Abstract
Metallothioneins (MTs) are short, cysteine-rich proteins for heavy metal homeostasis and detoxification; they bind a variety of heavy metals and also act as radical scavengers. Transcription of mammalian MT genes is activated by heavy metal load via the metal-responsive transcription factor 1 (MTF-1), an essential zinc finger protein whose elimination in mice leads to embryonic lethality due to liver decay. Here we characterize the Drosophila homolog of vertebrate MTF-1 (dMTF-1), a 791-amino-acid protein which is most similar to its mammalian counterpart in the DNA-binding zinc finger region. Like mammalian MTF-1, dMTF-1 binds to conserved metal-responsive promoter elements (MREs) and requires zinc for DNA binding, yet some aspects of heavy metal regulation have also been subject to divergent evolution between Drosophila and mammals. dMTF-1, unlike mammalian MTF-1, is resistant to low pH (6 to 6.5). Furthermore, mammalian MT genes are activated best by zinc and cadmium, whereas in Drosophila cells, cadmium and copper are more potent inducers than zinc. The latter species difference is most likely due to aspects of heavy metal metabolism other than MTF-1, since in transfected mammalian cells, dMTF-1 responds to zinc like mammalian MTF-1. Heavy metal induction of both Drosophila MTs is abolished by double-stranded RNA interference: small amounts of cotransfected double-stranded RNA of dMTF-1 but not of unrelated control RNA inhibit the response to both the endogenous dMTF-1 and transfected dMTF-1. These data underline an important role for dMTF-1 in MT gene regulation and thus heavy metal homeostasis.
Heavy metals, both essential and nonessential ones, pose a threat to every organism, and their concentration has to be carefully controlled. For example, copper, zinc, and cobalt have to be enriched if scarce but removed if abundant, while nonessential heavy metals, such as cadmium, have to be detoxified. Most species contain small, cysteine-rich proteins, called metallothioneins (MTs), to bind heavy metals for redistribution and/or detoxification (18, 28, 29, 52). In addition to their role in heavy metal homeostasis, MTs have been found to act as potent radical scavengers, thus contributing to antioxidant defense and cellular redox balance. Accordingly, transcription of the genes for MTs is induced not only by heavy metal load but also by a variety of other stress conditions.
Vertebrates contain four types of MTs, all about 60 to 70 amino acids long, of which MT-I and MT-II are stress induced and expressed in all organs, while MT-III and MT-IV are cell type specific and respond only moderately to heavy metal load (41, 55). The stress-induced MT-I and MT-II genes contain in their promoters multiple copies of so-called metal-responsive element (MRE) sequence motifs with the core consensus sequence TGCRCNC (50, 54). The transcription factor that binds to these MREs and confers metal inducibility was cloned in our laboratory and termed MTF-1 (for MRE-binding transcription factor 1 or metal-responsive transcription factor 1) (25, 43, 57). MTF-1, a protein with six characteristic zinc fingers of the C2H2 type, is essential for heavy metal response and for embryonic development, since homozygous knockout mouse embryos die on embryonic days 13 to 14 due to liver decay (23). MTF-1, which also contributes to the activation of genes other than MTs, in resting cells localizes to the cytoplasm and enters the nucleus under stress conditions (47, 53). Zinc induction works by direct metal binding to the MTF-1 zinc fingers (7, 11, 15, 25), but the exact signaling pathways for transcriptional activation remain to be established. Even though other metals, such as cadmium and copper, also activate transcription via MTF-1, activation must be indirect since these metals cannot replace zinc in zinc finger binding (6, 25; B. Zhang and W. Schaffner, unpublished). MTF-1 also mediates response to oxidative stress and hypoxia (16, 23, 38, 59). Recently, researchers isolated the MTF-1 gene from another vertebrate, the Japanese puffer fish Fugu rubripes. This MTF-1 turned out to be conserved to the ones of mouse and human, most notably in the DNA-binding zinc finger domain (3; see also reference 17). In order to gain more insights into the mechanisms of cellular defense against heavy metal and other stress, we decided to have a closer look at heavy metal homeostasis in Drosophila melanogaster, where a rapid genetic dissection is feasible. While transcriptional regulation of heavy metal homeostasis in yeast and the worm Caenorhabditis elegans differs from the one in vertebrates, the system in Drosophila promises to yield evolutionary insights of great relevance also to mammals.
In Drosophila, the two MT genes characterized to date, designated Mto (or MtnB) and Mtn (or MtnA), encode proteins of 43 and 40 amino acids, respectively. Mto, transcribed from a TATA-less promoter, is primarily active during embryogenesis, while Mtn, characteristic for late embryos, larvae, and adult flies, is strongly expressed in the gut, Malpighian tubules, and fat body and also in hemocytes (8, 9, 19, 35, 36, 37, 51). Even though the overall protein sequences of both Drosophila MTs deviate considerably from the ones of mammals, their promoters contain sequence motifs corresponding to mammalian MREs (20, 51). In support of this notion, a promoter fragment from Mtn containing two MREs was responsive to zinc after transfection into cultured hamster cells (39). This indicated to us that, if not the MTs themselves, at least the mechanism of their induction should be conserved, and prompted a search for a Drosophila homolog of the mammalian MTF-1. A Drosophila cDNA encoding a protein reminiscent of vertebrate MTF-1 was found in the expressed sequence tag database. Isolation of cDNAs and of the genomic locus, sequence comparisons, and functional tests reveal that Drosophila harbors, some differences in structure and properties notwithstanding, a functional homolog of the vertebrate stress regulator MTF-1. Drosophila MTF-1 (dMTF-1) can activate transcription from the promoters of MTs, Mtn and Mto, and in cultured Drosophila cells confers particularly strong transcriptional responses to copper and cadmium.
MATERIALS AND METHODS
Isolation of the phages containing the dMTF-1 gene.
A Drosophila λ phage genomic library was probed with an EST cDNA (BDGP clone identification, SD03560) containing a putative MTF-1 sequence labeled by random hexamer priming using [α-32P]dCTP. Filters were hybridized according to the method described by Church and Gilbert (12). Forty positive clones were obtained, and eight of them were chosen for rescreening and further analysis. Subsequently, all eight positive clones were mapped by standard restriction enzyme digestion and Southern blot analysis (46). The overlapping positive fragments were subcloned into pBluescript SK(−) (Stratagene) and sequenced.
cDNA library screening and isolation of cDNA for dMTF-1.
dMTF-1 cDNA was isolated from a cDNA library derived from 8- to 12-h-old Drosophila embryos (22), using the same probe as for the screening of the genomic library. To determine the 5′ end of dMTF-1 transcripts, rapid amplification of cDNA ends (RACE) was performed with RNA from 0- to 20-h-old Drosophila embryos using specific primers for dMTF-1 (SMART RACE kit; Clontech).
Poly(A) RNA isolation and Northern blot.
Poly(A) RNA was isolated with oligo(dT) cellulose, and Northern blot analyses were performed according to the method described by Sambrook et al. (46), using approximately 5 μg of poly(A) RNA per slot.
Generation of OVEC reporter constructs and expression vectors.
The reporter and reference plasmids used in the transient transfection assays were generated based on the OVEC-1 reporter system, which is based on quantitative S1 nuclease mapping of transcripts (58). The OVEC reporter constructs containing mouse MT-I (mMT-I) promoter or four copies of a mouse MRE (4× mMREd) used in this study are described by Radtke et al. (43). The OVEC reporter genes containing Drosophila MT Mtn promoter or Mto promoter were generated by PCR from the promoter of Mtn (−373 to −39 relative to transcription start site) or of Mto (−169 to +11 relative to transcription start site) and were inserted into the SacI/SalI-digested OVEC vector. The synthetic “minipromoter” construct was generated by insertion of a synthesized oligonucleotide containing the four MREs found in Mto promoter in a tandem array (−160 to −143, −133 to −116, −71 to −54, and −12 to +5) into the OVEC vector as done before (57). The human MTF-1 (hMTF-1) expression vector driven by the cytomegalovirus (CMV) promoter (used in mammalian cells) is described by Heuchel et al. (25). The hMTF-1 (with vesicular stomatitis virus tagged at its C-terminal) expression vector used in Drosophila Schneider-2 (S2) cells was generated by subcloning hMTF-1 cDNA-vesicular stomatitis virus fragment into the pAc5* vector containing the Drosophila actin 5C promoter or the pT vector containing the Drosophila tubulin α1 promoter. After reconstruction of the full-length dMTF-1 open reading frame in pBluescript SK(−), the entire cDNA was subcloned into pcDNA I (Invitrogen) containing the CMV promoter or pAc5* and pT vectors and was used as the expression vectors for dMTF-1 in mammalian or Drosophila cells, respectively.
dsRNA preparation.
Double-stranded RNA (dsRNA) was prepared according to Kennerdell and Carthew (31). Briefly, the templates for in vitro transcription of dsRNA were generated by PCR using primers which are flanked by a T7 promoter sequence at the 5′ end. These primers specifically amplified a cDNA fragment of 512 bp corresponding to the N-terminal part of dMTF-1 (including the first 104 amino acids) and of 498 bp corresponding to part of the coding region of lacZ (amino acids 6 to 171), respectively. The dsRNAs were synthesized using T7 RNA polymerase (New England BioLabs) followed by DNase I digestion, phenol treatment, and ethanol precipitation. The quality and amount of the dsRNA were controlled by using agarose gel electrophoresis.
Cell culture.
Human embryonic kidney 293 cells, as well as dko7 cell line derived from mouse embryonic stem cells from MTF-1 knockout embryos (42) and KO1-9 cells derived from MTF-1−/− mouse embryo fibroblasts, were grown in Dulbecco modified Eagle medium (GIBCO) supplemented with 5% fetal calf serum (GIBCO) and 5% newborn calf serum (GIBCO), 100 U of penicillin/ml, 100 U of streptomycin/ml, and 2 mM l-glutamine. Drosophila S2 cells (48) were grown at 24°C in Schneider's Drosophila medium (GIBCO) supplemented with 10% fetal calf serum (Eurobio) and 100 U of penicillin/ml and 100 U of streptomycin/ml. In the case of the transient transfection assay, 60- by 15-mm cell culture plates (Corning) containing 5 ml of medium were used.
Transient transfections and nuclease S1 mapping.
Transfections and nuclease S1 mapping of transcripts were performed as described previously (30, 43, 56). Various OVEC reporter constructs driven by either a mammalian or Drosophila promoter were transfected with or without respective expression plasmids using the calcium phosphate technique and together with dsRNA when necessary. For heavy metal induction, ZnCl2, CdCl2, or CuSO4 was added to the tissue culture medium to a final concentration as indicated in the figure legends and was incubated for 4 or 24 h before harvesting for mammalian or Drosophila S2 cells, respectively. S1 nuclease data were developed using a PhosphorImager (Molecular Dynamics) and quantified using ImageQuaNT software.
Preparation of nuclear extracts and EMSA.
The electrophoretic mobility shift assay (EMSA) was performed as described by Radtke et al. (43). Binding reactions were performed by incubating 25 fmol of end-labeled, 31-bp-long oligonucleotides containing the core MRE consensus sequence (MRE-s), TGCACAC, with nuclear extracts obtained according to the protocol of Schreiber et al. (49). For competition experiments, 5 pmol of unlabeled oligonucleotides was added to the reaction mixture prior to the addition of the extracts. The MRE-s oligonucleotide used for EMSA is as follows: 5′-CGAGGGAGCTCTGCACACGGCCCGAAAAGTG-3′ and 3′-TCGAGCTCCCTCGAGACGTGTGCCGGGCTTTTCACAGCT-5′.
Database searches and computer analyses of the sequences.
Database homology searches were carried out using the National Center for Biotechnology Information blast server (2). Splice sites and intron/exon boundaries were determined by alignment of the genomic sequence with the dMTF-1 cDNA. Sequence alignments were performed using the CLUSTALX program.
Nucleotide sequence accession number.
The GenBank accession numbers are AJ271817 for the dMTF-1 gene and AJ297844 for the cDNA.
RESULTS
Cloning and expression of dMTF-1.
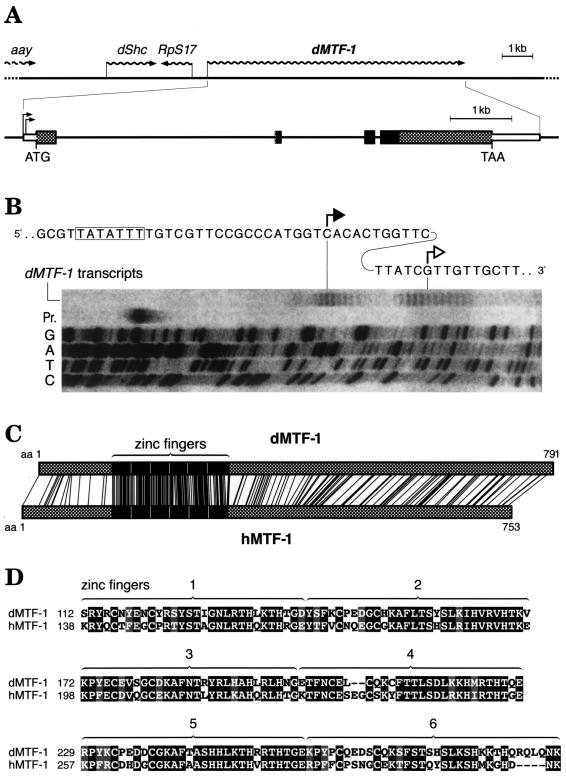
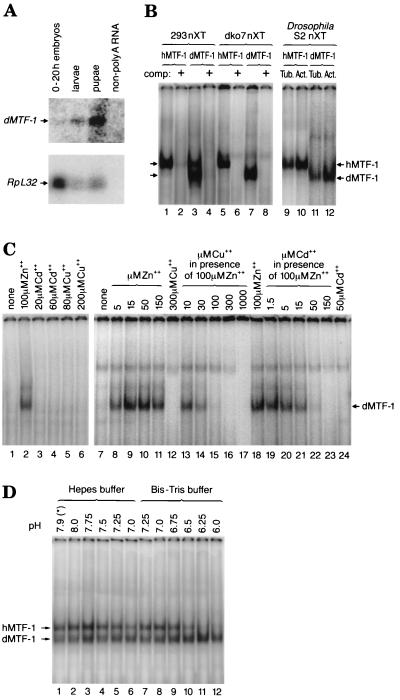
Searching through Drosophila databases, we found open reading frames related to MTF-1 on EST sequences derived from adult fly heads and from Schneider cells. However, the MTF-1 candidate sequence reconstructed from EST clones lacked the first zinc finger and also lacked any reasonable in-frame start codon. The complete N-terminal region could only be deduced after isolation and analysis of clones from an embryonic cDNA library provided by Fu and Noll (22) (GenBank accession no. AJ297844). The dMTF-1 locus was mapped to (3L) 67B using a digoxigenin-labeled cDNA hybridization probe in a salivary gland chromosome squash preparation (not shown). This chromosome region also contains a number of lethal and nonlethal mutations; however, none of them bears a phenotype that can easily be related to the loss of a putative regulator of heavy metal or other stress. Sequencing of a genomic insert in a lambda phage clone revealed the presence, besides dMTF-1, of three additional genes: the gene for ribosomal protein S17 (RpS17), in opposite orientation; the gene for SHC adapter protein (Shc), coding for a putative regulator of epidermal growth factor receptor signaling; and the 3′ end of the astray (aay) gene. This gene constellation was confirmed by the Drosophila genomic sequence (1). The intron-exon structure of dMTF-1 reveals three introns; the last one is at a position identical to that for the one in vertebrates (3, 4), while the other two intron positions are unique to Drosophila (Fig. 1A) (GenBank accession no. AJ271817). To determine more accurately the transcription start site, we performed an extension reaction (RACE) and also determined the 5′ end of transcripts independently by transcript mapping with nuclease S1 (Fig. 1B). These data are consistent and indicate a transcription start downstream of a putative TATA box and a 3.4-kb mRNA for dMTF-1. dMTF-1 cDNA encodes a protein of 791 amino acids (predicted molecular mass, 85 kDa), which is slightly larger than vertebrate MTF-1. A comparison of the protein sequences between the three vertebrate species and Drosophila reveals a particularly striking similarity in the region of all six zinc fingers (Fig. 1C and D). The similarity is 78% in the zinc finger region and 27% outside it, i.e., 39% in the total protein. In order to verify the data obtained by sequencing and transcript mapping (Fig. 1B), we also performed a Northern blot with samples of Drosophila poly(A) RNA (Fig. 2A), which indicates a steady increase of dMTF-1 mRNA in embryos, larvae, and pupae relative to the mRNA for ribosomal protein L32. In order to obtain some information on the expression of dMTF-1 in the adult, frozen tissue sections were subjected to in situ hybridization. Preliminary results indicate a strong expression in the fat body and in the gut (not shown), consistent with a major role in the control of MT (Mtn), which is expressed in the gut and also the fat body of the adult fly (8, 9, 19, 35).
FIG. 1.
Structure of the dMTF-1 locus. (A) Schematic view of the dMTF-1 locus and neighboring genes (GenBank accession no. AJ271817 for the gene, AJ297844 for cDNA). The dMTF-1 gene is preceded by the astray (aay), SHC adapter protein (dShc) (34), and ribosomal protein S17 (RpS17) genes. The dMTF-1 gene contains two large introns and one small intron of 3,571, 1,362, and 108 bp, respectively. The position of intron 3 coincides exactly with that of a vertebrate intron in MTF-1. The DNA-binding zinc finger region is indicated by black boxes, and the rest of the coding region is indicated by dotted boxes. Note that in the EST database and in the Drosophila Genome Annotation Database, only a cDNA that lacks exon 2 (zinc finger 1) is listed. (B) Mapping of the dMTF-1 gene transcription start sites. Cytoplasmic total RNA was isolated from Drosophila S2 cells after transfection with a dMTF-1 genomic clone. The RNA was digested with DNase I and then subjected to S1 nuclease mapping after hybridization to a 32P-labeled DNA oligonucleotide probe corresponding to the promoter region of dMTF-1 (electrophoresis is shown from left to right). The lane “dMTF-1 transcripts” shows two clusters of initiation sites downstream of a putative TATA box (framed), which coincide with the RACE extension product (solid arrowhead) and the 5′ end of the longest dMTF-1 cDNA clone (empty arrowhead), respectively. Pr., input oligonucleotide probe; G, A, T, and C, sequencing reaction of the dMTF-1 promoter region used as reference. (C) Similarity of dMTF-1 and hMTF-1 proteins. Identical amino acids (aa) are indicated by connecting lines. The zinc finger regions (black) are best conserved with 66% identical/78% similar amino acids. (D) Alignment of the amino acid sequences in the zinc finger region of dMTF-1 and hMTF-1. Zinc fingers 1 to 6 are indicated by numbered brackets above the corresponding sequences.
FIG. 2.
Expression of dMTF-1 in Drosophila. (A) Northern blot with poly(A) RNA of embryos, larvae, and pupae. Poly(A) RNA was isolated, and approximately 5 μg of the RNA was loaded, electrophoresed, blotted, and hybridized to a cDNA probe corresponding to dMTF-1 or ribosomal protein L32 (RpL32) according to standard procedures. The result shows a steady increase of dMTF-1 transcripts, following development of Drosophila from embryos (0 to 20 h old) to larvae to pupae, relative to those of RpL32. Non-poly(A) RNA in the last lane was used as a negative control. (B) EMSA of d- and hMTF-1. All the binding reactions were performed in the presence of 100 μM ZnCl2. Human embryonic kidney 293 cells were transfected with hMTF-1 (lanes 1 and 2) or dMTF-1 (lanes 3 and 4) cDNA expression plasmids, and 15 μg of nuclear protein extract (nXT) was used for bandshift. Lanes 1 and 3, bandshift with 32P-labeled MRE consensus oligonucleotide (MRE-s). Lanes 2 and 4, same conditions but also including a 200-fold excess of unlabeled MRE-s competitor oligonucleotide (comp +). The upper band in lane 3 is from endogenous hMTF-1. Mutant mouse cells (designated dko7) lacking MTF-1 due to targeted gene disruption were transfected with hMTF-1 (lanes 5 and 6) or with dMTF-1 (lanes 7 and 8) expression plasmids, and 12 μg of nuclear protein extract (nXT) was used for bandshift. Lanes 5 and 7, presence of labeled MRE-s probe; lanes 6 and 8, presence of 200-fold cold competitor. In the case of Drosophila S2 cells transfected with expression clones for hMTF-1 (lanes 9 and 10) or dMTF-1 (lanes 11 and 12), two different constitutive Drosophila promoters were used to drive MTF-1 expression, namely, D. tubulin α1 (Tub.) (lanes 9 and 11) and D. actin 5C (Act.) (lanes 10 and 12). Fifteen micrograms of nuclear protein extract (nXT) was used for bandshift. Note that hMTF-1 migrates unusually slowly compared to the similarly sized dMTF-1; this is attributed to the higher proline content of the former, a condition known to retard electrophoretic migration (e.g., reference 42). (C) DNA-binding activity of dMTF-1 is dependent on zinc but not on cadmium or copper. Mouse embryonic stem cells lacking endogenous MTF-1 (dko7 cells) (lanes 1 to 6) or mouse embryo fibroblasts derived from MTF-1 knockout embryos (KO1-9 cells) (lanes 7 to 24) were transfected with dMTF-1 expression plasmids. Nuclear extracts were prepared 40 h later, and 12 μg (lanes 1 to 6) or 8.5 μg (lanes 7 to 24) of protein was used for each EMSA. Different amounts of heavy metal ions (ZnCl2, CdCl2, or CuCl2) were included in the binding reactions as indicated in the figure. Arrow, position of MRE-s oligonucleotide bound by dMTF-1. (D) Species-specific difference of MTF-1 DNA binding at low pH. Eighteen micrograms of protein from dMTF-1-transfected human 293 cell nuclear extracts was used in an EMSA with binding buffers of different pH buffered by either HEPES (lanes 1 to 6) or bis-Tris (lanes 7 to 12). ZnCl2 (100 μM) was included in the binding reaction. Note that the reactions at pHs 7.25 and 7.0 were performed with each of the two binding buffers, to account for possible differences in buffer properties. The lane marked by an asterisk is with our standard EMSA binding buffer, i.e., pH 7.9 buffered by HEPES.
dMTF-1 activates MT promoters in vivo.
To assess the biological properties of dMTF-1, we cloned the full-length cDNA under the control of either the D. tubulin α1 or D. actin 5C promoter. After transfection into cultured Drosophila S2 cells, nuclear extract preparation, and EMSA, a characteristic band migrating slightly faster than the one with hMTF-1 became apparent (Fig. 2B). dMTF-1 protein could also be readily identified after transfection into mammalian cells (Fig. 2B). As was found with mammalian MTF-1, the binding of dMTF-1 to a consensus MRE was competed by an MRE sequence but not by a GC box oligonucleotide that specifically binds Sp1 factor (Fig. 2B and data not shown). dMTF-1, like mammalian MTF-1, depends on zinc for DNA binding in vitro, and other heavy metals like copper and cadmium cannot replace it but rather interfere with DNA binding (Fig. 2C). Nevertheless, there is also a difference in DNA-binding activity between h- and dMTF-1: when the reaction was performed at different pH values between 6.0 and 8.0, dMTF-1 showed a greater tolerance than did hMTF-1 towards low pH (6.0 to 6.5), perhaps reflecting the low optimum pH (6.5) of Drosophila cells (Fig. 2D). We also found that hMTF-1 is not damaged at low pH (6.0 at room temperature for 1 h), since binding could be fully restored upon shift to high pH (not shown).
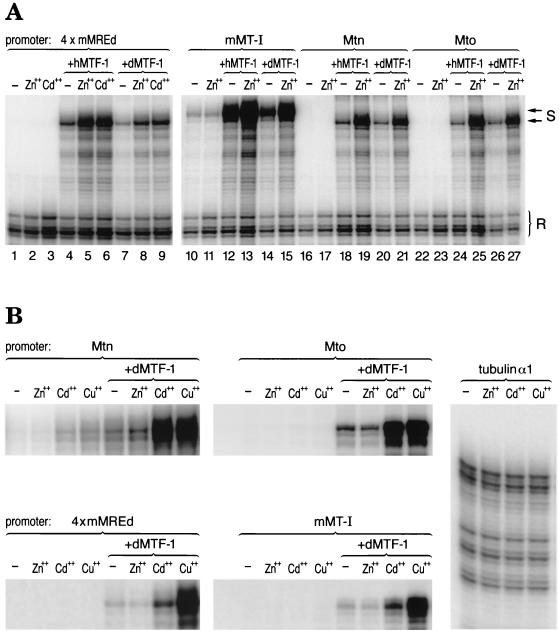
To test for biological activity of dMTF-1, it was first compared side by side with hMTF-1 in a transient transfection assay in a mammalian cell line which was derived from mouse embryonic stem cells lacking endogenous MTF-1 (dko7 cells) (Fig. 3A). In that comparison dMTF-1 showed a very similar, though somewhat less efficient, response to zinc and cadmium induction than did hMTF-1. As a control, the viral CMV promoter was not affected by heavy metal treatment and/or expression of extra MTF-1. Similar results were also obtained with dMTF-1 when transfected into another cell line derived from MTF-1−/− mouse embryo fibroblasts (designated KO 1-9 cells) (data not shown). For a test in fly cells, dMTF-1 cDNA driven by D. actin 5C promoter was transfected, together with reporter genes, into S2 cells. As shown in Fig. 3B and 4, dMTF-1 is able to confer very strong transcriptional activity to both Drosophila MT promoters (Mto and Mtn) in response to both copper and cadmium but hardly, if at all, to zinc even at a 500 μM concentration. A clear response of the Mtn promoter was achieved, however, when the cells were exposed to 2 mM zinc (Fig. 6A). Unfortunately, dMTF-1 and hMTF-1 could not be studied side by side in Drosophila, since transfected hMTF-1 was transcriptionally inactive in S2 cells (not shown; see also Discussion). As a control, the D. tubulin α1 promoter was neither affected by heavy metal treatment nor by expression of transfected dMTF-1 (Fig. 3B and data not shown).
FIG. 3.
Transcriptional activity of dMTF-1 in mammalian and Drosophila cells. (A) Comparison between the activity of hMTF-1 and dMTF-1 on both mammalian (mMT-I, 4× mMREd) and Drosophila (Mtn, Mto) MT promoters in a mammalian cell line lacking endogenous MTF-1 (dko7). Reporter gene constructs containing MT promoters were transfected into dko7 cells together with a reference gene construct driven by the CMV promoter, with or without cotransfection of an expression vector for hMTF-1 or dMTF-1. Cytoplasmic total RNA was extracted after metal induction for 4 h and was subjected to nuclease S1 mapping. With the mMT-I promoter (lanes 10 to 15) and the synthetic promoter 4× mMREd (lanes 1 to 9), dMTF-1 conferred metal-inducible transcription, though not as efficiently as hMTF-1 (compare lanes 12 and 14 and 4 and 7 for basal activation; lanes 13 and 15 and 5 and 8 for Zn2+ response; and lanes 6 and 9 for Cd2+ response). The Drosophila MT promoters (Mtn and Mto) responded equally well to both hMTF-1 and dMTF-1 in the same experiment. (In the case of the Mtn promoter, compare lanes 18 and 20 for basal activation and lanes 19 and 21 for Zn2+ induction. In the case of the Mto promoter, compare lanes 24 and 26 for basal activity and lanes 25 and 27 for Zn2+ induction.) Note that in all the cases, the CMV promoter was not affected by either cotransfection of MTF-1 (hMTF-1 or dMTF-1) or metal induction. Zn2+ induction, 100 μM ZnCl2; Cd2+ induction, 50 μM CdCl2; S, reporter gene signal (MT promoters); R, reference gene signal as an internal control (CMV promoter); −, control. (B) Drosophila Mto and Mtn promoters as well as mammalian MT (mMT-I, 4× mMREd) promoters are active in Drosophila S2 cells in response to heavy metals and dMTF-1. OVEC reporter gene constructs driven by the corresponding testing promoters were transfected into S2 cells; where indicated, a dMTF-1 expression vector under the control of the D. actin 5C promoter was cotransfected. Cytoplasmic total RNA was isolated after heavy metal induction for 24 h, and 160 μg of the RNA was subjected to nuclease S1 mapping. As a control, heavy metal treatment did not have any effect on the D. tubulin α1 promoter. Zn2+ induction, 100 μM ZnCl2 for mMT-I and Mto; 200 μM for 4× mMREd; 500 μM for Mtn and tubulin α1. Cd2+ induction, 20 μM CdCl2. Cu2+ induction, 500 μM CuSO4. −, control.
FIG. 4.
Induction of Drosophila MT Mto and Mtn promoters by dMTF-1 in response to different heavy metal concentrations. Reporter gene constructs driven by the promoters of Drosophila MT genes (Mto and Mtn) were transfected into Drosophila S2 cells. Induction by cadmium or copper was tested with different heavy metal concentrations for 24 h, with and without cotransfected dMTF-1 expression vector. To determine the activity of the Mto promoter, 100 μg of cytoplasmic RNA was used except for lanes 13 (50 μg), 19 (25 μg), and 23 (25 μg), where a fraction of cells was killed by high metal concentration. For the Mtn promoter, 160 μg of RNA was used except for lanes 6 (150 μg), 12 (100 μg), 13 (100 μg), 19 (40 μg), and 23 (40 μg) due to the same reason as above. Shown above the autoradiograms is the quantification, in which Mto and Mtn are represented and signals are normalized for total RNA content. Note that the TATA-less Mto promoter (top row of gel) has weak activity in S2 cells and even with cotransfected dMTF-1 expression vector has low basal activity; upon metal load, its activity is similar to that of the Mtn promoter.
FIG. 6.
Inhibition of MT Mtn and Mto transcription by RNAi specific for dMTF-1. (A) dsRNA of dMTF-1 inhibits heavy metal-induced transcription of the Mtn reporter gene. The Mtn promoter-reporter gene construct was transfected into Drosophila S2 cells, and various amounts (ranging from 0.3 to 2.7 μg per 5 ml of culture medium) of dsRNA of either dMTF-1 or lacZ were included. Cytoplasmic RNA was isolated after 24 h of heavy metal induction (0.5 to 2 mM ZnCl2 as indicated, 60 μM CdCl2, or 500 μM CuSO4) and the reporter gene transcripts were quantified by S1 nuclease mapping. Lanes 1 and 13, basal expression level of transfected Mtn reporter; lanes 2 to 4, 6 to 8, and 10 to 12, induction by zinc; lanes 14 to 20, induction by cadmium; lanes 21 to 27, induction by copper. LacZ dsRNA was included in lanes 5 to 8, 15 to 17, and 22 to 24 (lanes 5 to 8, 1 μg; lanes 15 to 17 and 22 to 24, 0.3 to 2.7 μg as indicated). dMTF-1 dsRNA was added in the transfections shown in lanes 9 to 12, 18 to 20, and 25 to 27 (lanes 9 to 12, 1 μg; lanes 18 to 20 and 25 to 27, 0.3 to 2.7 μg as indicated). −, control. F, free probe. (B) Inhibition of heavy metal (Cd2+ or Cu2+) induction in the presence of cotransfected (cotransf.) dMTF-1 by dsRNA of dMTF-1. The Mtn or Mto reporter construct was introduced into S2 cells together with a dMTF-1 expression vector, and results very similar to those for Mtn reporter transfection alone (described for panel A) were obtained. Lanes 1 and 10, basal transcription of the Mtn and Mto reporters, respectively. Lanes 2, 5, 8, 11, 14, and 17, cadmium induction (60 μM CdCl2, 24 h); lanes 3, 6, 9, 12, 15, and 18, copper induction (500 μM CuSO4, 24 h). Lanes 4 to 6 and 7 to 9, cotransfection with 10 μg of lacZ dsRNA and dMTF-1 dsRNA, respectively; lanes 13 to 15 and 16 to 18, cotransfection with 5 μg of LacZ dsRNA and dMTF-1 dsRNA, respectively. −, control. (C) dsRNA of dMTF-1 inhibits endogenous MT gene (Mtn) transcription. Various amounts of dsRNA were transfected into S2 cells. Endogenous Mtn transcripts were detected by S1 mapping after 24 h of induction with 60 μM CdCl2 or 500 μM CuSO4. F, free S1 probe for detecting endogenous Drosophila Mtn transcripts. Lanes: 1, basal level of endogenous Mtn transcripts; 2 and 9, after Cd2+ and Cu2+ induction, respectively; 3 to 5 and 6 to 8, effects on cadmium induction of dsRNA corresponding to lacZ and dMTF-1, respectively; 10 to 12 and 13 to 15, effects on copper induction of dsRNA corresponding to lacZ and dMTF-1, respectively. Endogenous Mtn transcripts following Cd2+ or Cu2+ induction were reduced to 20 and 47%, respectively, of the control level in the presence of dMTF-1 dsRNA, whereas lacZ dsRNA was not inhibitory.
S2 cells contain only low amounts of dMTF-1 (not shown) unless transfected with a dMTF-1 expression vector. Accordingly, the TATA-less Mto promoter was hardly active in S2 cells but became strongly metal-inducible in the presence of transfected dMTF-1. Even the adult-type Mtn promoter, while responsive to heavy metals on its own, depended on extra transfected dMTF-1 for maximal activity (Fig. 3B and 4). Comparable results were obtained with lacZ as a reporter gene under the control of either Mto or Mtn promoter when transfected together with dMTF-1 expression vector (data not shown), which is consistent with previous findings that MT levels in Drosophila are primarily regulated at the level of transcription (9, 10, 35, 39) and suggests that dMTF-1 plays a critical role in heavy metal homeostasis.
dMTF-1 activates transcription via MRE sequence motifs.
To determine whether the four putative MREs of the embryonic Mto promoter were indeed responsive to heavy metals, we made a synthetic minipromoter which merely contained these MRE motifs in a tandem array (Fig. 5). The activity of this promoter shows that the Drosophila MREs alone are sufficient to confer heavy metal inducibility to a corresponding reporter gene.
FIG. 5.
Drosophila MRE-like motifs can confer heavy metal response. The promoter of the embryonic MT gene (Mto) of Drosophila contains four MRE-like motifs (51), which, however, were found by EMSA to bind relatively poorly to MTF-1 under our standard experimental conditions. (A) To evaluate their potential, a minipromoter was assembled into a OVEC reporter construct as shown (for details, see Materials and Methods) and was transfected into S2 cells with or without a dMTF-1 expression vector. a to d indicate four MRE-like sequence motifs. Numbering refers to nucleotide position relative to transcription start. (B) Cytoplasmic RNA was isolated after metal induction for 24 h, and 160 μg of the RNA was subjected to nuclease S1 mapping as shown. Zn2+ induction, 100 μM ZnCl2; Cd2+ induction, 20 μM CdCl2; Cu2+ induction, 500 μM CuSO4; F, free S1 oligonucleotide probe; S, correctly initiated reporter gene transcripts.−, absence of dMTF-1.
RNAi demonstrates dependence of MT promoters on dMTF-1.
To further analyze the role of dMTF-1 in the transcriptional regulation of MT genes, we also applied the technique of dsRNA-mediated interference (RNAi) (21, 24, 31). A 512-bp segment of the dMTF-1 cDNA was flanked on either side by a T7 phage promoter and transcribed in a cell-free reaction by T7 RNA polymerase. The dsRNA was purified by phenol extraction followed by ethanol precipitation, and various amounts of dsRNA were cotransfected with a constant amount of reporter gene. As seen in Fig. 6A, RNAi eliminated all transcripts from a reporter gene with an MT (Mtn) promoter that was driven by the Drosophila host cell's transcription factors, indicating that MT transcription depends on dMTF-1. dsRNA interference was highly effective: even when dMTF-1 was overexpressed from a cotransfected gene, there was still a strong inhibition of reporter gene transcription (Fig. 6B). The transfected Mtn as well as the Mto promoter constructs were inhibited, suggesting that both MT genes rely on MTF-1 for transcriptional induction. Furthermore, the inhibition was specific: dsRNA of the lacZ gene blocked β-galactosidase activity (not shown) but failed to block reporter genes with Mtn or Mto promoters (Fig. 6A and B), and transcripts from the D. tubulin α1 gene promoter were not inhibited by either of the two dsRNAs (not shown). Quite contrary to the specific RNAi effect, transfection of an unrelated dsRNA even boosted the signals, perhaps due to an inhibition of cellular nucleases by excess substrate (Fig. 6A). In agreement with such an interpretation, nonspecific stimulation was also observed with cotransfected yeast tRNA (not shown). Finally, we found that dMTF-1 dsRNA inhibited not only the activation of a transfected MT promoter but also significantly reduced the transcripts from the endogenous MT Mtn gene (Fig. 6C). Under standard transfection conditions, not all cells can be transfected; thus, our finding that the transfected dsRNA reduced (but did not eliminate) the signal with cadmium and copper, while lacZ dsRNA was ineffective, is yet more evidence for the role of dMTF-1 in MT gene regulation.
DISCUSSION
In Drosophila, the two genes encoding MTs, Mto and Mtn, have distinct but partially overlapping expression patterns, with Mto being primarily expressed in early embryogenesis and Mtn being expressed in late embryogenesis/adulthood. In addition, Mtn is expressed in hemocytes, possibly to regulate copper supply to hemocyanin (8). It was postulated that Drosophila Mto is important for copper homeostasis during embryogenesis, while Mtn, in particular due to its very strong expression in the gut, Malpighian tubules, and fat body, is thought to balance the toxic effects of copper and other metals, such as cadmium and mercury. Thus, Mtn seems to play a role that in the snail is delegated to two different MTs, one active in hemocytes, the other one in the gut (14).
Here we have isolated a cDNA and dMTF-1, the corresponding gene from Drosophila encoding a transcription factor related to vertebrate MTF-1. The expression pattern of dMTF-1 is compatible with a role in MT gene activation/heavy metal detoxification and homeostasis. Furthermore, in transfection experiments both in mammalian cells and Drosophila S2 cells, dMTF-1 is able to confer strong cadmium induction to MT promoters of both Drosophila and mammalian origins. As in mammals, metal induction seems to involve characteristic MRE sequence motifs. In support of this notion, the promoters of both Drosophila MT genes, Mto and Mtn, contain multiple sequence motifs resembling mammalian MREs. Their role in metal response was further corroborated by a synthetic minipromoter where the four MREs from the Mto gene were juxtaposed in a tandem array and conferred strong heavy metal response to the reporter gene (Fig. 5). In spite of some striking similarities, it should also be pointed out that significant differences exist between dMTF-1 and vertebrate MTF-1. First of all, the strongly conserved zinc finger domain notwithstanding, there is limited, albeit significant, protein sequence similarity between vertebrate MTF-1 and dMTF-1. Specifically, the three hallmark activation domains of mammalian MTF-1, namely, acidic, proline-rich, and serine/threonine-rich, do not have obvious counterparts in Drosophila. The functional equivalents of one or all of them remain to be identified. Secondly, dMTF-1 is quite forgiving towards low pH (6.0 to 6.5) while mammalian MTF-1 loses its DNA-binding capacity under these conditions (Fig. 2D). This property may explain why mammalian MTF-1 in our hands was inactive in transfected Drosophila Schneider cells, which are grown at pH 6.5 (not shown). Conversely, dMTF-1 performed well in mammalian cells grown in Dulbecco modified Eagle medium at pH 7.4 (Fig. 3A). A third difference may concern the tissue distribution: while mammalian MTF-1 is expressed in all tissues analyzed so far (4), dMTF-1 is expressed in a few tissues, notably gut and fat body, although further experiments will have to probe into this issue. This is compatible with a major role in the activation of MT and possibly other toxicity/cell stress-related genes. Finally, there is a difference between mammals and Drosophila concerning heavy metal metabolism. In mammalian cells, MTF-1 mediates MT gene transcription primarily in response to zinc and cadmium and, to a lesser extent, to copper and nickel (25). We were surprised to find that dMTF-1 was unable to induce transcription in response to zinc at concentrations which readily induce transcription in mammalian cells. However, at 2 mM zinc, a concentration that is lethal to mammalian cells, dMTF-1 activated transcription from the Mtn promoter (Fig. 6A). This finding is in agreement with earlier heavy metal studies of the fly: of all metals tested for Mto and Mtn transcription, Cd2+, Cu2+, and Hg2+ were found to be more efficient than Zn2+, which was required at concentrations of at least 1 mM (7, 9, 19, 35, 51). The high concentrations of zinc (and copper) tolerated by Schneider cells may reflect inefficient uptake or, more likely, particularly efficient export of these two essential heavy metals (13, 40). In fact, a search of the Drosophila genome revealed six homologs of mammalian zinc transporters (1). Whatever the reason for the poor response to zinc at submillimolar concentrations, it cannot be attributed to a species difference between MTF-1 factors themselves, since dMTF-1, upon transfection into mouse cells, mediates zinc-responsive transcription like hMTF-1 (Fig. 3A). Further evidence for functional similarity is that dMTF-1, like mammalian MTF-1, requires elevated zinc concentrations for DNA binding in vitro, while copper and cadmium interfere with zinc rather than replacing it (Fig. 2C) (6, 11, 25). At first sight, cadmium and copper induction of a transcription factor that strictly requires zinc appears paradoxical. However, we have recently been able to induce MTF-1-dependent transcription in vitro by cadmium and copper. Activation was dependent on the presence of MT, which has a particularly high affinity for these two metals and upon binding to them releases zinc on behalf of MTF-1 (B. Zhang, O. Georgiev, and W. Schaffner, unpublished). Another question concerns the regulation of the dMTF-1 gene itself. In mammals, the MTF-1 promoter does not contain MREs and transcripts are marginally, if at all, elevated by heavy metal treatment (4, 25). Although we have not quantified MTF-1 transcripts in Drosophila, we note that the MTF-1 promoter, like the mammalian one, lacks MRE-type sequence motifs, at least within a 2-kb segment around the site of transcription initiation.
Perhaps the best evidence for an important role of dMTF-1 in MT gene regulation is provided by our studies with RNAi. Without RNAi and in the absence of any Drosophila mutation in the dMTF-1 gene, one might have argued that the observed effect in transfected cells was due to a nonphysiological stimulation by a heterologous zinc finger factor. Ectopic activation effects have been observed before, when related family members of mammalian transcription factors could replace each other in transient transfection/overexpression conditions. This scenario is ruled out by the finding that dMTF-1 dsRNA completely blocked the activation of the Mtn promoter without any cotransfected MTF-1 expression plasmid, i.e., under conditions that relied entirely on the host cell's transcription factors. Furthermore, unlike many other transcription factors, MTF-1 is a unique protein without related family members in Drosophila and, apparently, in mammals. Taken together, the RNAi experiments corroborated the important role of dMTF-1 for MT transcription and thus heavy metal homeostasis.
It certainly will be of interest to study the effect of loss of dMTF-1 in vivo, for example, by screening deletion mutants at the MTF-1 locus or by the newly introduced techniques of inheritable RNAi (32) or targeted gene disruption in Drosophila (45). As mentioned, a targeted disruption of MTF-1 in the mouse results in embryonic lethality due to liver decay on embryonic days 13 to 14. We note that disruption of either of two other stress-associated transcription factors, namely, c-Jun (26) and NF-κB/RelA (5), also results in embryonic death from liver decay at about the same stage of mouse embryogenesis. In this context it is of interest that AP-1 sites, which bind jun-fos heterodimers, are present in the promoters of both Drosophila MTs, pointing to a possible interconnection of MTF-1 and AP-1 in the cellular stress response of insects. Thanks to the availability of jun (27, 33) and fos (1, 44, 60) mutants and the power of Drosophila genetics, these and other aspects of cellular stress response are amenable to analysis.
ACKNOWLEDGMENTS
We are indebted to Denise Nellen for chromosome mapping; to Zhaobing Ding for initial studies on in situ hybridization of adult fly sections; to Peter Lichtlen for providing mouse KO1-9 cells; to Markus Noll, Erich Frei, and Werner Boll for providing high-quality Drosophila embryonic cDNA and genomic libraries and for valuable discussions; and to Fritz Ochsenbein for the preparation of figures. We also thank Konrad Basler for critical reading of the manuscript.
This work was supported by the Schweizerischer Nationalfonds and by the Kanton Zürich.
REFERENCES
- 1.Adams M D, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auf der Maur A, Belser T, Elgar G, Georgiev O, Schaffner W. Characterization of the transcription factor MTF-1 from the Japanese pufferfish (Fugu rubripes) reveals evolutionary conservation of heavy metal stress response. Biol Chem. 1999;380:175–185. doi: 10.1515/BC.1999.026. [DOI] [PubMed] [Google Scholar]
- 4.Auf der Maur A, Belser T, Wang Y, Günes Ç, Lichtlen P, Georgiev O, Schaffner W. Characterization of the mouse gene for the heavy metal-responsive transcription factor MTF-1. Cell Stress Chaperon. 2000;5:196–206. doi: 10.1379/1466-1268(2000)005<0196:cotmgf>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beg A A, Sha W C, Bronson R T, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the relA component of NF-κB. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 6.Bittel D, Dalton T, Samson S L, Gedamu L, Andrews G K. The DNA binding activity of metal response element-binding transcription factor-1 is activated in vivo and in vitro by zinc, but not by other transition metals. J Biol Chem. 1998;273:7127–7133. doi: 10.1074/jbc.273.12.7127. [DOI] [PubMed] [Google Scholar]
- 7.Bittel D C, Smirnova I V, Andrews G K. Functional heterogeneity in the zinc fingers of the metalloregulatory transcription factor, MTF-1. J Biol Chem. 2000;275:37194–37201. doi: 10.1074/jbc.M003863200. [DOI] [PubMed] [Google Scholar]
- 8.Bonneton F, Wegnez M. Developmental variability of metallothionein Mtn gene expression in the species of the Drosophila melanogaster subgroup. Dev Genet. 1995;16:253–263. doi: 10.1002/dvg.1020160305. [DOI] [PubMed] [Google Scholar]
- 9.Bonneton F, Théodore L, Silar P, Maroni G, Wegnez M. Response of Drosophila metallothionein promoters to metallic, heat shock and oxidative stresses. FEBS Lett. 1996;380:33–38. doi: 10.1016/0014-5793(95)01544-2. [DOI] [PubMed] [Google Scholar]
- 10.Bunch T A, Grinblat Y, Goldstein L S. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Chu M, Giedroc D P. MRE-Binding transcription factor-1: weak zinc-binding finger domains 5 and 6 modulate the structure, affinity, and specificity of the metal-response element complex. Biochemistry. 1999;38:12915–12925. doi: 10.1021/bi9913000. [DOI] [PubMed] [Google Scholar]
- 12.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox D W. Disorders of copper transport. Br Med Bull. 1999;55:544–555. doi: 10.1258/0007142991902619. [DOI] [PubMed] [Google Scholar]
- 14.Dallinger R, Berger B, Hunziker P, Kägi J H. Metallothionein in snail Cd and Cu metabolism. Nature. 1997;388:237–238. doi: 10.1038/40785. [DOI] [PubMed] [Google Scholar]
- 15.Dalton T P, Bittel D, Andrews G K. Reversible activation of mouse metal response element-binding transcription factor 1 DNA binding involves zinc interaction with the zinc finger domain. Mol Cell Biol. 1997;17:2781–2789. doi: 10.1128/mcb.17.5.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalton T P, Li Q, Bittel D, Liang L, Andrews G K. Oxidative stress activates metal-responsive transcription factor-1 binding activity. Occupancy in vivo of metal response elements in the metallothionein-I gene promoter. J Biol Chem. 1996;271:26233–26241. doi: 10.1074/jbc.271.42.26233. [DOI] [PubMed] [Google Scholar]
- 17.Dalton T P, Solis W A, Nebert D W, Carvan M J. Characterization of the MTF-1 transcription factor from zebrafish and trout cells. Biochem Physiol. 2000;126:325–335. doi: 10.1016/s0305-0491(00)00182-6. [DOI] [PubMed] [Google Scholar]
- 18.DeMoor J M, Koropatnick D J. Metals and cellular signaling in mammalian cells. Cell Mol Biol (Noisy-le-Grand) 2000;46:367–381. [PubMed] [Google Scholar]
- 19.Durliat M, Bonneton F, Boissonneau E, Andre M, Wegnez M. Expression of metallothionein genes during the post-embryonic development of Drosophila melanogaster. Biometals. 1995;8:339–351. doi: 10.1007/BF00141608. [DOI] [PubMed] [Google Scholar]
- 20.Echalier G. Drosophila cells in culture. New York, N.Y: Academic Press; 1997. [Google Scholar]
- 21.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 22.Fu W, Noll M. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 1997;11:2066–2078. doi: 10.1101/gad.11.16.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Günes Ç, Heuchel R, Georgiev O, Müller K-H, Lichtlen P, Blüthmann H, Marino S, Aguzzi A, Schaffner W. Embryonic lethality and liver degeneration in mice lacking the metal-responsive transcriptional activator MTF-1. EMBO J. 1998;17:2846–2854. doi: 10.1093/emboj/17.10.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond S M, Bernstein E, Beach D, Hannon G J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 25.Heuchel R, Radtke F, Georgiev O, Stark G, Aguet M, Schaffner W. The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression. EMBO J. 1994;13:2870–2875. doi: 10.1002/j.1460-2075.1994.tb06581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilberg F, Aguzzi A, Howells N, Wagner E F. c-Jun is essential for normal mouse development and hepatogenesis. Nature. 1993;365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- 27.Hou X S, Goldstein E S, Perrimon N. Drosophila jun relays the jun amino-terminal kinase signal transduction pathway to the decapentaplegic signal transduction pathway in regulating epithelial cell sheet movement. Genes Dev. 1997;11:1728–1737. doi: 10.1101/gad.11.13.1728. [DOI] [PubMed] [Google Scholar]
- 28.Kägi J H, Kojima Y. Chemistry and biochemistry of metallothionein. Exper Suppl. 1987;52:25–61. doi: 10.1007/978-3-0348-6784-9_3. [DOI] [PubMed] [Google Scholar]
- 29.Kägi J H. Overview of metallothionein. Methods Enzymol. 1991;205:613–626. doi: 10.1016/0076-6879(91)05145-l. [DOI] [PubMed] [Google Scholar]
- 30.Kemler I, Bucher E, Seipel K, Müller-Immerglück M M, Schaffner W. Promoters with the octamer DNA motif (ATGCAAAT) can be ubiquitous or cell type-specific depending on binding affinity of the octamer site and Oct-factor concentration. Nucleic Acids Res. 1991;19:237–242. doi: 10.1093/nar/19.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennerdell J R, Carthew R W. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 32.Kennerdell J R, Carthew R W. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 33.Kockel L, Zeitlinger J, Staszewski L M, Mlodzik M, Bohmann D. Jun in Drosophila development: redundant and nonredundant functions and regulation by two MAPK signal transduction pathways. Genes Dev. 1997;11:1748–1758. doi: 10.1101/gad.11.13.1748. [DOI] [PubMed] [Google Scholar]
- 34.Lai K M, Olivier J P, Gish G D, Henkemeyer M, McGlade J, Pawson T A. Drosophila shc gene product is implicated in signaling by the DER receptor tyrosine kinase. Mol Cell Biol. 1995;15:4810–4818. doi: 10.1128/mcb.15.9.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lastowski-Perry D, Otto E, Maroni G. Nucleotide sequence and expression of a Drosophila metallothionein. J Biol Chem. 1985;260:1527–1530. [PubMed] [Google Scholar]
- 36.Maroni G, Otto E, Lastowski-Perry D. Molecular and cytogenetic characterization of a metallothionein gene of Drosophila. Genetics. 1986;112:493–504. doi: 10.1093/genetics/112.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mokdad R, Debec A, Wegnez M. Metallothionein genes in Drosophila melanogaster constitute a dual system. Proc Natl Acad Sci USA. 1987;84:2658–2662. doi: 10.1073/pnas.84.9.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy B J, Andrews G K, Bittel D, Discher D J, McCue J, Green C J, Yanovsky M, Giaccia A, Sutherland R M, Laderoute K R, Webster K A. Activation of metallothionein gene expression by hypoxia involves metal response elements and metal transcription factor-1. Cancer Res. 1999;59:1315–1322. [PubMed] [Google Scholar]
- 39.Otto E, Allen J M, Young J E, Palmiter R D, Maroni G. A DNA segment controlling metal-regulated expression of the Drosophila melanogaster metallothionein gene Mtn. Mol Cell Biol. 1987;7:1710–1715. doi: 10.1128/mcb.7.5.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmiter R D, Findley S D. Cloning and functional characterization of a mammalian zinc transporter that confers resistance to zinc. EMBO J. 1995;14:639–649. doi: 10.1002/j.1460-2075.1995.tb07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmiter R D, Findley S D, Whitmore T E, Durnam D M. MT-III, a brain-specific member of the metallothionein gene family. Proc Natl Acad Sci USA. 1992;89:6333–6337. doi: 10.1073/pnas.89.14.6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radtke F, Georgiev O, Muller H P, Brugnera E, Schaffner W. Functional domains of the heavy metal-responsive transcription regulator MTF-1. Nucleic Acids Res. 1995;23:2277–2286. doi: 10.1093/nar/23.12.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Radtke F, Heuchel R, Georgiev O, Hergersberg M, Gariglio M, Dembic Z, Schaffner W. Cloned transcription factor MTF-1 activates the mouse metallothionein I promoter. EMBO J. 1993;12:1355–1362. doi: 10.1002/j.1460-2075.1993.tb05780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riesgo-Escovar J R, Hafen E. Common and distinct roles of DFos and DJun during Drosophila development. Science. 1997;278:669–672. doi: 10.1126/science.278.5338.669. [DOI] [PubMed] [Google Scholar]
- 45.Rong Y S, Golic K G. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Saydam, N., O. Georgiev, M. Y. Nakano, U. Greber, and W. Schaffner. Nucleo-cytoplasmic trafficking of metal-responsive transcription factor MTF-1 is regulated by diverse stress signals. J. Biol. Chem., in press. [DOI] [PubMed]
- 48.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- 49.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts,’ prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Searle P F, Stuart G W, Palmiter R D. Metal regulatory elements of the mouse metallothionein-I gene. Exper Suppl. 1987;52:407–414. doi: 10.1007/978-3-0348-6784-9_39. [DOI] [PubMed] [Google Scholar]
- 51.Silar P, Theodore L, Mokdad R, Erraiss N E, Cadic A, Wegnez M. Metallothionein Mto gene of Drosophila melanogaster: structure and regulation. J Mol Biol. 1990;215:217–224. doi: 10.1016/S0022-2836(05)80340-7. [DOI] [PubMed] [Google Scholar]
- 52.Simpkins C O. Metallothionein in human disease. Cell Mol Biol (Noisy-le-Grand) 2000;46:465–488. [PubMed] [Google Scholar]
- 53.Smirnova I V, Bittel D C, Ravindra R, Jiang H, Andrews G K. Zinc and cadmium can promote rapid nuclear translocation of metal response element-binding transcription factor-1. J Biol Chem. 2000;275:9377–9384. doi: 10.1074/jbc.275.13.9377. [DOI] [PubMed] [Google Scholar]
- 54.Stuart G W, Searle P F, Palmiter R D. Identification of multiple metal regulatory elements in mouse metallothionein-I promoter by assaying synthetic sequences. Nature. 1985;317:828–831. doi: 10.1038/317828a0. [DOI] [PubMed] [Google Scholar]
- 55.Uchida Y, Takio K, Titani K, Ihara Y, Tomonaga M. The growth inhibitory factor that is deficient in the Alzheimer's disease brain is a 68 amino acid metallothionein-like protein. Neuron. 1991;7:337–347. doi: 10.1016/0896-6273(91)90272-2. [DOI] [PubMed] [Google Scholar]
- 56.Weaver R F, Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5′ termini of 15 S beta-globin mRNA precursor and mature 10 S beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979;7:1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westin G, Schaffner W. A zinc-responsive factor interacts with a metal-regulated enhancer element (MRE) of the mouse metallothionein-I gene. EMBO J. 1988;7:3763–3770. doi: 10.1002/j.1460-2075.1988.tb03260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Westin G, Gerster T, Müller M M, Schaffner G, Schaffner W. OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 1987;15:6787–6798. doi: 10.1093/nar/15.17.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoo H Y, Chang M S, Rho H M. Heavy metal-mediated activation of the rat Cu/Zn superoxide dismutase gene via a metal-responsive element. Mol Gen Genet. 1999;262:310–313. doi: 10.1007/s004380051088. [DOI] [PubMed] [Google Scholar]
- 60.Zeitlinger J, Kockel L, Peverali F A, Jackson D B, Bohmann D. Defective dorsal closure and loss of epidermal decapentaplegic expression in Drosophila fos mutants. EMBO J. 1997;16:7393–7401. doi: 10.1093/emboj/16.24.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]