As the basic building blocks of all living species, cells play a crucial role in sustaining life activities. Traditional bulk methods only give us molecular insight into tissue, organ, or/and individual. With the rapid development of sequencing technology, especially the 10× Genomics based on microfluidic omics, biological research has entered the era of single‐cell level, which enables us to gain insight into life activities from more microscopic perspective. With this cutting‐edge technology, it is now possible to mine heterogeneity between tissue types and within cells like never before. However, the preparation of single‐cell suspension (namely, protoplasm suspension) and the annotation of cell clusters are still two main obstacles faced by single‐cell research. A few marker gene databases are currently available, including CellMarker, PanglaoDB, and SignatureDB (https://lymphochip.nih.gov/signaturedb/). Single‐cell research in mouse and human is at the rapid development stage. However, in plant science, the research for the single‐cell profiling is still in its infancy and quite limited resource were available, such as in Arabidopsis thaliana (Gala et al., 2021; Zhang and Chen, 2021b; Zhang et al., 2019), Zea mays (Marand et al., 2021; Xu et al., 2021), Oryza sativa (Liu, Liang, et al., 2021; Wang et al., 2021; Zhang et al., 2021a), Solanum lycopersicum (Tian et al., 2020), and Arachis hypogaea (Liu, Hu, et al., 2021). The existence of cell wall makes the preparation of plant single‐cell suspension more difficult than that of animals. On the other hand, due to the lack of effective marker gene database, single‐cell research in plant was usually time‐consuming for performing large amount of in situ RNA hybridization or genetic transformation with reporter gene to provide reliable experimental evidence for the identification of tissue type of cell cluster. It is in urgent demands to establish a comprehensive database and exploit efficient tools to analyze such scattered over thousands marker genes of specific cell types from different plant species. Therefore, based on the Shiny and bootstrap frameworks, we developed the Plant Single Cell Transcriptome Hub (PsctH) (http://jinlab.hzau.edu.cn/PsctH/), aiming to provide a comprehensive and accurate resource of cell markers and web tool for various cell types in tissues of plant species (Figure 1a). All marker genes included in the PsctH must have been evidenced via RNA in situ hybridization or expression of GFP reporter. Based on this standard, we presented over 20 published plant single‐cell reports including 98 cell markers from 51 cell types in 9 plant tissues/sub‐tissues of five plant species (Arabidopsis thaliana, Zea mays, Oryza sativa, Arachis hypogaea, and Solanum lycopersicum), and the data were collected and deposited in PsctH (Figure 1b–d). There have been more than 2 marker genes for phloem parenchyma cells and veins from leaf, cortex, lateral root primordia, and stele from root; inflorescence meristem; and spikelet meristem from staminate primordia. Notably, the majority of the cell marker entries are derived from roots, involving 26 cell types, and the most abundant root cell type is lateral root primordia cell. All marker genes were displayed in page of ‘MarkerGeneDB’ page and can be searched via keywords (Figure 1e). Meanwhile, all experimental evidence derived from RNA in situ hybridization or GFP reporter were accompanied for each marker gene. In addition, all marker genes’ information can be download as a tab‐delimited file on the ‘Download’ page and can be easily used in the command line.
Figure 1.
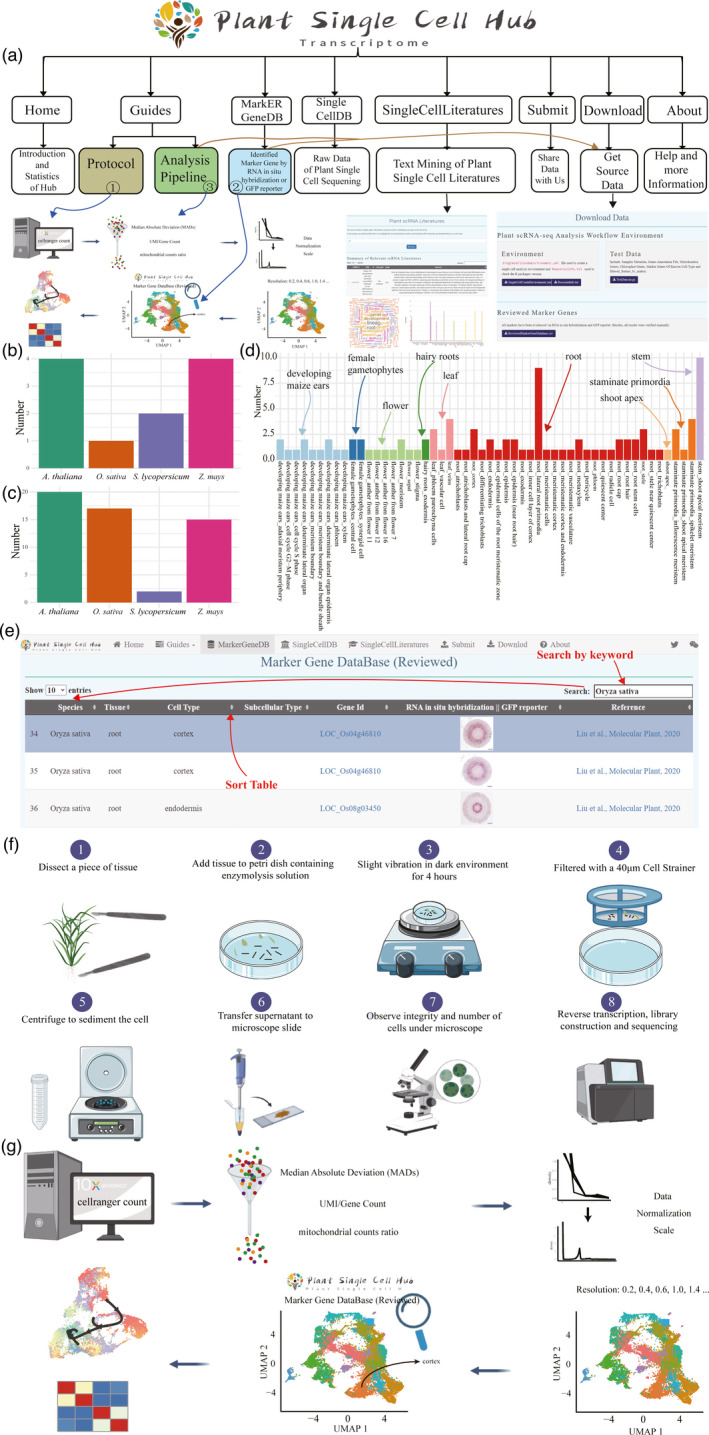
Statistics of cell markers and function interface in PsctH. (a) Overview of PsctH sitemap, which provided the information and relationship about the pages. Distribution of tissues (b) and cell types (c) for different species in PsctH. (d) Distribution of cell markers for different tissue types in PsctH. (e) The web images in the ‘MarkerGeneDB’ page and schematic diagram illustrate the process of browsing and keyword searching (such as gene id, species, tissue, and cell type) the database, which allows to quickly search for marker genes of cells in different tissues. (f) Schematic diagram highlighting the experimental setup for prepare protoplasts used for single‐cell RNA sequencing, which includes enzymatic hydrolysis of tissue samples and single‐cell quality detection under the microscope. (g) Illustration of the workflow used for performing plant single‐cell RNA data analysis, which includes filter out poor‐quality cells, and normalize and identify variable genes, cluster, and annotation cell type.
Another challenge of applying single‐cell analysis in plants is to prepare integrity and living single‐cell suspension. The first step of preparing plant single‐cell suspension usually starts with digestion of cell wall via cellulase and hemicellulase. In PsctH, we compared and evaluated different protocols used to prepare protoplasts and offer a feasible and efficient experimental pipeline for reference, which included preparation and isolation of tissue samples, enzyme digestion, purification, and the detection of the integrity of single cells (Figure 1f). To standardize and link the process of plant single‐cell data mining, a flexible pipeline of plant single‐cell transcriptome (scRNA‐seq) analysis was also provided, including the process of quality control, normalization and scale, clustering, and marker genes identification (Figure 1g). Users can configure key parameters to obtain a dedicated R analysis script. In addition, we also provide configuration files (SingleCellCondaEnvironment.yml) that can easily reproduce the analysis environment of single‐cell transcriptome through conda and run the R script obtained previously.
The scRNA‐Seq data are particularly powerful in resolving progressions in gene expression. Single‐cell sequencing data from different tissues of different species will help us to construct a complete plant single‐cell landscape, dissecting cellular heterogeneity, identify the key regulatory genes involved in life activities, and insight into specific biological mechanisms. In this case, we collected all published high throughput sequencing data of plant single‐cell and accessible in ‘SingleCellDB’ page, which were categorized by species and tissues. To best serve the community, we build a user interface that allows intuitive searching of plant single‐cell literature and performed text mining using natural language processing (NLP) to highlight the most important keywords and explore their associations. The results are compiled by searching for keywords in abstracts and titles using data from PubMed.
In summary, PsctH will be a comprehensive and valuable resource for researchers applying single‐cell experiments to plants. The isolation of protoplast, pipeline of data processing, and manually curated resource of cell markers collected from experimental researches will be expected to promote plant science research into a single‐cell resolution. Indeed, the rapidly growing field of single‐cell biology has given us an opportunity to constantly improve and enrich the database. Therefore, we will continue to track the single‐cell sequencing studies and update the database by frequent additions of new cell markers across all plant species. Meanwhile, we hope researchers can contribute PsctH by submitting data to us by ‘Submit’ page.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
S.J. and F.D. conceptualized and supervised the research. Z.X. designed and coded this database. Z.X. and G.W. wrote the manuscript. Q.W., X.Z., and G.W. help collected literature and manually curated data. Q.Y., L.T., X.Z., and H.D. provided constructive comments and suggestions on this research. All authors contributed to the writing of the final manuscript.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31872077) and Science and Technology Major Project of Guangxi (Gui Ke AA18118046) to Dr. Fang Ding and Fundamental Research Funds for the Central Universities (2021ZKPY003) to Dr. Shuangxia Jin.
Xu, Z. , Wang, Q. , Zhu, X. , Wang, G. , Qin, Y. , Ding, F. , Tu, L. , Daniell, H. , Zhang, X. and Jin, S. (2022) Plant Single Cell Transcriptome Hub (PsctH): an integrated online tool to explore the plant single‐cell transcriptome landscape. Plant Biotechnol. J., 10.1111/pbi.13725
Contributor Information
Fang Ding, Email: dinfany@mail.hzau.edu.cn.
Shuangxia Jin, Email: jsx@mail.hzau.edu.cn.
References
- Gala, H.P. , Lanctot, A. , Jean‐Baptiste, K. , Guiziou, S. , Chu, J.C. , Zemke, J.E. , George, W. et al. (2021) A single‐cell view of the transcriptome during lateral root initiation in Arabidopsis thaliana . Plant Cell, 33(7), 2197–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Hu, D. , Du, P. , Wang, L. , Liang, X. , Li, H. , Lu, Q. et al. (2021) Single‐cell RNA‐seq describes the transcriptome landscape and identifies critical transcription factors in the leaf blade of the allotetraploid peanut (Arachis hypogaea L.). Plant Biotechnol. J., 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Liang, Z. , Feng, D. , Jiang, S. , Wang, Y. , Du, Z. , Li, R. et al. (2021) Transcriptional landscape of rice roots at the single‐cell resolution. Mol. Plant, 14, 384–394. [DOI] [PubMed] [Google Scholar]
- Marand, A.P. , Chen, Z. , Gallavotti, A. and Schmitz, R.J. (2021) A cis‐regulatory atlas in maize at single‐cell resolution. Cell, 184(11), 3041–3055.e21. https://onlinelibrary.wiley.com/doi/full/10.1111/pbi.13656 [DOI] [PubMed] [Google Scholar]
- Tian, C. , Du, Q. , Xu, M. , Du, F. & Jiao, Y. (2020) Single‐nucleus RNA‐seq resolves spatiotemporal developmental trajectories in the tomato shoot apex. bioRxiv, 2020.09.20.305029. [Google Scholar]
- Wang, Y. , Huan, Q. , Li, K. and Qian, W. (2021) Single‐cell transcriptome atlas of the leaf and root of rice seedlings. J. Genet Genomics, 1–18. https://www.sciencedirect.com/science/article/pii/S1673852721001673 [DOI] [PubMed] [Google Scholar]
- Xu, X. , Crow, M. , Rice, B.R. , Li, F. , Harris, B. , Liu, L. , Demesa‐Arevalo, E. et al. (2021) Single‐cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Dev. Cell, 56, 557–568.e556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T.‐Q. , Chen, Y. , Liu, Y. , Lin, W.‐H. and Wang, J.‐W. (2021a) Single‐cell transcriptome atlas and chromatin accessibility landscape reveal differentiation trajectories in the rice root. Nat. Commun., 12, 2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T.‐Q. , Chen, Y. and Wang, J.‐W. (2021b) A single‐cell analysis of the Arabidopsis vegetative shoot apex. Dev. Cell, 56(7), 1056–1074.e8. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Xu, Z. , Shang, G. and Wang, J. (2019) A single‐cell RNA sequencing profiles the developmental landscape of Arabidopsis root. Mol. Plant, 12, 648–660. [DOI] [PubMed] [Google Scholar]


