Summary
Endosymbiosis is a relationship between two organisms wherein one cell resides inside the other. This affiliation, when stable and beneficial for the ‘host’ cell, can result in massive genetic innovation with the foremost examples being the evolution of eukaryotic organelles, the mitochondria and plastids. Despite its critical evolutionary role, there is limited knowledge about how endosymbiosis is initially established and how host–endosymbiont biology is integrated. Here, we explore this issue, using as our model the rhizarian amoeba Paulinella, which represents an independent case of primary plastid origin that occurred c. 120 million yr ago. We propose the ‘chassis and engine’ model that provides a theoretical framework for understanding primary plastid endosymbiosis, potentially explaining why it is so rare.
Keywords: endosymbiotic gene transfer, genome reduction, organellogenesis, primary endosymbiosis, photosynthetic eukaryotes, Rhizaria
I. Introduction
Two cases of primary plastid endosymbiosis are known in which a nonphotosynthetic protist engulfed a cyanobacterial prey that became stabilized within the cytoplasm of the cell and evolved into a photosynthetic organelle. The first of these is ancient (c. 1.5 billion yr old) and involved the uptake of a β-cyanobacterium, giving rise to the canonical plastid in the putative single common ancestor of the Archaeplastida (Yoon et al., 2004), which includes all land plants, red, green, and glaucophyte algae and other more recently discovered protist lineages (Schöon et al., 2021). This canonical plastid has been transferred independently multiple times into other eukaryotic lineages via secondary and tertiary endosymbiosis (Fig. 1), giving rise to a diverse range of photosynthetic eukaryotes that, although polyphyletic, contain plastids that can be traced back to the original, ancient engulfment. The second plastid primary endosymbiosis occurred more recently (c. 120 million yr ago) in the rhizarian amoeba Paulinella. This event involved a heterotrophic ancestor of Paulinella and an α-cyanobacterial prey, with the latter evolving into a photosynthetic organelle termed the ‘chromatophore’ (Lauterborn et al., 1895; Melkonian & Mollenhauer, 2005; Lhee et al., 2021a). Given the incomplete and fragmented genomic evidence regarding early events in plastid evolution that is available from the Archaeplastida (Gabr et al., 2020), there is a need for new models from nature or the application of synthetic biology approaches to understand this process (e.g. Mehta et al., 2018). We discuss here how analyses of photosynthetic Paulinella have advanced our understanding of primary plastid endosymbiosis.
Fig. 1.
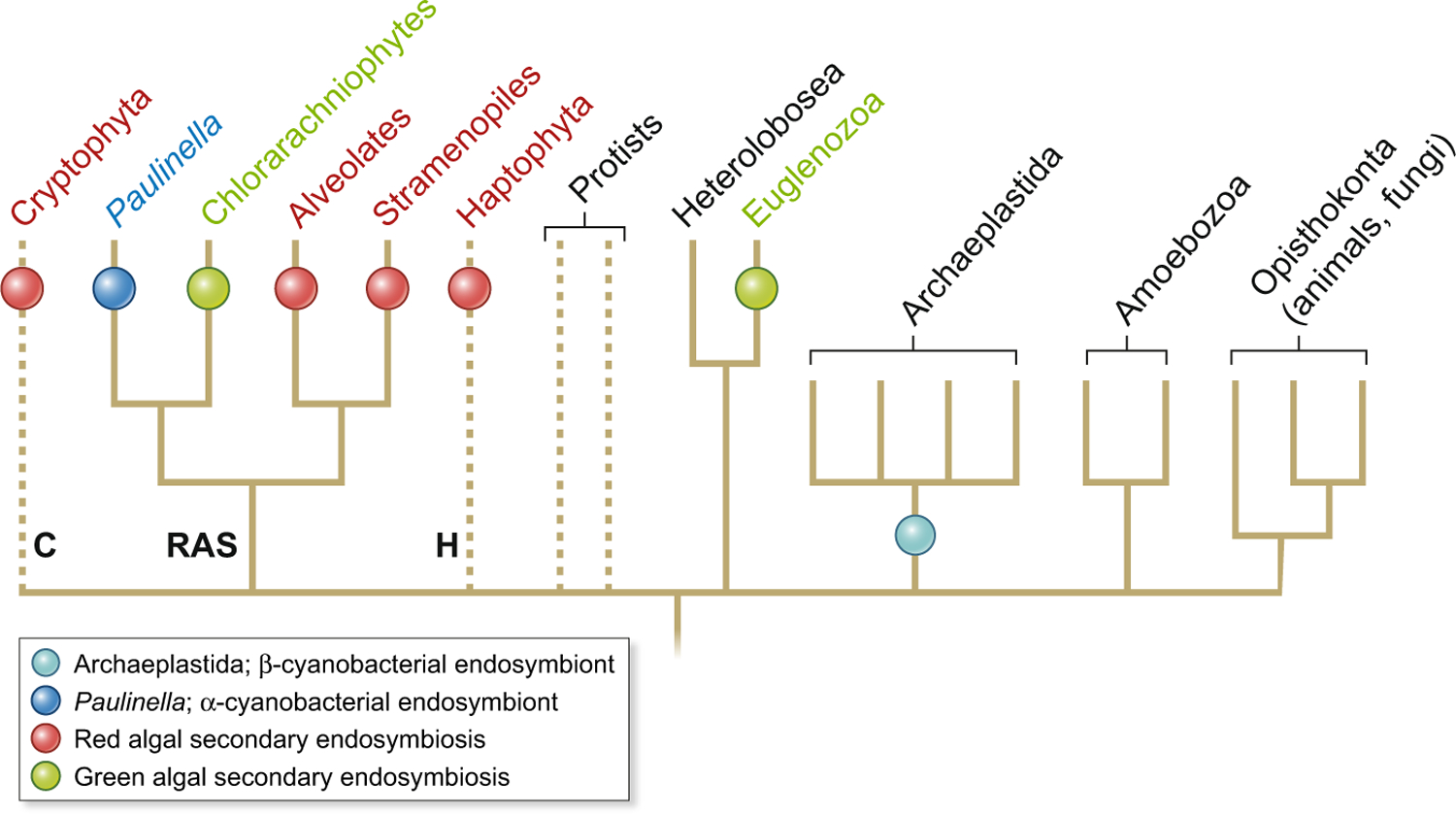
The history of plastid endosymbiosis in eukaryotes. This is a highly reduced tree that only shows the groups of interest (see also Ponce-Toledo et al., 2019). Primary plastid origin occurred in the ancestors of Archaeplastida and photosynthetic Paulinella (within Rhizaria). Green algal secondary endosymbiosis occurred independently in the chlorarachniophyte (Rhizaria) and Euglenozoa lineages. Red algal secondary endosymbiosis has occurred in the alveolates (e.g. dinoflagellates), stramenopiles, cryptophytes and haptophytes. These taxa are informally referred to as CRASH (i.e. cryptophytes, rhizarians, alveolates, stramenopiles and haptophytes) with phylogenetic evidence existing for the monophyly of SAR (stramenopiles, alveolates and rhizarians) taxa (Bhattacharya & Price, 2020; Fan et al., 2020). Broken lines indicate unclear phylogenetic affiliations that may impact the number of red algal endosymbiosis if some (or all, highly unlikely) of these lineages share a single event. There is a variety of plastid-lacking heterotrophic lineages, not shown here, such as ciliates, telonemids and katablepharids, that are sister to photosynthetic taxa in the CRASH. Multiple tertiary endosymbioses involving green algae, haptophytes and diatoms have occurred in the dinoflagellates that are not shown here (Gross et al., 2012).
II. Differing evolutionary trajectories of host and endosymbiont genomes
The available nuclear genomes of photosynthetic Paulinella lineages exhibit extensive divergence and uneven expansion with the genome sizes of sister species varying from 707 Mbp in P. micropora KR01 (hereafter, KR01; Lhee et al., 2021a) to 967 Mbp in P. micropora MYN1 (Matsuo et al., 2019), and c. 10 Gbp in the more distantly related P. chromatophora (Nowack et al., 2016). The chromatophore genome retains c. 35% of the ancestral cyanobacterial gene content (860–876 protein-coding genes; Lhee et al., 2019) with a size of c. 1 Mbp. In comparison, Synechococcus sp. WH5701, the cyanobacterial lineage most closely related to the chromatophore, has a c. 3 Mbp genome that contains 3346 protein-coding genes. In each chromatophore genome, > 90% of genes are shared among all strains (Lhee et al., 2019) and are under strong purifying selection (dN/dS ratios << 1) (Reyes-Prieto et al., 2010). In comparison, the canonical Archaeplastida-derived plastids have at most c. 250 genes (in some red algae) and as few as c. 7 genes (in holoparasites of legumes) (Vries & Archibald, 2018; Arias-Agudelo et al., 2019) with genome sizes varying between c. 10 kbp to > 1 Mbp. The chromatophore genome, while similar in size to the largest plastid genomes (Muñoz-Gómez et al., 2017), contains c. threefold more genes and retains most genes encoding proteins essential for photosynthetic electron transport, light harvesting and ATP synthesis (Nowack et al., 2008).
Mechanisms for endosymbiont integration with the host are primarily evolving from novel genetic elements
Despite an approximately two-thirds reduction in chromatophore genome size, only c. 50 endosymbiotic gene transfer (EGT) candidates have been found in the nuclear genome of Paulinella species (Nowack et al., 2016, Lhee et al., 2021a). By contrast, c. 600–1000 EGT-derived genes arose during the evolution of the Archaeplastida plastid (Nowack & Weber, 2018; Ponce-Toledo et al., 2019). The heterotrophic ancestor of Paulinella could have acquired genes via horizontal gene transfer (HGT) from its photosynthetic prey that allowed it to maintain cyanobacteria internally for increasingly longer periods of time (‘shopping bag’ model; Larkum et al., 2007) before lysis (Fig. 2a), potentially paving the way for plastid endosymbiosis.
Fig. 2.
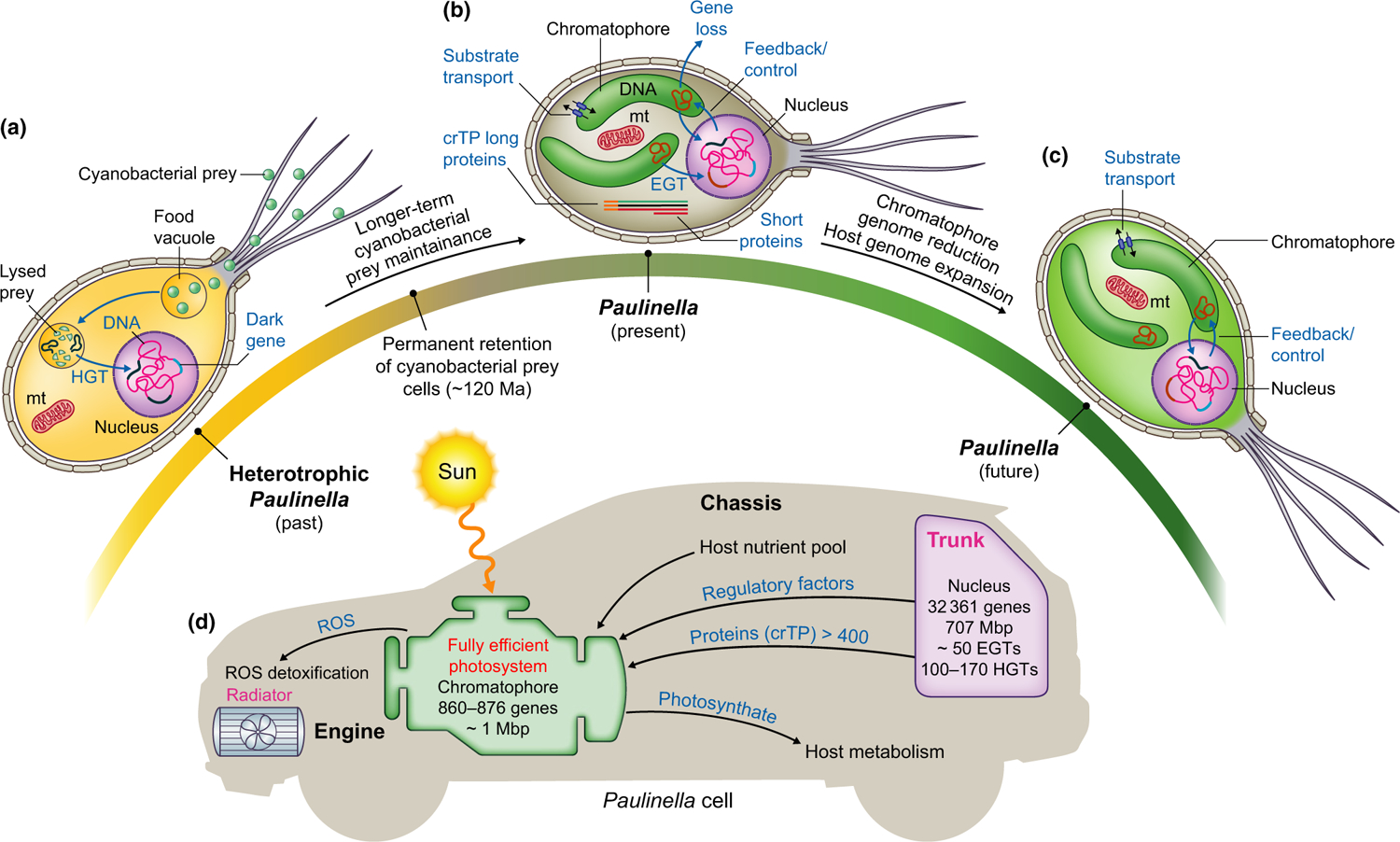
Putative timeline of plastid evolution in Paulinella encompassing both a historical and future perspective based on the chassis and engine model. (a) Heterotrophic Paulinella ingesting cyanobacterial cells likely using filose, feeding pseudopodia as a result of the presence of silica scales on the cell surface (Johnson et al., 1988). DNA from ingested bacterial cells can enter the nucleus and become integrated into the host genome via horizontal gene transfer (HGT). These foreign genes, as well as other novel ‘dark’ genes that evolved in this lineage, enabled the host to maintain the cyanobacteria in food vacuoles for increasingly longer periods of time until the association became permanent. (b) The status of extant photosynthetic Paulinella showing the two chromatophores and current evolutionary processes. (c) Putative future status of photosynthetic Paulinella. The chromatophore will encode only essential genes that cannot be transferred to the host genome because the encoded proteins either are problematic to transport into the chromatophore or require redox regulationwithin the organelle(e.g. co-locationfor redox regulation (CoRR hypothesis); Allen, 2017).The chromatophore may also lose its peptidoglycan layer, as observed in other lineages containing primary plastids (except Glaucophyta). Adaptations will probably increase the Paulinella growth rate and the ability to deal with high light, expanding its environmental niche. (d) The chassis and engine model of plastid endosymbiosis. The approximate size of the chromatophore genome and the number of genes it encodes across Paulinella species are shown inside the engine. Parts of the model where questions remain about the endosymbiosis in Paulinella are shown in red text (for more detail see Box 1). crTP, chromatophore targeting peptide; EGT, endosymbiotic gene transfer; mt, mitochondrion; ROS, reactive oxygen species.
We focus here on the evolution of Paulinella after its transition to permanent photoautotrophy, for which significant empirical results exist. These data suggest that in addition to numerous contributions from externally derived genes (i.e. c. 98 derived through HGT; Lhee et al., 2021a), there is strong selection for host-centered solutions to chromatophore maintenance and integration. The origin of novel genes and genetic elements plays a key role in endosymbiont maintenance, with > 51% of the predicted diurnally (day/night) rhythmic genes in KR01 lacking an annotation. In P. chromatophora, proteomic and in silico analyses identified 433 chromatophore-targeted proteins, of which 226 (52.19%) have unknown functions (Singer et al., 2017). These so-called ‘dark’ genes (of de novo origin and those shared more broadly but lacking annotation) are a major source of novelty in different lineages (Stephens et al., 2018). In comparison, among Archaeplastida, the GreenCut includes proteins that are encoded by the nuclear genomes of green algae, flowering and nonflowering plants, and can also occur in other algal groups, but are absent in heterotrophs (Grossman et al., 2019). Of the 597 GreenCut proteins, 189 are shared with cyanobacteria, whereas most of the remainder are lineage-specific (e.g. those present in green lineage organisms, in both the green lineage and red algae (PlantCut), and in the green lineage and diatoms (DiatomCut)). These lineage-specific GreenCut proteins are analogous to many of the Paulinella-specific dark genes that await study to uncover their roles in a novel context of photoautotrophy.
Paulinella has novel strategies for transporting proteins into chromatophores
Proteins encoded by nuclear genes essential to chromatophore function, such as those that have been transferred via EGT, acquired via HGT, or evolved de novo, must be targeted and transported across the chromatophore double membrane. Two classes of chromatophore-targeted proteins exist in Paulinella. The first comprises short (< 90 amino acid (aa)) proteins that enter the chromophore without the apparent need for a transit peptide, relying on the secretory pathway (Nowack & Grossman, 2012). The second are long proteins (> 268 aa) that contain a novel c. 200 aa N-terminal extension capable of functioning as a transit peptide for chromatophores (crTP), but that also enable protein targeting to plant chloroplasts (Singer et al., 2017). The efficiency of the chromatophore targeting systems, specifically the limitations imposed by the addition of the large crTP and the import of short proteins via a separate mechanism are unknown. Most solute transporters encoded on the chromatophore genome have been lost and, surprisingly, this has not been compensated by the retargeting of nuclear-encoded transporters. Instead, the amoeba appears to have evolved small, single transmembrane helix containing proteins that are chromatophore-targeted and may modulate membrane permeability (Oberleitner et al., 2020).
Inefficiencies of existing mechanisms for coping with light stress may impede chromatophore evolution
Excess light absorbed by photosynthetic organisms can cause extensive cellular damage through the generation of reactive oxygen species (ROS), which includes singlet oxygen, superoxides, hydrogen peroxide, hydroxyl radicals, and nitric and nitrous oxides (Khorobrykh et al., 2020). Accumulation of ROS can lead to inhibition of photosynthetic electron transport, denaturation of proteins, and peroxidation of lipids, which in turn can cripple various cellular functions and ultimately cause cell death. Under low or moderate light intensities (> 60 μmol photons m−2 s−1), Paulinella accumulates ROS and induces expression of genes encoding the ROS scavenger enzymes superoxide dismutase and glutathione-S-transferase, which are encoded on the chromatophore and nuclear genomes, respectively (Lhee et al., 2021b). These relatively low light conditions are sufficient to slow growth and cause cell bleaching, which suggests a limited ability of the amoeba to manage cellular redox and ROS generated in the chromatophore (Zhang et al., 2017). Transcriptional and physiological studies of Paulinella suggest inefficient integration of light-induced control of genes participating in photosynthesis and photoprotection. For instance, the induced expression of genes involved in photosynthesis in the light that are encoded on the chromatophore genome (core photosystem I and II (PSI and PSII) subunits psaA and psbA, respectively; phycobilisome linker polypeptide, cpcC) is weak when compared with induction in free-living cyanobacteria (Zhang et al., 2017; Tan et al., 2018). This diminished transcriptional regulation of chromatophore genes is also observed for plastid genes (Mettler et al., 2014). Despite their predominant localization to the chromatophore genome, some genes involved in photosynthesis and photoprotection are present on the nuclear genome, with others encoded on the genomes of both compartments. The genes encoding PSI subunit psaI and the carboxysome shell protein csoS4A are examples of the latter, with only one copy exhibiting a light intensity-dependent transcriptional response. In addition, some EGT-derived genes (e.g. PSI subunits psaE and psaK) in P. chromatophora are only present in the nuclear genome and are not light-responsive, in contrast to their differential expression in free-living cyanobacteria (Zhang et al., 2017; Tan et al., 2018). Differences in the regulation of chromatophore and nuclear genes might reflect an ongoing process of adaptation that will probably end with the loss of the organelle copy.
A highly duplicated gene family in the Paulinella nuclear genome encodes the chromatophore-derived high light-inducible (hli) genes (Zhang et al., 2017, Lhee et al., 2021a). In cyanobacteria, these genes are critical for growth under high light and stress conditions (He et al., 2001) and also regulate Chl biosynthesis and quenching of singlet oxygen (Komenda & Sobotka, 2016). The EGT-derived hli in photosynthetic Paulinella has undergone extensive expansion with 51 copies in KR01 and 64 in P. micropora MYN1 (Matsuo et al., 2019; Lhee et al., 2021a), compared with 13 copies in free-living Synechococcus sp. WH 5701. Hli gene family expansion also occurs in the high light ecotype of Prochlorococcus spp. (Bhaya et al., 2002). Growth of this Paulinella gene family may reflect selection to limit the generation of, or ameliorate the consequences of, photosynthetically derived ROS, which may lessen with increased metabolic integration of the host and endosymbiont.
III. The ‘chassis and engine’ model of endosymbiosis
Research on artificial endosymbiosis highlights the importance of several key aspects of biotic associations: compartmentalization, synchronized growth and cell division, processing of genetic and other types of information and, importantly, energy transduction and adaptability (Yewdall et al., 2018). In the case of Paulinella, these aspects have been accommodated with clear evidence of synchronized cell division of the chromatophore compartment and host cell (Hoogenraad, 1927; Melkonian & Mollenhauer, 2005), the finding that the ancestral pool of chromatophore-targeted long proteins are involved in genetic information processing (Lhee et al., 2021a), and the generation of energy in the chromatophore and its transfer to the amoeba (Sato et al., 2020), which is no longer capable of phagotrophic growth (Bhattacharya et al., 2012).
Based on our understanding of Paulinella, we suggest a simple analogy to explain plastid evolution that is broadly applicable and can guide understanding of the evolution of the Archaeplastida plastid. Here, the chromatophore is the ‘engine’ of a vehicle that must be supplied with ‘fuel’ and other resources to function (sunlight, H2O, proteins, and various nutrients including inorganic carbon). Excess heat generated by the engine is removed by the radiator (nonphotochemical quenching, photochemical quenching, ROS detoxification) and the speed is controlled (e.g. regulation of electron transport, reductant and ATP production, Calvin–Benson–Bassham cycle) by regulatory mechanisms associated with the ‘chassis’ (host cell) and through internal feedback controls (Fig. 2d). The power (photosynthate) produced by the engine generates motion and maintains other functions (e.g. metabolite and macromolecule biosynthesis), whereas some power is used to generate new engines and maintain existing ones.
Photosynthetic Paulinella appears to be in an intermediate stage of endosymbiosis (Fig. 2b). These species have evolved some key pathways needed for stable long-term maintenance of the endosymbiont such as the crTP and upregulation of nuclear hli genes under high light stress. However, there appear to be inefficiencies in fully harnessing the photosynthetic capacity of the chromatophore, probably explaining the extremely slow growth rate (5–7 d doubling time). The engine in photosynthetic Paulinella is, and has always been, capable of high performance, efficiently converting sunlight and H2O into energy, a process performed by oxygen-producing cyanobacteria for over 2 billion yr (Shih et al., 2017). By contrast, the chassis is not initially able to accommodate this powerful engine and is therefore forced to undergo significant remodeling. Paulinella may not efficiently transport resources (e.g. nutrients and inorganic carbon, proteins) to the engine and is still developing mechanisms to monitor and control engine speed, possibly through sensing energetic outputs (reductant, ATP, ROS) and needed inputs (light intensity and quality, nutrient concentrations). These sensing mechanisms need to couple with delivery of resources to the engine and modulate expression of nuclear-encoded proteins involved in photosynthesis and CO2 fixation, photoprotection and repair, and chromatophore replication. We speculate that because of the inefficiencies in using chromatophore products and controlling the ‘speed’ of this biological motor (which produces ROS and other damaging photoproducts), Paulinella is limited to low light environments. During the transitionary phase, the chassis is under selection to develop more efficient methods for dealing with the high-energy outputs of the engine and associated accumulation of toxic byproducts. Therefore, we postulate that the next major innovations in the evolution of photosynthetic Paulinella will not involve further reduction of the chromatophore genome into a more plastid-like structure, but rather will involve the development and optimization of novel or existing host systems to better accommodate the chromatophore in its current state. Once this process has occurred, continuation of chromatophore gene loss and additional EGT events, potentially resulting in a streamlined genome that more closely resembles the canonical Archaeplastida plastid, may occur (Fig. 2c). It is also conceivable, but highly unlikely, that the chromatophore will be lost outright in favor of the ancestral phagotrophic lifestyle. A more likely possibility is that the chromatophore loses photosynthetic function in some Paulinella species, as found in many algal lineages (e.g. diatoms; Kamikawa et al., 2017). We recognize, of course, that these ideas are speculative because the future of photosynthetic Paulinella will reflect currently unknowable, local biotic and abiotic conditions as well as the impact of population size that can strengthen or weaken selection on individuals.
IV. Why is secondary and tertiary endosymbiosis more common?
Whereas cases of primary endosymbiosis of a photosynthetic organism appear to be exceedingly rare, instances of secondary and tertiary endosymbiosis are more common. An explanation for this observation is the presence of existing host adaptations (e.g. transit peptides, plastid translocon, plastid metabolite transporters (Karkar et al., 2015), light sensing, photoacclimation (Duanmu et al., 2017), redox control and retrograde signaling) that optimize the metabolic and regulatory fit (interface) between the eukaryote host and bacterial symbiont (i.e. chassis and engine, respectively). In secondary and tertiary endosymbiosis, the presence of these genes in one or both eukaryote partners (e.g. from a previous plastid endosymbiosis, as observed for some dinoflagellates (Fig. 1)) facilitates the integration of the plastid into the metabolic systems of the new eukaryote host. This history alleviates the need to evolve novel genes and pathways, and contributes to the increased frequency of these events in nature.
V. Conclusions and perspectives
We believe that the chassis and engine model provides a theoretical framework to assist researchers, both established and new to the field, to better envision the complex evolutionary forces associated with primary endosymbiosis. As a testable hypothesis, it is meant to stimulate discussion and research within the community to advance the study of endosymbiosis and potentially provide guidance to synthetic biology efforts to design novel organelles. Although we focus on the Paulinella model, the ‘rules of engagement’ between a nonphotosynthetic protist and an oxygenic bacterial photoautotroph are likely to be universal, governing different types of host–symbiont interactions.
Box 1 Unanswered questions about evolution of the chromatophore in Paulinella.
What factors allowed Paulinella to integrate cyanobacterial photosynthate from the chromatophore into its metabolism, given that eukaryotic enzymes may not be able to utilize some of these substrates?
What role, if any, do chimeric or symbiogenetic genes play in establishing endosymbiosis (Meheust et al., 2016)?
What are the origins of proteins involved in coping with redox conditions, reactive oxygen species and the quenching of excess excitation?
Will the lost chromatophore-encoded transporters be ultimately compensated for by the retargeting of transporters encoded on the host nuclear genome, as found in Archaeplastida (Tyra et al., 2007)?
Does the Archaeplastida plastid represent the end-state of the chromatophore? Will Paulinella follow the same evolutionary trajectory? If not, then why and how do we expect these two events to differ?
If genes involved in the establishment of endosymbiosis were present before ingestion of the cyanobacterial cell, what role did they play in the biology of the heterotrophic Paulinella ancestor?
What biosynthetic processes have been gained by acquiring the chromatophore, are these pathways still encoded in the chromatophore, and are nuclear-encoded chromatophore pathways still maintained in the chromatophore? Or do different parts of the pathway occur in different cellular locations?
What metabolites are exchanged between the cytoplasm of the host and the chromatophore?
Acknowledgements
AG, TGS, VC and DB were supported by a grant from the National Aeronautics and Space Administration (80NSSC19K0462). DB was supported by a NIFA-USDA Hatch grant (NJ01180). ARG and VC were supported by the Carnegie Institution for Science.
References
- Allen JF. 2017. The CoRR hypothesis for genes in organelles. Journal of Theoretical 434: 50–57. [DOI] [PubMed] [Google Scholar]
- Arias-Agudelo LM, Gonzalez F, Isaza JP, Alzate JF, Pabon-Mora N. 2019. Plastome reduction and gene content in New World Pilostyles (Apodanthaceae) unveils high similarities to African and Australian congeners. Molecular Phylogenetics and Evolution 135: 193–202. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Price DC. 2020. The algal tree of life from a genomics perspective.In: Larkum AWD, Grossman AR, Raven JA, eds. Photosynthesis in algae: biochemical and physiological mechanisms. Cham, Switzerland: Springer International, 11–24. [Google Scholar]
- Bhattacharya D, Price DC, Yoon HS, Yang EC, Poulton NJ, Andersen RA, Das SP. 2012. Single cell genome analysis supports a link between phagotrophy and primary plastid endosymbiosis. Scientific Reports 2: 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D, Dufresne A, Vaulot D, Grossman A. 2002. Analysis of the hli gene family in marine and freshwater cyanobacteria. FEMS Microbiology Letters 215: 209–219. [DOI] [PubMed] [Google Scholar]
- Duanmu D, Rockwell NC, Lagarias JC. 2017. Algal light sensing and photoacclimation in aquatic environments. Plant, Cell & Environment 40: 2558–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Qiu H, Han W, Wang Y, Xu D, Zhang X, Bhattacharya D, Ye N. 2020. Phytoplankton pangenome reveals extensive prokaryotic horizontal gene transfer of diverse functions. Science Advances 6: eaba0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabr A, Grossman AR, Bhattacharya D. 2020. Paulinella, a model for understanding plastid primary endosymbiosis. Journal of Phycology 56: 837–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Bhattacharya D, Pelletreau KN, Rumpho ME, Reyes-Prieto A. 2012. Secondary and tertiary endosymbiosis and kleptoplasty. In: Bock R, Knoop V, eds. Genomics of chloroplasts and mitochondria. Dordrecht, the Netherlands: Springer, 31–58. [Google Scholar]
- Grossman A, Sanz-Luque E, Yi H, Yang W. 2019. Building the GreenCut2 suite of proteins to unmask photosynthetic function and regulation. Microbiology (Reading) 165: 697–718. [DOI] [PubMed] [Google Scholar]
- He Q, Dolganov N, Bjorkman O, Grossman AR. 2001. The high light-inducible polypeptides in Synechocystis PCC6803. Expression and function in high light. Journal of Biological Chemistry 276: 306–314. [DOI] [PubMed] [Google Scholar]
- Hoogenraad HR. 1927. Rhizopoden en Heliozoë uit het zoetwater van Nederland. Tijdschrift der Nederlandsche Dierkundige Vereeniging 2: 1–18. [Google Scholar]
- Johnson PW, Hargraves PE, Sieburth JM. 1988. Ultrastructure and ecology of Calycomonas ovalis Wulff, 1919, (Chrysophyceae) and its redescription as a testate rhizopod, Paulinella ovalis N. Comb. (Filosea: Euglyphina). Journal of Eukaryotic Microbiology 35: 618–626. [Google Scholar]
- Kamikawa R, Moog D, Zauner S, Tanifuji G, Ishida KI, Miyashita H, Mayama S, Hashimoto T, Maier UG, Archibald JM, Inagaki Y. 2017. A non-photosynthetic diatom reveals early steps of reductive evolution in plastids. Molecular Biology and Evolution 34: 2355–2366. [DOI] [PubMed] [Google Scholar]
- Karkar S, Facchinelli F, Price DC, Weber AP, Bhattacharya D. 2015. Metabolic connectivity as a driver of host and endosymbiont integration. Proceedings of the National Academy of Sciences, USA 112: 10208–10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorobrykh S, Havurinne V, Mattila H, Tyystjarvi E. 2020. Oxygen and ROS in photosynthesis. Plants 9: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J, Sobotka R. 2016. Cyanobacterial high-light-inducible proteins – protectors of chlorophyll-protein synthesis and assembly. Biochimica et Biophysica Acta (BBA) – Bioenergetics 1857: 288–295. [DOI] [PubMed] [Google Scholar]
- Larkum AW, Lockhart PJ, Howe CJ. 2007. Shopping for plastids. Trends in Plant Science 12: 189–195. [DOI] [PubMed] [Google Scholar]
- Lauterborn R 1895. Paulinella chromatophora nov. gen., nov. spec., ein beschalter Rhizopode des Siisswassers mit blaugriinen chromatophorenartigen Einschlussen. Zeitschrift für Wiss Zoologische 59: 537–544. [Google Scholar]
- Lhee D, Bhattacharya D, Yoon HS. 2021b. Independent evolution of the thioredoxin system in photosynthetic Paulinella species. Current Biology 31: R328–R329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhee D, Ha JS, Kim S, Park MG, Bhattacharya D, Yoon HS. 2019. Evolutionary dynamics of the chromatophore genome in three photosynthetic Paulinella species. Scientific Reports 9: 2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhee D, Lee J, Ettahi K, Cho CH, Ha J-S, Chan Y-F, Zelzion U, Stephens TG, Price DC, Gabr A, Nowack ECM, Bhattacharya D, Yoon HS. 2021a. Amoeba genome reveals dominant host contribution to plastid endosymbiosis. Molecular Biology and Evolution 38: 344–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M, Katahata A, Tachikawa M, Minakuchi Y, Noguchi H, Toyoda A, Fujiyama A, Suzuki Y, Hata T, Satoh S et al. 2019. Large DNA virus promoted the endosymbiotic evolution to make a photosynthetic eukaryote. bioRxiv. doi: 10.1101/809541. [DOI] [Google Scholar]
- Meheust R, Zelzion E, Bhattacharya D, Lopez P, Bapteste E. 2016. Protein networks identify novel symbiogenetic genes resulting from plastid endosymbiosis. Proceedings of the National Academy of Sciences, USA 113: 3579–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AP, Supekova L, Chen JH, Pestonjamasp K, Webster P, Ko Y, Henderson SC, McDermott G, Supek F, Schultz PG. 2018. Engineering yeast endosymbionts as a step toward the evolution of mitochondria. Proceedings of the National Academy of Sciences, USA 115: 11796–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkonian M, Mollenhauer D. 2005. Robert Lauterborn (1869–1952) and his Paulinella chromatophora. Protist 156: 253–262. [DOI] [PubMed] [Google Scholar]
- Mettler T, Muhlhaus T, Hemme D, Schottler MA, Rupprecht J, Idoine A, Veyel D, Pal SK, Yaneva-Roder L, Winck FV et al. 2014. Systems analysis of the response of photosynthesis, metabolism, and growth to an increase in irradiance in the photosynthetic model organism Chlamydomonas reinhardtii. Plant Cell 26: 2310–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Gomez SA, Mejia-Franco FG, Durnin K, Colp M, Grisdale CJ, Archibald JM, Slamovits CH. 2017. The new red algal subphylum Proteorhodophytina comprises the largest and most divergent plastid genomes known. Current Biology 27: 1677–1684. [DOI] [PubMed] [Google Scholar]
- Nowack EC, Melkonian M, Glockner G. 2008. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Current Biology 18: 410–418. [DOI] [PubMed] [Google Scholar]
- Nowack EC, Price DC, Bhattacharya D, Singer A, Melkonian M, Grossman AR. 2016. Gene transfers from diverse bacteria compensate for reductive genome evolution in the chromatophore of Paulinella chromatophora. Proceedings of the National Academy of Sciences, USA 113: 12214–12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack EC, Weber APM. 2018. Genomics-informed insights into endosymbiotic organelle evolution in photosynthetic eukaryotes. Annual Review of Plant Biology 69: 51–84. [DOI] [PubMed] [Google Scholar]
- Nowack ECM, Grossman AR. 2012. Trafficking of protein into the recently established photosynthetic organelles of Paulinella chromatophora. Proceedings of the National Academy of Sciences, USA 109: 5340–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberleitner L, Poschmann G, Macorano L, Schott-Verdugo S, Gohlke H, Stuhler K, Nowack ECM. 2020. The puzzle of metabolite exchange and identification of putative octotrico peptide repeat expression regulators in the nascent photosynthetic organelles of Paulinella chromatophora. Frontiers in Microbiology 11: 607182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce-Toledo RI, Lopez-Garcia P, Moreira D. 2019. Horizontal and endosymbiotic gene transfer in early plastid evolution. New Phytologist 224: 618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Prieto A, Yoon HS, Moustafa A, Yang EC, Andersen RA, Boo SM, Nakayama T, Ishida K, Bhattacharya D. 2010. Differential gene retention in plastids of common recent origin. Molecular Biology and Evolution 27: 1530–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Yoshitomi T, Mori-Moriyama N. 2020. Characterization and biosynthesis of lipids in Paulinella micropora MYN1: evidence for efficient integration of chromatophores into cellular lipid metabolism. Plant and Cell Physiology 61: 869–881. [DOI] [PubMed] [Google Scholar]
- Schön ME, Zlatogursky VV, Singh RP, Poirier C, Wilken S, Mathur V, Strassert JFH, Pinhassi J, Worden AZ, Keeling PK, et al. , 2021. Picozoa are archaeplastids without plastid. bioRxiv: 10.1101/2021.0414.439778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih PM, Hemp J, Ward LM, Matzke NJ, Fischer WW. 2017. Crown group Oxyphotobacteria postdate the rise of oxygen. Geobiol 15: 19–29. [DOI] [PubMed] [Google Scholar]
- Singer A, Poschmann G, Muhlich C, Valadez-Cano C, Hansch S, Huren V, Rensing SA, Stuhler K, Nowack ECM. 2017. Massive protein import into the early-evolutionary-stage photosynthetic organelle of the amoeba Paulinella chromatophora. Current Biology 27: 2763–2773. [DOI] [PubMed] [Google Scholar]
- Stephens TG, Ragan MA, Bhattacharya D, Chan CX. 2018. Core genes in diverse dinoflagellate lineages include a wealth of conserved dark genes with unknown functions. Scientific Reports 8: 17175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan XM, Hou SW, Song K, Georg J, Klahn S, Lu XF, Hess WR. 2018. The primary transcriptome of the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Biotechnology for Biofuels 11: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyra HM, Linka M, Weber AP, Bhattacharya D. 2007. Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biology 8: R212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Archibald JM. 2018. Plastid genomes. Current Biology 28: R336–R337. [DOI] [PubMed] [Google Scholar]
- Yewdall NA, Mason AF, van Hest JCM. 2018. The hallmarks of living systems: towards creating artificial cells. Interface Focus 8: 20180023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. 2004. A molecular timeline for the origin of photosynthetic eukaryotes. Molecular Biology and Evolution 21: 809–818. [DOI] [PubMed] [Google Scholar]
- Zhang R, Nowack ECM, Price DC, Bhattacharya D, Grossman AR. 2017. Impact of light intensity and quality on chromatophore and nuclear gene expression in Paulinella chromatophora, an amoeba with nascent photosynthetic organelles. The Plant Journal 90: 221–234. [DOI] [PubMed] [Google Scholar]


