Summary
Legume–rhizobia symbiosis enables biological nitrogen fixation to improve crop production for sustainable agriculture. Small heat shock proteins (sHSPs) are involved in multiple environmental stresses and plant development processes. However, the role of sHSPs in nodule development in soybean remains largely unknown. In the present study, we identified a nodule‐localized sHSP, called GmHSP17.9, in soybean, which was markedly up‐regulated during nodule development. GmHSP17.9 was specifically expressed in the infected regions of the nodules. GmHSP17.9 overexpression and RNAi in transgenic composite plants and loss of function in CRISPR‐Cas9 gene‐editing mutant plants in soybean resulted in remarkable alterations in nodule number, nodule fresh weight, nitrogenase activity, contents of poly β‐hydroxybutyrate bodies (PHBs), ureide and total nitrogen content, which caused significant changes in plant growth and seed yield. GmHSP17.9 was also found to act as a chaperone for its interacting partner, GmNOD100, a sucrose synthase in soybean nodules which was also preferentially expressed in the infected zone of nodules, similar to GmHSP17.9. Functional analysis of GmNOD100 in composite transgenic plants revealed that GmNOD100 played an essential role in soybean nodulation. The hsp17.9 lines showed markedly more reduced sucrose synthase activity, lower contents of UDP‐glucose and acetyl coenzyme A (acetyl‐CoA), and decreased activity of succinic dehydrogenase (SDH) in the tricarboxylic acid (TCA) cycle in nodules due to the missing interaction with GmNOD100. Our findings reveal an important role and an unprecedented molecular mechanism of sHSPs in nodule development and nitrogen fixation in soybean.
Keywords: biological nitrogen fixation, small heat shock proteins (sHSPs), soybean nodule, chaperone, sucrose synthase, yield
Introduction
Nitrogen, one of the key macronutrients in living beings, is abundant in the atmosphere and cannot be directly assimilated by crop plants. Available nitrogen deficiency in soil limits the productivity and production of crop plants. To achieve maximum crop yield, excessive industrial nitrogen fertilizer has been applied in the field (Mueller et al., 2012). Nevertheless, it is estimated that only 50% of the applied nitrogen fertilizer is captured by crop plants due to leaching into the underground water, which leads to environmental pollution (Erisman et al., 2011). Thus, it is important to find a natural and efficient approach to replace nitrogen fertilizer to increase crop production. Legume plants are of great interest for this purpose owing to their distinctive ability for biological nitrogen fixation (BNF). The prospect of increasing BNF importance in agriculture is currently a hot topic. BNF is the most efficient method of N‐fixation with amounts of 20–300 kg/ha/an, which accounts for approximately 60% of all fixed N on earth (Soumare et al., 2020). Researchers have attempted to explore genes that improve nodule formation and development in legume plants. For example, an SHR‐SCR module in the roots of the legume, Medicago truncatula, enabled cortical cell division and, consequently, de novo nodule organogenesis (Dong et al., 2021). The glycogen synthase kinase 3 (GSK3)‐like kinase, GmSK2‐8, inhibits symbiotic signalling and nodule formation in soybean under salt stress by phosphorylating GmNSP1 (He et al., 2021).
Soybean (Glycine max Merr.), the most widely cultivated legume plant in the world, symbiotically associates with nitrogen‐fixing rhizobia in the soil for root nodule formation. The fixed N products in nodules are in the form of ureides (allantoin and allantoic acids), which are transported to shoots through the xylem and used as nitrogen sources for plant growth and development (Collier and Tegeder, 2012; Ferguson et al., 2010; Oldroyd and Downie, 2008). Nodule formation and BNF are complicated and energy consuming, and soybean plants have developed strategies to regulate nodule numbers, such as long‐distance autoregulation feedback mechanisms, and achieve high BNF efficiency (Li et al., 2015; Magori and Kawaguchi, 2009; Marsh et al., 2007). Therefore, a better understanding of the molecular mechanisms and optimization of the process of BNF are urgently needed for crop growth and yield. In soybean, GmPT7, a phosphate transporter, was found to be localized in the fixation zone of nodules. The overexpression of GmPT7 promotes nodulation and shoot nitrogen, leading to increased soybean yield by up to 36% (Chen et al., 2019). In addition, with the inoculation of effective rhizobia in the field, nodule fresh weight and total N content of soybean were markedly improved, and soybean yield was increased by 92% compared to the non‐inoculated control, suggesting that effective nodulation enhances soybean yield through BNF (Qin et al., 2012).
Small heat shock proteins (sHSPs) are a large group of proteins present in all organisms that are induced by heat shock stress as well as other stress conditions (Carra et al., 2017; Eyles and Gierasch, 2010; Poulain et al., 2010; Waters, 2013; Waters and Rioflorido, 2007). sHSPs contain a conserved α‐crystallin domain (ACD) flanked by an N‐terminal domain (NTD) and a carboxy‐terminal extension domain (CTD) (Carra et al., 2017; Haslbeck and Vierling, 2015; Jacob et al., 2017; Sun and MacRae, 2005). Structural studies have demonstrated that sHSPs exist in the form of monomers, dimers and large multimers, which appear to be necessary for chaperone functions (Candido, 2002; McDonald et al., 2012). The sHSPs were first characterized and named based on their high expression during heat shock stress. Currently, sHSPs are known to be ubiquitous molecular chaperones that assist in protein folding and prevent irreversible protein aggregation (Basha et al., 2006; Haslbeck and Vierling, 2015; Karney‐Grobe et al., 2018; Smykal et al., 2000; Waters, 2013). Wheat sHSP16.9 and pea sHSP18.1 act as chaperones with specific substrates through their N‐terminal arm (Basha et al., 2006). Tobacco sHSP18 expressed in E. coli can form oligomers and exhibit ATP‐dependent chaperone activity (Smykal et al., 2000).
As chaperones for a wide range of clients, sHSPs participate in the regulation of many biological processes, such as oxidative stress, cell cycle and plant development (Basha et al., 2004a; Derham et al., 2003; Haslbeck et al., 2004; Reissig et al., 2018). Chloroplast sHSP21, which interacts with pTAC5 in Arabidopsis, is involved in chloroplast development under heat stress (Zhong et al., 2013). In ‘Micro‐Tom’ tomato fruits, sHSP23.6 is related to post‐harvest abiotic stress, such as hypoxia (Reissig et al., 2018). In addition, in tomato (Solanum lycopersicum), SlHSP17.7 is involved in sugar metabolism to improve fruit quality (Zhang et al., 2018b). However, little is known about the role of sHSPs in nodule formation and nitrogen fixation in legume plants. In this study, we reported a cytoplasmic‐localized sHSP in nodules, GmHSP17.9, which was required for nodule growth, nitrogen fixation and yield based on proteome analysis, transgenic assays, gene editing and yield‐trait genotyping. Furthermore, the results showed that GmHSP17.9 directly interacted with GmNOD100 to regulate symbiotic nitrogen fixation and seed yield. Our work illuminates an unprecedented molecular mechanism of sHSPs in plant nodule development and lays a foundation for cultivation of soybean varieties with high nitrogen‐fixing ability via biotechnology approach.
Results
GmHSP17.9 encodes a chaperone protein in soybean nodules
Through proteomic analysis of soybean nodules, we found several sHSPs, among which GmHSP17.9 showed markedly higher expression levels than any other sHSPs (Figure S1a). Sequence analysis revealed that the length of the predicted mature transcript of GmHSP17.9 was 878 bp with one exon and no intron, and had 160 bp corresponding to the 5’ untranslated region (UTR), 241 bp to the 3’ UTR and 477 bp to an open reading frame (ORF) (Figure 1a). The ORF of GmHSP17.9 encoded a predicted protein of 158 amino acid residues with a conserved ACD (50–139 aa) in the middle region flanked by less conserved N‐ (1–49 aa) and C‐ (140–158 aa) terminal sequences, similar to its homologs in other plant species (Figure 1b and Figure S1b). Phylogenetic analysis revealed that GmHSP17.9 belonged to the CII subfamily in the angiosperm kingdom, which was reported to be localized in the cytoplasm (Figure S1c) (Waters, 2013). To determine the subcellular distribution of GmHSP17.9, Arabidopsis protoplasts were transfected with a construct expressing GmHSP17.9‐GFP fusion or GFP only under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Fluorescence was detected in the cytoplasm of Arabidopsis protoplasts expressing GmHSP17.9‐GFP, similar to the GFP control (Figure 1c). In addition, protoplasts of roots expressing GmHSP17.9‐GFP or GFP constructs in transgenic composite soybean plants were isolated, and the fluorescence pattern of GmHSP17.9‐GFP or GFP was also observed in the cytoplasm (Figure 1d).
Figure 1.
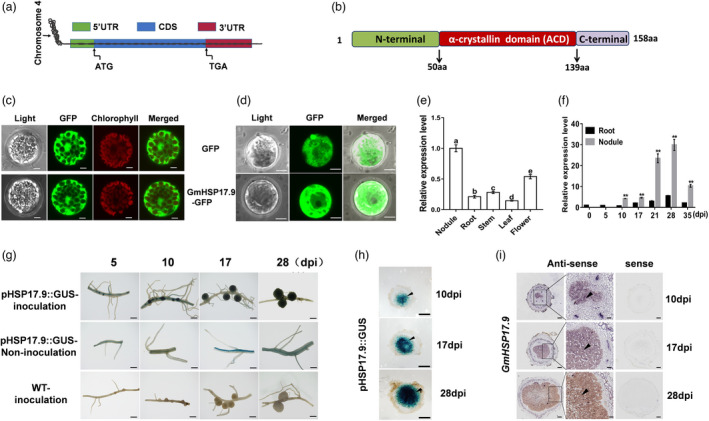
Expression pattern and localization of GmHSP17.9 in soybean nodules. (a) Schematic representation of gene structure showed that GmHSP17.9 was located on chromosome 4 with only one exon and no intron in the genomic sequence. (b) Domain structure of GmHSP17.9 protein. The conserved α‐crystallin domain (ACD) (50–139 aa) in the middle region flanked by less conserved N‐ (1–49 aa) and C‐ (140–158aa) terminal sequences are indicated. (c) Subcellular localization of GmHSP17.9 protein in Arabidopsis protoplasts. Fluorescence of GmHSP17.9‐GFP was observed in the cytoplasm of Arabidopsis protoplasts. The free GFP (empty vector) used as control was distributed in both nucleus and cytoplasm. GmHSP17.9‐GFP: GmHSP17.9 and GFP fusion protein. Scale bars = 10 μm. (d) Subcellular localization of GmHSP17.9 protein in soybean root protoplasts. Transgenic composite soybean plants overexpressing GmHSP17.9‐GFP were generated and protoplasts were isolated from positive roots expressing GmHSP17.9‐GFP. GFP fluorescence was observed by confocal fluorescence microscope and GFP vector alone was used as cytosolic marker. Scale bars = 5 μm. (e, f) Expression profiles of GmHSP17.9 in various tissues of soybean including root, nodule, stem, leaf, flower (e) and in rhizobia‐inoculated roots (0, and 5 dpi), nodules (10, 17, 28 and 35 dpi) and nodule‐detached roots (f). The relative expression value was normalized based on the expression of GmActin11 (Glyma.18g290800) used as reference gene. In (e), the different letters indicate significant differences, P < 0.05. Each error bar represents the mean of four biological replicates with ± SE. In (f), asterisks indicate significant difference within a P level in t‐tests. **, P < 0.01. (g) Histochemical localization analysis of GUS expression driven by the promoter of GmHSP17.9 (pHSP17.9::GUS) in transgenic composite soybean roots and nodules at different developmental stages. Three independent experiments were performed, and images from one representative experiment were shown here (n > 10). Scale bar = 1 mm. (h) Tissue‐specific expression of GmHSP17.9 in soybean nodules. Nodules were stained with X‐Gluc, and frozen sections were observed with a microscope. Black arrow head indicated the infected cells of nodules. Scale bar = 500 μm. (i) RNA in situ hybridization analysis of expression of GmHSP17.9 in soybean nodules. Digoxigenin (DIG) labelled antisense or sense probe was used for detection of the transcripts. Black arrow head indicated the infected cells of nodules. Section thickness was 5 μm. Scale bar =200 μm (left panel of antisense and sense) and 100 μm (right panel of antisense).
The molecular chaperone activity of GmHSP17.9 was monitored by measuring the thermal aggregation of malate dehydrogenase (MDH) and chemically induced aggregation of insulin as substrates. We found that the GmHSP17.9 proteins efficiently prevented the thermal aggregation of MDH at 45 °C and chemically induced aggregation of insulin by DTT (Figure S2a,b). When the GmHSP17.9 proteins were replaced in the reaction mixture with GFP as a control, no holdase activity was detected. Previously, the oligomerization status of sHSPs was reported to be associated with its chaperone activity (Carra et al., 2017; Klein et al., 2014; Mymrikov et al., 2020). Here, recombinant GmHSP17.9 proteins were treated with ß‐mercaptoethanol and only monomers were detected in reducing SDS‐PAGE stained by Coomassie Brilliant Blue (CBB) as well as GFP (Figure S2c). When recombinant GmHSP17.9 proteins were analysed on SDS‐PAGE under non‐reducing condition, the GmHSP17.9 proteins could predominantly form higher oligomeric complexes, while GFP proteins could not (Figure S2d). These findings demonstrate that GmHSP17.9 acts as a classical molecular chaperone in soybean nodules.
GmHSP17.9 is preferentially expressed in the infected cells of nodules in soybean
The spatiotemporal expression pattern of GmHSP17.9 was determined in different organs such as nodules, root, stem, leaves, and flowers. The results showed that GmHSP17.9 was mainly expressed in nodules, suggesting a role of GmHSP17.9 in nodulation (Figure 1e). The transcript abundance of GmHSP17.9 was enhanced at 10 dpi until it peaked at 28 dpi in the nodules, and then dramatically decreased at 35 dpi. In addition, the expression of GmHSP17.9 was markedly induced in soybean nodules than in nodule‐detached roots at all time points (Figure 1f). To further precisely determine the tissue‐specific expression of GmHSP17.9 in nodules, transgenic soybean roots and nodules containing β‐glucuronidase (GUS) reporter gene (pHSP17.9::GUS) were generated through hairy roots transformation. Histochemical GUS staining analysis was performed in transgenic hairy roots inoculated with rhizobia at different stages of nodule growth. The results showed that GmHSP17.9 was mainly expressed in roots and nodule primordium at 5 and 10 dpi, and was preferentially expressed in the nodules at 17 and 28 dpi compared to the uninoculated transgenic soybean hairy roots and wild‐type control (Figure 1g). In addition, GUS staining was visualized mainly in the infection zone of the transverse sections of the transgenic nodules during nodule development (Figure 1h).
To further confirm the expression specificity of GmHSP17.9 in soybean nodules, we performed RNA in situ hybridization analysis. Sections of nodules at different developmental stages were hybridized with sense and antisense probes specific for GmHSP17.9. As shown in Figure 1i, the transcript location of GmHSP17.9 was mainly found in the central infected zones during nodule development until mature and these results were consistent with GUS staining. Not surprisingly, no signals were found in any of the nodule sections hybridized with the sense probe. Taken together, these results indicate that GmHSP17.9 is involved in nodule development in soybean.
GmHSP17.9 plays a key role in nodulation, plant growth and seed yield in soybean
To elucidate the possible roles of GmHSP17.9 in nodule development, we generated transgenic composite soybean plants overexpressing (OX) or suppressing (RNAi) GmHSP17.9. We found that the altered expression of GmHSP17.9 in transgenic composite plants significantly affected soybean nodulation and plant growth (Figure 2a,b). qRT‐PCR showed that the transcript accumulation of GmHSP17.9 was threefold higher in the OX lines and 50% lower in the RNAi lines than in the wild‐type control (Figure 2c). The total nodule number and nodule fresh weight increased in the OX lines by 30.2% and 65.9%, respectively, and decreased in the RNAi lines by 26.1% and 42.6% respectively (Figure 2d,e). In addition, the OX lines showed increased nitrogenase activity by 28.2%, while the RNAi lines showed a 33.3% reduction compared to the control (Figure 2f). Importantly, the OX lines exhibited significantly enhanced soybean growth, with increases of 48.9%, 26.1%, 17.1% and 12.5% in plant height, shoot fresh weight, shoot dry weight and N content, respectively, compared with the control. In contrast, the RNAi lines displayed reduced soybean growth, with decreases of 25.9%, 15.6%, 22.4% and 25.1% in plant height, shoot fresh weight, shoot dry weight and N content respectively (Figure 2g–j).
Figure 2.
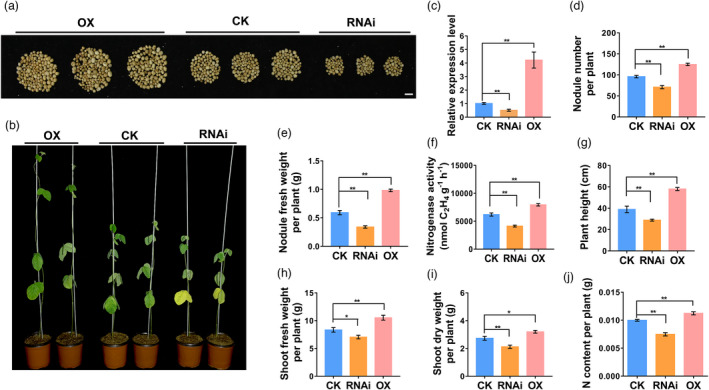
Phenotypic analysis of nodulation of transgenic composite soybean plants overexpressing (OX) or suppressing (RNAi) GmHSP17.9. (a, b) Growth performance of nodules (a) and composite transgenic soybean plants (b) at 28 dpi. Scale bar in (a) =1 cm. (c) Relative expression level of GmHSP17.9 in nodules at 28 dpi. Relative expression level was calculated as the fold change of the expression value of GmHSP17.9 in transgenic nodules to that of in wild type and was normalized based on the expression of GmActin11 used as reference gene. (d) Nodule number. (e) Nodule fresh weight. (f) Nitrogenase activity measured by the acetylene reduction assay. (g) Plant height. (h) Shoot fresh weight. (i) Shoot dry weight. (j) N content. CK refers to transgenic plants carrying empty vector. These experiments were repeated at least three times and similar results were obtained. Data are means with SE from three independent replicates (n = 10).
To further evaluate the role of GmHSP17.9 on nodule development and yield in soybean, hsp17.9 mutant lines were generated using the CRISPR‐Cas9 technology. Two target sites in the ORF region of GmHSP17.9 were selected and introduced into a binary vector under the control of the GmU6 promoter, followed by Cas9 recognition and cleavage of gRNA sites (Figure S3a,b). Two homozygous hsp17.9 mutant lines were obtained by genomic PCR and sequencing analysis of target sites using specific primers (Figure S3c,d). The mutant line, hsp17.9‐1, had a 1‐bp insertion in Target 1 and a 5‐bp deletion in Target 2 while the mutant line, hsp17.9‐2, had a 98‐bp insertion and a 6‐bp deletion in Target 2. The insertion and deletion in the two mutant lines led to frameshift mutations and the production of premature stop codons in the ORF region (Figure S3e,f). We tested these insertions and deletions in each generation until two independent stable mutant lines (T3 generation) were obtained. Six sHSP proteins containing GmHSP17.9 were previously reported to belong to the CII subfamily according to their amino acid sequences in soybeans (Figure S4a) (Lopes‐Caitar et al., 2013). Here, through PCR and sequencing, we found that there was no change in the genomic DNA sequences of the other five sHSP genes, except GmHSP17.9, in the two mutant lines compared with wild‐type plants. When the transcript accumulation of the six sHSP genes was examined, no expression of GmHSP17.9 was detected, whereas the transcripts of the other five sHSPs remained unchanged in the two hsp17.9 lines compared with wild‐type plants (Figure S4b). The two hsp17.9 lines were inoculated with rhizobia under N‐free nutrient conditions, and the nodule phenotypes were assessed at 28 dpi. The formation of root nodules in the two hsp17.9 lines was markedly reduced (Figure 3a,b). Compared with wild‐type plants, the two hsp17.9 lines displayed 39.8% and 43.1% decreases in nodule numbers, and 48.8% and 53.6% decreases in nodule fresh weight per plant, respectively, followed by a 33.6% and 38.3% decrease in nodule size (Figure 3c–e). Aligning with the defective nodule fresh weight and nodule size, nitrogenase activity in the hsp17.9 lines was decreased by 29.2% and 37.5%, respectively, compared with those in wild‐type plants (Figure 3f). Consistently, the expression of GmLbc3, which encodes leghemoglobin for nitrogen fixation, was also decreased in the hsp17.9 lines (Figure 3g). The ultrastructure of nodules showed that most bacteria in the nodules of the two hsp17.9 lines were not enclosed by the symbiosome membrane, indicating damages to symbiosome formation. Interestingly, we also found a markedly lower accumulation of poly β‐hydroxybutyrate bodies (PHBs) in the bacteroids of the nodules of the two hsp17.9 lines, with decreases of 32.7% and 45.7% compared with that of the wild‐type nodules (Figure 3h,i). PHB was synthesized in the bacteroids from acetyl‐CoA, implying the involvement of the tricarboxylic acid (TCA) cycle in soybean nodulation (Trainer and Charles, 2006).
Figure 3.
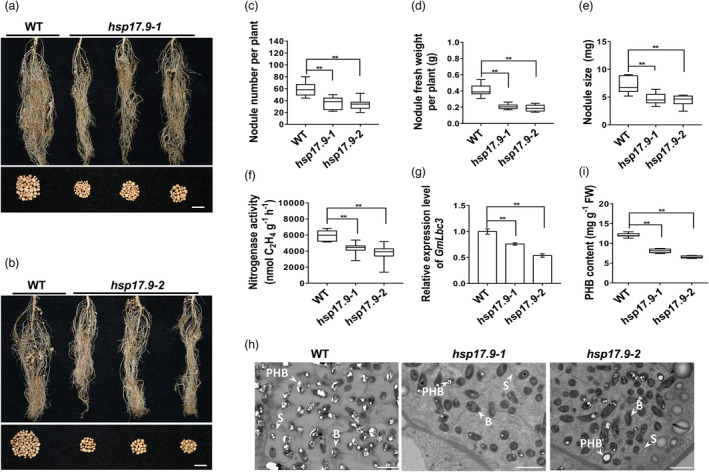
Effect of altered expression of GmHSP17.9 on nodulation of hsp17.9 plants at 28 dpi. (a, b) Nodule growth performance of hsp17.9‐1 (a) and hsp17.9‐2 (b) at 28 dpi. (c) Nodule number per plant. (d) Nodule fresh weight per plant. (e) Nodule size (single nodule weight). (f) Nitrogenase activity of nodules measured by the acetylene reduction assay. Wild‐type (WT) Williams 82 and hsp17.9 plants were grown in the vermiculite with rhizobia inoculation. (g) Quantitative real‐time RT‐PCR analysis of GmLbc3 expression in nodules. (h) Ultrastructure of nodules of hsp17.9 plants. The ultrastructure was observed by transmission electron microscopy (TEM) (n = 5). PHB: poly‐β‐hydroxybutyrate bodies; S: symbiosome; B: bacteroids. (i) Content of PHB in nodules. PHB was extracted by sodium hypochlorite in chloroform, and the turbidity was monitored at 530 nm using a fluorescence spectrophotometer. FW: fresh weight. Data presented are averaged from three independent experiments (n = 10) and similar data were obtained. For each box plot, the centre black line represents the median; the box limits are the upper and lower quartiles; the whiskers represent the lowest or highest data point within the 1.5 interquartile range of the lower or upper quartile; and the dot outliers are the data points outside the 1.5 interquartile range of the lower or upper quartile. (a,b) Scale bar = 1 cm.
Significantly reduced growth was observed in the two hsp17.9 lines after inoculation with rhizobia under N‐free nutrient condition at 28 dpi (Figure 4a,b), with decreases of 38.5% and 43.6% in plant height, 37.8% and 47.2% in plant fresh weight and 39.4% and 46.5% in dry weight, respectively, compared to those of wild‐type plants (Figure 4c–e). Furthermore, we evaluated the soybean yield traits of the two hsp17.9 lines in the field and found that they showed a significant decrease of 50.5% and 52.1% in pod number, 40.8% and 46.3% in seed number, and 41.4% and 46.0% in seed weight per plant, respectively, compared with the wild‐type lines (Figure 4f–k). Taken together, these results indicate that GmHSP17.9 plays a key role in nodulation, plant growth and yield in soybean.
Figure 4.
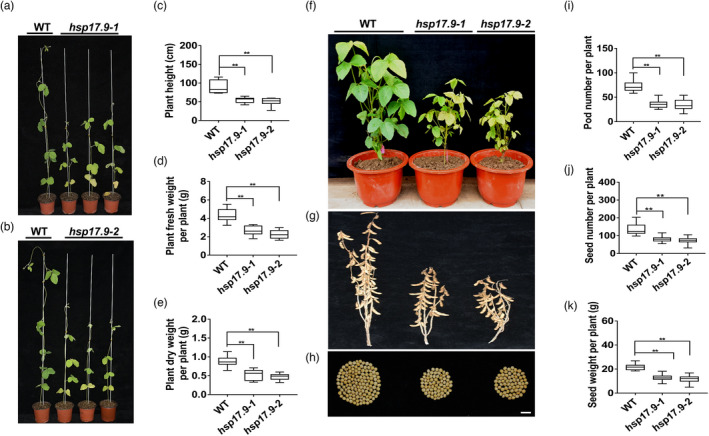
Phenotype and yield analysis of hsp17.9 plants. (a, b) Growth performance of the hsp17.9 plants. Box plots of plant height (c), plant fresh (d) and dry weight (e) per plant. Soybean seeds were germinated for 3 days and then incubated with Bradyrhizobium diazoefficiens USDA110 in vermiculite watered with a nitrogen‐free nutrient solution. The phenotype of the mutants was analysed at 28 dpi. (f–h) Growth performance of hsp17.9 plants in the field condition. Growth phenotype (f), pods (g) and seeds (h) of the two mutant plants. (i‐k) Box plots of pod number (i), seed number (j) and seed weight (k) per plant. Soybean seeds were planted in the pots containing vermiculite in the field condition. Fifty millilitres of USDA110 solution (OD600 = 0.08) was applied to each pots. The pod number, seed number and weight were determined at soybean maturation stage (n = 20). For each box plot in c‐e, i‐k: the centre black line represents the median; the box limits are the upper and lower quartiles; the whiskers represent the lowest or highest data point within the 1.5 interquartile range of the lower or upper quartile; and the dot outliers are the data points outside the 1.5 interquartile range of the lower or upper quartile. (h) Scale bar = 1 cm.
GmHSP17.9 interacts with GmNOD100 in the nodules of soybean
To uncover the molecular mechanism by which GmHSP17.9 regulates nodule development, we sought to identify the interacting proteins of GmHSP17.9 from soybean nodules by liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) (Figure S5a). A subunit of sucrose synthase, encoded by Glyma.17G045800, was identified as top candidate of GmHSP17.9 because its largest number of matches of MS/MS spectra and was called GmNOD100 (Table S1). To confirm the physical interaction between GmHSP17.9 and GmNOD100 in the cytoplasm in vivo, a bimolecular fluorescence complementation (BiFC) assay was carried out using Arabidopsis protoplasts. A strong yellow fluorescence (YFP) signal was observed in the cytoplasm when GmHSP17.9‐YFPN and GmNOD100‐YFPC or GmHSP17.9‐YFPC and GmNOD100‐YFPN were co‐expressed in Arabidopsis protoplasts, and a YFP signal was observed when the combination of GmHSP17.9‐YFPN and GmHSP17.9‐YFPC was expressed, suggesting that GmHSP17.9 may form homodimers or oligomers in vivo (Figure 5a). To identify which region in GmHSP17.9 is responsible for GmNOD100 binding, three truncated GmHSP17.9s (1–49, 50–139 and 140–158 amino acids) were generated. Based on the BiFC assay, only the ACD region could interact with GmNOD100 (Figure S5b). The interaction between GmHSP17.9 and GmNOD100 in either direction (BD‐GmHSP17.9 and AD‐NOD100; BD‐GmNOD100 and AD‐GmHSP17.9) was further tested in the yeast two‐hybrid assay followed by a co‐immunoprecipitation (Co‐IP) assay (Figure 5b,c). Finally, the interaction between these two proteins was further demonstrated by an in vitro pull‐down assay using recombinant proteins purified from E. coli. (Figure 5d). These findings illustrate that GmHSP17.9 directly interacts with GmNOD100.
Figure 5.
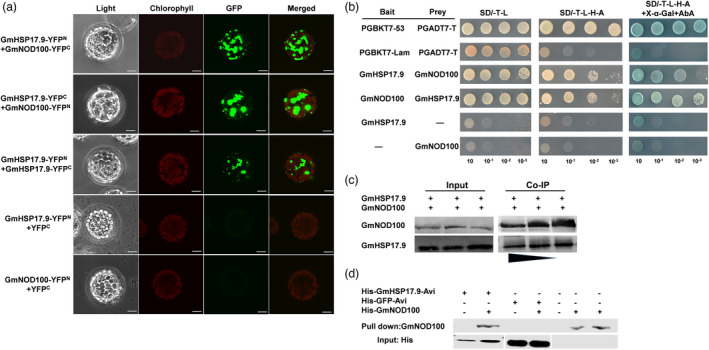
Interaction of GmHSP17.9 and GmNOD100. (a) BiFC analysis of interaction between GmHSP17.9 and GmNOD100 in Arabidopsis protoplasts. GmHSP17.9‐YFPN and GmNOD100‐YFPC or GmHSP17.9‐YFPC and GmNOD100‐YFPN were co‐transformed into Arabidopsis protoplasts. The YFP fluorescent signal was specifically detected in the cytoplasm. When GmHSP17.9‐YFPN or GmNOD100‐YFPN co‐expressed with empty vector YFPC, no fluorescence signal was detected. (b) Y2H assay showed the interaction between GmHSP17.9 and GmNOD100. Positive yeast strains were selected on SD/‐T‐L medium and further verified on SD/‐T‐L‐H‐A medium containing 125 ng/mL AbA and 40 µg/mL X‐α‐Gal. (c) Interaction of GmHSP17.9 and GmNOD100 revealed by CoIP assay. Anti‐GmHSP17.9 and anti‐GmNOD100 antibodies were used to detect GmHSP17.9 and GmNOD100 proteins respectively. These experiments were repeated three times with similar results. (d) GmHSP17.9 directly interacted with GmNOD100 in vitro by pull‐down assay. Pull‐down assay was performed using recombinant His‐GmHSP17.9‐Avi protein purified by streptavidin agarose resin and the total cell lysates of His‐GmNOD100. Input and pull‐down proteins were separated by 12% SDS‐PAGE and detected by Western blot with anti‐his antibody. (a) Scale bars = 10 μm.
GmNOD100 is responsible for nodule development in soybean
The expression pattern of GmNOD100 in various organs of soybean was determined by analysing the RNA‐Seq data in SoyBase as well as our qRT‐PCR results, which revealed that GmNOD100 was mainly expressed in soybean nodules and its transcript abundance gradually increases during nodulation (Figure S6a,b and Figure 6a). RNA in situ hybridization analysis revealed that GmNOD100 was specifically expressed in the infection regions of nodules, similar to GmHSP17.9 (Figure 6b). Furthermore, we found that GmNOD100 was localized in the cytoplasm of Arabidopsis protoplasts (Figure 6c). To explore the function of GmNOD100 in soybean nodulation, composite transgenic soybean plants were generated by either overexpression or suppression of GmNOD100 (Figure 6d). The expression level of GmNOD100 was 4.3‐fold higher in overexpressing lines and 65.8% lower in suppressing lines than in the control (Figure 6e). The nodule number, nodule fresh weight and nitrogenase activity were increased by 51.8%, 29.1% and 26.7% in overexpressed transgenic plants and decreased by 30.3%, 67.9% and 27.9% in RNAi plants of GmNOD100 respectively (Figure 6f–h). Consequently, the sucrose synthase activity, UDP‐glucose, acetyl coenzyme A (acetyl‐CoA) and activity of succinic dehydrogenase (SDH) in the tricarboxylic acid (TCA) cycle increased by 54.1%, 36.9%, 27.7% and 15.1% in nodules of overexpressed lines, while decreased by 51.9%, 38.4%, 26.3% and 38.6% in RNAi lines respectively (Figure 6i–l). Overexpression of GmNOD100 resulted in an increase of 26.8%, 55.6%, 34.7% and 67.6% while suppression of GmNOD100 caused a decrease of 30.9%, 22.5%, 21.6% and 32.2% in plant height, shoot fresh and dry weight and total N content, respectively, compared with those of the control (Figure S7). These findings indicate that GmNOD100 plays an important role in nodule development and nitrogen fixation, promoting soybean growth and development.
Figure 6.
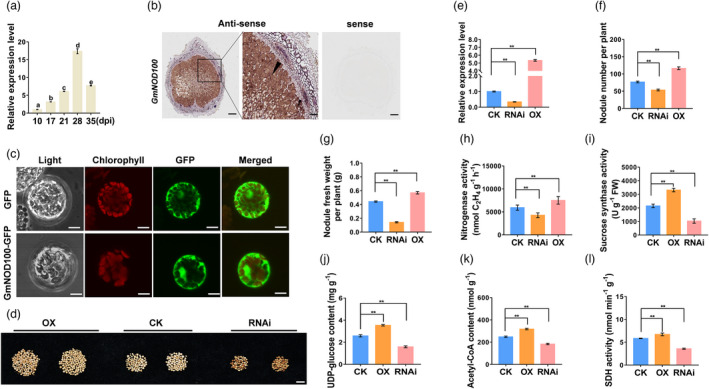
Phenotypic analysis of nodulation of transgenic composite soybean plants overexpressing (OX) or suppressing (RNAi) GmNOD100. (a) Expression patterns of GmNOD100 in nodules at 10, 17, 28 and 35 dpi. The different letters indicate significant differences, P < 0.05. (b) RNA in situ hybridization analysis of expression of GmNOD100 in nodules. Black arrow head indicated the infected cells of nodules. Scale bar = 200 μm (left panel of antisense and sense) and 100 μm (right panel of antisense). (c) Subcellular localization of GmNOD100‐GFP in Arabidopsis protoplasts. GmNOD100‐GFP signal was distributed in the cytoplasm. GmNOD100‐GFP: GmNOD100 and GFP fusion protein. Scale bars = 10 μm. (d) Phenotype of nodules at 28 dpi. Scale bar = 1 cm. (e) Relative expression level of GmNOD100 in nodules at 28 dpi. Relative expression level was calculated as the fold change of the expression value of GmNOD100 in transgenic nodules to that of in wild type and was normalized based on the expression of GmActin11. (f) Nodule number. (g) Nodule fresh weight. (h) Nitrogenase activity. (i) Sucrose synthase activity. (j) Content of UDP‐glucose. (k) Content of acetyl‐CoA. (l) Succinate dehydrogenase (SDH) activity. CK refers to transgenic nodules carrying empty vector. These experiments were repeated at least three times and similar results were obtained. Data are means with SE from three independent replicates (n = 10).
GmHSP17.9 promotes nitrogen fixation by interacting with GmNOD100 in soybean
To explore whether GmHSP17.9 is a chaperone for GmNOD100, recombinant purified GmNOD100 was denatured in vitro at 55 °C in the presence or absence of recombinant purified GmHSP17.9 and the sucrose synthase activity of denatured GmNOD100 was monitored. In the presence of recombinant GmHSP17.9, the activity of denatured GmNOD100 was rescued more than that of the GFP control (Figure 7a). In nodules of the hsp17.9 lines, transcript abundance of GmNOD100 did not change; however, the sucrose synthase activity decreased by 14.3% and 20.9%, respectively, compared to that of wild‐type plants. Such findings indicate that GmHSP17.9 regulates GmNOD100 at the protein level (Figure 7b,c). Owing to the reduced sucrose synthase activity, the content of UDP‐glucose, a product of sucrose synthase, decreased by 39.8% and 46.7%, respectively, in the two hsp17.9 lines (Figure 7d). In nodules, UDP‐glucose is catalysed via glycolysis, and in the form of acetyl‐CoA, enters the TCA cycle, providing a carbon source and energy for nitrogen fixation in nodules (Liu et al., 2018). As expected, the acetyl‐CoA content and activity of succinic dehydrogenase (SDH), a key enzyme in the TCA cycle, decreased by 23.4% and 12.4%, and 41.8% and 51.0% in nodules of the two hsp17.9 lines respectively (Figure 7e,f). Consequently, the ureide and N contents of the whole plant in the two hsp17.9 lines were significantly reduced by 24.6% and 23.6%, and 50.3% and 59.0% respectively (Figure 7g,h). Taken together, we conclude that GmHSP17.9 acts as a chaperone in nodules for GmNOD100 to regulate nodule development and nitrogen fixation.
Figure 7.
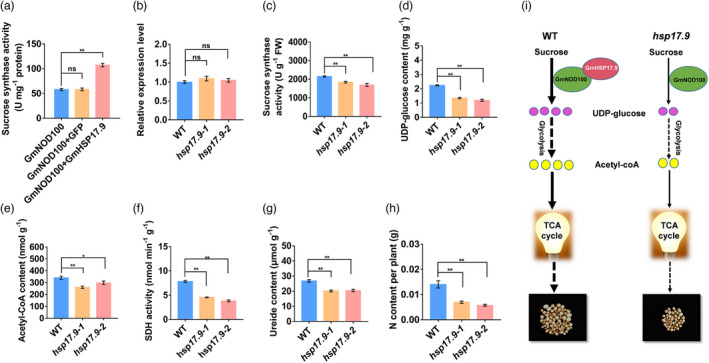
Effect of GmHSP17.9 and GmNOD100 interaction on nodulation and plant growth. (a) Sucrose synthase activity of recombinant GmNOD100. Recombinant GmNOD100 proteins were incubated in the presence or absence of GmHSP17.9, and then the sucrose synthase activity was measured. (b) Expression of GmNOD100 in nodules of hsp17.9 plants at 28 dpi. The relative expression value was normalized based on the expression of GmActin11 (Glyma.18g290800) used as reference gene. ns: not significant. Sucrose synthase activity (c), content of the UDP‐glucose (d), content of acetyl‐CoA (e) and activity of succinate dehydrogenase (SDH) (f) in nodules at 28 dpi. Content of ureide (g) and N (h) of the whole plants at 28 dpi. Values are means of 10 independent plants. Error bars represent means ± SE values. Statistics data were derived from three biological replicates (P < 0.05, t‐test). (i) A proposed model of the regulatory pathway of the interaction between GmHSP17.9 and GmNOD100 in soybean nodulation. When GmHSP17.9 interacts with GmNOD100, sucrose synthase activity of GmNOD100 is elevated, and the product UDP‐glucose converts into acetyl‐CoA through glycolysis pathway. Acetyl‐CoA then enters into the TCA cycle, which provides energy for nodule development. While in hsp17.9 plants, the contents of UDP‐glucose, acetyl‐CoA and the activity of SDH were remarkably decreased. Pink solid circle: UDP‐glucose. Yellow solid circle: acetyl‐CoA.
Discussion
Previous studies have shown that sHSPs are not only involved in defence responses against diverse stresses but also in plant development, such as in pollen, chloroplast and seed development (Dafny‐Yelin et al., 2008; Sun et al., 2020; Volkov et al., 2005; Zhong et al., 2013). A mitochondrial matrix‐localized HSP, GhHSP24.7, regulates cotton seed germination in a temperature‐dependent manner (Ma et al., 2019). AsHSP17, a creeping bentgrass sHSP and sHSP21 in Arabidopsis, modulates chloroplast development and photosynthesis (Sun et al., 2016; Zhong et al., 2013). Five HSPs with high molecular weights over 60 kDa were implicated in the symbiosis interactome between soybean and rhizobia, as revealed by proteome analysis (Zhang et al., 2018a). However, the function and molecular mechanisms of sHSPs in nodule development are less known in legume plants, with only a non‐canonical sHSP, PvNod22, known for infection thread progression during nodule formation in common bean (Rodriguez‐Lopez et al., 2019). In the present study, the role of GmHSP17.9, an sHSP in nodule development and nitrogen fixation, was first characterized in soybean. This research was prompted by the preferential expression of GmHSP17.9 in infected cells of soybean nodules from GUS section and in situ hybridization of wild type, while no hybridization signal was detected in the hsp17.9 mutant nodules (Figure 1f, h‐i, Figure S8). Our results were similar with the PHR‐PHT1 modules reported recently, which displayed specific expression in the infected tissues of soybean nodules (Lu et al., 2020). Loss of GmHSP17.9 function in soybean nodules led to the retardation of nodule development and nitrogen fixation based on the reduction in nodule number, nodule fresh weight and nitrogenase activity. Furthermore, the ultrastructure of nodules in the hsp17.9 lines showed damaged symbiosomes and less accumulation of PHBs (Figures 2 and 3). In addition, to know whether GmHSP17.9 functions in other organs of soybean, hsp17.9 and wild‐type plants were planted in full nutrition condition for 28 days under non‐nitrogen fixing condition. The results showed that there was no significant differences in the plant height and shoot and root fresh weight between hsp17.9 and wild type at different growing stages 10, 17, 21 and 28 day after planting (Figure S9). Based on these findings, we speculate that GmHSP17.9 is required for proper nodule development and nitrogen fixation.
sHSPs usually act as molecular chaperones that can form oligomers to prevent the aggregation of their target proteins to maintain protein homeostasis under various stress conditions (Carra et al., 2017; Haslbeck and Vierling, 2015; Jacob et al., 2017; Sun and MacRae, 2005). Here, we examined the holdase chaperone activity of GmHSP17.9 using MDH and insulin as substrates (Basha et al., 2004b). Recombinant GmHSP17.9 could effectively prevent denatured MDH and insulin from aggregation; the holdase chaperone activity was found to be related to its oligomer status, and recombinant GmHSP17.9 could protect the sucrose synthase activity of GmNOD100 in vitro under heat shock stress conditions (Figure 7a and Figure S2). The sucrose synthase activity in the nodules of the hsp17.9 lines was found to decrease compared to that of the wild‐type plants (Figure 7c). These findings suggest that GmHSP17.9 may associate with its target proteins as a chaperone in nodules to regulate nodule development.
Small heat shock proteins range from 15 to 22 kDa in size and are referred to as the HSP20 subfamily. In plants, sHSPs have 11 subfamilies according to their diverse localizations, including cytoplasmic, nuclear, chloroplast, mitochondria, peroxisomes and endoplasmic reticulum (ER), suggesting that they may play various roles in different organs (Haslbeck and Vierling, 2015; Siddique et al., 2008; Waters, 2013; Waters and Rioflorido, 2007). Moreover, sHSPs acting as chaperones do not function alone, but rather interact with partners. For example, chloroplast sHSP21 in Arabidopsis regulates chloroplast development by modulating its partner plastid protein, pTAC5, under heat stress (Zhong et al., 2013). Here, a subunit of sucrose synthase, GmNOD100, was isolated as a potential target of GmHSP17.9 in soybean nodules, purified previously in soybean nodules and named nodulin‐100 (Table S1) (Zhang et al., 1999). Sucrose synthase has been isolated from several legume plants, including soybean, M. truncatula, P. sativum and L. japonicus. Loss of function of MtSucS1 (encoding sucrose synthase) in M. truncatula impairs plant growth and nodulation and reduces amino acid content (Baier et al., 2007). The double mutants of sucrose synthase isoform, sus1‐1/sus3‐1, in L. japonicus nodules exhibited significantly decreased sucrose synthase activity and reduced nodule growth compared to wild‐type plants under the same growth conditions (Horst et al., 2007). In this study, we found that GmNOD100 was localized in the cytoplasm and was mainly expressed in infected tissues of soybean nodules, similar to GmHSP17.9 (Figure 6a–c). The overexpression and suppression of GmNOD100 in composite transgenic plants showed increased and decreased nodule performance respectively (Figure 6d–i). These results indicate that GmHSP17.9 and GmNOD100 contribute to nodulation in soybean. In the whole genome of soybean, 11 additional homolog proteins of GmNOD100 were identified, among which Glyma.13G114000 (named as GmSS1 in this study) was preferentially expressed in nodules and showed the highest homology with GmNOD100 (Figure S6). Not surprisingly, GmSS1 could also interact with GmHSP17.9 (Figure S10). These findings indicate that GmHSP17.9, acting as a chaperone in nodules, not only interacts with GmNOD100 but also with GmSS1.
In the present study, the reduced protein accumulation of GmNOD100 in hsp17.9 lines led to a markedly lower sucrose synthase activity than that obtained in wild‐type nodules, indicating that GmHSP17.9 is responsible for the stability and enzyme activity of GmNOD100 in soybean nodules. Subsequently, the contents of UDP‐glucose, ureide, total N and yield were markedly decreased in the hsp17.9 lines (Figure 7i). GmHSP17.9 as a chaperone interacts with GmNOD100 in nodules, which result in the production of UDP‐glucose. Then, UDP‐glucose converts into acetyl‐CoA via glycolysis, which then acting as carbon source enters into TCA cycle. TCA cycle as a result can provide energy and inorganic ions for nitrogen fixation process and finally plant growth and yield is promoted (Figures 4 and 7i). These results expand our understanding of the essential role of sHSPs as molecular chaperones in legume–rhizobia symbiosis. The novel functional genes, GmHSP17.9 and GmNOD100 obtained in this study, not only could be applied in soybean symbiosis and nitrogen fixation via transgenic breeding but also offer new insights for exploring the molecular mechanism of nitrogen fixation and engineering symbiotic systems into non‐leguminous crops, such as maize, wheat and rice in the future.
Experimental procedures
Plant materials and growth conditions
Soybean ecotype Williams 82 was used as the wild‐type material in all experiments in this study. Two hsp17.9 mutant lines in the Williams 82 background generated by the CRISPR‐Cas9 system through Agrobacterium‐mediated transformation were used with the wild‐type material for phenotypic analysis.
For phenotypic analysis, sterilized soybean seeds were germinated in Petri dishes with wet filter papers in the dark for 3 days. Thereafter, the seedlings were inoculated with rhizobia Bradyrhizobium diazoefficiens USDA110 suspension media and then transferred to pots with vermiculite and grown in a controlled chamber under a 16 h light: 8 h dark cycle at 28 °C. The plants were watered with nitrogen‐free nutrient solution containing 2.5 mM K2SO4, 2 mM MgSO4·7H2O, 1 mM KH2PO4, 0.15 mM FeCl2, 1.5 mM CaSO4·2H2O, 4.6×10−2 mM H3BO3, 9.1×10−3 mM MnCl2·4H2O, 7.5×10−4 mM ZnSO4, 5×10−4 mM CuSO4, 1.1×10−4 mM MoO3, 9.4×10−5 mM CoCl2·6H2O. [Correction added on 11 November 2021, after first online publication: the composition and concentration of the nitrogen‐free nutrient solution are updated in this version.] Images of the soybean plants were captured and the plants were harvested at day 28 post‐inoculation (dpi) with rhizobia to determine plant height, fresh and dry weight, total N content, ureide content, nitrogenase activity, nodule number and fresh weight, sucrose synthase activity and the contents of UDP‐glucose, PHB, acetyl‐CoA and SDH. For spatial expression analysis, roots, shoots, leaves and flowers were harvested separately at different developmental stages. Nodules were harvested at 10, 17, 28 and 35 dpi. All tissues were frozen in liquid nitrogen and stored at −80 °C for further mRNA and protein analyses.
Gene expression analysis by qRT‐PCR
Total RNA from different soybean tissues was isolated separately using the RNeasy Plant Mini Kit (Tiangen, Beijing, China), according to the manufacturer’s instructions. First‐strand cDNA synthesis was performed with anchored oligo dT primers using the PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, Ōtsu, Shiga Prefecture, Japan). Quantitative real‐time PCR was run on a CFX96TM real‐time system (Bio‐Rad) using SYBR Premix EX TagTM (Takara, Ōtsu, Shiga Prefecture, Japan). The specific primers used are listed in Table S2. The fold change in expression was normalized to the reference housekeeping gene, GmActin11. The experiments were performed three times, each with three replicates. Statistically significant differences in gene expression were determined using the Student’s t‐test.
Histochemical GUS staining analysis
For the analysis of the GmHSP17.9, a 2‐kb fragment upstream of ATG was cloned into the pBI121 vector to generate the pHSP::GUS construct. The construct was transformed into the Agrobacterium rhizogenes strain, K599, to obtain transgenic hairy roots which were screened by GUS staining. GUS staining was performed as described previously (Luo et al., 2013; Zhong et al., 2013). Fresh root tips were incubated in 5‐bromo‐4‐chloro‐3‐indolyl‐b‐D‐glucuronic acid (X‐Gluc) containing solution at 37 °C for 12 h, washed with 70% ethanol, and examined under a microscope. After GUS staining, nodules were embedded and frozen, then sectioned transversely to a thickness of 25 µm with a microtome (CRYOSTARNX50). GUS activity was observed with a light microscope (OLYMPUS U‐TV0.5XC‐3).
RNA in situ hybridization analysis
RNA in situ hybridization was performed as previously described (Kim et al., 2013). Soybean nodules were harvested at 10, 17 and 28 dpi and fixed in formaldehyde–acetic acid solution for 24 h at 4 °C. After embedding in paraffin, 5 µm sections were prepared using a microtome (RM2016). The samples were hybridized with digoxigenin‐labelled antisense and sense RNA probes (GmHSP17.9 antisense: 5’‐DIG‐GGCGAUCUUAACCUCUAUGGUCUUGGGCUUUUUG‐3’ and sense: 5’‐DIG‐CAAAAAGCCCAAGACCAUAGAGGUUAAGAUCGCC‐3’; GmNOD100 antisense: 5’‐DIG‐CCUACGGCUAGUUUCGGUGGGGGGGAA‐3’ and 5’‐DIG‐UUCCCCCCCACCGAAACUAGCCGUAGG‐3’). Hybridization signals were detected by alkaline phosphatase‐catalysed colour reaction with 5‐bromo‐4‐chloro‐3‐indolyl phosphate/nitroblue tetrazolium (Roche; catalogue no. 11175041910). Images were captured with a scanner (Pannoramic DESK, P‐MIDI, P250).
Vector construction for the CRISPR‐Cas9 system
The target sequences for CRISPR‐Cas9‐mediated targeted mutagenesis of GmHSP17.9 were designed using the web tool, CRISPR‐P (http://cbi.hzau.edu.cn/crispr/) (Lei et al., 2014). Two target sequences of GmHSP17.9 were subcloned into different single‐guide RNA (sgRNA) expression cassettes to construct GmU6 promoter‐target1 sgRNA–GmU6 promoter‐target2 sgRNA–GmU6 terminator cassettes, which were built into the pCBSG015/Cas9P35S‐B vector. The resulting positive construct was introduced into the Agrobacterium tumefaciens strain, EHA105, for stable soybean transformation. The transgenic plants were screened for bar resistance, and gene editing was confirmed by specific genomic PCR and sequencing.
Agrobacterium rhizogene K599‐mediated transformation of soybean hairy roots
The overexpression constructs harbouring the full‐length ORF of GmHSP17.9 and GmNOD100 in pCAMBIA1304 were generated. For the RNAi constructs, approximately 230 bp of sense and antisense fragments targeting GmHSP17.9 were cloned into the pTCK‐303 vector under the CaMV 35S promoter, respectively, as described previously (Du et al., 2016; Wang et al., 2020). Composite transgenic soybean hairy roots were generated by Agrobacterium rhizogene K599‐mediated transformation as described previously (Kim et al., 2013). Positive transgenic hairy roots were screened using GUS staining analysis.
Identification of the target proteins associated with GmHSP17.9
GmHSP17.9 was cloned into a modified pET‐28a (+) vector containing an additional Avi‐tag at the C‐terminal end (His‐GmHSP17.9‐Avi) (Du et al., 2015). The His‐GmHSP17.9‐Avi construct was introduced into the Escherichia coli strain, BL21 (DE3) (EMD Chemicals, Gibbstown, NJ), pretransformed with birA (encoding biotin protein ligase) for biotinylation (Tirat et al., 2006). GmHSP17.9 proteins were induced by 0.5 mM isopropyl‐β‐d‐thiogalactoside for 4 h at 28 °C and purified by affinity chromatography using streptavidin agarose resin (Thermo Fisher Scientific, Waltham, MA). The purified GmHSP17.9 proteins were detected by 12% SDS‐PAGE followed by Western blotting using anti‐His monoclonal antibody (Invitrogen). GFP was cloned in the same manner as GmHSP17.9 and used as a negative control. Protein concentration was measured using the Bradford protein assay kit (TaKaRa) according to the manufacturer’s instructions. Target proteins in the nodules were isolated using recombinant GmHSP17.9 affinity chromatography. The purified target proteins were resolved on a 12% SDS‐PAGE gel and further analysed by liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) (Table S1). Purified GFP proteins from E. coli DE3 were used as negative controls to exclude nonspecific targets that directly bound to the streptavidin beads.
Chaperone activity of GmHSP17.9
The chaperone activity of GmHSP17.9 was monitored by measuring the thermal aggregation of malate dehydrogenase (MDH; from porcine heart; Sigma‐Aldrich, St Louis, USA) and chemically induced aggregation of insulin (from bovine pancreas; Sigma‐Aldrich, St Louis, USA). MDH was incubated at 45 °C in 70 mM Tris‐HCl buffer containing 1 mM DTT in the presence or absence (used as control) of GmHSP17.9. Thermal aggregation of MDH was determined by monitoring the increase in turbidity at 340 nm using a spectrophotometer. Insulin was incubated at 37 °C in the presence of 20 mM DTT in 70 mM Tris‐HCl buffer with or without GmHSP17.9, and the aggregation of insulin was monitored by recording the absorbance at 360 nm.
To protect GmNOD100 by GmHSP17.9 in vitro, the recombinant GmNOD100 protein was incubated at 55 °C in the presence or absence of GmHSP17.9. The sucrose synthase activity was then measured by recording the absorbance at 480 nm.
Subcellular localization and bimolecular fluorescence complementation (BiFC) analysis
For subcellular localization, the coding domain sequences (CDSs) of GmHSP17.9 and GmNOD100 were cloned into the 326‐GFP vector to generate GmHSP17.9‐GFP and GmNOD100‐GFP respectively. GFP fluorescence was observed using a confocal microscope. For BiFC, the CDSs of GmHSP17.9 and GmNOD100 were cloned into vectors, p326YFPN and p326YFPC, respectively, to generate GmHSP17.9‐YFPN, GmHSP17.9‐YFPC, GmNOD100‐YFPC and GmNOD100‐YFPN (Du et al., 2015). The resulting constructs were then co‐transformed into Arabidopsis protoplasts via polyethylene glycol (PEG)‐mediated transformation, as described previously (Yoo et al., 2007). For specific fragments of GmHSP17.9, the CDSs for the N‐terminal (amino acid No. 1 to 50), conserved ACD (amino acid No. 51 to 139) and C‐terminal region (amino acid No. 140 to 158) of GmHSP17.9 were cloned into p326YFPN to generate GmHSP17.9‐N‐YFPN, GmHSP17.9‐ACD‐YFPN and GmHSP17.9‐C‐YFPN respectively.
Yeast two‐hybrid assay
To verify the protein interaction between GmHSP17.9 and GmNOD100 in yeast cells, the Matchmaker Gold Yeast Two‐Hybrid System (Clontech, Mountain View CA, USA) was used according to the manufacturer’s instructions. The ORFs of GmHSP17.9 as bait and GmNOD100 as prey were cloned into the pGBKT7 and pGADT7 vectors respectively. The resulting bait and prey constructs were co‐transformed into the yeast strain, Y2H Gold, and then incubated on SD/Leu‐Trp plates at 30 °C for 3 days. Single fresh positive clones were transferred to SD/His‐Ade‐Leu‐Trp‐ plates containing 125 ng/mL of the antibiotic, aureobasidin A (AbA), and 40 µg/mL X‐α‐Gal and incubated at 30 °C for 3 to 5 days to confirm the interaction. pGBKT7‐53 and pGADT7‐T were used as positive controls while pGBKT7‐Lam and pGADT7‐T were used as negative controls. The Y2H assay was repeated three times. The primers used are shown in Table S2.
Pull‐down assay
For the pull‐down assay, the CDS of GmNOD100 was cloned into the pET‐28a (+) vector to generate His‐tag fusion protein (His‐GmNOD100) and transformed into the E. coli strain, BL21. His‐GmHSP17.9‐Avi was purified using a streptavidin agarose resin as bait, and incubated with the total proteins isolated from BL21‐expressing His‐GmNOD100. The precipitates were washed three times and visualized on 12% SDS‐PAGE using an anti‐His antibody (GE Healthcare, 27‐4710‐01, UK).
Co‐immunoprecipitation (Co‐IP) Assay
Total proteins from wild‐type soybean nodules were isolated and incubated with protein G‐immobilized agarose and anti‐GmHSP17.9 antibody at 4 °C for 2 h with gentle continual inversion. The beads were washed with the protein extraction buffer, and the eluted protein was analysed using the anti‐GmNOD100 antibody.
Conflict of interest
The authors have no conflict of interests to declare.
Author contributions
C.Z. and H.D. designed the research; Z.Y. and H.D. conducted most of the experiments and analysed the data; X.X. carried out the Y2H assays; H.D. and Z.Y. wrote the manuscript; C.Z. corrected the manuscript and W.L., Y.K. and X.L. provided suggestions during all the processes of experiments.
Supporting information
Figure S1 Expression pattern and phylogenetic analysis of GmHSP17.9.
Figure S2 Chaperone activity of GmHSP17.9.
Figure S3 Analysis of the mutagenesis of GmHSP17.9 generated by CRISPR‐Cas9 technology.
Figure S4 Alignment and transcript accumulation of homologs of GmHSP17.9 in soybean.
Figure S5 Identification of targets of GmHSP17.9 in soybean nodules.
Figure S6 Expression pattern of GmNOD100 in various organs of soybean.
Figure S7 Phenotypic analysis of composite transgenic soybean plants overexpressing (OX) or suppressing (RNAi) GmNOD100.
Figure S8 RNA in situ hybridization analysis of expression of GmHSP17.9 in hsp17.9 mutant nodules.
Figure S9 Phenotypic analysis of hsp17.9 plants under full nutrient condition.
Figure S10 Interaction of GmHSP17.9 and GmSS1 in Arabidopsis protoplasts.
Table S1 Protein targets of GmHSP17.9 isolated from soybean nodules of 28 dpi.
Table S2 All primers used in this study.
Acknowledgements
We thank Zhiying Ma professor (Hebei Agricultural University, China) for making constructive suggestions on this work and helpful comments on the manuscript. This research was funded by the Project of Hebei Province Science and Technology Support Program (17927670H and 16227516D‐1) and Natural Science Foundation of Hebei Province (C2019204199).
Yang, Z. , Du, H. , Xing, X. , Li, W. , Kong, Y. , Li, X. and Zhang, C. (2022) A small heat shock protein, GmHSP17.9, from nodule confers symbiotic nitrogen fixation and seed yield in soybean. Plant Biotechnol. J., 10.1111/pbi.13698
References
- Baier, M.C. , Barsch, A. , Kuster, H. and Hohnjec, N. (2007) Antisense repression of the Medicago truncatula nodule‐enhanced sucrose synthase leads to a handicapped nitrogen fixation mirrored by specific alterations in the symbiotic transcriptome and metabolome. Plant Physiol. 145, 1600–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha, E. , Friedrich, K.L. and Vierling, E. (2006) The N‐terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J. Biol. Chem. 281, 39943–39952. [DOI] [PubMed] [Google Scholar]
- Basha, E. , Lee, G.J. , Breci, L.A. , Hausrath, A.C. , Buan, N.R. , Giese, K.C. & Vierling, E. (2004a) The identity of proteins associated with a small heat shock protein during heat stress in vivo indicates that these chaperones protect a wide range of cellular functions. J. Biol. Chem. 279, 7566–7575. [DOI] [PubMed] [Google Scholar]
- Basha, E. , Lee, G.J. , Demeler, B. and Vierling, E. (2004b) Chaperone activity of cytosolic small heat shock proteins from wheat. Eur. J. Biochem. 271, 1426–1436. [DOI] [PubMed] [Google Scholar]
- Candido, E.P. (2002) The small heat shock proteins of the nematode Caenorhabditis elegans: structure, regulation and biology. Prog. Mol. Subcell Biol. 28, 61–78. [DOI] [PubMed] [Google Scholar]
- Carra, S. , Alberti, S. , Arrigo, P.A. , Benesch, J.L. , Benjamin, I.J. , Boelens, W. , Bartelt‐Kirbach, B. et al. (2017) The growing world of small heat shock proteins: from structure to functions. Cell Stress Chaperones, 22, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Qin, L. , Zhou, L. , Li, X. , Chen, Z. , Sun, L. , Wang, W. et al. (2019) A nodule‐localized phosphate transporter GmPT7 plays an important role in enhancing symbiotic N2 fixation and yield in soybean. New Phytol. 221, 2013–2025. [DOI] [PubMed] [Google Scholar]
- Collier, R. and Tegeder, M. (2012) Soybean ureide transporters play a critical role in nodule development, function and nitrogen export. Plant J. 72, 355–367. [DOI] [PubMed] [Google Scholar]
- Dafny‐Yelin, M. , Tzfira, T. , Vainstein, A. and Adam, Z. (2008) Non‐redundant functions of sHSP‐CIs in acquired thermotolerance and their role in early seed development in Arabidopsis. Plant Mol. Biol. 67, 363–373. [DOI] [PubMed] [Google Scholar]
- Derham, B.K. , Ellory, J.C. , Bron, A.J. and Harding, J.J. (2003) The molecular chaperone alpha‐crystallin incorporated into red cell ghosts protects membrane Na/K‐ATPase against glycation and oxidative stress. Eur. J. Biochem. 270, 2605–2611. [DOI] [PubMed] [Google Scholar]
- Dong, W. , Zhu, Y. , Chang, H. , Wang, C. , Yang, J. , Shi, J. , Gao, J. et al. (2021) An SHR‐SCR module specifies legume cortical cell fate to enable nodulation. Nature, 589, 586–590. [DOI] [PubMed] [Google Scholar]
- Du, H. , Kim, S. , Hur, Y.S. , Lee, M.S. , Lee, S.H. and Cheon, C.I. (2015) A cytosolic thioredoxin acts as a molecular chaperone for peroxisome matrix proteins as well as antioxidant in peroxisome. Mol. Cells, 38, 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y. , He, W. , Deng, C. , Chen, X. , Gou, L. , Zhu, F. , Guo, W. et al. (2016) Flowering‐Related RING Protein 1 (FRRP1) regulates flowering time and yield potential by affecting histone H2B monoubiquitination in rice (Oryza sativa). PLoS One, 11, e0150458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erisman, J.W. , Galloway, J. , Seitzinger, S. , Bleeker, A. and Butterbach‐Bahl, K. (2011) Reactive nitrogen in the environment and its effect on climate change. Curr. Opin. Env. Sust. 3, 281–290. [Google Scholar]
- Eyles, S.J. and Gierasch, L.M. (2010) Nature's molecular sponges: small heat shock proteins grow into their chaperone roles. Proc. Natl Acad. Sci. USA, 107, 2727–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, B.J. , Indrasumunar, A. , Hayashi, S. , Lin, M.H. , Lin, Y.H. , Reid, D.E. and Gresshoff, P.M. (2010) Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52, 61–76. [DOI] [PubMed] [Google Scholar]
- Haslbeck, M. , Braun, N. , Stromer, T. , Richter, B. , Model, N. , Weinkauf, S. and Buchner, J. (2004) Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae . Embo J. 23, 638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck, M. and Vierling, E. (2015) A first line of stress defense: small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 427, 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C. , Gao, H. , Wang, H. , Guo, Y. , He, M. , Peng, Y. and Wang, X. (2021) GSK3‐mediated stress signaling inhibits legume‐rhizobium symbiosis by phosphorylating GmNSP1 in soybean. Mol. Plant, 14, 488–502. [DOI] [PubMed] [Google Scholar]
- Horst, I. , Welham, T. , Kelly, S. , Kaneko, T. , Sato, S. , Tabata, S. , Parniske, M. et al. (2007) TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule‐enhanced sucrose synthase. Plant Physiol. 144, 806–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, P. , Hirt, H. and Bendahmane, A. (2017) The heat‐shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 15, 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karney‐Grobe, S. , Russo, A. , Frey, E. , Milbrandt, J. and DiAntonio, A. (2018) HSP90 is a chaperone for DLK and is required for axon injury signaling. Proc. Natl Acad. Sci. USA, 115, E9899–E9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.K. , Kim, S. , Um, J.H. , Kim, K. , Choi, S.K. , Um, B.H. , Kang, S.W. et al. (2013) Functional implication of β‐carotene hydroxylases in soybean nodulation. Plant Physiol. 162, 1420–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, R.D. , Chidawanyika, T. , Tims, H.S. , Meulia, T. , Bouchard, R.A. and Pett, V.B. (2014) Chaperone function of two small heat shock proteins from maize. Plant Sci. 221–222, 48–58. [DOI] [PubMed] [Google Scholar]
- Lei, Y. , Lu, L. , Liu, H.Y. , Li, S. , Xing, F. and Chen, L.L. (2014) CRISPR‐P: a web tool for synthetic single‐guide RNA design of CRISPR‐system in plants. Mol. Plant, 7, 1494–1496. [DOI] [PubMed] [Google Scholar]
- Li, X. , Zhao, J. , Tan, Z. , Zeng, R. & Liao, H. (2015) GmEXPB2, a cell wall β‐expansin, affects soybean nodulation through modifying root architecture and promoting nodule formation and development. Plant Physiol. 169, 2640–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, A. , Contador, C.A. , Fan, K. and Lam, H.M. (2018) Interaction and regulation of carbon, nitrogen, and phosphorus metabolisms in root nodules of legumes. Front. Plant Sci. 9, 1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes‐Caitar, V.S. , de Carvalho, M.C. , Darben, L.M. , Kuwahara, M.K. , Nepomuceno, A.L. , Dias, W.P. , Abdelnoor, R.V. et al. (2013) Genome‐wide analysis of the Hsp20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genom. 14, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, M. , Cheng, Z. , Zhang, X.M. , Huang, P. , Fan, C. , Yu, G. , Chen, F. et al. (2020) Spatial divergence of PHR‐PHT1 modules maintains phosphorus homeostasis in soybean nodules. Plant Physiol. 184, 236–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y.J. , Wang, Z.J. , Ji, H.T. , Fang, H. , Wang, S.F. , Tian, L.N. and Li, X. (2013) An Arabidopsis homolog of importin β1 is required for ABA response and drought tolerance. Plant J. 75, 377–389. [DOI] [PubMed] [Google Scholar]
- Ma, W. , Guan, X. , Li, J. , Pan, R. , Wang, L. , Liu, F. , Ma, H. et al. (2019) Mitochondrial small heat shock protein mediates seed germination via thermal sensing. Proc. Natl Acad. Sci. USA, 116, 4716–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magori, S. and Kawaguchi, M. (2009) Long‐distance control of nodulation: molecules and models. Mol. Cells, 27, 129–134. [DOI] [PubMed] [Google Scholar]
- Marsh, J.F. , Rakocevic, A. , Mitra, R.M. , Brocard, L. , Sun, J. , Eschstruth, A. , Long, S.R. et al. (2007) Medicago truncatula NIN is essential for rhizobial‐independent nodule organogenesis induced by autoactive calcium/calmodulin‐dependent protein kinase. Plant Physiol. 144, 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, E.T. , Bortolus, M. , Koteiche, H.A. and McHaourab, H.S. (2012) Sequence, structure, and dynamic determinants of Hsp27 (HspB1) equilibrium dissociation are encoded by the N‐terminal domain. Biochemistry, 51, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, N.D. , Gerber, J.S. , Johnston, M. , Ray, D.K. , Ramankutty, N. and Foley, J.A. (2012) Closing yield gaps through nutrient and water management. Nature, 490, 254–257. [DOI] [PubMed] [Google Scholar]
- Mymrikov, E.V. , Riedl, M. , Peters, C. , Weinkauf, S. , Haslbeck, M. and Buchner, J. (2020) Regulation of small heat‐shock proteins by hetero‐oligomer formation. J. Biol. Chem. 295, 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd, G.E. and Downie, J.A. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59, 519–546. [DOI] [PubMed] [Google Scholar]
- Poulain, P. , Gelly, J.C. and Flatters, D. (2010) Detection and architecture of small heat shock protein monomers. PLoS One, 5, e9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, L. , Zhao, J. , Tian, J. , Chen, L.Y. , Sun, Z.A. , Guo, Y.X. , Lu, X. et al. (2012) The high‐affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol. 159, 1634–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissig, G.N. , Posso, D.A. , Borella, J. , da Silveira, R.V.D. , Rombaldi, C.V. and Bacarin, M.A. (2018) High MT‐sHSP23.6 expression increases antioxidant system in 'Micro‐Tom' tomato fruits during post‐harvest hypoxia. Sci. Hortic‐Amsterdam, 242, 127–136. [Google Scholar]
- Rodriguez‐Lopez, J. , Lopez, A.H. , Estrada‐Navarrete, G. , Sanchez, F. and Diaz‐Camino, C. (2019) The noncanonical heat shock protein PvNod22 Is essential for infection thread progression during rhizobial endosymbiosis in common bean. Mol. Plant Microbe Interact. 32, 939–948. [DOI] [PubMed] [Google Scholar]
- Siddique, M. , Gernhard, S. , von Koskull‐Doring, P. , Vierling, E. and Scharf, K.D. (2008) The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones, 13, 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smykal, P. , Masin, J. , Hrdy, I. , Konopasek, I. and Zarsky, V. (2000) Chaperone activity of tobacco HSP18, a small heat‐shock protein, is inhibited by ATP. Plant J. 23, 703–713. [DOI] [PubMed] [Google Scholar]
- Soumare, A. , Diedhiou, A.G. , Thuita, M. , Hafidi, M. , Ouhdouch, Y. , Gopalakrishnan, S. and Kouisni, L. (2020) Exploiting biological nitrogen fixation: a route towards a sustainable agriculture. Plants, 9, 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. , Sun, C. , Li, Z. , Hu, Q. , Han, L. and Luo, H. (2016) AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA‐dependent and independent signalling to attenuate plant response to abiotic stress. Plant Cell Environ. 39, 1320–1337. [DOI] [PubMed] [Google Scholar]
- Sun, X. , Zhu, J. , Li, X. , Li, Z. , Han, L. and Luo, H. (2020) AsHSP26.8a, a creeping bentgrass small heat shock protein integrates different signaling pathways to modulate plant abiotic stress response. BMC Plant Biol. 20, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. and MacRae, T.H. (2005) Small heat shock proteins: molecular structure and chaperone function. Cell Mol. Life Sci. 62, 2460–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirat, A. , Freuler, F. , Stettler, T. , Mayr, L.M. and Leder, L. (2006) Evaluation of two novel tag‐based labelling technologies for site‐specific modification of proteins. Int. J. Biol. Macromol. 39, 66–76. [DOI] [PubMed] [Google Scholar]
- Trainer, M.A. and Charles, T.C. (2006) The role of PHB metabolism in the symbiosis of rhizobia with legumes. Appl. Microbiol. Biotechnol. 71, 377–386. [DOI] [PubMed] [Google Scholar]
- Volkov, R.A. , Panchuk, I.I. and Schoffl, F. (2005) Small heat shock proteins are differentially regulated during pollen development and following heat stress in tobacco. Plant Mol. Biol. 57, 487–502. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Yang, Z. , Kong, Y. , Li, X. , Li, W. , Du, H. and Zhang, C. (2020) GmPAP12 is required for nodule development and nitrogen fixation under phosphorus starvation in soybean. Front. Plant Sci. 11, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, E.R. (2013) The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 64, 391–403. [DOI] [PubMed] [Google Scholar]
- Waters, E.R. and Rioflorido, I. (2007) Evolutionary analysis of the small heat shock proteins in five complete algal genomes. J. Mol. Evol. 65, 162–174. [DOI] [PubMed] [Google Scholar]
- Yoo, S.D. , Cho, Y.H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Liu, J.Y. , Gu, H. , Du, Y.F. , Zuo, J.F. , Zhang, Z.B. , Zhang, M.L. et al. (2018a) Bradyrhizobium diazoefficiens USDA 110–Glycine max Interactome Provides Candidate Proteins Associated with Symbiosis. J. Proteome. Res. 17, 3061–3074. [DOI] [PubMed] [Google Scholar]
- Zhang, N. , Shi, J. , Zhao, H. and Jiang, J. (2018b) Activation of small heat shock protein (SlHSP17.7) gene by cell wall invertase inhibitor (SlCIF1) gene involved in sugar metabolism in tomato. Gene, 679, 90–99. [DOI] [PubMed] [Google Scholar]
- Zhang, X.Q. , Lund, A.A. , Sarath, G. , Cerny, R.L. , Roberts, D.M. and Chollet, R. (1999) Soybean nodule sucrose synthase (nodulin‐100): further analysis of its phosphorylation using recombinant and authentic root‐nodule enzymes. Arch. Biochem. Biophys. 371, 70–82. [DOI] [PubMed] [Google Scholar]
- Zhong, L. , Zhou, W. , Wang, H. , Ding, S. , Lu, Q. , Wen, X. , Peng, L. et al. (2013) Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell, 25, 2925–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Expression pattern and phylogenetic analysis of GmHSP17.9.
Figure S2 Chaperone activity of GmHSP17.9.
Figure S3 Analysis of the mutagenesis of GmHSP17.9 generated by CRISPR‐Cas9 technology.
Figure S4 Alignment and transcript accumulation of homologs of GmHSP17.9 in soybean.
Figure S5 Identification of targets of GmHSP17.9 in soybean nodules.
Figure S6 Expression pattern of GmNOD100 in various organs of soybean.
Figure S7 Phenotypic analysis of composite transgenic soybean plants overexpressing (OX) or suppressing (RNAi) GmNOD100.
Figure S8 RNA in situ hybridization analysis of expression of GmHSP17.9 in hsp17.9 mutant nodules.
Figure S9 Phenotypic analysis of hsp17.9 plants under full nutrient condition.
Figure S10 Interaction of GmHSP17.9 and GmSS1 in Arabidopsis protoplasts.
Table S1 Protein targets of GmHSP17.9 isolated from soybean nodules of 28 dpi.
Table S2 All primers used in this study.


