Summary
Plant ethylene‐responsive factors (ERFs) play essential roles in cold stress response, but the molecular mechanisms underlying this process remain poorly understood. In this study, we characterized PtrERF9 from trifoliate orange (Poncirus trifoliata (L.) Raf.), a cold‐hardy plant. PtrERF9 was up‐regulated by cold in an ethylene‐dependent manner. Overexpression of PtrERF9 conferred prominently enhanced freezing tolerance, which was drastically impaired when PtrERF9 was knocked down by virus‐induced gene silencing. Global transcriptome profiling indicated that silencing of PtrERF9 resulted in substantial transcriptional reprogramming of stress‐responsive genes involved in different biological processes. PtrERF9 was further verified to directly and specifically bind with the promoters of glutathione S‐transferase U17 (PtrGSTU17) and ACC synthase1 (PtrACS1). Consistently, PtrERF9‐overexpressing plants had higher levels of PtrGSTU17 transcript and GST activity, but accumulated less ROS, whereas the silenced plants showed the opposite changes. Meanwhile, knockdown of PtrERF9 decreased PtrACS1 expression, ACS activity and ACC content. However, overexpression of PtrERF9 in lemon, a cold‐sensitive species, caused negligible alterations of ethylene biosynthesis, which was attributed to perturbed interaction between PtrERF9, along with lemon homologue ClERF9, and the promoter of lemon ACS1 gene (ClACS1) due to mutation of the cis‐acting element. Taken together, these results indicate that PtrERF9 acts downstream of ethylene signalling and functions positively in cold tolerance via modulation of ROS homeostasis by regulating PtrGSTU17. In addition, PtrERF9 regulates ethylene biosynthesis by activating PtrACS1 gene, forming a feedback regulation loop to reinforce the transcriptional regulation of its target genes, which may contribute to the elite cold tolerance of Poncirus trifoliata.
Keywords: Trifoliata orange, ERF, cold tolerance, ROS homeostasis, ethylene biosynthesis, glutathione S‐transferase, ACC synthase
Introduction
Citrus is an economically important fruit crop in the world and occupies a key position in international fruit trade. Being sessile organism, citrus production is constantly subjected to a myriad of environment stresses, among which low temperature is a major limiting factor for plant growth, development, geographical distribution and productivity (Ding et al., 2019; Zhu, 2016). Thus, improving the cold resistance of citrus is regarded as an important breeding programme. Trifoliate orange (Poncirus trifoliata (L.) Raf.), closely related to citrus, exhibits desirable resistance to extremely low temperature when fully acclimated (Peng et al., 2014). It is usually used as a citrus rootstock to incorporate its elite tributes. In addition, it is widely accepted that trifoliate orange is a good germplasm source for exploring cold‐responsive genes that can be employed for genetic engineering for manipulating cold tolerance. To this end, great efforts have been devoted to elucidating molecular mechanisms underlying the cold tolerance of trifoliate orange and to identifying valuable genes that are tightly involved in this process and play key roles in modulating the cold response.
Plant perceived stress signals through various signalling pathways mediated by different signalling molecules, including phytohormones, reactive oxygen species (ROS) and Ca2+ (Guo et al., 2018; Hu et al., 2017; Peng et al., 2017). These signalling pathways play crucial roles in orchestration of stress signal perception, signal transduction and magnification, and finally trigger a large spectrum of stress responses at physiological, biochemical, metabolic and molecular levels (Ding et al., 2020; Zhang et al., 2019a). Among the phytohormones, ethylene has been increasingly regarded as an important member helping the plants adapt to adverse environmental stresses. This argument is supported by the findings that ethylene synthesis is prominently altered by diverse stresses, such as drought, cold, wounding and pathogen attack (Wang et al., 2021; Müller and Munné‐Bosch, 2015). Interestingly, ethylene has been shown to act as either a positive or negative regulator of cold stress response. Some studies showed that increased endogenous ethylene level was accompanied by enhanced cold tolerance of grapevine and apple (Sun et al., 2016; Wang et al., 2021). By contrast, ethylene levels were negatively correlated with freezing tolerance of Arabidopsis thaliana and Medicago truncatula (Shi et al., 2012; Zhao et al., 2014). In addition, treatment with 1‐methylcyclopropene (1‐MCP), an inhibitor of ethylene signal perception, increased the chilling injury of tomato fruit (Zhao et al., 2009). Despite the contrasting conclusions obtained in different plant species, these findings suggested that ethylene may be tightly implicated in cold tolerance. However, the molecular network pertinent to ethylene biosynthesis in response to cold stress remains largely uncharacterized in most plants.
Plants have evolved a sophisticated adaptation mechanism so as to maintain growth and development and to survive under the harsh environmental cues. One of the mechanisms is the transcriptional reprogramming of a vast number of stress‐responsive molecular modules composed of transcription factors (TFs) and their corresponding target genes. The TFs serve as the indispensable links connecting the external environmental stimuli by relaying the stress signal to the downstream gene expression. The APETALA2/ethylene‐responsive factors (AP2/ERFs) superfamily is one of the largest plant‐specific TF families and functions as the major regulatory factors for ethylene signalling pathway in plant stress responses. The AP2/ERF proteins, defined by containing one or two conserved AP2 domains, are divided into three separate subfamilies, ERF, AP2 and RAV (Licausi et al., 2013). Accumulating evidence has demonstrated that the AP2/ERF TFs are involved in a wide range of environmental stresses, including drought (Zhao et al., 2020, 2020,2020, 2020), heavy metal (Lin et al., 2017), salinity (Schmidt et al., 2013), high light (Vogel et al., 2014) and cold (Wang et al., 2019; Zhuo et al., 2018), in a variety of plant species. However, it is worth mentioning that the roles of most ERF members in cold tolerance remain far from being clearly characterized. In addition, the target genes by which the ERFs regulate to orchestrate the cold tolerance remain largely ambiguous, which hampers our integrative understanding of the cold stress response at molecular levels.
Cold stress influences plant growth by causing several negative effects. One of them is increased production of ROS, which, at high concentrations, result in membrane lipid peroxidation, protein and nucleic acids oxidation, leading to cell structure damages or ultimately cell death (Considine and Foyer, 2014). Plants have developed non‐enzymatic and enzymatic antioxidant defence systems for removing excessive ROS produced under cold conditions. Glutathione S‐transferases (GST) is one of the enzymes that are mobilized by plants to scavenge ROS (Cui et al., 2019). Earlier studies have shown that increased GST activity or expression conferred abiotic stress tolerance, whereas loss‐of‐function of GST in rice mutant osgst4 was found to exhibit reduced salt tolerance (Jha et al., 2011; Xu et al., 2018). The GST family is grouped into seven classes, tau (GSTU), phi (GSTF), lambda (GSTL), dehydroascorbate reductase (DHAR), zeta (GSTZ), theta (GSTT) and tetrachlorohydroquinone dehydrogenase (TCHQD), in which the first four GSTs are specific to plants (Rezaei et al., 2013). Currently, substantial evidence has suggested that GSTUs can scavenge H2O2 and O2 ·− to modulate ROS homeostasis under various stresses (Horváth et al., 2020; Zhang et al., 2019b, 2019a). Although the GSTs have been well reported to participate in plant abiotic stress tolerance by elimination of ROS, the upstream regulatory proteins of the GSTs are still unclear. A raised question is whether the GSTs, in particular the plant‐specific ones, are regulated by the ERFs under cold stress.
In an earlier study, we carried out a global transcriptome analysis in Poncirus trifoliata subjected to cold treatment in order to identify cold‐inducible genes (Wang et al., 2015). A number of genes annotated as ERFs were induced to different extent by cold, and one of them (Pt2g011090), designated as PtrERF9 in this study, was strongly induced. We thus hypothesize that PtrERF9 may act as a critical regulator of cold tolerance in trifoliate orange by modulation of pertinent metabolic pathways through its regulating target genes. Here, we demonstrated that PtrERF9 plays a positive role in cold tolerance based on overexpression and virus‐induced gene silencing (VIGS) assays. By using RNA‐sequencing (RNA‐seq), we found that PtrERF9 may act to regulate a myriad of stress‐associated genes involved in various signalling and metabolic pathways. Furthermore, we demonstrate that PtrERF9 functions in cold tolerance by modulation of ROS homeostasis via directly targeting PtrGSTU17, a GSTU gene. Interestingly, ethylene participates in cold‐induced up‐regulation of PtrERF9, which was further demonstrated to regulate ethylene synthesis by interacting with the promoter of PtrACS1, but not with the promoter of ClACS1 from lemon, implying that PtrERF9 undergoes a feedback regulation of ethylene to orchestrate cold stress response in trifoliate orange.
Results
Identification of PtrERF9 based on a transcriptome dataset
The transcript profiles of ERF genes in cold‐treated trifoliate orange were examined using a heat map based on our previous RNA‐seq data (Wang et al., 2015). Among the ERF genes, Pt2g011090 was induced to the greatest extent by cold (Figure 1a). A full‐length cDNA sequence of Pt2g011090 was obtained by RT‐PCR from trifoliate orange. The 696‐bp ORF encodes a predicted protein of 231 amino acids (aa), with an estimated molecular mass of 24.85 kDa and an isoelectric point of 9.15. Phylogenetic tree analysis revealed that Pt2g011090 was most closely related to AtERF9 (Figure S1A & B), which allowed us to rename Pt2g011090 as PtrERF9 in this study. Protein structure analysis showed that PtrERF9 contained a conserved AP2 domain of 64 aa. Since the residues at the 14th and 19th positions in the AP2 domain were alanine (A) and aspartic acid (D), respectively (Figure S1C), PtrERF9 was categorized into the ERF subfamily.
Figure 1.
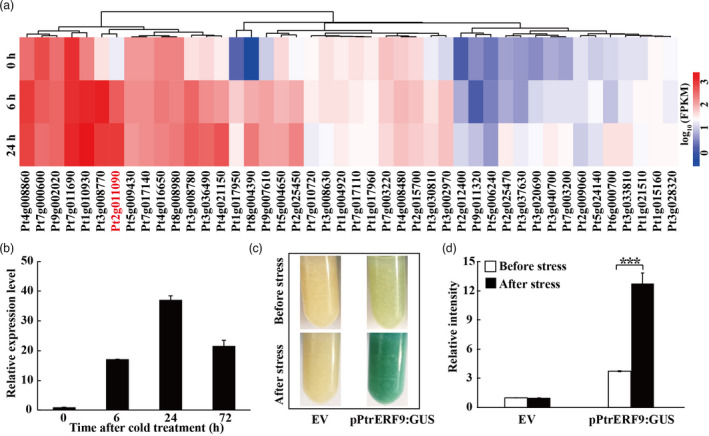
The expression of ERF genes analysis under cold treatment in Poncirus trifoliata. (a) Expression profiles of 44 ERF genes in trifoliate orange under cold stress. (b) Relative expression of PtrERF9 was quantitated by qPCR in response to low temperature. Error bars represent ± SE (n = 3). (c, d) GUS staining (c) and relative GUS intensity (d) analysis in sweet orange callus transformation with empty vector (EV) and pPtrERF9: GUS in response to cold treatment. Error bars represent ± SE (n = 3). Asterisks indicate significant difference between the vectors under before and after the cold treatment (***P < 0.001).
Cold induction of PtrERF9 is dependent on ethylene
Expression pattern of PtrERF9 under cold stress was checked by qPCR. PtrERF9 transcript level was up‐regulated after low‐temperature treatment and peaked at 24 h (37‐fold of that at 0 h), followed by a steady decrease to 21‐fold of the initial level at the last time point (Figure 1b). In order to confirm this pattern, a transient expression assay was performed by expressing the GUS gene driven by the promoter of PtrERF9 (pPtrERF9) in citrus callus. Histochemical staining showed that the GUS expression level in the calli expressing pPtrERF9:GUS construct was drastically increased following the cold treatment, justifying that PtrERF9 is a cold‐responsive gene (Figures 1c and d).
Ethylene has been shown to play a vital role in plant response to various abiotic stresses, including cold (Sun et al., 2019). We also checked dynamic changes of ethylene in trifoliate orange exposed to cold condition. Ethylene production displayed a slow elevation at 12 h of cold treatment, followed by a sharp and steady increase to the highest level at 48 h (Figure 2a). Next, we explored whether ethylene contributed to cold tolerance by comparing plants treated with ACC and AVG, the precursor and inhibitor of ethylene biosynthesis, respectively, using water treatment as a control. No difference in plant morphology was observed between seedlings pretreated with ACC or AVG and water. However, upon exposure to cold treatment at freezing temperature, less serious necrotic phenotype, lower electrolyte leakage (EL) level and MDA content were noticed in the ACC‐treated seedlings, whereas the AVG‐treated seedlings plant exhibited more serious damages, concurrent with higher EL and MDA levels, in comparison with the water‐treated plants (Figure 2b–d). These results suggest that cold promoted ethylene production and that ethylene plays a positive role in cold tolerance of trifoliate orange.
Figure 2.
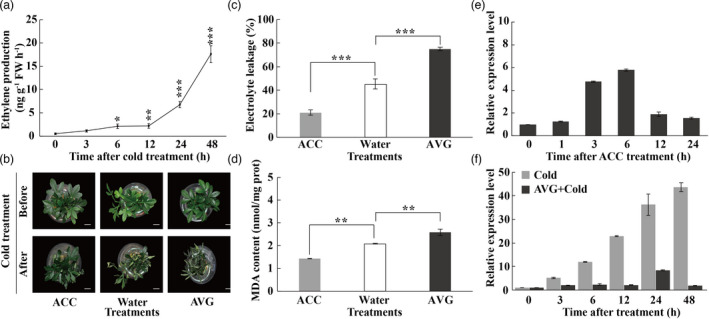
Ethylene enhanced trifoliate orange cold tolerance and PtrERF9 expression was increased association with ethylene production under cold stress. (a) Cold induced ethylene production in trifoliate orange. (b) Phenotype of ACC‐, water‐ or AVG‐pretreated wild‐type plants in response to freezing treatment. Scale bars = 1 cm. (c, d) Electrolyte leakage (c) and MDA levels (d) in ACC‐, water‐, AVG‐pretreated wild‐type plants after cold treatment. Error bars indicate ± SE (n = 3). Asterisks indicate significant differences between different groups (**P < 0.01, ***P < 0.001). (e, f) The expression of PtrERF9 under ACC treatment (e), cold and cold with AVG treatment (f) in trifoliate orange. Error bars indicate ± SE (n = 3).
It has been suggested that ERFs act downstream of ethylene signalling pathway to regulate abiotic stress response. This compels us to investigate how PtrERF9 expression was influenced by ethylene under cold stress. To answer this question, we first determined PtrERF9 expression in the presence of ACC. Interestingly, transcript level of PtrERF9 was prominently increased by exogenous ACC treatment (Figure 2e). However, AVG application substantially compromised the cold induction of PtrERF9 (Figure 2f). All of these data demonstrated that up‐regulation of PtrERF9 by cold is dependent on synthesis ethylene.
PtrERF9 is localized to the nucleus
To determine the subcellular location of PtrERF9, a construct was generated by fusing the PtrERF9 cDNA to the YFP gene, driven by the CaMV 35S promoter. Then, the 35S:PtrERF9‐YFP fusion construct or 35S:YFP empty vector was transiently expressed in N. benthamiana leaves via agroinfiltration. Confocal microscopic observation showed that the YFP signal was ubiquitously distributed in the cytoplasm and the nucleus of epidermal cells transformed with the control vector. However, when the fusion construct was examined, the YFP signal was detected exclusively in the nucleus, precisely co‐locating with the nuclear marker (mCherry, red fluorescence) or indicator (DAPI, blue fluorescence), implying that PtrERF9 is a nuclear protein (Figure 3).
Figure 3.
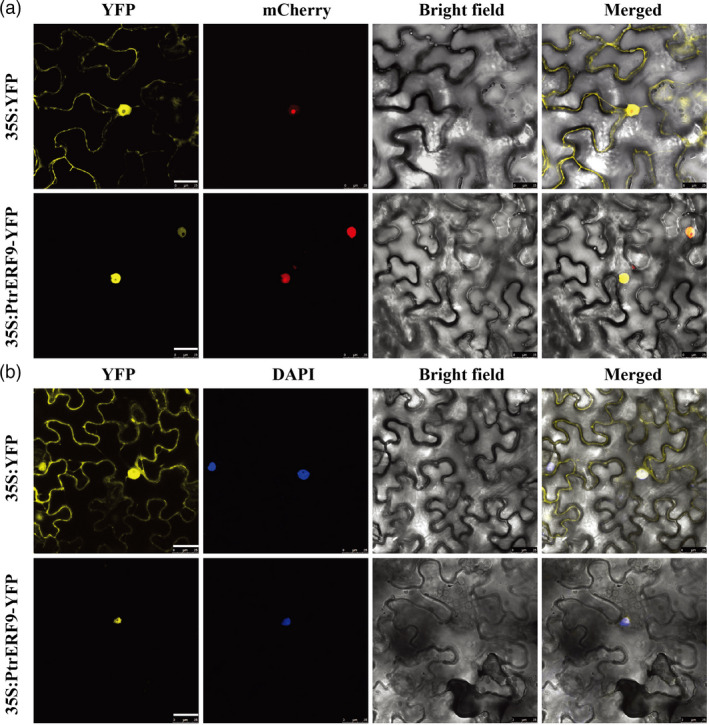
PtrERF9 is localized in the nucleus. (a) The fusion construct (35S: PtrERF9‐YFP) or an empty vector (35S: YFP) was co‐transformed with a nucleus marker gene VirD2NLS fused to mCherry in tobacco (Nicotiana benthamiana) leaves. Confocal microscopic images of the epidermal cells were taken under bright filed and yellow (for YFP), red (for mCherry) fluorescence signals. (b) DAPI staining, shown in blue, was used to stain the nucleus. The overlapped images are shown on the right. Scale bars = 25 µm.
Overexpression of PtrERF9 improves cold tolerance in transgenic plants
Since PtrERF9 was dramatically induced by cold stress, we speculated that PtrERF9 might play a pivotal role in regulation of cold tolerance. To verify this hypothesis, we generated PtrERF9‐overexpressing transgenic tobacco plants. We selected two independents lines (#2 and #4) with high transcript levels of PtrERF9 for cold tolerance assay (Figure S2). No morphological differences were observed between the wild‐type (WT) and transgenic tobacco plants under normal growth conditions. After exposure to the cold condition at freezing temperature (−4°C for 6 h), the WT plants exhibited severe water‐soaking phenotype in comparison with the transgenic counterparts (Figure 4a). After recovery for 3 d at ambient temperature, 80%–85% of the transgenic plants survived, while the survival rate of WT was only 20% (Figure 4b). EL, MDA, chlorophyll fluorescence and Fv/Fm ratios between the WT and the transgenic plants were similar to each other without stress, while significantly lower EL and MDA contents, together with stronger chlorophyll fluorescence and higher F v/F m ratios, were measured in the transgenic tobacco lines than in the WT after the freezing treatment (Figure 4c–f).
Figure 4.
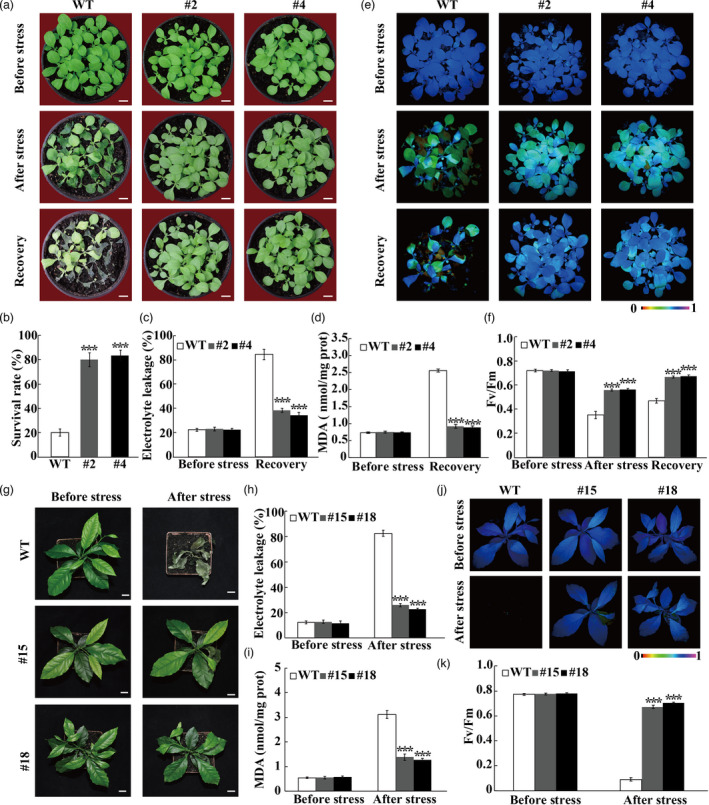
Overexpression of PtrERF9 improved cold tolerance of transgenic plants. (a) Phenotypes of wild‐type (WT) and PtrERF9 overexpression transgenic tobacco lines (#2 and #4) in response to cold treatment. Scale bars = 1 cm. (b) Survival rate of transgenic tobacco plants after the growth recovery for 3 d at ambient environment. (c, d) Electrolyte leakage (c), MDA content (d) of transgenic tobacco plants before and after the growth recovery for 3 d at 25 °C. (e, f) Chlorophyll fluorescence (e) and F v/F m ratios (f) of transgenic tobacco plants before and after cold treatment and the growth recovery for 3 d at 25 °C. (g) Phenotypes of WT and PtrERF9 overexpression transgenic lemon plants (#15 and #18) before and after cold treatment. Scale bars = 1 cm. (h‐k) Electrolyte leakage (h), MDA content (i), chlorophyll fluorescence (j) and F v/F m (k) of WT and transgenic lemon before and after the cold treatment. Error bars represent ± SE (n = 3). Asterisks indicate significant difference between WT and the transgenic lines in response to cold treatment (***P < 0.001).
To further investigate the role of PtrERF9 in cold tolerance, PtrERF9 was overexpressed in lemon, one of the most cold‐sensitive citrus species; two transgenic lines (#15 and #18) were used for assessing cold tolerance (Figure S3). Both the wild‐type (WT) lemon and transgenic plants grew well under normal growth environment. Upon exposure to freezing at −4 °C for 8 h and a subsequent growth recovery for 5 d at room temperature, the WT plants withered and died, whereas the transgenic plants were still alive despite existence of some wilted leaves (Figure 4g). Consistent with the phenotype, the transgenic plants had lower EL and MDA levels, better chlorophyll fluorescence and higher Fv/Fm ratios in comparison with the WT after the cold stress (Figure 4h–k). All of these results demonstrated that overexpression of PtrERF9 significantly enhanced cold tolerance in transgenic plants.
Knockdown of PtrERF9 in Poncirus trifoliata leads to elevated cold sensitivity
To further investigate the function of PtrERF9 in cold tolerance, VIGS was used to knock down PtrERF9 in Poncirus trifoliata. The mRNA abundance of PtrERF9 in the VIGS plants (defined as TRV‐PtrERF9) was sharply reduced compared with the control plants (defined as TRV, Figure S4). Under normal growth condition, there was no difference in plant phenotype between the VIGS plants and the control. After the freezing treatment, the VIGS plants displayed more serious growth inhibition and leaf wilting, whereas the control plants just exhibited slight water‐soaking in the leaves (Figure 5a). In line with the morphology observation, substantially higher EL and MDA content, along with reduced chlorophyll fluorescence and Fv/Fm ratio, were detected in the TRV‐PtrERF9 plants relative to the control (Figure 5b–e). Moreover, we checked the expression pattern of six other homologous ERF genes in the TRV‐PtrERF9 plants and found that their mRNA abundance underwent negligible changes when PtrERF9 was silenced (Figure S5). These results illustrated that VIGS‐mediated knockdown of PtrERF9 led to enhanced cold sensitivity in trifoliate orange. Taken together, these findings demonstrated that PtrERF9 plays a positive role in mediating cold tolerance.
Figure 5.
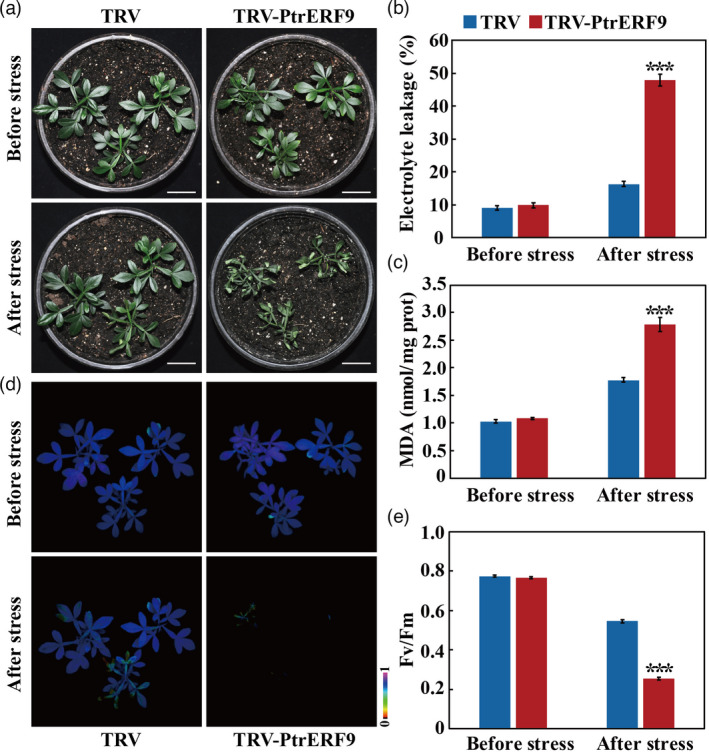
Silencing of PtrERF9 reduced the cold tolerance in trifoliate orange. (a) Phenotypes of TRV plants and TRV‐PtrERF9 plants before and after cold treatment. Scale bars = 5 cm. (b–e) Electrolyte leakage (b), MDA content (c), chlorophyll fluorescence (d) and F v/F m (e) of TRV control and TRV‐PtrERF9 plants before and after the cold treatment. Error bars indicate ± SE (n = 3). Asterisks indicate significant difference between the VIGS line and the TRV control plants under same conditions (***P < 0.001).
Knockdown of PtrERF9 causes dramatic alteration of transcriptome
For further elucidation of the molecular mechanism and identification of downstream target genes of PtrERF9, we performed RNA‐seq using the VIGS and control plants grown under normal conditions. A total of 3864 differentially expressed genes (DEGs), 1087 up‐regulated and 2777 down‐regulated, with significant changes in transcript levels (fold change ≥ 2, P ≤ 0.05) were revealed in the VIGS line relative to the control (Figure 6a). To verify the transcriptome profile, expression levels of 12 randomly selected down‐regulated DEGs were analysed by qPCR. The results demonstrated that there is a good correlation between the qPCR and the RNA‐seq data (Figure S6).
Figure 6.
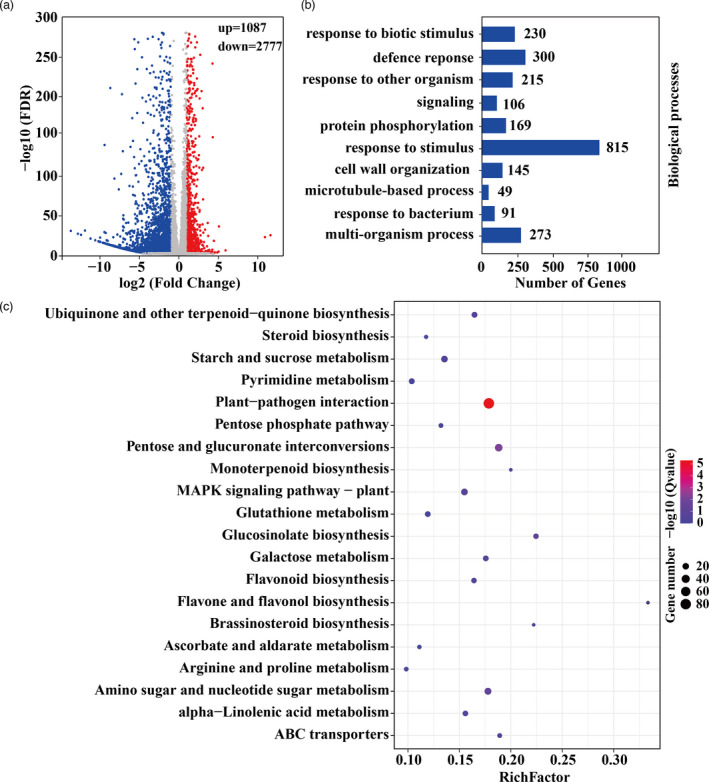
Silencing of PtrERF9 leads to transcriptional reprogramming of a larger number of genes in trifoliate orange. (a) Scatterplots of gene expression patterns in the VIGS plants compared with TRV control under normal condition. Blue and red circles represent down‐regulated and up‐regulated genes, respectively. (b) GO analysis of enrichment of down‐regulated genes in terms of biological process. (c) The top 20 enriched KEGG pathways among the down‐regulated DEGs.
We then performed GO and KEGG enrichment analyses to gain deeper insight into the down‐regulated DEGs in the VIGS line. GO analysis showed that the enriched DEGs were involved in various biological processes, with the largest number being grouped into ‘response to stimulus’ (Figure 6b). In addition, 15 out of the top 20 enriched KEGG pathways were related to metabolism or biosynthesis of a range of metabolites, including starch and sucrose, galactose, flavonoid, arginine and proline and glutathione (Figure 6c). In addition, MAPK signalling pathway was also included in the enriched KEGG pathway. An in‐depth analysis of the DEGs showed that some of them are well‐known stress‐related genes, including transcription factors (ZAT12 and NAC2), kinases or phosphatases (CDPK2 and PP2C2), enzymes involved in ROS scavenging (POD14, POD30 and AO1) and metabolism (FAD8, UDT76C4, GA2ox2 and PAL). Collectively, all these results demonstrated that PtrERF9 possibly regulated a large number of stress‐responsive genes at transcriptional levels to orchestrate the cold stress response.
PtrERF9 binds to the promoter of PtrGSTU17 and activates its expression
Among the DEGs, PtrGSTU17 (Pt4g003440) was found to be tremendously down‐regulated in the VIGS line. In addition, we found that mRNA abundance of PtrGSTU17 was steadily and progressively increased in the presence of cold treatment, suggesting that PtrGSTU17 was a cold‐responsive gene (Figure S7). Further analysis demonstrated that the PtrGSTU17 promoter contained one GCC‐box that is potentially recognized by ERF proteins (Figure 7a). To explore whether PtrERF9 interacts with PtrGSTU17 promoter, yeast one‐hybrid (Y1H) assay was performed. The yeast cells transformed with the prey and the bait containing P1 fragment harbouring the original GCC‐box, along with the positive control, grew well on the selection medium supplemented with Aureobasidin A (AbA), while mutation of the GCC‐box element (mP1) completely inhibited the yeast growth (Figure 7b,c), indicating that PtrERF9 could interact with the promoter of the PtrGSTU17 via the GCC‐box core sequence.
Figure 7.
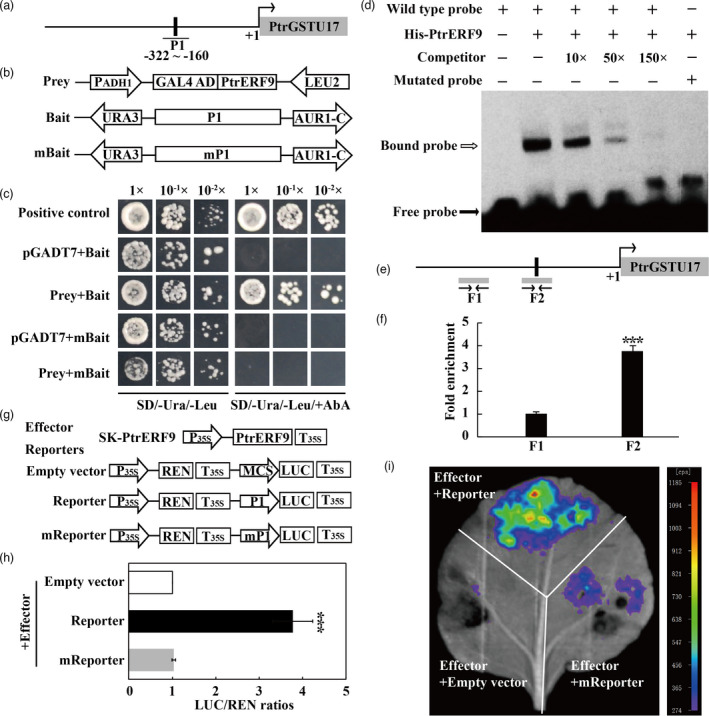
PtrERF9 binds to PtrGSTU17 promoter and positively regulates its expression. (a) Schematic diagram of the PtrGSTU17 promoter. The black rectangle indicates the position of the partial promoter fragment (P1) within GCC‐box elements. (b) The prey and bait vectors were used for yeast one‐hybrid assay (Y1H). mP1 is mutated form P1 fragment as the GCCGCC was changed to TCCTCC. (c) Growth of yeast cells co‐transformed with baits (P1 or mP1) and prey combinations (pGADT7‐PtrERF9), positive control (p53‐AbAi + pGAD‐p53), negative control (bait + pGADT7) on SD/‐Ura/‐Leu medium without (left) or with (right) AbA. (d) EMSA assay of specific binding of PtrERF9 to the GCC‐box of the PtrGSTU17 promoter. The purified His‐PtrERF9 protein was incubated with the biotin‐labelled probe containing the wild‐type or mutated GCC‐box element, with or without unlabelled probe was used as a competitor. Open and closed arrows indicate bound and free probes, respectively. +, presence; −, absence. (e) Schematic diagram of two PtrGSTU17 promoter fragments (F1, F2), shown as grey bars, used for ChIP‐qPCR analysis. GCC‐box element is marked by black bar. (f) ChIP‐qPCR assays revealed the enrichment of PtrERF9 in the promoter of PtrGSTU17 by using specific primers as indicated by arrows below the grey bars in (e). (g) Schematic diagrams of the effector and reporter constructs used for dual‐LUC transient expression assay. P35S and T35S, the promoter and terminator of CaMV 35S, respectively. MCS, multiple cloning sites. LUC, firefly luciferase. REN, Renilla luciferase. (h) Dual‐LUC transient expression assays of the promoter activity were measured by the LUC/REN ratios in tobacco (Nicotiana benthamiana) protoplasts. LUC/REN ratio of the negative control (SK‐PtrERF9 + pGreen0800 empty vector) was considered as 1 for normalization. (i) Representative bioluminescence image of the capability of PtrERF9 to regulate the PtrGSTU17 promoter activity in tobacco leaves. Error bars indicate ± SE (n = 3). Asterisks indicate that the value is significantly different from that of the control (***P < 0.001).
In order to further confirm this interaction, we carried out an electrophoretic mobility shift assay (EMSA) using His‐PtrERF9 fusion protein. Incubation of the fusion protein and biotin‐labelled probe containing the wild‐type GCC‐box led to shift of the protein‐DNA complex, which was reduced by adding the unlabelled competitor probe in a dosage‐dependent manner. In addition, no bind shift was detected when the mutated probe was incubated with the fusion protein (Figure 7d). These findings indicate that PtrERF9 directly and specifically bound in vitro to the GCC‐box element within the promoter of PtrGSTU17.
To examine the in vivo binding of PtrERF9 to the PtrGSTU17 promoter, chromatin immunoprecipitation (ChIP) assay with GFP antibody was performed (Figure S8). The ChIP‐qPCR results revealed that PtrERF9 was significantly enriched in the F2 region of PtrGSTU17 promoter containing a GCC‐box, but not in the F1 region without the GCC‐box (Figures 7e and f). In addition, we also performed a dual luciferase (LUC) assay to examine whether PtrERF9 could activate the PtrGSTU17 promoter. When the effector (SK‐PtrERF9) and the reporter containing the original GCC‐box sequence were transiently co‐expressed in N. benthamiana protoplasts, promoter activity, expressed by the LUC/REN ratio, was significantly elevated relative to the control. However, when the GCC‐box was mutated the LUC/REN ratio was resumed to the control level (Figure 7g,h). Quantitative measurement was also supported by microscopic visualization of the LUC fluorescence (Figure 7i). These results demonstrated that PtrERF9 could activate PtrGSTU17 expression by interacting with the GCC‐box within its promoter.
Alteration of GST enzymes activity and ROS levels in the transgenic and VIGS plants
In consideration of interaction and activation of PtrGSTU17 promoter by PtrERF9, we analysed GSTU17 expression and GST activity in the plants with overexpression and knockdown of PtrERF9. Transcript levels of GSTU17 and GST enzyme activity were increased in transgenic lemon plants overexpressing PtrERF9, but decreased in the VIGS line, relative to the corresponding WT or control in the absence or presence of cold treatment. It is noticed that cold‐induced expression of PtrGSTU17 and GST activity was more prominent in the PtrERF9‐overexpressing lines, but noticeably suppressed in the VIGS plants, when compared to the WT or control (Figures 8a–d).
Figure 8.
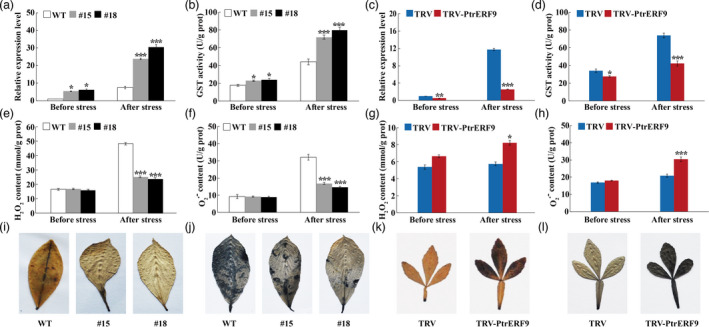
PtrERF9 regulates PtrGSTU17 expression to control ROS levels in response to cold stress. (a–d) PtrGSTU17 expression (a, c) and GST activity (b, d) in lemon wild‐type (WT) and transgenic plants (a, b) and TRV control plants and trifoliate orange VIGS (TRV‐PtrERF9) plants (c, d) before and after cold stress. (e–h) Levels of H2O2 (e, g) and O2 ·− content (f, h) in transgenic lemon and WT (e, f) and TRV control and VIGS plants (g, h) before and after cold treatment. (i–l) In situ detection of H2O2 (i, k) and O2 ·− (j, l) in the WT and transgenic lemons (i, j) and TRV control and VIGS plants (k, l) after cold treatment, as revealed by histochemical staining with DAB and NBT, respectively. Error bars indicate ± SE (n = 3). Asterisks indicate significant differences between different groups under the same growth condition (*P < 0.05; ***P < 0.001).
GST is known to play an essential role in scavenging ROS, including H2O2 and O2 ·−, which are known to be toxic or detrimental to cellular organelles. Therefore, we analysed ROS levels in the tested line. Under normal growth conditions, no profound difference in both H2O2 and O2 ·− levels was observed between the transgenic or VIGS lines and the WT or control. When subjected to the freezing treatment, H2O2 and O2 ·− levels were significantly lower in the PtrERF9‐overexpressing plants than in the WT. By contrast, the VIGS line contained significantly higher levels of H2O2 and O2 ·− than the control (Figure 8e–h). Histochemical staining with DAB and NBT is an effective way to detect in vivo accumulation of H2O2 and O2 ·−, respectively (Ming et al., 2021). The leaves from transgenic plants were stained by DAB and NBT to a much lighter extent, whereas staining of the VIGS line was evidently stronger, when compared to the WT or the control counterparts after the cold treatment (Figures 8i–l). These results showed that accumulation of ROS was dramatically alleviated in the PtrERF9‐overexpressing plants, but exacerbated when PtrERF9 was silenced.
PtrERF9 binds to promoter of PtrACS1 and activates its expression
Based on the results that ethylene is necessary for cold induction of PtrERF9, PtrERF9 was proposed to be possibly involved in ethylene signalling pathway. Examination of the RNA‐seq database showed that two ACC synthase (ACS)‐encoding genes (Pt3g038000 and Pt3g002280) were substantially repressed in the VIGS line in comparison with the control. However, promoter analysis of the two genes showed that only Pt3g038000 contained a canonical GCC‐box core sequence within its promoter. Pt3g03800 and its homologue of Arabidopsis thaliana were annotated as the ACS1 gene in the released genomes of these two plant species, so Pt3g03800 was named as PtrACS1 hereafter. In order to illustrate whether PtrERF9 could regulate PtrACS1 expression, we investigated interaction between PtrERF9 and the promoter of PtrACS1. Y1H assay showed that the yeast cells co‐transformed with prey and the bait containing the original PtrACS1 promoter fragment P2 with a genuine GCC‐box sequence grew normally on the selection medium containing AbA, whereas the growth was completely inhibited when the GCC‐box was mutated from GGCGGC to GGTGGC (Figure 9a–c). In addition, EMSA assay showed that an apparent band shift was observed when His‐PtrERF9 fusion protein was incubated with the labelled probe derived from the P2 sequence, whereas inclusion of the competitor DNA in the reaction resulted in a dosage‐dependent suppression of band migration. Furthermore, mutation of the GCC‐box completely abolished the band shift, suggesting that PtrERF9 bound directly and specifically to the GCC‐box within the PtrACS1 promoter (Figure 9d). Quantitative dual LUC assay and microscopic observation of LUC fluorescence demonstrated that PtrERF9 activated PtrACS1 though interacting with the GCC‐box, within the promoter, while mutation of the GCC‐box impaired the activation (Figure 9e‐g). In addition, the ChIP‐qPCR assay demonstrated that PtrERF9 was conspicuously enriched in the F3 region of the PtrACS1 promoter having a GCC‐box sequence, whereas no enrichment was observed in the F4 region without the cis‐acting element (Figures 9h,i). All of these results indicate that PtrERF9 regulated PtrACS1 expression through interacting with the GCC‐box core sequence.
Figure 9.
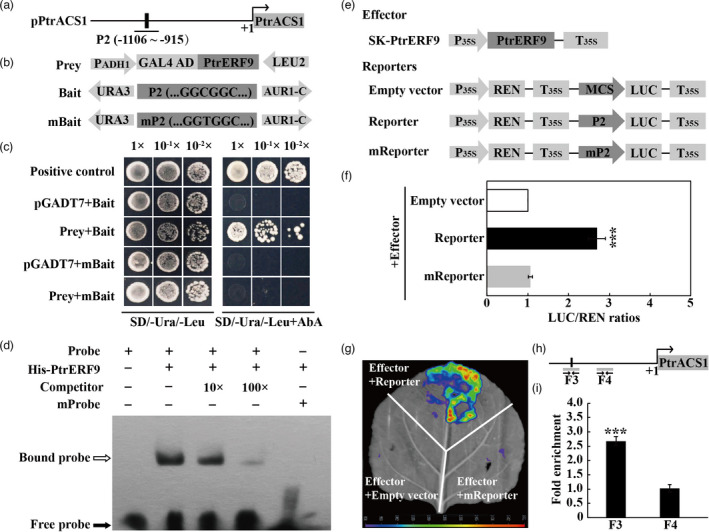
PtrERF9 binds to PtrACS1 promoter and positively regulates its expression. (a) Schematic diagram of PtrACS1 promoter, black rectangle indicates the GCC‐box element in the partial promoter fragment (P2). (b) Prey and bait constructs were used for Y1H analysis, the mutated bait mP2 is a mutated form P2 by changing ‘GGCGGC’ into ‘GGTGGC’. (c) Growth of yeast cells co‐transformed with the prey and bait on selective medium with or without AbA. Positive control: pGADT7‐Rec‐p53 + p53‐AbAi; Negative control: pGADT7 + bait. (d) EMSA assay using purified His‐PtrERF9 fusion protein incubated with biotin‐labelled probe containing wild‐type or mutated probe, along with or without unlabelled competitor DNA. Open and closed arrows indicate bound and free probes, respectively. +, presence; −, absence. (e) Schematic diagrams of effector and reporter constructs used for dual‐LUC transient assay. (f) The various combinations of vectors were performed on Dual‐LUC transient expression assays in tobacco protoplasts. (g) Representative bioluminescence image of PtrERF9 activation on the PtrACS1 promoter in tobacco leaves. (h) Schematic diagrams of F3 and F4 regions, shown as grey bars, in the PtrACS1 promoter, in which only F3 contains a GCC‐box sequence (black bar). (i) Enrichment of PtrERF9 in the PtrACS1 promoter based on ChIP‐qPCR assays using an anti‐GFP antibody. The arrows below the grey bars in (h) show the specific primers used for ChIP‐qPCR. Error bars indicate ± SE (n = 3). Asterisks indicate that the value is significantly different from that of the control (***P < 0.001).
Analysis with qPCR showed that transcript level of PtrACS1 was drastically down‐regulated in the VIGS line relative to the control (Figure 10a). Consistently, the ACS activity and ACC content were significantly decreased in the VIGS line as compared to the control (Figures 10b,c). Surprisingly, no difference in ACS1 gene expression, ACS activity and ACC content was detected between transgenic lemon lines overexpressing PtrERF9 and WT (Figure 10d–f). It seems that PtrERF9 could not regulate ACS1 gene of lemon (ClACS1). In order to confirm this assumption, we first compared the promoter sequences of ClACS1 and PtrACS1 as well as the AP2 domains of lemon ERF9 (ClERF9) and PtrERF9. The promoter sequence of ClACS1 was largely like that of PtrACS1 except presence of a mutation in the GCC‐box sequence, from GGCGGC in the PtrACS1 promoter to GGTGGC in the ClACS1 promoter (Figure S9). By contrast, the sequences of AP2 domains were completely identical between ClERF9 and PtrERF9 (Figure S10). This raised the question of whether the mutation of GCC‐box in the ClACS1 promoter impeded interaction between ClERF9 or PtrERF9 and ClACS1 promoter. To answer this question, we carried out Y1H assay to examine interaction between PtrERF9 or ClERF9 and the ClACS1 promoter. Yeast cells co‐transformed with either ClERF9 or PtrERF9 and the bait constructed using the ClACS1 promoter fragment P3 containing the GGTGGC sequence could not grow on the selection medium (Figure 10i), implying that PtrERF9 and ClERF9 failed to interact with the ClACS1 promoter. These findings indicate that PtrERF9 and ClERF9 could not regulate the ClACS1 gene due to mutation of the GCC‐box element within the promoter region.
Figure 10.
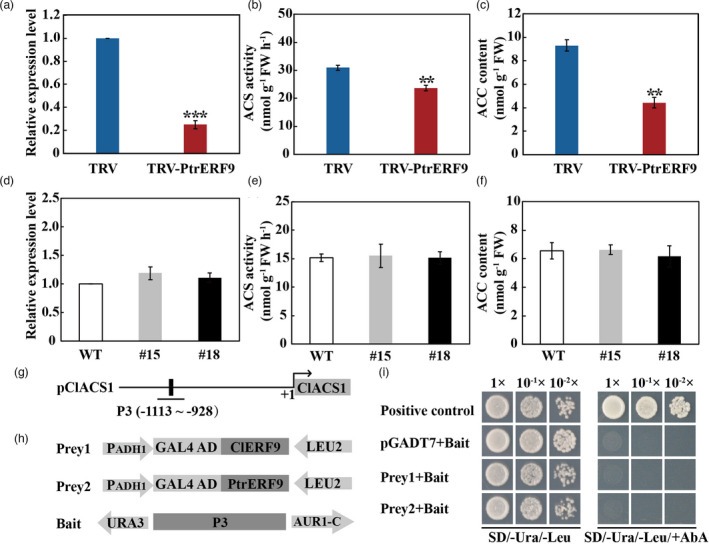
PtrERF9 regulates ethylene synthesis via controlling ACS expression in trifoliate orange for cold response. (a–c) ACS1 expression (a), ACS activity (b) and ACC content (c) in TRV control and PtrERF9‐silencing trifoliate orange. (d–f) ACS1 expression (d), ACS activity (e) and ACC content (f) in wild‐type and PtrERF9 overexpression transgenic lemon plants. Error bars indicate ± SE (n = 3). Asterisks indicate significant differences between different groups under the same growth condition (**P < 0.01; ***P < 0.001). (g) Schematic diagram of ClACS1 promoter, black rectangle indicated the GGTGGC motif in the partial promoter fragment (P3). (h) Prey and bait constructs were used for yeast one‐hybrid analysis. pGADT7‐PtrERF9 and pGADT7‐ClERF9 were used as prey. P3 fragment was used as bait. (i) Growth of yeast cells co‐transformed with prey (PtrERF9, ClERF9) and bait (P3) on selective medium added with or without AbA. positive control (p53‐AbAi + pGAD‐p53), negative control (bait + pGADT7).
Discussion
Plant AP2/ERFs have been known to be involved in plant adaption to various extreme environmental cues, including low temperature, oxidative stress, drought, heavy metals and nutrient starvation (Lin et al., 2017; Yao et al., 2017; Zhao et al., 2020a,b; Zhuo et al., 2018). However, the physiological and molecular mechanisms accounting for the functions of most ERFs in cold stress are not well characterized, particularly in perennial plants like Poncirus trifoliata. In this study, we characterized an ERF gene, PtrERF9, from P. trifoliata. PtrERF9 was explored through bioinformatics analysis of the ERF genes based on an earlier RNA‐seq dataset. The cold‐inducible silhouette was further confirmed by analysing expression patterns of the gene and the promoter. In line with the cold‐responsive tribute of PtrERF9, we demonstrated that PtrERF9 functioned as a positive regulator of cold tolerance, as overexpression and knockdown of the gene conferred enhanced and impaired cold tolerance, respectively. Our work adds PtrERF9 as a valuable member for the cold‐responsive gene repository for genetic engineering in an attempt to improve cold tolerance.
The most direct way for transcription factors to exert their functions is the regulation of downstream target genes. In this study, we analysed transcriptome profiles of the VIGS line and the control plants in order to know how knockdown of PtrERF9 influenced gene expression atlas. We predominantly focussed on the down‐regulated genes in the VIGS line, as PtrERF9 plays a positive role in modulation of cold tolerance. It showed that silencing of PtrERF9 led to alteration of numerous genes (DEGs) involved in a variety of biological processes and molecular functions. Many transcription factors and important components in the signalling network, such as CDPK and PP2C (Yang et al., 2018; Zhu, 2016), were down‐regulated in the VIGS line relative to the control. Of note, GO analysis showed that the biological process ‘response to stimulus’ contained the largest number of DEGs, implying that PtrERF9 acts as a significant regulator of cold stress response. Moreover, KEGG analysis revealed a large number of down‐regulated genes were associated with diverse metabolic pathways, implying that PtrERF9 may function in cold tolerance by modulating the levels of different metabolites, including fatty acids, flavonoid and phenol, via regulating the relevant genes. In the transcriptome data, we found that FAD, PAL and UDPG genes were substantially down‐regulated in the VIGS line. FAD (fatty acid desaturase) is the key enzyme responsible for biosynthesis of unsaturated fatty acids, which are implicated in modulation of cold tolerance by impacting the membrane fluidity and stabilization (Chen and Thelen, 2013). PAL (phenylalanine ammonia‐lyase) is a rate‐limiting enzyme in the phenylpropanoid pathway associated with the synthesis of secondary metabolites, such as flavonoids and phenols, which have been shown to function in protecting plants from environmental stress (Habibollahi et al., 2020). UDPG (UDP‐glycosyltransferase) plays an important role in the biosynthesis of eugenol glucoside or ABA glycosylation, which has been reported to be crucial for coping with abiotic stress (Chen et al., 2020; Zhao et al., 2020, 2020,2020, 2020). Therefore, PtrERF9 may possibly regulate biosynthesis of these metabolites to impart cold tolerance. However, more extra work is required to clarify this assumption in the future.
It has been documented that plants are well equipped with a highly complex dynamic ROS scavenging system composed of non‐enzymatic and enzymatic antioxidants to combat abiotic stresses, including cold. Thus, ROS homeostasis and relevant antioxidant metabolism are crucial for plant performance and survival under the cold stress. Based on this scenario, we screened the DEGs pertinent to the ROS scavenging and found that expression level of PtrGSTU17 was tremendously repressed in the VIGS line. GSTU is a plant‐specific GST that has been known to play a critical role in ROS scavenging. Integrative analyses indicated that PtrERF9 directly and specifically bound to the GCC‐box element within the promoter region of PtrGSTU17 and activated its transcription. This indicates that PtrGSTU17 is a direct downstream target gene of PtrERF9. To our knowledge, our work is the first to justify transcriptional regulation of a GSTU gene by an ERF protein. Although GST is activated by cold stress, the associated molecular mechanism remains unclear. The illustration of PtrERF9‐PtrGSTU17 module in this study may provide evidence to explain this physiological phenomenon, shedding light on the transcriptional regulation of GST activation in plants exposed to cold stress. In agreement with this regulatory pattern, expression level of GSTU17 and enzyme activity of GST were increased in the transgenic plants overexpressing PtrERF9, but reduced in the PtrERF9‐silenced line, in comparison with the WT or control plants. Consistent with these alterations, the transgenic plants accumulated significantly less ROS, whereas ROS level was noticeably increased in the VIGS plants, suggesting that ROS scavenging capacity was elevated by overexpression of PtrERF9, but repressed when PtrERF9 was knocked down. Hence, one of the physiological roles of PtrERF9 in cold tolerance is the modulation of ROS homeostasis through activating PtrGSTU17 to exert a more robust ROS scavenging capacity.
It is known that ethylene serves as an essential phytohormone involved in cold stress response. Previous studies have demonstrated that ethylene acted as either a positive or a negative regulator of cold tolerance (Shi et al., 2012; Sun et al., 2016, 2019; Zhang et al., 2009; Zhao et al., 2014), implying that the role of ethylene in cold tolerance differs among various plant species. In this study, we found cold treatment promoted ethylene production in trifoliate orange. Moreover, exogenous supply of ACC, the precursor of ethylene synthesis, resulted in enhanced cold tolerance, whereas the cold tolerance capacity was greatly impaired by AVG, the inhibitor of ethylene synthesis. These results showed that ethylene plays a positive role in modulation of cold tolerance in trifoliate orange. Numerous studies have demonstrated that ethylene participates in stress response by triggering the well‐defined ethylene signalling pathway (Argueso et al., 2007; Tao et al., 2015). A few important components are crucial for orchestrating the ethylene‐mediated signalling network, in which the ERFs are important players by relaying the stress signal to downstream stress response. Ethylene‐responsive ERFs involved in cold tolerance have been previously reported in various plants, such as ERF057 of grape (Sun et al., 2016), ERF1 of Medicago falcata (Zhuo et al., 2018) and ERF2 of tomato (Zhang and Huang, 2010). Herein, we also showed that exogenous ACC treatment increased mRNA abundance of PtrERF9, and up‐regulation of PtrERF9 under cold stress was dependent on ethylene. These findings indicated that PtrERF9 acts downstream of ethylene signalling to regulate its target genes for adaptation to the cold stress.
Ethylene biosynthesis involves two enzymes, ACS and ACC oxidase (ACO), in which ACS has been widely accepted as the rate‐limiting enzyme for ethylene production. In this study, two ACS‐encoding genes, PtrACS1 and PtrACS2, were down‐regulated in the PtrERF9‐silencing plants. Further bioinformatics analysis, along with Y1H, EMSA, LUC and ChIP‐qPCR assays, demonstrated that PtrERF9 could directly bind to the GCC‐box element within the promoter of PtrACS1 and activated its expression. These data indicate that PtrACS1 served as a direct target gene of PtrERF9. In accordance with this, expression level of PtrACS1 was dramatically reduced in the PtrERF9‐silenced plants, accompanied by lower ACS activity and ACC content, relative to control plants. Earlier studies have proposed that the ACS genes were regulated at transcriptional and posttranscriptional levels to control ethylene production under normal and stressful conditions (Catalá et al., 2014). Except PtrERF9, several other transcription factors, such as AtWRKY33, AtERF11 of Arabidopsis thaliana (Datta et al., 2015; Li et al., 2011; Zheng et al., 2019), ERF1B of apple (Wang et al., 2021), have been illustrated as the regulators of ACS gene expression and ethylene synthesis in response to various stresses, including pathogen infection, high salinity and cold stress. The facts that ethylene induced expression of PtrERF9 and that PtrERF9 regulated ethylene synthesis by targeting the ACS1 gene indicate that PtrERF9 might undergo a positive feedback regulation by ethylene under cold stress. In this regard, PtrERF9 could be fine‐tuned by ethylene to reinforce its transcriptional regulation of target genes for counteracting the cold stress. Trifoliate orange is extremely cold tolerant, while lemon is one of the most cold‐sensitive species in Citrus. Although ClERF9 of lemon had the same AP2 domain sequence as that of PtrERF9, surprisingly, ClERF9 and PtrERF9 could not interact with the promoter of ClACS1 due to mutation of the GCC‐box sequence. Absence of interaction between ClERF9 and ClACS1 indicates that ethylene synthesis of lemon is possibly weakened under cold stress. As a result, the feedback regulation of ethylene on ClERF9, as observed on PtrERF9, was alleviated. This may provide explanation for the slight induction of ClERF9 under cold stress (Figure S11). Interestingly, the discrepancy in the interaction of ERF9 and ACS1 between trifoliate orange and lemon is largely congruent with cold tolerance capacity of the two species. Therefore, we can assume that cold tolerance may be ascribed, at least partly, to the existence of a regulatory module consisting of ERF9‐ACS1, although other undetermined regulatory modules might also work in this process.
In conclusion, we have demonstrated that PtrERF9 of trifoliate orange, a cold‐responsive ERF transcription factor, plays a positive role in cold tolerance. By using RNA‐seq, we revealed that knockdown of PtrERF9 led to transcriptome reprogramming of a vast number of genes involved in various biological processes or molecular functions, in which PtrGSTU17 and PtrACS1 were confirmed as two downstream target genes of PtrERF9. PtrERF9 functioned in cold tolerance by modulation of ROS homeostasis through regulating PtrGSTU17‐mediated ROS scavenging. In addition, PtrERF9 was responsive to ethylene induction and could regulate ethylene biosynthesis via regulating PtrACS1, forming a feedback regulation loop. Based on our findings, we put forward a working model for PtrERF9‐mediated cold tolerance (Figure 11). Cold increases the ethylene production and induces the expression of PtrERF9. PtrERF9, on the one hand, binds to the promoter of PtrGSTU17 and activates its expression to promote ROS scavenging. On the other hand, PtrERF9 induces PtrACS1 expression and stimulates synthesis of ethylene, which is integrated into the existing ethylene pool to trigger further up‐regulation and activation of PtrERF9 for transcriptionally regulating downstream target genes to promote cold tolerance.
Figure 11.
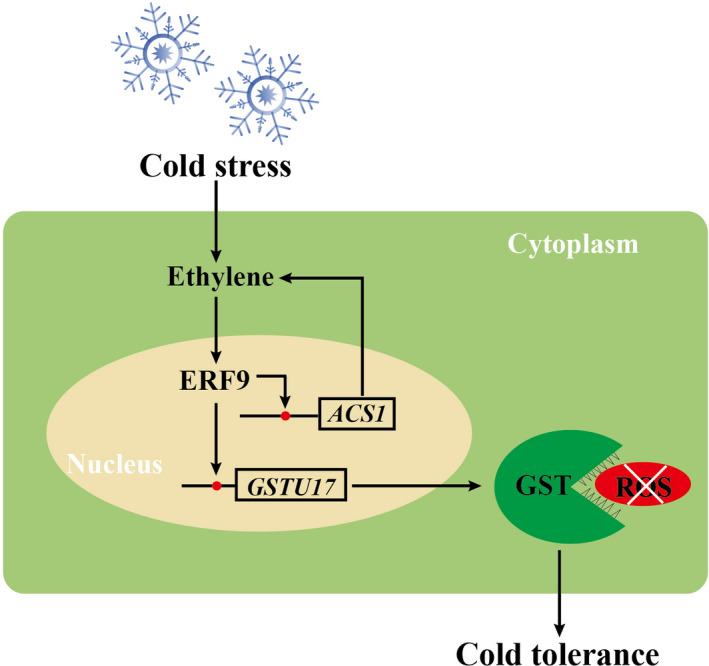
A proposed model for the regulatory function of PtrERF9 under cold stress. Cold stress induces ethylene production and activates the expression of PtrERF9. PtrERF9 binds to the GCC‐box in the promoter of PtrGSTU17, improves the plant ROS scavenging captivity. Additionally, PtrERF9 involves in the signalling pathway and acts as a feedback enhancer to elevate ethylene biosynthesis gene (PtrACS1) expression and ethylene level, and finally increases plant cold tolerance. The GCC‐box elements are shown by red circles.
Materials and methods
Plant materials and stress treatments
To verify the expression patterns of the PtrERF9, we performed qPCR via using the original materials kept at the −80 °C fridge which was used for high‐throughput sequencing as previously described (Wang et al., 2015). Wild‐type seeds of trifoliate orange (Poncirus trifoliata) and lemon (Citrus limon) were obtained from the Citrus Breeding Center at Huazhong Agricultural University. The seeds were grown in soil pots and kept in growth room (16‐h light/8‐h dark cycle) at 25 °C. Two‐month‐old seedlings were used for the following experiments. For ACC treatment, the seedlings were treated with 100 μM ACC, and leaves were collected at 0, 1, 3, 6, 12 and 24 h. For cold treatment, the seedlings were transferred into the growth chamber at 4 °C for 0, 3, 6, 12, 24 and 48 h, and the leaf samples were collected at the designated time points. For the ethylene inhibitor AVG treatment, 100 μM AVG was applied to plants under low‐temperature conditions at 4 °C and the leaves were collected at 0, 3, 6, 12, 24 and 48 h. All samples were immediately frozen in liquid nitrogen and stored at −80 °C until further analyses.
RNA extraction and qPCR analysis
Total RNA was isolated from the leaves using a TRIzol kit (TaKaRa, Ostu, Shiga, Japan), and the cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo, Waltham, MA, USA) according to the manufacturer’s instruction. Quantitative real‐time PCR (qPCR) was carried out using a QuantStudio 7 Flex instrument (Applied Biosystems, Foster City, CA, USA) with the SYBR® Green PCR kit (Vazyme, Nanjing, China). PCR reactions were performed in triplicate using 0.2 μM of forward and reverse primers, 5 μL of SYBR qPCR master mix and 200 ng of cDNA in a final volume of 10 μL. Cycling conditions were as follows: 5 min of denaturation at 95 °C, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. Actin and ubiquitin were used as the normalizing internal controls for citrus and tobacco, respectively. Relative gene expression levels were calculated using the 2–ΔΔCt method, and three replicates were used for all quantitative experiments. The primers for qPCR analysis are listed in Table S1.
Histochemical assay of GUS activity
A 1431bp promoter fragment upstream of the translation start site (TSS) of PtrERF9 was amplified with the primers and inserted into DX2181 vector within β‐glucuronidase (GUS) gene to generate pPtrERF9: GUS. The construct was transformed into sweet orange (Citrus sinensis) embryogenic callus as described in previous report (Dai et al., 2018). After culturing at ambient temperature for 2 d in the dark, the callus was treated at 4 °C for 2 d. Histochemical staining of GUS was performed by the histochemical assay kit (Zhongkeruitai, China) according to the manufacturer's protocol.
Cloning and sequence analysis of PtrERF9
An earlier transcriptome dataset (Wang et al., 2015) was analysed to unravel PtrERFs involved in cold response. PtrERF9 (Pt2g011090) was selected for further analysis. The full‐length coding sequence of PtrERF9 was amplified from Poncirus trifoliata leaf cDNA with the primers Pt2g011090‐F/R (Table S1) by PCR. The sequence analysis was performed to construct phylogenetic tree and multiple alignments by using MEGA X and GENEDOC software, respectively.
Subcellular localization analysis
The PtrERF9 full‐length CDS without the stop codon was amplified with primers containing EcoRI and BamHI restriction sites and then fused to the 101LYFP vector, under the control of the CaMV 35S promoter. The construct (35S: PtrERF9‐YFP) or the control vector (35S: YFP) was co‐transformed with the nuclear marker VirD2NLS‐mCherry into N. benthamiana leaves by Agrobacterium tumefaciens‐mediated transformation as described previously (Sparkes et al., 2006). DAPI was also used for staining of nucleus. The infiltrated plants were kept in growth chamber for another two days, and the fluorescent signals were observed with a laser scanning confocal microscope (Leica TCS‐SP8, Wetzlar, Germany).
Vector construction and plant transformation
For the PtrERF9 overexpression lines, the full‐length PtrERF9 CDS sequence was cloned into the pGWB411 vector according to the gateway recombination technology (Invitrogen). The resulting construct was transformed into tobacco (Nicotiana nudicaulis) and lemon (C. limon) via Agrobacterium. tumefaciens‐mediated transformation as previously reported (Fu et al., 2011). And then the transgenic explants were grown on MS (for tobacco; Murashige and Skoog, 1962) or MT (for lemon; Murashige and Tucker, 1969) medium containing 50 μg/mL kanamycin. Kanamycin‐resistant plants were selected which were further confirmed by qPCR assay. The transgenic tobacco lines at T2 generation and lemon lines in the vegetative period were used for phenotype analysis. Similarly, transient expression of PtrERF9 in trifoliate orange was performed according to Acanda et al. (2021) with minor modifications. To this end, full‐length cDNA of PtrERF9 was cloned into the pH7WG2D expression vector with a GFP tag. The 35S: PtrERF9‐GFP construct was transformed into Agrobacterium strain (EHA105) and then used for injection into the young tender leaves of three‐month‐old trifoliate orange seedlings. After 12 days of agroinfiltration, the infected leaves were subjected to fluorescence observation using a hand‐held lamp providing dual‐wavelength fluorescent protein excitation light source (LUYOR‐3415RG, USA). The leaves emitting the green fluorescence were used for Western blotting and ChIP‐qPCR assays.
Virus‐induced gene silencing
The VIGS system with the tobacco rattle virus was used to decrease PtrERF9 expression in trifoliate orange plants. A 390‐bp fragment of the PtrERF9 was amplified and inserted into pTRV2 vector to generate the pTRV2‐PtrERF9 construct. The vectors pTRV1, pTRV2 and pTRV2‐PtrERF9 were transformed into Agrobacterium tumefaciens strain GV3101. The Agrobacterium mixture containing pTRV1, pTRV2 or pTRV2‐PtrERF9 (v: v, 1: 1) was infiltrated into the germinating seeds of trifoliate orange with shoots (around 2 cm) as described previously (Dai et al., 2018; Wang et al., 2019). After infiltration, the plants were maintained at 25 °C in a growth room for 3 days under darkness and then the plants were washed with distilled water and transplanted into soil. After four weeks, the leaves of seedlings were subjected to qPCR assay to select the silenced plants for further analyses.
Cold tolerance assays
For cold stress, one‐month‐old transgenic tobacco and wild‐type (WT) seedlings were directly exposed at −4 °C for 6 h, followed by recovery at 25 °C for 3 d. In addition, for lemon, the wild‐type plans and transgenic lines were treated for 8 h at −4 °C, and then recovered for 5 d at ambient temperature. As for the VIGS trifoliate orange plants, two‐month‐old TRV control plants and the TRV‐PtrERF9‐VIGS lines were subjected to −4 °C treatment for 12 h and then recovered for 3 d at ambient temperature. In addition, two‐month‐old health wild‐type seedlings of trifoliate orange were transferred into triangle flasks containing water, or 100 μM ACC, or 100 μM AVG solution for 2 h prior to exposure at −5 °C for 9 h, followed by growth recovery for 2 d at 25 °C. The leaves of tested plants were harvested before or after cold treatment and then used for physiological analysis.
Physiological measurement and histochemical staining
Electrolyte leakage was measured as described in previous report (Dahro et al., 2016). MDA content, H2O2 content, O2 ·− level and GST activity were measured by using the relevant detection kits (A003‐1 for MDA; A064 for H2O2; A052 for O2 ·−, A004‐1 for GST activity, Nanjing Jiancheng Bioengineering Institute, China) according to manufacturer’s instructions. Total protein levels were tested using the Coomassie Brilliant Blue G‐250 staining method as previously described (Bradford, 1976). H2O2 and O2.− were also checked by histochemical staining with DAB and NBT as previously reported (Huang et al., 2013). The chlorophyll fluorescence was detected by using an IMAGING‐PAM chlorophyll fluorometer (Walz, Germany), and the Imaging WinGege software was used to measure the F v/F m ratio.
RNA‐seq analysis
High‐throughput RNA‐sequencing was used to identify downstream target genes of PtrERF9. Total RNA was extracted by TRIzol kit (TaKaRa, Japan) from leaves of PtrERF9‐silenced and TRV control plants according to the user manual. Three biological replicates were used for each genotype sample, and the individual total RNA quality and concentration were checked by Agilent 2100 Bioanalyzer and NanoDrop 2000 (Thermo Scientific). The cDNA libraries were constructed and then sequenced with BGISEQ‐500 platform, which was processed by the BGI Company (Shenzhen, China). The raw reads were filtered to remove reads with > 5% unknown nucleotides and low‐quality reads (Q‐value < 10). Then, the clean reads were mapped to the Poncirus trifoliata genome (Huang et al., 2021) by using HISAT and Bowtie2 software. The gene expression levels were calculated by FPKM method and the NOISeq software was used to identify differentially expressed genes in pair‐wise comparisons as described by Tarazona (Tarazona et al., 2015). Genes with expression fold change ≥ 2 and false discovery rate (FDR) < 0.05 were defined as DEGs. Gene ontology categories represented by DEGs were performed by the GOseq analysis (Young et al., 2010), and Kyoto Encyclopedia of Genes and Genomes (KEGG) classifications were performed using KABAS software as previously described (Mao et al., 2005).
Yeast one‐hybrid assay
Yeast one‐hybrid assay was used to check the binding of PtrERF9 to the PtrGSTU17/PtrACS1/ClACS1 promoter and was also used for testing the interaction between ClERF9 with the ClACS1 promoter. The PtrERF9/ClERF9 coding sequence was cloned into the NdeI‐EcoRI site of the pGADT7 vector as preys. The promoter fragment within GCC‐box (P1 for PtrGSTU17, −160 to −322, P2 for PtrACS1, −915 to −1106, P3 for ClACS1, −928 to −1113) and mP1 and mP2 mutated forms of P1 (GCCGCC mutated to TCCTCC) and P2 (GGCGGC mutated GGTGGC) was amplified and inserted into the pAbAi vector as baits. These preys and baits were combined and co‐transformed into yeast Y1H Gold strain using the Matchmaker Y1H library screening system (Clontech, Mountain View, CA, USA) following by the manufacturer’s protocol. Positive control (pGAD‐p53 + p53‐AbAi) and negative control (pGADT7‐AD + bait) control were also handled in the same method.
Electrophoretic mobility shift assay
The full‐length PtrERF9 was cloned into the expression vector pHMGWA; then, the construct was transformed into Escherichia coli strain Rosetta (DE3). The fused protein (His‐PtrERF9) was induced by 0.05 mM isopropyl‐β‐D‐1‐thiogalactopyranoside and purified using Ni‐NTA Agarose according to the manufacturer’s protocol (Qiagen, Germany). The probes containing GCC‐box or mutated elements were synthesized and labelled with biotin by Tsingke Biological Technology (Beijing, China). Unlabelled probe was used as the competitor. EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Pierce). All DNA‐binding reactions were carried out in 1× binding buffer containing 5 mM MgCl2, 50 mM KCl, 10 mM EDTA, 2.5% glycerol, 50 ng/μL Poly (dI‐dC) and 0.05% NP‐40. The reaction mixtures were separated on 6.5% nondenaturing polyacrylamide gel; then, gel was electroblotted onto the nylon membrane (Biosharp, Hefei, China), followed by chemiluminescence detection.
Chromatin immunoprecipitation‐qPCR assay
Immunoblot assays were performed as described by Acanda et al. (2021). The protein was incubated with anti‐GFP antibody (Abbkine, a02020, Wuhan, China) or actin antibody (ABclonal, ac009, Wuhan, China) and was then detected with HRP goat anti‐mouse IgG (H + L) antibody (ABclonal, aS003). ChIP‐qPCR assay was performed following the procedure as described previously (Ming et al., 2021). In brief, approximately 1 g of leaves collected from the PtrERF9‐GFP‐overexpressing or wild‐type plants was cross‐linked in 1% (w/v) formaldehyde, and the reaction was stopped by adding 125 mM glycine. After sonication, the anti‐GFP mAb‐magnetic agarose beads (MBL, Nagoya, Japan) were used to immunoprecipitate the chromatin. The recovered DNA was used to performed qPCR by using specific primers (Table S1). The fold enrichment of DNA fragments was calculated according to Chen et al. (2018).
Dual luciferase assay
The CDS of PtrERF9 was inserted into pGreen 62‐SK vector to generate the effector, and the original and mutated promoter fragments of PtrGSTU17 or PtrACS1 were recombined to the pGreen 0800‐LUC vector to generate reporters (Hellens et al., 2005). The recombinant plasmids were separately transformed into N. benthamiana leaves via the A. tumefaciens‐mediated transformation. The isolation of tobacco protoplasts was used to analyse transient expression. Luciferase activities were measured with Dual‐Luciferase® Reporter Assay Kit (Promega, Madison, WI, USA) according to the manufacturer’s instruction. Fluorescence was measured using an Infinite 200 Pro microplate reader (Tecan, Mannedorf, Switzerland) and NightSHADE LB 985 (Berthold, Germany).
Measurement of ethylene production
Ethylene was measured following previous description (Sun et al., 2016) with some minor modifications. Two‐month‐old trifoliate orange seedlings were incubated in beakers containing sterile deionized water under normal growth condition (16‐h light/8‐h dark cycle, 25 °C) for 3 d. Then, the plants were transferred to a growth chamber set at 4 °C. The seedlings were taken out at 0, 3, 6, 12, 24 and 48 h for leaves collection. Each plant leaves with petioles were weighed and put into 20 mL sealed bottle containing wet cottons. After 24 h room temperature incubation, 1 mL of gas was withdrawn with a syringe from each bottle and injected into a gas chromatograph (Agilent 7890B, Palo Alto, CA, USA) to quantify the ethylene production. ET production (μg/g/h) was determined in comparison with a standard curve of ethylene. All measurements were gained in triplicate from three independent samples.
Measurement of ACC content
Determination of ACC content was performed according to protocols described previously (Li et al., 2020) with some modifications. The leaf of plants was ground with liquid nitrogen, and about 0.05 g of tissues powder was placed into 2 mL tube and then vortexed with 1.5 mL 80% ethanol. The mixture was incubated at 80 °C for 15 min, and then was centrifuged for 15 min at 8000 g . The supernatant was transferred to a new 2 mL centrifuge tube with 0.5 mL 80% ethanol and then vortexed for 1 min. The content was incubated at 80 °C for 15 min and then centrifuged at 8000 g for 15 min. The supernatant was merged and dried in a Speed Vac (Labconco, Kansas City, MO, USA), and then 2 mL double‐distilled H2O with 0.5 mL CHCl3 was added to the tube to resuspend the dried product for 5 min. Finally, the mixture was centrifuged at 4000 g for 15 min to extract ACC in aqueous phase. Transfer 0.8 mL of suspended in two separate 20 mL vials (one of them to be used for the reaction, the other was added 20 μL 50 μM ACC as an internal standard), then 0.4 mL 50 mM HgCl2 was added into both vials. The vials were sealed with a serum cap. Then, 0.2 mL of 5% NaClO and saturated NaOH (2 : 1, v/v) were injected into the vials, which were vortexed for 10 s and incubated on ice for 15 min. One mL of the head space gas was sampled and assayed for ethylene as described above. The ACC content was calculated from the reaction efficiency of ACC converted into ethylene.
Measurement of ACS activity
ACS activity was measured according to previous protocol (Zhao et al., 2013) with minor modifications. Plants were frozen in liquid nitrogen, and the 0.05 g tissue was ground with 2 mL 0.1 M KBS buffer (pH = 8) and then was gently shaken 30 min at 4 °C. The homogenate was centrifuged for 30 min at 4000 g at 4 °C, and the supernatant was collected for ACS activity measurement. The 0.5 mL extract was mixed with 1.5 mL reaction buffer containing 0.05 M KBS (pH = 8), 250 μM SAM, 1 mM PLP; then, the mixture was placed into a 20 mL bottle with a rubber stopper and incubated 1 h at 30 °C. The purified extract was reacted with chlorinated SAM, and the amount of ACC formed was quantified by chemically converting it to ethylene. The following step of ACC measurement was performed according to the method described above.
Statistical analysis
All the experiments were repeated at least three times for each study. Statistical differences were evaluated using one‐way analysis of variance (ANOVA) method in SPSS (IBM, NY) based on a t test, taking * P < 0.05, ** P < 0.01 and *** P < 0.001 as significant.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
J.H.L. and Y.Z. conceived and designed the research. Y.Z. performed the experiments. Y.W. assisted bioinformatics analysis. Y.Z., R.M., M.K., B.D. and W.X. analysed the data. Y.Z. and C.L. wrote the manuscript draft, J.H.L. finalized writing and revision of the manuscript. All authors have read and approved the final version of the manuscript.
Supporting information
Figure S1 Phylogenetic analysis and sequence alignments of PtrERF9 and ERFs from Poncirus trifoliata and other plants.
Figure S2 Generation and molecular identification of transgenic tobacco plants overexpressing PtrERF9.
Figure S3 Generation and molecular Identification of transgenic lemon plants overexpressing PtrERF9.
Figure S4 Molecular characterization of the PtrERF9‐VIGS plants.
Figure S5 Expression levels of six ERF genes in TRV control and PtrERF9‐silencing trifoliate orange.
Figure S6 Validation of differentially expressed genes by qPCR analysis.
Figure S7 Relative expression of PtrGSTU17 at the designated time points of cold treatment by qPCR.
Figure S8 Analysis of PtrERF9‐GFP protein level in the Poncirus trifoliata leaves transiently expressing PtrERF9 by western blot.
Figure S9 Comparison and analysis of PtrACS1 and ClACS1 promoters.
Figure S10 Sequence alignments of PtrERF9 and ClERF9.
Figure S11 Expression levels of ClERF9 from Citrus limon under cold treatment.
Table S1 List of primers used in this study.
Table S2 Summary of RNA‐seq results.
Table S3 Up‐ and down‐regulated DEGs in RNA‐seq.
Acknowledgements
This work was supported by National Key Research and Development Program of China (2018YFD1000302), and the National Natural Science Foundation of China (31772273, 31972377).
Zhang, Y. , Ming, R. , Khan, M. , Wang, Y. , Dahro, B. , Xiao, W. , Li, C. and Liu, J.‐H. (2022) ERF9 of Poncirus trifoliata (L.) Raf. undergoes feedback regulation by ethylene and modulates cold tolerance via regulating a glutathione S‐transferase U17 gene. Plant Biotechnol. J., 10.1111/pbi.13705
Contributor Information
Chunlong Li, Email: cl2444@mail.hzau.edu.cn.
Ji‐Hong Liu, Email: liujihong@mail.hzau.edu.cn.
References
- Acanda, Y. , Welker, S. , Orbović, V. and Levy, A. (2021) A simple and efficient agroinfiltration method for transient gene expression in Citrus . Plant Cell Rep. 40, 1171–1179. [DOI] [PubMed] [Google Scholar]
- Argueso, C.T. , Hansen, M. and Kieber, J.J. (2007) Regulation of ethylene biosynthesis. J. Plant Growth Regul. 26, 92–105. [Google Scholar]
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Catalá, R. , López‐Cobollo, R. , Mar Castellano, M. , Angosto, T. , Alonso, J.M. , Ecker, J.R. and Salinas, J. (2014) The Arabidopsis 14‐3‐3 protein RARE COLD INDUCIBLE 1A links low‐temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell, 26, 3326–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Zhao, Y. , Xu, S. , Zhang, Z. , Xu, Y. , Zhang, J. and Chong, K. (2018) OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol., 218, 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M. and Thelen, J.J. (2013) ACYL‐LIPID DESATURASE2 is required for chilling and freezing tolerance in Arabidopsis. Plant Cell, 25, 1430–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T.T. , Liu, F.F. , Xiao, D.W. , Jiang, X.Y. , Li, P. , Zhao, S.M. et al. (2020) The Arabidopsis UDP‐glycosyltransferase75B1, conjugates abscisic acid and affects plant response to abiotic stresses. Plant Mol. Biol., 102, 389–401. [DOI] [PubMed] [Google Scholar]
- Considine, M.J. and Foyer, C.H. (2014) Redox regulation of plant development. Antioxid. Redox Signal. 21, 1305–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, X.Y. , Gao, Y. , Guo, J. , Yu, T.F. , Zheng, W.J. , Liu, Y.W. et al. (2019) BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1 . Plant Physiol. 180, 605–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahro, B. , Wang, F. , Peng, T. and Liu, J.H. (2016) PtrA/NINV, an Alkaline/neutral invertase gene of Poncirus trifoliata, confers enhanced tolerance to multiple abiotic stresses by modulating ROS levels and maintaining photosynthetic efficiency. BMC Plant Biol. 16, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, W.S. , Wang, M. , Gong, X. and Liu, J.H. (2018) The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol. 219, 972–989. [DOI] [PubMed] [Google Scholar]
- Datta, R. , Kumar, D. , Sultana, A. , Hazra, S. , Bhattacharyya, D. and Chattopadhyay, S. (2015) Glutathione regulates 1‐aminocyclopropane‐1‐carboxylate synthase transcription via WRKY33 and 1‐aminocyclopropane‐1‐carboxylate oxidase by modulating messenger RNA stability to induce ethylene synthesis during stress. Plant Physiol. 169, 2963–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y. , Shi, Y. and Yang, S. (2019) Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222, 1690–1704. [DOI] [PubMed] [Google Scholar]
- Ding, Y. , Shi, Y. and Yang, S. (2020) Molecular regulation of plant responses to environmental temperatures. Mol. Plant. 13, 544–564. [DOI] [PubMed] [Google Scholar]
- Fu, X.Z. , Khan, E.U. , Hu, S.S. , Fan, Q.J. and Liu, J.H. (2011) Overexpression of the betaine aldehyde dehydrogenase gene from Atriplex hortensis enhances salt tolerance in the transgenic trifoliate orange (Poncirus trifoliata L. Raf.). Environ. Exp. Bot. 74, 106–113. [Google Scholar]
- Guo, X. , Liu, D. and Chong, K. (2018) Cold signaling in plants: insights into mechanisms and regulation. J. Integr. Plant Biol. 60, 745–756. [DOI] [PubMed] [Google Scholar]
- Habibollahi, M. , Kavousi, H.R. , Lohrasbi‐Nejad, A. and Rahpeyma, S.A. (2020) Cloning, characterization and expression of a phenylalanine ammonia‐lyase gene (CcPAL) from cumin (Cuminum cyminum L.). J. Appl. Res. Med. Aromat. Plants 18, 100253. [Google Scholar]
- Hellens, R.P. , Allan, A.C. , Friel, E.N. , Bolitho, K. , Grafton, K. , Templeton, M.D. , Karunairetnam, S. et al. (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods, 1, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth, E. , Bela, K. , Gallé, Á. , Riyazuddin, R. , Csomor, G. , Csenki, D. and Csiszár, J. (2020) Compensation of mutation in arabidopsis glutathione transferase (AtGSTU) genes under control or salt stress conditions. Int. J. Mol. Sci. 21, 2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Jiang, Y. , Han, X. , Wang, H. , Pan, J. and Yu, D. (2017) Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot. 68, 1361–1369. [DOI] [PubMed] [Google Scholar]
- Huang, X.S. , Wang, W. , Zhang, Q. and Liu, J.H. (2013) A basic helix‐loop‐helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase‐mediated scavenging of hydrogen peroxide. Plant Physiol. 162, 1178–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. , Xu, Y. , Jiang, X. , Yu, H. , Jia, H. , Tan, C. et al. (2021) Genome of a citrus rootstock and global DNA demethylation caused by heterografting. Hortic. Res. 8, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha, B. , Sharma, A. and Mishra, A. (2011) Expression of SbGSTU (tau class glutathione S‐transferase) gene isolated from Salicornia brachiata in tobacco for salt tolerance. Mol. Biol. Rep. 38, 4823–4832. [DOI] [PubMed] [Google Scholar]
- Li, D. , Flores‐Sandoval, E. , Ahtesham, U. , Coleman, A. , Clay, J.M. , Bowman, J.L. and Chang, C. (2020) Ethylene‐independent functions of the ethylene precursor ACC in Marchantia polymorpha . Nat. Plants 6, 1–10. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Zhang, L. , Yu, Y. , Quan, R. , Zhang, Z. , Zhang, H. and Huang, R. (2011) The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J. 68, 88–99. [DOI] [PubMed] [Google Scholar]
- Licausi, F. , Ohme‐Takagi, M. and Perata, P. (2013) APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. [DOI] [PubMed] [Google Scholar]
- Lin, T. , Yang, W. , Lu, W. , Wang, Y. and Qi, X. (2017) Transcription factors PvERF15 and PvMTF‐1 form a cadmium stress transcriptional pathway. Plant Physiol., 173, 1565–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, X. , Cai, T. , Olyarchuk, J.G. and Wei, L. (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics, 21, 3787–3793. [DOI] [PubMed] [Google Scholar]
- Ming, R. , Zhang, Y. , Wang, Y. , Khan, M. , Dahro, B. and Liu, J.H. (2021) The JA‐responsive MYC2‐BADH‐like transcriptional regulatory module in Poncirus trifoliata contributes to cold tolerance by modulation of glycine betaine biosynthesis. New Phytol. 229, 2730–2750. [DOI] [PubMed] [Google Scholar]
- Müller, M. and Munné‐Bosch, S. (2015) Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T. and Skoog, F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plantarum. 15, 473–497. [Google Scholar]
- Murashige, T. and Tucker, D.P.H. (1969) Growth factor requirement of citrus tissue culture. Proc 1st Int Citrus Symp. 3, 1155–1161. [Google Scholar]
- Peng, L. , Lang, S. , Wang, Y. , Pritchard, H.W. and Wang, X. (2017) Modulating role of ROS in re‐establishing desiccation tolerance in germinating seeds of Caragana korshinskii Kom. J. Exp. Bot. 68, 3585–3601. [DOI] [PubMed] [Google Scholar]
- Peng, T. , Zhu, X. , Duan, N. and Liu, J.H. (2014) PtrBAM1, a β‐amylase‐coding gene of Poncirus trifoliata, is a CBF regulon member with function in cold tolerance by modulating soluble sugar levels. Plant Cell Environ. 37, 2754–2767. [DOI] [PubMed] [Google Scholar]
- Rezaei, M.K. , Shobbar, Z.S. , Shahbazi, M. , Abedini, R. and Zare, S. (2013) Glutathione S‐transferase (GST) family in barley: Identification of members, enzyme activity, and gene expression pattern. J. Plant Physiol. 170, 1277–1284. [DOI] [PubMed] [Google Scholar]
- Schmidt, R. , Mieulet, D. , Hubberten, H.‐M. , Obata, T. , Hoefgen, R. , Fernie, A.R. , Fisahn, J. et al. (2013) Salt‐responsive ERF1 regulates reactive oxygen species‐dependent signaling during the initial response to salt stress in rice. Plant Cell, 25, 2115–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. , Tian, S. , Hou, L. , Huang, X. , Zhang, X. , Guo, H. and Yang, S. (2012) Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type‐A ARR genes in Arabidopsis . Plant Cell, 24, 2578–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes, I.A. , Runions, J. , Kearns, A. and Hawes, C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. [DOI] [PubMed] [Google Scholar]
- Sun, X. , Zhang, L. , Wong, D.C.J. , Wang, Y. , Zhu, Z. , Xu, G. et al. (2019) The ethylene response factor VaERF092 from Amur grape regulates the transcription factor VaWRKY33, improving cold tolerance. Plant J. 99, 988–1002. [DOI] [PubMed] [Google Scholar]
- Sun, X. , Zhao, T. , Gan, S. , Ren, X. , Fang, L. , Karungo, S.K. et al. (2016) Ethylene positively regulates cold tolerance in grapevine by modulating the expression of ETHYLENE RESPONSE FACTOR 057. Sci. Rep. 6, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, J.J. , Chen, H.W. , Ma, B. , Zhang, W.K. , Chen, S.Y. and Zhang, J.S. (2015) The role of ethylene in plants under salinity stress. Front. Plant Sci. 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona, S. , Furió‐Tarí, P. , Turrà, D. , Di Pietro, A. , Nueda, M.J. , Ferrer, A. and Conesa, A. (2015) Data quality aware analysis of differential expression in RNA‐seq with NOISeq R/Bioc package. Nucleic Acids Res., 43, e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, M.O. , Moore, M. , König, K. , Pecher, P. , Alsharafa, K. , Lee, J. and Dietz, K.J. (2014) Fast retrograde signaling in response to high light involves metabolite export, MITOGEN‐ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell, 26, 1151–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Dai, W. , Du, J. , Ming, R. , Dahro, B. and Liu, J.H. (2019) ERF109 of trifoliate orange (Poncirus trifoliata (L.) Raf.) contributes to cold tolerance by directly regulating expression of Prx1 involved in antioxidative process. Plant Biotechnol. J. 17, 1316–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Zhang, X. and Liu, J.H. (2015) Deep sequencing‐based characterization of transcriptome of trifoliate orange (Poncirus trifoliata (L.) Raf.) in response to cold stress. BMC Genom. 16, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Jiang, H. , Mao, Z. , Liu, W. , Jiang, S. , Xu, H. , Su, M. et al. (2021) Ethylene increases the cold tolerance of apple via the MdERF1B–MdCIbHLH1 regulatory module. Plant J. 106, 379–393. [DOI] [PubMed] [Google Scholar]
- Xu, N. , Chu, Y. , Chen, H. , Li, X. , Wu, Q.I. , Jin, L. , Wang, G. et al. (2018) Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA‐mediated regulatory pathway and ROS scavenging. PLoS Genet, 14, 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q.I. , Liu, K. , Niu, X. , Wang, Q.I. , Wan, Y. , Yang, F. , Li, G. et al. (2018) Genome‐wide identification of PP2C genes and their expression profiling in response to drought and cold stresses in Medicago truncatula . Sci. Rep, 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Y. , He, R.J. , Xie, Q.L. , Zhao, X.H. , Deng, X.M. , He, J.B. , Song, L. et al. (2017) ETHYLENE RESPONSE FACTOR 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog D (RbohD)‐dependent mechanism in response to different stresses in Arabidopsis. New Phytol. 213, 1667–1681. [DOI] [PubMed] [Google Scholar]
- Young, M.D. , Wakefield, M.J. , Smyth, G.K. and Oshlack, A. (2010) Gene ontology analysis for RNA‐seq: accounting for selection bias. Genome Biol. 11, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Yang, J. , Li, W. , Chen, Y. , Lu, H. , Zhao, S. et al. (2019b) PuHSFA4a enhances tolerance to excess zinc by regulating reactive oxygen species production and root development in Populus . Plant Physiol. 180, 2254–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Li, X.M. , Lin, H.X. and Chong, K. (2019a) Crop improvement through temperature resilience. Annu. Rev. Plant Biol. 70, 753–780. [DOI] [PubMed] [Google Scholar]
- Zhang, Z.J. and Huang, R.F. (2010) Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. Biol. 73, 241–249. [DOI] [PubMed] [Google Scholar]
- Zhang, Z.J. , Zhang, H.W. , Quan, R.D. , Wang, X.C. and Huang, R.F. (2009) Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol. 150, 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D. , Shen, L. , Fan, B. , Yu, M. , Zheng, Y. , Lv, S. and Sheng, J. (2009) Ethylene and cold participate in the regulation of LeCBF1 gene expression in postharvest tomato fruits. FEBS Lett. 583, 3329–3334. [DOI] [PubMed] [Google Scholar]
- Zhao, M. , Cai, B. , Jin, J. , Zhang, N. , Jing, T. , Wang, J. et al. (2020) Cold stress‐induced glucosyltransferase CsUGT78A15 is involved in the formation of eugenol glucoside in Camellia sinensis . Hortic. Plant J. 6, 439–449. [Google Scholar]
- Zhao, M. , Liu, W. , Xia, X. , Wang, T. and Zhang, W.H. (2014) Cold acclimation‐induced freezing tolerance of Medicago truncatula seedlings is negatively regulated by ethylene. Physiol. Plant. 152, 115–129. [DOI] [PubMed] [Google Scholar]
- Zhao, Q. , Hu, R.S. , Liu, D. , Liu, X. , Wang, J. , Xiang, X.H. and Li, Y.Y. (2020) The AP2 transcription factor NtERF172 confers drought resistance by modifying NtCAT . Plant Biotechnol. J. 18, 2444–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, R. , Xie, H. , Lv, S. , Zheng, Y. , Yu, M. , Shen, L. and Sheng, J. (2013) LeMAPK4 participated in cold‐induced ethylene production in tomato fruit. J. Sci. Food Agric. 93, 1003–1009. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , Xing, J. , Zhang, K. , Pang, X. , Zhao, Y. , Wang, G. et al. (2019) Ethylene response factor ERF11 activates BT4 transcription to regulate immunity to Pseudomonas syringae . Plant Physiol. 180, 1132–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2016) Abiotic stress signaling and responses in plants. Cell, 167, 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo, C. , Liang, L. , Zhao, Y. , Guo, Z. and Lu, S. (2018) A cold responsive ethylene responsive factor from Medicago falcata confers cold tolerance by up‐regulation of polyamine turnover, antioxidant protection, and proline accumulation. Plant Cell Environ. 41, 2021–2032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Phylogenetic analysis and sequence alignments of PtrERF9 and ERFs from Poncirus trifoliata and other plants.
Figure S2 Generation and molecular identification of transgenic tobacco plants overexpressing PtrERF9.
Figure S3 Generation and molecular Identification of transgenic lemon plants overexpressing PtrERF9.
Figure S4 Molecular characterization of the PtrERF9‐VIGS plants.
Figure S5 Expression levels of six ERF genes in TRV control and PtrERF9‐silencing trifoliate orange.
Figure S6 Validation of differentially expressed genes by qPCR analysis.
Figure S7 Relative expression of PtrGSTU17 at the designated time points of cold treatment by qPCR.
Figure S8 Analysis of PtrERF9‐GFP protein level in the Poncirus trifoliata leaves transiently expressing PtrERF9 by western blot.
Figure S9 Comparison and analysis of PtrACS1 and ClACS1 promoters.
Figure S10 Sequence alignments of PtrERF9 and ClERF9.
Figure S11 Expression levels of ClERF9 from Citrus limon under cold treatment.
Table S1 List of primers used in this study.
Table S2 Summary of RNA‐seq results.
Table S3 Up‐ and down‐regulated DEGs in RNA‐seq.


