Summary
Increasing grain yield has always been the primary goal of crop breeding. KLUH/CYP78A5 has been shown to affect seed size in several plant species, but the relevant molecular mechanism is still unclear and there are no reports of this gene contributing to yield. Here, we demonstrate that modified expression of TaCYP78A5 can enhance wheat grain weight and grain yield per plant by accumulating auxin. TaCYP78A5 is highly expressed in maternal tissues, including ovary and seed coat during wheat development. The constitutive overexpression of TaCYP78A5 leads to significantly increased seed size and weight but not grain yield per plant due to the strengthening of apical dominance. However, localized overexpression of TaCYP78A5 in maternal integument enhances grain weight and grain yield per plant by 4.3%–18.8% and 9.6%–14.7%, respectively, in field trials. Transcriptome and hormone metabolome analyses reveal that TaCYP78A5 participates in auxin synthesis pathway and promotes auxin accumulation and cell wall remodelling in ovary. Phenotype investigation and cytological observation show that localized overexpression of TaCYP78A5 in ovary results in delayed flowering and prolonged proliferation of maternal integument cells, which promote grain enlargement. Moreover, naturally occurring variations in the promoter of TaCYP78A5‐2A contribute to thousand‐grain weight (TGW) and grain yield per plant of wheat;TaCYP78A5‐2A haplotype Ap‐HapII with higher activity is favourable for improving grain weight and grain yield per plant and has been positively selected in wheat breeding. Then, a functional marker of TaCYP78A5 haplotype Ap‐HapII is developed for marker‐assisted selection in wheat grain and yield improvement.
Keywords: wheat, TaCYP78A5, grain weight, grain yield, auxin, maternal integument cell proliferation, haplotype, functional marker
Introduction
Wheat (Triticum aestivum L.) is one of the most important staple crops worldwide, providing more than 20% calories and protein for humans. Increasing wheat yield is critical for global food and nutrition security (FAO, http://faostat.fao.org). Wheat yield is composed of the number of panicles per unit area, the number of grains per panicle and grain weight, and among which the grain weight has high heritability and stability, with great potential for improvement (Li et al., 2019b). In practice, attempts to enhance grain yield through enlarging grain size/weight have always been impeded by the trade‐off between grain weight and grain number. Increasing grain weight without changing grain number has become a major goal of high‐yield wheat breeding (Bustos et al., 2013). Therefore, increasing grain weight and understanding the mechanism underlying grain size/weight control are pivotal to increase yield of wheat.
Seed is composed of embryo, endosperm and the seed coat from the maternal tissue, which together determine the size and weight of the seed (Shewry et al., 2012). It was demonstrated that KLUH/CYP78A5, which encodes cytochrome P450 monooxygenase, plays an important role in controlling grain size. In Arabidopsis, KLUH increases seed size by non‐cell autonomously stimulating maternal integument cell proliferation (Adamski et al., 2009). The rice KLUH homolog OsCYP78A13 affects seed size through regulating the balance of resources for cell between embryo and endosperm (Xu et al., 2015). In tomato, SiKLUH controls fruit weight by increasing cell layer and delaying fruit ripening, as well regulating plant architecture by adjusting the number and the length of branches (Chakrabarti et al., 2013). Previous studies in Arabidopsis suggest that CYP78A5 is involved in the production of downstream mobile signal molecule (Anastasiou et al., 2007). Although KLUH has been shown to affect seed size in several species, there are no reports of this gene increasing yield. The molecular mechanism of KLUH controlling seed size remains elusive.
Auxin, the first discovered plant growth hormone, plays an important role in plant growth and development, including cell proliferation and expansion at the cytological level, embryogenesis, apical dominance and flowering at the macroscopic level (Pagnussat et al., 2009; Sauer et al., 2013; Shimizu‐Sato et al., 2009). Appropriately increasing auxin can increase crop yield (Shao et al., 2017). Recent studies showed that increasing the expression of PLA1/CYP78A1 in maize and CYP78A9 in rapeseed can increase seed weight and yield by affecting auxin metabolism (Shi et al., 2019; Sun et al., 2017), but a recent study in Arabidopsis showed that CYP78A5 mainly affects cytokinin rather than auxin metabolism (Jiang et al., 2021).
In this study, we find that TaCYP78A5 is highly expressed in ovaries and seed coat and locates within the QTLs for grain weight and yield‐related traits in wheat. Modified expression of TaCYP78A5 in maternal integument enhances grain weight and grain yield per plant by 4.3%–18.8% and 9.6%–14.7%, respectively, in field trials. Transcriptome and hormone metabolome analyses reveal that TaCYP78A5 participates in auxin synthesis pathway and promotes auxin accumulation and cell wall remodelling in ovary. Phenotype investigation and cytological observation show that localized overexpression of TaCYP78A5 in ovary results in delayed flowering, which prolongs proliferation of maternal integument cells, increases the number of seed coat cell and eventually promotes grain enlargement. Association analysis demonstrates that TaCYP78A5 haplotype Ap‐HapII with higher activity is favourable for increasing grain weight and grain yield per plant and has been positively selected in wheat breeding in China. These findings reveal that TaCYP78A5 can serve as a valuable gene for improving wheat yield.
Results
TaCYP78A5 is highly expressed in maternal tissues during grain development
Based on the sequence of KLUH/CYP78A5 in Arabidopsis, we obtained TaCYP78A5, a homologous gene of KLUH/CYP78A5 in wheat. TaCYP78A5 had three homoeologs (TaCYP78A5‐2A/‐2B/‐2D; TraesCS2A01G175700.1, TraesCS2B01G201900.1 and TraesCS2D01G183000.1 respectively) in the wheat genome (Figure S1a). Evolutionary analysis exhibited that TaCYP78A5 in wheat is most closely related to those in Thinopyrum and Hordeum vulgare (Figure S1b). Domain analysis showed that TaCYP78A5 is conserved among different species, all containing the typical N‐terminal hydrophobic region, oxygen and heme‐binding domains (Figure S1b). The subcellular localization of TaCYP78A5 in wheat protoplast showed that it located in the endoplasmic reticulum (Figure 1a). Spatiotemporal expression profile analysis showed that TaCYP78A5 was ubiquitously expressed in wheat root, stem, leaf and spike, with most abundant in ovary and young seed (Figure 1b). Further investigation of the spatial expression of TaCYP78A5 in developmental grains showed that it was highly expressed in maternal integument (seed coat) but not in embryo and endosperm (Figure 1c). This indicates that TaCYP78A5 may be involved in grain development through maternal pathways.
Figure 1.
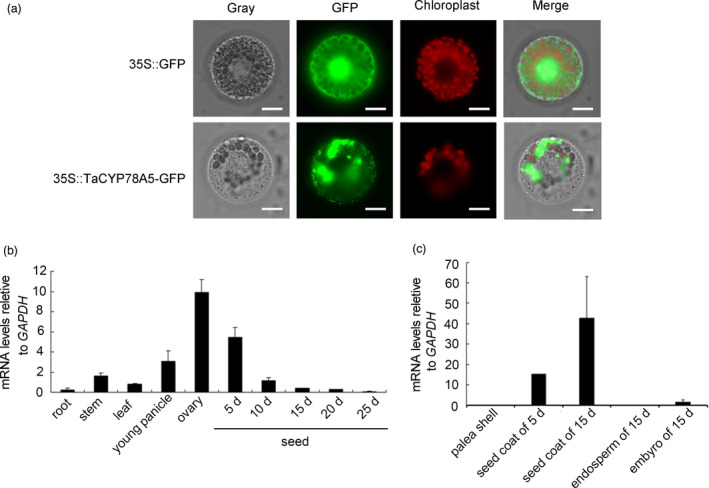
Expression profile and subcellular localization of TaCYP78A5 in wheat. (a) Subcellular localization of TaCYP78A5 in wheat. The vector control (35S::GFP) and fusion protein vector (35S::TaCYP78A5‐GFP) were each introduced into wheat protoplasts. GFP was observed with laser scanning confocal microscope. Scale bar = 10 µm. (b, c) Expression profiles of TaCYP78A5 across developmental cycle of wheat (b) and developmental grains (c). GADPH (TraesCS6B01G243700.1) was used as a reference gene. Data are shown as the mean ± SE (n = 3).
To reveal if TaCYP78A5 contributes to yield, we integrated the physical location of three homoeologs of TaCYP78A5 and the known genetic loci of yield component traits on the short arms of chromosome 2A, 2B and 2D in wheat (Table S1) based on the known genetic maps, physical map and wheat genome reference sequence IWGSC Ref v1.0 (IWGSC, 2018). The results showed that TaCYP78A5‐2A locates within the QTLs associated with grain thickness (GT), grain length (GL) and thousand‐grain weight (TGW) in wheat (Figure S2). The above results suggest that TaCYP78A5 may play an important role in regulating grain weight and yield.
Constitutive overexpression of TaCYP78A5 enhances grain weight but not grain yield per plant
To verify if TaCYP78A5 affects grain weight of wheat, we knocked down the expression of TaCYP78A5 in developing grains of wheat cultivar Shaan 512 that has large‐size/heavy weight kernel (with TGW 52 g) by using barley stripe mosaic virus‐induced gene silencing (BSMV‐VIGS) technique as reported previously (Ma et al., 2012). The result showed that the grain size and weight of TaCYP78A5‐knockdown plants (BSMV:TaCYP78A5) were significantly reduced, compared with those of the control plants (BSMV:00) (Figure S3a–e). We further investigated the cellular characteristics of seed coat, and found that the number of seed coat cells of BSMV:TaCYP78A5 plants was significantly decreased, but the size of the seed coat cell was not altered, compared with those of the control plants (Figure S3f–j). These results suggest that TaCYP78A5 regulates grain weight by promoting proliferation of seed coat cells.
To obtain increased yield of transgenic wheat and further verify the biological effect of TaCYP78A5, we generated transgenic wheat lines constitutively overexpressing TaCYP78A5‐2A under the control of maize ubiquitin promoter (named as UBI lines for simplicity). Nine independent transgenic events were obtained; of which two single‐locus transgenic events (UBI‐1 and UBI‐4) with higher expression levels of TaCYP78A5 compared to wild‐type plants (WT) are shown as representatives of UBI lines (Figure 2). The grain length, width and thickness of the UBI lines increased by 9.3%–10.3%, 9.4%–10.0% and 3.5%–4.0%, respectively, (Figure 2a–c), which resulted in significantly increased grain weight (by 26.9%–30.7%), compared with that of WT (Figure 2d). Further cytological analysis of grains at 15 days after fertilization (DAF) indicated that both the number and the length of seed coat cells of UBI lines were significantly higher than those of WT (Figure 2e,f). Taken together, TaCYP78A5 has a positive role in increasing grain weight of wheat.
Figure 2.
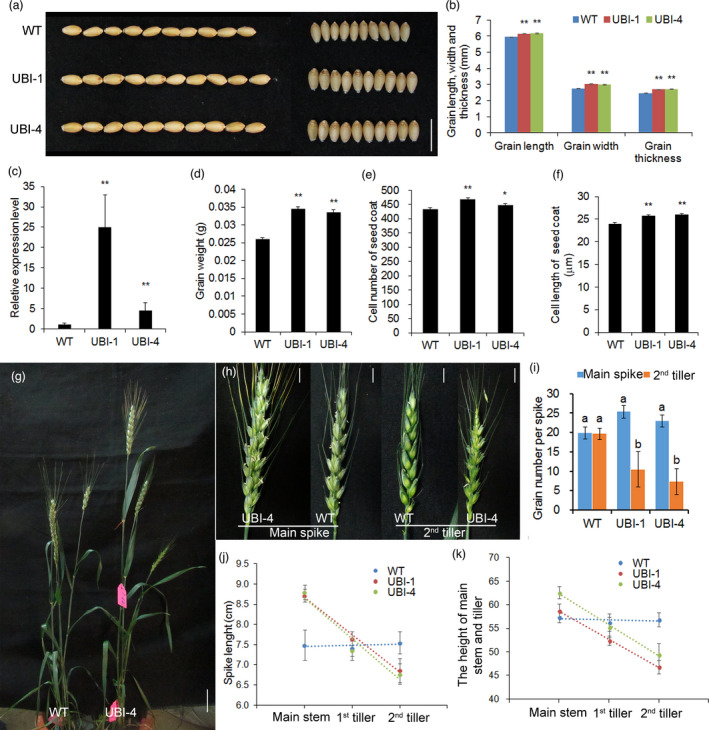
The phenotypes of UBI::TaCYP78A5‐transgenic wheat lines (UBI lines) and wild‐type wheat plants (WT). (a) The phenotypes of grain length and width of UBI lines and WT. Bar = 1 cm. (b) Grain length, width and thickness of UBI lines and WT (n ≥ 8). (c) Relative expression level of TaCYP78A5 in UBI lines and WT, GADPH as a reference gene (n = 3). (d) Grain weight of UBI lines and WT (n ≥ 8). (e–f) Cell number (e) and cell length (f) of outer seed coat of wheat grain 15 days after fertilization (n ≥ 8). (g) The plant architectures of UBI line‐4 and WT. Bar = 5 cm. (h) The phenotypes of main spike and the 2nd tiller spike of UBI lines and WT. Bar = 1 cm. (i) The grain number per main spike and per 2nd tiller spike of UBI lines and WT (n ≥ 8). (j) The spike length of the main stem and the tillers of UBI lines and WT (n ≥ 8). (k) The height of the main stem and the tillers of UBI lines and WT (n ≥ 8). Data are shown as the mean ± SE, *P < 0.05, **P < 0.01 by Student’s t‐test. Different lowercase letters on the bar chart indicate a significant level of difference.
Unfortunately, UBI lines showed apical dominance, the tiller growth was inhibited and hysteresis (Figures 2g–k and S4). Consequently, the grain yield per plant of the UBI lines did not increase (Figure S5), because the grain weight effects at the whole plant level were offset by significantly reduced grain number per tiller spike (Figure 2i).
Localized overexpression of TaCYP78A5 in maternal integument increases grain weight and grain yield per plant
In view of the fact that constitutive overexpression of TaCYP78A5 in wheat enhances apical dominance and that TaCYP78A5 is preferentially expressed in ovary and seed coat, we generated wheat transgenic lines overexpressing TaCYP78A5 driven by Arabidopsis INNER NO OUTER promoter (pINO), a maternal integument‐specific expression promoter (Villanueva et al., 1999), in order to reveal the biological effects of TaCYP78A5 on grain development and avoid enhancing apical dominance. More than 20 independent transgenic lines (called pINO lines for simplicity) were obtained, and three independent single‐locus transgenic lines (pINO‐1, pINO‐13 and pINO‐24) with various transgene expression levels were used for subsequent analysis (Figure 3a). The grain length and thickness of the pINO lines were not significantly different from those of WT, but the grain width was increased by 1.2%–3.4% (P < 0.01) (Figure 3b,c), this led to an increase in grain size and grain weight by 7.3%–8.9% and 5.7%–8.6%, respectively, compared with WT (Figure 3d,e). There was no difference in quality traits between the pINO lines and WT (Table S2). We further investigated the numbers of seed coat cells of the pINO lines and WT by cross‐cutting the grains 15 DAF (Figure 3f,g). The result showed that the pINO lines had more outer layer cells than WT (Figure 3h); however, the cell lengths of the pINO lines were similar as those of WT (Figure 3i). These indicate that localized overexpression of TaCYP78A5 in maternal integument causes an increase in the number of seed coat cells, which leads to enlargement in grain size and grain weight.
Figure 3.
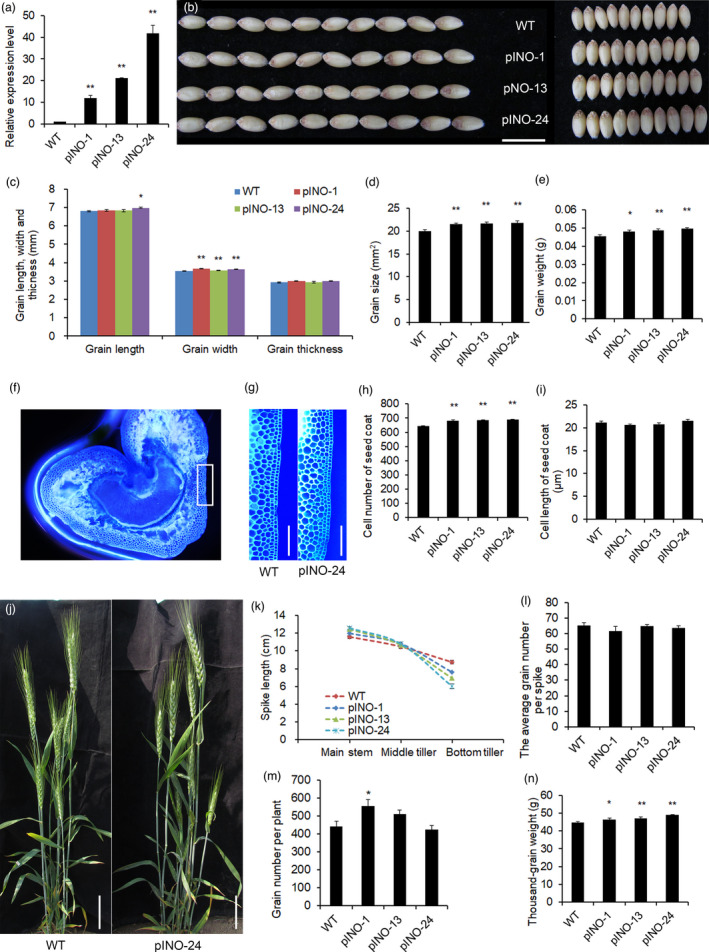
The phenotypes of pINO::TaCYP78A5‐transgenic wheat lines (pINO lines) and wild‐type plant (WT). (a) Relative expression of TaCYP78A5 in pINO lines and WT (n = 3). (b) Grain phenotypes of pINO lines and WT. Bar = 1 cm. (c) Grain length, width and thickness of pINO lines and WT (n ≥ 10). (d, e) Grain size (d) and grain weight (e) of pINO lines and WT (n ≥ 10). (f) A representative cross section of the grain 15 days after fertilization (DAF) stained with Fluorescent Brightener. (g) Enlarged view of the seed coat cells of pINO lines and WT. Bar = 200 µm. (h, i) Cell number (h) and cell length (i) of the outer seed coat of grain 15 DAF (n ≥ 20). (j) The plant architecture of pINO‐24 and WT. (k) The spike length of the main stem and the tillers of pINO line and WT (n > 10). (l, m) The average grain number per spike (l) and grain number per plant (m) of pINO lines and WT (n = 20). (n) Thousand‐grain weight of pINO lines and WT (n > 10). Data are shown as the mean ± SE, *P < 0.05, **P < 0.01 by Student’s t‐test.
As we expected, the pINO lines had no obvious apical dominance (Figure 3j). The main spikes and the middle tiller spikes of the pINO lines had similar spike length with those of WT, except the smallest tiller spike (bottom tiller spike) of the pINO lines that had decreased spike length, compared to WT (Figure 3k). There was no difference in the average grain number per spike and grain number per plant between the pINO lines and WT, except for pINO‐1 which had significantly increased grain number per plant (Figure 3m). The TGW of the pINO lines was increased by 4.0%–9.5% (Figure 3n).
To further explore the effects of TaCYP78A5 on grain yield per plant, we investigated the grain yield per plant of the pINO lines for 3 consecutive years, that is, the pINO lines grown in the greenhouse in 2017 and grown in the Transgenic Plant Experimental Station of Northwest A & F University, Yangling (108°4′E, 34°17′N) in natural growth seasons in 2018–2019 and 2019–2020. The results showed that the TGW of the pINO lines significantly increased in all three years (4.9%, 4.3% and 18.8% respectively), compared with those of WT (Figure 4a–c). The grain yield per plant of the pINO lines increased by 11.1% and 14.7% in 2017 and 2018–2019, respectively, and the biomass per plant increased only in 2018–2019 (9.6%), compared with those of WT (Figure 4d–f). There were no differences in other yield‐related traits between the pINO lines and WT (Figure S6). Taken together, localized overexpression of TaCYP78A5 in maternal integument enhances grain size, weight and yield per plant of wheat.
Figure 4.
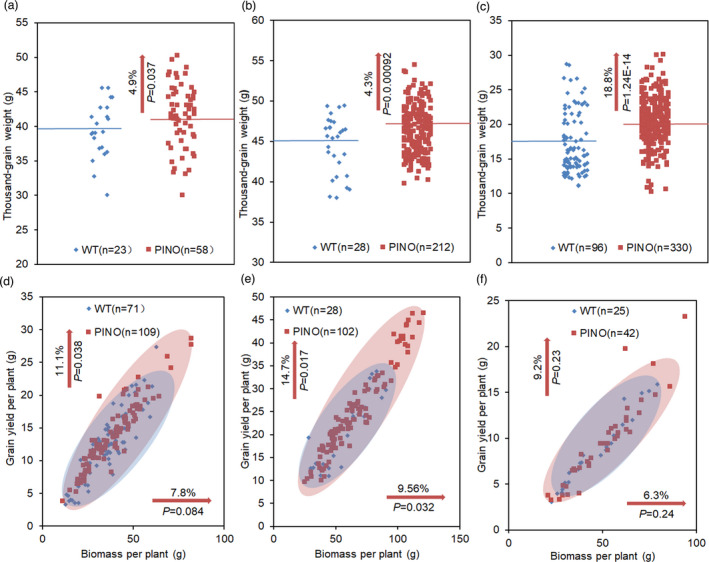
TaCYP78A5 affects thousand‐grain weight and yield of wheat. (a–c) Thousand‐grain weight of all transgenic wheat lines overexpressing TaCYP78A5 in integument under the control of INO promoter (pINO lines) and wild‐type wheat plant (WT) at green house in 2017 (a) and at field in 2018 (b) and in 2019 (c), the line represents the mean. (d–f) Grain yields per plant and biomass per plant of all pINO lines and WT at green house in 2017 (d) and at field in 2018 (e) and in 2019 (f). The percentages in the vertical of direction in each pane indicate the increase of grain yield per plant, and the percentages in the horizontal direction show the increase of biomass per plant. P‐values by Student’s t‐test.
The growth‐promoting effect of TaCYP78A5 on plant organs is limited by the travel distance of a mobile factor
In above study, we noticed that localized overexpression of TaCYP78A5 in ovaries resulted in a significant increase in biomass, implying extensive growth‐promoting effects of TaCYP78A5 on other organs. Histochemical observations using β‐glucuronidase (GUS) staining showed that the fusion protein TaCYP78A5‐GUS only aggregated in the ovaries of the pINO lines (Figure 5a), which resulted in enlarged glumes and lengthen spikes in the pINO lines, compared with those in WT (Figure 5b–i). The flag leaves of the pINO‐13 line are also significantly longer than WT (Figure 5j). Cytological observation showed that the cell sizes of the glume outer integument of the pINO lines were similar as those of WT, but the cell numbers of the pINO lines were significantly increased, compared with those of WT (Figure 5d,e). These results suggested that the growth‐promoting effect of TaCYP78A5 may depend on a mobile growth‐promoting factor. This is consistent with the previous inference that CYP78A5 may promote the growth of reproductive organs through a mobile molecule in Arabidopsis (Adamski et al., 2009; Anastasiou et al., 2007).
Figure 5.
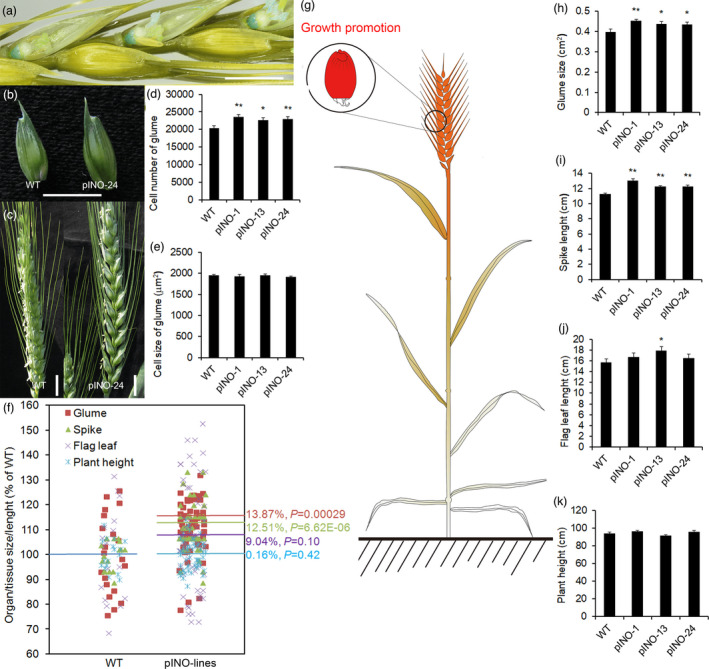
Growth‐promoting effects of TaCYP78A5 overexpressing in ovaries on other tissues/organs of wheat. (a) Histochemical β‐glucuronidase (GUS) staining of heading stage spike from transgenic wheat lines overexpressing fusion protein TaCYP78A5‐GUS in ovary (named as pINO lines for simplicity). Bar = 1 cm. (b, c) The glumes (b) and the main spike (c) of transgenic line pINO‐24 and wild‐type wheat plant (WT). Bar = 1 cm. (d, e) Cell number (d) and cell size (e) of the glumes of pINO lines and WT (n = 20). (f) The data of all the glume size, spike length, flag leaf length and plant height of pINO lines and WT, the mean of WT was set to 100%. The horizontal line represents the mean (n > 10). (g) Schematic diagram of the growth‐promoting effects of TaCYP78A5 overexpressing in ovary on other tissues/organs and its distance limitation. (h–k) Glume size (h), spike length (i), flag leaf length (j) and plant height (k) of pINO lines and WT (n > 10). Data are shown as the mean ± SE, *P < 0.05, **P < 0.01 by Student’s t‐test.
Interestingly, it can be seen that the growth‐promoting effects of TaCYP78A5 on tissues/organs were obviously related to the physical distance where the organ is from the ovary/grain tissues. Glumes and spikes had the closest physical distance to grains, and their enlargement effects were obvious and significant, with an increase of 13.9% and 12.5% respectively (P = 0.00029 for glume, P = 6.62E‐06 for spike). However, the growth‐promoting effects on flag leaf and plant height gradually decreased with increasing distance from the grains (Figure 5f–k). Collectively, overexpression of TaCYP78A5 only in ovaries can extend the growth‐promoting effects beyond the grains of wheat, and this promotion effects may be through a mobile molecule downstream.
TaCYP78A5 promotes grain enlargement through auxin accumulation
To investigate the mechanism underlying TaCYP78A5 affects grain size and the possible downstream mobile molecule, we conducted transcriptome analysis using the 1‐mm size ovaries of pINO‐24 and WT. Consequently, a total of 3328 differentially expressed genes (DEGs) were detected between pINO‐24 and WT, with 903 up‐regulated and 2425 down‐regulated (q value <0.05, fold change >2; Table S3‐1). Ten DEGs were randomly selected to perform real‐time quantitative RT‐PCR (qRT‐PCR) and the results from qRT‐PCR and from RNA‐Seq data were highly consistent (Figure S7), suggesting that RNA‐Seq data are reliable. To explore the functions, these DEGs involved in Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of the DEGs were performed, and the result showed that the DEGs involved in hormone signal transduction and cell wall metabolic process were significantly enriched (Figures S8 and 6a, Tables S4‐1 and S4‐2). Further analysis of the hormone signal transduction‐related DEGs revealed that 49% (30/61) were involved in auxin signalling (Table S3‐2) and that the genes associated with auxin metabolism, transport and response underwent were significantly differentially expressed (Figure 6b, Table S3‐3). Considering that the changes in auxin metabolism may affect cell wall process (Pacheco‐Villalobos et al., 2016), we further analysed the cell wall metabolism‐related genes regulated by auxin and found that the genes involved in cell wall remodelling were prominently differentially expressed (Figure 6c, Table S4‐3). This is in line with above conclusion that TaCYP78A5 overexpression led to an increase in the number of seed coat cells. Interestingly, cell wall expansin‐related genes were consistently down‐regulated (Figure 6c, Table S4‐3), suggesting that cell expansion might be inhibited. Taken together, these results are consistent with above conclusion that TaCYP78A5 regulates grain size by promoting maternal integument cell proliferation.
Figure 6.
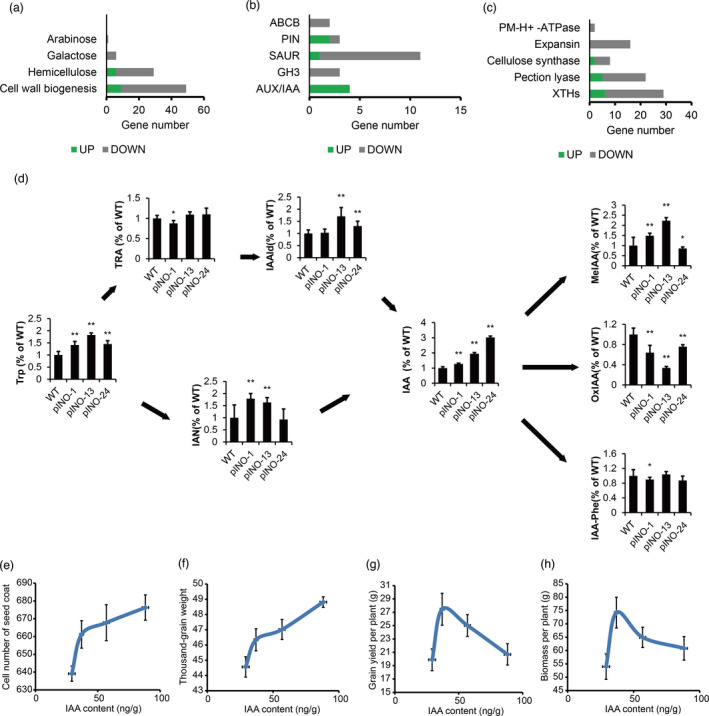
TaCYP78A5 overexpression affects auxin‐related pathways and causes auxin accumulation. WT, wild‐type wheat; pINO‐1,‐13,‐24, transgenic wheat lines overexpressing TaCYP78A5 driven by INO promoter. (a–c) Expression changes of genes associated with cell wall metabolism (a), IAA transduction and signalling (b), cell wall metabolism regulated by auxin(c). (d) The relative levels of auxin, auxin precursors and auxin conjugates in pINO lines compared to those of WT. The mean of WT was set to 100%. The contents of these substances were measured by ESI‐HPLC‐MS/MS, auxin/IAA, indole‐3‐acetic acid; IAAld, indole‐3‐acetaldehyde; IAA‐Phe, IAA‐phenylalanine; IAN, indole‐3‐acetonitrile; MeIAA, methyl‐IAA; oxIAA, 2‐oxoindole‐3‐acetic acid; TRA, tryptamine; Trp, tryptophan. (e–h) The correlation between the auxin concentration and the number of seed coat cells (e), Thousand‐grain weight (f), grain yield per plant (g) and biomass per plant (h) in pINO lines. Data are shown as the mean ± SE (n > 3), *P < 0.05, **P < 0.01 by Student’s t‐test.
As described above, all the auxin transport, auxin response and cell wall metabolism affected by auxin were significantly changed in the pINO line, so we speculated that overexpression of TaCYP78A5 might lead to changes in auxin metabolism, although a recent study in Arabidopsis showed that CYP78A5 mainly affects cytokinin rather than auxin metabolism (Jiang et al., 2021). In order to confirm whether TaCYP78A5 overexpression affects cytokinin or auxin metabolism in transgenic wheat, the levels of auxin, auxin precursors, auxin conjugates and cytokinin were determined in the ovaries (≥1 mm in length) and the spikes of the pINO lines and WT at heading stage through liquid chromatography–tandem mass spectrometry (LC‐MS/MS). Interestingly, the cytokinin levels in the pINO lines were not changed significantly, compared with that in WT, with the exception of pINO‐1 (Figure S9). However, the auxin levels of the pINO lines were significantly higher than that of WT and were positively correlated with the expression levels of TaCYP78A5 (Figures 3a, 6d and S10). Moreover, the levels of auxin biosynthesis precursors, such as tryptophan (TRP), tryptamine (TRA), indole‐3‐acetaldehyde (IAAld) and auxin conjugates MeIAA in the pINO lines were also higher than those in WT, while the levels of auxin inactivation product (oxIAA) in the pINO lines were much lower than that in WT (Figure 6d). Therefore, we deduced that TaCYP78A5 might function by affecting the auxin pathway in wheat. Auxin is an essential hormone for plant growth, seed development, cell division and yield. In order to explore the possible influence of TaCYP78A5 on yield‐related traits via auxin, we analysed the correlations between the TaCYP78A5 activity and the auxin concentration, the number of seed coat cells, TGW, grain yield per plant and biomass per plant of the pINO lines. The results showed that the concentration of auxin in the ovary was positively correlated with the expression levels of TaCYP78A5 (Figures 3a and 6d). The number of seed coat cell and TGW were continuously increased with the increase of the auxin concentration and the TaCYP78A5 activity in the pINO lines, while the grain yield and biomass per plant were first increased and then decreased with the increase of the auxin concentration and the TaCYP78A5 activity in the pINO lines (Figure 6e–h). These results suggest that grain size and TGW increased with the increase of the auxin concentration in the pINO lines, but an optimal auxin concentration existed to maximize grain yield and biomass per plant. This may explain the reason that the UBI lines did not increase grain yield per plant.
In order to further verify that auxin accumulation plays an essential role in enhancing grain weight, we treated wheat (JW1) plants at the booting stage with auxin or auxin synthesis inhibitor 5‐methyl‐tryptophan (5‐MT) every 3 days till the plants at 15 days post flowering, and then measured grain weight after maturity. The results showed that 100 μmol/L of auxin treatment led to increased grain weight, while 50 μmol/L of 5‐MT treatment caused reduced grain weight (Figure S11), indicating that auxin accumulation enhances grain weight.
Taken together, transcriptome and hormone metabolome analyses revealed the involvement of TaCYP78A5 in auxin synthesis pathway and auxin accumulation in the pINO lines to enhance grain weight and grain yield per plant of wheat.
TaCYP78A5 promotes grain enlargement by auxin‐mediated prolongation of maternal epidermal cell proliferation
Flowering time and ripening time have important effects on biomass of crops by affecting duration of basic vegetative growth (Andres and Coupland, 2012; Gao et al., 2014). In the present study, heading and flowering time of the pINO lines were delayed by 1 and 2–3 days, respectively, compared with those of WT; however, the maturity time of the pINO lines is the same as that of WT (Figure S12a,b). The delayed heading and flowering of the pINO lines may attribute to the increased auxin level, because wheat plants at booting stage treated with exogenous auxin, naphthylacetic acid (NAA), exhibited delayed flowering (Figure S13). This is in line with previous reports that high concentration of auxin can delay flowering and fruit ripening (Dal Santo et al., 2020; Zhao et al., 2013). Then, we questioned if there is any relationship between auxin‐mediated delayed flowering and the enlarged grains due to the increased number of seed coat cells. To answer this question, we selected six time points throughout the period from heading to ripening to observe proliferation of maternal integument/seed coat cells of pINO line‐24 and WT, and the results showed that proliferation of maternal integument/seed coat cells mainly occurred during ovary development stage (Figure S12c). A similar phenomenon also appeared in barley (Radchuk et al., 2011). Delayed flowering resulted in extending proliferation time of maternal integument cells of the pINO lines, which ultimately led to the increased number of seed coat cells (Figure S12d). Thus, we conclude that TaCYP78A5 promotes grain enlargement through auxin‐mediated delayed flowering, which prolongs proliferation of maternal integument cells and enhances the number of seed coat cell.
Genetic variations in TaCYP78A5‐2A promoter affect wheat grain weight and the favourable haplotype Ap‐HapII has been positively selected in wheat breeding
To uncover the naturally allelic variations of TaCYP78A5 in wheat, we compared the DNA sequences of the coding regions and the promoters of three homoeologs of TaCYP78A5 in 30 wheat cultivars with various genetic backgrounds (Table S5). Two haplotypes of TaCYP78A5‐2A were characterized by five single‐nucleotide polymorphisms (SNPs) in the promoter region (named as TaCYP78A5‐Ap for simplicity), that is, TaCYP78A5 Ap‐HapI and TaCYP78A5 Ap‐HapII (named as Ap‐HapI and Ap‐HapII, respectively, for simplicity) (Figure 7a). A cleaved amplified polymorphic sequence (CAPS) marker was developed based on −1191 bp (C/T) in TaCYP78A5‐Ap to distinguish these two haplotypes (Figure 7b). This CAPS marker was further verified in wheat population with 323 accessions (Table S6). Since the two haplotypes have SNPs in the promoter region of TaCYP78A5‐2A, we speculated that these SNPs might lead to changes in promoter activity. Therefore, we tested the promoter activity of these two haplotypes, and the results showed that Ap‐HapII has higher promoter activity than Ap‐HapI (Figure 7c). In order to investigate if the two haplotypes affect wheat yield potential, we conducted association analysis between the two haplotypes and TGW and grain yield per plant of the 323 accessions in 16 environmental sites. The results showed that Ap‐HapII had significantly higher TGW and grain yield per plant than Ap‐HapI in most environments (Figure 7d,e). These suggested that Ap‐HapII with higher promoter activity was a favourable haplotype for TGW and grain yield per plant in wheat.
Figure 7.
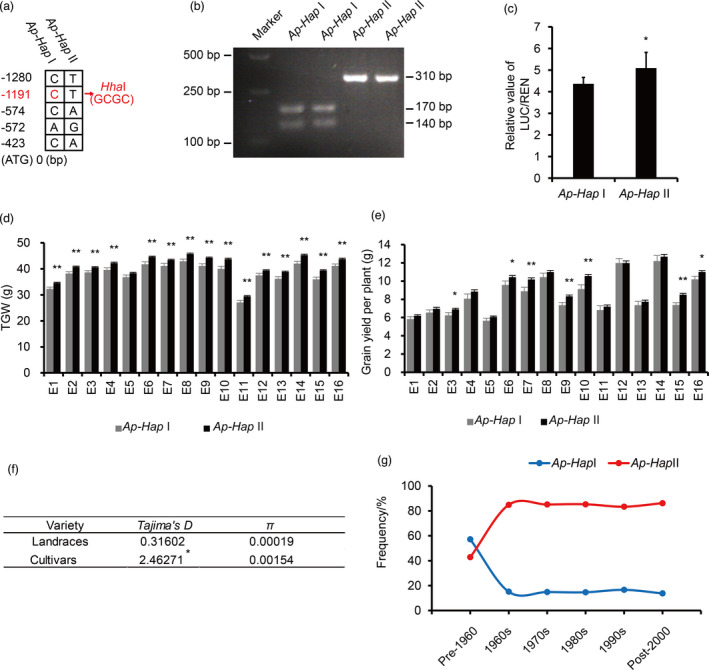
Sequence variations of TaCYP78A5‐2A and their associations with grain yield‐related traits. (a) Two haplotypes (Ap‐HapI and Ap‐HapII) based on the sequence variation in the promoter region of TaCYP78A5‐2A. (b) A cleaved amplified polymorphic sequence (CAPS) marker developed based on −1191 bp (C/T) with restriction endonuclease HhaI showed in (a). After enzyme digestion, the Ap‐HapI be cleaved into 170 and 140 bp, but Ap‐HapII could not be cleaved. (c) The relative activity of TaCYP78A5 promoters with haplotype Ap‐HapI and Ap‐HapII. The two haplotypes (Ap‐HapI and Ap‐HapII) of TaCYP78A5‐2A were separately cloned into the pGreenII 0800‐LUC (firefly luciferase) to generate expression vectors Ap‐HapI::LUC and Ap‐HapII::LUC, and the two constructs were separately infiltrated into the leaves of 30‐day‐old Nicotiana benthamiana, with the expression of renilla luciferase (REN) as an internal control. The LUC activity and REN activity were measured with GloMax‐multi‐luminescence reader. The ration of LUC activity to REN activity (LUC/REN) indicates the relative expression level of TaCYP78A5 Ap‐HapI or Ap‐HapII. Data are shown as the mean ± SE (n = 3). (d, e) Association of Ap‐HapI and Ap‐HapII with thousand‐grain weight (TGW) (d) and yield per plant (e) of 323 wheat accessions at 16 environmental sites. E1–E16 indicate the environments at Shunyi in 2015 under drought stressed (DS), DS + heat stress (HS), well‐watered (WW), WW + HS; Shunyi in 2016 under DS, DS + HS, WW, WW + HS; Changping in 2016 under DS, WW; Shunyi in 2017 under DS, DS + HS, WW, WW + HS; Changping in 2017 under DS and WW. (f) The nucleotide diversity (π) and Tajima’s D of TaCYP78A5‐2A promoter regions of 43 landraces and 42 cultivars. (g) Frequency changes of the two haplotypes of TaCYP78A5‐2A over decades in modern cultivars (total of 311 accessions). 14, 33, 47, 34, 60 and 123 accessions were released in pre‐1960s, 1960s, 1970s, 1980s, 1990s and post‐2000 respectively. Ap‐HapI and Ap‐HapII indicate two haplotypes of TaCYP78A5‐2A.
Breeding selection leaves intense footprints in genomes, showing progressive accumulation of favourable haplotypes (Barrero et al., 2011). To examine the evolutionary history of TaCYP78A5‐Ap, the Tajima’s D and diversity (π) analysis of TaCYP78A5‐Ap (1.5 kb of promoter region) were investigated in 43 landraces and 42 cultivars (Table S7). Tajima’s D of the cultivars showed significant values and was higher than that of the landraces, and the diversity (π) in the cultivars was also higher than that in the landraces, this suggesting that allelic variations of TaCYP78A5‐Ap were strongly artificially selected during wheat domestication (Figure 7f). To determine whether favourable haplotype Ap‐HapII was selected during wheat breeding programs, we evaluated frequency changes of the two haplotypes in wheat population with 311 accessions released in different decades in China (Table S6). The results exhibited the frequency of the two haplotypes of TaCYP78A5‐2Ap in modern cultivars changed over decades. The frequencies of favourable haplotype Ap‐HapII sharply increased from 42.9% to 84.8% in the cultivars released from pre‐1960 to 1960s, and subsequently remained stable in the decades from 1970s to post‐2000, while the frequencies of haplotype Ap‐HapI showed the opposite trend (Figure 7g), this implying that favourable haplotype Ap‐HapII was positively selected in wheat breeding in China.
Discussion
Altered expression of TaCYP78A5 in wheat integument promotes grain weight with yield potential
In plant, seed size is a key factor affecting yield. Larger seeds have greater seed weight and offer the potential to increase yield, but larger seeds usually tend to be accompanied by a decrease in seed number, which counteract the increase in seed yield caused by enlarged seeds (Bustos et al., 2013; Foulkes et al., 2011; Molero et al., 2019). KLUH/CYP78A5 and its homologous genes have been shown to affect seed/fruit size in Arabidopsis, rice, tomato and other plants (Anastasiou et al., 2007; Chakrabarti et al., 2013; Nagasawa et al., 2013; Zhao et al., 2016); but overexpression of KLUH/CYP78A5 in Arabidopsis did not increase seed yield per plant, because the increase in seed size was offset by the decrease in seed number (Adamski et al., 2009). Here, we show that constitutive overexpression of TaCYP78A5 in wheat leads to enlarged seeds and increased seed weight, but not increased grain yield per plant due to enhanced apical dominance and reduced grain number of tillers (Figure 2g–k). In order to avoid this problem, we generated wheat transgenic lines overexpressing TaCYP78A5 specifically in integument. Consequently, unlike UBI lines, pINO lines had no obvious apical dominance and normal grain number (Figure 3j–m). Therefore, grain weight and grain yield per plant of the pINO lines were increased significantly compared with those of WT (Figures 3n and 4). The trade‐off between grain size and grain number has been reported in wheat, and enhancing grain yield through enlarging grain size had always been impeded by the trade‐off between grain weight and grain number (Bustos et al., 2013; Foulkes et al., 2011; Molero et al., 2019). A recent study raised one solution to overcome this problem by ectopic expression of α‐expansin in developing seeds, which can lead to grain enlargement but does not reduce the grain number in wheat (Calderini et al., 2021). Here, we provide another solution to overcome this problem by localized overexpression of TaCYP78A5 in wheat integument, which had the potential for grain enlargement by increasing the number of maternal integument /seed coat cells, and ultimately led to the increase in grain size/weight without affecting grain number (Figure 3m,n).
TaCYP78A5 promotes grain weight and grain yield per plant through auxin accumulation
A previous study in Arabidopsis demonstrated that KLU/CYP78A5 is involved in generating a mobile growth‐promoting signal molecule different from known classic hormones (Anastasiou et al., 2007). A study in rice indicated that GE/CYP78A13 does not participate in the biosynthesis of auxin (Xu et al., 2015). But studies in maize and rapeseed showed that overexpression of PLA1/CYP78A1 and BnaA9.CYP78A9, both belonging to CYP78A family, could affect auxin pathway (Shi et al., 2019; Sun et al., 2017). More recently, a study in Arabidopsis reported that KLUH participates in the cytokinin rather than auxin pathway (Jiang et al., 2021). In this study, we find that overexpression of TaCYP78A5 in integument promotes the growth of organs surrounding, suggesting that TaCYP78A5 involved in the production of a mobile growth‐promoting signalling molecule (Figure 5). Overexpression of TaCYP78A5 in wheat could enhance apical dominance (Figure 2g–k). Conversely, Arabidopsis cyp78a5/klu mutant exhibits reduced apical dominance phenotype (Anastasiou et al., 2007), which corresponds to the phenotype of the classic auxin synthesis gene mutant yucca in Arabidopsis (Cheng et al., 2006). Correspondingly, auxin‐synthesis‐related gene MEE45 can promote the cell division of the seed coat, leading to seed enlargement (Li et al., 2021), and here overexpression of TaCYP78A5 in wheat can also promote proliferation of the seed coat cell by prolongation of maternal epidermal cell proliferation, which increases the number of seed coat cell and causes seed enlargement (Figures 2b–i, 3b–h and S12). These findings suggested that TaCYP78A5 may be involved in auxin‐related pathway. The transcriptome and hormone metabolome analyses further confirmed that overexpression of TaCYP78A5 affects auxin synthesis pathway and causes auxin accumulation in wheat (Figure 6d). However, which step of auxin synthesis pathway TaCYP78A5 participates in needs further to explore.
Genetic variations of TaCYP78A5‐2A affect grain yield‐related traits and has been selected in wheat domestication and breeding
As one of the most successful crops on the earth, wheat has expanded from the small core area within the Fertile Crescent to all parts of the world in <10 000 years (Lev‐Yadun et al., 2000; Salamini et al., 2002). The genetic diversity of its genome and the convergent adaptation to human selection are one of the important reasons for its evolutionary success (Zhou et al., 2020). In the course of evolution, genotypes controlling favourable agronomic traits were preserved. In this study, we found that TaCYP78A5‐2A locates within QTLs for TGW and yield‐related traits by integrating the physical location of TaCYP78A5 homoeologs with the known QTL maps of group 2 chromosomes (2A, 2B and 2D) in wheat (Figure S2, Table S1), suggesting that TaCYP78A5‐2A might contribute to grain yield of wheat. Further analysis of naturally genetic variations in TaCYP78A5‐2A identified two haplotypes, haplotype Ap‐HapII exhibiting higher promoter activity than Ap‐HapI (Figure 7c). Association analysis between the two haplotypes and the agronomic traits of 323 wheat accessions in 16 environments revealed that haplotype Ap‐HapII exhibited significantly higher TGW and grain yield per plant than haplotype Ap‐HapI in most environments (Figure 7d,e). This is consistent with the result that overexpression of TaCYP78A5‐2A leads to an increase in grain size and grain yield per plant (Figure 3). Tajima’s D and the diversity (π) analysis of TaCYP78A5‐2A promoter sequences in the 43 landraces and the 42 cultivars showed the genetic variations of TaCYP78A5‐Ap strongly artificially being selected during wheat domestication and breeding (Figure 7f). Further, the frequency of haplotype Ap‐HapII increased rapidly in wheat breeding in China in 1960s and kept stable high level after 1970s (Figure 7g), and this time period is consistent with the time of the wheat green revolution, indicating that favourable haplotype Ap‐HapII of TaCYP78A5‐2A may have been strongly artificially selected during the wheat green revolution in China. Application of marker‐assisted selection (MAS) can significantly accelerated wheat breeding (Gupta et al., 2010). In this study, a CAPS marker developed to identify Ap‐HapI and Ap‐HapII (Figure 7b) provides an important functional marker for MAS for improving TGW and grain yield in future wheat breeding.
Materials and methods
Winter wheat cultivar Xiaoyan 6 was used to clone cDNA of TaCYP78A5 and to analyse its spatiotemporal expression profile. Wheat cultivar Shaan 512 with high thousand‐grain weight (52 g) was used to conduct BSMV‐VIGS to rapid identification of TaCYP78A5 function in wheat grain development. The 30 wheat cultivars with various genetic backgrounds were used to detect SNPs of three homoeologs of TaCYP78A5 (Table S5). The 323 wheat accessions described previously (Li et al., 2019a) were used for association analysis (Table S6). Spring wheat accession JW1 was used as a receptor material for wheat transformation.
The growth conditions of the wheat cultivars, wheat accessions and transgenic wheat lines are described in Appendix S1.
Total RNA isolation and cDNA synthesis
Wheat tissue/organ samples kept at −80 were used for RNA isolation. Total RNA was extracted using the Steady Pure Plant RNA Extraction Kit (Accurate Biotechnology, Changsha, China) according to the manufacturer’s protocol. The quality of RNA samples was determined by agarose gel electrophoresis and RNA concentration was measured using a Nanodrop 2000 spectrophotometer (ND2000; Thermo Scientific, Wilmington, NC). The cDNA was synthesized using the Evo M‐MLVRT Premix (Accurate Biotechnology, Changsha, China).
cDNA cloning and sequence alignment of TaCYP78A5 in wheat
To clone wheat TaCYP78A5, the mRNA sequence of Arabidopsis KLU/CYP78A5 (AT1G13710) was used to blast against wheat genome reference sequences to acquire the putative TaCYP78A5. Specific primers were designed based on the putative sequences of TaCYP78A5, and the cDNA synthesized from the mixed RNA samples of roots, leaves, young panicles and grains of Xiaoyan 6 was used as the template for TaCYP78A5 amplification. The primers used are shown in Table S8. The amplified products were constructed into pMD19T vector (Takara, Japan) and more than 10 clones were randomly selected for sequencing. The resulted sequences were compared with IWGSC Ref v 1.0 to obtain full‐length cDNA of three homoeologs of TaCYP78A5 in wheat. DNAMAN8 (www.lynnon.com) was used to compare these sequences. The tree plot of TaCYP78A5 and its homologues were investigated in Triticeae‐Gene Tribe (http://wheat.cau.edu.cn/TGT/) (Chen et al., 2020).
Subcellular localization of TaCYP78A5 in wheat
The coding region of TaCYP78A5‐2A from +1 to +1017 bp that contains the hydrophobic domain and oxygen‐binding motif was inserted into 35S::GFP of expression vector p16318 by using restriction endonuclease HindIII and SalI site to generate the construct 35S::TaCYP78A5‐GFP. The resulted construct and the vector control (35S::GFP) were transferred into wheat protoplast prepared according to previous study (Yoo et al., 2007). GFP was observed by inverted fluorescence microscope (DMi8, Leica, Germany). The primers used for developing the constructs are listed in Table S8.
Genetic mapping of TaCYP78A5 in wheat
The physical locations of three homoeologs of TaCYP78A5 were localized based on physical maps and wheat genome reference sequence IWGSC Ref v1.0 (IWGSC, 2018). The known genetic loci associated with yield‐related traits on the short arms of chromosome 2A, 2B and 2D in wheat (Table S1) were integrated to the physical maps of the short arms of group 2 chromosomes to obtain the genetic maps of TaCYP78A5 in wheat.
Detection of genetic variations of TaCYP78A5 in wheat
Single‐nucleotide polymorphism (SNP) detection of three homoeologs of TaCYP78A5 in the 30 wheat cultivar and functional marker development based on the SNPs are described in Appendix S1.
The sequences of TaCYP78A5‐2A of 43 landraces and 42 cultivars (Table S7) were obtained from GVM database (https://bigd.big.ac.cn/gvm, GVM000082; Zhou et al., 2020), and the molecular diversity π values and Tajima’s D were computed using DnaSP5.0 (Librado and Rozas, 2009).
Association analysis between TaCYP78A5 haplotypes and agronomic traits of wheat accessions
The genotypic data based on genetic variations of TaCYP78A5‐2A and phenotypic data of agronomic traits of the 323 wheat accessions at 16 environmental sites (E1–E16) over 3 years were analysed with the TASSEL5.1 software (Bradbury et al., 2007). The Details of the 16 environmental sites and agronomic trait measurement of wheat accessions are described in Appendix S1.
Determination of TaCYP78A5‐2A promoter activity
The promoter activity was detected according to the instruction of Dual‐Luciferase Reporter Assay System (Promega), and the procedure is shown in Appendix S1.
BSMV‐mediated TaCYP78A5 gene silencing in wheat
The BSMV‐VIGS technique used for knockdown the expression of TaCYP78A5 in developmental grains of wheat was performed as previously reported (Ma et al., 2012). The protocol of BSMV‐derived vector construction and BSMV‐mediated TaCYP78A5 gene silencing is described in Appendix S1.
Construction of TaCYP78A5 overexpression vectors and development of transgenic wheat lines
The protocol for constructing TaCYP78A5 overexpression vectors UBI::TaCYP78A5‐2A‐GFP/GUS and pINO::TaCYP78A5‐GUS is described in Appendix S1. The constructed vectors were, respectively, introduced into Agrobacterium tumefaciens strain EHA105. The embryogenic calli of the immature embryo 15 days after fertilization (DAF) of wheat line JW1 were used as recipient materials. Wheat transformation was conducted by A. tumefaciens‐mediated method as described previously (Ishida et al., 2015; Zhang et al., 2018). The positive transgenic plants were identified by leaf daubing with 0.2% glufosinate (BASTA), and transgenic lines were obtained by continuous self‐crossing of single‐locus transgenic plants and screening by Basta.
RNA sequencing and data analysis
The RNA samples isolated from the 1‐mm size ovaries of transgenic wheat line pINO‐24 and WT were used for RNA sequencing. The differentially expressed genes were identified as described in Appendix S1.
All the genes detected were subjected to GO (http://geneontology.org/) to obtain GO annotations, GO and KEGG enrichment were conducted as previously described (Chi et al., 2019).
Determination of auxin and cytokinin metabolite levels in wheat
The 1‐mm size ovaries of the pINO lines and WT were used to detect auxin levels, and auxin was determined by electrospray ionization–high‐performance liquid chromatography–tandem mass spectrometry (ESI‐HPLC‐MS/MS) method. The spikes of the pINO lines and WT at heading stage were used for testing relative quantification of auxin precursors, auxin conjugates and cytokinin. The protocols of auxin and cytokinin metabolite determination are shown in Appendix S1.
Quantitative real‐time reverse transcriptase–polymerase chain reaction (qRT‐PCR)
The qRT‐PCR was performed using the SYBR Green premix Pro Taq HS qPCR Kit (Accurate Biotechnology) on a CFX96™ Real‐time PCR Detection System (Bio‐Rad, Hercules, USA), wheat GAPDH gene (TraesCS6B01G243700.1) being used as a reference gene. The comparative Ct (threshold cycle) method was used to calculate the relative quantity (Livak and Schmittgen, 2001). All date were obtained from three biological replicates. The primers used are listed in Table S8.
Morphological characterization of transgenic wheat lines
The transgenic wheat lines of T2 and T3 generations and wild‐type wheat (WT) grown in the Transgenic Plant Experiment Station (108°4′E, 34°17′N) of Northwest A & F University, Yangling, China, were used for assessment of agronomic trait, including plant height, tiller number, spike length, grain number per main spike, thousand‐grain weight, grain yield per plant and biomass per plant. At least 15 plants were investigated for each transgenic line. For grain length, width and thickness determination, the grains from the middle spikelets were measured with a digital calliper (Pro’sKit, PD‐151).
Cytological characterization of ovary, seed coat and glume cells of wheat
To measure the number and area of outer integument/seed coat cells, the ovaries of the transgenic wheat plants and WT were collected and transected with frozen slicer (POLAR DM; Sakura, Tokyo, Japan), the grains at 15 DAF were rapidly frozen in liquid nitrogen and then cross‐cut. The sliced samples were stained with 0.01% solution of Fluorescent Brightener 28 (Sigma, Hongkong, China) for 10 min, and photographed under fluorescent microscope (Olympus, System Microscope BX53) to determine the number and the size of the outermost layer cells of integument/seed coat. At least 10 ovaries or grains from individual plants of each transgenic line or WT were analysed.
Histochemical observations using β‐glucuronidase (GUS) staining
Histochemical observations of GUS activity was determined as described previously (Tittarelli et al., 2007). The spikes of the pINO lines and WT at heading stage were incubated in the GUS staining solution (50 mm Phosphate buffer (pH = 7.2), 0.01% Triton X‐100, 2 mm K3Fe(CN)6, 2 mm K4[Fe(CN)6]·3H2O, 10 mm EDTA, 0.2 mm X‐Gluc) at 37 °C for 6 h, followed by gradient decolourization, then photographed by stereomicroscope (SMZ25; Nikon, Tokyo, Japan).
Statistical analysis
All data obtained were processed in SPSS, and statistical analysis was conducted by one‐way ANOVA or unpaired Student’s t‐test. Significant differences were considered if P‐values <0.05.
Conflict of interest
Authors claim that there is no conflict of interest.
Author contributions
H.Z., R.J., M.M. and L.G. conceived and designed the experiments. L.G., M.M., L.W., M.Z., M.L., B.W. and L.L. conducted the experiments. X.L. helped to construct expression vector. L.G. and M.M. analysed the data and wrote the draft. H.Z., R.J. and W.C. revised the manuscript.
Supporting information
Figure S1 The sequence characteristics and phylogenetic tree of TaCYP78A5 in wheat.
Figure S2 The genetic loci of yield‐related traits on the short arms of chromosome 2A, 2B and 2D in wheat.
Figure S3 TaCYP78A5 silencing resulted in decreased seed size and seed coat cell number.
Figure S4 The ripening time of UBI::TaCYP78A5‐transgenic wheat line (UBI‐4) and wild‐type plants (WT).
Figure S5 Grain yield per plant of UBI::TaCYP78A5‐transgenic wheat lines (UBI‐1 and UBI‐2) and wild‐type plants (WT).
Figure S6 The yield‐related traits of transgenic wheat lines and wild‐type wheat.
Figure S7 Validation of RNA‐seq data by quantitative real‐time RT‐PCR (qPCR).
Figure S8 KEGG enrichment (a) and GO enrichment (b) of differentially expressed genes (DEGs) between wild‐type wheat and transgenic line pINO‐24.
Figure S9 Relative levels of cytokinin in the ovaries of wild‐type wheat and transgenic lines.
Figure S10 The correlation analysis between the auxin concentration and the relative expression of pINO lines.
Figure S11 Auxin affects grain weight of wheat.
Figure S12 Localized overexpression of TaCYP78A5 in integument causes delayed flowering and prolongs proliferation of maternal integument/seed coat cells.
Figure S13 Exogenous auxin treatment causes delayed flowering in wheat.
Table S1 The known genetic loci of yield‐related traits on the short arms of chromosome 2A, 2B and 2D in wheat
Table S2 The quality traits of pINO lines and WT
Table S3‐1 The differentially expressed genes between wild‐type wheat (WT) and transgenic line pINO‐24
Table S3‐2 The differentially expressed genes involved in hormone signal
Table S3‐3 The differentially expressed genes associated with IAA metabolism, transport and signalling
Table S4‐1 The signalling genes that response for AUXIN
Table S4‐2 The cell‐wall‐related process
Table S4‐3 The cell‐wall‐related genes regulated by AUXIN
Table S5 Thirty cultivars used for detecting the natural allelic variations of TaCYP78A5 in wheat
Table S6 Basic information of 323 wheat accessions used for association analysis between TaCYP78A5‐2A haplotypes and agronomic traits
Table S7 Basic information of wheat landraces and cultivars used for analysis of evolutionary history
Table S8 The information of the primers used in this study
Appendix S1 Supplemental Methods
Acknowledgements
We thank Jinan Bondi Biotechnology Co., Ltd and Wheat Transformation Platform, State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A & F University, for assistance in generation of transgenic wheat plants. We thank Dr. Li Huang, Department of Plant Sciences & Plant Pathology, Montana State University, for kindly providing us with BSMV vector used in this study. We also thank Prof. Wanquan Ji, College of Agronomy, Northwest A & F University, for kindly providing with the Experimental Base of Transgenic Plants for us to grow transgenic wheat used in this study. Finally, we thank Prof. Rudi Appels, Honorary Professor, University of Melbourne, for improving the draft of this manuscript. This study was financially supported by the Natural Science Foundation of China (32072003 and 32072059), and Key Research and Development Program of Shaanxi Province (2021NY‐079) and the Natural Science Foundation of Shaanxi province (2017JQ3032).
Guo, L. , Ma, M. , Wu, L. , Zhou, M. , Li, M. , Wu, B. , Li, L. , Liu, X. , Jing, R. , Chen, W. and Zhao, H. (2022) Modified expression of TaCYP78A5 enhances grain weight with yield potential by accumulating auxin in wheat (Triticum aestivum L.). Plant Biotechnol. J., 10.1111/pbi.13704
Contributor Information
Ruilian Jing, Email: jingruilian@caas.cn.
Wei Chen, Email: chenwei0609@mail.hzau.edu.cn.
Huixian Zhao, Email: hxzhao212@nwafu.edu.cn.
Data availability statement
The raw data of the transcriptome in this study were deposited in NCBI’s Sequence Read Archive under BioProject number PRJNA718479.
References
- Adamski, N.M. , Anastasiou, E. , Eriksson, S. , O'Neill, C.M. and Lenhard, M. (2009) Local maternal control of seed size by KLUH/CYP78A5‐dependent growth signaling. Proc. Natl Acad. Sci. USA, 106, 20115–20120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou, E. , Kenz, S. , Gerstung, M. , MacLean, D. , Timmer, J. , Fleck, C. and Lenhard, M. (2007) Control of plant organ size by KLUH/CYP78A5‐dependent intercellular signaling. Dev. Cell, 13, 843–856. [DOI] [PubMed] [Google Scholar]
- Andres, F. and Coupland, G. (2012) The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. [DOI] [PubMed] [Google Scholar]
- Barrero, R.A. , Bellgard, M. and Zhang, X. (2011) Diverse approaches to achieving grain yield in wheat. Funct. Integr. Genomics, 11, 37–48. [DOI] [PubMed] [Google Scholar]
- Bradbury, P.J. , Zhang, Z. , Kroon, D.E. , Casstevens, T.M. , Ramdoss, Y. and Buckler, E.S. (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics, 23, 2633–2635. [DOI] [PubMed] [Google Scholar]
- Bustos, D.V. , Hasan, A.K. , Reynolds, M.P. and Calderini, D.F. (2013) Combining high grain number and weight through a DH‐population to improve grain yield potential of wheat in high‐yielding environments. Field Crop Res. 145, 106–115. [Google Scholar]
- Calderini, D.F. , Castillo, F.M. , Arenas, M.A. , Molero, G. , Reynolds, M.P. , Craze, M. , Bowden, S. et al. (2021) Overcoming the trade‐off between grain weight and number in wheat by the ectopic expression of expansin in developing seeds leads to increased yield potential. New Phytol. 230, 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti, M. , Zhang, N. , Sauvage, C. , Munos, S. , Blanca, J. , Canizares, J. , Diez, M.J. et al. (2013) A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc. Natl Acad. Sci. USA, 110, 17125–17130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.M. , Song, W.J. , Xie, X.M. , Wang, Z.H. , Guan, P.F. , Peng, H.R. , Jiao, Y.N. et al. (2020) A collinearity‐incorporating homology inference strategy for connecting emerging assemblies in the Triticeae tribe as a pilot practice in the plant pangenomic era. Mol. Plant, 13, 1694–1708. [DOI] [PubMed] [Google Scholar]
- Cheng, Y.F. , Dai, X.H. and Zhao, Y.D. (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, Q. , Guo, L. , Ma, M. , Zhang, L. , Mao, H. , Wu, B. , Liu, X. et al. (2019) Global transcriptome analysis uncovers the gene co‐expression regulation network and key genes involved in grain development of wheat (Triticum aestivum L.). Funct. Integr. Genomics, 19, 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Santo, S. , Tucker, M.R. , Tan, H.T. , Burbidge, C.A. , Fasoli, M. , Bottcher, C. , Boss, P.K. et al. (2020) Auxin treatment of grapevine (Vitis vinifera L.) berries delays ripening onset by inhibiting cell expansion. Plant Mol. Biol. 103, 91–111. [DOI] [PubMed] [Google Scholar]
- Foulkes, M.J. , Slafer, G.A. , Davies, W.J. , Berry, P.M. , Sylvester‐Bradley, R. , Martre, P. , Calderini, D.F. et al. (2011) Raising yield potential of wheat. III. Optimizing partitioning to grain while maintaining lodging resistance. J. Exp. Bot. 62, 469–486. [DOI] [PubMed] [Google Scholar]
- Gao, H. , Jin, M. , Zheng, X.M. , Chen, J. , Yuan, D. , Xin, Y. , Wang, M. et al. (2014) Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl Acad. Sci. USA, 111, 16337–16342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, P.K. , Langridge, P. and Mir, R.R. (2010) Marker‐assisted wheat breeding: present status and future possibilities. Mol. Breeding, 26, 145–161. [Google Scholar]
- Ishida, Y. , Tsunashima, M. , Hiei, Y. and Komari, T. (2015) Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol. Biol. 1223, 189–198. [DOI] [PubMed] [Google Scholar]
- IWGSC (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science, 361, 661. [DOI] [PubMed] [Google Scholar]
- Jiang, L. , Yoshida, T. , Stiegert, S. , Jing, Y. , Alseekh, S. , Lenhard, M. , Perez‐Alfocea, F. et al. (2021) Multi‐omics approach reveals the contribution of KLU to leaf longevity and drought tolerance. Plant Physiol. 185, 352–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev‐Yadun, S. , Gopher, A. and Abbo, S. (2000) Archaeology. The cradle of agriculture. Science, 288, 1602–1603. [DOI] [PubMed] [Google Scholar]
- Li, L. , Peng, Z. , Mao, X.G. , Wang, J.Y. , Chang, X.P. , Reynolds, M. and Jing, R.L. (2019a) Genome‐wide association study reveals genomic regions controlling root and shoot traits at late growth stages in wheat. Ann. Bot. 124, 993–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. , Xu, R. and Li, Y. (2019b) Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 70, 435–463 [DOI] [PubMed] [Google Scholar]
- Li, Y.J. , Yu, Y. , Liu, X. , Zhang, X.S. and Su, Y.H. (2021) The Arabidopsis MATERNAL EFFECT EMBRYO ARREST45 protein modulates maternal auxin biosynthesis and controls seed size by inducing AINTEGUMENTA. Plant Cell, 33, 1907–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado, P. and Rozas, J. (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451–1452. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2‐ΔΔ CT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma, M. , Yan, Y. , Huang, L. , Chen, M.S. and Zhao, H.X. (2012) Virus‐induced gene‐silencing in wheat spikes and grains and its application in functional analysis of HMW‐GS‐encoding genes. BMC Plant Biol. 12, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molero, G. , Joynson, R. , Pinera‐Chavez, F.J. , Gardiner, L.J. , Rivera‐Amado, C. , Hall, A. and Reynolds, M.P. (2019) Elucidating the genetic basis of biomass accumulation and radiation use efficiency in spring wheat and its role in yield potential. Plant Biotechnol. J. 17, 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa, N. , Hibara, K.I. , Heppard, E.P. , Vander Velden, K.A. , Luck, S. , Beatty, M. , Nagato, Y. et al. (2013) GIANT EMBRYO encodes CYP78A13, required for proper size balance between embryo and endosperm in rice. Plant J. 75, 592–605. [DOI] [PubMed] [Google Scholar]
- Pacheco‐Villalobos, D. , Diaz‐Moreno, S.M. , van der Schuren, A. , Tamaki, T. , Kang, Y.H. , Gujas, B. , Novak, O. et al. (2016) The effects of high steady state auxin levels on root cell elongation in Brachypodium. Plant Cell, 28, 1009–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat, G.C. , Alandete‐Saez, M. , Bowman, J.L. and Sundaresan, V. (2009) Auxin‐dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science, 324, 1684–1689. [DOI] [PubMed] [Google Scholar]
- Radchuk, V. , Weier, D. , Radchuk, R. , Weschke, W. and Weber, H. (2011) Development of maternal seed tissue in barley is mediated by regulated cell expansion and cell disintegration and coordinated with endosperm growth. J. Exp. Bot. 62, 1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamini, F. , Ozkan, H. , Brandolini, A. , Schafer‐Pregl, R. and Martin, W. (2002) Genetics and geography of wild cereal domestication in the near east. Nat. Rev. Genet. 3, 429–441. [DOI] [PubMed] [Google Scholar]
- Sauer, M. , Robert, S. and Kleine‐Vehn, J. (2013) Auxin: simply complicated. J. Exp. Bot. 64, 2565–2577. [DOI] [PubMed] [Google Scholar]
- Shao, A. , Ma, W. , Zhao, X. , Hu, M. , He, X. , Teng, W. , Li, H. et al. (2017) The auxin biosynthetic TRYPTOPHAN AMINOTRANSFERASE RELATED TaTAR2.1‐3A increases grain yield of wheat. Plant Physiol. 174, 2274–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry, P.R. , Mitchell, R.A.C. , Tosi, P. , Wan, Y.F. , Underwood, C. , Lovegrove, A. , Freeman, J. et al. (2012) An integrated study of grain development of wheat (cv. Hereward). J. Cereal Sci. 56, 21–30. [Google Scholar]
- Shi, L.L. , Song, J.R. , Guo, C.C. , Wang, B. , Guan, Z.L. , Yang, P. , Chen, X. et al. (2019) A CACTA‐like transposable element in the upstream region of BnaA9.CYP78A9 acts as an enhancer to increase silique length and seed weight in rapeseed. Plant J. 98, 524–539. [DOI] [PubMed] [Google Scholar]
- Shimizu‐Sato, S. , Tanaka, M. and Mori, H. (2009) Auxin‐cytokinin interactions in the control of shoot branching. Plant Mol. Biol. 69, 429–435. [DOI] [PubMed] [Google Scholar]
- Sun, X. , Cahill, J. , Van Hautegem, T. , Feys, K. , Whipple, C. , Novak, O. , Delbare, S. et al. (2017) Altered expression of maize PLASTOCHRON1 enhances biomass and seed yield by extending cell division duration. Nat. Commun. 8, 14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittarelli, A. , Milla, L. , Vargas, F. , Morales, A. , Neupert, C. , Meisel, L.A. , Salvo‐G, H. et al. (2007) Isolation and comparative analysis of the wheat TaPT2 promoter: identification in silico of new putative regulatory motifs conserved between monocots and dicots. J. Exp. Bot. 58, 2573–2582. [DOI] [PubMed] [Google Scholar]
- Villanueva, J.M. , Broadhvest, J. , Hauser, B.A. , Meister, R.J. , Schneitz, K. and Gasser, C.S. (1999) INNER NO OUTER regulates abaxial–adaxial patterning in Arabidopsis ovules. Genes Dev. 13, 3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F. , Fang, J. , Ou, S. , Gao, S. , Zhang, F. , Du, L. , Xiao, Y. et al. (2015) Variations in CYP78A13 coding region influence grain size and yield in rice. Plant Cell Environ. 38, 800–811. [DOI] [PubMed] [Google Scholar]
- Yoo, S.D. , Cho, Y.H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang, S.J. , Zhang, R.Z. , Song, G.Q. , Gao, J. , Li, W. , Han, X.D. , Chen, M.L. et al. (2018) Targeted mutagenesis using the Agrobacterium tumefaciens‐mediated CRISPR‐Cas9 system in common wheat. BMC Plant Biol. 18, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B. , Dai, A. , Wei, H. , Yang, S. , Wang, B. , Jiang, N. and Feng, X. (2016) Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 90, 33–47. [DOI] [PubMed] [Google Scholar]
- Zhao, Z. , Zhang, Y. , Liu, X. , Zhang, X. , Liu, S. , Yu, X. , Ren, Y. et al. (2013) A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev. Cell, 27, 113–122. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Zhao, X.B. , Li, Y.W. , Xu, J. , Bi, A.Y. , Kang, L.P. , Xu, D.X. et al. (2020) Triticum population sequencing provides insights into wheat adaptation. Nat. Genet. 52, 1412–1422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The sequence characteristics and phylogenetic tree of TaCYP78A5 in wheat.
Figure S2 The genetic loci of yield‐related traits on the short arms of chromosome 2A, 2B and 2D in wheat.
Figure S3 TaCYP78A5 silencing resulted in decreased seed size and seed coat cell number.
Figure S4 The ripening time of UBI::TaCYP78A5‐transgenic wheat line (UBI‐4) and wild‐type plants (WT).
Figure S5 Grain yield per plant of UBI::TaCYP78A5‐transgenic wheat lines (UBI‐1 and UBI‐2) and wild‐type plants (WT).
Figure S6 The yield‐related traits of transgenic wheat lines and wild‐type wheat.
Figure S7 Validation of RNA‐seq data by quantitative real‐time RT‐PCR (qPCR).
Figure S8 KEGG enrichment (a) and GO enrichment (b) of differentially expressed genes (DEGs) between wild‐type wheat and transgenic line pINO‐24.
Figure S9 Relative levels of cytokinin in the ovaries of wild‐type wheat and transgenic lines.
Figure S10 The correlation analysis between the auxin concentration and the relative expression of pINO lines.
Figure S11 Auxin affects grain weight of wheat.
Figure S12 Localized overexpression of TaCYP78A5 in integument causes delayed flowering and prolongs proliferation of maternal integument/seed coat cells.
Figure S13 Exogenous auxin treatment causes delayed flowering in wheat.
Table S1 The known genetic loci of yield‐related traits on the short arms of chromosome 2A, 2B and 2D in wheat
Table S2 The quality traits of pINO lines and WT
Table S3‐1 The differentially expressed genes between wild‐type wheat (WT) and transgenic line pINO‐24
Table S3‐2 The differentially expressed genes involved in hormone signal
Table S3‐3 The differentially expressed genes associated with IAA metabolism, transport and signalling
Table S4‐1 The signalling genes that response for AUXIN
Table S4‐2 The cell‐wall‐related process
Table S4‐3 The cell‐wall‐related genes regulated by AUXIN
Table S5 Thirty cultivars used for detecting the natural allelic variations of TaCYP78A5 in wheat
Table S6 Basic information of 323 wheat accessions used for association analysis between TaCYP78A5‐2A haplotypes and agronomic traits
Table S7 Basic information of wheat landraces and cultivars used for analysis of evolutionary history
Table S8 The information of the primers used in this study
Appendix S1 Supplemental Methods
Data Availability Statement
The raw data of the transcriptome in this study were deposited in NCBI’s Sequence Read Archive under BioProject number PRJNA718479.


