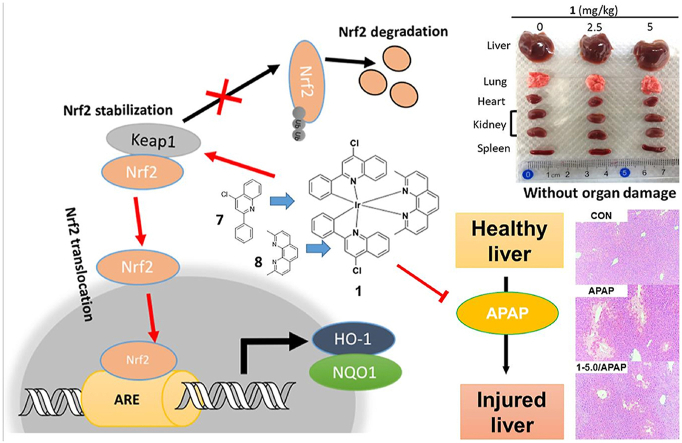
Abstract
Hepatotoxicity caused by an overdose of acetaminophen (APAP) is the leading reason for acute drug-related liver failure. Nuclear factor erythroid-2-related factor 2 (Nrf2) is a protein that helps to regulate redox homeostasis and coordinate stress responses via binding to the Kelch-like ECH-associated protein 1 (Keap1). Targeting the Keap1-Nrf2 interaction has recently emerged as a potential strategy to alleviate liver injury caused by APAP. Here, we designed and synthesized a number of iridium (III) and rhodium (III) complexes bearing ligands with reported activity against oxidative stress, which is associated with Nrf2 transcriptional activation. The iridium (III) complex 1 bearing a bioactive ligand 2,9-dimethyl-1,10-phenanthroline and 4-chloro-2-phenylquinoline, a derivative of the bioactive ligand 2-phenylquinoline, was identified as a direct small-molecule inhibitor of the Keap1–Nrf2 protein-protein interaction. 1 could stabilize Keap1 protein, upregulate HO-1 and NQO1, and promote Nrf2 nuclear translocation in normal liver cells. Moreover, 1 reversed APAP-induced liver damage by disrupting Keap1–Nrf2 interaction and without inducing organ damage and immunotoxicity in mice. Our study demonstrates the identification of a selective and efficacious antagonist of Keap1–Nrf2 interaction possessed good cellular permeability in cellulo and ideal pharmacokinetic parameters in vivo, and, more importantly, validates the feasibility of conjugating metal complexes with bioactive ligands to generate metal-based drug leads as non-toxic Keap1–Nrf2 interaction inhibitors for treating APAP-induced acute liver injury.
Keywords: Iridium(III) complex, Kelch-like ECH-Associated protein 1, Nuclear factor E2-related factor 2, Inhibitor, Liver injury
Graphical abstract
A bioactive ligand-conjugated iridium (III) metal-based complex 1 reverses acetaminophen-induced liver damage by disrupting Keap1–Nrf2 interaction and without inducing organ damage and immunotoxicity in mice.
Highlights
-
•
1 reversed APAP-induced liver damage by disrupting Keap1–Nrf2 interaction without inducing organ damage or immunotoxicity.
-
•
Complex 1 possessed good cellular permeability in cellulo and ideal pharmacokinetic parameters in vivo.
-
•
Conjugating metal complexes with bioactive ligands opens a novel avenue for the treatment of APAP-induced liver damage.
Abbreviations
- APAP
Acetaminophen
- ARE
Antioxidant response element
- NAPQI
N-acetyl-p-benzoquinone imine
- Co-IP
Co-immunoprecipitation
- DMSO
Dimethyl sulfoxide
- CETSA
Cellular thermal shift assay
- FBS
Fetal bovine serum
- HO-1
Heme oxygenase-1
- Keap1
Kelch-like ECH-associated protein 1
- NQO1
NAD(P)H: quinone oxidoreductase
- Nrf2
Nuclear factor E2-related factor 2
1. Introduction
Acetaminophen (APAP) is one of the most frequently used analgesics in the world [1]. Overdose of APAP is a leading factor for drug-related acute liver failure in developed nations, and is the second-most responsible reason for liver replacement [2]. The liver metabolizes APAP to generate the toxic quinone metabolite, N-acetyl-p-benzoquinone imine (NAPQI) [3,4]. Excess NAPQI adducts to biomolecules in the cell to produce NAPQI-protein complexes, leading to mitochondrial aberration, oxidative stress, and nuclear DNA damage, which drives hepatocyte damage and liver injury [5]. Currently, N-acetylcysteine (NAC) is the only FDA-approved treatment for hepatotoxicity induced by APAP [6]. However, some serious side effects of NAC include chest tightness, bronchoconstriction, and bleeding [7].
The transcription factor nuclear factor E2-related factor 2 (Nrf2) is activated in the presence of oxidative tension, where it binds to the antioxidant response element (ARE) within the promoter regions of genes coding for a range of antioxidant products [8,9], including quinone oxidoreductase (NQO1) and heme oxygenase-1 (HO-1) [10,11]. NQO1 utilizes NADH and NADPH to reduce quinones to hydroquinones and to decrease NAPQI production [12]. HO-1 induction reduced pro-inflammatory cytokine levels, and limited liver damage in acute hepatic inflammation rodent models [13]. Normally, Nrf2 interacts with Kelch-like ECH-associated protein 1 (Keap1), a substrate adaptor, in the cytoplasm, forming a complex with a CUL3-containing E3 ubiquitin ligase [14]. This CUL3-Keap1-E3 ligase complex adds a polyubiquitin chain to Nrf2, targeting it for proteasomal degradation to maintain a low base level of Nrf2 [15]. The loss of Keap1 activity in the liver leads to constitutive activation of Nrf2 and the expression of its target antioxidant genes [16]. Numerous natural (e.g. isothiocyanate, sulforaphane, and curcumin) and synthetic (e.g. dimethyl fumarate, bardoxolone methyl, and oltipraz) small molecules have been reported to activate ARE-dependent antioxidant gene transcription [17,18]. However, these reported activators of Nrf2 may have a higher potential for affecting multiple signaling pathways. ML334 (9), the first direct small-molecule antagonist of the Keap1–Nrf2 interaction, promotes Nrf2 nuclear translocation and induces ARE activity in vitro [19]. Following the discovery of ML334, several other direct small-molecule antagonists of Keap1–Nrf2 interaction have been reported (Supplementary Table 1 and Fig. S1) [[20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37]], including tetrahydroisoquinoline, 1,4-diaminonaphthalene, and its analogs, phenyl pyrrole, phenyl pyrazole, phenyl triazole, phenyl furan, and miscellaneous scaffolds. However, while some of this small-molecule the Keap1–Nrf2 protein-protein interaction (PPI) inhibitors exhibit nanomolar potency in vitro, they usually possess aliphatic acid groups, such as carboxylic acid (e.g. A1, B2–B4, B6–B8, B10–B11, B13, B18–B19, B23, B26–B29, C2–C3, D2), and hydroxamic acid (e.g. B14) groups. Although these aliphatic acid groups may play an important role for the recognition and interaction with Keap1 [33], they may also decrease membrane permeability in cellulo [33]. Moreover, while some studies have described the use of Keap1-Nrf2 inhibitors for treating APAP-induced liver injury have been reported [21,38], concerns regarding pharmacokinetic parameters and relatively low in vivo efficacy still remain [24]. Several small-molecule antagonists without aliphatic acid groups have been reported [24,31,32], but they are all derived from the 1,4-diaminonaphthalene scaffold and none of these inhibitors, to our knowledge, have been investigated for treating APAP-induced hepatotoxicity in cellulo and in vivo. Moreover, assessments of toxicity, including organ damage and immunotoxicity, for these compounds are still limited.
While conventional inorganic anticancer drugs such as cisplatin and its derivatives act through binding to DNA, recent advantages in chemical biology have enabled the concept of selectively focusing on medicinally relevant proteins or protein-protein interactions (PPIs) using transition-metal complexes [[39], [40], [41], [42], [43]]. It is now recognized that metal compounds can be fine-tuned for non-covalent interactions, due to their ability to incorporate a flexible array of ligands in different oxidation states [44,45], structural configurations [46,47], and stereochemical variations [[48], [49], [50]]. Many of these are based on Group 9 metal centers such as iridium and rhodium, which have the advantages of structural diversity [51,52], kinetic inertness [39,53], and tunable photophysical properties [[54], [55], [56], [57], [58]].
2-Phenylquinoline, an alkaloid, has potential antioxidant activity owing to its ability to inhibit detoxification enzyme depletion and as well as increase antioxidant levels [[59], [60], [61]]. Meanwhile, 2,9-dimethyl-1,10-phenanthroline has been described as a neuronal cell protector in oxidative stress, as it selectively chelates oxidative stress inducers such as copper ion [62]. Furthermore, metal complexes can possess various geometries depending on their oxidation status as well as the nature of both the auxiliary ligands and central metal ion [[63], [64], [65]]. Such compounds can be readily adapted to specifically recognize the binding regions of proteins through complementary interactions [66]. Specifically, conjugative bioactive ligands to metal scaffolds is an emerging design strategy for developing metal-based drug leads [54,63].
In this study, we synthesized a series of iridium (III) and rhodium (III) bearing ligands (2-phenylquinoline, 2,9-dimethyl-1,10-phenanthroline, and their derivatives) with reported activity against oxidative stress, and evaluated their impact on the regulation of the Keap1-Nrf2 pathway. This work led to the identification of the lead complex 1 as the first metal-based Keap1–Nrf2 inhibitor in the literature with the protective effect against APAP-induced hepatotoxicity. Thus, conjugating bioactive motifs to a metal scaffold to generate small-molecule inhibitors of Keap1–Nrf2 PPI is a powerful alternative strategy to obtain drug candidates against oxidative stress compared to conventional approaches.
2. Experimental section
2.1. Reagents
All compounds were analyzed by 1H and 13C NMR and high-resolution mass spectrometry. The purity of all complexes were validated by combustion using automatic analyzers (Atlantic Microlab Inc., Norcross, GA) to confirm the purity is ≥ 95%. Human liver LO2 cells were purchased from Shanghai institute of biochemistry and cell biology (Shanghai, China). A549, HUVEC, PC3, HeLa, and DU145 cells were obtained from ATCC (Manassas, USA). Fetal bovine serum (FBS) and Dulbecco's modified Eagle's medium (DMEM) were obtained from Gibco BRL (Gaithersburg, MD, USA). The Keap1–Nrf2 Inhibitor Screening Assay Kit was purchased from BSP Bioscience (San Diego, CA). The positive control ML334 was purchased from Merck KGaA (Darmstadt, Germany).
2.2. General experimental
Mass spectrometry was performed at the Mass Spectroscopy Unit at the Department of Chemistry, Hong Kong Baptist University, Hong Kong (China). Deuterated solvents for NMR purposes were obtained from Armar and used as received.1H and 13C NMR were recorded on a Bruker Advance 400 spectrometer operating at 400 MHz (1H) and 100 MHz (13C). 1H and 13C chemical shifts were referenced internally to solvent shift (Acetonitrile-d3: 1H, δ1.94, 13C, δ118.7; Acetone-d6: 1H, δ2.05, 13C, δ29.7). Chemical shifts (d) are quoted in ppm, the downfield direction being defined as positive. Uncertainties in chemical shifts are typically ±0.01 ppm for 1H and ±0.05 for 13C. Coupling constants are typically ±0.1 Hz for 1H–1H and ±0.5 Hz for 1H–13C couplings. The following abbreviations are used for convenience in reporting the multiplicity of NMR resonances: s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet. All NMR data were acquired and processed using standard Bruker software (Topspin).
2.3. Synthesis of iridium(III) and rhodium(III) complexes
2.3.1. Preparation of the precursor complexes [M2(C^N)4Cl2]
A solution of rhodium (III)/iridium (III) chloride (200 mg, 0.88 mmol) and corresponding C^N ligands (1.93 mmol, 2.2 eq.) in a mixture of methoxyethanol: water (3:1, 48 mL) was heated under reflux overnight under a nitrogen atmosphere. The reaction mixture was cooled to room temperature. The solid was collected by filtration, washed with additional portions of water (2 × 100 mL) and diethyl ether (2 × 50 mL), and dried to yield the corresponding precursor complexes.
2.3.2. Preparation of the final complexes
A suspension of [Rh2(C^N)4Cl2] (0.2 mM) and corresponding N^N ligands (0.24 mmol) in a mixture of dichloromethane: methanol (1:1, 20 mL) was refluxed overnight under a nitrogen atmosphere. The resulting solution was then allowed to cool to room temperature, and filtered to remove unreacted cyclometalated dimer. To the filtrate, an aqueous solution of ammonium hexafluorophosphate (excess) was added and the filtrate was reduced in volume by rotary evaporation until precipitation of the crude product occurred. The precipitate was then filtered and washed with several portions of water (2 × 50 mL) followed by diethyl ether (2 × 50 mL). The product was recrystallized by acetonitrile: diethyl ether vapor diffusion to yield the titled compounds.
Complex 1: (Yield 61%) 1H NMR (400 MHz, Acetone) δ 8.57 (d, J = 9.2 Hz, 4H), 8.14 (t, J = 8 Hz, 4H), 7.83 (d, J = 8.1 Hz, 4H), 7.57–7.48 (m, 4H), 7.14–7.07 (m, 4H), 6.85 (td, J = 7.7, 1.3 Hz, 2H), 6.55 (d, J = 7.6 Hz, 2H), 2.06 (s, 6H). 13C NMR (101 MHz, Acetone-d6) δ 170.8, 164.6, 148.3, 148.5, 147.7, 146.0, 145.2, 138.7, 133.2, 131.6, 130.8, 129.2, 127.7, 127.6, 127.6, 126.5, 125.1, 125.1, 124.4, 122.8, 117.7, 24.4. HRMS: calcd. For C44H30Cl2IrN4 [M − PF6]+: 877.1459, found: 877.1421; elemental anal. (C44H30Cl2N4IrPF6) C, H, N: calcd: 51.67, 2.96, 5.48; found: 51.66, 3.06, 5.59.
Complex 2: Reported [67].
Complex 3: Reported [68].
Complex 4: Reported [68].
Complex 5: (Yield: 49%) 1H NMR (400 MHz, acetone-d6) δ 8.51 (d, J = 8.3 Hz, 2H), 8.41 (d, J = 8.7 Hz, 2H), 8.28 (d, J = 8.8 Hz, 2H), 7.95 (d, J = 8.1 Hz, 2H), 7.85 (dd, J = 8.4, 1.6 Hz, 2H), 7.82–7.74 (m, 4H), 7.41 (d, J = 8.8 Hz, 2H), 7.34 (t, J = 7.5 Hz, 2H), 6.89–7.01 (m, 4H), 6.36 (s, 2H), 2.33–2.22 (m, 4H), 2.03 (s, 6H), 0.85 (t, J = 7.6 Hz, 6H). 13C NMR (101 MHz, Acetone) δ 171.8, 165.4, 149.7, 148.9, 148.6, 147.5, 144.8, 140.7, 139.4, 133.5, 131.2, 130.0, 129.9, 128.4, 128.3, 128.0, 127.3, 127.1, 124.7, 123.4, 118.1, 55.0, 25.1, 15.4. HRMS: calcd. For C48H40IrN4 [M − PF6]+: 865.2880, found: 865.2881. Anal. (C48H40IrN4PF6) C, H, N: calcd: 57.08, 3.89, 5.55; found: 55.61, 3.87, 5.51.
Complex 6: (Yield: 39%) 1H NMR (400 MHz, Acetone-d6) δ 8.58 (s, 2H), 8.52 (d, J = 8.3 Hz, 2H), 8.21 (dd, J = 7.9, 1.5 Hz, 2H), 8.14 (dd, J = 8.4, 1.4 Hz, 2H), 7.83–7.72 (m, 6H), 7.51 (t, J = 8.3 Hz, 2H), 7.22–7.11 (m, 4H), 6.93 (t, J = 7.9 Hz, 2H), 6.61 (dt, J = 7.9, 1.0 Hz, 2H), 2.14 (s, 6H). 13C NMR (101 MHz, Acetone) δ 168.0, 164.6, 148.4, 146.9, 146.8, 146.0, 139.6, 135.3, 132.2, 131.6, 129.5, 128.7, 128.4, 128.4, 127.0, 126.3, 125.8, 125.5, 124.8, 119.0, 119.0, 24.9. HRMS: calcd. For C44H30RhCl2N4 [M − PF6]+:787.0892, found: 787.0882; elemental anal. (C44H30Cl2N4RhPF6+1.5H2O) C, H, N: calcd: 55.02, 3.46, 5.83; found: 54.87, 3.59, 6.28.
Complex 1a: (Yield: 46%) 1H NMR (400 MHz, CDCl3) δ 8.82 (d, J = 5.4 Hz, 2H), 8.76 (s, 2H), 8.41 (d, J = 7.1 Hz, 2H), 8.08 (m, 4H), 8.01 (s, 2H), 7.63–7.52 (m, 12H), 7.457 (t, J = 7.6 Hz, 2H), 7.27 (t, J = 7.2 Hz, 2H), 7.11 (t, J = 7.2 Hz, 2H), 6.93 (t, J = 7.5, 2H), 6.78 (dd, J = 7.7, 0.8 Hz, 2H), 2.79 (s, 4H). 13C NMR (101 MHz, CDCl3) δ 171.8, 164.9, 151.5, 149.5, 149.3, 149.0, 146.8, 146.0, 136.6, 134.4, 132.6, 131.6, 130.6, 130.4, 130.0, 128.6, 128.6, 128.4, 127.7, 126.1, 126.1, 125.7, 124.9, 123.7, 118.6, 58.2, 54.9, 25.1. MALDI-TOF-HRMS: Calcd. For C56H38N4Cl2Ir [M − PF6]+: 1029.2103, Found: 1029.2114. Anal.: (C56H38IrCl2PF6N4) C, H, N: calcd. 57.24, 3.26, 4.77; found 57.41, 3.43, 4.94.
Complex 1b: (Yield: 61%) 1H NMR (400 MHz, Acetone-d6) δ 8.83 (d, J = 5.4 Hz, 2H), 8.76 (s, 2H), 8.42 (dd, J = 7.8, 1.2 Hz, 2H), 8.12–8.04 (m, 4H), 8.02 (s, 2H), 7.65–7.52 (m, 12H), 7.46 (ddd, J = 8.2, 6.9, 1.0 Hz, 2H), 7.28 (ddd, J = 8.1, 7.2, 1.2 Hz, 2H), 7.12 (ddd, J = 8.7, 6.9, 1.5 Hz, 2H), 6.94 (td, J = 7.4, 1.3 Hz, 2H), 6.78 (dd, J = 7.8, 1.1 Hz, 2H). 13C NMR (101 MHz, Acetone) δ 171.4, 152.3, 151.8, 149.6, 149.1, 148.1, 147.0, 146.3, 136.3, 135.7, 132.7, 132.0, 130.7, 129.9, 129.5, 128.9, 128.8, 128.2, 126.7, 126.1, 126.1, 125.8, 124.1, 119.3. MALDI-TOF-HRMS: Calcd. For C54H34Cl2IrN4 [M − PF6]+: 1001.1790 Found: 1001.1834; Anal.: (C54H34Cl2IrN4PF6) C, H, N: calcd. 56.55, 2.99, 4.88; found 56.06, 2.66, 4.94.
Complex 1c: (Yield: 51%) 1H NMR (400 MHz, CDCl3) δ 8.73 (s, 2H), 8.33 (d, J = 8.0 Hz, 2H), 8.26 (d, J = 8.3 Hz, 2H), 8.17 (dd, J = 8.3, 1.1 Hz, 2H), 8.10 (s, 2H), 7.96–7.89 (m, 2H), 7.62–7.52 (m, 4H), 7.30–7.17 (m, 4H), 6.86 (td, J = 7.6, 1.3 Hz, 2H), 6.62 (dd, J = 8.0, 0.8 Hz, 2H), 2.28 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 171.4, 166.5, 152.3, 151.4, 149.0, 148.6, 148.5, 147.1, 146.9, 146.8, 146.6, 146.5, 146.3, 137.8, 136.5, 135.9, 135.7, 134.5, 133.3, 132.7, 132.6, 132.4, 132.1, 132.0, 132.0, 129.2, 128.8, 128.8, 128.7, 128.2, 127.6, 126.7, 126.0, 125.8, 125.7, 125.6, 124.1, 121.0, 119.3, 15.3. MALDI-TOF-HRMS: Calcd. For C42H30Cl2IrN4 [M − PF6]+: 853.1477 Found: 853.1459; Anal.: (C42H30Cl2IrN4PF6+H2O) C, H, N: calcd. 49.61, 3.17, 5.51; found 49.72, 3.09, 5.71.
Complex 1d: (Yield: 58%) 1H NMR (400 MHz, CDCl3) δ 8.87 (dd, J = 8.6, 1.2 Hz, 2H), 8.73 (s, 2H), 8.69 (dd, J = 5.1, 1.2 Hz, 2H), 8.38 (d, J = 7.0 Hz, 2H), 8.10–8.01 (m, 4H), 7.41 (t, J = 8.4 Hz, 4H), 7.29–7.21 (m, 2H), 6.96 (td, J = 8.4, 1.6 Hz, 2H), 6.90 (td, J = 7.5, 1.3 Hz, 2H), 6.73 (dd, J = 7.7, 0.7 Hz, 2H), 2.70 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 171.4, 166.5, 152.3, 151.4, 149.0, 148.6, 148.5, 147.1, 146.9, 146.8, 146.6, 146.4, 146.3, 137.8, 136.5, 135.9, 135.7, 134.5, 133.3, 132.7, 132.6, 132.4, 132.1, 132.0, 132.0, 129.2, 128.8, 128.8, 128.7, 128.2, 127.6, 126.7, 126.0, 125.8, 125.7, 125.6, 124.1, 121.0, 119.3, 15.3. MALDI-TOF-HRMS: Calcd. For C44H30Cl2IrN4 [M − PF6]+: 877.1459 Found: 877.1472; Anal.: (C44H30Cl2IrN4PF6+H2O) C, H, N: calcd. 50.77, 3.10, 5.38; found 50.97, 3.06, 5.58.
Complex 1e: (Yield: 53%) 1H NMR (400 MHz, CDCl3) δ 8.54 (s, 2H), 8.30 (dd, J = 8.4, 1.1 Hz, 2H), 8.10 (dd, J = 7.9, 1.2 Hz, 2H), 7.95 (t, J = 7.8 Hz, 2H), 7.83 (t, J = 7.9 Hz, 4H), 7.68 (d, J = 8.2 Hz, 2H), 7.50 (d, J = 7.6 Hz, 2H), 7.34 (t, J = 7.2, 2H), 7.07 (t, J = 8.0 Hz, 2H), 6.83 (t, J = 7.2 Hz, 2H), 6.50 (dd, J = 7.8, 0.8 Hz, 2H), 1.81 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 171.5, 164.1, 159.2, 148.9, 148.5, 147.1, 145.8, 140.5, 133.8, 132.8, 131.7, 129.2, 128.8, 128.5, 126.3, 126.3, 126.2, 123.7, 122.5, 118.6, 24.5. MALDI-TOF-HRMS: Calcd. For C42H30N4Cl2Ir [M − PF6]+: 853.1477, Found: 853.1488. Anal.: (C42H30IrCl2PF6N4+ 2H2O) C, H, N: calcd. 48.75, 3.31, 5.41; found 48.29, 3.05, 5.51.
Complex 10: (Yield: 39%) 1H NMR (400 MHz, Acetone-d6) δ 8.52 (d, J = 5.3 Hz, 2H), 8.50–8.38 (m, 4H), 8.24–8.16 (m, 4H), 7.89 (d, J = 5.2 Hz, 2H), 7.77 (dd, J = 8.1, 1.4 Hz, 2H), 7.36 (d, J = 8.9 Hz, 2H), 7.25 (t, J = 8.0 Hz, 2H), 7.07 (dd, J = 8.0, 1. Hz, 2H), 6.87 (dt, J = 8.7, 1.6 Hz, 2H), 6.55 (d, J = 1.6 Hz, 2H), 2.87 (s, 6H), 2.28 (q, J = 7.6 Hz, 4H), 0.82 (td, J = 7.6, 1.3 Hz, 6H). 13C NMR (101 MHz, Acetone) δ 206.3, 206.1, 205.95, 171.2, 152.7, 149.8, 148.9, 148.5, 147.6, 147.3, 144.8, 140.7, 135.0, 131.2, 131.0, 129.9, 128.3, 128.3, 128.1, 127.2, 125.1, 125.0, 123.7, 118.7, 29.3, 18.9, 15.3. MALDI-TOF-HRMS: Calcd. For C48H40IrN4 [M − PF6]+: 865.2882 Found: 864.9313. Anal.: (C48H40IrN4PF6) C, H, N: calcd. 57.08, 3.99, 5.55; found 56.66, 3.97, 5.60.
Complex 11: Reported [69].
Complex 12: Reported [70].
Complex 13: (Yield: 42%) 1H NMR (400 MHz, CDCl3) δ 8.19 (d, J = 8.0 Hz, 2H), 7.91 (d, J = 8.0 Hz, 2H), 7.79 (m, 4H), 7.64 (d, J = 8.0 Hz, 2H), 7.57 (d, J = 8.0 Hz, 2H), 7.50 (d, J = 8.0 Hz, 2H), 7.43 (t, J = 6.0 Hz, 2H), 7.15 (d, J = 8.0 Hz, 2H), 7.08 (t, J = 8.0 Hz, 2H), 6.84 (d, J = 8.0 Hz, 2H), 2.27 (q, J = 8.0 Hz, 4H), 1.67 (s, 6H), 0.87 (t, J = 8.0 Hz, 6H). 13C NMR (101 MHz, CDCl3) δ 170.3, 162.5, 158.2, 147.7, 147.5, 147.1, 142.9, 139.6, 139.4, 132.5, 130.5, 129.1, 127.3, 126.6, 126.5, 124.4, 122.5, 121.6, 116.6, 28.8, 24.0, 14.9. HRMS: Calcd. For C46H40IrN4 [M − PF6]+: 841.2879 Found: 841.2893.
Complex 14: Reported [71].
Complex 15: Reported [72].
Complex 16: Reported [49].
Complex 17: (Yield: 42%) 1H NMR (400 MHz, CDCl3) δ 8.19 (d, J = 8.0 Hz, 2H), 7.95 (d, J = 8.0 Hz, 2H), 7.76 (d, J = 8.0 Hz, 2H), 7.70 (d, J = 8.0 Hz, 2H), 7.55 (m, 9H), 7.46 (s, 2H), 7.43 (d, J = 6.0 Hz, 2H), 7.37 (m, 5H), 6.97 (t, J = 6.0 Hz, 2H), 6.88 (d, J = 8.0 Hz, 2H), 6.31 (s, 2H), 2.30 (q, J = 8.0 Hz, 4H), 1.96 (s, 6H), 0.89 (t, J = 8.0 Hz, 6H). 13C NMR (101 MHz, CDCl3) δ 167.1, 160.8, 159.6, 150.6, 149.6, 148.0, 141.6, 137.8, 135.9, 131.2, 129.7, 128.8, 128.0, 122.3, 121.8, 118.6, 116.3, 107.9, 54.8. HRMS: Calcd. For C60H48IrN4 [M − PF6]+: 1017.3507, Found: 1017.3554.
Complex 18: (Yield: 42%) 1H NMR (400 MHz, CDCl3) δ 8.50 (d, J = 8.0 Hz, 2H), 8.17 (d, J = 8.0 Hz, 1H), 8.06 (t, J = 10.0 Hz, 2H), 7.84 (s, 2H), 7.71 (d, J = 8.0 Hz, 2H), 7.64 (m, 4H), 7.47 (d, J = 8.0 Hz, 2H), 7.33 (t, J = 8.0 Hz, 2H), 7.11 (t, J = 8.0 Hz, 4Hz, 2H), 6.99 (t, J = 8.0 Hz, 2H), 6.90 (m, 2H), 6.49 (s, 7H), 6.43 (d, J = 8.0 Hz, 2H), 5.61 (s, 2H), 2.00 (q, J = 8.0 Hz, 4H), 0.80 (t, J = 8.0 Hz, 6H). 13C NMR (101 MHz, CDCl3) δ 170.3, 163.3, 147.4, 146.5, 146.4, 144.8, 140.8, 139.0, 137.6, 136.8, 136.3, 131.2, 129.8, 129.1, 128.9, 128.3, 128.1, 127.5, 126.8, 126.7, 126.4, 126.0, 125.8, 125.7, 125.5, 125.4, 124.9, 122.6, 120.8, 115.7, 27.5, 13.4. HRMS: Calcd. For C58H44IrN4 [M − PF6]+: 989.3194 Found: 989.3167.
Complex 19: (Yield: 43%) 1H NMR (400 MHz, DMSO‑d6) δ 13.88 (s, 2H), 8.44 (s, 2H), 8.13 (s, 2H), 7.91 (d, J = 8.0 Hz, 2H), 7.54 (d, J = 8.0 Hz, 2H), 7.10 (m, 4H), 6.88 (t, J = 8.0 Hz, 2H), 6.74 (t, J = 8.0 Hz, 2H), 6.34 (d, J = 8.0 Hz, 2H), 5.35 (d, J = 8.0 Hz, 2H), 2.83 (s, 6H), 2.36 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 164.2, 151.1, 150.4, 146.2, 139.2, 134.7, 134.2, 133.0, 132.7, 130.3, 128.8, 124.4, 124.2, 123.4, 123.0, 121.9, 113.0, 112.6, 17.7, 14.8.
Complex 20: Reported [73].
Complex 21: (Yield: 52%) 1H NMR (400 MHz, CDCl3) δ 9.06 (s, 2H), 8.93 (d, J = 8.0 Hz, 2H), 8.25 (d, J = 8.0 Hz, 2H), 8.00 (d, J = 8.0 Hz, 2H), 7.77 (m, 4H), 7.60 (d, J = 8.0 Hz, 2H), 7.50 (d, J = 8.0 Hz, 2H), 7.42 (d, J = 8.0 Hz, 2H), 7.34 (d, J = 8.0 Hz, 2H), 7.10 (t, J = 8.0 Hz, 2H), 6.88 (t, J = 6.0 Hz, 2H), 6.26 (d, J = 8.0 Hz, 2H), 1.49 (s, 18H). 13C NMR (101 MHz, CDCl3) δ 169.0, 164.6, 155.7, 154.2, 149.7, 145.5, 140.4, 137.0, 132.2, 131.6, 130.8, 130.6, 128.6, 127.7, 126.9, 126.3, 124.9, 123.1, 122.1, 36.0, 30.5. HRMS: Calcd. For C48H44IrN4 [M–PF6]+: 869.3195 Found: 869.1659.
Complex 22: Reported [74].
Complex 23: Reported [75].
2.4. Fluorescence polarization assay
The disruption of the Keap1/Nrf2 peptide interaction by the lead compounds was tested following the manufacturer's instruction (BPS Bioscience, San Diego, CA). Briefly, the tested compounds were dissolved in the kit assay buffer. To each test well, 24 μL of the diluted binding assay buffer, 0.5 μL Nrf2 peptide (1 μM), 0.5 μL BSA (10 mg/mL) and 5 μL tested inhibitor (50 μM) and 20 μL diluted Keap1 (15 ng/mL) were then added and incubated for 0.5 h at ambient temperature. The change in fluorescent polarization can then be measured using a fluorescence reader capable of excitation at 485 nm and detection at 535 nm. Competitive assays were performed using a Corning 384-well plate as described in the procedures above. Wells were treated with biotinylated Nrf2 and with 1 at the indicated concentrations.
2.5. Protein thermal shift assay using purified recombinant Keap1 and Nrf2
Purified recombinant Keap1 and Nrf2 were firstly treated with 1 (5 μM) at room temperature for 30 min. The proteins were then heated at different temperatures ranging from 25 °C to 75 °C for 5 min. The collected protein samples were then detected by Western blotting using either Keap1 or Nrf2 antibodies. Densitometry analysis of Keap1 and Nrf2 levels was performed.
2.6. Cellular thermal shift assay
Cellular thermal shift assay was performed to monitor the target engagement of 1 in LO2 cell lysates. Briefly, cell lysates from 2 × 106 LO2 cells were collected, diluted in PBS, and separated in the same aliquots. Each aliquot was treated with 1 (5.0 μM) or DMSO. 30 min after incubation at room temperature, the compound-treated lysates were divided into 50 μL in each of the PCR tubes and heated individually at different temperatures (Veriti thermal cycler, Applied Biosystems/Life Technologies). The heated lysates were centrifuged and the supernatants were analyzed by SDS-PAGE followed by immunoblotting analysis by probing with the indicated antibody.
2.7. Isothermal titration calorimetry
ITC experiments were carried in a MicroCal PEAQ-ITC Isothermal Titration Calorimeter (Malvern Panalytical) as previously described with minor modification [[76], [77], [78]]. Briefly, complex 1 and recombinant proteins (Keap1 and Nrf2) were dialyzed into ITC buffer (20 mM Bis-Tris, 150 mM NaCl, 2 mM DTT). Complex 1 (200 μM) was titrated against 20 μM of proteins, over 19 injections of 2 μL complex 1 solution at a rate of 2 s/μL at 150 s time intervals. The assay was performed out at 25 °C with agitation at 750 rpm. The generated data was analyzed using the Setup MicroCal PEAQ-ITC Analysis Software. Three control titrations, in which (1) 1 is titrated into the buffer; (2) buffer is titrated into proteins; (3) buffer is titrated into the buffer, were also analyzed by using the composite model.
2.8. Expression and purification of Keap1 and Nrf2 proteins
The plasmids pET28a-His6-Keap1 (Addgene plasmid #62454) and pET28a-His6-Nrf2 (Addgene plasmid #62455) were extracted using TIANprep Rapid Mini Plasmid Kit (DP105), and then transformed into the expression strain E. coli (BL21) (DE3), respectively. The transformed E. coli (BL21) (DE3) cells were cultured in lysogeny broth (LB) medium with 50 μg/mL kanamycin and grew at 37 °C until the OD600 reached 0.5–0.8. Then, cells were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37 °C for 4 h. Cells were harvested by centrifugation at 4000g for 20 min and then homogenized by sonication in buffer (20 mM sodium phosphate, 500 mM NaCl, 5 mM imidazole, pH 7.4) and precleared lysates were applied to His GraviTrap columns (GE Healthcare), following kit protocols for purifications. The protein was assayed by SDS-PAGE by staining with Coomassie brilliant blue R250.
2.9. Mitochondrial dysfunction
Evaluation of mitochondrial depolarization was performed by rhodamine 123 staining, as described previously [79]. LO2 cells were cultured in the 6-well plates and allowed to attach overnight. After cellular adhesion, were treated for 8 h with compounds. For microscopic observation, after treatment with the compounds, cells were incubated in complete medium containing 5 μg/mL rhodamine 123 for 30 min and washed twice with PBS, and then analyzed using an inverted fluorescence microscope. For spectrofluorometer measurement, cells were harvested and adjusted to 1.0 × 105 cells/mL, washed twice with PBS, and then incubated with 5 μg/mL rhodamine 123 staining solution at 37 °C in the dark for 30 min. Afterward, cells were washed twice with PBS and centrifuged at 500×g for 10 min. Finally, absorbance was determined using a spectrofluorometer at an excitation wavelength of 505 nm and an emission wavelength of 534 nm.
2.10. ROS detection
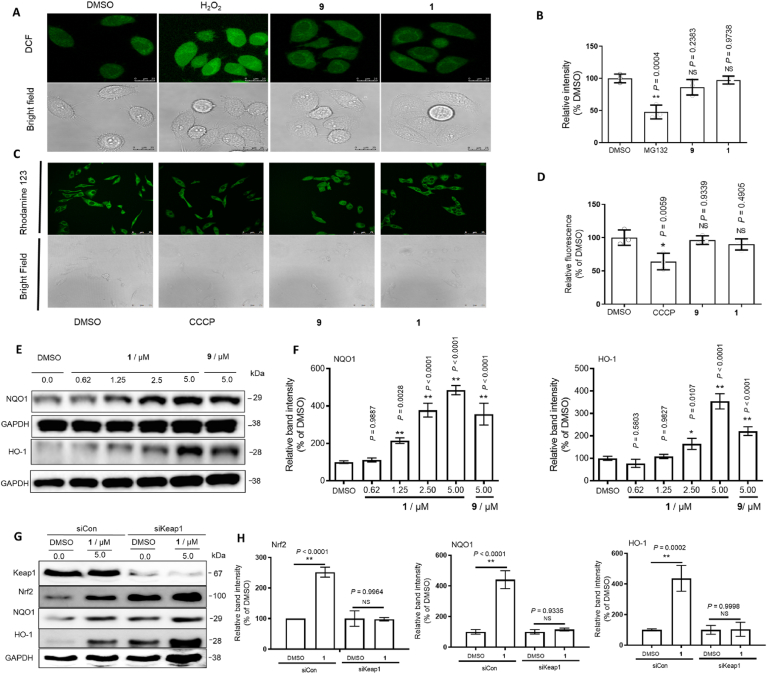
LO2 cells seeded into 6-well plates were treated with H2O2 (800 μM) for 3 h, and 1 (5 μM) or 9 (5 μM) for 8 h. Cells were collected and labeled with 5 μM of H2DCF-DA in serum-free DMEM at 37 °C for 30 min. Cells were washed twice with PBS, and the fluorescence intensity of the cells was measured a confocal microscopy with excitation at 488 nm and emission at 525 nm.
2.11. Measurement of proteasome activity
The effect of complex 1 on proteasome activity was determined using a commercial Proteasome Activity Fluorometric Assay Kit (BioVision Incorporated, CA, USA).
2.12. Animals and treatments
C57BL/6 male mice (8–10 weeks old) were obtained from the Faculty of Health Science, University of Macau (Macau, China). Mice were housed under identical conditions in a controlled environment with a 12:12 h light/dark cycle and fed with water and a standard laboratory rodent diet (Teklad 2018SX, Envigo) as following a previous study [80]. After a week acclimation, mice were randomly assigned to four groups (n = 10) as following: CON, APAP, APAP + 1–2.5 mg kg−1, APAP + 1–5.0 mg kg−11 treatment groups were administrated with 2.5 mg kg−1 or 5.0 mg kg−1 (dissolved in PEG 400: distilled water = 4:6, v/v) intraperitoneally (i.p.) once daily for five days. APAP was dissolved in warm phosphate buffer at 55–60 °C and cooled to 37 °C before injection. Mice fasted overnight before 400 mg kg−1 APAP injection. After the last dose of 1 pretreatment for 1 h, all the mice except the CON group were injected i.p. With APAP at a dose of 400 mg kg−1. The CON group received the same volume of vehicle solution (PEG 400: distilled water = 4:6, v/v). All mice were anesthetized after 6 h after the APAP challenge, and samples including blood and liver were immediately harvested and stored at −80 °C for further analysis. The experimental procedures (ICMS-AEC-031-2017) were performed in accordance with the animal care guidelines of the University of Macau and were approved by the ethics committee.
2.13. Measurement of serum aminotransferase levels
Serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT), the makers for liver injury [81], were determined using colorimetric kits according to the manufacture's protocols (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
2.14. Histopathological analysis
The identical lobe of the liver and other organs (spleen, heart, lung, and kidney) in each group were fixed in 4% phosphate-buffered polyoxymethylene for 24 h. The fixed organ tissues were processed routinely by being dehydrated in alcohol with progressively increased concentrations and embedded in paraffin. Sections (4 μm thickness) were stained with hematoxylin and eosin (H&E) according to the manufacturer's instructions. The pathological changes of organ sections were photographed and analyzed by an Olympus CX-31 light microscope (Olympus Corp., Tokyo, Japan). Necrotic area analysis was performed using ImageJ software (version 1.53e, National Institutes of Health).
2.15. Immunofluorescence assay
To determine whether the protection of complex 1 against APAP hepatotoxicity was involved in Nrf2 nuclear translocation, an immunofluorescence assay was performed. In brief, the cryostat sections (8 μm) of the liver were fixed in ice-cold 4% paraformaldehyde for 15 min and rehydrated with PBS three times. After permeabilization with 0.1% Triton-X-100 in PBS for 20 min, followed by blocking endogenous peroxidase with 5% goat serum in PBS at room temperature for 1 h, the slides were incubated with rabbit anti-mouse Nrf2 (1:50, Santa Cruz Biotechnology Inc., Santa Cruz, CA) overnight at 4 °C, then incubated with Alexa Fluor® 568 goat anti-rabbit IgG (1:200, Life Technologies, Carlsbad, CA, USA) for 2 h at 37 °C in dark. Nuclear were counterstained with DAPI. All fluorescent images were visualized and captured by a Leica TCS SP8 confocal microscope (Leica Microsystems).
2.16. Drug release experiments
After fasting overnight prior to the test, mice were intraperitoneally administered with complex 1 (5.0 mg kg−1), and blood was collected at the indicated time points (0, 0.5, 1, 2, 6, 12, 24, and 48 h) and placed into tubes containing EDTA-2Na. After centrifugation, plasma samples were used to determine the complex 1 concentration for the appropriate administration time using inductively coupled plasma mass spectrometry (ICP-MS).
2.17. Toxicity evaluation
Mice were intraperitoneally administrated with complex 1 (2.5 mg kg−1 or 5.0 mg kg−1) for 6 h, anesthetized, and body weight was recorded. Fresh organs (liver, kidney, lung, spleen, and heart) were weighed and photographed. Organ index was calculated by the following formula: organ index (%) = organ wet weight/body weight × 100% respectively. Approximately 100 mg organs were ground in liquid nitrogen and were digested by nitric acid. The concentration of complex 1 in each organ was determined using ICP-MS.
2.18. Flow cytometry determination
Mice were administrated with or without complex 1 (5.0 mg kg−1) for 6 h, anesthetized, and T cells were extracted from the spleen as previously described [82]. T cells were incubated with antibodies for surface markers, including anti-mouse-CD3-PE-Cy7, anti-mouse-CD4-FITC, and anti-mouse-CD8-PE antibodies (BioLegend, Shanghai, China) for 20 min, then the cells were accounted 104 cells and determined using flow cytometry analysis. The acquisition was performed with FACSpCanto II using the FACSDiva software (BD Biosciences) and data was analyzed with FlowJo software (Tree Star, Ashland, OR, USA).
2.19. Statistical analysis
All data were analyzed by using GraphPad Prism 6.0. Error bars represent the standard deviations of the results from three independent experiments unless otherwise noted. Significant differences between groups were determined using a one-way analysis of variance (ANOVA) unless otherwise noted. P < 0.05 was considered statistically significant throughout the study.
3. Results and discussion
3.1. Synthesis and screening of metal complexes as Keap1–Nrf2 interaction inhibitors
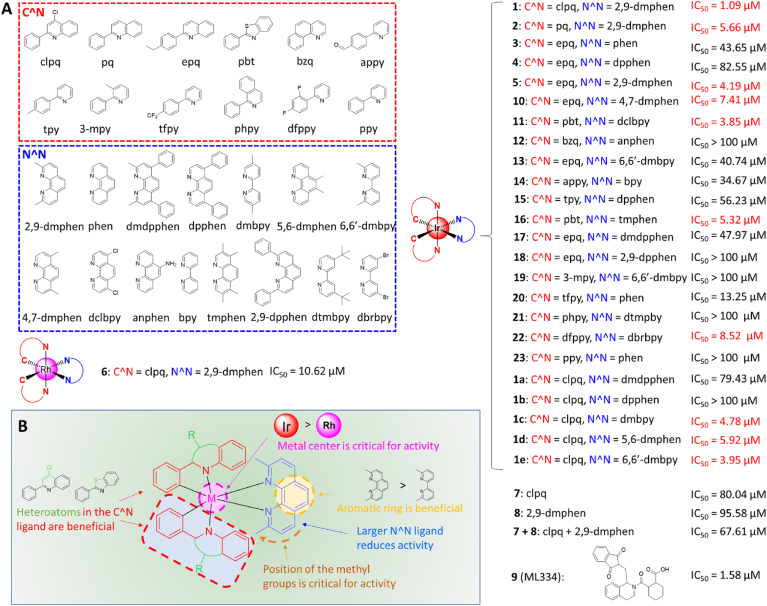
Fluorescence polarization (FP) is commonly used to investigate the interactions between various biomacromolecules in solution. We employed fluorescent Nrf2 tracer peptides carrying the ETGE motif to track Keap1–Nrf2 interaction inhibition. We synthesized iridium (III) and rhodium (III) complexes (1–6) with diverse C^N and N^N ligands (Fig. 1A). From the screening results, the iridium (III) complex 1 [Ir (clpq)2 (2,9-dmphen)](PF6) (where clpq = 4-chloro-2-phenylquinoline (7) and 2,9-dmphen = 2,9-dimethyl-1,10-phenanthroline (8)) was identified as the most effective compound at disrupting the Keap1–Nrf2 interaction, with an IC50 value of 1.09 μM. In contrast, the positive control ML334 (9) showed an IC50 value of 1.58 μM (Fig. 1A).
Fig. 1.
(A) Chemical structures and Keap1–Nrf2 binding inhibition IC50 values of the cyclometalated iridium (III) and rhodium (III) complexes 1–6, 1a–1e, 10–23, the ligands 7 and 8, and positive control 9 (ML334) evaluated in this study using a FP assay. (B) Summary of SAR for these complexes.
3.2. Structure-activity relationships
From the FP results, preliminary structure-activity relationships (SARs) could be drawn. Comparing iridium (III) complexes 1 and 2 suggests that having chlorine groups on the C^N ligand is desirable for activity since complex 1 bearing the clpq ligand showed higher potency (IC50 = 1.09 μM) than complex 2 (IC50 = 5.66 μM) with the 2-phenylquinoline (pq) ligand or complex 5 (IC50 = 4.19 μM) with the 2-(4-ethylphenyl)quinoline (epq) ligand. Interestingly, complexes containing the 2-phenylbenzothiazole (pbt) C^N ligand (11 and 16) and fluorine groups (20 and 22) showed quite potent inhibition of Keap-Nrf2 binding. Together with the lead complex 1, which bears the clpq ligand, this result suggests that having heteroatoms in the C^N ligand could be beneficial for Keap1-Nrf2 potency. Changing from 2,9-dmphen (as in 5) to 1,10-phenanthroline (phen, as in 3) or 4,7-diphenyl-1,10-phenanthroline (dpphen, as in 4) led to drastically reduced activity.
To test the importance of the 2,9-dmphen N^N ligand, a focused library of five cyclometalated iridium (III) complexes bearing clpq (1a–1e) C^N ligands were designed and synthesized. These were tested against for Keap1–Nrf2 inhibitory activity using the FP assay as above. However, none of these complexes (1a-1e) exceeded the potency of complex 1 in the second round of screening, indicating that the combination of the clpq C^N ligand and 2,9-dmphen N^N ligand were optimal for activity. Interesting SAR trends could be deduced from this focused library. For example, increasing the steric bulk of the N^N ligand by appending phenyl rings (1a and 1b) led to drastic decreases in activity, which is also demonstrated when considering the complexes 15, 17, and 18. On the other hand, reducing the size of the N^N ligand from 2,9-dmphen to 5,5′-dimethyl-2,2′-bipyridine (1c) or 6,6′-dimethyl-2,2′-bipyridine (6,6′-dmbpy, as in 1e) only slightly reduced activity, indicating that having small substituents at the N^N ligand may be important for Keap1-Nrf2 PPI inhibition. Taken together, these data suggest that the size of the complex may be an important determinant of activity, with larger complexes being less effective. Adding methyl groups at other positions (as in 1d, 10, and 16) reduced activity, which showed the position of the methyl groups is also important for inhibition of Keap-Nrf2 binding. Moreover, the identity of the metal center is critical for activity, since the rhodium (III) congener (6) of the lead iridium (III) complex 1 showed substantially weaker activity. Finally, the isolated ligands 7 and 8 showed poor activity at inhibiting the Keap1–Nrf2 interaction, even when incubated together simultaneously as a mixture (IC50 = 67.61 μM) with ligand 7 being raised to twice the concentration as in the initial screen (Fig. 1A). These data highlight the key role of the iridium (III) ion in organizing the co-ligands into a medicinally potent configuration. The overall SAR trends of these complexes are summarized in Fig. 1B.
3.3. 1 possesses good cellular permeability in cellulo
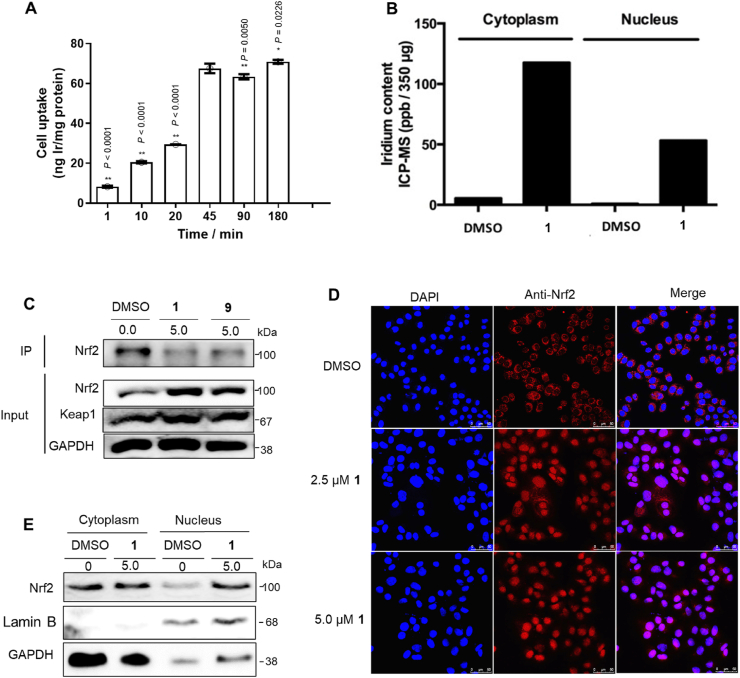
Complex 1 showed good stability in 4:1 acetonitrile/aqueous medium for seven days as revealed by UV–Vis (Supplementary Fig. S2A) and 1H NMR (Supplementary Fig. S2B). In 80% acetonitrile/20% buffer solution, the complex had an excitation peak at 365 nm and an emission peak at 591 nm, respectively, giving a Stokes shift of approximately 226 nm (Supplementary Fig. S2C). The luminescence lifetime was 1.16 μs (Supplementary Fig. S2D). Subsequently, the cellular uptake of 1 in LO2 cells was investigated by ICP-MS. As shown in Fig. 2A and B, cells treated with complex 1 displayed higher iridium levels in the nucleus and cytoplasm, indicating that 1 possesses good cellular permeability.
Fig. 2.
Effects of 1 on Nrf2 translocation. (A) ICP-MS quantification of cell uptake for 1. LO2 cells were incubated with 5 μM of 1 at different times. Error bars represent the standard deviations of the results from three independent experiments. P values were calculated using a one-way ANOVA with Tukey's multiple comparison test. **P < 0.01 vs. 45 min group. NS (not significant, P > 0.05) vs. 45 min group. (B) ICP-MS comparison of 1 in the nucleus and cytoplasm from LO2 cells. (C) Interactions between Keap1 and Nrf2 in LO2 cells were examined by Western blotting and co-IP. This experiment was repeated twice with similar results. (D) 1 induces nuclear translocation of Nrf2 in LO2 cells. (E) Western blot analysis of Nrf2 in the nucleus and cytoplasm in LO2 cells. This experiment was repeated twice with similar results.
3.4. 1 enhances the nuclear accumulation of Nrf2 in cellulo
A co-immunoprecipitation experiment was conducted to test the ability of 1 to disrupt the Keap1–Nrf2 interaction in cellulo. Treatment of LO2 cells with complex 1 led to a reduction of Nrf2 co-precipitated with Keap1, indicating that the complex could inhibit the Keap1–Nrf2 interaction in living cells (Fig. 2C). Inhibition of the Keap1–Nrf2 interaction in response to pro-oxidant stimuli allows the liberated Nrf2 to move into the nuclear region and bind to the ARE, thereby activating antioxidant genes. As shown in Fig. 2D, 1 induced Nrf2 accumulation in the nucleus of LO2 cells after 1 h has been observed using immunofluorescence. This result was further supported by Western blotting data which showed that complex 1 enhances the nuclear accumulation of Nrf2 in the LO2 cell line (Fig. 2E).
3.5. 1 engages Keap1 in cellulo
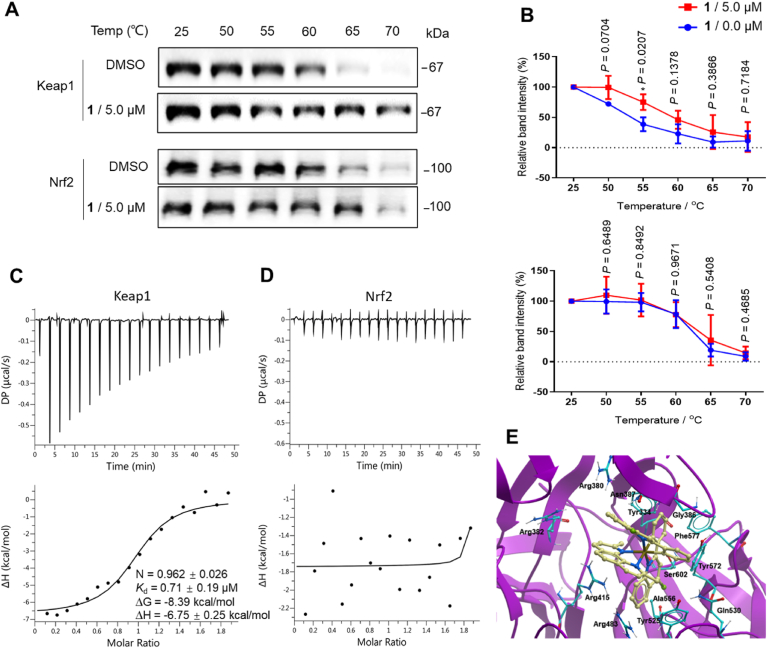
To investigate the ability of 1 to target Keap1 and Nrf2 in cellulo, the cellular thermal shift assay (CETSA) was conducted (Fig. 3A) [83,84]. Complex 1 significantly stabilized Keap1 and shifted the Keap1 melting curve by ca. 6 °C, whereas no significant shift for Nrf2 was detected (Fig. 3B), This result indicates that 1 can target Keap1 even within cell lysates, and further suggests that the ability of complex 1 to antagonize the Keap1–Nrf2 interaction in the FP assay could be attributed to its ability to bind to Keap1.
Fig. 3.
Complex 1 engages Keap1 in cellulo. (A and B) Thermal stabilization of Keap1 and Nrf2 by 1 and densitometry analysis. Proteins were detected by Western blotting using the corresponding antibodies. Error bars represent the standard deviations of the results from three independent experiments. *P < 0.05 vs. DMSO group. (C and D) ITC thermograms showing titration of 200 μM of 1 into (C) Keap1 (20 μM) and (D) Nrf2 (20 μM). (E) Docking pose of complex 1 with Keap1 (PDB code: 4L7B) which is depicted as a space-filling representation showing carbon (yellow), nitrogen (blue), chlorine (green) atoms. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To exclude the influence of cellular factors, human recombinant Keap1 and Nrf2 proteins were firstly expressed from E. coli (BL21) (DE3) (Supplementary Fig. S3). To understand the nature of inhibition of Keap1 and Nrf2 binding by complex 1, a competitive assay was also performed (Supplementary Fig. S4). The double-reciprocal plot showed that 1 was a competitive inhibitor of Keap1 and Nrf2 binding with a Ki value of 1.03 μM. Isothermal titration calorimetry (ITC) was used to characterize the thermodynamic parameters of complex 1 binding to Keap1 or Nrf2. The results showed that complex 1 bound to Keap1 with an approximate 1:1 stoichiometry (N = 0.962 ± 0.026) and with a dissociation constant of Kd = 0.71 ± 0.79 μM (Fig. 3C). Moreover, the binding between 1 and Keap1 could be driven by enthalpy/entropy compensation (ΔG = −8.39 kcal/mol, ΔH = −6.75 kcal/mol, -TΔS = −1.64 kcal/mol), suggesting that noncovalent interactions may play a key role in the binding. However, no binding was observed between complex 1 and Nrf2 by ITC (Fig. 3D).
To further investigate the mechanism of action of complex 1, we evaluated the binding mode of complex 1 to Keap1 via molecular docking using the X-ray co-crystal structure of the Keap1 dimer with ML334 9 (PDB code: 4L7B). Docking was performed using the Molsoft ICM method (ICM version 3.9-1b molecular docking software). In the lowest-energy binding pose of 1 to the Keap1 dimer, 1 is observed to occupy the same binding pocket as the positive control 9 (Fig. 3E and Supplementary Fig. S5A). The predicted binding mode of between complex 1 and Keap1 is expected to be stabilized by noncovalent interactions, including van der Waals, π-π stacking, and hydrophobic interactions. The quinoline moiety of one C^N ligand forms partial π-π interactions with Tyr334, van der Waals interactions with Arg 380, and hydrophobic interactions with Asn387. Another C^N ligand forms van der Waals interactions with Arg 483, and multiple hydrophobic interactions with Gly 386, Tyr525, Gln 530, Ala556, Tyr572, Phe 577, and Ser602. The phenanthroline N^N ligand interacts with Arg415, Asn387, and Arg 382 through van der Waals interactions. Interestingly, amino acids such as Tyr334, Arg415, Ala556, Tyr572, and Ser602 that are important for the binding of positive control 9 [85] are also critical for maintaining the interactions between complex 1 and Keap1 dimer. One of the clpq C^N ligands of 1 is located at the same position of the tetrahydroisoquinoline and phthalimide moieties of 9. Meanwhile, the other 4-clpq ligand and the 2,9-dmphen N^N ligand of 1 overlap with the cyclohexane ring of 9. In contrast, the inactive complex 4 occupies only part of the binding area engaged by 9 (Supplementary Fig. S5B), probably due to its larger size and incorrect geometry.
3.6. 1 exerts its cellular effects via blocking the Keap1–Nrf2 interaction and activating HO-1 and NQO1
Mitochondria generated higher ROS levels during oxidation damage, which activates and releases Nrf2 from the Keap1-Nrf2 complex in the cytoplasm [86]. The released Nrf2 then enters into the nuclear region to enhance ARE-directed transcription, thus driving antioxidant activity as well as mitochondrial biogenesis. Moreover, the impairment of the ubiquitin-proteasome system has been reported to lead to the transcriptional activation of Nrf2 [87]. In this study, we monitored the effect of 1 on the mitochondrial membrane potential, ROS levels, and proteasomal activity in LO2 cells. The results showed that 5 μM of complex 1 had no significant effect on ROS elevation (Fig. 4A), proteasomal inhibition (Fig. 4B), and/or mitochondrial dysfunction (Fig. 4C and D) [79]. This suggests that the relevant concentration of complex 1 is unlikely to exert its biological effects through proteasomal inhibition, ROS elevation, and/or mitochondrial dysfunction.
Fig. 4.
Complex 1 exerts its cellular effects via blocking the Keap1–Nrf2 interaction and activating HO-1 and NQO1. (A) Detection of intracellular ROS by confocal microscopy with excitation at 488 nm. LO2 cells seeded into 6-well plates were treated with H2O2 (800 μM) for 3 h, and 1 (5 μM) or 9 (5 μM) for 8 h. Complex 1 and ML334 9 (5 μM) had no significant effect on ROS elevation and proteasomal inhibition. (B) Measurement of proteasome activity using a commercial fluorometric assay kit after 1 (5 μM) or 9 (5 μM) treatment for 8 h. MG132 was used as a positive control. (C) Effect of 1 on mitochondrial membrane potential in LO2 cells as measured via rhodamine 123 staining. Cells seeded into 6-well plates were treated with 10 μM of positive control CCCP for 1 h, 5 μM of 9, and 1 for 8 h. Cells were imaged by confocal microscopy with excitation at 488 nm. (D) Fluorescence intensity was determined using a spectrofluorometer at an excitation wavelength of 505 nm and an emission wavelength of 534 nm. (E and F) Effects of 1 and ML334 on HO-1 and NQO1 levels in LO2 cells after 6 h treatment and densitometry analysis of Western blotting results. (G and H) Keap1 siRNA treatment produces efficient target knockdown in LO2 cells. Keap1, Nrf2, HO-1, and NQO1 were blotted to control for total protein levels. SiCon: control siRNA. siKeap1: Keap1 siRNA. Error bars represent the standard deviations of the results from three independent experiments. P values were calculated using a one-way ANOVA with Tukey's multiple comparison test. **P < 0.01 vs. DMSO group. NS (not significant, P > 0.05) vs. DMSO group.
Nuclear Nrf2 forms hetero-dimers with Maf to bind to ARE and activate the expression of antioxidant proteins, including HO-1 and NQO1. As shown in Fig. 4E and F, 1 increased the levels of HO-1 and NQO1 in treated LO2 cells. A Keap1 knockdown assay was performed to further confirm the relationship between Keap1 activity and HO-1 and NQO1 in LO2 cells [88]. The results of Western blotting showed that complex 1 was less effective at increasing HO-1 and NQO1 levels in Keap1 knockdown cells versus control cells (Fig. 4G and H). Similarly, the increase of the levels of HO-1 and NQO1 induced by complex 1 was muted after Nrf2 knockdown (Supplementary Fig. S6). Taken together, the data show that complex 1 exerts its cellular effects via blocking the Keap1–Nrf2 interaction and activating HO-1 and NQO1 [89].
3.7. 1 displays cytoprotective effects against APAP-induced toxicity in cellulo
It has been suggested that Nrf2 activators should be tested for two opposing effects: cytoprotection versus cytotoxicity [90]. The cytoprotective effects are due to the activation of Nrf2-dependent antioxidant proteins by complex 1, while cytotoxicity can result from other in vitro toxicity mechanisms of 1. After 48 h of treatment, complex 1 showed IC50 values of between 10.6 and 109.0 μM in seven different cell lines, including LO2 cells with an IC50 of 48.0 μM (Supplementary Fig. S7). The isolated ligands 7 and 8 also showed low cytotoxicity to LO2 cells, with IC50 > 100 μM (Supplementary Fig. S8). Importantly, complex 1 showed dose-dependent restoration of cell viability of LO2 cells after treatment with APAP (Supplementary Fig. S9). This suggests complex 1 displays cytoprotective effects against APAP-induced toxicity in LO2 cells.
3.8. The effect of metal complex 1 on APAP-induced liver injury in vivo
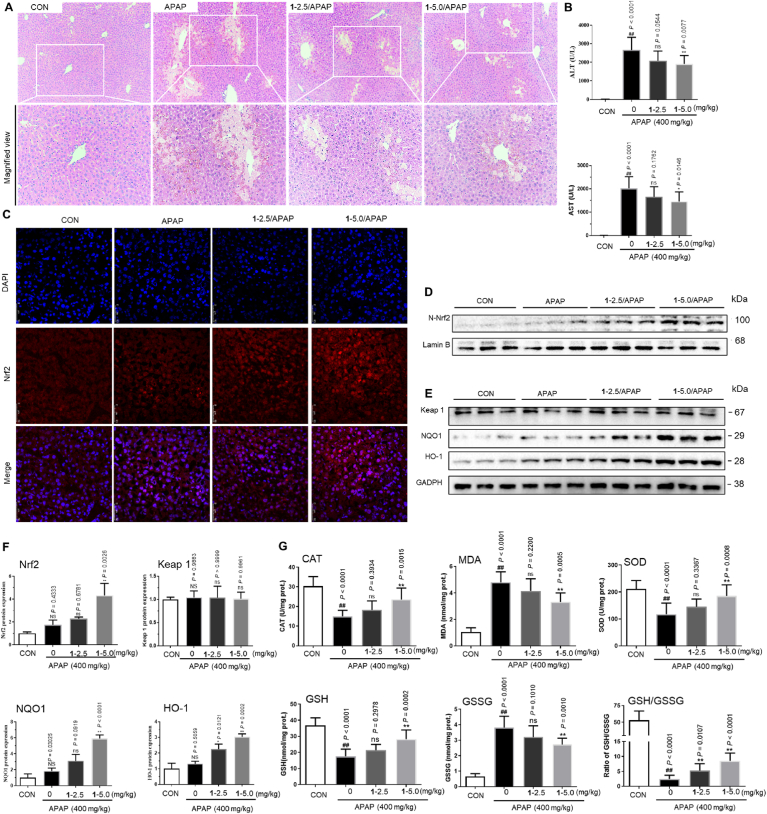
Inspired by the effective activity of complex 1 in vitro and in cellulo, the protective ability of 1 versus acute liver damage induced by APAP was next studied in an animal model. To evaluate the hepatoprotective ability of 1 on APAP injury, mice were pretreated with 1 (2.5 mg kg−1 or 5.0 mg kg−1) once daily for five days before the APAP challenge. The results of histopathological examination revealed that the APAP challenge caused extensive liver damage including destruction of liver architecture, hepatocyte ballooning degeneration, and centrilobular hepatocyte death. APAP-induced histological changes were attenuated significantly in 1 treatment group. After 2.5 mg kg−1 or 5.0 mg kg−1 of 1 treatment, the percentage of necrotic liver areas induced APAP 21.01% was reduced to 16.04% and 8.99%, respectively (Fig. 5A). Moreover, mice treated with APAP (400 mg kg−1) showed marked increases in serum ALT and AST by 188-fold and 216-fold, respectively compared with the control group, indicating significant liver injury. However, the increases of ALT and AST levels in serum were attenuated in the 1 treatment group in a dose-dependent fashion compared with the control group, with 5.0 mg kg−1 1 decreasing serum ALT and AST by 23.5% and 25.2%, respectively (Fig. 5B). To evaluate the degree of protection of 1 against APAP toxicity, serum ALT and AST levels at various time points (e.g. 12 h, 24 h, and 48 h) after APAP challenge were also detected. Time course analysis demonstrated that the enzymatic activities of ALT and AST increased in a time-dependent fashion before 12 h, reached maximum levels (3334.98 U/L for ALT and 2002.31 U/L for AST) at 12 h, and returned to lower levels (608.41 U/L for ALT and 488.20 U/L for AST) by 48 h after APAP challenge (Supplementary Fig. S10A). The elevation of serum ALT and AST levels in APAP-induced mice was markedly reduced after 1 treatment at different time intervals (6, 12, 24, and 48 h), as evidenced by the 31.45–51.77% reductions of ALT levels and 32.29–53.62% reductions of AST levels. In parallel with serum activities of ALT and AST, the elevation of APAP-induced liver necrosis area was also significantly inhibited after 1 treatment at the indicated time intervals (Supplementary Fig. S10B). Collectively, these findings indicate that 1 administration could alleviate APAP-induced acute liver injury. Moreover, complex 1 treatment induced Nrf2 nuclear translocation (Fig. 5C), as well as the immunofluorescence results in vitro. To investigate whether 1 protects against APAP-induced acute liver injury via targeting Nrf2, we next determined the levels of hepatic Nrf2 and its target antioxidant proteins, HO-1 and NQO1, in the liver cells. As shown in Fig. 5D–E and densitometry analysis Fig. 5F, 1 treatment significantly increased Nrf2 levels in the nucleus in comparison to the APAP group, which showed only a slight increase in Nrf2 levels. However, the protein expression of Keap1, which increases the proteasomal degradation of Nrf2, remained basal after APAP administration in 1-treated mice, indicating that 1-induced Nrf2 activation was likely not due to inhibition of Keap1 expression. As shown in Fig. 5G, the APAP challenge significantly decreased the activities of catalase (CAT) and superoxide dismutase (SOD), while decreasing the GSH/GSSG glutathione ratio in the liver compared with the control group. However, 1 administration (2.5 mg kg−1 or 5.0 mg kg−1) reversed these changes in a dose-dependent manner. Moreover, the ability of 1 to increase the in vivo expression of Nrf2 was also verified in liver tissue sections (Supplementary Fig. S11). In short, our data demonstrates that 1 can ameliorate APAP-induced liver injury, likely via inhibiting the degradation of Nrf2 and reversing the decrease of Nrf2-dependent hepatic antioxidative parameters.
Fig. 5.
Complex 1 alleviates APAP-induced acute liver injury in mice. (A) Representative hematoxylin and eosin (H&E) staining of liver tissue sections. (B) Serum levels of alanine aspartate transaminase (AST) and aminotransferase (ALT). 1 induces nuclear translocation of Nrf2. Immunofluorescence staining of Nrf2 (C) and Western blot analysis of Nrf2 in the nucleus (D). (E) Effect of 1 and ML334 on the HO-1 and NQO1 levels by Western Blotting. (F) 1 reduced APAP-induced liver injury was involved in the upregulation of Nrf2-mediated antioxidative protein. Immunoblots analysis of nuclear Nrf2 (A) and Keap1 (B) expressions respectively, and Nrf2 downstream target proteins NQO1 (C) and HO-1 (D). Lamin B was used as the loading control. (G) Complex 1 alleviated APAP-induced hepatic oxidative stress. Hepatic levels of MDA, GSH, GSSG, GSH/GSSG, and enzyme activities of CAT and SOD were determined after the APAP challenge for 6 h. P values were calculated using a one-way ANOVA with Tukey's multiple comparison test. Data are presented as mean ± SD (n = 3 mice). ##P < 0.01 vs. Control (CON) group. *P < 0.05, **P < 0.01 vs. APAP-induced model group.
3.9. Pharmacokinetic and toxicity of metal complex 1 in mice
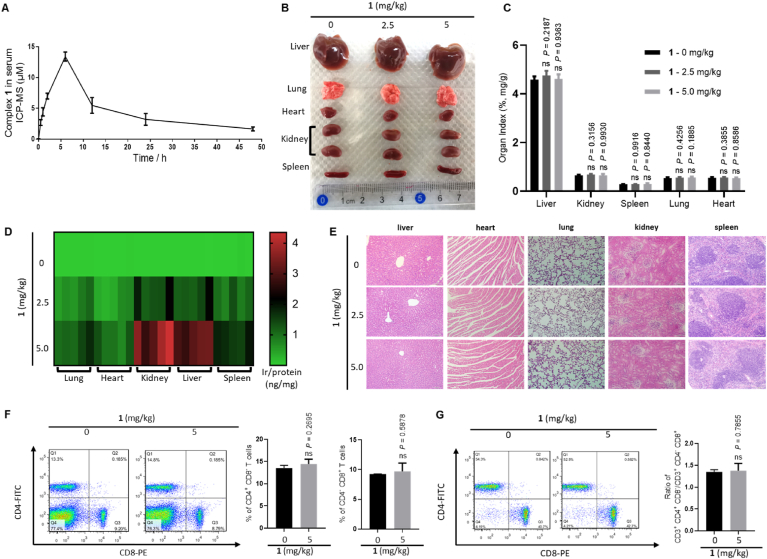
We next evaluated the pharmacokinetic parameters of metal complex 1 and its potential toxicity in vivo. The pharmacokinetic profile of complex 1 was studied after intraperitoneal injection of the complex (5.0 mg kg−1) in mice by monitoring plasma concentration at various time points (0, 0.5, 1, 2, 6, 12, 24, and 48 h) using ICP-MS. The Cmax value occurred between 2 and 12 h, and the maximum mean peak area (13.41 μM or 13.70 μg/mL) of complex 1 in plasma was obtained at 6 h (Fig. 6A). Moreover, administration with complex 1 (5.0 mg kg−1) did not cause organ damage at various time points (6, 12, and 24 h), as evidenced by lack of significant changes of organ morphology (e.g. liver, lung, heart, kidney, and spleen) and organ index (Fig. 6B and C, Supplementary Figs. S12A and S12B). In addition, the concentration of complex 1 in various organs was determined using ICP-MS after intraperitoneal injection with different concentrations of 1 for 6 h. The results indicated that complex 1 was mainly concentrated in the liver and kidney at Cmax (Fig. 6D). H&E staining of various organs also showed normal tissue structures without any obvious inflammatory lesions or organ damage after complex 1 treatment (Fig. 6E and Supplementary Fig. S12C). Moreover, immunotoxicity of complex 1 was assessed by counting CD4+ CD8− and CD4− CD8+ T lymphocytes in mice spleen. Consistently, T lymphocytes extracted from mice spleen showed no significant difference between the populations of CD4+ CD8− and CD4− CD8+ T lymphocytes, as well as in the ratio between CD3+ CD4+ CD8− and CD3+ CD4− CD8+ T lymphocytes, in treated and untreated mice as analyzed by flow cytometry (Fig. 6F and G). Taken together, these data suggest that 1 is non-toxic in mice without inducing organ damage and immunotoxicity.
Fig. 6.
Pharmacokinetics and toxicity of complex 1 in mice. (A) Pharmacokinetic profile of complex 1. Mean peak area of 1 in plasma at different time points. C57BL/6J mice were intraperitoneally injected with complex 1 at 5 mg kg−1. The concentrations of complex 1 in the plasma samples were measured at 0, 0.5, 1, 2, 6, 12, 24, and 48 h after administration. Data are expressed as means ± SD (n = 4). (B) Images of representative liver, lung, heart, kidney, and spleen of mice sacrificed at 6 h after intraperitoneal injection with different concentrations of 1. (C) Liver, lung, heart, kidney, and spleen indexes of complex 1-treated mice at 12 h after intraperitoneal injection. Data are expressed as means ± SD (n = 5), P values were calculated using a one-way ANOVA with Tukey's multiple comparison test. nsP > 0.05 vs. control group. (D) Heat map generated from ICP-MS for quantitative iridium element reflecting absorption values of the complex 1. (E) Histological H&E staining of main organs from mice at 6 h after intraperitoneal injection. (F) Representative density plots of splenic CD4+ CD8− and CD4− CD8+ cells. The numbers indicate the percentage of CD4+ CD8− and CD4− CD8+ cells. FITC, fluorescein isothiocyanate; PE, phycoerythrin. (G) Representative density plots of splenic CD3+ CD4+ CD8− and CD3+ CD4− CD8+ cells. The numbers indicate the ratio change of the percentage of CD3+ CD4+ CD8− and CD3+ CD4− CD8+ cells. Data are expressed as means ± SD (n = 3), P values were calculated using a two-sided t-test. nsP > 0.05 vs. 0 mg kg−1 group.
4. Conclusions
2-Phenylquinoline and 2,9-dimethyl-1,10-phenanthroline have been reported as regulators against oxidative stress by inhibiting detoxification enzyme depletion and chelating oxidative stress inducers, respectively [59,62]. Herein, we have developed an effective direct Keap1–Nrf2 inhibitor (iridium (III) complex 1) by coupling an iridium (III) core with 4-chloro-2-phenylquinoline (7), a derivative of the bioactive ligand 2-phenylquinoline, and 2,9-dimethyl-1,10-phenanthroline (8), which have previously shown activity against oxidative stress, a key activator of the Nrf2 transcriptional pathway. 1 inhibited the interaction Keap1–Nrf2 interaction in vitro by targeting Keap1 directly. The SAR study indicated the important determinants for Keap1-Nrf2 inhibitory activity, which are (1) having a heteroatom (e.g. chlorine) on the C^N ligand situated in the correct arrangement, (2) having C^N and N^N ligands that are not excessively bulky, and (3) having the appropriate number and position methyl groups on the auxiliary ligands. In terms of mechanism, 1 could stabilize Keap1 in LO2 cell lysates, increase Nrf2 nucleus translocation and upregulate HO-1 and NQO1. Additionally, complex 1 showed high cytoprotective activity and low cytotoxicity in vitro. In a murine model of APAP-induced liver injury, 1 decreased serum ALT and AST and alleviated the destruction of liver architecture caused by APAP. Paralleling the in vitro results, complex 1 increased Nrf2 signaling and activated downstream protein expression in vivo and without inducing organ damage and immunotoxicity.
Although complex 1 does not bear any aliphatic acid groups, it showed strong activity at disrupting the Keap1–Nrf2 interaction in vitro, with an IC50 value of 1.09 μM, which is more potent than ML334 (IC50 = 1.58 μM) bearing a carboxylic acid group. In cellulo, 1 possessed good cellular permeability with high iridium levels in the nucleus and cytoplasm after 45 min treatment (Fig. 2A and B). Moreover, the pharmacokinetic data for complex 1 shows that it is well absorbed, with peak plasma concentrations reached within 6 h after administration at 5 mg kg−1, and an ideal elimination half-life of 12 h in vivo (Fig. 6A), which is an important parameter for clinical drugs with regards to accumulation. Finally, our study demonstrated that metal-based complex 1, as the first metal-based Keap1–Nrf2 inhibitor in the literature, could reverse APAP-induced liver damage by disrupting the Keap1–Nrf2 interaction without inducing organ damage and immunotoxicity in mice (Fig. 6B–G). These results demonstrate that conjugating metal complexes with bioactive ligands to generate metal-based drug leads as and non-toxic Keap1–Nrf2 interaction inhibitors is an effective strategy to improve the relatively low in vivo efficacy of currently reported inhibitors for treating APAP-induced acute liver injury.
Declaration of competing interest
The authors declare no conflict of interest.
Author contributions
G.L. and T.S.K. carried out the in vitro and in cellulo experiments. H.L. and C·N.K. carried out synthesis and optimization of complexes. R.F. carried out the in vivo experiments. W.W. performed the molecular docking. G.L. and T.S.K. analyzed the data and wrote the paper; D.L.M., J.B·W., and C·H.L. designed the research study; M.Y. contributed essential reagents or tools.
Declaration of competing interest
All authors have approved the final version of the manuscript.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (21575121 and 21775131), the Hong Kong Baptist University Century Club Sponsorship Scheme 2020, the Science and Technology Development Fund, Macau SAR (File no. 0072/2018/A2, 0007/2020/A1 and 0034/2019/A1), SKL-QRCM(UM)-2020-2022, and the University of Macau (MYRG2019-00002-ICMS).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102129.
Contributor Information
Dik-Lung Ma, Email: edmondma@hkbu.edu.hk.
Jian-Bo Wan, Email: JBWan@um.edu.mo.
Chung-Hang Leung, Email: duncanleung@um.edu.mo.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ghosh A., Sil P.C. Anti-oxidative effect of a protein from Cajanus indicus L against acetaminophen-induced hepato-nephro toxicity. J. Biochem. Mol. Biol. 2007;40(6):1039. doi: 10.5483/bmbrep.2007.40.6.1039. [DOI] [PubMed] [Google Scholar]
- 2.Fontana R.J. Acute liver failure including acetaminophen overdose. Med. Clin. 2008;92(4):761–794. doi: 10.1016/j.mcna.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jadeja R.N., Urrunaga N.H., Dash S., Khurana S., Saxena N.K. Withaferin-A reduces acetaminophen-induced liver injury in mice. Biochem. Pharmacol. 2015;97(1):122–132. doi: 10.1016/j.bcp.2015.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng R., Wang Y., Liu C., Yan C., Zhang H., Su H., Kang J.X., Shang C.-Z., Wan J.-B. Acetaminophen-induced liver injury is attenuated in transgenic fat-1 mice endogenously synthesizing long-chain n-3 fatty acids. Biochem. Pharmacol. 2018;154:75–88. doi: 10.1016/j.bcp.2018.04.019. [DOI] [PubMed] [Google Scholar]
- 5.Jaeschke H., Bajt M.L. Intracellular signaling mechanisms of acetaminophen-induced liver cell death. Toxicol. Sci. 2005;89(1):31–41. doi: 10.1093/toxsci/kfi336. [DOI] [PubMed] [Google Scholar]
- 6.Jadeja R.N., Upadhyay K.K., Devkar R.V., Khurana S. Naturally occurring Nrf2 activators: potential in treatment of liver injury. Oxid. Med. Cell. Longev. 2016;2016:3453926. doi: 10.1155/2016/3453926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dodd S., Dean O., Copolov D.L., Malhi G.S., Berk M. N-acetylcysteine for antioxidant therapy: pharmacology and clinical utility. Expet Opin. Biol. Ther. 2008;8(12):1955–1962. doi: 10.1517/14728220802517901. [DOI] [PubMed] [Google Scholar]
- 8.Vomund S., Schäfer A., Parnham M.J., Brüne B., von Knethen A. Nrf2, the master regulator of anti-oxidative responses. Int. J. Mol. Sci. 2017;18(12):2772. doi: 10.3390/ijms18122772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ursini F., Maiorino M., Forman H.J. Redox homeostasis: the Golden Mean of healthy living. Redox Biol. 2016;8:205–215. doi: 10.1016/j.redox.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nam E., Derrick J.S., Lee S., Kang J., Han J., Lee S.J.C., Chung S.W., Lim M.H. Regulatory activities of dopamine and its derivatives towards metal-free and metal-induced amyloid-β aggregation, oxidative stress, and inflammation in alzheimer’s disease. ACS Chem. Neurosci. 2018;9(11):2655–2666. doi: 10.1021/acschemneuro.8b00122. [DOI] [PubMed] [Google Scholar]
- 11.Ke B., Shen X.-D., Zhang Y., Ji H., Gao F., Yue S., Kamo N., Zhai Y., Yamamoto M., Busuttil R.W. KEAP1-NRF2 complex in ischemia-induced hepatocellular damage of mouse liver transplants. J. Hepatol. 2013;59(6):1200–1207. doi: 10.1016/j.jhep.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srijiwangsa P., Na-Bangchang K. Roles of NAD (P) H-quinone oxidoreductase 1 (NQO1) on cancer progression and chemoresistance. J. Clin. Exp. Oncol. 2017;6(4):1–6. 2017. [Google Scholar]
- 13.Karatop-Cesur İ., Yildiz S., Uzun G., Oztas Y., Sabuncuoglu S., Ilgaz Y., Kutlu A., Dogan E., Cebi G., Oztas E. Effects of hyperbaric oxygen therapy on acetaminophen induced nephrotoxicity and hepatotoxicity: the role of heme oxygenase-1. Dis. Mol. Med. 2016;4(3):37–42. [Google Scholar]
- 14.Bo C., Lu Y., Chen Y., Cheng J. The role of Nrf2 in oxidative stress-induced endothelial injuries. J. Endocrinol. 2015;225(3):83–99. doi: 10.1530/JOE-14-0662. [DOI] [PubMed] [Google Scholar]
- 15.Villeneuve N.F., Lau A., Zhang D.D. Regulation of the Nrf2–Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxidants Redox Signal. 2010;13(11):1699–1712. doi: 10.1089/ars.2010.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paneni F., Volpe M., Lüscher T.F., Cosentino F. SIRT1, p66(shc), and set 7/9 in vascular hyperglycemic memory: bringing all the strands together. Diabetes. 2013;62(6):1800–1807. doi: 10.2337/db12-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eggler A.L., Gay K.A., Mesecar A.D. Molecular mechanisms of natural products in chemoprevention: induction of cytoprotective enzymes by Nrf2. Mol. Nutr. Food Res. 2008;52(S1):S84–S94. doi: 10.1002/mnfr.200700249. [DOI] [PubMed] [Google Scholar]
- 18.Hur W., Gray N.S. Small molecule modulators of antioxidant response pathway. Curr. Opin. Chem. Biol. 2011;15(1):162–173. doi: 10.1016/j.cbpa.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Hu L., Magesh S., Chen L., Wang L., Lewis T.A., Chen Y., Khodier C., Inoyama D., Beamer L.J., Emge T.J. Discovery of a small-molecule inhibitor and cellular probe of Keap1–Nrf2 protein–protein interaction. Bioorg. Med. Chem. Lett. 2013;23(10):3039–3043. doi: 10.1016/j.bmcl.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang C.-S., Zhuang C.-L., Zhu K., Zhang J., Muehlmann L.A., Figueiro Longo J.P., Azevedo R.B., Zhang W., Meng N., Zhang H. Identification of a novel small-molecule Keap1–Nrf2 PPI inhibitor with cytoprotective effects on LPS-induced cardiomyopathy. J. Enzym. Inhib. Med. Chem. 2018;33(1):833–841. doi: 10.1080/14756366.2018.1461856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu L.-L., Wu Y.-F., Wang L., Li C.-C., Li L., Di B., You Q.-D., Jiang Z.-Y. Structure-activity and structure-property relationships of novel Nrf2 activators with a 1, 2, 4-oxadiazole core and their therapeutic effects on acetaminophen (APAP)-induced acute liver injury. Eur. J. Med. Chem. 2018;157:1376–1394. doi: 10.1016/j.ejmech.2018.08.071. [DOI] [PubMed] [Google Scholar]
- 22.Sun H.-P., Jiang Z.-Y., Zhang M.-Y., Lu M.-C., Yang T.-T., Pan Y., Huang H.-Z., Zhang X.-J., You Q.-d. Novel protein–protein interaction inhibitor of Nrf2–Keap1 discovered by structure-based virtual screening. MedChemComm. 2014;5(1):93–98. [Google Scholar]
- 23.Zhuang C., Narayanapillai S., Zhang W., Sham Y.Y., Xing C. Rapid identification of Keap1–Nrf2 small-molecule inhibitors through structure-based virtual screening and hit-based substructure search. J. Med. Chem. 2014;57(3):1121–1126. doi: 10.1021/jm4017174. [DOI] [PubMed] [Google Scholar]
- 24.Lu M., Zhang X., Zhao J., You Q., Jiang Z. A hydrogen peroxide responsive prodrug of Keap1-Nrf2 inhibitor for improving oral absorption and selective activation in inflammatory conditions. Redox Biol. 2020;34 doi: 10.1016/j.redox.2020.101565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazzara P.R., Jain A.D., Maldonado A.C., Richardson B., Skowron K.J., David B.P., Siddiqui Z., Ratia K.M., Moore T.W. Synthesis and evaluation of noncovalent naphthalene-based KEAP1-NRF2 inhibitors. ACS Med. Chem. Lett. 2020;11(4):521–527. doi: 10.1021/acsmedchemlett.9b00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.You Q., Jiang Z., Mengchen L., Chen Z., Sun H., Zhang X., Xiaoke G., Xu X. Google Patents; 2019. 1-sulfonamido-4-aryloxy Compounds, and Preparation Method and Medicinal Application Thereof. [Google Scholar]
- 27.Meng N., Tang H., Zhang H., Jiang C., Su L., Min X., Zhang W., Zhang H., Miao Z., Zhang W. Fragment-growing guided design of Keap1-Nrf2 protein-protein interaction inhibitors for targeting myocarditis. Free Radic. Biol. Med. 2018;117:228–237. doi: 10.1016/j.freeradbiomed.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Hu L., Magesh S., Chen L., Lewis T., Munoz B., Wang L. Google Patents; 2014. Direct Inhibitors of Keap1-Nrf2 Interaction as Antioxidant Inflammation Modulators. [Google Scholar]
- 29.Yasuda D., Nakajima M., Yuasa A., Obata R., Takahashi K., Ohe T., Ichimura Y., Komatsu M., Yamamoto M., Imamura R. Synthesis of Keap1-phosphorylated p62 and Keap1-Nrf2 protein-protein interaction inhibitors and their inhibitory activity. Bioorg. Med. Chem. Lett. 2016;26(24):5956–5959. doi: 10.1016/j.bmcl.2016.10.083. [DOI] [PubMed] [Google Scholar]
- 30.Winkel A.F., Engel C.K., Margerie D., Kannt A., Szillat H., Glombik H., Kallus C., Ruf S., Güssregen S., Riedel J. Characterization of RA839, a noncovalent small molecule binder to Keap1 and selective activator of Nrf2 signaling. J. Biol. Chem. 2015;290(47):28446–28455. doi: 10.1074/jbc.M115.678136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain A.D., Potteti H., Richardson B.G., Kingsley L., Luciano J.P., Ryuzoji A.F., Lee H., Krunic A., Mesecar A.D., Reddy S.P. Probing the structural requirements of non-electrophilic naphthalene-based Nrf2 activators. Eur. J. Med. Chem. 2015;103:252–268. doi: 10.1016/j.ejmech.2015.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu M.-C., Tan S.-J., Ji J.-A., Chen Z.-Y., Yuan Z.-W., You Q.-D., Jiang Z.-Y. Polar recognition group study of Keap1-Nrf2 protein–protein interaction inhibitors. ACS Med. Chem. Lett. 2016;7(9):835–840. doi: 10.1021/acsmedchemlett.5b00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang Z.-Y., Lu M.-C., Xu L.L., Yang T.-T., Xi M.-Y., Xu X.-L., Guo X.-K., Zhang X.-J., You Q.-D., Sun H.-P. Discovery of potent Keap1–Nrf2 protein–protein interaction inhibitor based on molecular binding determinants analysis. J. Med. Chem. 2014;57(6):2736–2745. doi: 10.1021/jm5000529. [DOI] [PubMed] [Google Scholar]
- 34.Jnoff E., Albrecht C., Barker J.J., Barker O., Beaumont E., Bromidge S., Brookfield F., Brooks M., Bubert C., Ceska T. Binding mode and structure–activity relationships around direct inhibitors of the Nrf2–Keap1 complex. ChemMedChem. 2014;9(4):699–705. doi: 10.1002/cmdc.201300525. [DOI] [PubMed] [Google Scholar]
- 35.Abed D.A., Goldstein M., Albanyan H., Jin H., Hu L. Discovery of direct inhibitors of Keap1–Nrf2 protein–protein interaction as potential therapeutic and preventive agents. Acta Pharm. Sin. B. 2015;5(4):285–299. doi: 10.1016/j.apsb.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu M.-C., Zhao J., Liu Y.-T., Liu T., Tao M.-M., You Q.-D., Jiang Z.-Y. CPUY192018, a potent inhibitor of the Keap1-Nrf2 protein-protein interaction, alleviates renal inflammation in mice by restricting oxidative stress and NF-κB activation. Redox Biol. 2019;26 doi: 10.1016/j.redox.2019.101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng R.-B., Wang Y., He C., Yang Y., Wan J.-B. Gallic acid, a natural polyphenol, protects against tert-butyl hydroperoxide-induced hepatotoxicity by activating ERK-Nrf2-Keap1-mediated antioxidative response. Food Chem. Toxicol. 2018;119:479–488. doi: 10.1016/j.fct.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 38.Lu M.-C., Zhang X., Wu F., Tan S.-J., Zhao J., You Q.-D., Jiang Z.-Y. Discovery of a potent Kelch-like ECH-associated protein 1-nuclear factor erythroid 2-related factor 2 (Keap1–Nrf2) protein–protein interaction inhibitor with natural proline structure as a cytoprotective agent against acetaminophen-induced hepatotoxicity. J. Med. Chem. 2019;62(14):6796–6813. doi: 10.1021/acs.jmedchem.9b00818. [DOI] [PubMed] [Google Scholar]
- 39.Leung C.-H., Zhong H.-J., Chan D.S.-H., Ma D.-L. Bioactive iridium and rhodium complexes as therapeutic agents. Coord. Chem. Rev. 2013;257(11–12):1764–1776. [Google Scholar]
- 40.Bruijnincx P.C.A., Sadler P.J. New trends for metal complexes with anticancer activity. Curr. Opin. Chem. Biol. 2008;12(2):197–206. doi: 10.1016/j.cbpa.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporale C., Massi M. Cyclometalated iridium (III) complexes for life science. Coord. Chem. Rev. 2018;363:71–91. [Google Scholar]
- 42.Chen F., Huang X., Wu M., Gou S., Hu W. A CK2-targeted Pt (IV) prodrug to disrupt DNA damage response. Canc. Lett. 2017;385:168–178. doi: 10.1016/j.canlet.2016.10.026. [DOI] [PubMed] [Google Scholar]
- 43.Li G., Ko C.-N., Li D., Yang C., Wang W., Yang G.-J., Di Primo C., Wong V.K.W., Xiang Y., Lin L., Ma D.-L., Leung C.-H. A small molecule HIF-1α stabilizer that accelerates diabetic wound healing. Nat. Commun. 2021;12(1):3363. doi: 10.1038/s41467-021-23448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang P., Sadler P.J. Redox‐active metal complexes for anticancer therapy. Eur. J. Inorg. Chem. 2017;12:1541–1548. 2017. [Google Scholar]
- 45.Guo L., Zhang H., Tian M., Tian Z., Xu Y., Yang Y., Peng H., Liu P., Liu Z. Electronic effects on reactivity and anticancer activity by half-sandwich N, N-chelated iridium (III) complexes. New J. Chem. 2018;42(19):16183–16192. [Google Scholar]
- 46.Yam V.W.-W., Wong K.M.-C. Luminescent metal complexes of d 6, d 8 and d 10 transition metal centres. Chem. Commun. 2011;47(42):11579–11592. doi: 10.1039/c1cc13767k. [DOI] [PubMed] [Google Scholar]
- 47.Liu Z., Sadler P.J. Organoiridium complexes: anticancer agents and catalysts. Acc. Chem. Res. 2014;47(4):1174–1185. doi: 10.1021/ar400266c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.OuYang J., Crassous J. Chiral multifunctional molecules based on organometallic helicenes: recent advances. Coord. Chem. Rev. 2018;376:533–547. [Google Scholar]
- 49.Kang T.-S., Mao Z., Ng C.-T., Wang M., Wang W., Wang C., Lee S.M.-Y., Wang Y., Leung C.-H., Ma D.-L. Identification of an iridium (III)-based inhibitor of tumor necrosis factor-α. J. Med. Chem. 2016;59(8):4026–4031. doi: 10.1021/acs.jmedchem.6b00112. [DOI] [PubMed] [Google Scholar]
- 50.Liu L.-J., Wang W., Huang S.-Y., Hong Y., Li G., Lin S., Tian J., Cai Z., Wang H.-M.D., Ma D.-L. Inhibition of the Ras/Raf interaction and repression of renal cancer xenografts in vivo by an enantiomeric iridium (III) metal-based compound. Chem. Sci. 2017;8(7):4756–4763. doi: 10.1039/c7sc00311k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sluijter S.N., Elsevier C.J. Synthesis and reactivity of heteroditopic dicarbene rhodium (I) and iridium (I) complexes bearing chelating 1, 2, 3-triazolylidene–imidazolylidene ligands. Organometallics. 2014;33(22):6389–6397. [Google Scholar]
- 52.Nejman P.S., Morton-Fernandez B., Moulding D.J., Arachchige K.S.A., Cordes D.B., Slawin A.M., Kilian P., Woollins J.D. Structural diversity of bimetallic rhodium and iridium half sandwich dithiolato complexes. Dalton Trans. 2015;44(38):16758–16766. doi: 10.1039/c5dt02542g. [DOI] [PubMed] [Google Scholar]
- 53.Messori L., Marcon G., Orioli P., Fontani M., Zanello P., Bergamo A., Sava G., Mura P. Molecular structure, solution chemistry and biological properties of the novel [ImH][trans-IrCl4 (Im)(DMSO)],(I) and of the orange form of [(DMSO) 2H][trans-IrCl4 (DMSO) 2],(II), complexes. J. Inorg. Biochem. 2003;95(1):37–46. doi: 10.1016/s0162-0134(03)00069-2. [DOI] [PubMed] [Google Scholar]
- 54.Nam J.S., Kang M.-G., Kang J., Park S.-Y., Lee S.J.C., Kim H.-T., Seo J.K., Kwon O.-H., Lim M.H., Rhee H.-W. Endoplasmic reticulum-localized iridium (III) complexes as efficient photodynamic therapy agents via protein modifications. J. Am. Chem. Soc. 2016;138(34):10968–10977. doi: 10.1021/jacs.6b05302. [DOI] [PubMed] [Google Scholar]
- 55.Tian X., Zhu Y., Zhang M., Luo L., Wu J., Zhou H., Guan L., Battaglia G., Tian Y. Localization matters: a nuclear targeting two-photon absorption iridium complex in photodynamic therapy. Chem. Commun. 2017;53(23):3303–3306. doi: 10.1039/c6cc09470h. [DOI] [PubMed] [Google Scholar]
- 56.Li G., Wu C., Ma D.-L., Leung C.-H. Drug screening strategies using metal-based luminescent probes. Trends Anal. Chem. 2021;139 [Google Scholar]
- 57.Ma D.L., Wu C., Dong Z.Z., Tam W.S., Wong S.W., Yang C., Li G., Leung C.H. The development of G‐quadruplex‐based assays for the detection of small molecules and toxic substances. Chem. Asian J. 2017;12(15):1851–1860. doi: 10.1002/asia.201700533. [DOI] [PubMed] [Google Scholar]
- 58.Liu J., Dong Z.-Z., Yang C., Li G., Wu C., Lee F.-W., Leung C.-H., Ma D.-L. Turn-on luminescent probe for hydrogen peroxide sensing and imaging in living cells based on an iridium (III) complex–silver nanoparticle platform. Sci. Rep. 2017;7(1):1–7. doi: 10.1038/s41598-017-09478-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Breviglieri E., da Silva L.M., Boeing T., Somensi L.B., Cury B.J., Gimenez A., Cechinel Filho V., de Andrade S.F. Gastroprotective and anti-secretory mechanisms of 2-phenylquinoline, an alkaloid isolated from Galipea longiflora. Phytomedicine. 2017;25:61–70. doi: 10.1016/j.phymed.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 60.Souza M., Gushiken L., Pereira F., Pellizzon C. Evaluation of the gastroprotective and antioxidant effects of caffeine and caffeic acid on ethanol-induced gastric ulcer. JSM Hepatitis. 2017;2:1008. [Google Scholar]
- 61.Verbanac D., Malik R., Chand M., Kushwaha K., Vashist M., Matijašić M., Stepanić V., Perić M., Paljetak H.Č., Saso L. Synthesis and evaluation of antibacterial and antioxidant activity of novel 2-phenyl-quinoline analogs derivatized at position 4 with aromatically substituted 4 H-1, 2, 4-triazoles. J. Enzym. Inhib. Med. Chem. 2016;31(2):104–110. doi: 10.1080/14756366.2016.1190714. [DOI] [PubMed] [Google Scholar]
- 62.Nunes E.A., Manieri T.M., Matias A.C., Bertuchi F.R., da Silva D.A., Lago L., Sato R.H., Cerchiaro G. Protective effects of neocuproine copper chelator against oxidative damage in NSC34 cells. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2018;836:62–71. doi: 10.1016/j.mrgentox.2018.06.019. [DOI] [PubMed] [Google Scholar]
- 63.Yang G.J., Wang W., Mok S.W.F., Wu C., Law B.Y.K., Miao X.M., Wu K.J., Zhong H.J., Wong C.Y., Wong V.K.W. Selective inhibition of lysine‐specific demethylase 5A (KDM5A) using a rhodium (III) complex for triple‐negative breast cancer therapy. Angew. Chem. Int. Ed. 2018;130(40):13275–13279. doi: 10.1002/anie.201807305. [DOI] [PubMed] [Google Scholar]
- 64.Ko C.-N., Li G., Leung C.-H., Ma D.-L. Dual function luminescent transition metal complexes for cancer theranostics: the combination of diagnosis and therapy. Coord. Chem. Rev. 2019;381:79–103. [Google Scholar]
- 65.Ma D.-L., Chan D.S.-H., Leung C.-H. Group 9 organometallic compounds for therapeutic and bioanalytical applications. Acc. Chem. Res. 2014;47(12):3614–3631. doi: 10.1021/ar500310z. [DOI] [PubMed] [Google Scholar]
- 66.Kilpin K.J., Dyson P.J. Enzyme inhibition by metal complexes: concepts, strategies and applications. Chem. Sci. 2013;4(4):1410–1419. [Google Scholar]
- 67.Leung K.-H., He H.-Z., He B., Zhong H.-J., Lin S., Wang Y.-T., Ma D.-L., Leung C.-H. Label-free luminescence switch-on detection of hepatitis C virus NS3 helicase activity using a G-quadruplex-selective probe. Chem. Sci. 2015;6(4):2166–2171. doi: 10.1039/c4sc03319a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ko C.-N., Yang C., Lin S., Li S., Dong Z., Liu J., Lee S.M.-Y., Leung C.-H., Ma D.-L. A long-lived phosphorescence iridium (III) complex as a switch on-off-on probe for live zebrafish monitoring of endogenous sulfide generation. Biosens. Bioelectron. 2017;94:575–583. doi: 10.1016/j.bios.2017.03.050. [DOI] [PubMed] [Google Scholar]
- 69.Lin S., Gao W., Tian Z., Yang C., Lu L., Mergny J.-L., Leung C.-H., Ma D.-L. Luminescence switch-on detection of protein tyrosine kinase-7 using a G-quadruplex-selective probe. Chem. Sci. 2015;6(7):4284–4290. doi: 10.1039/c5sc01320h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu L., Zhong H.-J., Wang M., Ho S.-L., Li H.-W., Leung C.-H., Ma D.-L. Inhibition of beta-amyloid fibrillation by luminescent iridium (III) complex probes. Sci. Rep. 2015;5:14619. doi: 10.1038/srep14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiong L., Zhao Q., Chen H., Wu Y., Dong Z., Zhou Z., Li F. Phosphorescence imaging of homocysteine and cysteine in living cells based on a cationic iridium (III) complex. Inorg. Chem. 2010;49(14):6402–6408. doi: 10.1021/ic902266x. [DOI] [PubMed] [Google Scholar]
- 72.Lu L., Chan D.S.-H., Kwong D.W., He H.-Z., Leung C.-H., Ma D.-L. Detection of nicking endonuclease activity using a G-quadruplex-selective luminescent switch-on probe. Chem. Sci. 2014;5(12):4561–4568. [Google Scholar]
- 73.Yang X.H., Li M., Peng H., Zhang Q., Wu S.X., Xiao W.Q., Chen X.L., Niu Z.G., Chen G.Y., Li G.N. Highly luminescent mono‐and dinuclear cationic iridium (III) complexes containing phenanthroline‐based ancillary ligand. Eur. J. Inorg. Chem. 2019;6:847–855. 2019. [Google Scholar]
- 74.Cavazzini M., Quici S., Scalera C., Puntoriero F., La Ganga G., Campagna S. Synthesis, characterization, absorption spectra, and luminescence properties of multinuclear species made of Ru (II) and Ir (III) chromophores. Inorg. Chem. 2009;48(17):8578–8592. doi: 10.1021/ic9006108. [DOI] [PubMed] [Google Scholar]
- 75.You Y., Cho S., Nam W. Cyclometalated iridium (III) complexes for phosphorescence sensing of biological metal ions. Inorg. Chem. 2014;53(4):1804–1815. doi: 10.1021/ic4013872. [DOI] [PubMed] [Google Scholar]
- 76.Kraft L., Serpell L.C., Atack J.R. A biophysical approach to the identification of novel ApoE chemical probes. Biomolecules. 2019;9(2):48. doi: 10.3390/biom9020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu Y., Miyakawa T., Nosaki S., Nakamura A., Lyu Y., Nakamura H., Ohto U., Ishida H., Shimizu T., Asami T. Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga. Nat. Commun. 2018;9(1):3947. doi: 10.1038/s41467-018-06452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Montanier C., Flint J.E., Bolam D.N., Xie H., Liu Z., Rogowski A., Weiner D.P., Ratnaparkhe S., Nurizzo D., Roberts S.M. Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules. J. Biol. Chem. 2010;285(41):31742–31754. doi: 10.1074/jbc.M110.142133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He L., Wang K.-N., Zheng Y., Cao J.-J., Zhang M.-F., Tan C.-P., Ji L.-N., Mao Z.-W. Cyclometalated iridium (iii) complexes induce mitochondria-derived paraptotic cell death and inhibit tumor growth in vivo. Dalton Trans. 2018;47(20):6942–6953. doi: 10.1039/c8dt00783g. [DOI] [PubMed] [Google Scholar]
- 80.Jeong Y., Hoang N.T., Lovejoy A., Stehr H., Newman A.M., Gentles A.J., Kong W., Truong D., Martin S., Chaudhuri A., Heiser D., Zhou L., Say C., Carter J.N., Hiniker S.M., Loo B.W., Jr., West R.B., Beachy P., Alizadeh A.A., Diehn M. Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiation resistance. Canc. Discov. 2017;7(1):86–101. doi: 10.1158/2159-8290.CD-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li D., Liu Q., Lu X., Li Z., Wang C., Leung C.-H., Wang Y., Peng C., Lin L.J.A. Vol. 11. 2019. p. 11084. (α-Mangostin Remodels Visceral Adipose Tissue Inflammation to Ameliorate Age-Related Metabolic Disorders in Mice). 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Siddiqui A.J., Bhardwaj J., Goyal M., Prakash K., Adnan M., Alreshidi M.M., Patel M., Soni A., Redman W. Immune responses in liver and spleen against Plasmodium yoelii pre-erythrocytic stages in Swiss mice model. J. Adv. Res. 2020;24:29–41. doi: 10.1016/j.jare.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li G., Boyle J.W., Ko C.-N., Zeng W., Wong V.K.W., Wan J.-B., Chan P.W.H., Ma D.-L., Leung C.-H. Aurone derivatives as Vps34 inhibitors that modulate autophagy. Acta Pharm. Sin. B. 2019;9(3):537–544. doi: 10.1016/j.apsb.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li G., Henry S.A., Liu H., Kang T.S., Nao S.C., Zhao Y., Wu C., Jin J., Zhang J.T., Leung C.H., Wai Hong Chan P., Ma D.L. A robust photoluminescence screening assay identifies uracil-DNA glycosylase inhibitors against prostate cancer. Chem. Sci. 2020;11(7):1750–1760. doi: 10.1039/c9sc05623h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhuang C., Wu Z., Xing C., Miao Z. Small molecules inhibiting Keap1–Nrf2 protein–protein interactions: a novel approach to activate Nrf2 function. MedChemComm. 2017;8(2):286–294. doi: 10.1039/c6md00500d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.He L., He T., Farrar S., Ji L., Liu T., Ma X. Antioxidants maintain cellular redox homeostasis by elimination of reactive oxygen species. Cell. Physiol. Biochem. 2017;44(2):532–553. doi: 10.1159/000485089. [DOI] [PubMed] [Google Scholar]
- 87.Kageyama S., Sou Y.-s., Uemura T., Kametaka S., Saito T., Ishimura R., Kouno T., Bedford L., Mayer R.J., Lee M.-S. Proteasome dysfunction activates autophagy and the Keap1-Nrf2 pathway. J. Biol. Chem. 2014;289(36):24944–24955. doi: 10.1074/jbc.M114.580357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Devling T.W., Lindsay C.D., McLellan L.I., McMahon M., Hayes J.D. Utility of siRNA against Keap1 as a strategy to stimulate a cancer chemopreventive phenotype. Proc. Natl. Acad. Sci. U. S. A. 2005;102(20):7280–7285. doi: 10.1073/pnas.0501475102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gaikwad A., Long D.J., Stringer J.L., Jaiswal A.K. In Vivo Role of NAD (P) H: quinone oxidoreductase 1 (NQO1) in the regulation of intracellular redox state and accumulation of abdominal adipose tissue. J. Biol. Chem. 2001;276(25):22559–22564. doi: 10.1074/jbc.M101053200. [DOI] [PubMed] [Google Scholar]
- 90.Satoh T., McKercher S.R., Lipton S.A. Nrf2/ARE-mediated antioxidant actions of pro-electrophilic drugs. Free Radic. Biol. Med. 2013;65:645–657. doi: 10.1016/j.freeradbiomed.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.