Abstract
The Fur4p uracil permease, like most yeast plasma membrane proteins, undergoes ubiquitin-dependent endocytosis and is then targeted to the vacuole (equivalent to the mammalian lysosome) for degradation. The cell surface ubiquitination of Fur4p is mediated by the essential Rsp5p ubiquitin ligase. Ubiquitination of Fur4p occurs on two target lysines, which receive two ubiquitin moieties linked through ubiquitin Lys63, a type of linkage (termed UbK63) different from that involved in proteasome recognition. We report that pep4 cells deficient for vacuolar protease activities accumulate vacuolar unubiquitinated Fur4p. In contrast, pep4 cells lacking the Doa4p ubiquitin isopeptidase accumulate ubiquitin-conjugated Fur4p. These data suggest that Fur4p undergoes Doa4p-dependent deubiquitination prior to vacuolar degradation. Compared to pep4 cells, pep4 doa4 cells have huge amounts of membrane-bound ubiquitin conjugates. This indicates that Doa4p plays a general role in the deubiquitination of membrane-bound proteins, as suggested by reports describing the suppression of some doa4 phenotypes in endocytosis and vacuolar protein sorting mutants. Some of the small ubiquitin-linked peptides that are a hallmark of Doa4 deficiency are not present in rsp5 mutant cells or after overproduction of a variant ubiquitin modified at Lys 63 (UbK63R). These data suggest that the corresponding peptides are degradation products of Rsp5p substrates and probably of ubiquitin conjugates carrying UbK63 linkages. Doa4p thus appears to be involved in the deubiquitination of endocytosed plasma membrane proteins, some of them carrying UbK63 linkages.
The covalent modification of proteins by the 76-residue ubiquitin (Ub) polypeptide is involved in many aspects of cell function. Ub molecules are transferred to lysine residues of target proteins via an E1/E2/E3 (Ub-activating enzyme/Ub-conjugating enzyme/Ub ligase) enzyme cascade. Ubiquitination is known mainly as a signal targeting substrate proteins for recognition and degradation by the 26S proteasome (23, 82). The proteasome preferentially degrades multiubiquitinated substrates (chains at least four Ub long) in which the carboxy terminus of one Ub is ligated to Lys48 of the previously attached Ub (50). Ub conjugation is also involved in the down-regulation of membrane receptors, transporters, and channels (reviewed in references 6, 24, 57, and 67). In Saccharomyces cerevisiae, this modification serves to trigger the internalization of most plasma membrane proteins, followed by their degradation in the vacuole (equivalent to the mammalian lysosome) without involvement of the proteasome (reviewed in references 24 and 57). The existence of a receptor that recognizes Ub as an internalization signal was thus postulated (61). Many mammalian receptors, and some channels, are also modified by Ub in response to stimulation. In some cases, this modification also appears to play a role in internalization followed by lysosomal degradation, but some receptors appear to undergo proteasomal degradation, and it is not clear how many features are shared between the yeast and mammalian systems (6, 67). The ubiquitination of most yeast plasma membrane proteins seems to be mediated by a single Ub ligase, Rsp5p (57), which belongs to the Nedd4 HECT domain superfamily of E3 Ub ligases (21). Ubiquitination of two channels in higher eucaryotes is mediated by Ub ligases of the same family, but it was recognized for a growing list of receptors that their ubiquitination is mediated by another Ub ligase, c-Cbl (32, 37, 38, 81). The Rsp5-dependent ubiquitination of two yeast plasma membrane transporters leads to the addition of short Ub chains on two target Lys residues. These chains are two to three residues long and are linked through Ub Lys63, i.e., different from the type of Ub chains recognized by the proteasome (17, 66). Several other yeast plasma membrane proteins also undergo either primarily monoubiquitination (addition of one Ub molecule to one or several target lysines) (25, 71) or the addition of several short (two- to three-residue-long) Ub chains (20, 54), but Ub linkages involved in the latter cases have not been not reported.
The observation of a Ub-dependent endocytosis raises many questions. The way in which ubiquitination is involved in the internalization of membrane proteins remains to be determined. An important issue that also needs to be addressed is the fate of the Ub moieties after internalization of plasma membrane proteins. The ubiquitination of proteins is a versatile covalent protein modification. Ubiquitinated protein substrates of the proteasome undergo deubiquitination prior to breakdown by the proteasome (50). Since a constant Ub concentration is crucial for normal cell function, internalized ubiquitinated plasma membrane proteins are probably deubiquitinated prior their breakdown in the vacuole. Deubiquitination is catalyzed by processing proteases called deubiquitinating (DUB) enzymes (84). As a group, these enzymes comprise the largest known family in the Ub system. DUBs fall into two classes, the Ub C-terminal hydrolases and the Ub-specific processing proteases (UBPs). The former cleave primarily peptide bonds in poly-Ub precursor proteins or isopeptide bonds in small free Ub chains. UBPs, the larger class of DUB, cleave isopeptide bonds between two Ub residues or between Ub and another protein. There are 16 potential UBPs encoded by the S. cerevisiae genome (28), 14 of which were shown to have ubiquitin-cleaving activity (1). These proteins all have similar Cys and His domains, which form the catalytic region of the protein. The roles of UBPs are thought to be diverse but remain poorly understood in both mammals and yeast. Surprisingly, none of the yeast UBPs is essential for viability (1). Specific functions have been assigned to very few UBPs (reviewed in reference 84), and the specific substrates have been even more rarely identified. Among the few documented cases, it has been proposed on the basis of genetic data that the Drosophila Liquid facets (homolog of the vertebrate epsin) is the critical substrate of the UBP Fat facets (7). Fam (Fat facets in the mouse) would deubiquitinate the Ras effector AF-6 (70) and β-catenin (69). In S. cerevisiae, two UBPs, Ubp10p (Dot4p) (33) and Ubp3p (43), regulate silencing, in latter case by interacting with Sir4p. The yeast Ubp14p, like its mammalian homolog IsoT, specifically disassembles unanchored Ub chains (3).
One of the first yeast UBPs identified, and probably the most widely studied, is Ubp4p (Doa4p). DOA4 was identified in a genetic screen for mutants that stabilize an unstable MATα2–β-galactosidase fusion protein Deg1–β-galactosidase (30). In doa4 mutant cells, many substrates of the Ub proteasome pathway are stabilized (48, 49) and many plasma membrane proteins are protected against endocytosis (reviewed in reference 57). This is probably due to the depletion of Ub in doa4 cells (66, 68) (3- to 4-fold reduction for cells in the exponential growth phase and 10-fold reduction for cells in the stationary phase). This depletion results from more rapid Ub degradation (68). Hence, some phenotypes of doa4 cells, such as loss of viability in the stationary phase, impaired ubiquitination and degradation of some proteasome substrates, or deficient ubiquitination and endocytosis of some plasma membrane proteins, can be overcome by providing additional Ub. But overproduction of Ub cannot suppress all doa4 phenotypes. Indeed, doa4Δ cells accumulate low-molecular-mass Ub-containing species that cluster most prominently above free Ub, di-Ub, and tri-Ub (49) and do not disappear upon Ub overproduction (68). These small polyubiquitinated peptides are thought to be Ub remnants resulting from the extensive degradation of ubiquitinated substrates by the proteasome occurring in the absence of Doa4p-dependent deubiquitination (49). This is supported by the findings that a mutation in one of the proteasome subunit genes, DOA3, partly suppresses their generation, and that Doa4p partially copurifies with the proteasome (48). However, recent studies on Ub homeostasis led to extension of the potential role of Doa4p to other cellular processes. Introducing a mutation in END3, a gene involved in the internalization step of endocytosis, reestablished partially normal Ub concentrations in doa4Δ cells and led to the disappearance of the low-molecular-mass Ub-containing species. These two doa4 phenotypes were even better corrected after deletion of VPS24 or VPS27 (68), two genes required for later steps of endocytosis, namely, late endosome-to-vacuole sorting. The link between Doa4p and endocytosis was further strengthened by the observation that a green fluorescent protein (GFP)-tagged version of Doa4p partially relocated to late endosome/prevacuolar compartment in some vps mutants. These overall data suggested that one important function of Doa4p is to recover Ub from ubiquitinated plasma membrane proteins (2).
This report analyzes the potential involvement of Doa4p in the vacuolar/endocytic pathway by monitoring the fate of a plasma membrane protein, uracil permease (Fur4p). Fur4p has been shown to undergo basal endocytosis and degradation in actively growing cells and accelerated endocytosis in various stress conditions, such as heat shock, nutrient starvation, and inhibition of protein synthesis (18, 76), or in the presence of excess substrate (60). Fur4p undergoes cell surface phosphorylation on serine residues (75) in a PEST-like N-terminal sequence (42). This posttranslational modification, in turn, triggers Rsp5p-dependent Fur4p ubiquitination (18, 42) on two target lysines (17) lying just before the PEST sequence (41). These residues accept up to two Ubs, linked through Ub Lys63 (17). Permease ubiquitination is required for subsequent internalization of the protein, an event that is followed by vacuolar targeting and degradation (18, 76). We have monitored the ubiquitination status of Fur4p after the internalization step of endocytosis. We find that the protein undergoes deubiquitination before its vacuolar degradation. This process is strongly inhibited in doa4Δ cells, as evidenced by the massive accumulation of Fur4p-Ub conjugates in doa4Δ cells deficient for vacuolar protease activities. More generally, these cells were found to accumulate membrane-bound Ub conjugates. These experiments support the idea that Doa4p is involved in the deubiquitination of plasma membrane proteins prior to their vacuolar degradation. We also report the disappearance of the accumulation of both Ub-Fur4p conjugates and membrane-bound Ub conjugates as a result of introducing a vps mutation (vps27Δ) in cells lacking Doa4p.
MATERIALS AND METHODS
Materials.
The enzymes used in DNA manipulations were from Roche Diagnostic Life Technology. Monoclonal antibodies against Ub were from Zymed, and monoclonal antibodies against Pep12p and Vat2p were from Molecular Probes. The polyclonal antibody directed against the last 10 residues of Fur4p was a gift from M. R. Chevallier. The polyclonal antibody against H+-ATPase was a gift from R. Serrano, and the polyclonal antibody against carboxypeptidase S (CPS) was a gift from D. Katzman and S. Emr.
Yeast strains and growth conditions.
The strains used are listed in Table 1. Open reading frame replacement cassettes with long flanking homology regions were used to disrupt the PEP4 and DOA4 genes (77). PCR amplification using Pwo DNA polymerase (Boehringer Mannheim) from the genomic DNA of strain W3031B/D with the four oligonucleotide primers PEP4-1, PEP4-2, PEP4-3, and PEP4-4 (Table 2) generated two DNA products corresponding to the PEP4 promoter and terminator, respectively, with 25-bp extensions homologous to the KanMX4 marker (78) containing the geneticin (G418) resistance gene. In a second PCR amplification experiment, one strand of each of these molecules served as a long primer using KanMX4 as the template. The linear fragment was used to transform W3031B/D, thus generating strain MOB100, which is resistant to geneticin. Correct integration at the PEP4 locus was confirmed by whole-cell PCR using PEP4- and KanMX4-specific primers. The same technique was used to disrupt the DOA4 gene, with the HIS3 marker instead of KanMX4, using oligonucleotide primers DOA4-1, DOA4-2, DOA4-3, and DOA4-4 (Table 2). Yeast strains were transformed as described by Gietz et al. (19). Cells were grown at 30°C in minimal medium containing 0.67% yeast nitrogen base (YNB) without amino acids (Difco) and supplemented with appropriate nutrients. The carbon source was either 2% glucose, or 4% galactose plus 0.05% glucose, as indicated in the figure legends. Overproduction of Ub from the CUP1 promoter was obtained by growing the cells for 1 h in the presence of 0.1 mM CuSO4, unless otherwise stated.
TABLE 1.
S. cerevisiae strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference(s) or source |
|---|---|---|
| Strains | ||
| W3031B/D | MATα ade2-1 ura3-1 his3-11 leu2-3,112 trp1-1 can1-100 | 72 |
| MOB100 | MATα ade2-1 ura3-1 his3-11 leu2-3,112 trp1-1 can1-100 PEP4::KanMX4 (isogenic to W3031B/D) | Gift from M.-O. Blondel |
| SD20 | MATα ade2-1 ura3-1 his3-11 leu2-3,112 trp1-1 can1-100 DOA4::HIS3 (isogenic to W3031B/D) | This study |
| SD21 | MATα ade2-1 ura3-1 his3-11 leu2-3,112 trp1-1 can1-100 PEP4::KanMX4 DOA4::HIS3 (isogenic to W3031B/D) | This study |
| YMW82 | MATα ade2-101 his3-Δ200 lys2-801 trp1Δ63 ura3-52 arp2-1 | 45 |
| SD22 | MATα ade2-101 his3-Δ200 lys2-801 trp1-Δ63 ura3-52 arp2-1 DOA4::HIS3 (isogenic to YMW82) | This study |
| MHY1269 | matα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 vps27Δ::LEU2 | 2 |
| SD23 | matα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 vps27Δ::LEU2 PEP4::KanMX4 (isogenic to MHY1269) | |
| MHY1275 | matα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 vps27Δ::LEU2 doa4-Δ1::LEU2 | 2 |
| SD24 | matα his3-Δ200 leu2-3,112 ura3-52 lys2-801trp1-1 vps27Δ::LEU2 doa4-Δ1::LEU2 PEP4::KanMX4 (isogenic to MHY1275) | This study |
| 27061b | MATaura3 trp1 | 66 |
| 27064b | MATaura3 trp1 npi1 (isogenic to 27061b) | 66 |
| 27071b | MATaura3 trp1 npi2 (isogenic to 27061b) | 66 |
| 33276d | MATaura3 trp1 npil npi2 gap1 (isogenic to 27061b) | Gift from J. Decraene and B. Andre |
| Plasmids | ||
| YEp352fF | 2μm URA3 FUR4 | 18 |
| YEp96 | 2μm TRP1 CUP-UB | 12, 14 |
| YEp112 | 2μm TRP1 CUP-UB-HA | 29 |
| YEp96-UbK63R | 2μm TRP1 CUP-UBK63R | 4 |
| YEp96-UbK29, 48R | 2μm TRP1 CUP1-UBK29 K48R | Gift from J.-M. Galan |
| pFL38-FUR4-GFP | ARS CEN URA3 GAL10-FUR4-GFP | This study |
| YEp96fF | 2μm URA3 FUR4-CUP1-UB | This study |
| pUG35 | 2μm URA3 MET35-GFP-tCYC1 | Gift from J. Hegeman |
TABLE 2.
Oligonucleotides used in this study
| Name | Sequencea |
|---|---|
| PEP4-1 | 5′GCGGGTGTCGATGGATTAAGGG3′ |
| PEP4-2 | 5′GGGGATCCGTCGACCTGCAGCGTACGGGATTAAATACTAGGTCACTAGGC3′ |
| PEP4-3 | 5′GTCGAAAACGAGCTCGAATTCATCGATCGATTACACGCTTGAAGTTTCAGG3′ |
| PEP4-4 | 5′GGGCAGCAGCATAGAACAATGG3′ |
| DOA4-1 | 5′GAATCCAGATTATAAGCAAC3′ |
| DOA4-2 | 5′GGGGATCCGTCGACCTGCAGCGTACCATAACTTAAGCATGTATC3′ |
| DOA4-3 | 5′AACGAGCTCGAATTCATCGATGATAGTTTTATCACCGCGTCTACG3′ |
| DOA4-4 | 5′GAAAGTTGCTGTGTCTCAAGAGG3′ |
| GFP1 | 5′AACAACATTATAAAGGACAAGGGCTGGTTCC3′ |
| GFP2 | 5′CACGAACACGAAAAGACTTTCATTGCGGCCGCTATGTCTAAAGGTGAAGAATTATTCACTGGTGTTGTCC3′ |
| GFP3 | 5′GGACAACACCAGTGAATAATTCTTCACCTTTAGACATAGCGGCCGCAATGAAAGTCTTTTCGTGTTCGTG3′ |
| GFP4 | 5′CGGGATCCTTAAAGCCTTCGAGCGTCCCAAAAC3′ |
| CUP1 | 5′CGGGATCCCATTACCGACATTTG3′ |
| CUP2 | 5′CGGGATCCAAGCTTGCAAATTAAAGCCTTCGAGCG3′ |
Boldface denotes 25-bp extensions.
Plasmid constructions.
Plasmid YEp96 (2μm TRP1UB) contains a synthetic yeast UB gene under the control of the copper-inducible CUP1 promoter (12, 14). YEp112 (2μm TRP1 HA-UB) is identical to YEp96 except that it encodes a hemagglutinin (HA)-tagged version of Ub (29). The plasmid encoding a mutant Ub in which Lys63 is replaced by arginine (UbK63R) was a derivative of YEp96 (4). The plasmid encoding a mutant Ub in which Lys29 and Lys48 were replaced by arginine was constructed from UbK29R (4) by using a Chameleon double-stranded site-directed mutagenesis kit. The resulting gene encoding mutant Ub was sequenced using double-stranded DNA and a Sequenase 2.0 kit (U.S. Biochemicals). The chromosome-encoded uracil permease was produced in very small amounts, and cells that produced the permease from a strong promoter or from multicopy plasmids were used for accurate measurements of permease activity and for immunodetection of the protein. All experiments were performed under conditions of overproduction of Ub, unless otherwise stated, to facilitate the detection of Ub-Fur4p conjugates and to correct the Ub deficiency of doa4Δ cells. The multicopy plasmid YEp96-fF (2μm URA3 CUP1-UB FUR4) was constructed as follows. The CUP1-UB gene (including CYC1 terminator) was amplified by PCR using YEp96 as a template and the oligonucleotides CUP1 and CUP2 (Table 2). The resulting PCR fragment was digested with BamHI and cloned at the unique BamHI site of plasmid YEp352fF (2μm URA3 FUR4) (18). The centromeric plasmid pFL38gF (URA3 GAL-FUR4) was constructed as described elsewhere (60). We constructed a plasmid, pFL38gF-GFP, which expressed a GFP variant (S65G, S72A) (10) fused to wild-type permease at its C terminus to locate uracil permease in living cells. PCR with oligonucleotides GFP1 and GFP2 (Table 2) was used to generate the Fur4p C-terminus coding sequence from the unique HpaI site and excluding the stop codon. The entire GFP gene, excluding the initiator ATG, was amplified by PCR with oligonucleotides GFP3 and GFP4, resulting in the introduction in the 3′ region of a BamHI restriction site. A DNA fragment encoding the Fur4 C terminus fused in frame to GFP was then generated by fusion PCR using GFP1 and GFP4, digested with HpaI and BamHI, and cloned in pFL38gF that had been cut with the same enzymes. The constructions were checked at the fusion points by sequencing.
Measurement of uracil uptake.
Uracil uptake was measured in exponentially growing cells as previously described (62). Yeast culture (1 ml) was incubated with 5 μM [14C]uracil (I C/N) for 20 s at 30°C and then quickly filtered through Whatman GF/C filters, which were then washed twice with ice-cold water and counted for radioactivity.
Membrane preparation.
Yeast cells (40 A600 units) in the exponential growth phase were harvested by centrifugation in the presence of 10 mM sodium azide, washed once in distilled water plus 10 mM sodium azide, and used to prepare membrane-enriched fractions, essentially as previously described (18). Washed cells were transferred to a conical 1.5-ml Eppendorf tube and suspended in 0.2 ml of lysis buffer (0.1 M Tris-HCl [pH 7.5]–0.15M NaCl–5 mM EDTA plus a mixture of protease inhibitors [Complete; Roche] and 25 mM freshly prepared N-ethylmaleimide to prevent artifactual deubiquitination). All subsequent steps were carried out at 4°C. Chilled glass beads (0.2 ml) were added, and the cells were lysed by vigorous vortex mixing for three times for 3 min each, separated by 30 s on ice. The homogenate was transferred to a new tube, and the glass beads were washed three times with 0.2 ml of lysis buffer. The resulting homogenate was centrifuged at 3,000 rpm for 3 min to remove unbroken cells and debris. Small aliquots of the total protein extracts were withdrawn for Western blot analysis (referred as total protein extracts, glass beads technique). The total protein extract was centrifuged for 45 min at 12,000 rpm. The resulting supernatant was centrifuged for 1 h at 100,000 × g in a Beckman TLA-100 rotor. Both the 12,000 × g and 100,000 × g pellets were washed by suspension in 0.4 ml of lysis buffer plus 5 M urea, incubation at 0°C for 30 min, and sedimentation as above. The resulting pellets were suspended in 0.4 ml of lysis buffer, and trichloroacetic acid (TCA) was added to 10% to precipitate proteins. This step was needed to prevent proteolysis by residual endogenous proteases. The precipitates were neutralized and dissolved in 40 μl of 1 M Tris base plus 80 μl of 2× sample buffer (100 mM Tris-HCl [pH 6.8], 4 mM EDTA, 4% sodium dodecyl sulfate [SDS], 20% glycerol, 0.002% bromophenol blue) containing 2% 2-mercaptoethanol and heated at 37°C for 15 min. Aliquots of these final 12,000 × g and 100,000 × g pellets, enriched respectively in plasma membrane, vacuole, and endoplasmic reticulum and in endosomal markers (plus Golgi cisternae), were analyzed by Western blotting as described below.
Yeast cell extracts, SDS-polyacrylamide gel electrophoresis, and Western immunoblotting.
Total protein extracts were prepared as described above or by the NaOH-TCA lysis technique (76). Proteins were separated by SDS-polyacrylamide gel electrophoresis on 12% Tricine gels (59) and transferred to nitrocellulose. The membranes were probed with the various antisera. For anti-Ub immunoblotting, cell extracts were prepared as described above except that they were boiled in sample buffer instead of incubated at 37°C. Proteins were then transferred to Immobilon-P membranes and treated as described elsewhere (66). Primary antibodies were detected with a horseradish peroxidase-conjugated anti-rabbit (or anti-mouse) immunoglobulin G secondary antibody (Sigma) detected by enhanced chemiluminescence (Amersham).
Equilibrium density centrifugation.
Cell organelles were fractionated on equilibrium density gradients essentially as described elsewhere (36, 53). Exponentially growing cultures (40 A600 units) were arrested by adding 10 mM sodium azide, washed once in 10 mM sodium azide, and broken by vigorous shaking with 0.2 ml of glass beads and 0.2 ml of STET (10% [wt/wt] sucrose, 10 mM Tris-HCl [pH 7.6], 10 mM EDTA, protease inhibitors, and 25 mM N-ethylmaleimide). The mixture was centrifuged at 3,000 rpm for 3 min, and 0.4-ml aliquots of the cleared extracts were layered on top of 5-ml 20 to 60% linear sucrose gradients made up in buffer A (10 mM Tris-HCl [pH 7.6], 10 mM EDTA). Samples were centrifuged for 18 h at 100,000 × g in an SW50.1 rotor (Beckman). Fractions were collected from the top of the gradient, and proteins were precipitated by 10% TCA. The proteins were incubated for 30 min on ice, pelleted by centrifugation, and suspended in 2× sample buffer (plus 1 M Tris base, as described above). The proteins in each gradient fraction were analyzed by Western blotting as described above.
Fluorescence microscopy.
Cells were grown to the mid-logarithmic phase in galactose minimal medium. To monitor GFP fluorescence, 5 × 106 cells were collected by centrifugation in the presence of 10 mM sodium azide and resuspended in 50 μl of Citifluor plus 10 mM sodium azide. Microscopic observations were done in a Leitz microscope equipped with fluorescent optics. Direct image acquisitions were made with a charge-coupled device Princeton cooled camera equipped with the Metaview imaging system.
RESULTS
Fur4p is deubiquitinated prior to vacuolar degradation.
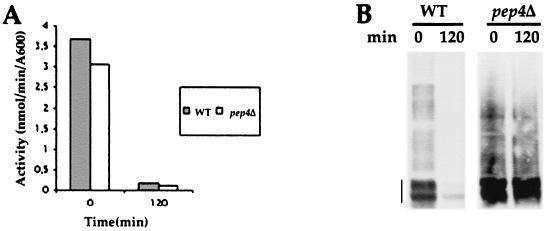
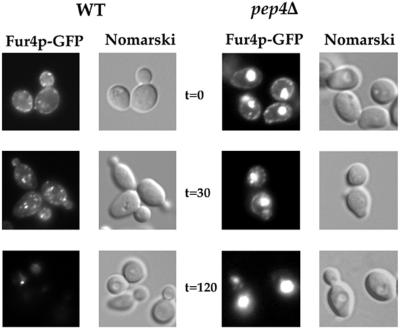
Fur4p has been shown to undergo basal endocytosis in actively growing cells and accelerated endocytosis under various stress conditions, such as inhibition of protein synthesis. We followed the fate of Fur4p in wild-type (WT) cells after adding cycloheximide (CHX) and in pep4Δ cells, deficient for the maturation of several vacuolar proteases, to determine whether Fur4p was deubiquitinated prior to vacuolar degradation. Uracil uptake was measured, and protein extracts were prepared and analyzed for uracil permease by Western immunoblotting before and after 2 h of CHX treatment (Fig. 1). We also monitored the fate of a version of Fur4p tagged at its C terminus with a brilliant version of GFP (Fig. 2). Fur4p-GFP was detected as punctate staining at the cell periphery in exponentially growing WT cells, as often described for plasma membrane proteins (27, 53). Small dots were also found throughout the cytoplasm. The plasma membrane signal gradually disappeared from cells incubated with CHX; intracellular dots, probably corresponding to endosomes, formed transiently, and the GFP signal disappeared after incubation for 2 h with CHX (Fig. 2). In agreement with these observations, uracil uptake, a measure of cell surface active permease, dropped to almost zero after 2 h in CHX, and immunodetectable Fur4p was degraded (Fig. 1), as was Fur4p-GFP (not shown). Growing pep4Δ cells had the same permease activity as WT cells but about threefold more immunodetectable protein, as previously described (76). This is probably explained by basal permease endocytosis. Growing pep4Δ cells indeed displayed vacuolar fluorescence in addition to small dots and plasma membrane Fur4p-GFP staining. The PEP4 deletion did not prevent permease internalization, as judged by the disappearance of uracil uptake and plasma membrane fluorescence after incubation for 2 h, as for WT cells, but involved strong protection against permease degradation (Fig. 1). Identical results were obtained on immunoblots with Fur4p or Fur4p-GFP (data not shown). There was massive vacuolar staining at the end of CHX treatment. The fluorescence signal corresponded only to the fusion protein Fur4p-GFP; no fluorescence was observed for cells expressing Fur4p which was not fused to GFP, and no GFP was formed by proteolysis of Fur4p-GFP, as evidenced by Western immunoblotting with anti-GFP antibody (data not shown).
FIG. 1.
Fur4p turnover is dependent on vacuolar proteases. Strains W3031B/D (WT) and MOB100 (pep4Δ) transformed with YEp96fF (2μm URA3 FUR4 CUP1-UB) were grown at 30°C in YNB with glucose as the carbon source. CuSO4 (100 μM) was added for 1 h to induce Ub synthesis from the CUP1 promoter. CHX (100 μg/ml) was added to cells in exponential growth (A600 = 0.8). At the times indicated after addition of CHX, uracil uptake was measured (A), and total protein extracts (glass beads technique) were prepared and analyzed for uracil permease by Western immunoblotting (B). Permease (vertical line) appeared as several bands (here mostly two) corresponding to the various phosphorylated states of uracil permease (75).
FIG. 2.
Fur4p-GFP trafficks from the plasma membrane to the vacuole lumen in pep4Δ cells. Strains W3031B/D (WT) and MOB100 (pep4Δ) cotransformed with pFL38-FUR4-GFP (2μm URA3 GAL10-FUR4) and YEp96 (2μm TRP1 CUP1-UB) were grown at 30°C in YNB with galactose as the carbon source. CuSO4 (100 μM) was added for 1 h. CHX was added to cells in exponential growth. Aliquots were withdrawn at the times indicated after addition of CHX, washed with ice-cold azide, and examined by Nomarski optics and for GFP fluorescence.
Additional experiments were needed to define the ubiquitination status of the protein before and after CHX treatment, since Fur4p Ub conjugates are not easily detected in total protein extracts. We therefore compared the electrophoretic patterns of Fur4p in subcellular fractions of WT and pep4Δ cells in the experimental conditions defined above. Glass bead extracts of cells withdrawn before and after incubation for 2 h with CHX were fractionated on sucrose density gradients (Fig. 3). In exponentially growing WT cells, the Fur4p that fractionated with the plasma membrane Pma1p (essentially fractions 12 to 14) appeared mostly as a ladder of four distinct bands with slower mobility than the Fur4p main signal (Fig. 3a). These bands corresponded to Ub-Fur4p conjugates containing one to four Ubs (17, 18) (Fig. 4). This plasma membrane Fur4p appeared mostly enriched in conjugates carrying four Ubs. Some Fur4p also fractionated partly with the endosomal marker Pep12p. Most of the Fur4p in these intracellular fractions was in a nonubiquitinated form, with small amounts of Fur4p containing one Ub and only trace amounts of higher-molecular-mass Ub-Fur4p. These intracellular fractions probably corresponded to endosomal Fur4p, rather than to Fur4p en route to the plasma membrane. First, uracil permease is delivered rather rapidly (in <20 min) to the plasma membrane (44), whereas its vacuolar trafficking is far slower. Second, the same profile was observed on sucrose gradients prepared from cells expressing Fur4p under the control of the GAL10 promoter and submitted to 20 min of glucose repression to produce complete plasma membrane delivery of Fur4p (D. Urban- Grimal, unpublished data). These observations suggest that deubiquitination occurs in the course of the trafficking of Fur4p from the plasma membrane to endosomal compartments. Incubation with CHX for 2 h caused the plasma membrane Fur4p to disappear completely, and only trace amounts of Fur4p (nonubiquitinated and probably monoubiquitinated forms) were still detected in internal fractions (Fig. 3).
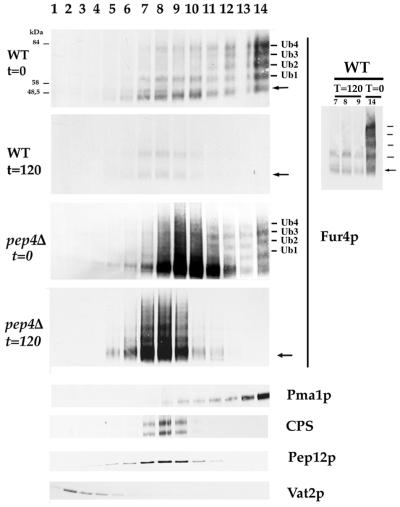
FIG. 3.
Fur4p is deubiquitinated prior vacuolar degradation. Strains W3031B/D (WT) and MOB100 (pep4Δ) transformed with YEp96fF (2μm URA3 FUR4 CUP1-UB) were grown and treated with CHX as described in the legend to Fig. 1. At the times indicated after the addition of CHX, cells were lysed, cleared of unbroken cells, and fractionated on equilibrium sucrose density gradients as described in Materials and Methods. Aliquots of the various fractions were analyzed by Western immunoblotting for uracil permease, Pma1p (plasma membrane marker), Pep12p (late endosome marker), Vat2p (vacuolar marker), and CPS, a vacuolar marker in WT cells and a marker of internal dense vacuolar vesicles in pep4 cells. All immunoblots were probed for the various protein markers, giving identical positions of these markers, except for CPS. Markers are shown in only one case (pep4 cells, time 120 min [t = 120]). Unubiquitinated Fur4p is indicated by an arrow. The Ub-Fur4p conjugates carrying one to four Ubs are indicated as Ub1 to Ub4. The positions of size standards are indicated on the left. Unprocessed CPS appears in pep4 cells as two bands, corresponding to two different precursor glycoforms of the protein, both found at steady state (65). To improve the identification of the ubiquitinated forms of Fur4p still detected in internal fractions of WT cells after CHX treatment, aliquots of these fractions were migrated close to aliquots from plasma membrane fractions, time zero (insert on the right). The apparent shift of Fur4p to lighter fractions after CHX treatment is probably attributable to the peak of Fur4p at time zero being the sum of plasma membrane and internal Fur4p.
FIG. 4.
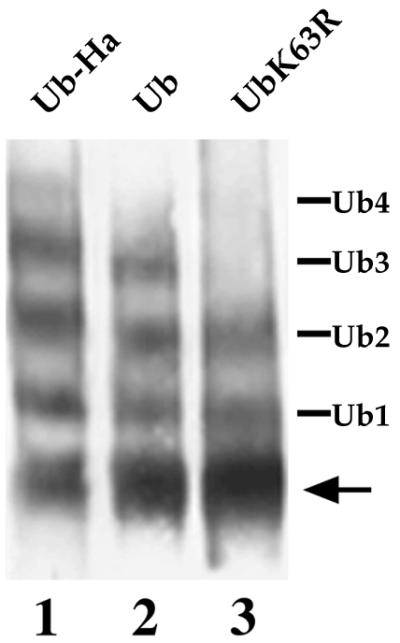
Effect of overexpression of WT and mutant Ubs on the ubiquitination pattern of uracil permease in doa4Δ cells. SD20 (doa4Δ) cells cotransformed with YEp352fF (2μm URA3 FUR4) and either YEp96 (2μm TRP1 CUP1-UB), YEp96-UB-HA, or YEp96-UBK63R were grown in YNB plus glucose and induced for 1 h with CuSO4. Cells were then collected and used to prepare membrane-enriched fractions (P13) as described in Materials and Methods. Aliquots were analyzed for uracil permease by Western immunoblotting. The various Fur4p forms are as indicated in the legend to Fig. 3.
Growing pep4Δ cells contained some plasma membrane Fur4p with the same ubiquitinated pattern as in WT cells, but most of the Fur4p was in intracellular fractions (Fig. 3). Some of this intracellular Fur4p fractionated with the endosomal marker Pep12p and not with the membrane-bound vacuolar marker Vat2p, although most of the Fur4p-GFP fluorescence was in the entire vacuole lumen. The subcellular fractions of Fur4p from pep4Δ cells incubated for 2 h with CHX shifted slightly toward lighter fractions that comigrated clearly with Pep12p, whereas all of the Fur4-GFP was in the vacuole. As for growing pep4Δ cells, the major fraction of intracellular Fur4p was unubiquitinated, although some Ub-Fur4p conjugates were still detected.
Thus, Fur4p targeted from the plasma membrane to the vacuoles in pep4Δ cells deficient for vacuolar protease activity was recovered in fractions whose density was clearly distinct from that containing the vacuolar membrane-bound Vat2p. Although, to our knowledge, similar fractionation data have not been reported for endocytosed plasma membrane proteins, it has been reported that the vacuolar carboxypeptidase CPS also fractionates with endosomes and Golgi cisternae in pep4 cells, despite being in the vacuole lumen in these cells (46). This protein is synthesized as a type II integral membrane protein and targeted via late endosomes to the vacuole, where it undergoes lumenal PEP4-dependent cleavage from its transmembrane anchor. These studies are consistent with the sorting of CPS at a prevacuolar endosome into membrane vesicles that invaginate and bud into the endosome (multivesicular bodies [MVBs]) (46), as in mammalian cells (16). One possible explanation for the membranes from MVBs having a different density from lysosomal membrane in mammals is their different lipid composition (35). The yeast endocytic pathway converges with the Golgi to vacuole trafficking at the late endosome-prevacuolar compartment. Hence, the finding that lumenal vacuolar Fur4p fractionates differently from vacuolar membrane-bound proteins in protease-deficient cells appears to be congruent with the CPS data, and we did indeed observe complete cofractionation of intracellular, vacuolar Fur4p and CPS in pep4 cells (Fig. 3). Like CPS, Fur4p is probably located in pep4Δ cells in intravacuolar vesicles whose density is distinct from that of the vacuolar membrane, which were clearly seen in ultrastructural studies (46).
In addition to these observations on the fractionation of plasma membrane proteins delivered to the vacuole of pep4Δ cells, the above experiments clearly indicate that Fur4p, in ubiquitinated form at the plasma membrane, undergoes deubiquitination leading mostly to unubiquitinated forms upon arrival at internal compartments and then at the vacuoles.
Doa4p dependence of uracil permease deubiquitination.
We reasoned that preventing vacuolar degradation might help to decipher the fate of Ub conjugates. We therefore compared the fate of Fur4p in pep4Δ and pep4Δ doa4Δ cells and, as a control, that in WT and doa4Δ cells. We used experimental conditions in which the deficiency of doa4Δ cells for free Ub was corrected by overproduction of Ub. Cells were thus all transformed with a multicopy plasmid carrying the UB gene under the control of the regulatable CUP1 promoter and induced by incubation for 1 h in the presence of Cu2+ before incubation with CHX. We have previously observed that Ub overproduction does not modify the rate of Fur4p internalization or degradation or the overall pattern of Fur4p ubiquitination in WT cells (17, 18). We also checked that Fur4p deubiquitination was not modified by Ub overproduction; identical results were obtained before and after CHX treatment, using sucrose gradients prepared from pep4Δ cells overproducing Ub (Fig. 2) or not (data not shown). All cells displayed identical permease internalization (Fig. 1A and 5A). PEP4 deletion provided strong protection against degradation in DOA4 and doa4Δ genetic contexts. The ubiquitination status of Fur4p in the various mutants was analyzed by differential centrifugation. We separated the 13,000 × g membrane pellet (P13), enriched for plasma membrane, endoplasmic reticulum and vacuole, and the 100,000 × g pellet (P100), enriched for membranes of the Golgi apparatus, endosomes, and transport vesicles. As expected, the plasma membrane Pma1p was entirely in P13 (Fig. 5C). In agreement with the observations for sucrose gradients, both unubiquitinated Fur4p and mono- to tetraubiquitinated forms, which fractionate mostly with P13, were present in exponentially growing WT cells. Only a minor fraction was recovered in P100. Degradation was complete after 2 h of CHX treatment, whereas the stable Pma1p remained unchanged (Fig. 5B and C). As previously reported, doa4Δ cells overproducing Ub displayed the same internalization and the same Fur4p ubiquitination pattern as WT cells (17). Fur4p ubiquitination was mainly dependent on plasmid-encoded Ub in these cells: the Ub-Fur4p conjugates shifted in doa4Δ cells producing Ub extended by an HA tag (Fig. 4). The residual Fur4p in P13 of cells incubated for 2 h with CHX was mostly unubiquitinated, with some Ub conjugates still detectable.
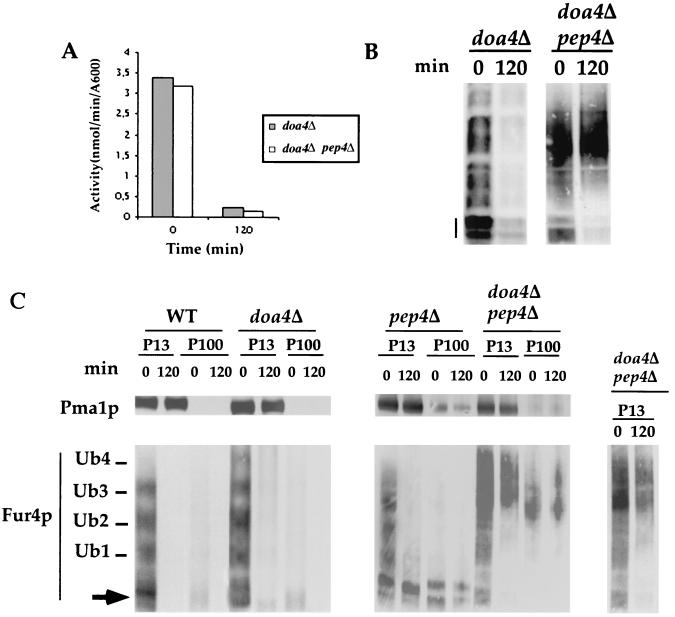
FIG. 5.
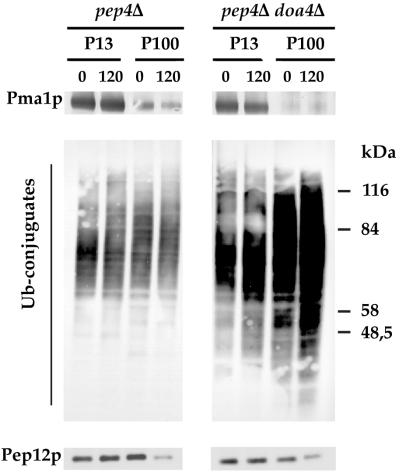
Accumulation of Ub-Fur4p conjugates in doa4Δ pep4Δ cells. W3031B/D (WT), SD20 (doa4Δ), MOB100 (pep4Δ), and SD21 (doa4Δ pep4Δ) cells transformed with YEp96fF (2μm URA3 FUR4 CUP1-UB) were grown and induced for Ub by CuSO4 as described in the legend to Fig. 1. At the times indicated after addition of CHX, uracil uptake was measured (panel A and Fig. 1A), total protein extracts (glass beads technique) were withdrawn (panel B and Fig. 1B), and membrane-enriched fractions (P13 and P100) (C) were prepared and analyzed by Western immunoblotting for uracil permease and Pma1p. The various Fur4p forms are as indicated in the legend to Fig. 3 except for unubiquitinated Fur4p in panel A, indicated by a dash (the various phosphorylated bands of Fur4p are well separated). To obtain better characterization of the forms of Ub-Fur4p accumulated in doa4Δ pep4Δ cells, a shorter exposure of the material in P13 is shown (inset).
There was a striking difference between pep4Δ and pep4Δ doa4Δ cells. The PEP4 deletion protected strongly against degradation, and permease was recovered mostly unubiquitinated from cells incubated with CHX (Fig. 5C). Some higher-molecular-mass Ub-Fur4p conjugates (>4 Ubs) were detected in growing pep4Δ doa4Δ cells, in addition to the species usually observed. Unubiquitinated Fur4p was entirely lost from cells incubated for 2 h with CHX, and Ub-Fur4p conjugates (enriched in forms with three to four Ubs) were recovered in P13 and P100 (Fig. 5C). The accumulation of Fur4p-Ub conjugates was so great that they were clearly detectable far above any background in total protein extracts both before and after CHX treatment (Fig. 5B).
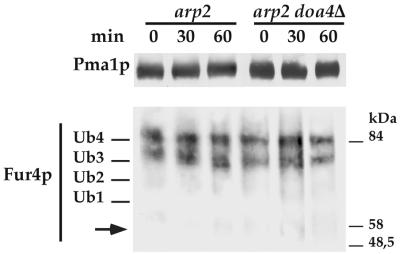
It therefore seems that ubiquitinated Fur4p undergoes a Doa4p-dependent deubiquitination step. This processing is probably a postinternalization event. Whether or not DOA4 was deleted, only Fur4p species with three to four Ubs were found in arp2 (actin-related protein 2) cells deficient for the internalization step of endocytosis because of an altered actin cytoskeleton (44) that had been kept for over 1 h at the restrictive temperature (Fig. 6).
FIG. 6.
Uracil permease accumulates as Ub conjugates in arp2 cells lacking or carrying DOA4. YMW82 (arp2) and SD22 (arp2 doa4Δ) cells transformed with YEp96fF (2μM URA3 FUR4 CUP1-UB) were grown at 24°C with glucose as the carbon source, induced for 1 h with CuSO4, and incubated for 10 min at 37°C. CHX was then added for the times indicated. Membrane-enriched fractions (P13) were prepared and analyzed for uracil permease and Pma1p by Western immunoblotting. The positions of size standards are indicated on the left.
Accumulation of membrane-bound Ub conjugates in pep4Δ cells lacking Doa4p.
We compared the patterns of total Ub conjugates in membrane-enriched fractions from growing pep4Δ and pep4Δ doa4Δ cells (Fig. 7) to see whether Doa4p had a more general impact on membrane-bound proteins. Some membrane-bound Ub conjugates were found in pep4Δ cells. These conjugates appeared to be rather stable and fractionated almost equally between P13 and P100. In pep4Δ doa4Δ cells, Ub conjugates were clearly more abundant. Serial dilution of the membrane-bound extracts prepared from pep4Δ and pep4Δ doa4Δ cells analyzed on the same gel indicated an about fourfold enrichment of Ub conjugates in the latter cells (not shown). The pep4Δ doa4Δ cells were further enriched in Ub conjugates after CHX treatment, with conjugates in both P13 and P100. The conjugates in P100 were more abundant than those in P13. The newly ubiquitinated proteins would reach the vacuole in an ubiquitinated state in these CHX-treated cells and then accumulate as a result of the inhibition of vacuolar proteases. The characteristic fractionation pattern of Ub conjugates in these cells could also indicate a delay in intracellular targeting of these conjugates that would transiently accumulate in some endosomal compartments. Whatever the precise intracellular location of these membrane-bound conjugates, it seems that Doa4p plays a central role in the deubiquitination of membrane-bound proteins.
FIG. 7.
doa4Δ pep4Δ cells accumulate huge amounts of Ub conjugates. MOB100 (pep4Δ) and SD21 (doa4Δ pep4Δ) cells transformed with YEp96fF (2μm URA3 FUR4 CUP1-UB) were grown and induced for Ub with CuSO4 as described in the legend to Fig. 1. They were treated with CHX for the times indicated. Membrane-enriched fractions (P13 and P100) were prepared. Aliquots were analyzed by Western immunoblotting for Ub conjugates, Pma1p, and Pep12p. The position of size standards are indicated on the left.
Deleting VPS27 restores the deubiquitination of Fur4p and other membrane-bound Ub conjugates in cells lacking Doa4p.
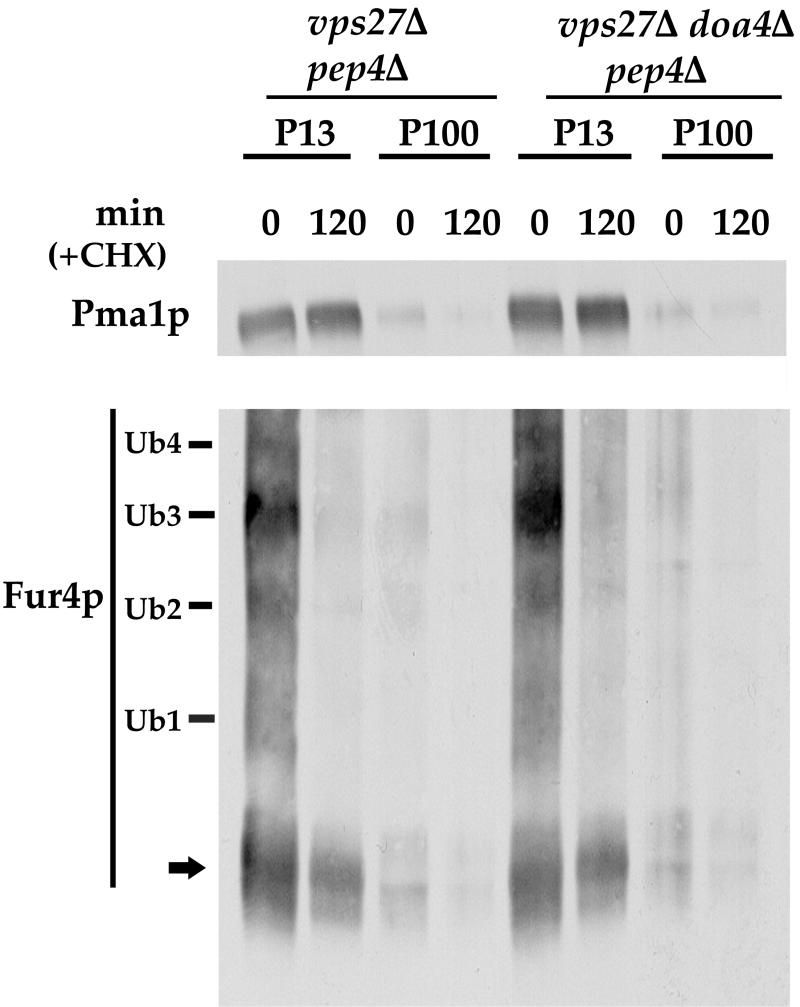
When we first detected the involvement of Doa4p in the deubiquitination of Fur4p, we attempted to identify the step in the endocytic pathway where deubiquitination occurs. The basic idea was that any mutation blocking the endocytic pathway before the place where deubiquitination occurs would prevent deubiquitination, as observed in arp2 cells (Fig. 6). Surprisingly, we observed that deubiquitination occurs in vps27 cells, in which Fur4p late endosome-to-vacuole traffic was blocked. Moreover, we also observed deubiquitination in doa4 vps27 cells (data not shown). Americk et al. then reported the isolation of numerous extragenic did (doa4-independent degradation) suppressors of the doa4-1 allele, all of which correspond to class E VPS genes, which function in the maturation of the late endosome into MVBs (2). Our preliminary result seemed to fit with these genetic data. To determine whether deletion of VPS27 influences Doa4p-dependent deubiquitination of Fur4p, we monitored the fate of Fur4p in vps27Δ pep4Δ cells and in the vps27Δ pep4Δ doa4Δ triple mutant. We used the same experimental conditions as we had used to observe Doa4p-dependent deubiquitination of Fur4p; i.e., we compared the pattern of Fur4p in P13 and P100 before and 2 h after CHX treatment. At time zero, we observed the usual pattern of ubiquitinated and unubiquitinated Fur4p (Fig. 8) in both types of cells. The combined VPS27 and PEP4 deletions inhibited Fur4p degradation, as revealed by the remaining Fur4p (unubiquitinated) after 2 h of CHX treatment. Only small amounts of Ub-Fur4p conjugates were still present in the triple mutant, which contrasted with the accumulation of these conjugates in the pep4Δ doa4Δ double mutant (compare Fig. 5B and 8). Consistent with this observation, vps27Δ pep4Δ doa4Δ cells did not display any particular accumulation of membrane-bound Ub conjugates (data not shown), which contrasts with what we found for doa4Δ pep4Δ cells.
FIG. 8.
Deletion of VPS27 restores the deubiquitination of Fur4p in pep4Δ doa4Δ cells. Membrane-enriched fractions were prepared from SD23 (vps27Δ pep4Δ) and SD24 (vps27Δ pep4Δ doa4Δ) cells transformed with YEp96fF (2μm URA3 FUR4 CUP1-UB), grown and induced for Ub and treated as described in the legend to Fig. 1. Aliquots were analyzed by Western immunoblotting for Fur4p, Pma1p, and Pep12p.
These data provide evidence that deleting VPS27 restores the deubiquitination of Fur4p and other membrane-bound Ub conjugates in cells lacking Doa4p. One hypothesis to interpret these data would be that impairment of the VPS pathway allows another deubiquitinating enzyme(s) to reach endocytic Doa4p targets, as suggested by Amerik et al. to explain the phenotype of did doa4 mutants (2).
Deficiency in the Ub-ligase Rsp5p suppresses the accumulation of small Ub-peptides in doa4 cells.
Papa and Hochstrasser demonstrated long ago that doa4Δ cells accumulate small Ub-linked peptides (Ub peptides) (49). They suggested recently that these peptides could at least in part result from vacuolar degradation of plasma membrane proteins targeted to the vacuole without prior Doa4p-dependent deubiquitination (2, 68). This was partly based on the observation that a mutation in the END3 gene involved in the internalization step of endocytosis suppressed the accumulation of these peptides (68). Since most of the plasma membrane proteins in yeast undergo Ub-dependent endocytosis, we wondered whether these peptides would also disappear from cells deficient in ubiquitination of plasma membrane proteins. Many plasma membrane proteins now appear to undergo ubiquitination and/or endocytosis controlled by the Ub ligase Rsp5p (reviewed in reference 57). We thus investigated the occurrence of the small doa4 Ub peptides in cells deficient for Rsp5p. Several rsp5 thermosensitive mutants have been described, but they did not appear appropriate for this investigation, since the small doa4 Ub peptides accumulate more specifically during exponential growth (49). We therefore used npil cells, which have reduced (<10-fold) amounts of Rsp5p as a result of the insertion of a Ty1 element in the 5′ region of RSP5 gene (22). These cells grow normally but are severely deficient in ubiquitination/endocytosis of a dozen plasma membrane proteins (reviewed in reference 57). We compared the patterns of Ub and small Ub conjugates in WT, npi1, doa4, and npi1 doa4 cells (Fig. 9A). To compare cells of the same genetic background, we used a specific doa4 allele, npi2. npi2 cells carry a point mutation in a conserved residue of the DOA4 gene that results in the same phenotype as a complete DOA4 deletion (66). As reported previously, the amount of free Ub in npi2 cells was much lower than that in WT cells (66, 68) but was normal in npi1 cells (Fig. 9A). npi2 cells had greater amounts of di-Ub than did WT (2, 68) or npi1 cells and the signature consisting of small peptides that migrated slightly above di-Ub, absent from both wild-type and npi1 cells. Such peptides were not observed in the npi1 npi2 double mutant. This suggests that these peptides are degradation products of Rsp5p substrates.
FIG. 9.
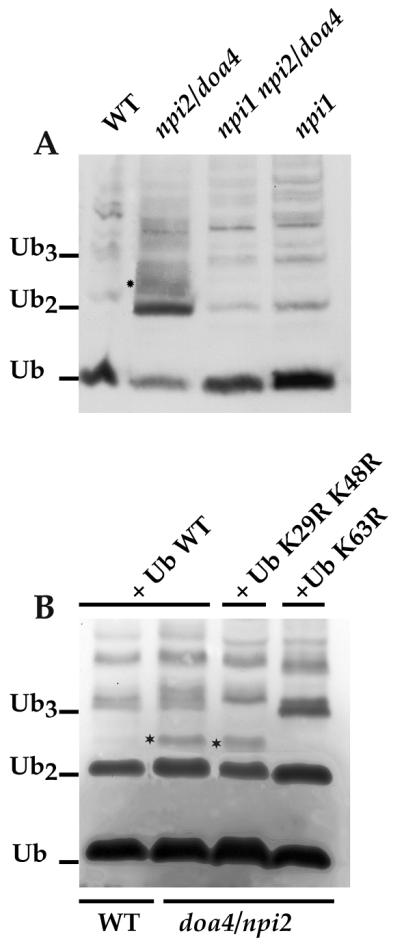
Suppression of the small Ub-containing peptides of doa4 cells by mutation in NPI1/RSP5 or overexpression of UbK63R. (A) 27061b (WT), 27064b (npi1), 27081a (npi2), or 33276d (npi1 npi2) cells were grown in YNB plus glucose. Cells were collected in exponential growth phase and used to prepare total protein extracts (NaOH-TCA technique). Aliquots were analyzed by Western immunoblotting using an antibody against Ub. The positions of Ub, di-Ub (Ub2), and tri-Ub (Ub3) are indicated. Small Ub peptides above di-Ub characteristic of doa4 cells are indicated (∗). (B) 27061b (WT) transformed with YEp96 (2 μm TRP1 CUP1-UB), and 27081a (npi2) cells transformed with YEp96, YEp96-UbK29, 48R, or YEp96-UbK63R were grown in YNB with glucose as the carbon source and induced for 2 h with CuSO4. Cells were collected and used to prepare protein extracts analyzed as in panel A.
Possible involvement of the UbK63 linkage in some of the small Ub peptides that accumulate in doa4Δ cells.
Two Rsp5p plasma membrane substrates were described to carry short Ub chains linked through Ub Lys63 (UbK63 linkage) (17, 66). The type of Ub linkage in other plasma membrane proteins has not yet been reported, but other examples of plasma membrane proteins carrying short Ub chains, two to three residues long, have been described (20, 54). If the doa4 Ub peptides result from vacuolar degradation of plasma membrane proteins, then what is the nature of linkage between Ub residues in these peptides? Three lysines in Ub are the in vivo acceptors of additional Ubs, Lys29, Lys48, and Lys63 (4). We investigated the type of Ub linkages in the small doa4 Ub peptides by overexpressing in doa4Δ cells several variant Ubs, in which these three lysines were mutated to arginine, preventing formation of Ub chains by elongation at each of these points. This resulted in doa4Δ cells overexpressing these variant Ubs, incorporating them statistically more frequently in Ub conjugates, notably in plasma membrane proteins (17, 66). For instance, Ub-Fur4p conjugates exhibited an up-shift in doa4 cells overproducing Ub extended by an HA tag, and the ubiquitination pattern in doa4 cells overproducing UbK63R showed two bands, instead of four, corresponding to the addition of only one Ub to the two Fur4p lysine acceptor sites (reference 17 and Fig. 4). Overexpression of wild-type Ub in doa4Δ cells corrected their Ub level, but small Ub-containing species still accumulated, notably above di-Ub and in the region of tri-Ub. Overproduction of UbK29 K48R led to disappearance of bands in the region of tri-Ub, whereas overproduction of UbK63R led to the disappearance of some of the small species above di-Ub and an accumulation of species in the region of tri-Ub. Hence, it is likely that some of the small peptides that accumulate in doa4 cells are degradation products of Ub conjugates carrying UbK63 linkages, whereas others are degradation products of Ub conjugates carrying UbK29 or UbK48 linkages.
DISCUSSION
We have determined whether the Ub isopeptidase Doa4p is involved in deubiquitination of yeast plasma membrane proteins by using the yeast plasma membrane transporter Fur4p, a well-characterized substrate of the Ub-dependent endocytic pathway. We show that this protein undergoes deubiquitination prior to vacuolar degradation, since the vacuoles of pep4 cells accumulated Fur4p mostly in an unubiquitinated form. In contrast, doa4 pep4 cells accumulated large amounts of Ub-Fur4p conjugates, strongly suggesting the involvement of Doa4p in the deubiquitination of this protein. This accumulation was suppressed when VPS27 was deleted, providing biochemical data supporting the hypothesis that impairment of MBV formation allows another UBP(s) to reach proteins that are usually Doa4p targets (2). At least some of the small Ub-containing peptides that accumulated in doa4Δ cells carried a UbK63-type linkage, a characteristic feature of Ub-Fur4p conjugates, supporting the idea that Doa4p is involved in the deubiquitination of plasma membrane proteins that display UbK63-type linkage like Ub-Fur4p conjugates.
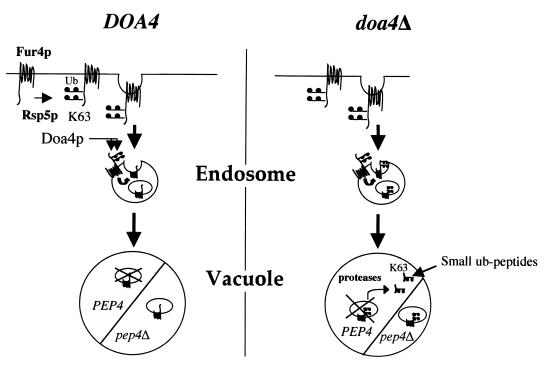
These data suggest that the events can be summarized in the following model (Fig. 10). Ub-Fur4p conjugates formed at the surface of WT cells in an Rsp5p-dependent fashion undergo Doa4p-dependent deubiquitination before vacuolar targeting. Doa4p is partly relocated in the late endosome of class E vps cells and thus probably lies partly at the cytoplasmic surface of late endosomes (2). Hence, deubiquitination probably occurs at the level of late endosome, releasing Ub into the cytosol prior to vesicle invagination toward the interior of late endosome, which gives rise to MVBs (16, 46). Deubiquitinated Fur4p is degraded in the vacuole by vacuolar proteases. Consequently, pep4 cells accumulate mostly unubiquitinated Fur4p. In agreement with our data, the a-factor receptor, which also undergoes ubiquitin-dependent endocytosis, accumulates in pep4 cells mainly in an unubiquitinated form, suggesting postinternalization deubiquitination (55). Ub-Fur4p conjugates are delivered to the vacuole in doa4Δ pep4Δ cells, where they accumulate, as do Ub conjugates presumably derived from many other plasma membrane proteins. Ub-Fur4p conjugates are delivered to the interior of late endosomes in doa4Δ cells without deubiquitination and are subsequently targeted to the vacuole. The vacuolar proteases then degrade them to small peptides. As Ub is rather resistant to proteolysis, small peptides carrying one to three Ub residues still attached to small pieces of Fur4p accumulate in the vacuoles. The sequestration of Ub in the vacuoles would ultimately result in its degradation, explaining the shorter half-life of Ub in doa4Δ cells (68). In agreement with this model, which fits with the hypothesis of Swaminathan et al. (68), Ub is greatly stabilized in doa4Δ pep4Δ cells.
FIG. 10.
Model of the involvement of Doa4p in the Fur4p endocytic pathway. Fur4p undergoes Rsp5-dependent ubiquitination at the plasma membrane, with the binding of di-Ub to two target lysines. Doa4p-dependent deubiquitination occurs prior to invagination into the late endosome. The absence of deubiquitination does not prevent the delivery of Ub-Fur4p conjugates to the vacuole. They accumulate in pep4Δ cells or are degraded by vacuolar proteases in PEP4 cells, giving rise to small Ub-containing peptides. Ub, plain circles.
The huge accumulation of Ub-Fur4p conjugates in doa4Δ pep4Δ cells indicates the involvement of Doa4p in the deubiquitination of Fur4p. We cannot definitely conclude from our data whether Doa4p is directly or indirectly involved in the cleavage between Ub and Fur4p or between Ub residues carried by this transporter. Among the relevant observations is the fact that the deubiquitination of Fur4p seems to be a processive event, since monoubiquitinated Fur4p is still detected in endosomal fractions after higher-molecular-mass Ub-Fur4p conjugates have disappeared. Hence, at least for some conjugates, the cleavage between Ub and Fur4p, catalyzed by Doa4p or by another UBP, probably occurs after cleavage between two Ubs.
The observation that overexpression of UbK63R led to the disappearance of some of the small Ub peptides that accumulate in doa4Δ cells suggests that the corresponding peptides carry UbK63 linkages and that Doa4p may be directly or indirectly involved in the cleavage of these linkages. Only two plasma membrane transporters have so far been shown to carry UbK63-linked short Ub chains. Our data suggest that this is a more general feature of yeast plasma membrane proteins. For instance, this is probably the case for the arginine permease and cadmium transporter, for the reasons detailed below. doa4Δ cells are hypersensitive to cadmium and to canavanine, a toxic compound that enters yeast cells via the arginine permease (68). The hypersensitivity of doa4Δ cells to these compounds is probably due to defective endocytic down-regulation of the corresponding two transporters because of Ub depletion. Hypersensitivity to cadmium and canavanine was also reported in cells carrying a point mutation in the RSP5 gene, in agreement with the hypothesis of a Ub-dependent endocytosis of the two transporters (34). In doa4Δ cells, hypersensitivity to cadmium and canavanine is shown to be complemented by the overexpression of Ub and UbK48R, but not of UbK63R (68), suggesting that UbK63-linked Ub chains must be formed for efficient endocytosis of cadmium transporter and arginine permease, as for Fur4p and Gap1p. UbK63 linkages are involved in mitochondrial inheritance (15), DNA repair (31, 64, 73), and modification of ribosomal proteins (63), in addition to their role in endocytosis. Whether Doa4p is involved in deubiquitination of the corresponding targets remains to be determined.
A lack of Doa4p in cells overproducing Ub did not seem to have a dramatic effect on the trafficking of uracil permease from the plasma membrane to the vacuole. Internalization occurs at a normal rate (reference 17 and Fig. 5), there is no accumulation of Ub-Fur4p conjugates, and there is only a small reduction in the rate of degradation. But the slowing of the postinternalization steps of endocytosis may vary from protein to protein, since doa4Δ cells accumulate more Ub conjugates than WT cells (not shown), and the Ub conjugates that accumulate in doa4Δ pep4Δ cells have distinct fractionation characteristics. These conjugates may be proteins en route to vacuolar degradation, and there may be blockage prior to late endosome delivery. An overaccumulation of ubiquitinated plasma membrane proteins prior or at the late endosome might result in the recycling of endocytosed proteins to the plasma membrane, as reported for a variety of mutants in which endosomal transport to the vacuole is blocked (11, 51). Preliminary data suggest that doa4Δ cells indeed show limited recycling under some experimental conditions. Recycling resulting from an absence of deubiquitination might be artifactual, being due to a backup resulting from a downstream block rather than a normal physiological event. Intuitively, a recycling event in a Ub-dependent endocytic pathway is more likely to be due to a deubiquitination step. A rare example of deubiquitination described in an endocytic pathway is that of the high-affinity immunoglobulin E receptor (FcɛRI) (47). Antigen-induced engagement of FcɛRI results in the immediate multiubiquitination of FcɛRI β and γ chains. Disengagement of the receptors is accompanied by massive, rapid deubiquitination without any receptor degradation (47); hence, receptors may be recycled. A recycling pathway was found recently in yeast by using a lipophilic dye, FM4-64 (83). Recycling in yeast has been described for a limited number of proteins, including chitin synthases (9) and the v-SNAREs Snc1p and Snc2p (39). Whether the internalization of chitin synthases and Snc proteins is or not Ub dependent has not been reported. Recycling was also recently reported for the a-factor receptor (Ste3p) internalized via a ligand-dependent pathway (8). The constitutive, ligand-independent internalization of Ste3p is Ub dependent (55) and has been extensively characterized (54, 56). Adding the ligand also causes the ubiquitination of a truncated form of Ste3p, lacking the sequence involved in constitutive internalization (55). Whether deubiquitination is involved in the recycling of Ste3p has not been reported. A potential role for Doa4p in recycling thus remains an open question.
The ubiquitination of uracil permease appears primarily to be a plasma membrane event. Ub-Fur4p conjugates are found in sucrose gradients in plasma membrane fractions. Conjugates accumulate in arp2 cells after the inhibition of internalization; the pool of plasma membrane Ub-Fur4p is very stable and unchanged in cells also lacking DOA4. Ub conjugates of several other plasma membrane proteins also accumulate after inhibition of the internalization step of endocytosis (13, 26, 55). In addition to this apparent general involvement of ubiquitination as a signal-triggering internalization, it was also proposed that ubiquitination may occur in internal compartments in yeast. Ubiquitination may be required for diverting intracellular pools of the tryptophan permease Tat2p, from the Golgi complex directly to the vacuole upon nutrient limitation (5). Deubiquitination might have other regulating roles in such situations. Both Tat2p and Ste6p, the transporter of the a-factor, are found at the vacuolar membrane (5) or as a ring delimiting the vacuole (40) in doa4 cells not complemented with Ub. Whether this unusual localization results from a lack of Ub, characteristic of doa4 cells, or a deficiency in Doa4-dependent deubiquitination remains to be elucidated. One possibility is that a ubiquitination event is required for the invagination to the interior of late endosomes. Nedd4, the human homolog of the Ub ligase Rsp5p, is found both at the plasma membrane and in endosome-like structures, suggesting that it may be involved in ubiquitination in internal compartments (52). Rsp5p is also found at these two sites (79).
Ubiquitination-deubiquitination events may also regulate the trafficking machinery. The ubiquitin ligase c-Cbl is involved in the multiubiquitination of several tyrosine kinases receptors associated with their endocytosis and itself undergoes plasma membrane multiubiquitination, followed by rapid deubiquitination before its return to the cytoplasm (80). Eps15, a protein required for the correct formation of clathrin-coated vesicles in mammalian cells (58), undergoes monoubiquitination in response to epidermal growth factor binding to its receptor (74). While the UBPs involved in deubiquitination of these proteins have not yet been identified, genetic data suggest that epsin, one of the partners of Eps15 (58), is a key target of the Drosophila Fat facets deubiquitinating enzyme (7).
Thus, this study indicates that the yeast Ub isopeptidase Doa4p is involved in the deubiquitination of endocytosed plasma membrane proteins. The other UBP(s) able to replace Doa4p in vps class E mutants remains to be identified. Further investigations are also needed to determine whether Doa4p has other roles in trafficking events associated with the late steps of the endocytic pathway.
ACKNOWLEDGMENTS
We are grateful to B. André, M.-O. Blondel, J. Decraene, M. Ellison, J.-M. Galan, J. Hegeman, S. Amerik, and M. Hochstrasser for providing plasmids and strains and to M.-R. Chevallier, R. Serrano, D. Katzman, and S. Emr for gifts of antibodies. We thank M. Hochstrasser for encouragment during this work. We are indebted to members of the laboratory for stimulating discussions. Special thanks are also due to G. Castillon, D. Urban-Grimal, and C. Volland for critical reading of the manuscript and to O. Parkes and M. Ghosh for editorial assistance.
This work was supported by grant 5681 from the Association pour la Recherche contre le Cancer.
ADDENDUM IN PROOF
While this paper was in press, Kölling and coworkers reported that the accumulation of the Ste6p tarnsporter at the vacuolar membrane in doa4Δ cells results from the decrease in the Ub pool in these cells (S. Losko, F. Kopp, and R. Kölling, Mol. Biol. Cell 12:1047–1059, 2001).
REFERENCES
- 1.Amerik A Y, Li S J, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000;381:981–992. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- 2.Amerik A Y, Nowak J, Swaminathan S, Hochstrasser M. The doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amerik A Y, Swaminathan S, Krantz B A, Wilkinson K D, Hochstrasser M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 1997;16:4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnason T, Ellison M J. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol. 1994;14:7876–7883. doi: 10.1128/mcb.14.12.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck T, Schmidt A, Hall M N. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol. 1999;146:1227–1238. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifacino J S, Weissman A. Ubiquitin and the control of protein fate in the scretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cadavid A L M, Ginzel A, Fischer J A. The function of the Drosophila Fat facets deubiquitinationg enzyme in limiting photoreceptor cell number is intimately associated with endocytosis. Development. 2000;127:1727–1736. doi: 10.1242/dev.127.8.1727. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Davis N G. Recycling of the yeast a-factor receptor. J Cell Biol. 2000;151:731–738. doi: 10.1083/jcb.151.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuang J S, Schekman R W. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cormack B P, Bertram G, Egerton M, Gow N A, Falkow S, Brown A J. Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 11.Davis N G, Horecka J L, Sprague J G F. Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ecker D J, Ishaq Khan M, Marsh J, Butt T R, Crooke S T. Chemical synthesis and expression of a cassette adapted ubiquitin gene. J Biol Chem. 1987;262:3524–3527. [PubMed] [Google Scholar]
- 13.Egner R, Kuchler K. The yeast multidrug transporter Pdr5 of the plasma membrane is ubiquitinated prior to endocytosis and degradation in the vacuole. FEBS Lett. 1996;378:177–181. doi: 10.1016/0014-5793(95)01450-0. [DOI] [PubMed] [Google Scholar]
- 14.Ellison M, Hochstrasser M. Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J Biol Chem. 1991;266:21150–21157. [PubMed] [Google Scholar]
- 15.Fisk H A, Yaffe M P. A role for ubiquitination in mitochondrial inheritance in S. cerevisiae. J Cell Biol. 1999;145:1199–1208. doi: 10.1083/jcb.145.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Futter C E, Pearse A, Hewlett L J, Hopkins C R. Multivesicular endosomes containing internlized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J Cell Biol. 1996;132:10011–1023. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galan J-M, Haguenauer-Tsapis R. Ubiquitin Lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galan J M, Moreau V, André B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 19.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gitan R S, Eide D J. Zinc-regulated ubiquitin conjugation signals endocytosis of the yeast ZRT1 zinc transporter. Biochem J. 2000;346:329–336. [PMC free article] [PubMed] [Google Scholar]
- 21.Harvey K F, Kumar S. Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 1999;9:166–169. doi: 10.1016/s0962-8924(99)01541-x. [DOI] [PubMed] [Google Scholar]
- 22.Hein C, Springael J Y, Volland C, Haguenauer-Tsapis R, André B. NPI1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 23.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 24.Hicke L. Gettin' down with ubiquitin: turning off cell-surface receptors transporters and channels. Trends Cell Biol. 1999;9:107–112. doi: 10.1016/s0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- 25.Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 26.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 27.Hicke L, Zanolari B, Pypaert M, Rohrer J, Riezman H. Transport through the yeast endocytic pathway occurs through morphologically distinct compartments and requires an active secretory pathway and Sec18p/N-ethylmaleimide-sensitive fusion protein. Mol Biol Cell. 1997;8:13–31. doi: 10.1091/mbc.8.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 29.Hochstrasser M, Ellison M J, Chau V, Varshavsky A. The short-lived MATalpha2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hochstrasser M, Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast alpha2 repressor. Cell. 1990;61:697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- 31.Hofmann R, Pickart C M. Noncanonical MMS2-encoded ubiquitin-conjugating enzymes functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 32.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2- dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 33.Kahana A, Gottschling D E. DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6608–6620. doi: 10.1128/mcb.19.10.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanda T. A ubiquitin-protein ligase (E3) mutation of Saccharomyces cerevisiae suppressed by co-overexpression of two ubiquitin-specific processing proteases. Genes Genet Syst. 1996;71:75–83. doi: 10.1266/ggs.71.75. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi T, Stang E, Fang K S, de Moerloose P, Parton R G, Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- 36.Kölling R, Hollenberg C P. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee P S, Wang Y, Dominguez M G, Yeung Y G, Murphy M A, Bowtell D D, Stanley E R. The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J. 1999;18:3616–3628. doi: 10.1093/emboj/18.13.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon W A, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis M J, Nichols B J, Prescianotto-Baschong C, Riezman H, Pelham H. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loayza D, Michaelis S. Role for the ubiquitin-proteasome system in the vacuolar degradation of Ste6p, the a-factor transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:779–789. doi: 10.1128/mcb.18.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchal C, Haguenauer-Tsapis R, Urban-Grimal D. Casein kinase I-dependent phosphorylation within a PEST sequence and ubiquitination at nearby lysines, signal endocytosis of yeast uracil permease. J Biol Chem. 2000;275:23608–23614. doi: 10.1074/jbc.M001735200. [DOI] [PubMed] [Google Scholar]
- 42.Marchal C, Haguenauer-Tsapis R, Urban-Grimal D. A PEST-like sequence mediates phosphorylation and efficient ubiquitination of the yeast uracil permease. Mol Cell Biol. 1998;18:314–321. doi: 10.1128/mcb.18.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moazed D, Johnson A D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 44.Moreau V, Galan J-M, Devilliers G, Haguenauer-Tsapis R, Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol Biol Cell. 1997;8:1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreau V, Madania A, Martin R P, Winsor B. The Saccharomyces cerevisiae actin-related protein Arp2 is involved in the actin cytoskeleton. J Cell Biol. 1996;134:117–132. doi: 10.1083/jcb.134.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Odorizzi G, Babst M, Emr S D. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- 47.Paolini R, Kinet J-P. Cell surface control of the multiubiquitination and deubiquitination of high-affinity immunoglobulin E receptors. EMBO J. 1993;12:779–786. doi: 10.1002/j.1460-2075.1993.tb05712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papa F R, Amerik A Y, Hochstrasser M. Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome. Mol Biol Cell. 1999;10:741–756. doi: 10.1091/mbc.10.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papa F R, Hochstrasser M. The yeast doa4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- 50.Pickart C M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 51.Piper R C, Cooper A A, Yang H, Stevens T H. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plant P J, Lafont F, Lecat S, Verkade P, Simons K, Rotin D. Apical membrane targeting of Nedd4 is mediated by an association of its C2 domain with annexin XIIIb. J Cell Biol. 2000;149:1473–84. doi: 10.1083/jcb.149.7.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberg K J, Rowley N, Kaiser C A. Physiological regulation of membrane sorting late in the secretory pathway of Saccharomyces cerevisiae. J Cell Biol. 1997;137:1469–1482. doi: 10.1083/jcb.137.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roth A F, Davis N G. Ubiquitination of the PEST-like endocytosis signal of the yeast a-factor receptor. J Biol Chem. 2000;275:8143–8153. doi: 10.1074/jbc.275.11.8143. [DOI] [PubMed] [Google Scholar]
- 55.Roth A F, Davis N G. Ubiquitination of the yeast a-factor receptor. J Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roth A F, Sullivan D M, Davis N G. A large PEST-sequence directs the ubiquitination, endocytosis, and vacuolar degradation of the yeast a-factor receptor. J Cell Biol. 1998;142:949–961. doi: 10.1083/jcb.142.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rotin D, Staub O, Haguenauer-Tsapis R. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J Membr Biol. 2000;176:1–17. doi: 10.1007/s00232001079. [DOI] [PubMed] [Google Scholar]
- 58.Santolini E, Salcini A E, Kay B K, Yamabhai M, Di Fiore P P. The EH network. Exp Cell Res. 1999;253:186–209. doi: 10.1006/excr.1999.4694. [DOI] [PubMed] [Google Scholar]
- 59.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Ann Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 60.Séron K, Blondel M-O, Haguenauer-Tsapis R, Volland C. Uracil-induced down regulation of the yeast uracil permease. J Bacteriol. 1999;181:1793–1800. doi: 10.1128/jb.181.6.1793-1800.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shih S C, Sloper-Mould K E, Hicke L. Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J. 2000;19:187–198. doi: 10.1093/emboj/19.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silve S, Volland C, Garnier C, Jund R, Chevallier M R, Haguenauer-Tsapis R. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol Cell Biol. 1991;11:1114–1124. doi: 10.1128/mcb.11.2.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spence J, Gali R R, Dittmar G, Sherman F, Finley D. Cell cycle regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- 64.Spence J, Sadis S, Haas A L, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spormann D O, Heim J, Wolf D H. Biogenesis of the yeast vacuole (lysosome)—the precursor forms of the soluble hydrolase carboxypeptidase yscY are associated with the vacuolar membrane. J Biol Chem. 1992;267:8021–8029. [PubMed] [Google Scholar]
- 66.Springael J-Y, Galan J-M, Haguenauer-Tsapis R, André B. NH4+-induced down-regulation of the Saccharomyces cerevisiae Gaplp permease involves its ubiquitination with lysine-63-linked chains. J Cell Sci. 1999;112:1375–1383. doi: 10.1242/jcs.112.9.1375. [DOI] [PubMed] [Google Scholar]
- 67.Strous G J, Govers R. The ubiquitin-proteasome system and endocytosis. J Cell Sci. 1999;112:1417–1423. doi: 10.1242/jcs.112.10.1417. [DOI] [PubMed] [Google Scholar]
- 68.Swaminathan S, Amerik A Y, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol Biol Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taya S, Yamamoto T, Kanai-Azuma M, Wood S A, Kaibuchi K. The deubiquitinating enzyme Fam interacts with and stabilize β-catenin. Genes Cells. 1999;4:757–767. doi: 10.1046/j.1365-2443.1999.00297.x. [DOI] [PubMed] [Google Scholar]
- 70.Taya S, Yamamoto T, Kano K, Kawano Y, Iwamatsu A, Tsuchiya T, Tanaka K, Kanai-Azuma M, Wood S A, Mattick J S, Kaibuchi K. The Ras target AF-6 is a substrate of the fam deubiquitinating enzyme. J Cell Biol. 1998;142:1053–1062. doi: 10.1083/jcb.142.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Terrell J, Shih S, Dunn R, Hicke L. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- 72.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 73.Ulrich H D, Jentsch S. Two ring finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Delft S, Govers R, Strous G J, Verkleij A J, van Bergen en Henegouwen P M. Epidermal growth factor induces ubiquitination of Eps15. J Biol Chem. 1997;272:14013–14016. doi: 10.1074/jbc.272.22.14013. [DOI] [PubMed] [Google Scholar]
- 75.Volland C, Garnier C, Haguenauer-Tsapis R. In vivo phosphorylation of the yeast uracil permease. J Biol Chem. 1992;267:23767–23771. [PubMed] [Google Scholar]
- 76.Volland C, Urban-Grimal D, Géraud G, Haguenauer-Tsapis R. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J Biol Chem. 1994;269:9833–9841. [PubMed] [Google Scholar]
- 77.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 78.Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 79.Wang G, McCaffery J M, Wendland B, Dupré S, Haguenauer-Tsapis R, Huibregtse J M. Localization of the Rsp5p ubiquitin-protein ligase at multiple sites within the endocytic pathway. Mol Cell Biol. 2001;21:3564–3575. doi: 10.1128/MCB.21.10.3564-3575.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Y, Yeung Y-G, Stanley E R. CSF-1 stimulated multiubiquitination of the CSF-1 receptor and of Cbl follows their tyrosine phosphorylation and association with other signaling proteins. J Cell Biochem. 1999;72:119–134. [PubMed] [Google Scholar]
- 81.Waterman H, Yarden Y. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 2001;490:142–152. doi: 10.1016/s0014-5793(01)02117-2. [DOI] [PubMed] [Google Scholar]
- 82.Weissman A M. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 83.Wiederkehr A, Avaro S, Prescianotto-Bashong C, Haguenauer-Tsapis R, Riezman H. The F-box Rcylp is involved in endocytic membrane traffic and recycling out of an early endosome in S. cerevisiae. J Cell Biol. 2000;149:397–410. doi: 10.1083/jcb.149.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilkinson K D. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]