Abstract
The precise assembly of the kinetochore complex at the centromere is epigenetically determined by substituting histone H3 with the centromere-specific histone H3 variant CENH3 in centromeric nucleosomes. A chromosome 1B reconstructed in wheat by centric misdivision from two wheat-rye centric translocations is known to carry a hybrid wheat-rye centromere. The resulting hybrid (dicentric)centromere is composed of both wheat and rye centromeric repeats. As CENH3 is a marker for centromere activity, we applied Immuno-FISH followed by ultrastructural super-resolution microscopy to address whether both or only parts of the hybrid centromere are active. Our study demonstrates that only the rye-derived centromere part incorporates CENH3 of wheat in the 1BL/1RS hybrid centromere. This finding supports the notion that one centromere part of a translocated chromosome undergoes inactivation, creating functional monocentric chromosomes to maintain chromosome stability.
Keywords: CENH3, 1BL/1RS, Robertsonian translocation, wheat, dicentric, rye, hybrid centromere
Introduction
The centromere is the assembly site for the proteinaceous kinetochore complex, which enables the proper segregation of sister chromatids and their transmission to the daughter cells during mitosis and meiosis. Despite the conserved function of the centromere, the primary constriction of monocentric species is generally enriched in highly diverged gene-free repetitive sequences. These repeats include satellite DNAs and retrotransposons (Barra and Fachinetti, 2018; Talbert and Henikoff, 2020). The centromeric DNA sequence is not sufficient to maintain the centromere position and function. The precise assembly of the kinetochore complex at the centromere is epigenetically determined by substituting a centromere-specific histone H3 variant CENH3 for histone H3 in centromeric nucleosomes (Henikoff and Dalal, 2005; Allshire and Karpen, 2008). Besides some diploid species encoding multiple functional CENH3 variants, e.g., barley (Sanei et al., 2011; Ishii et al., 2015), rye (Evtushenko et al., 2017, 2021), Fabeae species (Neumann et al., 2015), and cowpea (Ishii et al., 2020), most diploid eukaryotes encode only one variant of CENH3. In contrast, in allopolyploids species like wheat (Yuan et al., 2015), each parental subgenome possesses at least one CENH3 variant operating in the context of multiple species-specific centromeric sequences.
Defects in the pathways that regulate centromere assembly and function affect the proper chromosome segregation and lead to chromosome instability and lethality in various organisms (Maheshwari et al., 2015; Britt and Kuppu, 2016; Barra and Fachinetti, 2018). Also, the alteration of CENH3 that impairs its loading and maintenance results in chromosome missegregation (Au et al., 2008; Pidoux et al., 2009; Ravi and Chan, 2010; Karimi-Ashtiyani et al., 2015; Kuppu et al., 2015, 2020; Shrestha et al., 2017; Karimi-Ashtiyani, 2021).
Due to the high density of repetitive sequences and high similarity to non-homologous centromeres within a species, the primary constriction is vulnerable to breakage and recombination (Sullivan et al., 1996; Friebe et al., 2005; Barra and Fachinetti, 2018). Therefore, chromosome rearrangements and breaks often involve (peri)centromeric regions (Friebe et al., 2005; Lukaszewski, 2010; Barra and Fachinetti, 2018). Centric misdivisions, followed by the fusion of the broken arms from different chromosomes, result in whole-arm Robertsonian translocations (Robertson, 1961), which usually possess centromeric sequences of both chromosomes (Zhang et al., 2001; Song et al., 2016). However, to ensure mitotic stability, these chromosomes having a “hybrid (dicentric)centromere” convert to functionally monocentric chromosomes where only one centromere part remains active (Friebe et al., 2005; Han et al., 2006). It is suggested that centromere inactivation involves genomic and/or epigenomic reprogramming (Stimpson et al., 2012). Chromosome breaks and fusions occurred at centromeric and pericentromeric regions in wheat-rye translocation lines (Lukaszewski, 1997; Zhang et al., 2001; Friebe et al., 2005). The most common centric translocation is the 1BL/1RS translocation, in which the short arm of wheat chromosome 1B is replaced by the short arm of rye chromosome 1R (Lukaszewski, 2010; Moskal et al., 2021). Various 1BL/1RS translocations have served as valuable stocks in wheat breeding programs as chromosome 1R contains insect and disease resistance genes (Kim et al., 2004)1.
To address whether both or only one part of the 1BL/1RS hybrid centromere is active Wang et al. (2017) applied a combination of immuno-FISH, chromatin immunoprecipitation (ChIP)-qPCR, and RT-PCR. They concluded that all analyzed 1BL/1RS translocations contain hybrid centromeres and that the wheat-derived CENH3 is incorporated into both, the wheat- and rye-derived components of the hybrid centromere. The application of wheat and rye centromere-specific repeats, like the CRW clone 6C6 (Wang et al., 2017) and pAWRC.1/Bilby (Francki, 2001), respectively, allowed the visualization of the 1BL/1RS hybrid centromere. Both repeats are part of the CENH3-positive chromatin in either wheat (Liu et al., 2008; Li et al., 2013) or rye (Houben et al., 2011). None of the rye CENH3 variants is encoded by the short arm chromosome 1R (Evtushenko et al., 2021; Rabanus-Wallace et al., 2021). Hence, the 1BL/1RS-located CENH3 derives from wheat.
We extended the analysis and applied ultrastructural super-resolution microscopy to analyze the arrangement of the 1BL/1RS centromere at a resolution of ∼120 nm. Surprisingly, our findings demonstrate that only the rye-derived centromere incorporates CENH3 of wheat into the hybrid centromere of 1BL/1RS.
Materials and Methods
Plant Material and Cultivation
The 1Brec-1 line of cv. Pavon 76 carrying a reconstructed chromosome 1B (Lukaszewski, 1993, 1997) was grown in a greenhouse at 16 h light, 22°C day/16°C night conditions. Chromosome 1Brec-1 was reconstructed by centric misdivision from two centric wheat-rye translocations, 1RS.1BL and 1BS.1RL. In essence, the chromosome itself is a centric translocation, composed of a wheat chromosome arm 1BS from cv. Pavon 76 and 1BL arm from the translocation 1RS.1BL from the Aurora/Kavkaz source (Lukaszewski, 1993, 1997). As demonstrated by Zhang et al. (2001) this chromosome carries a hybrid centromere, composed in part of a wheat centromere and in part of a rye centromere.
Fluorescence in situ Hybridization and Indirect Immunostaining
Root tips and anthers of five individual plants of line 1BSPVN.1BLVEE were used for the preparation of mitotic and meiotic chromosomes, according to Gernand et al. (2003). Immunostaining followed by FISH was done as described by Ma et al. (2010). The rye centromere-specific repeat Bilby (Francki, 2001), and the wheat centromere-specific repeat CRW2 (Liu et al., 2008) labeled by nick translation with Alexa 488-dUTP (Invitrogen), and Cy5-dUTP (GE Healthcare Life Sciences), respectively, were used as FISH probes. The primary antibodies, rabbit anti-Os-CENH3 (Nagaki et al., 2004), and the secondary antibodies, rhodamine-conjugated anti-rabbit IgG (Dianova) were used for indirect immunostaining. Finally, the slides were mounted in antifade (Vectashield) containing 4,6-diamidino2-phenylindole (DAPI) as counterstain.
Microscopy
Imaging was performed by using an Olympus BX61 microscope and an ORCA-ER CCD camera (Hamamatsu). Deconvolution microscopy was employed for superior optical resolution of spherical structures. To investigate the centromeric chromatin ultrastructures at an optical resolution of ∼120 nm (super-resolution achieved with a 488 nm laser excitation), we applied spatial structural illumination microscopy (3D-SIM) using a 63/1.40 objective of an Elyra PS.1 super-resolution microscope system (Carl Zeiss GmbH), (Weisshart et al., 2016). 3D-SIM image stacks were used to perform surface rendering and to produce the movies via the Imaris 9.7 (Bitplane) software.
Results and Discussion
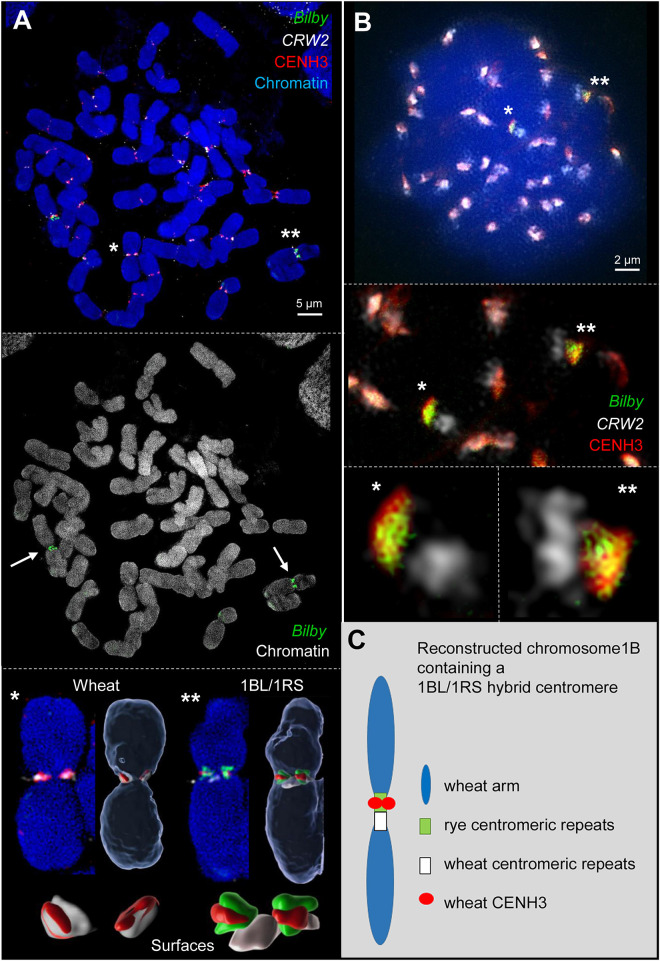
Fluorescence in situ hybridization with the rye- and wheat-specific centromere repeats pAWRC.1/Bilby (Francki, 2001) and CRW2 (Liu et al., 2008), respectively, demonstrated the hybrid structure of the 1BL/1RS centromere of the translocation line 1BSPVN.1BLVEE at somatic metaphase chromosomes (Figure 1A and Supplementary Figure 1). In line with previous publications (Lukaszewski, 1997; Zhang et al., 2001) one half of the hybrid centromere displayed rye and the other half wheat centromere-specific signals.
FIGURE 1.
Wheat CENH3 incorporates exclusively into rye centromere repeats containing chromatin of the 1BL/1RS hybrid centromere during mitosis (A) and meiosis (B) as revealed by 3D-SIM. (A) Somatic wheat metaphase cell containing two 1BL/1RS homologs labeled by rye-specific Bilby repeats (arrows). All wheat chromosomes incorporate wheat CENH3 into the wheat-specific centromeric CRW2 repeat-positive chromatin (*). Instead, wheat CENH3 loads only into the rye-specific Bilby repeat-positive chromatin of the translocated 1BL/1RS chromosomes (**). Surface rendering of the centromeric components (below) show clearly the different co-localizations (see also Supplementary Movies 1, 2). (B) Early meiotic metaphase I showing in addition to all labeled wheat centromeres both 1BL/1RS homologs labeled specifically by Bilby (asterisks). During meiosis wheat CENH3 places also exclusively into the rye derived centromeric chromatin as shown enlarged below. The co-localization of CENH3 and Bilby induces the yellow color. (C) Schematic representation of the reconstructed wheat chromosome 1B with a hybrid wheat-rye centromere. This centromere is composed of wheat and rye centromeric repeats. The wheat-derived CENH3 co-localized only with the rye-derived centromeric chromatin creating a functional chromosome.
As CENH3 is a mark for centromere activity (Talbert and Henikoff, 2020), the hybrid 1BL/1RS centromere activity was determined by immunostaining using antibodies that cross-reacts with the CENH3 proteins of cereals (Nagaki et al., 2004; Liu et al., 2008; Houben et al., 2011). Subsequent FISH with differentially labeled centromeric repeats Bilby (Francki, 2001) and CRW2 (Liu et al., 2008) was performed to address whether the entire hybrid centromere or only the wheat- or rye- derived centromere component is active. In all analyzed dividing cells (about 80) of five plants, CENH3 signals consistently co-localized with the wheat centromeric repeat CRW2 in all non-translocation chromosomes. In contrast, only the rye-derived centromere of 1BL/1RS co-localized with the CENH3 signals (Figure 1A and Supplementary Figure 1). To confirm this observation, naturally extended meiotic prophase I chromosomes were labeled and analyzed by super-resolution microscopy (Figure 1B). Again, the rye derived centromere component co-localized with CENH3 signals. The CRW2-positive wheat centromeric region of 1BL/1RS was free of CENH3. The results are summarized in Figure 1C.
Our observation differs from the findings described by Wang et al. (2017) who described that CENH3 binds to both components of the 1BL/1RS hybrid centromere. What might be the reason for this difference? The apparent difference between both studies might be caused by the different 1BL/1RS genotypes analyzed and the different microscopical resolution achieved (diffraction-limited versus super-resolution microscopy). In addition, the ChIP-qPCR experiment performed by Wang et al. (2017) confirmed the interaction of the rye centromeric repeat pAWRC.1/Bilby with CENH3-containing nucleosomes. Still, it could not address whether the wheat component of the 1BL/1RS hybrid centromere interacts with CENH3 too. Besides, in their colocalization study of centromeric repeats with CENH3, they solely used a rye centromeric-specific probe without including any wheat centromere-specific probe. As only a fraction of CENH3 signals co-localized with the rye-specific probe, the authors concluded that CENH3 interacts with wheat centromeric sequences as well. In our study, we differentially labeled both centromeric repeats of rye and wheat (Bilby and CRW2) together with CENH3 in a single immuno-FISH experiment.
Although the centromere position is epigenetically determined and does not depend on its underlying sequences, studies in maize (Zhong et al., 2002), rice (Nagaki et al., 2005), wheat-rye B addition lines (Banaei-Moghaddam et al., 2012) and many other species have shown that CENH3 containing nucleosomes bind differentially to the repeats of the centromeres. Why the rye- and not the wheat-derived centromere component of the 1BL/1RS chromosome is epigenetically marked by CENH3 is unknown. However, it is tempting to speculate that fusion of the chromosome arm 1BL with 1RS triggered centromere sequence changes and, therefore CENH3 preference. The centromeric sequences of hybrids between distantly related or translocated chromosomes may undergo rearrangements like expansion and reduction. For example, centromeric retrotransposons were strongly reduced or eliminated in wheat aneuploids and addition lines from wide hybrids (Guo et al., 2016). The presence of CENH3 signals at novel positions suggests a de novo centromere formation. In contrast, rye-specific centromeric sequences in wheat-rye hybrids expanded towards the chromosome arms, possibly causing centromere expansion (Guo et al., 2016).
It is also possible that differential methylation of centromeric repeats may underlay the inactivation of wheat centromeric repeats in the hybrid centromere of 1BL/1RS. Investigations of the epigenetic landscape for the centromeres in Arabidopsis and maize showed that the centromere repeats associated with CENH3 are hypomethylated compared to flanking centromere repeats (Zhang et al., 2008).
In summary, our analysis of a 1BL/1RS centromere provides an additional example that one component of a hybrid (dicentric)centromere undergoes inactivation creating functional monocentric chromosomes to maintain their stability. Further investigations are needed to determine whether the fusion of 1BL with 1RS triggered centromere sequence changes and or epigenetic modifications and consequently CENH3 preference.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
AH designed the study and provided editorial input to the manuscript. RK-A and VS performed the research and interpreted the results. RK-A wrote the first draft of the manuscript. All authors contributed to manuscript revision and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Adam Lukaszewski (UC, Riverside, CA, United States) for providing the translocation 1BSPVN.1BLVEE line and Jianyong Chen (IPK, Germany) for sequence analysis.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.802222/full#supplementary-material
Wheat CENH3 incorporates exclusively into rye centromere repeats containing chromatin of the 1BL/1RS hybrid centromere during mitosis (A–F) and meiosis (G–L) revealed by standard fluorescence microscopy. (A–F) Wheat metaphase cells containing 1BL/1RS translocations (arrows) after CENH3-immunostaining and FISH with rye (Bilby, in green) and wheat (CRW2, in white) centromere-specific repeats. CENH3 signals co-localized with Bilby (centromeric repeat of rye) and not with the centromeric repeats of wheat in 1BL/1RS. (G–L) Meiotic metaphase I showing in addition to all labeled wheat centromeres a 1BL/1RS bivalent (arrow). (C,I) Co-localization of CENH3 and CRW2. (D,J) Co-localization of CENH3 and Bilby. (E,K) FISH with rye and wheat centromere-specific repeats (Bilby and CRW2, respectively). (F,L) Further enlarged views of 1RS/1BL. Selected chromosomes are labeled by *.
Wheat CENH3 (red) co-localizes the wheat-specific CRW2 repeats (white) of a wheat centromere.
Wheat CENH3 (red) co-localizes with the rye-specific centromeric Bilby repeats (green), but not with the wheat-specific CRW2 repeats (white) of a hybrid centromere of a translocated 1BL/1RS chromosome.
References
- Allshire R. C., Karpen G. H. (2008). Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 9 923–937. 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au W.-C., Crisp M. J., Deluca S. Z., Rando O. J., Basrai M. A. (2008). Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics 179 263–275. 10.1534/genetics.108.088518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaei-Moghaddam A. M., Schubert V., Kumke K., Weiβ O., Klemme S., Nagaki K., et al. (2012). Nondisjunction in favor of a chromosome: the mechanism of rye B chromosome drive during pollen mitosis. Plant Cell 24 4124–4134. 10.1105/tpc.112.105270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra V., Fachinetti D. (2018). The dark side of centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA. Nat. Commun. 9:4340. 10.1038/s41467-018-06545-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt A. B., Kuppu S. (2016). Cenh3: an emerging player in haploid induction technology. Front. Plant Sci. 7:357. 10.3389/fpls.2016.00357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evtushenko E. V., Elisafenko E. A., Gatzkaya S. S., Lipikhina Y. A., Houben A., Vershinin A. V. (2017). Conserved molecular structure of the centromeric histone CENH3 in Secale and its phylogenetic relationships. Sci. Rep. 7:17628. 10.1038/s41598-017-17932-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evtushenko E. V., Elisafenko E. A., Gatzkaya S. S., Schubert V., Houben A., Vershinin A. V. (2021). Expression of two rye CENH3 variants and their loading into centromeres. Plants 10:2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francki M. G. (2001). Identification of Bilby, a diverged centromeric Ty1-copia retrotransposon family from cereal rye (Secale cereale L.). Genome 44 266–274. 10.1139/g00-112 [DOI] [PubMed] [Google Scholar]
- Friebe B., Zhang P., Linc G., Gill B. S. (2005). Robertsonian translocations in wheat arise by centric misdivision of univalents at anaphase I and rejoining of broken centromeres during interkinesis of meiosis II. Cytogenet. Genome Res. 109 293–297. 10.1159/000082412 [DOI] [PubMed] [Google Scholar]
- Gernand D., Demidov D., Houben A. (2003). The temporal and spatial pattern of histone H3 phosphorylation at serine 28 and serine 10 is similar in plants but differs between mono- and polycentric chromosomes. Cytogenet. Genome Res. 101 172–176. 10.1159/000074175 [DOI] [PubMed] [Google Scholar]
- Guo X., Su H., Shi Q., Fu S., Wang J., Zhang X., et al. (2016). De novo centromere formation and centromeric sequence expansion in wheat and its wide hybrids. PLoS Genet. 12:e1005997. 10.1371/journal.pgen.1005997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Lamb J. C., Birchler J. A. (2006). High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. U.S.A. 103 3238–3243. 10.1073/pnas.0509650103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Dalal Y. (2005). Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 15 177–184. 10.1016/j.gde.2005.01.004 [DOI] [PubMed] [Google Scholar]
- Houben A., Kumke K., Nagaki K., Hause G. (2011). CENH3 distribution and differential chromatin modifications during pollen development in rye (Secale cereale L.). Chromosome Res. 19 471–480. 10.1007/s10577-011-9207-6 [DOI] [PubMed] [Google Scholar]
- Ishii T., Juranić M., Maheshwari S., Bustamante F. D. O., Vogt M., Salinas-Gamboa R., et al. (2020). Unequal contribution of two paralogous CENH3 variants in cowpea centromere function. Commun. Biol. 3:775. 10.1038/s42003-020-01507-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T., Karimi-Ashtiyani R., Banaei-Moghaddam A. M., Schubert V., Fuchs J., Houben A. (2015). The differential loading of two barley CENH3 variants into distinct centromeric substructures is cell type- and development-specific. Chromosome Res. 23 277–284. 10.1007/s10577-015-9466-8 [DOI] [PubMed] [Google Scholar]
- Karimi-Ashtiyani R. (2021). Centromere engineering as an emerging tool for haploid plant production: advances and challenges. Methods Mol. Biol. 2289 3–22. 10.1007/978-1-0716-1331-3_1 [DOI] [PubMed] [Google Scholar]
- Karimi-Ashtiyani R., Ishii T., Niessen M., Stein N., Heckmann S., Gurushidze M., et al. (2015). Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc. Natl. Acad. Sci. U.S.A. 112 11211–11216. 10.1073/pnas.1504333112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Johnson J. W., Baenziger P. S., Lukaszewski A. J., Gaines C. S. (2004). Agronomic effect of wheat-rye translocation carrying rye chromatin (1R) from different sources. Crop Sci. 44 1254–1258. 10.2135/cropsci2004.1254 34798789 [DOI] [Google Scholar]
- Kuppu S., Ron M., Marimuthu M. P. A., Li G., Huddleson A., Siddeek M. H., et al. (2020). A variety of changes, including CRISPR/Cas9-mediated deletions, in CENH3 lead to haploid induction on outcrossing. Plant Biotechnol. J. 18 2068–2080. 10.1111/pbi.13365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppu S., Tan E. H., Nguyen H., Rodgers A., Comai L., Chan S. W., et al. (2015). Point mutations in centromeric histone induce post-zygotic incompatibility and uniparental inheritance. PLoS Genet. 11:e1005494. 10.1371/journal.pgen.1005494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Choulet F., Heng Y., Hao W., Paux E., Liu Z., et al. (2013). Wheat centromeric retrotransposons: the new ones take a major role in centromeric structure. Plant J. 73 952–965. 10.1111/tpj.12086 [DOI] [PubMed] [Google Scholar]
- Liu Z., Yue W., Li D., Wang R. R., Kong X., Lu K., et al. (2008). Structure and dynamics of retrotransposons at wheat centromeres and pericentromeres. Chromosoma 117 445–456. 10.1007/s00412-008-0161-9 [DOI] [PubMed] [Google Scholar]
- Lukaszewski A. J. (1993). Reconstruction in wheat of complete chromosomes 1B and 1R from the 1RS.1BL translocation of ‘Kavkaz’ origin. Genome 36, 821–824. 10.1139/g93-109 [DOI] [PubMed] [Google Scholar]
- Lukaszewski A. J. (1997). Further manipulation by centric misdivision of the 1RS.1BL translocation in wheat. Euphytica 94 257–261. 10.1023/A:1002916323085 [DOI] [Google Scholar]
- Lukaszewski A. J. (2010). Behavior of centromeres in univalents and centric misdivision in wheat. Cytogenet. Genome Res. 129 97–109. 10.1159/000314108 [DOI] [PubMed] [Google Scholar]
- Ma L., Vu G. T., Schubert V., Watanabe K., Stein N., Houben A., et al. (2010). Synteny between Brachypodium distachyon and Hordeum vulgare as revealed by FISH. Chromosome Res. 18 841–850. 10.1007/s10577-010-9166-3 [DOI] [PubMed] [Google Scholar]
- Maheshwari S., Tan E. H., West A., Franklin F. C., Comai L., Chan S. W. (2015). Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids. PLoS Genet. 11:e1004970. 10.1371/journal.pgen.1004970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskal K., Kowalik S., Podyma W., Łapiński B., Boczkowska M. (2021). The Pros and Cons of rye chromatin introgression into wheat genome. Agronomy 11:456. 10.3390/agronomy11030456 [DOI] [Google Scholar]
- Nagaki K., Cheng Z., Ouyang S., Talbert P. B., Kim M., Jones K. M., et al. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36 138–145. 10.1038/ng1289 [DOI] [PubMed] [Google Scholar]
- Nagaki K., Neumann P., Zhang D., Ouyang S., Buell C. R., Cheng Z., et al. (2005). Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol. Biol. Evol. 22 845–855. 10.1093/molbev/msi069 [DOI] [PubMed] [Google Scholar]
- Neumann P., Pavlíková Z., Koblížková A., Fuková I., Jedličková V., Novák P., et al. (2015). Centromeres off the hook: massive changes in centromere size and structure following duplication of CenH3 gene in fabeae species. Mol. Biol. Evol. 32 1862–1879. 10.1093/molbev/msv070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A. L., Choi E. S., Abbott J. K. R., Liu X., Kagansky A., Castillo A. G., et al. (2009). Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell. 33 299–311. 10.1016/j.molcel.2009.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabanus-Wallace M. T., Hackauf B., Mascher M., Lux T., Wicker T., Gundlach H., et al. (2021). Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat. Genet. 53 564–573. 10.1038/s41588-021-00807-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi M., Chan S. W. (2010). Haploid plants produced by centromere-mediated genome elimination. Nature 464 615–618. 10.1038/nature08842 [DOI] [PubMed] [Google Scholar]
- Robertson W. (1961). Chromosome studies. I. Taxonomic relationships shown in the chromosomes of Tettegidae and Acrididiae: V-shaped chromosomes and their significance in Acrididae, Locustidae and Grillidae:chromosomes and variations. J. Morphol. 27 179–331. [Google Scholar]
- Sanei M., Pickering R., Kumke K., Nasuda S., Houben A. (2011). Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc. Natl. Acad. Sci. U.S.A. 108 E498–E505. 10.1073/pnas.1103190108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R. L., Ahn G. S., Staples M. I., Sathyan K. M., Karpova T. S., Foltz D. R., et al. (2017). Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget 8 46781–46800. 10.18632/oncotarget.18108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Li X., Sun L., Xu S., Liu N., Yao Y., et al. (2016). A family with Robertsonian translocation: a potential mechanism of speciation in humans. Mol. Cytogenet. 9:48. 10.1186/s13039-016-0255-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson K. M., Matheny J. E., Sullivan B. A. (2012). Dicentric chromosomes: unique models to study centromere function and inactivation. Chromosome Res. 20 595–605. 10.1007/s10577-012-9302-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B. A., Jenkins L. S., Karson E. M., Leana-Cox J., Schwartz S. (1996). Evidence for structural heterogeneity from molecular cytogenetic analysis of dicentric Robertsonian translocations. Am. J. Hum. Genet. 59 167–175. [PMC free article] [PubMed] [Google Scholar]
- Talbert P. B., Henikoff S. (2020). What makes a centromere? Exp. Cell Res. 389:111895. 10.1016/j.yexcr.2020.111895 [DOI] [PubMed] [Google Scholar]
- Wang J., Liu Y., Su H., Guo X., Han F. (2017). Centromere structure and function analysis in wheat–rye translocation lines. Plant J. 91 199–207. 10.1111/tpj.13554 [DOI] [PubMed] [Google Scholar]
- Weisshart K., Fuchs J., Schubert V. (2016). Structured illumination microscopy (SIM) and photoactivated localization microscopy (PALM) to analyze the abundance and distribution of RNA polymerase II molecules on flow-sorted Arabidopsis nuclei. Bio Protocol 6:e1725. 10.21769/BioProtoc.1725 33656495 [DOI] [Google Scholar]
- Yuan J., Guo X., Hu J., Lv Z., Han F. (2015). Characterization of two CENH3 genes and their roles in wheat evolution. New Phytol. 206 839–851. 10.1111/nph.13235 [DOI] [PubMed] [Google Scholar]
- Zhang P., Friebe B., Lukaszewski A. J., Gill B. S. (2001). The centromere structure in Robertsonian wheat-rye translocation chromosomes indicates that centric breakage-fusion can occur at different positions within the primary constriction. Chromosoma 110 335–344. 10.1007/s004120100159 [DOI] [PubMed] [Google Scholar]
- Zhang W., Lee H.-R., Koo D.-H., Jiang J. (2008). Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and Maize. Plant Cell 20 25–34. 10.1105/tpc.107.057083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C. X., Marshall J. B., Topp C., Mroczek R., Kato A., Nagaki K., et al. (2002). Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14 2825–2836. 10.1105/tpc.006106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wheat CENH3 incorporates exclusively into rye centromere repeats containing chromatin of the 1BL/1RS hybrid centromere during mitosis (A–F) and meiosis (G–L) revealed by standard fluorescence microscopy. (A–F) Wheat metaphase cells containing 1BL/1RS translocations (arrows) after CENH3-immunostaining and FISH with rye (Bilby, in green) and wheat (CRW2, in white) centromere-specific repeats. CENH3 signals co-localized with Bilby (centromeric repeat of rye) and not with the centromeric repeats of wheat in 1BL/1RS. (G–L) Meiotic metaphase I showing in addition to all labeled wheat centromeres a 1BL/1RS bivalent (arrow). (C,I) Co-localization of CENH3 and CRW2. (D,J) Co-localization of CENH3 and Bilby. (E,K) FISH with rye and wheat centromere-specific repeats (Bilby and CRW2, respectively). (F,L) Further enlarged views of 1RS/1BL. Selected chromosomes are labeled by *.
Wheat CENH3 (red) co-localizes the wheat-specific CRW2 repeats (white) of a wheat centromere.
Wheat CENH3 (red) co-localizes with the rye-specific centromeric Bilby repeats (green), but not with the wheat-specific CRW2 repeats (white) of a hybrid centromere of a translocated 1BL/1RS chromosome.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.