Summary
High humidity during harvest season often causes pre‐harvest sprouting in barley (Hordeum vulgare). Prolonged grain dormancy prevents pre‐harvest sprouting; however, extended dormancy can interfere with malt production and uniform germination upon sowing. In this study, we used Cas9‐induced targeted mutagenesis to create single and double mutants in QTL FOR SEED DORMANCY 1 (Qsd1) and Qsd2 in the same genetic background. We performed germination assays in independent qsd1 and qsd2 single mutants, as well as in two double mutants, which revealed a strong repression of germination in the mutants. These results demonstrated that normal early grain germination requires both Qsd1 and Qsd2 function. However, germination of qsd1 was promoted by treatment with 3% hydrogen peroxide, supporting the notion that the mutants exhibit delayed germination. Likewise, exposure to cold temperatures largely alleviated the block of germination in the single and double mutants. Notably, qsd1 mutants partially suppress the long dormancy phenotype of qsd2, while qsd2 mutant grains failed to germinate in the light, but not in the dark. Consistent with the delay in germination, abscisic acid accumulated in all mutants relative to the wild type, but abscisic acid levels cannot maintain long‐term dormancy and only delay germination. Elucidation of mutant allele interactions, such as those shown in this study, are important for fine‐tuning traits that will lead to the design of grain dormancy through combinations of mutant alleles. Thus, these mutants will provide the necessary germplasm to study grain dormancy and germination in barley.
Keywords: Hordeum vulgare, seed dormancy, targeted genome modification, CRISPR, Cas9 nuclease, pre‐harvest sprouting
Introduction
Grains exhibit dormancy to prevent sprouting under adverse conditions and environments and only germinate when favourable conditions return. Grain dormancy in crops has been greatly shortened during domestication to allow repeated sowing and harvesting over a single growing season (Gubler et al., 2005). This brevity of dormancy can cause pre‐harvest sprouting in some temperate areas where the crop harvesting season coincides with high humidity. Pre‐harvest sprouting negatively affects agriculture and the economy by lowering grain yield and quality in cereal crops such as rice (Oryza sativa), maize (Zea mays), wheat (Triticum aestivum), barley (Hordeum vulgare), and sorghum (Sorghum bicolor). Barley has been domesticated for human consumption and is a widely cultivated cereal crop (Sato, 2020). Amongst others, barley is used as raw material for malting, which requires synchronized and rapid germination, but cultivated barley is more prone to pre‐harvest sprouting than its wild form (Li et al., 2004). The balance between dormancy and germinability is one of the main goals driving barley breeding programmes.
Grain dormancy is strongly affected by environmental conditions, for example temperature, humidity, light, and nutritional status during grain maturation and germination. For example, during grain maturation, high and low temperatures result in shorter and longer dormancy, respectively, in barley and wheat (Reddy et al., 1985; Schuurink et al., 1992). However, low temperatures prevent germination in rice cultivated in the summer season, while high temperatures repress germination in barley and wheat cultivated in the winter–spring season.
Grain dormancy is a quantitative trait regulated by many genes and metabolites. Among these factors, the endogenous levels of and sensitivity to two antagonistic phytohormones, abscisic acid (ABA) and gibberellic acid (GA), play critical roles in controlling dormancy and germination (Graeber et al., 2012). ABA is one of the most important regulators of grain dormancy and development, and it suppresses germination (McCarty, 1995). ABA levels increase and peak during grain maturation, leading to longer dormancy. Conversely, dormancy is gradually released after reaching full physiological maturity by the induction of reactive oxygen species (ROS) caused by higher levels of GA. Thus, mutants involved in grain dormancy have been used to study ABA and GA synthesis, metabolism, and sensitivity genes, which include those encoding the entire downstream signalling pathway originating from hormone receptors. However, due to the pleiotropic effects of hormones, these mutant alleles are difficult to use in crop breeding. Therefore, the use of genes for natural variation of grain dormancy and their precise control are required for future improvement.
The identification of natural polymorphisms may help improve pre‐harvest sprouting tolerance (Nakamura, 2018). Since the first report of a quantitative trait locus (QTL) for grain dormancy in barley (Ullrich et al., 1993), multiple additional QTL have been described that map across all chromosomes using several barley accessions as genetic test material (Hickey et al., 2012; Hori et al., 2007; Li et al., 2003; Nakamura et al., 2017; Sato et al., 2009). Of those, two major dormancy‐related loci were identified and named SEED DORMANCY 1 (SD1) and SD2 (Han et al., 1996). The causal gene for SD1 was identified by a map‐based cloning approach using barley cv. ‘Haruna Nijo’ and wild barley ‘OUH602’ and was renamed QTL FOR SEED DORMANCY 1 (Qsd1). Qsd1 is predicted to encode an alanine aminotransferase (Sato et al., 2016). An analysis of the association between genotype and phenotype across barley germplasm showed that the single nucleotide polymorphism G642C in exon 9 of Qsd1, causing the L214F amino acid change, is associated with grain dormancy; this polymorphism is thought to have originated in West Asia after domestication and helped usher in the transition from long dormancy in wild barley to short dormancy in cultivated barley (Sato et al., 2016).
The identity of Qsd1 was independently confirmed with transgenic techniques. First, grains from transgenic lines expressing an RNA interference (RNAi) construct that lowered Qsd1 mRNA in the cultivar ‘Golden Promise’, which carries a wild‐type functional Qsd1 allele, were suppressed in their germination (Sato et al., 2016). In addition, a backcross line harbouring a chromosome fragment derived from ‘OUH602’ and encompassing the qsd1 long dormancy allele in ‘Golden Promise’ background was generated; then the Qsd1 gene from cultivar ‘Haruna Nijo’ was transformed into this backcross line, resulting complementation of the short dormancy phenotype (Sato et al., 2016).
SD2 (renamed Qsd2) was also cloned by map‐based cloning using barley cvs. ‘Azumamugi’ and ‘Kanto Nakate Gold’ and was shown to encode MITOGEN‐ACTIVATED PROTEIN KINASE KINASE 3 (MAPKK3) (Nakamura et al., 2016). In the case of Qsd2, the N260T amino acid change in the dormant ‘Azumamugi’ allele was shown to be causal. Additional qsd2 mutants generated by ethyl methanesulfonate (EMS) mutagenesis in cv. ‘Barke’ were also described: premature termination of Qsd2 translation was associated with reduced germination. For Qsd2, there have been no functional confirmation experiments using genetic engineering. These cloning experiments of Qsd1 and Qsd2 were conducted in different genetic backgrounds, and it remains to be determined whether these genes interact, which would require a collection of mutants in the same cultivar.
A MAPKK3 gene was also identified as the causal gene of Phs1 for grain dormancy in common wheat (Torada et al., 2016), based on a map‐based cloning approach using cvs. ‘Haruyokoi’ and ‘Leader’ with short and long dormancy, respectively. The introduction of the Phs1 ‘Haruyokoi’ allele in the ‘Leader’ background led to relatively higher expression of the TaMAPKK gene in grains and shortened dormancy relative to that of ‘Leader’. These results indicated that the function of the MAPKK3 gene for grain dormancy is conserved among the Triticeae. These findings are critical for breeding of pre‐harvest sprouting tolerance in barley and wheat.
Targeted mutagenesis via ‘genome editing’ by means of customizable endonucleases is a new breeding technology that can efficiently produce desired mutants in a target gene and has been applied to generate mutants in multiple crop species, including barley (Gerasimova et al., 2020; Kapusi et al., 2017; Lawrenson et al., 2015; Thiel et al., 2021). Genome editing makes it possible to create single or multiple mutations at the target site(s) of choice without any alteration in the genetic background, allowing for powerful analyses of the effects of individual genes and their genetic interactions. Currently, RNA‐guided clustered regularly interspaced short palindromic repeats (CRISPR)‐associated (Cas) endonucleases are the molecular tools of choice for targeted mutagenesis in cereal crops (Hisano et al., 2021; Koeppel et al., 2019). Higher‐order gene‐edited mutants can be generated at once, as demonstrated by the simultaneous targeting of all three Qsd1 homeoalleles in the wheat ABD subgenomes, which produced prolonged grain dormancy (Abe et al., 2019). This result suggests that grain dormancy can be manipulated by deliberately introducing mutations. Here, we performed targeted mutagenesis of the barley Qsd1 and Qsd2 genes using RNA‐guided Cas9 endonuclease and analysed the consequences on dormancy in single and double mutants. The motivation for this study was fuelled by the need to fine‐tune barley grain dormancy for breeding by balancing pre‐harvest sprouting tolerance and uniform germination for malting.
Results
Generation and molecular analysis of qsd1 and qsd2 mutants in barley
We transformed ‘Golden Promise’ barley via Agrobacterium tumefaciens‐mediated DNA transfer to immature embryos using constructs expressing guide RNAs (gRNAs) designed against Qsd1 or Qsd2. We obtained 21 and 70 hygromycin‐resistant T0 plants for Qsd1 and Qsd2, respectively (Table 1). After PCR amplification of the gRNA target regions and Sanger sequencing, we identified nine and 21 independently mutated plants for Qsd1 and Qsd2, respectively (Figure 1, Figure S1). The obtained qsd1 alleles featured 1‐ to 4‐bp deletions or 1‐bp insertions (Figure S1). Likewise, qsd2 alleles comprised one 1‐bp insertion event, small deletions from 1 to 7 bp, and a slightly larger deletion of 17 bp (Figure S1). We did not observe any base substitutions in the plants analysed. We allowed T0 plants to self‐pollinate and collected M2 grains (T1 generation). We excised immature embryos from these grains and put them on the culture medium for precocious germination to shorten generation time.
Table 1.
Summary of targeted mutagenesis of Qsd1 and Qsd2 genes in barley
| target gene | No. of regenerated plants | No. of mutated plants | Efficiency of mutation events per regenerated plants (%) |
|---|---|---|---|
| Qsd1 | 21 | 9 | 42.9 |
| Qsd2 | 70 | 21 | 30.0 |
Figure 1.
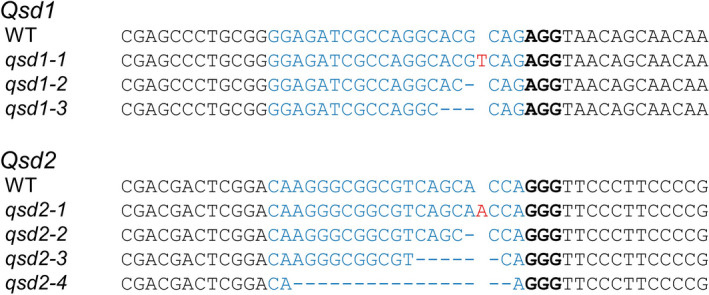
Comparison of DNA sequences for the Cas9/gRNA target sites in Qsd1 and Qsd2 in the wild type and mutants. Sanger sequencing was performed to detect mutations in Qsd1 and Qsd2. Wild‐type, WT; qsd1 mutants, qsd1‐1 to qsd1‐3; qsd2 mutants, qsd2‐1 to qsd2‐4. qsd1‐1, 1‐bp insertion; qsd1‐2, 1‐bp deletion; qsd1‐3, 3‐bp deletion; qsd2‐1, 1‐bp insertion; qsd2‐2, 1‐bp deletion; qsd2‐3, 6‐bp deletion; qsd2‐4, 17‐bp deletion. These representative mutants were used for further analysis; other mutants are listed in Figure S1. The sequence targeted by the gRNA (protospacer) is shown in blue, the protospacer‐adjacent motif (PAM, bound by the Cas9 enzyme) is in bold, dashes indicate deletions, and red nucleotides indicate insertions.
We tested for segregation of gene‐edited alleles at Qsd1 and Qsd2 by Sanger sequencing of the M2 generation. In addition, we determined whether individual plants harboured the T‐DNA by PCR on genomic DNA (Figure S2). We thus selected T‐DNA‐free and homozygous qsd1 mutants with a 1‐bp insertion (qsd1‐1), a 1‐bp deletion (qsd1‐2), or a 3‐bp deletion (qsd1‐3) and qsd2 mutants with a 1‐bp insertion (qsd2‐1), a 1‐bp deletion (qsd2‐2), a 6‐bp deletion (qsd2‐3), and a 17‐bp deletion (qsd2‐4) for further analysis (Figure 1). The deduced amino acid sequences of Qsd1 and Qsd2 from the wild‐type and the mutants are shown in Figure S3. While wild‐type Qsd1 contains 494 amino acids, the protein is shortened by one amino acid in qsd1‐3; the other mutants carry premature stop codons that lead to truncated proteins of 72 (qsd1‐1) or 67 (qsd1‐2) amino acids. Similarly, wild‐type Qsd2 is a 523‐amino acid protein that is shorter by two amino acids in qsd2‐3 and is truncated to 42 (qsd2‐1), 46 (qsd2‐2), or 36 (qsd2‐4) amino acids.
Grain dormancy tests in M3 (T2 generation) lines
We next determined grain dormancy in the wild type and mutants. The level of dormancy was evaluated as percentage of germination within a given period of time, where a shorter dormancy causes a higher percentage of germination. We used M2 progeny from T0 plants without a T‐DNA and with the wild‐type Qsd1 and Qsd2 alleles as controls. We performed germination tests on grains that had been after‐ripened for 6 weeks at 25 °C, which revealed that all controls started to germinate within 1 day after grain imbibition; almost all grains had germinated by 4 days (Figure S4). Seven days after grain imbibition, the controls as well as the in‐frame mutant qsd2‐3 displayed over 90% germination proportion (Figure 2), indicating that this in‐frame mutation does neither disrupt nor decrease Qsd2 function. By contrast, all other qsd1 and qsd2 mutants, including the in‐frame mutant qsd1‐3, did not germinate during the 7 days after grain imbibition (Figure 2). However, grains did germinate after treatment with 3% hydrogen peroxide (Figure S5), indicating that these mutant grains exhibit extremely long dormancy rather than grain lethality.
Figure 2.
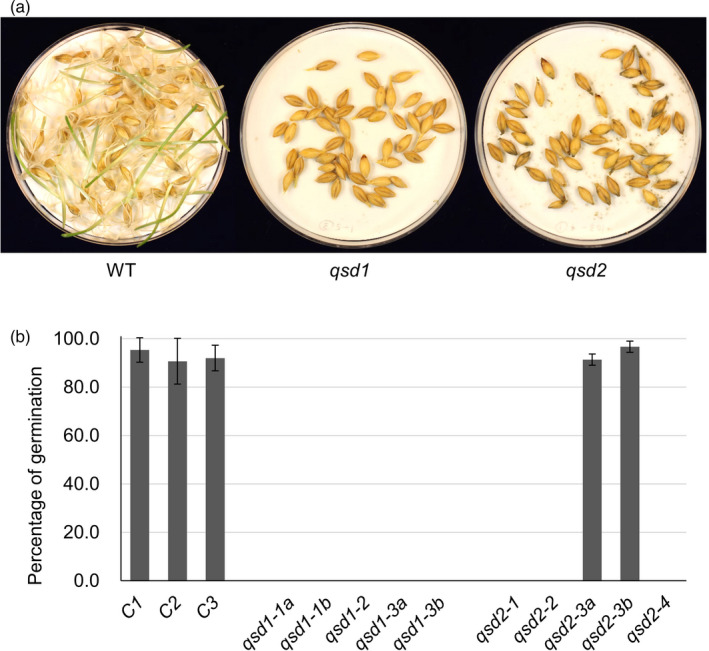
Loss of Qsd1 or Qsd2 function results in prolonged dormancy in barley. Germination test on wild‐type (WT) and genome‐edited barley qsd1 and qsd2 mutant grains. All grains were after‐ripened at 25 °C for 6 weeks under dry conditions to break dormancy. (a) Representation photographs of WT and mutant grains 7 days after grain imbibition. (b) Mean percentages of germination in wild‐type and mutant lines. Data are shown as means ± standard deviation (SD) from three replicates of 50 grains each. C1; non‐transgenic wild‐type Golden Promise control; C2 and C3, non‐edited control plants segregated from T0 plants for Qsd1 and Qsd2, respectively. qsd1‐1a, qsd1‐1b, qsd1‐3a, qsd1‐3b, qsd2‐3a, and qsd2‐3b were derived from individual M2 plants.
Generating and phenotyping of qsd1 and qsd2 double mutants
We generated two independent qsd1 and qsd2 double mutants by crossing a qsd2 mutant (qsd2‐4) with two mutants carrying different qsd1 mutant alleles (qsd1‐1 and qsd1‐3). We selected four genotypes of T‐DNA‐free F3 lines that were either wild‐type for Qsd1 and Qsd2, homozygous for qsd1 or qsd2 (single mutants), or homozygous for qsd1 and qsd2 (double mutant). The representative results of PCR analysis to check the presence or absence of the T‐DNA in F2 plants are shown in Figure S2. These results allowed us to confirm that the T‐DNA had been removed. Representative F2 plants are shown in Figure S6: we observed no obvious differences in their growth or flowering time. We then scored percentage of germination for all F3 lines after 6 weeks of after‐ripening at 25 °C and an additional 4 weeks at 40 °C. While wild‐type grains appeared to fully germinate within 7 days after grain imbibition, none of the single or double mutant lines did (Figure S7). A more quantitative analysis revealed that the controls germinated at proportions between 70% and close to 100% at 21 days after grain imbibition, whereas all single and double mutants showed similarly low germination proportions, with values spanning 1.3%–9.3% (for qsd1), 2%–5.3% (for qsd2), and 0.6%–6.7% (for qsd1qsd2) for the qsd2‐4×qsd1‐1 progeny; and 0%–15.3% (for qsd1), 0% (for qsd2), and 0%–0.7% (for qsd1qsd2) for the qsd2‐4×qsd1‐3 progeny (Figure 3).
Figure 3.
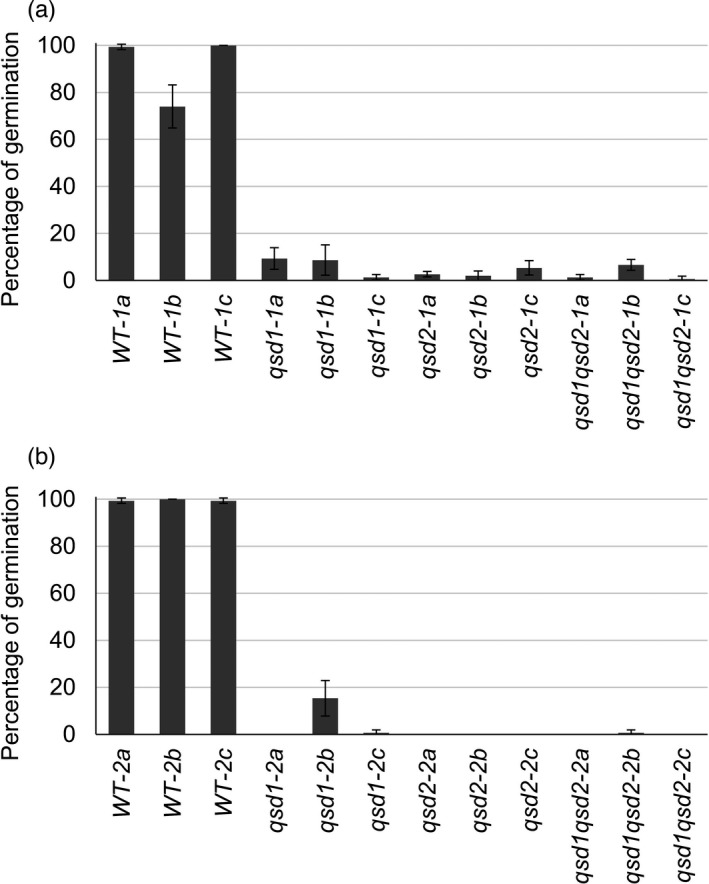
Percentage of germination of F3 lines harbouring the qsd1 and/or qsd2 mutations 21 days after grain imbibition. Percentage of germination of F3 lines resulting from crosses between qsd2‐4 as a seed parent and qsd1‐1 (a) or qsd1‐3 (b) as the pollen donors. All grains were after‐ripened at 25 °C for 6 weeks under dry conditions and an additional 4 weeks at 40 °C to reduce dormancy. Data are shown as means ± standard deviation (SD) from three replicates of 50 grains each. qsd1 mutant lines: qsd1‐1x, qsd1‐2x; qsd2 mutant lines: qsd2‐1x, qsd2‐2x; qsd1qsd2 double mutant lines: qsd1qsd2‐1x, qsd1qsd2‐2x; wild‐type siblings: WT‐1x, WT‐2x.
We also performed pre‐harvest sprouting tests using five unthreshed mature spikes of F3 progenies derived from qsd2‐4×qsd1‐1. All wild‐type spikes sprouted, but the qsd1, qsd2, and qsd1qsd2 double mutants did not sprout at all (Figure S8).
Germination of mutants at low temperature
As with other plants, grain dormancy in barley is alleviated by exposure to low temperature. We thus scored germination percentages of qsd1 and qsd2 grains maintained at 4 °C and in the dark from the start of grain imbibition. None of the genotypes germinated during the 8 days (Figure 4a). However, by 10 days after grain imbibition, wild‐type grains had reached over 90% germination, whereas the mutants exhibited delayed germination, as evidenced by their lower germination proportions: 38%–47% for qsd1, 6%–13% for qsd2, and 12%–20% for qsd1qsd2. Germination further increased slightly 12 days after grain imbibition, with proportions of 60%–73% (qsd1), 29%–35% (qsd2), and 42%–47% (qsd1qsd2). Twenty days after imbibition, qsd1 and qsd2 mutants showed 80%–90% and 70%–72% germination proportions, respectively. Notably, the qsd1qsd2 double mutants had a higher percentage of germination, at 81%–82%, compared with the qsd2 single mutant.
Figure 4.
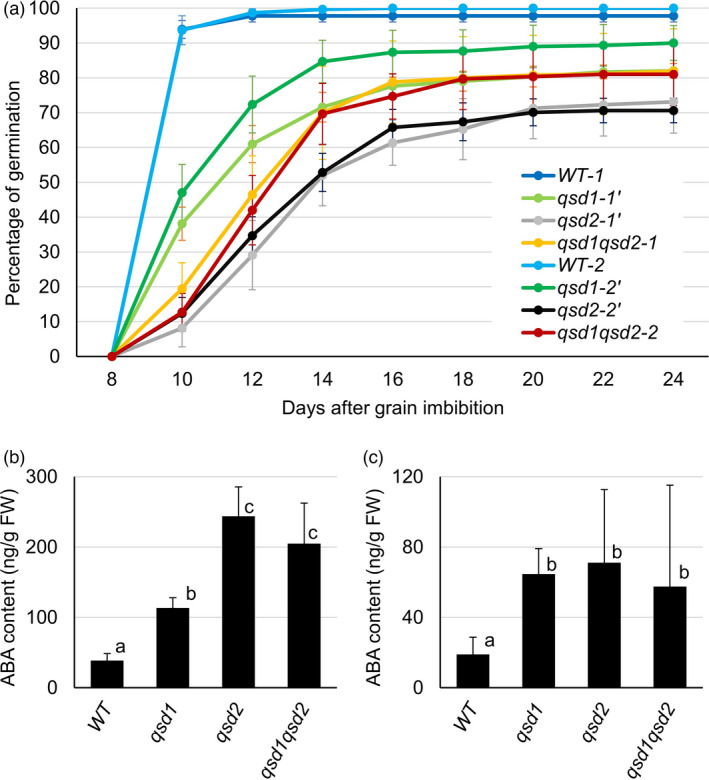
Germinations and ABA contents of F3 lines harbouring the qsd1 and/or qsd2 mutations at 4 °C. All grains were after‐ripened at 25 °C for 6 weeks under dry conditions and an additional 4 weeks at 40 °C to reduce dormancy. Grains were allowed to germinate at 4 °C. (a) Time course of percentages of germination from eight to 24 days after grain imbibition. Data are shown as means ± standard error (SE) from six replicates of 21–50 grains each. qsd1 mutant lines: qsd1‐1′, qsd1‐2′; qsd2 mutant lines: qsd2‐1′ qsd2‐2′; qsd1qsd2 double mutant lines: qsd1qsd2‐1, qsd1qsd2‐2; wild‐type siblings: WT‐1, WT‐2. qsd1‐1′, qsd2‐1′, qsd1qsd2‐1 and WT‐1 are segregating siblings derived from the qsd2‐4×qsd1‐1 progeny, and qsd1‐2′, qsd2‐2′, qsd1qsd2‐2 and WT‐2 are those from the qsd2‐4×qsd1‐3 progeny. (b, c) ABA content in embryos during germination at 4 °C 3 days (b) or seven days (c) after grain imbibition, respectively. Data are shown as means ± SD from six replicates. Different letters indicate significant differences, as determined by Tukey’s test (P < 0.05).
We concluded that exposure to low temperature following grain imbibition largely abrogates dormancy in mutant grains, although with a slight delay in the mutants compared to the wild type. To investigate the possible reason for this delay, we measured ABA contents in embryos after grain imbibition but before germination took place. ABA content was significantly higher in embryos from the qsd1 mutant compared with wild‐type embryos 3 days after grain imbibition, and with qsd2 and qsd1qsd2 embryos accumulating over twice as much ABA as the qsd1 mutant (P < 0.001, Figure 4b). Seven days after grain imbibition, ABA content was lower in all genotypes relative to levels measured at 3 days (Figures 4b,c). In addition, wild‐type embryos accumulated significantly less ABA than the mutants that had comparable ABA contents of approximately three times that of the wild type (P < 0.05, Figure 4c).
Germination test under different light conditions
Exposure to light also affects grain dormancy and germination. We therefore explored the effect of light exposure on germination using an immature embryo germination system, thus removing any influence from the endosperm and husks. Here, we considered an immature embryo as having germinated when the primary shoot and root grew more than 5 mm in length. Over 90% of immature embryos produced shoots and roots, regardless of the genotype, when immature embryos were incubated in the dark (Figure 5e–g, Table 2). In sharp contrast, qsd2 and qsd1qsd2 immature embryos rarely germinated when placed in the light; wild‐type and qsd1 immature embryos only exhibited a modest suppression of germination under the same conditions (Figure 5a–d, Table 2).
Figure 5.
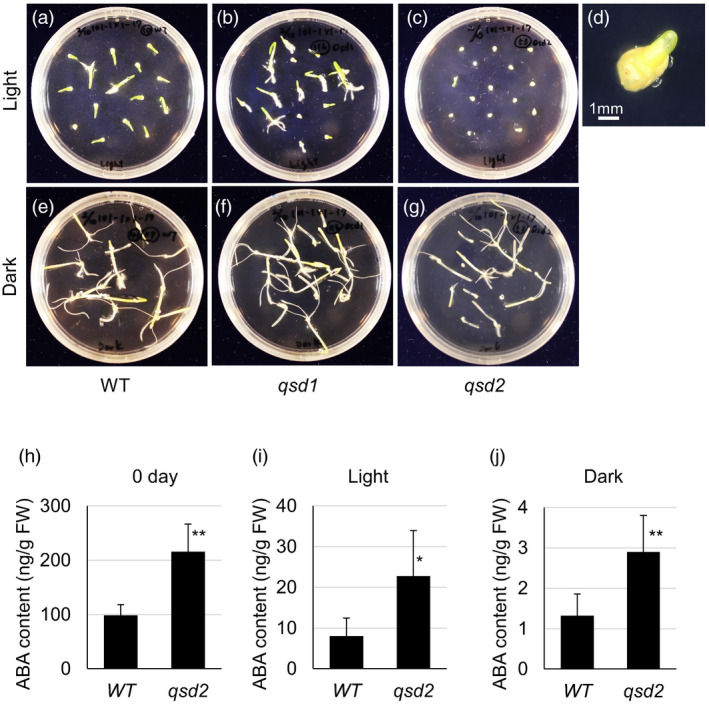
Germination of immature embryos incubated in the dark or in the light. Germination tests in the light (a–d) or in the dark (e–g) at 25 °C were conducted using immature embryos of the wild‐type (WT) (a, e), qsd1 (b, f), and qsd2 (c, d, g) mutants. Plates were photographed 7 days after the start of culture. d is a magnified view of a stagnating qsd2 embryo after 10 days in the light. (h–j) ABA contents in immature WT and qsd2 embryos at the start of the experiment (0 days, h) and after 7 days in the light (i) or in the dark (j). Statistical analysis was performed by Student's t‐test between the WT and qsd2. Asterisks indicate significant differences (*P < 0.05, **P < 0.01).
Table 2.
Germination proportion of barley immature embryos incubated in the dark or in the light
| Light | Dark | |||||
|---|---|---|---|---|---|---|
| n | Ave* | SD † | n | Ave* | SD † | |
| wild type | 4 | 15.2a | 4.9 | 4 | 18.7a | 1.2 |
| qsd1 | 4 | 13.7a | 2.8 | 4 | 18.4a | 1.1 |
| qsd2 | 4 | 4.9b | 3.1 | 4 | 18.1a | 1.3 |
| qsd1qsd2 | 3 | 5.7b | 1.0 | 3 | 18.4a | 1.5 |
Average of germination number per 20 immature embryos on the embryo culture medium at 25 °C. Different characters indicate significant differences (P < 0.05) as analysed by Tukey's test.
SD means standard deviation.
We finally measured the ABA content of immature embryos and cultured embryos. Immature embryos in qsd2 accumulated twice as much ABA as the wild type (P < 0.01, Figure 5h). Seven days after initiation of culture, the ABA content of both wild‐type and qsd2 cultured embryos had dropped by 90% in the light and by over 98% in the dark. Notably, qsd2 still had higher ABA levels than the wild type (P < 0.05, Figure 5i,j).
Discussion
Genetic variation in genes related to grain dormancy or germination are essential resources for breeding of pre‐harvest sprouting tolerance. Many QTL analyses have been performed for grain dormancy in barley, highlighting the two major QTL, Qsd1 and Qsd2 (Nakamura et al., 2016; Sato et al., 2016). However, each QTL and associated causal gene was characterized in different genetic backgrounds, making any comparison of their respective effects, or their interactions, difficult to interpret. Here, we employed Cas9‐mediated genome editing to specifically target the Qsd1 and Qsd2 loci in the same genetic background of the barley cultivar ‘Golden Promise’ (Figure 1, Figure S1, Table 1). Grains from these mutants exhibited long dormancy but were nonetheless viable (Figures 2 and 3, Figure S5). This prolonged dormancy must also contribute to tolerance for pre‐harvest sprouting (Figure S8). Our results suggest that targeted mutagenesis of Qsd1 and Qsd2 is a useful strategy to prolong grain dormancy in barley. This is the first report of controlling germination of barley grains using targeted mutagenesis.
The use of Cas9‐mediated gene editing was previously reported in barley, for instance, by targeting HvPM19, an ABA‐inducible gene that encodes a plasma membrane protein predicted to affect grain dormancy. However, no phenotypic characterization was conducted (Lawrenson et al., 2015). Our results show that mutations in Qsd1 and Qsd2 result in prolonged grain dormancy compared with wild‐type ‘Golden Promise’, observations that are in line with results previously obtained by RNAi‐mediated suppression of Qsd1 in barley cv. ‘Golden Promise’ and EMS mutants for Qsd2 in barley cv. ‘Barke’ (Nakamura et al., 2016; Sato et al., 2016). Like the qsd1 mutants, the Qsd1‐RNAi transformants showed little germination in almost the same environment as used in the present study (Sato et al., 2016). The EMS‐induced Qsd2 mutant has a different genetic background and the environmental conditions and methods used for its analysis were different from those used in our current work, so it is not possible to make an exact comparison, but the EMS mutant seems to have moderately prolonged dormancy compared with our qsd2 mutants in this study (Nakamura et al., 2016). We hypothesize that barley cultivation and breeding have selected alleles with enhanced function at Qsd1 and Qsd2 to promote rapid germination, from the pool of natural alleles represented among barley cultivars (Nakamura et al., 2016; Sato et al., 2016; Vetch et al., 2020). We attempted to break dormancy by using the high temperature of 40 °C for 4 weeks, with a dormant after‐ripening period of 10 weeks; however, even under these extreme conditions, the germination of all mutants was still significant delayed at 25 °C, thus preventing a comparative analysis of the individual contribution of Qsd1 and Qsd2 to dormancy (Figure 3). We therefore suspect that both genes are essential for germination under natural settings and, conversely, that loss‐of‐function alleles cause severely delayed germination and are therefore under strong selective pressure. This hypothesis should be tested on plants grown in the field, but the level of grain dormancy is expected to fluctuate due to temperature, wind, rain, and pests and diseases in a natural environment. We have conducted all of the current experiments in an environmentally controlled growth chamber, to exclude the possibility that the extent of dormancy might be shortened in barley grown in the field particularly due to higher temperatures that plants would experience during the ripening period. Mimicking natural growth conditions may help dissect the individual contributions of Qsd1 and Qsd2 to dormancy.
In barley breeding, rapid germination has been selected for in malting barley cultivars, while long dormancy confers tolerance to pre‐harvest sprouting. Creating weak mutant alleles is considered an effective strategy to modulate traits more subtly than loss‐of‐function alleles, for example, by targeting the 3′ end of genes or to delete just one or a few amino acids while retaining the translational reading frame. Gene editing systems with Cas endonucleases will allow such precise genomic modifications (Komor et al., 2016; Nishida et al., 2016; Schedel et al., 2017). In this study, we identified one in‐frame mutation each for Qsd1 (qsd1‐3) and Qsd2 (qsd2‐3), resulting in the removal of one (qsd1‐3) or two (qsd2‐3) amino acids near the N terminus of the proteins. Dormancy in qsd2‐3 was not affected, in contrast to all other qsd2 mutants isolated in this study. Thus two‐amino‐acid deletion may not alter the kinase activity of Qsd2, as its kinase domain is located between amino acids 100 and 300 (Figure S3) (Nakamura et al., 2016). Although the qsd1‐3 allele encodes a protein lacking a single amino acid, a histidine residue at position 32, this mutant showed prolonged dormancy. This suggests that qsd1‐3 is a loss‐of‐function mutation of Qsd1. The deletion of a histidine residue may have affected the secondary structure of Qsd1, leading to an inactive protein. We performed the F3 experiments assuming an in‐frame mutant allele would be milder for grain dormancy; however, the results were similar compared with frame‐shifted mutations. How these proteins perform their function for grain dormancy and germination remains to be investigated. The mutants produced in this study are in the same genetic background and will be instrumental in understanding these issues.
Under low‐temperature (4 °C) conditions, the qsd2 mutants exhibited a significant delay of germination than did qsd1 mutants (Figure 4). This observation suggests that the dormancy or germination defect in qsd2 might be stronger than that of qsd1 in the ‘Golden Promise’ genetic background. On the other hand, the germination percentage of the qsd2 single mutant was lower than that of the qsd1qsd2 double mutant and qsd1 single mutant. It cannot be ruled out that the qsd1 mutation might counteract the suppressive effect of qsd2 on germination under cold conditions, although the underlying molecular mechanism is unknown.
We also performed germination assays using cultured immature embryos incubated in the dark or in the light (Figure 5a–g, Table 2). In the dark, wild‐type and mutant immature embryos showed the same germination proportion. In sharp contrast, qsd2 mutants exhibited a strong suppression of germination when incubated in the light. The CRYPTOCHROME blue light photoreceptors perceive light and repress germination in barley grains (Barrero et al., 2014). Loss of Qsd1 did not affect embryo germination in the light, indicating that Qsd1 acts independently from light signalling related to germination or dormancy of barley grains. By contrast, germination of qsd2 mutant immature embryos was repressed by light. Qsd2 encodes MAPKK3, suggesting that MAPK cascades might be involved in photoreception during germination of barley grains. A phosphoproteomic analysis of freshly harvested and after‐ripened barley embryos treated with ABA revealed the involvement of different phosphorylation signalling networks in each set of embryos, suggesting that after‐ripening modulates phosphorylation signalling pathways, leading to the decay of ABA signalling (Ishikawa et al., 2019). Although it remains unclear whether Qsd2 is a key MAPKK in this phosphorylation pathway linked to ABA regulation, we did notice an effect of qsd2 mutants on ABA accumulation in immature embryos (Figure 5).
Although different plant species have different dormancy mechanisms, reflecting their physiology and morphology (Baskin and Baskin, 2004), Arabidopsis (Arabidopsis thaliana) and barley share core conserved mechanisms. For example, nitric oxide can break dormancy in both Arabidopsis and barley (Bethke et al., 2004), as do GA and ROS. The cytochrome P450 gene ABA 8ʹ‐HYDROXYLASE is involved in ABA catabolism and thus regulates dormancy or germination in both Arabidopsis and barley (Millar et al., 2006). When measuring ABA contents, we discovered that ABA accumulates to higher levels in the mutants compared to the wild type (Figures 4b,c, 5h–j). The qsd2 mutants initially had higher ABA levels than qsd1 mutants or the wild type 3 days after grain imbibition, but these levels dropped to become comparable to those of qsd1 mutants after 7 days at 4 °C (Figure 5h–j). Likewise, both qsd1 and qsd2 mutants germinated later than the wild type under cold conditions, suggesting that while ABA may delay the initial germination in the mutants, it does not contribute to long‐term dormancy of the barley grain. In addition, ABA levels in qsd2 immature embryos were higher than those of wild‐type embryos after 7 days of incubation in the light or in the dark; notably, ABA levels in dark‐incubated qsd2 embryos reached only ~2% of non‐cultured qsd2 embryos, suggesting the possibility that such ABA content allowed germination in qsd2. Although Wang et al. (1995) reported that ABA contents are higher in dormant barley embryos compared with non‐dormant embryos, they concluded that ABA content is not related to grain germination. Further detailed analysis is needed to clarify whether ABA directly affects germination in the qsd2 mutants.
Genes involved in seed dormancy have been isolated from forward genetic studies and studies of natural variation in Arabidopsis and other species. DELAY OF GERMINATION 1 (DOG1) was isolated from a major QTL for seed dormancy in Arabidopsis accessions, with the dog1 mutant having reduced dormancy (Bentsink et al., 2006). DOG1 is expressed at higher levels in Arabidopsis seeds ripened at low temperatures (Kendall et al., 2011). DOG1 physically interacts with the protein phosphatases ABA HYPERSENSITIVE GERMINATION 1 (AHG1) and AHG3 (Nishimura et al., 2018); however, the function of DOG1 has not fully been elucidated. DOG1‐like genes have been identified in barley and wheat, and surprisingly they exhibited distinct expression patterns from Arabidopsis DOG1 (Ashikawa et al., 2010). Although these genes share low sequence identity with Arabidopsis DOG1, their ectopic overexpression in Arabidopsis demonstrated their conserved function in seed dormancy. In another example, the rice Seed dormancy 4 (Sdr4) QTL was identified by map‐based cloning using near isogenic lines derived from crosses between the short dormancy japonica‐type rice cv. ‘Nipponbare’ and long dormancy indica‐type rice cv. ‘Kasalath’ (Sugimoto et al., 2010). Sdr4 expression was positively controlled by VIVIPAROUS‐1 (VP1), the rice ortholog to Arabidopsis ABA INSENSITIVE 3 (ABI3), a master regulator of seed dormancy as well as seed maturation (Hattori et al., 1994; McCarty et al., 1991). A rice sdr4 mutant was also shown to be insensitive to ABA, that is the germination of the mutant was not blocked by ABA, although ABA does block germination of wild‐type grains. Rice Sdr4 is therefore thought to play an intermediate regulator role in dormancy during grain maturation; in agreement, the positive regulators of germination GA20 OXIDASE 1 (OsGA20ox‐1) and PLASMA MEMBRANE INTRINSIC PROTEIN 1;3 (PIP1;3) were highly expressed in the sdr4 mutant after grain imbibition. The loss of DOG1 and Sdr4 functions were associated with shortened dormancy (Bentsink et al., 2006; Sugimoto et al., 2010). On the other hand, loss of Qsd1 and Qsd2 might result in tolerance to pre‐harvest sprouting due to longer dormancy. However, too long of a dormancy period could be difficult for malting. So, the use of multiple genes for natural variation of seed/grain dormancy and their precise control will be required for future breeding.
In this study, we used targeted mutagenesis to introduce mutations in the barley genes Qsd1 and Qsd2, in which loss of function resulted in prolonged grain dormancy. The introduction of mutations in the same genetic background further allowed us to investigate their genetic interactions. qsd2 dominated dormancy responses slightly more than qsd1, although qsd1 did appear to mitigate the effect of qsd2 on suppressing germination under cold conditions. For a practical use for barley breeding, we had hoped for mutants with shorter dormancy, but all mutants showed long dormancy phenotypes. However, this experiment demonstrated that grain dormancy can be regulated by mutations in Qsd1 and Qsd2, paving the way for the creation of additional alleles by genome editing, for instance by the use of base editors like artificial deaminases (Nishida et al., 2016).
Experimental procedures
Plant materials
Barley cv. ‘Golden Promise’ was grown in growth chambers for 2 months in daily cycles of 12 h light at 15 °C and 12 h darkness at 13 °C before transfer to a long‐day photoperiod (16 h light at 16 °C and 8 h darkness at 13 °C) until flowering (ca. 1 month in long days) to collect immature embryos for Agrobacterium‐mediated transformation. Transgenic and mutant barley plants were grown in growth chambers in long days (16 h light at 16 °C and 8 h darkness at 13 °C).
Design of guide RNAs and vector construction
Guide RNAs (gRNAs) were designed to target each exon of barley Qsd1 and Qsd2 with the gRNA online design tool WU‐CRISPR (http://crisprdb.org/wu‐crispr/; Wong et al., 2015). The predicted secondary structures of gRNAs were generated by RNAfold (http://rna.tbi.univie.ac.at/cgi‐bin/RNAWebSuite/RNAfold.cgi), and the most suitable targets were selected largely considering the criteria described by Liang et al. (2016) and Kumlehn et al. (2018). Consequently, Qsd1‐gRNA1 (5′‐GGAGATCGCCAGGCACGCAG‐3′) and Qsd2‐gRNA1 (5′‐AAGGGCGGCGTCAGCACCA‐3′) were chosen for this study (Figure S9).
The synthetic oligonucleotides (5′‐tggcGGAGATCGCCAGGCACGCAG‐3′ and 5′‐aaacCTGCGTGCCTGGCGATCTCC‐3′ for Qsd1, 5′‐tggcAAGGGCGGCGTCAGCACCA‐3′ and 5′‐aaacTGGTGCTGACGCCGCCCTT‐3′ for Qsd2) were annealed and ligated between the OsU3 promoter and the gRNA scaffold in the intermediate vector pSH121 (Gerasimova et al., 2018) predigested with the restriction enzyme BsaI. The lowercase sequences in the synthetic oligonucleotides are the overhang sticky ends for ligation with BsaI‐cleaved plasmid DNA. The expression units for gRNAs and Cas9 were introduced into the binary vector p6i‐2×35S‐TE9 (DNA Cloning Service e.K., Germany) using the SfiI restriction sites. The T‐DNA of p6i‐2×35S‐TE9 confers resistance to hygromycin in plants. The components of the T‐DNA region of the final vector used for transformation are depicted in Figure S10.
Production and molecular analysis of transgenic barley plants
Transformation vectors were introduced into Agrobacterium strain AGL1. The resulting agrobacteria were then used for transformation via co‐cultivation with barley immature embryos as previously described (Hisano et al., 2017; Hisano and Sato, 2016).
Total DNA was extracted from regenerated plantlets with Kaneka Easy DNA Extraction Kit version 2 (Kaneka, Japan) and used as a template for PCR validation of the presence of transgenes; PCR products spanning the genomic region targeted by each gRNA were PCR‐amplified and sequenced to identify mutations. PCR was performed following the method described by Hisano and Sato (2016). Mutations were detected by visual inspection of sequencing data from PCR products using the ‘Assemble Sequence to Reference’ module in CLC Main Workbench software (QIAGEN, Germany). The primers used in this study are listed in Table S1.
Immature embryo culture
For rapid generation and to prevent germination delays due to dormancy, immature embryo culture was performed on half‐strength Murashige and Skoog (MS) medium adjusted to pH 5.8, with 15 g/L sucrose and 3 g/L Phytagel (Sigma‐Aldrich). To this end, immature embryos were isolated from immature grains, ca. 20 days after pollination, that had been surface sterilized with a solution of sodium hypochlorite (1% effective chlorine concentration), and incubated on the above growth medium for 2 days in the dark and for 2–3 days under long days (16 h light/8 h dark) at 25 °C. Germinated embryos were then transferred to a growth chamber and sown in soil. To analyse the response of embryo germination to light, immature embryos were incubated in the dark or exposed to light in a long‐day photoperiod under fluorescent lights (17.7 μmol m−2 s−1 PPF in 400–500 nm blue region of the spectrum, and 53.0 μmol m−2 s−1 PPF in 400–700 nm) at 25 °C.
Generation of double mutants
Double mutants were generated by conventional crossing of an M2 mutant line carrying a 17‐bp deletion in Qsd2 with two independent M2 mutant lines with either a 1‐bp insertion or a 3‐bp deletion in Qsd1.
Grain germination test
The spikes of each plant were harvested at physiological maturity when the colour of rachis had changed to a straw yellow. Spikes were desiccated at 25 °C for 10 days in a drying cabinet (Tolihan Co., Japan) and then stored at –20 °C until all samples were collected. The spikes of M2 plants were hand‐threshed at the same time, and M3 grains were after‐ripened at 25 °C and 10%–15% relative humidity for 6 weeks. Spikes from F2 plants were treated similarly, with an additional 4 weeks at 40 °C after 6 weeks of after‐ripening at 25 °C. Fifty grains were assessed for germination on moistened filter paper (ADVANTEC, Japan) in 90‐mm disposable dishes with triplication at 25 °C for normal germination tests. For low‐temperature response germination tests at 4 °C, 21–50 grains per dish with six replications were used. The percentage of germination was calculated by counting seedlings with primary shoots and roots that had elongated more than 5 mm.
For pre‐harvest sprouting tests, unthreshed mature spikes of F3 progenies of qsd2‐4×qsd1‐1 were maintained on wet soil for 11 days with daily cycles of 16 h light at 25 °C and 8 h darkness at 15 °C.
Analysis of ABA content
Embryos from four or five grains for each sample representing 11–33 mg fresh weight were collected with five replicates and lyophilized in a freeze dryer. The embryos were sampled at 3 days or 7 days after grain imbibition for the test under 4 °C conditions, and at 7 days after the start of incubation for the test under light/dark conditions. Five immature embryos were collected from each spike as controls. Dried embryos were then ground and used for extraction and analysis of phytohormones according to Hisano et al. (2016) and Tsukahara et al. (2015).
Conflict of interest
The authors declare no conflicts of interest associated with this manuscript.
Author contributions
HH, FA, SN, JK, and KS designed experiments, HH, RH, HM, and TM performed experiments, ME, MM, and SN provided vectors, sequences of target genes and supported experiments, HH, FA, SN, JK, and KS drafted the manuscript, and RH, ME, and MM revised the manuscript. All authors looked through the final version of the manuscript and approved the submission.
Supporting information
Figure S1 Partial DNA sequence of the gRNA target sites at the Qsd1 and Qsd2 loci in mutants.
Figure S2 PCR analysis for detection of T‐DNA region in T1 and T‐DNA‐free mutant plants.
Figure S3 Partial deduced amino acid sequences from the wild‐type and qsd1 and qsd2 mutants.
Figure S4. Germination of the wild‐type and M3 lines of genome‐edited barley 4 days after grain imbibition.
Figure S5 Germination of the M3 qsd1‐2 mutant 1 month after treatment with 3% hydrogen peroxide.
Figure S6 T‐DNA‐free F2 generation of wild‐type, qsd1 and qsd2 single mutant, and qsd1qsd2 double mutant plants.
Figure S7 Germination from segregating progeny with no mutation in Qsd1 or Qsd2 (wild‐type) or homozygous for qsd1, qsd2, or qsd1qsd2 double mutations (photographed 7 days after grain imbibition).
Figure S8 Pre‐harvest sprouting test of F3 progenies derived from qsd2‐4×qsd1‐1 (photographed 11 days after place spikes on the soil).
Figure S9 Target positions and 20‐nt sequence of gRNAs in Qsd1 and Qsd2 genes.
Figure S10 Structure of the T‐DNA region in the vector used in this study.
Table S1. PCR primers used in this study
Acknowledgements
The authors thank Drs. S. Toki, S. Hirose (NARO) and T. Hirayama (IPSR, Okayama U.) for valuable discussion, and M. Yamane, H. Saito and M. Miyake (IPSR, Okayama U.) for technical assistance. This work was supported by the Cabinet Office, Government of Japan, Cross‐ministerial Strategic Innovation Promotion Program (SIP), ‘Technologies for Creating Next‐Generation Agriculture, Forestry and Fisheries’ (funding agency: Bio‐oriented Technology Research Advancement Institution, NARO, Japan), JSPS KAKENHI, Japan, grant 19H00943 and 19H02930, and the Joint Usage/Research Center, IPSR, Okayama U. (FY2017). The grain sample of cv. Golden Promise was provided by the National BioResource Project‐Barley, Japan.
Hisano, H. , Hoffie, R. E. , Abe, F. , Munemori, H. , Matsuura, T. , Endo, M. , Mikami, M. , Nakamura, S. , Kumlehn, J. and Sato, K. (2022) Regulation of germination by targeted mutagenesis of grain dormancy genes in barley. Plant Biotechnol. J., 10.1111/pbi.13692
References
- Abe, F. , Haque, E. , Hisano, H. , Tanaka, T. , Kamiya, Y. , Mikami, M. , Kawaura, K. et al. (2019) Genome‐edited triple‐recessive mutation alters seed dormancy in wheat. Cell Rep. 28, 1362–1369. [DOI] [PubMed] [Google Scholar]
- Ashikawa, I. , Abe, F. and Nakamura, S. (2010) Ectopic expression of wheat and barley DOG1‐like genes promotes seed dormancy in Arabidopsis. Plant Sci. 179, 536–542. [DOI] [PubMed] [Google Scholar]
- Barrero, J.M. , Downie, A.B. , Xu, Q. and Gubler, F. (2014) A role for barley CRYPTOCHROME1 in light regulation of gain dormancy and germination. Plant Cell, 26, 1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin, C.C. and Baskin, J.M. (2004) Germinating seeds of wildflowers, an ecological perspective. HortTechnology Horttech, 14, 467. [Google Scholar]
- Bentsink, L. , Jowett, J. , Hanhart, C.J. and Koornneef, M. (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis . Proc. Natl Acad. Sci. 103, 17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke, P.C. , Gubler, F. , Jacobsen, J.V. and Jones, R.L. (2004) Dormancy of Arabidopsis seeds and barley grains can be broken by nitric oxide. Planta, 219, 847–855. [DOI] [PubMed] [Google Scholar]
- Gerasimova, S.V. , Hertig, C. , Korotkova, A.M. , Kolosovskaya, E.V. , Otto, I. , Hiekel, S. , Kochetov, A.V. et al. (2020) Conversion of hulled into naked barley by Cas endonuclease‐mediated knockout of the NUD gene. BMC Plant Biol. 20, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova, S.V. , Korotkova, A.M. , Hertig, C. , Hiekel, S. , Hoffie, R. , Budhagatapalli, N. , Otto, I. et al. (2018) Targeted genome modification in protoplasts of a highly regenerable Siberian barley cultivar using RNA‐guided Cas9 endonuclease. Vavilov Journal of Genetics and Breeding, 22, 1033–1039. [Google Scholar]
- Graeber, K. , Nakabayashi, K. , MiattonI, E. , Leubner‐Metzger, G. and Soppe, W.J.J. (2012) Molecular mechanisms of seed dormancy. Plant, Cell Environ. 35, 1769–1786. [DOI] [PubMed] [Google Scholar]
- Gubler, F. , Millar, A.A. and Jacobsen, J.V. (2005) Dormancy release, ABA and pre‐harvest sprouting. Curr. Opin. Plant Biol. 8, 183–187. [DOI] [PubMed] [Google Scholar]
- Han, F. , Ullrich, S.E. , Clancy, J.A. , Jitkov, V. , Kilian, A. and Romagosa, I. (1996) Verification of barley seed dormancy loci via linked molecular markers. Theor. Appl. Genet. 92, 87–91. [DOI] [PubMed] [Google Scholar]
- Hattori, T. , Terada, T. and Hamasuna, S.T. (1994) Sequence and functional analyses of the rice gene homologous to the maize Vp1. Plant Mol. Biol. 24, 805–810. [DOI] [PubMed] [Google Scholar]
- Hickey, L.T. , Lawson, W. , Arief, V.N. , Fox, G. , Franckowiak, J. and Dieters, M.J. (2012) Grain dormancy QTL identified in a doubled haploid barley population derived from two non‐dormant parents. Euphytica, 188, 113–122. [Google Scholar]
- Hisano, H. , Abe, F. , Hoffie, R.E. and Kumlehn, J. (2021) Targeted genome modifications in cereal crops. Breed. Sci. 10.1270/jsbbs.21019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano, H. , Matsuura, T. , Mori, I.C. , Yamane, M. and Sato, K. (2016) Endogenous hormone levels affect the regeneration ability of callus derived from different organs in barley. Plant Physiol. Biochem. 99, 66–72. [DOI] [PubMed] [Google Scholar]
- Hisano, H. , Meints, B. , Moscou, M.J. , Cistue, L. , Echávarri, B. , Sato, K. and Hayes, P.M. (2017) Selection of transformation‐efficient barley genotypes based on TFA (transformation amenability) haplotype and higher resolution mapping of the TFA loci. Plant Cell Rep. 36, 611–620. [DOI] [PubMed] [Google Scholar]
- Hisano, H. and Sato, K. (2016) Genomic regions responsible for amenability to Agrobacterium‐mediated transformation in barley. Sci. Rep. 6, 37505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, K. , Sato, K. and Takeda, K. (2007) Detection of seed dormancy QTL in multiple mapping populations derived from crosses involving novel barley germplasm. Theor. Appl. Genet. 115, 869–876. [DOI] [PubMed] [Google Scholar]
- Ishikawa, S. , Barrero, J.M. , Takahashi, F. , Nakagami, H. , Peck, S.C. , Gubler, F. , Shinozaki, K. et al. (2019) Comparative phosphoproteomic analysis reveals a decay of ABA signaling in barley embryos during after‐ripening. Plant Cell Physiol. 60, 2758–2768. [DOI] [PubMed] [Google Scholar]
- Kapusi, E. , Corcuera‐Gómez, M. , Melnik, S. and Stoger, E. (2017) Heritable genomic fragment deletions and small indels in the putative ENGase gene induced by CRISPR/Cas9 in barley. Front Plant Sci. 8, 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall, S.L. , Hellwege, A. , Marriot, P. , Whalley, C. , Graham, I.A. and Penfield, S. (2011) Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell, 23, 2568–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppel, I. , Hertig, C. , Hoffie, R. and Kumlehn, J. (2019) Cas endonuclease technology ‐ a quantum leap in the advancement of barley and wheat genetic engineering. Int. J. Mol. Sci. 20, 2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor, A.C. , Kim, Y.B. , Packer, M.S. , Zuris, J.A. and Liu, D.R. (2016) Programmable editing of a target base in genomic DNA without double‐stranded DNA cleavage. Nature, 533, 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumlehn, J. , Pietralla, J. , Hensel, G. , Pacher, M. and Puchta, H. (2018) The CRISPR/Cas revolution continues: from efficient gene editing for crop breeding to plant synthetic biology. J. Integr. Plant Biol. 60, 1127–1154. [DOI] [PubMed] [Google Scholar]
- Lawrenson, T. , Shorinola, O. , Stacey, N. , Li, C. , Østergaard, L. , Patron, N. , Uauy, C. et al. (2015) Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA‐guided Cas9 nuclease. Genome Biol. 16, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Ni, P. , Francki, M. , Hunter, A. , Zhang, Y. , Schibeci, D. , Li, H. et al. (2004) Genes controlling seed dormancy and pre‐harvest sprouting in a rice‐wheat‐barley comparison. Funct. Integr. Genomics. 4, 84–93. [DOI] [PubMed] [Google Scholar]
- Li, C.D. , Tarr, A. , Lance, R.C.M. , Harasymow, S. , Uhlmann, J. , Westcot, S. , Young, K.J. et al. (2003) A major QTL controlling seed dormancy and pre‐harvest sprouting/grain α‐amylase in two‐rowed barley (Hordeum vulgare L.). Aust. J. Agric. Res. 54, 1303–1313. [Google Scholar]
- Liang, G. , Zhang, H. , Lou, D. and Yu, D. (2016) Selection of highly efficient sgRNAs for CRISPR/Cas9‐based plant genome editing. Sci. Rep. 6, 21451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, D.R. (1995) Genetic control and integration of maturation and germination pathways in seed development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 71–93. [Google Scholar]
- McCarty, D.R. , Hattori, T. , Carson, C.B. , Vasil, V. , Lazar, M. and Vasil, I.K. (1991) The Viviparous‐1 developmental gene of maize encodes a novel transcriptional activator. Cell, 66, 895–905. [DOI] [PubMed] [Google Scholar]
- Millar, A.A. , Jacobsen, J.V. , Ross, J.J. , Helliwell, C.A. , Poole, A.T. , Scofield, G. , Reid, J.B. et al. (2006) Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′‐hydroxylase. Plant J. 45, 942–954. [DOI] [PubMed] [Google Scholar]
- Nakamura, S. (2018) Grain dormancy genes responsible for preventing pre‐harvest sprouting in barley and wheat. Breed. Sci. 68, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, S. , Pourkheirandish, M. , Morishige, H. , Kubo, Y. , Nakamura, M. , Ichimura, K. , Seo, S. et al. (2016) Mitogen‐activated protein kinase kinase 3 regulates seed dormancy in barley. Curr. Biol. 26, 775–781. [DOI] [PubMed] [Google Scholar]
- Nakamura, S. , Pourkheirandish, M. , Morishige, H. , Sameri, M. , Sato, K. and Komatsuda, T. (2017) Quantitative trait loci and maternal effects affecting the strong grain dormancy of wild barley (Hordeum vulgare ssp. spontaneum). Frontiers . Plant Sci. 8, 1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, K. , Arazoe, T. , Yachie, N. , Banno, S. , Kakimoto, M. , Tabata, M. , Mochizuki, M. et al. (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science, 353, aaf8729. [DOI] [PubMed] [Google Scholar]
- Nishimura, N. , Tsuchiya, W. , Moresco, J.J. , Hayashi, Y. , Satoh, K. , Kaiwa, N. , Irisa, T. et al. (2018) Control of seed dormancy and germination by DOG1‐AHG1 PP2C phosphatase complex via binding to heme. Nat. Commun. 9, 2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, L.V. , Metzger, R.J. and Ching, T.M. (1985) Effect of temperature on seed dormancy of wheat1 . Crop Sci. 25, 455–458. [Google Scholar]
- Sato, K. (2020) History and future perspectives of barley genomics. DNA Res. 27, dsaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, K. , Matsumoto, T. , Ooe, N. and Takeda, K. (2009) Genetic analysis of seed dormancy QTL in barley. Breed. Sci. 59, 645–650. [Google Scholar]
- Sato, K. , Yamane, M. , Yamaji, N. , Kanamori, H. , Tagiri, A. , Schwerdt, J.G. , Fincher, G.B. et al. (2016) Alanine aminotransferase controls seed dormancy in barley. Nat. Commun. 7, 11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedel, S. , Pencs, S. , Hensel, G. , Müller, A. , Rutten, T. and Kumlehn, J. (2017) RNA‐guided Cas9‐induced mutagenesis in tobacco followed by efficient genetic fixation in doubled haploid plants. Front Plant Sci. 7, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurink, R.C. , Sedee, N.J.A. and Wang, M. (1992) Dormancy of the barley grain is correlated with gibberellic acid responsiveness of the isolated aleurone layer. Plant Physiol. 100, 1834–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, K. , Takeuchi, Y. , Ebana, K. , Miyao, A. , Hirochika, H. , Hara, N. , Ishiyama, K. et al. (2010) Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl Acad. Sci. 107, 5792–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel, J. , Koppolu, R. , Trautewig, C. , Hertig, C. , Kale, S. , Erbe, S. , Mascher, M. et al. (2021) Transcriptional landscapes of floral meristems in barley. Sci. Adv. 7, eabf0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torada, A. , Koike, M. , Ogawa, T. , Takenouchi, Y. , Tadamura, K. , Wu, J. , Matsumoto, T. et al. (2016) A causal gene for seed dormancy on wheat chromosome 4A encodes a MAP kinase kinase. Curr. Biol. 26, 782–787. [DOI] [PubMed] [Google Scholar]
- Tsukahara, K. , Sawada, H. , Kohno, Y. , Matsuura, T. , Mori, I.C. , Terao, T. , Ioki, M. et al. (2015) Ozone‐induced rice grain yield loss is triggered via a change in panicle morphology that is controlled by ABERRANT PANICLE ORGANIZATION 1 gene. PLoS One, 10, e0123308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich, S.E. , Hayes, P.M. , Dyer, W.E. , Blake, T.K. and Clancy, J.A. (1993) Quantitative trait locus analysis of seed dormancy in “Steptoe” barley. In Pre‐harvest Sprouting in Cereals 1992 ( Walker‐Simmons, M.K. and Reid, J.L. , eds.), pp. 136–145. St Paul, Minnesota: American Association of Cereal Chemists. [Google Scholar]
- Vetch, J.M. , Walling, J.G. , Sherman, J. , Martin, J.M. and Giroux, M.J. (2020) Mutations in the HvMKK3 and HvAlaAT1 genes affect barley preharvest sprouting and after‐ripened seed dormancy. Crop Sci. 60, 1897–1906. [Google Scholar]
- Wang, M. , Heimovaara‐Dijkstra, S. and Van Duijn, B. (1995) Modulation of germination of embryos isolated from dormant and nondormant barley grains by manipulation of endogenous abscisic acid. Planta, 195, 586–592. [Google Scholar]
- Wong, N. , Liu, W. and Wang, X. (2015) WU‐CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 16, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Partial DNA sequence of the gRNA target sites at the Qsd1 and Qsd2 loci in mutants.
Figure S2 PCR analysis for detection of T‐DNA region in T1 and T‐DNA‐free mutant plants.
Figure S3 Partial deduced amino acid sequences from the wild‐type and qsd1 and qsd2 mutants.
Figure S4. Germination of the wild‐type and M3 lines of genome‐edited barley 4 days after grain imbibition.
Figure S5 Germination of the M3 qsd1‐2 mutant 1 month after treatment with 3% hydrogen peroxide.
Figure S6 T‐DNA‐free F2 generation of wild‐type, qsd1 and qsd2 single mutant, and qsd1qsd2 double mutant plants.
Figure S7 Germination from segregating progeny with no mutation in Qsd1 or Qsd2 (wild‐type) or homozygous for qsd1, qsd2, or qsd1qsd2 double mutations (photographed 7 days after grain imbibition).
Figure S8 Pre‐harvest sprouting test of F3 progenies derived from qsd2‐4×qsd1‐1 (photographed 11 days after place spikes on the soil).
Figure S9 Target positions and 20‐nt sequence of gRNAs in Qsd1 and Qsd2 genes.
Figure S10 Structure of the T‐DNA region in the vector used in this study.
Table S1. PCR primers used in this study


