman S, Shapira M. A regulatory role for the 5′ and 3′ untranslated regions in differential expression of hsp83 in Leishmania. Nucleic Acids Res. 1994;22:2922–2929. doi: 10.1093/nar/22.15.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berberof M, Vanhamme L, Tebabi P, Pays A, Jefferies D, Welburn S, Pays E. The 3′-terminal region of the mRNAs for VSG and procyclin can confer stage specificity to gene expression in Trypanosoma brucei. EMBO J. 1995;14:2925–2934. doi: 10.1002/j.1460-2075.1995.tb07292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogenhagen D, Clayton D A. Mouse L cell mitochondrial DNA molecules are selected randomly for replication throughout the cell cycle. Cell. 1977;11:719–727. doi: 10.1016/0092-8674(77)90286-0. [DOI] [PubMed] [Google Scholar]
- 4.Brown G W, Melendy T E, Ray D S. Conservation of structure and function of DNA replication protein A in the trypanosomatid Crithidia fasciculata. Proc Natl Acad Sci USA. 1992;89:10227–10231. doi: 10.1073/pnas.89.21.10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown L M, Ray D S. Cell cycle regulation of RPA1 transcript levels in the trypanosomatid Crithidia fasciculata. Nucleic Acids Res. 1997;25:3281–3289. doi: 10.1093/nar/25.16.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosgrove W B, Skeen M J. The cell cycle in Crithidia fasciculata.Temporal relationships between synthesis of deoxyribonucleic acid in the nucleus and in the kinetoplast. J Protozool. 1970;17:172–177. doi: 10.1111/j.1550-7408.1970.tb02350.x. [DOI] [PubMed] [Google Scholar]
- 7.Coughlin B C, Teixeira S M, Kirchhoff L V, Donelson J E. Amastin mRNA abundance in Trypanosoma cruzi is controlled by a 3′-untranslated region position-dependent cis-element and an untranslated region-binding protein. J Biol Chem. 2000;275:12051–12060. doi: 10.1074/jbc.275.16.12051. [DOI] [PubMed] [Google Scholar]
- 8.Guttes E W, Hanawalt P C, Guttes S. Mitochondrial DNA synthesis and the mitotic cycle in Physarum polycephalum. Biochim Biophys Acta. 1967;142:181–194. doi: 10.1016/0005-2787(67)90526-6. [DOI] [PubMed] [Google Scholar]
- 9.Hines J C, Ray D S. Periodic synthesis of kinetoplast DNA topoisomerase II during the cell cycle. Mol Biochem Parasitol. 1997;88:249–252. doi: 10.1016/s0166-6851(97)00076-5. [DOI] [PubMed] [Google Scholar]
- 10.Hines J C, Ray D S. Tandem arrangement of two genes encoding kinetoplast-associated H1 histone-like proteins. Mol Biochem Parasitol. 1997;89:41–49. doi: 10.1016/s0166-6851(97)00099-6. [DOI] [PubMed] [Google Scholar]
- 11.Hotz H R, Hartmann C, Huober K, Hug M, Clayton C. Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 1997;25:3017–3026. doi: 10.1093/nar/25.15.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jefferies D, Tebabi P, Pays E. Transient activity assays of the Trypanosoma bruceivariant surface glycoprotein gene promoter: control of gene expression at the posttranscriptional level. Mol Cell Biol. 1991;11:338–343. doi: 10.1128/mcb.11.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson P J, Kooter J M, Borst P. Inactivation of transcription by UV irradiation of T. bruceiprovides evidence for a multicistronic transcription unit including a VSG gene. Cell. 1987;51:273–281. doi: 10.1016/0092-8674(87)90154-1. [DOI] [PubMed] [Google Scholar]
- 14.Mahmood R, Hines J C, Ray D S. Identification of cis and trans elements involved in the cell cycle regulation of multiple genes in Crithidia fasciculata. Mol Cell Biol. 1999;19:6174–6182. doi: 10.1128/mcb.19.9.6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mair G, Shi H, Li H, Djikeng A, Aviles H O, Bishop J R, Falcone F H, Gavrilescu C, Montgomery J L, Santori M I, Stern L S, Wang Z, Ullu E, Tschudi C. A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA. 2000;6:163–169. doi: 10.1017/s135583820099229x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noel D, Nikaido K, Ames G F. A single amino acid substitution in a histidine-transport protein drastically alters its mobility in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochemistry. 1979;18:4159–4165. doi: 10.1021/bi00586a017. [DOI] [PubMed] [Google Scholar]
- 17.Pasion S G, Brown G W, Brown L M, Ray D S. Periodic expression of nuclear and mitochondrial DNA replication genes during the trypanosomatid cell cycle. J Cell Sci. 1994;107:3515–3520. doi: 10.1242/jcs.107.12.3515. [DOI] [PubMed] [Google Scholar]
- 18.Pasion S G, Hines J C, Aebersold R, Ray D S. Molecular cloning and expression of the gene encoding the kinetoplast-associated type II DNA topoisomerase of Crithidia fasciculata. Mol Biochem Parasitol. 1992;50:57–67. doi: 10.1016/0166-6851(92)90244-e. [DOI] [PubMed] [Google Scholar]
- 19.Pasion S G, Hines J C, Ou X, Mahmood R, Ray D S. Sequences within the 5′ untranslated region regulate the levels of a kinetoplast DNA topoisomerase mRNA during the cell cycle. Mol Cell Biol. 1996;16:6724–6735. doi: 10.1128/mcb.16.12.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramamoorthy R, Swihart K G, McCoy J J, Wilson M E, Donelson J E. Intergenic regions between tandem gp63 genes influence the differential expression of gp63 RNAs in Leishmania chagasi promastigotes. J Biol Chem. 1995;270:12133–12139. doi: 10.1074/jbc.270.20.12133. [DOI] [PubMed] [Google Scholar]
- 21.Siegel L M, Monty K J. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 22.Teixeira S M, Kirchhoff L V, Donelson J E. Trypanosoma cruzi: suppression of tuzin gene expression by its 5′-UTR and spliced leader addition site. Exp Parasitol. 1999;93:143–151. doi: 10.1006/expr.1999.4446. [DOI] [PubMed] [Google Scholar]
- 23.Vanhamme L, Pays E. Control of gene expression in trypanosomes. Microbiol Rev. 1995;59:223–240. doi: 10.1128/mr.59.2.223-240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williamson D H, Moustacchi E. The synthesis of mitochondrial DNA during the cell cycle in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1971;42:195–201. doi: 10.1016/0006-291x(71)90087-8. [DOI] [PubMed] [Google Scholar]