Double haploid (DH) technology, based on in vivo haploid induction, enables the fixation of recombinant haplotypes within two generations, thereby greatly increasing crop breeding efficiency (Jacquier et al., 2020). Although haploid plants can be produced from some legumes via an in vitro anther/microspore culture approach (Croser et al., 2006), an in vivo (seed‐based) haploid induction system has not yet been established for this family, hindering the application of DH technology. Here, we report the successful generation of haploid plants through seeds by editing DMP (DOMAIN OF UNKNOWN FUNCTION 679) homologues in Medicago truncatula, a well‐characterized model legume.
Mutations in ZmDMP were shown to enhance haploid induction in maize (Zea mays) when combined with mutations in MTL/NLD/ZmPLA1 (Gilles et al., 2017; Kelliher et al., 2017; Liu et al., 2017; Zhong et al., 2019). Although MTL/NLD/ZmPLA1 is not conserved in dicots, DMP is conserved in both monocots and dicots (including legumes), and loss of function ZmDMP orthologues in the dicot Arabidopsis (Arabidopsis thaliana) trigger maternal haploid induction (Zhong et al., 2020), opening the possibility of applying the DMP‐triggered in vivo haploid induction system to leguminous plants. In agreement with previous reports (Zhong et al., 2019, 2020), phylogenetic analysis showed that ZmDMP has homologues in several legumes, including soybean (Glycine max), alfalfa (Medicago sativa) and M. truncatula (Figure 1a). Using M. truncatula, we explored whether the mutation of DMP homologues might be used for haploid induction in legumes.
Figure 1.
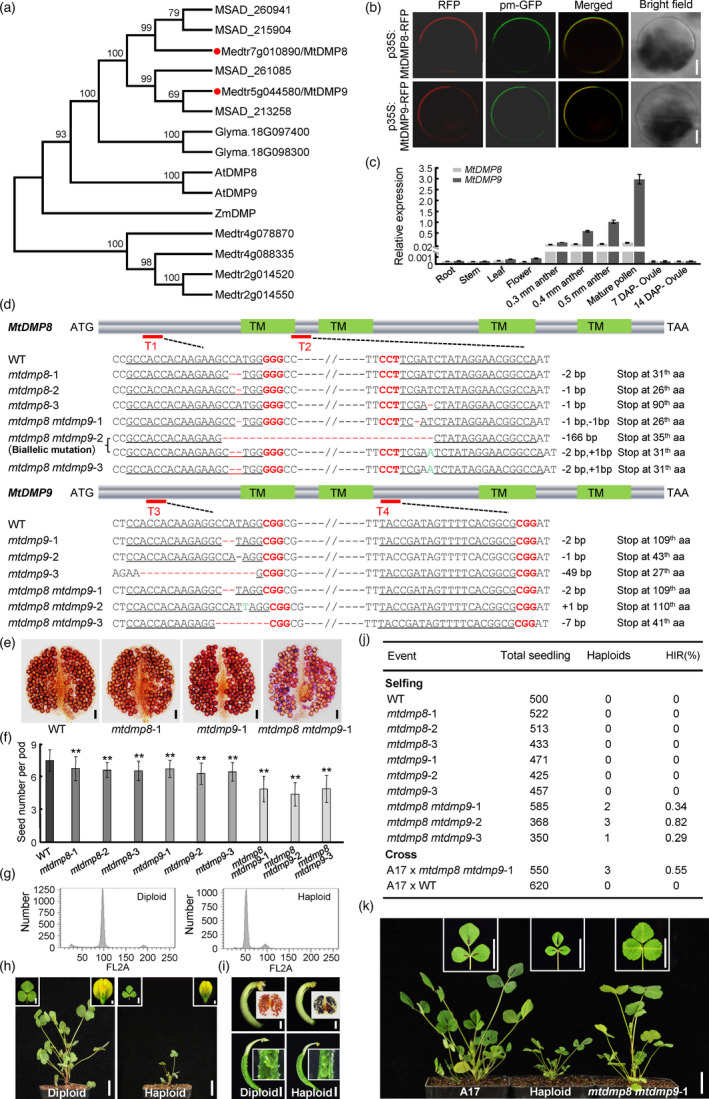
Inactivation of DMP homologues triggers haploid induction in Medicago truncatula. (a) Phylogenetic analysis of ZmDMP and its homologues in M. truncatula (Medtr or Mt), Arabidopsis (At), alfalfa (MSAD) and soybean (Glyma.). MtDMP8 and MtDMP9 are highlighted with red dots. Full‐length protein sequences were aligned using ClustalW, and a neighbour‐joining phylogenetic tree was constructed using MEGA6 software. Numbers on branches indicate bootstrap percentages for 1000 replicates. (b) Subcellular localization of MtDMP8‐RFP and MtDMP9‐RFP proteins in Arabidopsis leaf protoplasts; pm‐GFP was used as a plasma membrane marker. Bars, 5 μm. (c) Relative transcript levels of MtDMP8 and MtDMP9 in the indicated tissues, as determined by RT‐qPCR. MtActin was used as an internal control. Values are means ± SD of three technical replicates. Three independent experiments were performed, with similar results. (d) Schematic representation of MtDMP8 and MtDMP9 gene structures and genome editing experimental design. Filled blocks indicate the coding region. Green blocks correspond to the regions encoding the four predicted transmembrane domains (TMs). Red lines indicate the four regions (T1–4) targeted by sgRNAs. The relevant sequences from the wild‐type (WT) and mutant alleles are shown below the gene structure schematics. (e) Pollen viability assays with Alexander’s stain in the T1 progeny of selfed WT and mtdmp mutants. Bars, 50 μm. (f) Comparison of seed number per pod in the T1 progeny of selfed WT and mtdmp mutants. Bars represent means ± SD (n = 30); asterisks indicate significant differences from the WT (**P < 0.01, Student’s t‐test). (g) Confirmation of ploidy by flow cytometry analysis. (h) Phenotypic differences between M. truncatula haploid and diploid plants (whole plant, leaf and flower). Bars, 2 cm for whole plant; 5 mm for leaf; and 1 mm for flower. (i) Comparison of anther and pollen viability, as well as carpels and ovules between haploid and diploid M. truncatula plants. Bars, 1 mm. (j) Haploid induction rate (HIR) determined by self‐pollination or crossing. For crossing, the M. truncatula ecotype A17 was used as the female parent and was pollinated with mtdmp8 mtdmp9‐1. (k) Representative haploid plant from crossing. Bars, 2 cm.
We searched the M. truncatula genome (v4.0) using a Basic Local Alignment Sequence Tool for Protein (BLASTP) analysis and ZmDMP as query. When using a minimum protein sequence identity of 40%, we identified six putative DMP‐like proteins. Phylogenetic analysis showed that MtDMP8 (Medtr7g010890) and MtDMP9 (Medtr5g044580), which are most similar to ZmDMP (63.9% and 62.8% sequence identity, respectively), cluster together with ZmDMP in a separate subclade that includes Arabidopsis DMP8 and DMP9 (Figure 1a). MtDMP8 and MtDMP9 both contained four putative transmembrane domains. Consistent with this prediction, both proteins colocalized with the PIP2A (At3g53420)‐based plasma membrane marker pm‐GFP (Zhu et al., 2020) when MtDMP8 and MtDMP9 were transiently expressed as red fluorescent protein (RFP) fusions in Arabidopsis leaf protoplasts (Figure 1b). RT‐qPCR analysis revealed that both MtDMP8 and MtDMP9 are highly expressed in mature anthers and pollen, with MtDMP9 being more highly expressed, suggesting that MtDMP8 and MtDMP9 function during the late stages of gametophyte development (Figure 1c).
To assess the role of MtDMP8 and MtDMP9 in haploid induction in M. truncatula, we generated single and double knockout mutants in MtDMP8 or MtDMP9 (Figure 1d) using the pDIRECT_22C vector of the CRISPR‐Cas9 toolkit (Cermak et al., 2017) and two pairs of specific guide RNA sequences (gRNAs, each pair targeting one gene). After Agrobacterium (Agrobacterium tumefaciens)‐mediated transformation of M. truncatula accession R108 (Zhu et al., 2020), CRISPR mutants with deletions and insertions that led to translational frame shifts were found at MtDMP8 and/or MtDMP9 in the T0 generation (Figure 1d). Pollen development was normal in the T1 progeny of mtdmp8 and mtdmp9 single mutants, but pollen viability was reduced in mtdmp8 mtdmp9 double mutants (Figure 1e). Furthermore, seed set was slightly reduced in both mtdmp8 and mtdmp9 single mutants, but mtdmp8 mtdmp9 double mutants showed drastically reduced seed set (Figure 1f), confirming previously reported defects in seed set and putative roles for MtDMP8 and MtDMP9 in fertilization. Haploid M. truncatula plants, which exhibit typical haploid characteristics of reduced stature, as well as small ovules and sterile pollen, were identified amongst the self‐pollinated progenies of mtdmp8 mtdmp9 mutants (Figure 1g–i). The average haploid induction rate (HIR) ranged from 0.29% to 0.82% among the T2 progeny of mtdmp8 mtdmp9 mutant lines (Figure 1j). However, not a single haploid plant was identified among the T2 progeny from selfing mtdmp8 and mtdmp9 single mutants or wild‐type plants (Figure 1j). To investigate whether mtdmp8 mtdmp9 mutants could induce haploid embryos in different female parents, the M. truncatula ecotype Jemalong A17 was pollinated with pollen from mtdmp8 mtdmp9‐1. We identified three haploids among 550 plants from this crossing, whereas no haploids were found among the 620 plants resulting from the cross using wild‐type R108 as pollen donor (Figure 1j). The haploid plants were morphologically similar to the female parent A17 (Figure 1k). Thus, the simultaneous inactivation of MtDMP8 and MtDMP9 can trigger in vivo maternal haploid induction in M. truncatula.
Our successful haploid induction in M. truncatula provides a promising starting point for legume haploid gene editing and mechanistic studies of haploid induction in legumes. Future work will extend the range of applications of DMP‐triggered in vivo haploid induction to crops and forages such as soybean and alfalfa, paving the way for the deployment of DH technology in legume breeding.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
N.W., L.N. and H.L. designed the research. N.W., X.X, T.J., L.L. and P.Z. performed the experiments and analysed the data. H.C. and K.W. provided technical support. H.L. wrote the manuscript.
Acknowledgements
This work was supported by grants from the Fundamental Research Funds for Central Non‐profit Scientific Institution (Y2020YJ12 and No.1610392020005), and the Agricultural Science and Technology Innovation Program of CAAS (CAAS‐ZDRW202009 and CAAS‐ZDXT2019004).
Wang, N. , Xia, X. , Jiang, T. , Li, L. , Zhang, P. , Niu, L. , Cheng, H. , Wang, K. and Lin, H. (2022) In planta haploid induction by genome editing of DMP in the model legume Medicago truncatula . Plant Biotechnol. J., 10.1111/pbi.13740
References
- Čermák, T. , Curtin, S.J. , Gil‐Humanes, J. , Čegan, R. , Kono, T.J.Y. , Konečná, E. , Belanto, J.J. et al. (2017) A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell, 29, 1196–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croser, J.S. , Lulsdorf, M.M. , Davies, P.A. , Clarke, H.J. , Bayliss, K.L. , Mallikarjuna, N. and Siddique, K.H.M. (2006) Toward doubled haploid production in the Fabaceae: progress, constraints, and opportunities. Crit Rev Plant Sci. 25, 139–157. [Google Scholar]
- Gilles, L.M. , Khaled, A. , Laffaire, J.‐B. , Chaignon, S. , Gendrot, G. , Laplaige, J. , Bergès, H. et al. (2017) Loss of pollen‐specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 36, 707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier, N.M.A. , Gilles, L.M. , Pyott, D.E. , Martinant, J.P. , Rogowsky, P.M. and Widiez, T. (2020) Puzzling out plant reproduction by haploid induction for innovations in plant breeding. Nat Plants, 6, 610–619. [DOI] [PubMed] [Google Scholar]
- Kelliher, T. , Starr, D. , Richbourg, L. , Chintamanani, S. , Delzer, B. , Nuccio, M.L. , Green, J. et al. (2017) MATRILINEAL, a sperm‐specific phospholipase, triggers maize haploid induction. Nature, 542, 105–109. [DOI] [PubMed] [Google Scholar]
- Liu, C. , Li, X. , Meng, D. , Zhong, Y. , Chen, C. , Dong, X. , Xu, X. et al. (2017) A 4 bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol Plant, 10, 520–522. [DOI] [PubMed] [Google Scholar]
- Zhong, Y.U. , Chen, B. , Li, M. , Wang, D. , Jiao, Y. , Qi, X. , Wang, M. et al. (2020) A DMP‐triggered in vivo maternal haploid induction system in the dicotyledonous Arabidopsis . Nat Plants, 6, 466–472. [DOI] [PubMed] [Google Scholar]
- Zhong, Y.U. , Liu, C. , Qi, X. , Jiao, Y. , Wang, D. , Wang, Y. , Liu, Z. et al. (2019) Mutation of ZmDMP enhances haploid induction in maize. Nat Plants, 5, 575–580. [DOI] [PubMed] [Google Scholar]
- Zhu, B. , Li, H. , Xia, X. , Meng, Y. , Wang, N.A. , Li, LuLu , Shi, J. et al. (2020) ATP‐binding cassette G transporters SGE1 and MtABCG13 control stigma Exsertion. Plant Physiol. 184, 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]


