Summary
Pathogenic fungi cause major postharvest losses. During storage and ripening, fruit becomes highly susceptible to fungi that cause postharvest disease. Fungicides are effective treatments to limit disease. However, due to increased public concern for their possible side effects, there is a need to develop new strategies to control postharvest fungal pathogens. Botrytis cinerea, a common postharvest pathogen, was shown to uptake small double‐stranded RNA (dsRNA) molecules from the host plant. Such dsRNA can regulate gene expression through the RNA interference system. This work aimed to develop a synthetic dsRNA simultaneously targeting three essential transcripts active in the fungal ergosterol biosynthesis pathway (dsRNA‐ERG). Our results show initial uptake of dsRNA in the emergence zone of the germination tube that spreads throughout the fungus and results in down‐regulation of all three targeted transcripts. Application of dsRNA‐ERG decreased B. cinerea germination and growth in in vitro conditions and various fruits, leading to reduce grey‐mould decay. The inhibition of growth or decay was reversed by the addition of ergosterol. While dual treatment with dsRNA‐ERG and ergosterol‐inhibitor fungicide reduced by 100‐fold the required amount of fungicide to achieve the same protection rate. The application of dsRNA‐ERG induced systemic protection as shown by decreased decay development at inoculation points distant from the treatment point in tomato and pepper fruits. Overall, this study suggests that dsRNA‐ERG can effectively control B. cinerea growth and grey‐mould development suggesting its efficacy as a future method for postharvest control of fungal pathogens.
Keywords: double‐stranded RNA, ergosterol biosynthesis, Botrytis cinerea, grey mould, postharvest
Introduction
Globally, postharvest fruit and vegetable loss are estimated to impact more than 40% of yield. Pathogenic fungi are responsible for a major part of that loss (Abiad and Meho, 2018; Lipinski et al., 2013). Botrytis cinerea represents one of the most predominant and common necrotrophic fungal pathogen promoting postharvest decay of fresh fruit and vegetables (Romanazzi and Feliziani, 2014). B. cinerea has a wide range of hosts and can infect over 200 plant species, causing grey mould disease (Kumar et al., 2020; Schumacher, 2012). Infections by B. cinerea usually occur in the field, where after initial colonization, the fungi remain latent until harvest, storage and ripening. During ripening, fruits lose their innate resistance and become susceptible to the dormant fungal pathogens that switch from a latent to an active necrotrophic stage and cause grey mould (Romanazzi and Feliziani, 2014).
The most effective treatment to date against postharvest diseases is the application of fungicides (Nerva et al., 2020; Rani et al., 2020). However, fungicides hold disadvantages such as the risk of the development of fungicide‐resistant fungi (Elmer and Reglinski, 2006). Furthermore, due to their lack of specificity, they are a potential threat to beneficial micro‐organisms in the environment including their impact on human health (Rani et al., 2020). Therefore, there is a need to develop eco‐friendly approaches to control fungal pathogen and postharvest diseases (Wisniewski et al., 2016).
Recent progress in the control of pathogenic fungi takes advantage of a natural RNA interference (RNAi) process that occurs in eukaryotic cells (Mbengue et al., 2016; Mitter et al., 2017; Nerva et al., 2020). RNAi is based on the cellular recognition of double‐stranded RNA (dsRNA) molecules that leads to sequence‐specific degradation or interference of translation of a target messenger RNA (mRNA; Hua et al., 2018; Voinnet, 2008). In plants, dsRNA regulatory systems were found to be involved in endogenous gene regulation as well as in plant–pathogen interactions. Thus, plants control virus replication using this mechanism, and can also transfer inhibitory RNA from the host plant cell to fungal pathogens (Hua et al., 2018; Llave, 2010; Weiberg et al., 2013), which provides new possibilities for plant protection.
In a procedure called host‐induced gene silencing, transgenic plants produce small RNA molecules that induce post‐transcriptional gene silencing (Koch et al., 2013; Nunes and Dean, 2012). Despite its high potential, this method is limited by regulatory restrictions for genetically modified organisms and by difficulties in generating transgenic plants with stable expression. Alternatively, spray‐induced gene silencing (SIGS), where the dsRNA is externally applied on the plant parts such as leaves, branches or fruit was also shown to be effective (Koch et al., 2016; Wang et al., 2016). SIGS also has the clear advantage of being potentially rapidly adapted to a dynamic pathogenic environment. Several studies have demonstrated how direct application of dsRNA, using SIGS targeting virulence genes (e.g. dcl1 and dcl2) or essential genes for fungal colonization (e.g. erg11, chs1 and EF2), can offer plant protection (Nerva et al., 2020; Wang et al., 2016).
Ergosterol is a C28 sterol found particularly in fungal cell membranes (Lees et al., 1995). It has been shown that ergosterol plays a vital role in the cell membrane structure and function of fungi. Ergosterol is responsible for maintaining membrane fluidity (Lees et al., 1979), regulates membrane permeability (Bard et al., 1978), influences the activity of membrane‐related enzymes (Rottem et al., 1973) and alters fungal cell growth (Lees et al., 1980). Due to the importance of ergosterol for fungal growth and survival, the ergosterol biosynthesis pathway was found to be a good candidate for the development of antifungal agents (Bhattacharya et al., 2018; Yan et al., 2011). Indeed, various fungicides target the ergosterol biosynthesis of fungi (Ahmad et al., 2010). Furthermore, a gene, erg11 that encodes for a crucial step in the synthesis of ergosterol was shown to be effective when targeted together with additional genes, for example chitinase, and elongation factor (chs1 and EF2; Nerva et al., 2020).
The current study has the following aims. First, to test the efficacy of multiple targeting of essential transcripts solely in the ergosterol biosynthesis pathway. Second, to investigate the ability of the designed dsRNA to decrease and control fungal germination, growth and decay development in postharvest products. Third to evaluate a possible systemic mechanism. Fourth, to explore the use of SIGS in combination with a commercially used fungicide to reduce their needed concentrations.
Results
dsRNA penetrates the emergence zone of the hyphae and inhibit B. cinerea germination and growth
Three different genes in the ergosterol biosynthesis pathway of B. cinerea were chosen as targets. Sequences from erg13, erg11 and erg1 were joined to yield a construct in a total length of 751 bp (dsRNA‐ERG; Figure 1, see Materials and methods). For positive control, another construct targeting B. cinerea DCL1/DCL2 dicer (dsRNA‐Dicer) was cloned in a similar manner. To verify that B. cinerea is capable of taking up external dsRNA from the environment, a fluorescent dsRNA‐ERG construct was prepared by incorporation of Cy5‐labelled nucleotides into the sequence of dsRNA‐ERG or dsRNA‐Dicer. The results showed that the dsRNA was taken up and internalized near the emergence zone of the hyphae from the conidia in a punctate manner (Figure 1).
Figure 1.
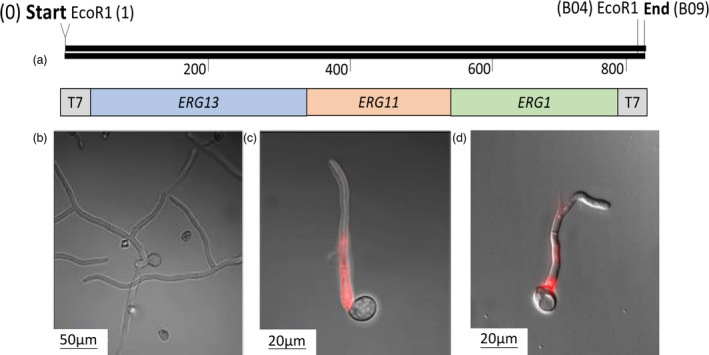
Map of the dsRNA‐ERG template and penetration of labelled dsRNA into conidia germination of B. cinerea. (a) Map of dsRNA‐ERG template made up of three essential genes in the ergosterol pathway: erg13, erg11 and erg1, flanked by T7 promoter and unique restriction enzymes sites (total 751 bases). (b–d) Conidia of the B. cinerea were germinated in the presence of Cy5‐labelled dsRNA‐Dicer (c) or Cy5‐labelled dsRNA‐ERG (d). The spores were germinated on a PDA substrate for 12 h. The germinating conidia were observed under a confocal microscope. (b) Germination of control after 12 h without dsRNA. (c and d) Conidia germination after 12 h in the presence of fluorescent dsRNA‐Dicer or dsRNA‐ERG, respectively.
B. cinerea growth from conidia was monitored over 50 h in the presence of different amounts of dsRNA‐ERG and showed a gradual decrease in fungal growth in a dose‐dependent manner in the concentration ranges of 200–800 ng (1–4 ng/μL) of dsRNA‐ERG (Figure S1). In vitro germination assay showed a significant reduction in germination rate and germination tube length compared to control when dsRNA‐ERG was added to the growth media at the beginning of the incubation (Figure 2a–c). The addition of dsRNA‐ERG had a slight effect on the percent of germination when the dsRNA was applied after 8 h of conidia incubation (Figure 2a). However, the germination tube length remained significantly lower (Figure 2b,c). To evaluate the effect on germination in vivo, B. cinerea was inoculated on grapes treated with water (control) or dsRNA‐ERG and then stained with lactophenol blue. B. cinerea treated with dsRNA‐ERG on grapes exhibited a shorter germination tube (Figure 2d) compared to B. cinerea which grew on water‐treated grapes. These results suggest that dsRNA‐ERG is more effective in inhibiting fungal germination tube elongation than the percent of germination.
Figure 2.
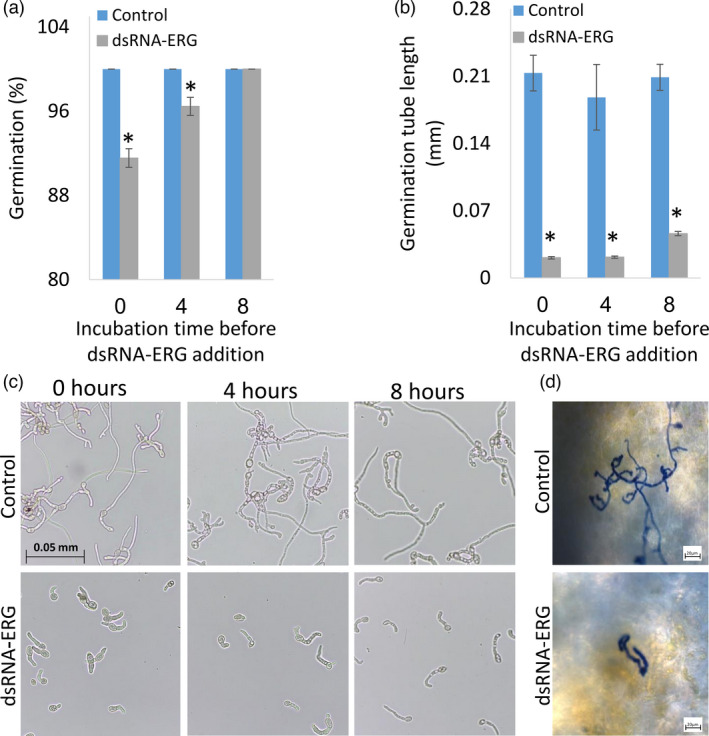
dsRNA‐ERG reduces germination and hyphal growth in vitro and in vivo. B. cinerea conidia were germinated in 0.2% SMB on glass slides. dsRNA‐ERG was added to the growth media at time 0 or after 4 or 8 h of incubation. Conidia germination was evaluated microscopically after 20 h of incubation. (a) Germination percentage. (b) Germ tube length. The presented data are mean and standard errors. Asterisks indicate statistically significant differences (P < 0.05). (c) Microscopic images (40× magnification) of B. cinerea conidia germination. (d) Representative pictures of B. cinerea conidia germination and growth on grapes treated with water (control; upper picture) or dsRNA‐ERG (lower picture).
dsRNA targeting ergosterol biosynthesis inhibits grey mould development on fruits
dsRNA‐ERG was applied externally to various crops following inoculation with B. cinerea conidia and monitored for decay development. Treated tissue exhibited a slower decay development rate compared to the control (water treated) as well as a smaller decay diameter around the inoculation site (Figure 3). Calculation of the area under the disease progress curve (AUDPC) showed a significant reduction in decay development of approximately fivefold in onion scale, eightfold in rose petals and ninefold in strawberries, compared to control (Figure 3). Moreover, dsRNA‐ERG displayed a similar reduction or better efficacy in the reduction in rot development as dsRNA targeting Dicer encoding genes (dcl1 and dcl22; dsRNA‐Dicer; Figure 3), which was shown previously to reduce B. cinerea decay development (Wang et al., 2016).
Figure 3.
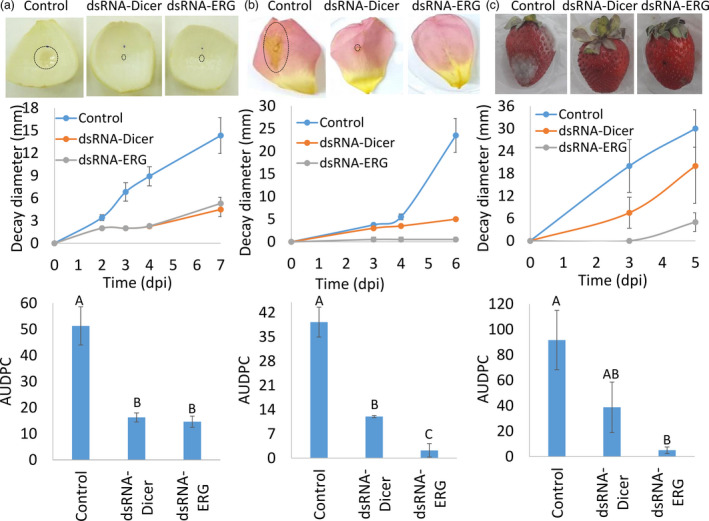
dsRNA‐ERG reduces decay development caused by Botrytis cinerea. Onion skin (a), rose petals (b) and strawberry (c) were treated with water (control), Bc‐DCL1/2‐dsRNA (dsRNA‐Dicer) or Bc‐ERG1/13/11‐dsRNA (dsRNA‐ERG) following infection by B. cinerea. Decay diameter was measured and the AUDPC was calculated for each experiment (7 dpi for onion skin, 6 dpi for rose petals and 5 dpi for strawberry). The top panel shows representative pictures were taken 3 dpi. The data presented are mean and standard errors. Values followed by different letters are statistically significant (P < 0.05).
The effect of dsRNA‐ERG on decay development was tested on various fruits as well. Inoculation of various fruits including tomato by B. cinerea without prior wounding led to very minor decay development in the control (Figure S2). This was probably due to poor penetration of B. cinerea through the waxy epidermal layer. To achieve consistent wounding, the fruit inoculation area was pre‐treated by rubbing with hydrocarborundum powder to establish micro‐injuries. This facilitated conidia inoculation and promoted uniform mould development on the fruit. The fruit was then sprayed by dsRNA‐ERG or water (control), followed by spraying of B. cinerea conidia. In all tested fruits (bell‐pepper, cherry, mango and grape) there was a decrease in decay development rate and decay area (Figure 4). AUDPC analysis showed a significant reduction in mould development of approximately fivefold in bell‐pepper, mango and grapes, while in cherry fruits the decrease in decay development was not statistically significant (Figure 4b).
Figure 4.
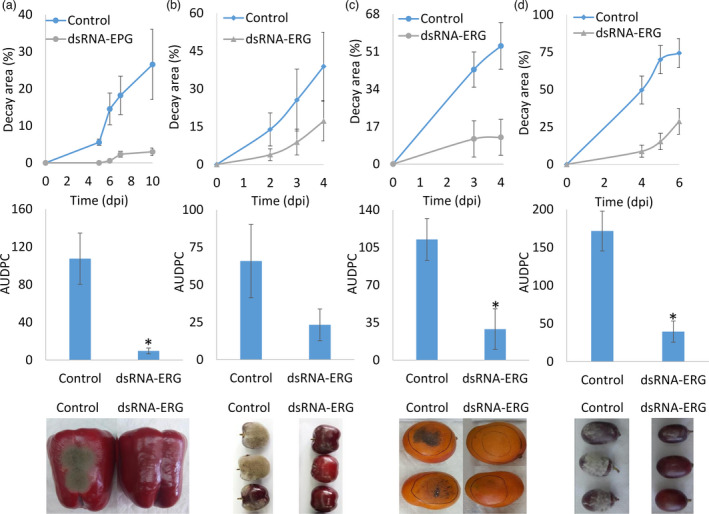
dsRNA‐ERG reduces fruit decay caused by Botrytis cinerea. Hydrocarborundum powder was used to create micro‐injuries in fruit cuticles of bell pepper (a), cherry (b), mango (c) and grape (d). The fruits were sprayed with water (control) or Bc‐ERG1/13/11‐dsRNA (dsRNA‐ERG) following spray infection by B. cinerea. The decayed area was measured and the AUDPC was calculated for each experiment (10 dpi for bell‐pepper, 4 dpi for cherry and mango and 6 dpi for grape). The lower panel shows representative pictures taken 4 days post‐inoculation (dpi). The presented data are mean and standard errors. Values followed by an asterisk indicate statistically significant differences (P < 0.05).
Systemic fruit protection by application of dsRNA‐ERG
Next, we aimed to evaluate the capability of dsRNA‐ERG to disperse to other fruit parts and provide protection. Bell‐peppers and tomatoes were used to evaluate the systemic protection of dsRNA‐ERG by applying dsRNA‐ERG at one point on the fruit and inoculate along the horizontal and vertical axes (Figure S4). The decayed area was measured every day and the AUDPC of the disease curve was calculated. The AUDPC in dsRNA‐ERG‐treated bell‐peppers and tomatoes were surprisingly lower, albeit not significantly, at all the inoculation points when inoculation was conducted on the same day of dsRNA‐ERG application (Figure 5a,b). However, inoculation with B. cinerea 1‐day post‐treatment resulted in a significant fourfold reduction in decay development in both bell peppers and tomatoes compared to control fruits (Figure 5c,d). It is worth mentioning that the decrease in decay development was similar at all inoculation points, close and far from the dsRNA‐ERG treatment point (Figure 5c,d).
Figure 5.
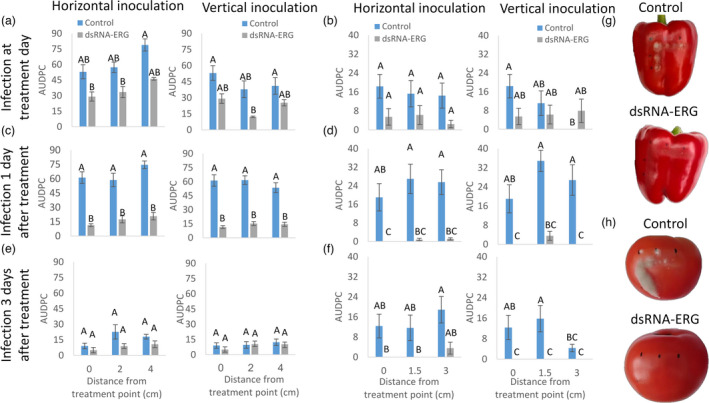
Systemic effect of dsRNA‐ERG. Bell‐peppers (a, c, e) and tomato (b, d, f) were treated with water (control) or dsRNA‐ERG at the upper‐left corner of the fruit, following inoculation with B. cinerea at the dsRNA‐ERG treatment point and two other points along the horizontal and vertical axes (Figure S4). The infection was conducted on the day of the treatment (a, b), or 1 (c, d), or 3 days post‐treatment (e, f). Decay area was measured and the AUDPC was calculated. (g) and (h) are representative pictures of bell peppers and tomatoes (respectively) which were inoculated 1 day post‐treatment. Representative pictures were taken 3 (for bell peppers) or 6 (for tomatoes) days post‐inoculation (dpi). The presented data are mean and standard errors. Values followed by different letters are statistically significantly different (P < 0.05).
The ability of dsRNA‐ERG treatment to provide lasting systemic protection was evaluated by inoculation of B. cinerea 3 days after treatment. Bell‐peppers that were infected 3 days after treatment did not exhibit any differences between the dsRNA‐ERG‐treated fruits and the control group (Figure 5e). Nonetheless, the decayed area in both groups was markedly lower compared to bell peppers that were inoculated at treatment day or 1 day after treatment (Figure 5a,c,e). This might be due to the healing of the micro‐injuries on the bell pepper's cuticle. In tomatoes that were inoculated 3 days after dsRNA‐ERG treatment, dsRNA‐ERG‐treated fruits showed notably lower decay development at all the inoculation points compared to control (Figure 5f). Furthermore, a significant reduction in decay development was observed in the vicinity of the treatment point. These results suggest that dsRNA‐ERG can be systemically transferred in the fruit and reduce decay development.
To evaluate penetration and action of the dsRNA‐ERG, micro‐injuries were generated on the grape peel using hydrocarborundum powder, then the grapes were sprayed with water (control) or with dsRNA‐ERG. Next, the fruits were spray‐inoculated with B. cinerea conidia, and peel (exocarp) or pulp (mesocarp) samples were collected after 8, 24 and 48 h for RNA analysis (Figure 6). The late time point was included as initial symptoms of decay appear 48 h post‐inoculation. Eight hours post‐treatment of dsRNA‐ERG on the fruit peel, the dsRNA‐ERG moved to the pulp and was found in the grape pulp in lesser quantities (Figure 6a). While the relative abundance of dsRNA‐ERG on the peel remained high during 48 h post‐treatment, the relative quantity in the pulp was reduced by 10‐fold after 24 h and remained almost unchanged after 48 h (Figure 6a). The effectiveness to down‐regulate erg11 transcript over time was tested in both peel and pulp, using primers from the transcript sequence region that was not included in the dsRNA construct. In the grape peel, down‐regulation of erg11 transcript has been observed at all tested time points (Figure 6b). In the grape pulp, at 8 and 24 h post‐infection, B. cinerea actin and erg11 were not detected in dsRNA‐ERG‐treated grapes, while it was detected in the inoculated control, probably due to fungal inhibition. These results show that the chosen erg11 primers are specific to the transcript and cannot detect the dsRNA‐ERG construct. At 48 h, the expression of erg11 transcript levels was significantly reduced in the pulp of dsRNA‐ERG‐treated grape compared to the control (Figure 6b).
Figure 6.
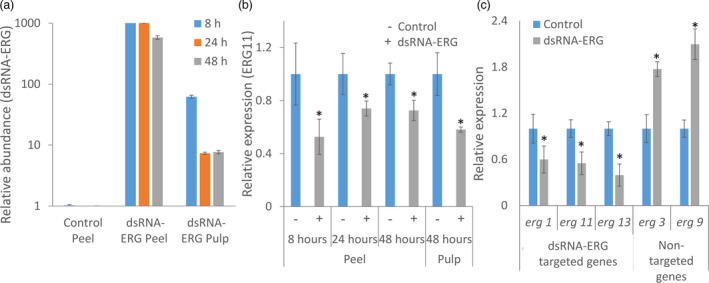
Penetration of dsRNA‐ERG to fruit pulp and down‐regulation of gene expression. Grapes were treated with water (control) or dsRNA‐ERG followed by spray inoculation with B. cinerea. Peel and pulp samples for RNA extraction were taken 8, 24 and 48 h post‐inoculation. (a) Relative abundance of the applied dsRNA‐ERG construct was evaluated by qPCR and normalized to the actin gene of grape at 8, 24 and 48 h post‐infection. (b) Relative expression of erg11 gene at 8, 24 and 48 h post‐infection, normalized to actin gene of B. cinerea in the grape peel or pulp. (c) Relative expression of the three targeted genes by the dsRNA‐ERG and two non‐targeted genes in the ergosterol biosynthesis pathway, 48 h post‐inoculation in peel tissue. The data shown are means and standard errors. Asterisks indicate statistically significant differences (P < 0.05).
The dsRNA‐ERG construct was designed to impact the post‐transcriptional expression of three transcripts in the ergosterol biosynthesis pathway. Therefore, the expression of the three targeted transcripts; erg13, erg11 and erg1 (Figure 6c) were quantified by qPCR as well as two non‐targeted transcripts erg3 and erg9. The relative expression of the mRNA transcripts was measured by qPCR and normalized to the expression of the fungal actin. The relative expression of the three targeted transcripts was reduced on average by more than 50% in the dsRNA‐ERG‐treated fruit, 48 h post‐inoculation, compared to control (Figure 6c). However, the two non‐targeted transcripts were significantly up‐regulated, 48 h post‐inoculation, at the treated grapes compared to control indicating an attempt of compensation (Figure 6c).
dsRNA‐ERG synergism with ergosterol‐inhibitors and complementation by ergosterol
In an attempt to complement the effect of dsRNA‐ERG on the growth of B. cinerea, external ergosterol was added to the dsRNA‐ERG treatments. Fungal growth kinetics in vitro was determined using optical density (O.D) measurements correlating to fungal biomass. The fungal growth curves showed a major reduction when dsRNA‐ERG was added to the media, compared to control (Figure 7a). However, when B. cinerea conidia were grown in media containing external ergosterol in addition to the dsRNA‐ERG, the fungal growth curve displayed a similar growth pattern as the untreated B. cinerea conidia, reaching the same final O.D levels (Figure 7a). Importantly, the addition of ergosterol to control did not alter fungal growth (Figure S3). The effect of the addition of external ergosterol on fruit decay development in vivo was evaluated on infected grapes (Figure 7b,c). Micro‐injuries were generated on the grape peel using hydrocarborundum powder, then the grapes were spray treated with water (control), dsRNA‐ERG or dsRNA‐ERG combined with external ergosterol. The fruits were inoculated by spraying with B. cinerea conidia and assessed daily for decay severity. dsRNA‐ERG treatment reduced B. cinerea mould development rate and decay severity by more than 50% during 6 days post‐inoculation (dpi), compared to control (Figure 7b,c). Conversely, the addition of external ergosterol complemented the decay development and restored decay progression to a similar level to control (Figure 7b,c). The results of reduced mRNA accumulation and complementation by exogenous ergosterol suggest that the inhibition in rot development is a result of the lack of ergosterol synthesis.
Figure 7.
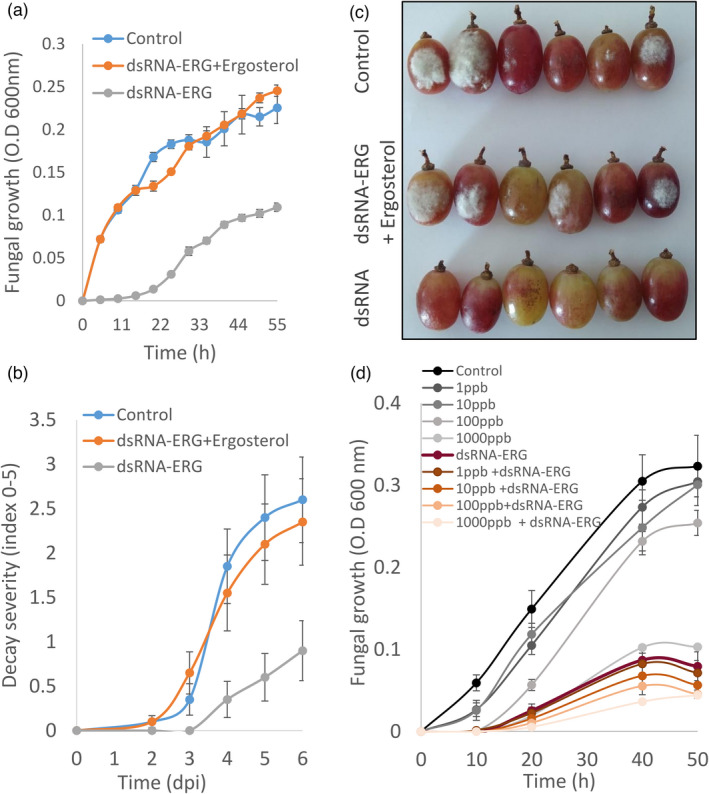
dsRNA application reduces fungicide needed concentration for growth inhibition of B. cinerea. (a) B. cinerea conidia were grown at room temperature in 1% SMB, or 1% SMB supplemented with dsRNA‐ERG, or dsRNA‐ERG and ergosterol as described in the Materials and methods. O.D measurements at 600 nm were taken every hour. (b) Grapes were treated with water (control), or dsRNA‐ERG or dsRNA‐ERG and ergosterol, following spray inoculation with B. cinerea. Decay severity was recorded every day. (c) Representative picture of grapes treated with water (control), or dsRNA‐ERG or dsRNA‐ERG and ergosterol, following spray inoculation with B. cinerea were taken 4 days post‐infection (dpi). (d) B. cinerea conidia were incubated at room temperature in 1% SMB, or 1% SMB supplemented with various concentrations of Prochloraz, with or without dsRNA‐ERG. O.D measurements at 600 nm were taken every hour. The presented data are mean and standard errors.
It was of interest to explore the use of SIGS in combination with a commercially used fungicide. The fungicide, prochloraz is an ergosterol‐inhibitor of the enzyme lanosterol 14α‐demethylase (CYP51A1; encoded by erg11), which is necessary for the production of ergosterol. However, the fungicide also has multiple targets in animal systems that indicate a need for restricting its use (Vinggaard et al., 2006). Increasing concentrations of ‘Prochloraz’ in the growth media (1–1000 ppb) resulted in a dose‐dependent reduction of fungal growth as measured by O.D (Figure 7d). When 4 ng/μL dsRNA‐ERG were added, the inhibition of fungal growth was augmented (Figure 7d; Table S1). The combination of dsRNA‐ERG with multiple concentrations of ‘Prochloraz’ showed a synergistic decline in fungal growth compared to each fungicide treatment or dsRNA‐ERG treatment alone (Figure 7d; Table S1). Another fungicide, Fludioxonil has recently been shown to act on triosephosphate isomerase activity to induce methylglyoxal stress that alters sensing in a histidine kinase inducing fungal death (Brandhorst and Klein, 2019). ‘Fludioxonil’ reduced B. cinerea growth in a non‐linear manner, a greater and non‐synergistic reduction in fungal growth was achieved by the addition of 4 ng/μL dsRNA‐ERG (Figure S5, Table S1). These results indicate that the application of dsRNA‐ERG could significantly reduce the amount of fungicide needed to inhibit fungal growth.
Discussion
Non‐specific fungicidal toxic side effects have an impact on the environment and human health. Public concerns have motivated the need for the reduction or replacement of fungicides with eco‐friendly alternatives especially in harvested fresh produce (Rani et al., 2020; Wisniewski et al., 2016). The RNAi method of fungal control represents a promising alternative due to its specificity (Koch et al., 2016; Nerva et al., 2020; Wang et al., 2016). The current research was aimed to; test the efficacy of a dsRNA construct targeting three essential genes in the ergosterol biosynthesis pathway on harvested fresh produce, to examine the systemic response and synergism with fungicides.
Uptake of dsRNA and small RNA molecules was shown to occur through the hyphae of fungi (Koch et al., 2016; Morozov et al., 2019; Wang and Dean, 2020a). While it was suggested that the uptake of dsRNA is likely by clathrin‐mediated endocytosis (Wytinck et al., 2020), different fungi might possess different dsRNA uptake mechanisms and more studies will need to be conducted to better understand dsRNA uptake (Qiao et al., 2021). In our work, the application of dsRNA to ungerminated conidia arrested the elongation of the germination tube; however, the initial germination process was less inhibited (Figure 2). This difference could result from the preferential uptake of dsRNA. As shown in Figure 1, dsRNA appears to accumulate near the site of germ‐tube emergence. This is also consistent with a previous study showing the uptake of dsRNA in B. cinerea when applied at the post‐germination stage (Wang et al., 2016). The punctate appearance in the accumulation of fluorescently labelled dsRNA may imply that the dsRNA is less permeable to vacuolar regions as has been noted in other fungal pathogens (Qiao et al., 2021). Rapid uptake by germinating conidia might be due to changes in the cell wall permeability and disruption of the protective layer of the conidia (Beauvais and Latgé, 2018). Thus, initial uptake of the dsRNA in the emergence zone or by hypha may be limited by changes in the cell wall viscoelastic properties, membrane endocytic capabilities or cell membrane transporters (Qiao et al., 2021; Walker et al., 2018; Wang and Dean, 2020b). Nonetheless as shown in Figure 2, the time sequence needed for dsRNA manifest inhibition; or for endogenous ergosterol to deplete to levels that limit growth, occurs only later during hyphal elongation.
Application of dsRNA‐ERG significantly decreased decay development in various crops such as onion scale, rose petals, strawberries, mango, cherry, bell pepper and grape (Figures 3 and 4). The reduction in rot development caused by B. cinerea by application of dsRNA‐ERG was similar or superior to the reduction that was observed by dsRNA‐Dicer designed to target different virulence genes of B. cinerea (Nerva et al., 2020; Wang et al., 2016). The infection method adopted to obtain coherent infection of fruits necessitated pre‐wounding the cuticle with mild hydrocarborundum treatment. Abrasion by hydrocarborundum powder that emulates natural environmental wounding allowed for consistent inoculation by B. cinerea without releasing cell sap. Thus, intact fruit peel serves as an effective barrier against B. cinerea infection (Figure S2; Coertze et al., 2001). As shown here, the abrasion‐assisted infection by B. cinerea was mitigated by dsRNA treatments (Figures 3 and 4).
The dsRNA‐ERG was comprised of short sequences from the coding regions of three transcripts in the ergosterol biosynthesis pathway: erg11, erg1 and erg13. In the cell, it is expected to be processed into short inhibitory RNA fragments by the Dicer‐like enzyme where inhibition is hypothesized to be achieved through down‐regulation of the targeted RNA or by inhibition of mRNA translation (Morozov et al., 2019; Voinnet, 2008). Transcript expression analysis showed significant down‐regulation in all three transcripts (Figure 6). A result that favours the former hypothesis. This result is also consistent with observations made in Fusarium graminearum inhibited by dsRNA application (Koch et al., 2016). Interestingly, transcripts of other genes in the ergosterol biosynthesis pathway, which are not targeted by dsRNA‐ERG (such as erg3 and erg9; Figure 6c), were significantly enhanced. Our results are consistent with a previous study that demonstrated the relation between a deletion mutation or experimental interference with erg11 and subsequent up‐regulation in erg3 transcript expression (Geber et al., 1995). In addition, erg9 encodes squalene synthase, the first enzyme responsible for a specific step of ergosterol biosynthesis (Malmierca et al., 2013). It is, therefore, possible that erg9 up‐regulation like erg3 may be due to activation of a futile compensatory response in this highly regulated biosynthetic pathway.
When the application of dsRNA‐ERG and inoculation by B. cinerea were collocated, it may be assumed that dsRNA was taken up and processed to siRNA by the fungus. Here we show that the application of dsRNA‐ERG to a singular point on bell‐peppers or tomatoes was able to reduce decay development at other more distant points. A degree of systemic protection was already observed even when B. cinerea inoculation was conducted on the treatment day, suggesting rapid movement of either dsRNA or likely processed RNAi to other parts of the fruit. In bell‐pepper and tomato, systemic protection was detected at least 2 and 3 days post‐treatment respectively. When the level of the non‐processed dsRNA‐ERG was monitored in the grape berry it was detected in distal regions of the mesocarp pulp at 1/10 to 1/100 of the amounts found at the point of inoculation. Presumably, vascular bundles located below the epidermis participate in such dsRNA transport. Interestingly, distal levels of the whole dsRNA‐ERG were far below the inhibitory concentrations that were found in the application zone; yet, erg11 transcript expression was significantly down‐regulated in the fruit pericarp (Figure 6). An attractive possibility to explain this discrepancy is that alongside the movement of dsRNA‐ERG, cell‐to‐cell movement is facilitated by dicer‐processed dsRNA in the plant to siRNA (Melnyk et al., 2011). The resultant potent siRNA, which also undergoes transport, is taken up by the fungus as has been shown to occur in other plant–fungal interactions (Hua et al., 2018; Llave, 2010; Weiberg et al., 2013). Thus, resistance to infection may be achieved by siRNA sourced from the fungus in co‐localized infection and sourced from the plant in distal resistance.
External ergosterol was shown here to abrogate the effect of dsRNA‐ERG treatment; restoring the severity level of grey mould to the decay severity in water‐treated control (Figure 7). Similarly, when RNAi was used to inhibit the ergosterol pathway in Trypanosoma brucei, growth was restored by exogenous ergosterol. Relatedly, exogenous co‐application of ergosterol with tebuconazole (inhibitor of ergosterol biosynthesis) alleviated the fungicidal effects in B. cinerea (Dauchy et al., 2016; Zhang et al., 2010). Taken together, these results show that dsRNA‐ERG induced rapid gene‐specific RNA turnover, which impacted on ergosterol biosynthesis needed for fungal growth and colonization.
A further aim of the study was to compare the dsRNA‐ERG treatment to commercially used fungicides and test for possible synergism. Two fungicides were chosen due to their different modes of action. 'Prochloraz', an inhibitor of ergosterol biosynthesis, specifically inhibits the cytochrome P450 sterol 14α‐demethylase, a critical enzyme in ergosterol biosynthesis, encoded by erg11, that is one of the targeted genes in the dsRNA‐ERG construct (Zhang et al., 2020). However, as an imidazole‐based compound, it is of limited specificity and has numerous off‐targets. ‘Fludioxonil’, impacts indirectly on a class lll hybrid histidine kinase (Brandhorst and Klein, 2019; Diskin et al., 2019). The growth inhibition of B. cinerea by ‘Prochloraz’ was found to be dose‐dependent (Figure 7d), while Fludioxonil was toxic to the fungi in a non‐linear manner (Figure S5). In both cases, a combination of dsRNA‐ERG and fungicide displayed a greater reduction in fungal growth compared to the fungicide alone. The required concentration of 'Prochloraz' to reach B. cinerea growth inhibition decreased by more than 100‐fold in the combined treatment with dsRNA‐ERG and exhibited a synergistic effect (Table S1). Fludioxonil and dsRNA‐ERG inhibited B. cinerea growth better than the fungicide alone, yet the combined inhibition was not synergistic (Table S1). Synergism between 'Prochloraz' and dsRNA‐ERG can result from the reduced flux of sterol biosynthesis due to RNAi that is further exacerbated by the inhibition of the residual activity. The combination could rapidly lower threshold levels of ergosterol to critical levels. In all, dsRNA was found to significantly reduce the needed fungicide concentration for controlling postharvest decay development. The synergistic effect of the combined treatment indicates that both treatments can in the future be applied together as they have a similar mode of action. The chemical fungicide is a competitive inhibitor acting quickly to inhibit cytochrome P‐450 enzymes (Henry and Sisler, 1984). In contrast, the maturation of dsRNA first reduces transcripts levels and ultimately reduces enzyme levels. Based on those differences, the impact of such fungicides may be enhanced if they were applied following dsRNA applications. In contrast, prior application of fungicide may enhance the levels of ergosterol transcripts and interfere with dsRNA treatment. Hence, the development of application procedures to further optimize synergy should be sought.
The main advantages of SIGS are their eco‐friendly nature and high specificity making them potential alternatives to conventional fungicides. Recent progress showed the pre‐and postharvest application of dsRNA to protect grapes from the pathogenic fungi B. cinerea under conditions that mimic the vineyard (Nerva et al., 2020). The work here shows the efficacy in targeting multiple components of the same biosynthetic pathway that results in systemic protection and potential synergy with fungicides. SIGS can also obviate the need to focus only on pathogen‐specific metabolism in an effort to produce fungicidal agents that have specificity (e.g. ergosterol pathway). Instead, many additional critical hubs of metabolism or development can be targeted in the pathogen where specificity is achieved by the evolutionary diversification in gene sequence between species. Thus, dsRNA construct that comprises several genes could be more effective, as it targets several essential genes. Due to novel gene junctions, the hybrid siRNAs could potentially create off‐target effects. To evaluate possible off‐targets in the designed dsRNA‐ERG, the whole dsRNA sequence was analysed against the genome data of three fungi (B. cinerea (our model), Penicillium expansum and Alternaria alternata), grape and human. While the software identified the target genes in B. cinerea genome, no hits were found to the other tested organisms. Nonetheless, several problems need to be solved before commercial applications will be affordable, such as cost‐efficient dsRNAs production, modified application tools and stabilizing agents for field conditions that will maintain the potency of dsRNA (Nerva et al., 2020).
To summarize, the results of the current study suggest that a combination of several main genes of an essential pathway, as ergosterol biosynthesis, could be united into one construct and serve as an efficient SIGS treatment that inhibits the expression of all the targeted transcripts. We showed that the dsRNA penetrates to the emergence zone of the hyphae and then spreads throughout the fungi. In this manner, dsRNA‐ERG could inhibit fungal germ tube elongation and hyphal growth and significantly inhibit the B. cinerea pathogenicity and colonization in various crops and harvested fruits. We also show that dsRNA moves systemically in the fruit peel and pulp to inhibit decay development. Additionally, dsRNA‐ERG treatments lowered by 100‐fold the necessary effective concentration of commonly used fungicides that have an ergosterol‐related mode of action.
Experimental procedures
Construction and production of dsRNA
To silence the ergosterol pathway, dsRNA construct was designed using SnapGene (SnapGene® software, GSL Biotech) for three transcripts in the ergosterol biosynthesis pathway, erg11, erg1 and erg13 yielding 791 bp. Each sequence was chosen from an expressed region of 250–300 bp (Supplemental Experimental procedures). The full sequence including flanking EcoR1 restriction sites and T7 transcriptional promoters was synthesized by GeneScript Inc as shown in Supplemental M&M. The DCL1/DCL2 dicer using the sequence for DCL1 and DCL2 was cloned in a similar manner as shown in the Supplemental M&M. For RNA synthesis, a high yield transcription reaction was carried out based on Ambion MEGAscript protocol (Thermo Fisher Scientific). The plasmid templates were linearized by EcoR1 restriction where complete linearization was monitored by gel fractionation and purified by Wizard SV clean‐up system (Promega). After RNA synthesis to produce two self‐hybridized complementary RNA transcripts, DNA and single‐stranded RNA were removed by nuclease digestion. The remaining dsRNA was purified on a solid‐phase adsorption system to remove proteins and oligonucleotides. The integrity and efficiency of duplex formation of dsRNA were examined by agarose gel and spectrophotometry. Pure dsRNA was stored at −20 °C until usage. The dsRNA construct was tested for off‐target sequences using siFi21 (Licensed by CC‐BY‐SA‐4.0) as described previously by Lück et al. (2019). Briefly, siFi software was used to analyse the whole dsRNA‐ERG sequence against all the genome sequences of B. cinerea (RefSeq: GCF_000143535.2), Alternaria alternata (RefSeq: GCF_001642055.1), Penicillium expansum (RefSeq: GCF_000769745.1), Vitis venifera (RefSeq: GCF_000003745.3) and human (RefSeq: GCF_000001405.39; all data were downloaded from NCBI database) using the default parameters to achieve higher sensitivity for off‐targets. Additional off‐target searches used the ERG construct from B. cinerea as a DNA gene query for BLAST technique. Some fungal pathogens closely related to Botrytis were detected at 90% identity and above. These include ERG13, Monilinia and Botryotinia sp.; ERG11, Botryotinia and Sclerotinia sp.; and ERG1, Botryotinia sp. No significant homologies were detected to plant or animal DNA.
Fungal strains and growth media
Cultures of B. cinerea B05‐10 were routinely grown on potato dextrose agar (PDA; Difco, New Jersey) medium for 14 days at 22 °C. Conidia were gently collected by suspension in sterile distilled water, filtered through four layers of sterile cheesecloth and diluted to a concentration of 105 conidia mL−1 for in vitro experiments or 104 conidia mL−1 for in vivo experiments. Conidial concentration in the suspension was microscopically determined using a haemocytometer.
dsRNA uptake by B. cinerea
To evaluate dsRNA uptake by B. cinerea cells, fluorescent dsRNA was prepared by incorporation of cy5‐labelled nucleotides. Aminoallyl‐UTP‐Cy5 (Jena Bioscience, cat. NU‐821‐X‐CY5) 4 μL of a 5 mm solution was added directly to the Ambion MEGA‐script transcription assay to yield a final volume of 20 μL. The incorporation of UTP‐Cy5 was monitored by the NanoDrop One (Thermo Fisher Scientific) spectrophotometer. B. cinerea conidia suspension (10 μL of 105 conidia mL−1) was seeded on a PDA glass slide with water (control) or 800 ng cy5‐labelled dsRNA, and incubated in the dark in a humid petri dish for 12–16 h at room temperature. The conidia were examined under a confocal laser scanning microscope (CLSM; Olympus IX81, Center Valley, PA).
Effect of dsRNA on decay development
All fruit and crops were purchased from a local grower. For non‐wounding inoculation assays, yellow onion (Allium cepa) scale, a cultivated pink rose (Rosa) petals and strawberries (Fragaria ananassa) cv. Gilli was treated with water (control) or dsRNA‐ERG (15 ng/μL) following inoculation by a 7 μL drop of B. cinerea at a concentration of 104 conidia mL−1. The scales, petals and fruit were incubated at 25 °C for 5–7 days. Decay diameter was measured daily. To measure the irregular shape of the decay, for each rot, the decay diameter was determined as the average of three diameters in different directions. Each experiment was conducted using at least 10 repeats for treatment, the experiment was repeated twice.
For wound inoculation assays red bell‐peppers (Capsicum annuum), cherry cv. Sweet‐Hart (Prunus cerasus), mango cv. Shelly (Mangiferae indica) or grapes cv. Autumn Royal and Scarlotta (Vitis vinifera) were pre‐treated with hydrocarborundum powder (Fisher Scientific, Loughborough, UK) by gentle abrasing the fruit until the fruit peel lost its smooth texture without releasing cell sap, to create micro‐injuries on the fruit cuticle which mimic natural wounding. Without such uniform infection, one cannot get reliable quantitative measurements of protection. Spray treatment of water (control) or dsRNA (15 ng/μL) was applied up to drainage following spray inoculation of B. cinerea conidia suspension at a concentration of 104 conidia mL−1. Fruits were stored at 25 °C for 6 days in humid chambers and either decay area or severity (index 0‐5; 0‐no decay, 1‐mild decay, 5‐severe decay) were measured.
Evaluation of systemic protection
Red bell peppers (Capsicum annuum) and tomatoes (Solanum lycopersicum) were pre‐treated with hydrocarborundum powder to create micro‐injuries. 10 μL drop of water (control) or dsRNA‐ERG (15 ng/μL) was placed on the top‐left point of the fruit (T; Figure S5). Inoculation was conducted at three time points: treatment day, 1 day after treatment, or 3 days after treatment using a 7 μL drop of B. cinerea at a concentration of 104 conidia mL−1. B. cinerea conidia were seeded at five inoculation points: the treatment point (T) and four additional points in a distance of 2 and 4 cm for bell‐peppers or 1.5 and 3 cm for tomatoes, positioned vertically (D1, D2) and horizontally (S1, S2; Figure S5). Fruits were stored at 25 °C for 7 days post‐inoculation, in humid chambers and the decayed area was measured.
Effect of dsRNA on conidia germination
The effect of dsRNA‐ERG on B. cinerea conidia germination was tested using an in vitro micro‐assay (Galsurker et al., 2020; Gatto et al., 2011). Conidia suspension (10 μL) was seeded on a glass slide, containing 0.2% Sabouraud maltose broth (SMB; 10 g Peptone (Difco), 40 g Maltose (Caisson labs, Smithfield, Utah, United States) per 1 L) and incubated in a humid petri dish for 24 h at room temperature. dsRNA‐ERG (40 μL dsRNA at a concentration of 20 ng/μL) was added at time zero, after 4 h or 8 h of incubation. To test the effect of dsRNA‐ERG on B. cinerea conidia germination in vivo, B. cinerea was inoculated on Vitis vinifera cv. Superior with water (control) or dsRNA‐ERG as described above. The inoculated fruits were incubated in a humid chamber at 25 °C for 24 h, then a thin layer of the inoculated grape peel was subjected to lactophenol blue staining. The conidia were examined under a microscope (Leica DM500 equipped with a Leica ICC50 HD camera) to assess both germination percentage and germ tube elongation. Conidia germination was scored when the germ tube's length was equal to, or exceeded, the conidia diameter. The germ tube length was evaluated using ImageJ software (http://rsb.info.nih.gov/ij) in three microscopic fields in each droplet, six droplets for each treatment. In each field examined, at least 80 conidia were counted.
RNA extraction and transcript expression (quantitative PCR; qPCR)
Vitis vinifera cv. Superior or Scarlota inoculated with B. cinerea and treated with water (control) or dsRNA‐ERG as described above. The fruit peels or pulps were collected 8, 24 and 48 h post‐inoculation to liquid nitrogen. For RNA extraction, the peels or pulps were grounded to a fine powder using mortar and pestle and total RNA was extracted using Spectrum™ plant total RNA kit (Sigma‐Aldrich, St. Louis, Missouri) according to the manufacturer's instructions, following DNase treatment (TURBO DNA‐free Kit, Ambion Life Technologies). Total RNA (1 μg) was used for cDNA construction using the RevertAid First‐Strand cDNA Synthesis kit (Thermo Scientific) according to the manufacturer's instructions. cDNA samples were diluted at 1 : 10 and used for qRT‐PCR. The relative expression of the three ergosterol targeted genes (erg11, erg1 and erg13) and two ergosterol non‐targeted genes (erg3 and erg9) was evaluated by a qRT‐PCR analysis conducted with a Step One Plus Real‐Time PCR (Applied Biosystems, Waltham, Massachusetts, US). PCR amplification was performed with 3 μL of diluted cDNA template in a 10 μL reaction mixture containing 5 μL Syber Green (Applied Biosystems) and 250 nm primers. The qRT‐PCR analysis was conducted with the corresponding primer sets of the selected genes. All the primers were chosen from the transcript sequence region that was not part of the dsRNA‐ERG construct to avoid cross‐contamination. The primers that were used in the current study are as follows: forward, 5′‐GCTGATCTCCCTGCTCTCAAGTA‐3′ and reverse, 5′‐TGTGGAGGCGTAGAGTTTCCTT‐3′ for erg11. Forward, 5′‐GAGGAAAGCGTGTGCAAGCT‐3′ and reverse, 5′‐TTCGCCCGTAGATGGATTGG‐3′ for erg1. Forward, 5′‐AAGAAGCGTTTCAACGAGCG‐3′ and reverse, 5′‐AGGGCGGCACTATCAACATT‐3′ for erg13, forward, 5′‐TGGAAGTGACCGACAAATAC‐3′ and reverse, 5′‐GAGGGTTGTGCCATTGAT‐3′ for erg3, forward, 5′‐TCACCCATTTCACCCAGTTT‐3′ and reverse, 5′‐GTTGATCCGAGTCCGTCTATTG‐3′ for erg9. And to evaluate the dsRNA‐ERG construct we used forward 5′‐TACCTCCTTTGCCTCTCTACC‐3′ located on erg13 sequence and reverse 5′‐GTATTGCTTGCGGAATTTGGAG‐3′ located on erg 11 sequence. PCR cycling program included: 10 min at 94 °C, followed by 40 cycles of 94 °C for 10 s, 60 °C for 15 s and 72 °C for 20 s. The expression of the selected genes was normalized using Ct values of B. cinerea actin (forward, 5′‐TGCTCCAGAAGCTTTGTTCCAA‐3′ and reverse, 5′‐TCGGAGATACCTGGGTACATAG‐3′) or grape actin (forward, 5′‐CTTGCATCCCTCAGCACCTT‐3′ and reverse, 5′‐TCCTGTGGACAATGGATGGA‐3′) as reference gene, and expression values were calculated relatively to control sample (water treated) using Step One software v2.2.2 (Applied Biosystems). Each treatment consisted of three biological repeats and three technical replicates.
In vitro fungal growth
To evaluate fungal growth, 6 μL conidia suspension was seeded in a 96‐well plate (Bar‐Naor Ltd., Petah Tikva, Israel), containing 200 μL of 1% SMB. For dsRNA‐ERG treatment, 4 ng/μL of dsRNA‐ERG (total 800 ng) was added to each treatment well. Ergosterol complementation assay wells contained 50 μm ergosterol (Sigma‐Aldrich), which was initially dissolved in chloroform to obtain a 125 mm stock solution (Dauchy et al., 2016). To test the effect of commonly used fungicide on B. cinerea growth, 'Procloraz' (Sportak, Bayer Cropscience LLC, Monheim am Rhein, Germany) or 'fludioxonil' (scholar, Hod‐Hasharon, Israel) fungicide were diluted in 1% SMB to final concentrations of 1–1000 ppb or 0.01–1000 ppb respectively.
The plates were incubated at room temperature and O.D measurement at 600 nm was taken every hour during 48–72 h using Synergy LX plate reader (BioTek, Winooski, Vermont). The absorbance of three wells of repeats was averaged together and was background‐corrected by subtracting the average absorbance of media alone at time zero (Galsurker et al., 2020; Langvad, 1999). Growth inhibition percentage was calculated (O.D in control well − O.D in treatment well)/O.D in control well × 100) (Meletiadis et al., 2003). Coefficient treatment interaction (CTI) was calculated as follows: CTI = AB/(A × B). AB is the ratio of the treatment combination to control; A, or B is the ratio of the single treatment to control. CTI is considered as synergistic when CTI < 1 (Chen et al., 2014).
Statistical analysis
The data presented are averages and standard errors. A T‐test was performed to compare two treatments. One‐way ANOVA analysis was performed by Tukey–Kramer HSD test using JMP Pro 15 software (SAS Institute, Cary, NC). Different letters or asterisks indicate significant differences (P ≤ 0.05).
Conflict of interest
We declare that we do not know of any conflict of interest.
Author contributions
DDA and OG coordinated, planned, carried out the experiments and data analysis, and wrote the manuscript draft. OD, OF and DM carried out experiments and data analysis. MS and EP wrote the proposal, planned some of the experiments and revised the manuscript. RF and NA supervised the study, the experiments and the data analysis and prepared the manuscript. All authors have read and agreed to the final version of the manuscript.
Supporting information
Method S1 Sequences of Bc‐DCL1/2‐dsRNA (dsRNA‐Dicer), or Bc‐ERG1/13/11‐dsRNA (dsRNA‐ERG) constructs.
Table S1 Coefficient treatment interaction and growth inhibition by dsRNA and fungicides
Figure S1 Dose effect of dsRNA‐ERG on fungal growth.
Figure S2 Infection of tomato fruit with no wounding.
Figure S3 In vitro fungal growth with external ergosterol addition.
Figure S4 Illustration of systemic evaluation experiment set‐up.
Figure S5 dsRNA application reduces fungicide‐needed concentration for growth inhibition of B. cinerea.
Acknowledgements
This research was supported by the Chief Scientist of the Israeli Ministry of Agriculture and Rural Development (Grant No. 20‐06‐0071). Danielle Duanis‐Assaf is a recipient of a scholarship in the memory of Yehoram and Naomi Gorodiski given by the Gorodiski Foundation.
Duanis‐Assaf, D. , Galsurker, O. , Davydov, O. , Maurer, D. , Feygenberg, O. , Sagi, M. , Poverenov, E. , Fluhr, R. and Alkan, N. (2022) Double‐stranded RNA targeting fungal ergosterol biosynthesis pathway controls Botrytis cinerea and postharvest grey mould. Plant Biotechnol. J., 10.1111/pbi.13708
References
- Abiad, M.G. and Meho, L.I. (2018) Food loss and food waste research in the Arab world: a systematic review. Food Secur. 10, 311–322. [Google Scholar]
- Ahmad, A. , Khan, A. , Manzoor, N. and Khan, L.A. (2010) Evolution of ergosterol biosynthesis inhibitors as fungicidal against Candida. Microb. Pathog. 48, 35–41. [DOI] [PubMed] [Google Scholar]
- Bard, M. , Lees, N. , Burrows, L. and Kleinhans, F. (1978) Differences in crystal violet uptake and cation‐induced death among yeast sterol mutants. J. Bacteriol. 135, 1146–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauvais, A. and Latgé, J.‐P. (2018) Fungal cell wall. Basel, Switzerland: Multidisciplinary Digital Publishing Institute. [Google Scholar]
- Bhattacharya, S. , Esquivel, B.D. and White, T.C. (2018) Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae . MBio, 9, e01291‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandhorst, T.T. and Klein, B.S. (2019) Uncertainty surrounding the mechanism and safety of the post‐harvest fungicide fludioxonil. Food Chem. Toxicol. 123, 561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Ye, H.‐L. , Zhang, G. , Yao, W.‐M. , Chen, X.‐Z. , Zhang, F.‐C. and Liang, G. (2014) Autophagy inhibition contributes to the synergistic interaction between EGCG and doxorubicin to kill the hepatoma Hep3B cells. PLoS One, 9, e85771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coertze, S. , Holz, G. and Sadie, A. (2001) Germination and establishment of infection on grape berries by single airborne conidia of Botrytis cinerea . Plant Dis. 85, 668–677. [DOI] [PubMed] [Google Scholar]
- Dauchy, F.‐A. , Bonhivers, M. , Landrein, N. , Dacheux, D. , Courtois, P. , Lauruol, F. , Daulouede, S. et al. (2016) Trypanosoma brucei CYP51: essentiality and targeting therapy in an experimental model. PLoS Negl. Trop Dis. 10, e0005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin, S. , Sharir, T. , Feygenberg, O. , Maurer, D. and Alkan, N. (2019) Fludioxonil—a potential alternative for postharvest disease control in mango fruit. Crop Prot. 124, 104855. [Google Scholar]
- Elmer, P. and Reglinski, T. (2006) Biosuppression of Botrytis cinerea in grapes. Plant. Pathol. 55, 155–177. [Google Scholar]
- Galsurker, O. , Diskin, S. , Duanis‐Assaf, D. , Doron‐Faigenboim, A. , Maurer, D. , Feygenberg, O. and Alkan, N. (2020) Harvesting mango fruit with a short stem‐end altered endophytic microbiome and reduce stem‐end rot. Microorganisms, 8, 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto, M.A. , Ippolito, A. , Linsalata, V. , Cascarano, N.A. , Nigro, F. , Vanadia, S. and Di Venere, D. (2011) Activity of extracts from wild edible herbs against postharvest fungal diseases of fruit and vegetables. Postharvest Biol. Technol. 61, 72–82. [Google Scholar]
- Geber, A. , Hitchcock, C.A. , Swartz, J.E. , Pullen, F.S. , Marsden, K.E. , Kwon‐Chung, K.J. and Bennett, J.E. (1995) Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob. Agents Chemother. 39, 2708–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, M.J. and Sisler, H.D. (1984) Effects of sterol biosynthesis‐inhibiting (Sbi) fungicides on cytochrome‐P‐450 oxygenations in fungi. Pestic Biochem. Phys. 22, 262–275. [Google Scholar]
- Hua, C. , Zhao, J.‐H. and Guo, H.‐S. (2018) Trans‐kingdom RNA silencing in plant–fungal pathogen interactions. Mol. Plant, 11, 235–244. [DOI] [PubMed] [Google Scholar]
- Koch, A. , Biedenkopf, D. , Furch, A. , Weber, L. , Rossbach, O. , Abdellatef, E. , Linicus, L. et al. (2016) An RNAi‐based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 12, e1005901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, A. , Kumar, N. , Weber, L. , Keller, H. , Imani, J. and Kogel, K.‐H. (2013) Host‐induced gene silencing of cytochrome P450 lanosterol C14α‐demethylase–encoding genes confers strong resistance to Fusarium species. Proc. Natl Acad. Sci. 110, 19324–19329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, V. , Hatan, E. , Bar, E. , Davidovich‐Rikanati, R. , Doron‐Faigenboim, A. , Spitzer‐Rimon, B. , Elad, Y. et al. (2020) Phenylalanine increases chrysanthemum flower immunity against Botrytis cinerea attack. Plant J. 104, 226–240. [DOI] [PubMed] [Google Scholar]
- Langvad, F. (1999) A rapid and efficient method for growth measurement of filamentous fungi. J. Microbiol. Methods, 37, 97–100. [DOI] [PubMed] [Google Scholar]
- Lees, N.D. , Bard, M. , Kemple, M.D. , Haak, R.A. and Kleinhans, F.W. (1979) ESR determination of membrane order parameter in yeast sterol mutants. Biochim. Biophys. Acta‐Biomembranes, 553, 469–475. [DOI] [PubMed] [Google Scholar]
- Lees, N.D. , Lofton, S.L. , Woods, R.A. and Bard, M. (1980) The effects of varied energy source and detergent on the growth of sterol mutants of Saccharomyces cerevisiae . Microbiology, 118, 209–214. [Google Scholar]
- Lees, N. , Skaggs, B. , Kirsch, D. and Bard, M. (1995) Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae—a review. Lipids, 30, 221–226. [DOI] [PubMed] [Google Scholar]
- Lipinski, B. , Hanson, C. , Lomax, J. , Kitinoja, L. , Waite, R. and Searchinger, T. (2013) Reducing food loss and waste. World Resources Institute Working Paper, vol. 1, pp. 1–40. [Google Scholar]
- Llave, C. (2010) Virus‐derived small interfering RNAs at the core of plant–virus interactions. Trends Plant Sci. 15, 701–707. [DOI] [PubMed] [Google Scholar]
- Lück, S. , Kreszies, T. , Strickert, M. , Schweizer, P. , Kuhlmann, M. and Douchkov, D. (2019) siRNA‐finder (si‐Fi) software for RNAi‐target design and off‐target prediction. Front. Plant Sci. 10, 1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmierca, M.G. , Cardoza, R.E. , Alexander, N.J. , McCormick, S.P. , Collado, I.G. , Hermosa, R. , Monte, E. et al. (2013) Relevance of trichothecenes in fungal physiology: disruption of tri5 in Trichoderma arundinaceum . Fungal Genet. Biol. 53, 22–33. [DOI] [PubMed] [Google Scholar]
- Mbengue, M. , Navaud, O. , Peyraud, R. , Barascud, M. , Badet, T. , Vincent, R. , Barbacci, A. et al. (2016) Emerging trends in molecular interactions between plants and the broad host range fungal pathogens Botrytis cinerea and Sclerotinia sclerotiorum . Front. Plant Sci. 7, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletiadis, J. , te Dorsthorst, D.T. and Verweij, P.E. (2003) Use of turbidimetric growth curves for early determination of antifungal drug resistance of filamentous fungi. J. Clin. Microbiol. 41, 4718–4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk, C.W. , Molnar, A. and Baulcombe, D.C. (2011) Intercellular and systemic movement of RNA silencing signals. EMBO J. 30, 3553–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter, N. , Worrall, E.A. , Robinson, K.E. , Li, P. , Jain, R.G. , Taochy, C. , Fletcher, S.J. et al. (2017) Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants, 3, 1–10. [DOI] [PubMed] [Google Scholar]
- Morozov, S.Y. , Solovyev, A. , Kalinina, N. and Taliansky, M. (2019) Double‐stranded RNAs in plant protection against pathogenic organisms and viruses in agriculture. Acta Naturae, 11, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerva, L. , Sandrini, M. , Gambino, G. and Chitarra, W. (2020) Double‐stranded RNAs (dsRNAs) as a sustainable tool against gray mold (Botrytis cinerea) in grapevine: effectiveness of different application methods in an open‐air environment. Biomolecules, 10, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes, C.C. and Dean, R.A. (2012) Host‐induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 13, 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, L. , Lan, C. , Capriotti, L. , Ah‐Fong, A. , Sanchez, J.N. , Hamby, R. , Heller, J. et al. (2021) Spray‐induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol. J. 19, 1756–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani, T.S. , Nadendla, S.R. , Bardhan, K. , Madhuprakash, J. and Podile, A.R. (2020) Chitosan conjugates, microspheres, and nanoparticles with potential agrochemical activity. In Agrochemicals Detection, Treatment and Remediation, pp. 437–464. Oxford: Butterworth‐Heinemann. [Google Scholar]
- Romanazzi, G. and Feliziani, E. (2014) Botrytis cinerea (gray mold). In Postharvest Decay, pp. 131–146. Cambridge, US: Academic Press. [Google Scholar]
- Rottem, S. , Yashouv, J. , Ne'Eman, Z. and Razin, S. (1973) Cholesterol in mycoplasma membranes. Composition, ultrastructure and biological properties of membranes from Mycoplasma mycoides var. capri cells adapted to grow with low cholesterol concentrations. Biochim. Biophys. Acta‐Biomembranes, 323, 495–508. [DOI] [PubMed] [Google Scholar]
- Schumacher, J. (2012) Tools for Botrytis cinerea: new expression vectors make the gray mold fungus more accessible to cell biology approaches. Fungal Genet. Biol. 49, 483–497. [DOI] [PubMed] [Google Scholar]
- Vinggaard, A.M. , Hass, U. , Dalgaard, M. , Andersen, H.R. , Bonefeld‐Jørgensen, E. , Christiansen, S. , Laier, P. et al. (2006) Prochloraz: an imidazole fungicide with multiple mechanisms of action. Int. J. Androl. 29, 186–192. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2008) Post‐transcriptional RNA silencing in plant–microbe interactions: a touch of robustness and versatility. Curr. Opin. Plant Biol. 11, 464–470. [DOI] [PubMed] [Google Scholar]
- Walker, L. , Sood, P. , Lenardon, M.D. , Milne, G. , Olson, J. , Jensen, G. , Wolf, J. et al. (2018) The viscoelastic properties of the fungal cell wall allow traffic of Am Bisome as intact liposome vesicles. MBio, 9, e02383‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. and Dean, R.A. (2020a) Movement of small RNAs in and between plants and fungi. Mol. Plant Pathol. 21, 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M.Y. and Dean, R.A. (2020b) Movement of small RNAs in and between plants and fungi. Mol. Plant Pathol. 21, 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Weiberg, A. , Lin, F.‐M. , Thomma, B.P. , Huang, H.‐D. and Jin, H. (2016) Bidirectional cross‐kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants, 2, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg, A. , Wang, M. , Lin, F.‐M. , Zhao, H. , Zhang, Z. , Kaloshian, I. , Huang, H.‐D. et al. (2013) Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science, 342, 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski, M. , Droby, S. , Norelli, J. , Liu, J. and Schena, L. (2016) Alternative management technologies for postharvest disease control: the journey from simplicity to complexity. Postharvest Biol. Technol. 122, 3–10. [Google Scholar]
- Wytinck, N. , Sullivan, D.S. , Biggar, K.T. , Crisostomo, L. , Pelka, P. , Belmonte, M.F. and Whyard, S. (2020) Clathrin mediated endocytosis is involved in the uptake of exogenous double‐stranded RNA in the white mold phytopathogen Sclerotinia sclerotiorum . Sci. Rep. 10, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, X. , Ma, W.‐B. , Li, Y. , Wang, H. , Que, Y.‐W. , Ma, Z.‐H. , Talbot, N.J. et al. (2011) A sterol 14α‐demethylase is required for conidiation, virulence and for mediating sensitivity to sterol demethylation inhibitors by the rice blast fungus Magnaporthe oryzae . Fungal Genet. Biol. 48, 144–153. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Li, T. , Xiao, L. , Zhou, S. and Liu, X. (2020) Characterization of tebuconazole resistance in Botrytis cinerea from tomato plants in China. Phytopathol. Res. 2, 1–10. [Google Scholar]
- Zhang, Y.‐Q. , Gamarra, S. , Garcia‐Effron, G. , Park, S. , Perlin, D.S. and Rao, R. (2010) Requirement for ergosterol in V‐ATPase function underlies antifungal activity of azole drugs. PLoS Pathog. 6, e1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Method S1 Sequences of Bc‐DCL1/2‐dsRNA (dsRNA‐Dicer), or Bc‐ERG1/13/11‐dsRNA (dsRNA‐ERG) constructs.
Table S1 Coefficient treatment interaction and growth inhibition by dsRNA and fungicides
Figure S1 Dose effect of dsRNA‐ERG on fungal growth.
Figure S2 Infection of tomato fruit with no wounding.
Figure S3 In vitro fungal growth with external ergosterol addition.
Figure S4 Illustration of systemic evaluation experiment set‐up.
Figure S5 dsRNA application reduces fungicide‐needed concentration for growth inhibition of B. cinerea.


