Abstract
The interaction of interleukin-2 (IL-2) with its receptor (IL-2R) critically regulates the T-cell immune response, and the α chain CD25/IL-2Rα is required for the formation of the high-affinity receptor. Tissue-specific, inducible expression of the IL-2Rα gene is regulated by at least three positive regulatory regions (PRRI, PRRII, and PRRIII), but none responded to CD28 engagement in gene reporter assays although CD28 costimulation strongly amplifies IL-2Rα gene transcription. By DNase I hypersensitivity analysis, we have identified a novel TCR-CD3- and CD28-responsive enhancer (CD28rE) located 8.5 kb 5′ of the IL-2Rα gene. PRRIV/CD28rE contains a functional CRE/TRE element required for CD28 signaling. The T-cell-specific, CD28-responsive expression of the IL-2Rα gene appears controlled through PRRIV/CD28rE by cooperation of CREB/ATF and AP-1 family transcription factors.
Complete T-cell activation requires antigen-mediated signaling through the TCR-CD3 complex and costimulatory signals that can be provided by CD28 and its counterreceptor, B7. The B7-CD28 signaling pathways synergize with mitogenic signal from the TCR-CD3 complex to promote prolonged T-cell proliferation and increase interleukin-2 (IL-2) secretion (27, 33, 36). Although stimulation via CD28 alone usually cannot induce effector functions, its signaling pathways involve site-specific tyrosine phosphorylation of several effector proteins that are crucial for these functions (52). Triggering these cosignaling downstream pathways ultimately leads to the transcriptional activation and mRNA stabilization of genes encoding a number of cytokines critically involved in the regulation of Th1-Th2 differentiation (8). Among them, IL-2 plays a major role in the regulation of the magnitude and duration of T-cell activation in the immune responses (41). IL-2 promotes T-cell proliferation by interacting with its high-affinity receptor composed of three transmembrane polypeptides, the α, β, and γc chains. The α chain (CD25) is induced during T-cell activation, and its association with β and γc chains constitutes the high-affinity receptor. Most of the biologic effects of IL-2 are mediated through the high-affinity complex as evidenced by the almost identical phenotype of mice with targeted inactivation of the genes for IL-2 (53) and IL-2Rα (66). Furthermore, the importance of the IL-2Rα gene in the immune system is dramatically illustrated by the profound immunodeficiency observed in patient lacking CD25/IL-2Rα expression (12, 51).
Expression of IL-2Rα gene is tightly controlled at the transcriptional level (34, 61) and by mRNA stabilization (28). Previous studies have identified three positive regulatory regions, PRRI, PRRII, and PRRIII, within the 5′ noncoding region of the human IL-2Rα gene. Each regulatory region appears to play a well-defined and complementary role. PRRI, located between nucleotides −276 and −244, contributes to the inducibility of the IL-2Rα gene, whereas PRRII, located between nucleotides −137 and −64, is involved in basal promoter activity as well as in T-cell-specific expression (25). PRRIII, located between nucleotides −3786 and −3701, is a specific IL-2-responsive enhancer playing a crucial role in the response to the IL-2/IL-2R autoregulatory loop which amplifies IL-2Rα gene transcription in the late stage of T-cell activation (26, 32, 57). The transcriptional activity of PRRI can be regulated by the viral protein Tax (transcription activator protein of human T-cell leukemia virus type 1), and the transcription factors NF-κB and a serum response factor-related protein (2, 30, 35, 47). Elf-1, a member of the Ets family of proteins specific for lymphoid tissue, and the nonhistone chromatin-associated protein, HMG-I(Y), regulate PRRII (25). The transcriptional activity of PRRIII/IL-2rE is controlled by the functional interaction of Stat5, Ets-1, and Ets-2 in response to IL-2 stimulation (50).
While the molecular mechanisms linking the CD28-mediated signal transduction pathways (8, 52, 64) and the transcriptional machinery have yet to be fully characterized, different CD28 response elements (CD28RE) have been delineated within the promoters of several cytokines including IL-2 (9, 16, 62), IL-4 (38), IL-6 (10), IL-8 (65) IL-3, granulocyte-macrophage colony-stimulating factor and gamma interferon (17). Despite sequence similarity, at least two functionally distinct classes of CD28RE exist that coincide with differences in their NF-κB binding activity (10), and all characterized CD28RE to date were associated with this family of transcription factors. For instance, the well-characterized CD28-responsive complex (CD28RC) that binds to the IL-2 promoter contains at least three members of the Rel/NF-κB family, c-Rel, RelA(p65), and p50 (19, 31), but also unrelated transcription factors such as NF-ATp (39) and AP-1 proteins (54). Furthermore, the CD28 costimulatory signals can activates the cyclic AMP-responsive element binding proteins CREB/ATF-1 in T lymphocytes (24) and ATF-1/CREB2 in Jurkat leukemia T cells (5).
In marked contrast, none of the previously identified regulatory regions responds to CD28 signaling in gene reporter assays although CD28 stimulation strongly amplifies IL-2Rα gene transcription (7). To understand how CD28 can up-regulate IL-2Rα gene expression at the transcriptional level, we used an experimental strategy combining DNase I hypersensitivity analysis, in vivo footprinting, transient reporter gene assays, and electrophoretic mobility shift assays (EMSAs) to identify this important missing element. In the present study, we identified a novel inducible regulatory region, hereafter designated positive regulatory region IV (PRRIV). This region, located 8.5 kb upstream of the IL-2Rα gene, responds efficiently to CD28 stimulation and fulfills the requirements for a bona fide CD28 responsive enhancer (CD28rE). IL-2Rα PRRIV matches a T-cell receptor (TCR)–CD3- and/or CD28-inducible DNase I-hypersensitive (DH) site. Moreover, the inducible transcriptional activity of PRRIV is mediated by the functional cooperation between CREB/ATF and Jun/AP-1 family proteins and enhanced by CD28 costimulatory signals.
(This work partially fulfills the requirement for the doctoral thesis of J.-H. Yeh at the Université de la Méditerranée.)
MATERIALS AND METHODS
Cell culture and reagents.
The human leukemia Jurkat T-cell clone JH6.2 (43) and the CD28-deficient Jurkat cell clone JF3 were maintained in RPMI 1650 with 10% fetal calf serum containing 2 mM l-glutamine. Peripheral lymphocytes were isolated from blood donated by healthy volunteers and purified as previously described (6). Primary T cells were maintained in CHO-SFM II (Life Technologies). COS 1 cells were cultured in Dulbecco modified Eagle medium (DMEM) with 10% fetal calf serum containing 2 mM l-glutamine. Anti-CD28 248 (mouse immunoglobulin M [IgM]) and anti-CD3 289 (mouse IgG2a) were kindly provided by A. Moretta (Cancer Institute, Genoa, Italy) and used either as ascites fluid at a 1/4,000 dilution or as purified monoclonal antibody (MAb) (5 μg/ml). Anti-CD3 289 was used coated onto petri tissue culture dishes. Anti-CD28 BW828 (IgG2a) (56) was kindly provided by R. Schwinzer (Medizinische Hochschule, Hannover, Germany) and used at 10 μg/ml. T-cell activation was controlled by CD69 expression (40) and proliferation assays (13). Phorbol myristate acetate (PMA) (Calbiochem) was used at 20 ng/ml.
DNase I hypersensitivity analysis.
Isolation and DNase I digestion of nuclei and purification of genomic DNA were performed as described previously (11). Briefly, nuclei were isolated from 108 cells by lysis in 0.05% NP-40 and suspended at 2 × 107 and 5 × 107 nuclei per ml. Aliquots of nuclei were incubated for 3 min at room temperature. The amounts of DNase I (Amersham Life Science) used are indicated in the figure legends. DNA was purified by phenol-chloroform extraction, and 15 μg of DNA was digested with BgIII. Samples were resolved on an 0.8% agarose gel, transferred to a Hybond-N+ positively charged nylon membrane (Amersham Life Science), and hybridized with an IL-2Rα gene locus-specific probe labeled by random priming. Hybridized membranes were exposed to BioMax MR film (Kodak) for 5 to 7 days.
Plasmids and mutagenesis.
The different fragments of DNA presented in Fig. 1B were subcloned upstream of the TK gene minimal promoter into the pTK3-CAT reporter gene vector. The −481 to +110 (481) and −8942 to +110 (8942) EcoRI-PstI fragments were subcloned in a promoterless chloramphenicol acetyltransferase (CAT) reporter gene vector. The BcII-SphI 207-bp fragment containing PRRIV/CD28rE as defined in Results was subcloned upstream of its endogenous proximal promoter-enhancer region into 481.IIR and into the luciferase reporter gene pGL3 (Promega) at the SmaI site. pHβAPr-1-CD28 wt (neo) and pHβAPr-1-CD28 Δ30 (neo) have been previously described (44). The mutated pGL3/PRRIV (mCRE/TRE) was constructed by site-directed mutagenesis using the oligonucleotidic sequence CTCCTCTAGATT (substituted nucleotides are in boldface type). pcDNA3-c-Jun, pcDNA3Jun B, pcDNA3-Jun D and pCMV-ATF-1 were gifts of V. Coulon and J.-M. Blanchard (Centre National de la Recherche Scientifique, Montpellier, France). pcDNA3-CREB and the cylic AMP responsive element (CRE) binding (CREB) dominant negative form, pcDNA3-CREB (S133A) (49), were gifts of P. G. Quinn (Pennsylvania State University, Hershey, Pa.).
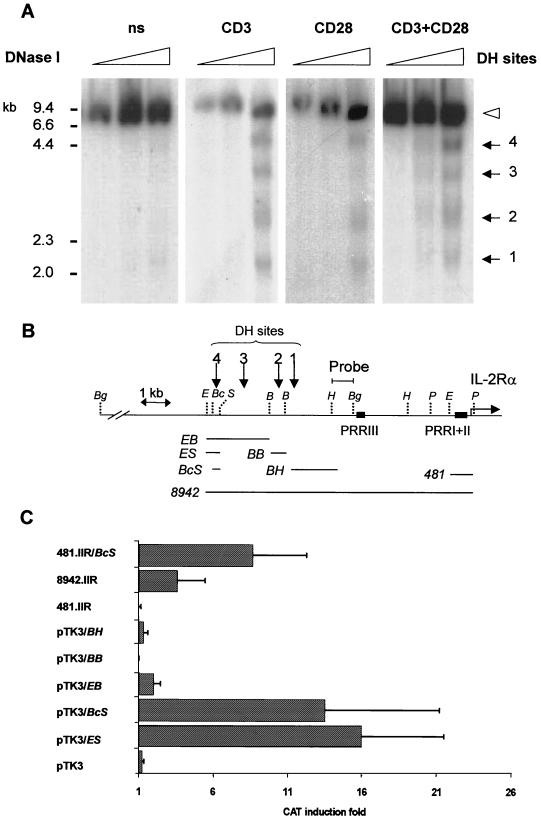
FIG. 1.
An inducible DH site exists upstream of the human IL-2Rα gene and corresponds to an inducible enhancer. (A) DNase I hypersensitivity pattern on the IL-2Rα locus in unstimulated (ns) and stimulated human primary T cells. Highly purified human primary T cells were maintained in serum-free medium and then stimulated with anti-CD3 289 and anti-CD28 248 for 16 h (see Materials and Methods); titration of DNase I (0 to 80 U/ml) is indicated above the panel. Southern blot analyses were performed with BglII-digested DNA samples probed with the HindIII-BglII genomic probe indicated in panel B. The sizes (in kilobases) of the molecular mass markers are indicated on the left and right sides, the open arrowhead indicates the DNase I-undigested genomic fragments cut by BglII; and arrows indicate the DH sites. (B) Map of the human IL-2Rα locus showing the locations of the DH sites. The previously characterized regulatory regions PRRI, PRRII, and PRRIII are indicated; Bc, BclI; Bg, BglII; E, EcoRI; H, HindIII; P, PstI; S, SphI. Horizontal lines below the map show the localization of the DNA fragments used in the reporter gene assays panel C, according to the GenBank sequence Z70243. EB, EcoRI-BamHI-digested 2,301-bp fragment; ES, EcoRI-SphI-digested 465-bp fragment; Bc/S, BclII-SphI-digested 207-bp fragment; BB, BamHI-digested 406-bp fragment; BH, BamHI-HindIII-digested 1,678-bp fragment; 481, EcoRI-PstI-digested 591-bp fragment containing the IL-2Rα gene PRRI-PRRII enhancer and promoter up to nucleotide −481; 8942, partial EcoRI-PstI-digested 9,052-bp fragment. (C) Transcriptional activity of the 5′ noncoding region of the human IL-2Rα gene in response to PMA-ionomycin. The DNA fragments covering the human IL-2Rα gene up to nucleotide −8942 presented in panel B were inserted upstream of the TK minimal promoter in the pTK3 CAT reporter, and 25 μg of each construct was transfected in Jurkat JH6.2 T cells. The constructs 481.IIR and 8942.IIR correspond to the −481 to +110 and −8942 to +110 EcoRI-PstI fragments, respectively, inserted upstream of a promoterless CAT reporter gene vector. The 481.IIR/BcS construct correspond to the insertion of the BcII-SphI 207-bp fragment upstream of the IL-2Rα-proximal promoter-enhancer region. The histogram bars represent the ratio of the CAT activity after PMA-ionomycin stimulation relative to the CAT activity in unstimulated cells.
Transient-transfection and reporter gene assays.
Jurkat T cells were transfected with 7.5 to 30 μg of the indicated plasmids at 250 V and 960 μF in a Bio-Rad (Hercules, Calif.) Gene Pulser. For COS 1 cells, transfections were performed using FuGENE 6 transfection reagent (Roche). Transfected cells were then activated with different stimuli as indicated in figure legends. Cells were harvested 16 h later for CAT assays and 6 h later for luciferase assays. Cell lysis and chloramphenicol acetyltransferase (CAT) assays were performed with the CAT enzyme-linked immunosorbent assay kit (Boehringer Mannheim), as specified by the manufacturer, with 50 μg of cell extracts. Luciferase assays were performed with the dual-luciferase reporter assay system (Promega) with 15 μg of cell extracts, and luciferase activity was measured with a Dynex MLX microplate luminometer. In Fig. 2 to 4 and 7, histogram and error bars represent the mean of at least 3 and up to 21 independent measurements, and the standard error of the mean, respectively.
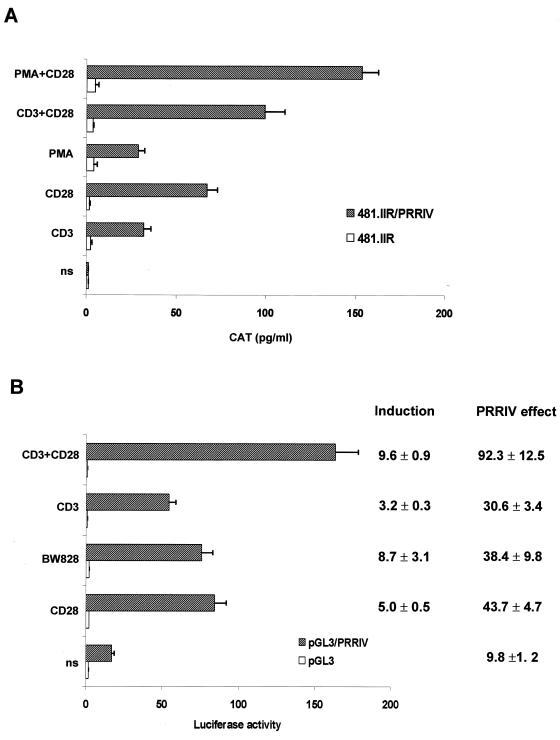
FIG. 2.
An inducible enhancer overlies the inducible DH site 4 upstream of the human IL-2Rα gene. (A) Transcriptional activity of PRRIV in its endogenous context. Jurkat JH6.2 T cells were transfected with 25 μg of 481.IIR and 481.IIR/PRRIV (identical to 481.IIR/BcS in Fig. 1) and treated with various stimuli as described in Materials and Methods. (B) JH6.2 T cells were transfected with the pGL3 luciferase vector and the pGL3/PRRIV construct containing the 207-bp BclI-SphI PRRIV element inserted in its natural orientation upstream of the SV40 promoter (7.5 μg). At 1 h after transfection, Jurkat T cells were treated with anti-CD3 289 and anti-CD28 248 or BW828, alone or in combination. Histogram bars represent the means of the CAT or luciferase activity (in relative light units) determined for each condition. Values on the right correspond to the induction ratio of each construct (Induction) and to the ratio of the luciferase activity of the pGL3/PRRIV construct relative to the luciferase activity of the pGL3 vector (PRRIV effect). ns, not stimulated.
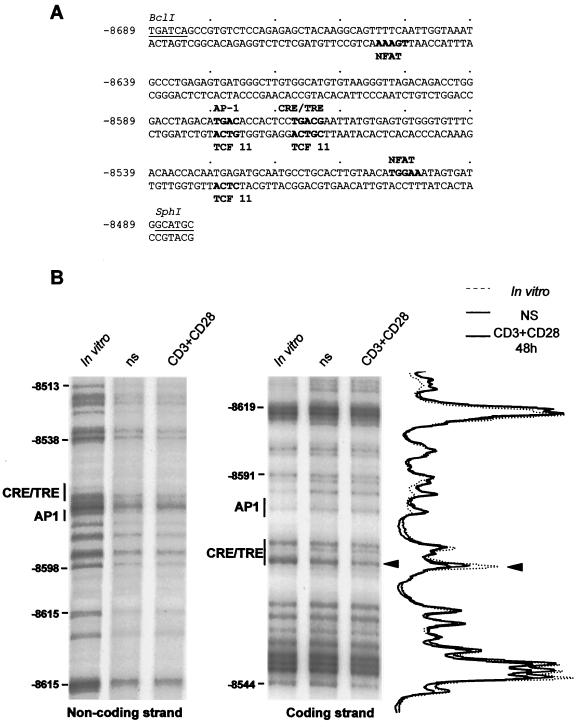
FIG. 4.
Characterization of the regulatory elements within PRRIV/CD28rE by in vivo footprinting. (A) DNA sequence of PRRIV/CD28rE in the human IL-2Rα gene between nucleotides −8689 and −8483. The putative regulatory elements are indicated in boldface. (B) DMS/LM-PCR was performed with in vitro-methylated DNA or DNA from human primary resting T cells or stimulated for 48 h with anti-CD3 289 and anti-CD28 248 using two sets of three specific primers for PRRIV/CD28rE noncoding and coding strands, respectively. ns, not stimulated.
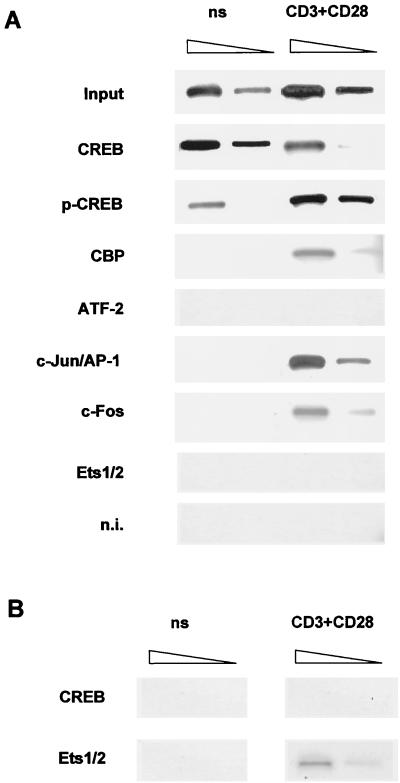
FIG. 7.
In vivo specific binding of CREB, CBP, and AP-1 family proteins to PRRIV in human primary T cells. (A) PCR analysis of chromatin-immunoprecipitated DNA with specific primers for PRRIV. PCR amplifications with 10% and 1% of input DNAs are used to evaluate the linear range of signal. DNA-protein complexes were immunoprecipitated with rabbit nonimmune serum (n.i.) or specific CREB, phospho-CREB (p-CREB), CBP, ATF-2, c-Jun/AP-1, c-Fos, or Ets1/2 antibodies. (B) As a control for the experiment in panel A, anti-Ets1/2 or anti-CREB chromatin-immunoprecipitated DNAs were analyzed by PCR with specific primers for PRRIII in the promoter region of the IL-2Rα gene. ns, not stimulated.
Reverse transcription-PCR (RT-PCR) and FACScan analysis.
CD28-deficient Jurkat JF3 T cells were transfected with 20 μg of pHβAPr-1-CD28 wt (neo) or pHβAPr-1-CD28 Δ30 (neo). Cells were harvested and washed 24 h after transfection, and RNA was extracted using Trizol. A 2-μg portion of total RNA was used for cDNA synthesis by random priming. PCR was used with primers 5′-hCD28 (GTGAAATGCTGCAGTCAGGA) and 3′-hCD28 (ACCTGAAGCTGCTGGGAGTA). All cell cultures were harvested 24 h after transfection and incubated at 4°C for 45 min with monoclonal antibodies (MAbs) in the dark. The cells were analyzed by flow cytometry (FACScan; Beckton Dickinson) using the CELLQuest software (Beckton Dickinson). The MAb used in our study was anti-human CD28 (phycoerythrin [PE]-conjugated clone CD28.2) from Immunotech (Marseille, France).
In vivo footprint analysis.
Genomic footprint analyses were performed as previously described (2) by the dimethylsulfate–ligation-mediated PCR (DMS/LM-PCR) method using in vitro and in vivo methylated genomic DNA purified from human primary T cells. The following oligonucleotide primers were used for the coding strand: primer 1, TCACTAGCACTGACTAGGC (−8408 to −8389; Tm, 60°C); primer 2, GGCAAGCACTGTGCTGAGAATGTTG (−8444 to −8419; Tm, 63°C); and primer 3, TGCTGAGAATGTTGCATGTCTCTGCCGCT (−8459 to −8430; Tm, 66°C); the following were used for the noncoding strand: primer 1, CAATGCTTAACGACTGAGC (−8804 to −8785; Tm, 58°C); primer 2, GCTAAGTACTGAACTCAGCACTAGG (−8774 to −8750; Tm, 61°C); and primer 3, CTCAGCACTAGGAATAAGAAGGCGACCTA (−8761 to −8733; Tm 64°C).
EMSAs and Western blot analyses.
Nuclear extracts of Jurkat JH6.2 T cells and purified T lymphocytes and EMSAs were performed as previously described (13). Synthetic oligonucleotide probes were end labeled with [γ-32P]ATP. The following probes were used: wt CRE/TRE, ACACCACTCCTGACGAATTATGTGAG; mCRE/TRE, ACACCACTCCTCTAGATTATGTGAG (the CRE/tetradecanoyl phorbol acetate responsive element (TRE) motif is underlined, and the substituted nucleotides to disrupt the consensus are in boldface); high-affinity CREa (a motif derived from the promoter region of the cyclin A gene [48]), CGCCTTGAATGACGTCAAGGC. Supershift and blocking assays of PRRIV CRE/TRE binding proteins were performed by incubating nuclear extract for 10 min on ice with different antibodies before the addition of 32P-labeled probe. Anti-c-Jun/AP-1 (D), anti-c-Fos (6-2H-2F), anti-ATF-1 (25C10G), anti-CREB-1 (X-12), anti-ATF-1 (C41-5.1), and anti-ATF-2 (C-19) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.). Anti-phospho-CREB was purchased from UBI. In competition experiments, unlabeled oligonucleotides were incubated for 15 min with nuclear extracts at 4°C before the addition of 32P-labeled probe. For Western blots, 5 μg of denatured nuclear extract was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide). Proteins were transferred to Immobilon-P membrane (Sigma), and phospho-CREB was detected with a 1/1,000 dilution of anti-phospho-CREB (Ser133) antibody (New England Biolabs) by using the enhanced chemiluminescence system (Amersham Life Science) with horseradish peroxidase-coupled goat anti-rabbit antibodies (Jackson ImmunoResearch Laboratories).
Chromatin immunoprecipitation (ChIP) assays.
Chromatin from unstimulated or stimulated (20 h with CD3 plus CD28) human primary T cells was extracted and immunoprecipitated using the chromatin immunoprecipitation ChIP assay kit (Upstate Biotechnology, Inc.) as recommended by the manufacturer. The purified chromatin was immunoprecipitated using 10 μg of anti-CREB or CBP (UBI); 15 μg of anti-phospho-CREB (UBI); 10 μg of anti-c-Jun/AP-1, c-Fos/AP-1, ATF-2, or Ets1/2 (Santa Cruz); and 5 μl of rabbit nonimmune serum. The input fraction corresponded to 0.1 and 0.01% of the chromatin solution before immunoprecipitation. Following DNA purification, the presence of selected DNA sequences was assessed by PCR. Fifteen cycles were performed with the first primer set in 50 μl with 2 μl of immunoprecipitated material. Then, using 2 μl of the first PCR product, 25 additional cycles of PCR were performed with the CRE/TRE 3′ primer set and 1 μCi of [α-32P]dCTP. The PCR products were resolved in 8% acrylamide gels. A twofold dilution series was typically used for the first PCR amplification. The primers used were as follows: PRRIV 5′ (5′-CTCCAGAGAGCTACAAGGCAGT-3′) and PRRIV 3′ (5′-GTTGTGAAACACCCACACTCAC-3′) (first PCR, 141-bp product); CRE/TRE 3′ (5′-CTCACATAATTCGTCAGGAGTGGTGT-3′) (second PCR, 125-bp product); PRRIII 5′ (5′-TTCACCCCACTGTACGTC-3′) and PRRIII 3′ (5′-GTCAACAGTGCAAGCTGAGTCT-3′) (first PCR, 173-bp product); and PRRIII 5′-2 (5′-TAAGGGAAGGCAGTCTAGGTCA-3′) (second PCR, 133-bp product).
RESULTS
Identification of PRRIV, a new positive regulatory region in the human IL-2Rα gene.
To further investigate the organization of the human IL-2Rα chain locus, we analyzed the chromatin structure accessibility of this gene using DNase I hypersensitivity analysis (67) in human primary T cells. The probe shown in Fig. 1B labeled the original genomic DNA restriction fragment in addition to the subbands created by the cutting of DNase I at the hypersensitivity sites (Fig. 1A). In human primary resting T cells, using high concentrations of DNase I, four weak sites of hypersensitivity were hardly detected. After stimulation via either TCR-CD3 or CD28, the chromatin structure appeared more sensitive to the nuclease since DH sites 1, 2, 3, and 4 were significantly reinforced. The potent mitogenic anti-CD3 and anti-CD28 combination resulted in a clear activation of the four sites and more particularly of DH site 4. DH sites 1, 2, 3, and 4 were located at kb −8.5, −6, −5, and −4.3 (Fig. 1B), respectively, relative to the major transcription initiation site (14). We excluded the possibility that the difference in DNase cleavage reflected differences in enzymatic activity in the different cell samples rather than an increased accessibility by analyzing the DNase I cleavage pattern of the IL-2Rα gene proximal promoter-enhancer region. No significant differences were observed when the same DNA preparations were used (data not shown).
On the basis of these results, the corresponding region was screened for putative regulatory elements by transient-transfection assays. For this purpose, a series of fragments derived from a 9-kb fragment between nucleotides −8942 and +110 were inserted upstream of the simian virus 40 (SV40) heterologous minimal promoter inserted either in the CAT reporter gene vector pTK3 or in the luciferase gene reporter vector pGL3. The transcriptional activity of these constructs was tested in the Jurkat JH6.2 T-cell line. These assays revealed that a BcII-SphI 207-nucleotide fragment, overlying DH site 4, conferred a positive transcriptional activity in response to PMA plus ionomycin that was much stronger than the activity conferred by a EcoRI-BamHI 2301-nucleotide fragment containing both DH sites 4 and 3, whereas the fragments containing DH sites 1 and 2 failed to respond (Fig. 1C). To exclude the possibility of aberrant function revealed by the heterologous pTK3 CAT construct, the activity was also tested for the endogenous promoter region. As previously reported (14), the proximal promoter-enhancer region containing PRRI plus PRRII failed to respond to PMA plus ionomycin (Fig. 1C, 481.IIR) whereas only a 3.5-fold induction was observed with the fragment from −8942 to +100 that included the four DH sites (Fig. 1C, 8942.IIR). Insertion of the BcII-SphI 207-nucleotide fragment upstream of the proximal promoter-enhancer region significantly increased CAT induction (Fig. 1C, 481.IIR/BcS). These results suggested the presence of some repressor sequences between the distal BcII-SphI 207-nucleotide fragment and the proximal promoter-enhancer region which were not further investigated. Furthermore, we have previously reported that a DNA fragment containing PRRIII does not respond to PMA plus ionomycin (32). Sequence analysis using the NIX package software (http://www.hgmp.mrc.ac.uk) confirmed that no significant open reading frame was present between this upstream putative enhancer and the previously characterized IL-2Rα PRRIII/IL-2rE (reference (32) and data not shown).
Collectively, our results strongly suggested that the BcII-SphI 207-bp fragment is a novel positive cis-acting regulatory region in the 5′-flanking region of the human IL-2Rα gene. It was designated PRRIV.
PRRIV can respond to TCR-CD3 and/or CD28 stimulatory signals.
Since TCR-CD3–CD28 costimulation potently up-regulates the transcription of the human IL-2Rα gene (7), we further investigated by transient transfection whether anti-CD3 and anti-CD28 activate the transcriptional activity of PRRIV in its endogenous context or in the heterologous pGL3/PRRIV construct. As illustrated in Fig. 2A, PRRIV inserted upstream of the IL-2Rα proximal promoter-enhancer region responded to various stimuli that mimicked TCR-CD3 and CD28 engagement. When Jurkat T cells were treated with a combination of anti-CD3 and anti-CD28, the 481.IIR construct was hardly activated whereas the transcriptional activity of 481.IIR/PRRIV was strongly increased (Fig. 2A). Interestingly, the CAT assay revealed that anti-CD28 248 alone stimulated the transcriptional activity of PRRIV, which was additive to the transcriptional effect of anti-CD3 289 alone. This observation led us to postulate that PRRIV can respond to signal 2 (costimulatory signal) alone without signal 1 (TCR-CD3 signal).
To exclude the effect of downstream signals from the TCR-CD3 complex, we used the CD28 MAb BW828 (56) which can induce resting human T-cell proliferation and cytokine synthesis without TCR-CD3 engagement. BW828 induced the transcriptional activity of PRRIV as effectively as did CD28 MAb 248 (Fig. 2B). Insertion of PRRIV upstream of the SV40 promoter led to a 90-fold increase of luciferase activity compared to pGL3 in response to the CD3-CD28 costimulation.
Collectively, our results strongly suggested that PRRIV is a CD28-responsive enhancer (CD28rE) in the human IL-2Rα gene.
CD28 can specifically induce PRRIV transcriptional activity.
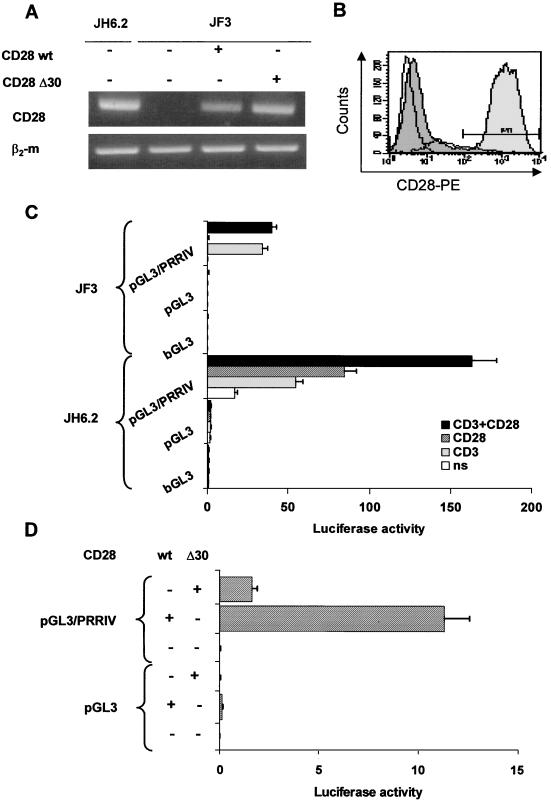
To demonstrate that PRRIV can specifically respond to CD28 downstream signals, we used the CD28-deficient mutant Jurkat T-cell clone, JF3. As the JH6.2 subclone, JF3 was selected by limiting dilution of Jurkat JA3.52 T cells (43). We used RT-PCR to control for the lack of CD28 expression in the JF3 subclone. As expected, no CD28 mRNA was detected in JF3 T cells (Fig. 3A). Fluorescence-activated cell sorter FACS analysis for CD28 expression confirmed the absence of protein at the cell surface (Fig. 3B). As a control, the Jurkat JH6.2 T cells expressed high levels of CD28 mRNA (Fig. 3A) and cell surface proteins (data not shown).
FIG. 3.
PRRIV is a bona fide CD28 response element. (A and B) Reconstitution of CD28 expression in CD28-deficient JF3 T cells transfected with CD28 wt or CD28 Δ30 expression vectors (20 μg). (A) Total RNA was isolated from CD28-deficient JF3 T cells 24 h after transfection, and specific fragments for CD28 or β2-microglobulin (β2m) mRNAs were amplified by RT-PCR. (B) FACS analysis of transfected JF3 T cells stained with anti-CD28–PE: nontransfected (white), CD28 wt (deep gray), and CD28 Δ30 (light gray) are shown. (C) The transcriptional activity of PRRIV is induced by anti-CD3 but not by anti-CD28 in JF3 T cells. JF3 T cells were transfected with a promoterless luciferase construct (bGL3) and an SV40 promoter-luciferase construct (pGL3) containing or not containing PRRIV (7.5 μg). (D) PRRIV transcriptional activity is restored by exogenous expression of CD28 wt but not by CD28 Δ30 in JF3 T cells. Luciferase activity was determined 24 h after transfection.
JF3 cells were transfected with pGL3/PRRIV and then treated with anti-CD3 and anti-CD28, alone or in combination. TCR-CD3 stimulation induced the activation of pGL3/PRRIV with a slightly lower efficiency than in Jurkat JH6.2 T cells (Fig. 3C). In sharp contrast, PRRIV failed to respond to anti-CD28 in JF3 cells, as expected for a bona fide CD28rE. In agreement, no further stimulatory effect was found in JF3 cells treated with both CD3 and CD28 MAbs.
To confirm the specificity of CD28 action on PRRIV/CD28rE, JF3 T cells were transiently cotransfected with pGL3/PRRIV and a wild-type CD28 cDNA expression vector or the truncated form CD28Δ30 (44). This nonfunctional form lacks almost all signal transduction motifs contained in the CD28 cytoplasmic domain. RT-PCR (Fig. 3A) and FACS analysis (Fig. 3B) confirmed reconstitution of CD28 wt and Δ30 expression. Cell surface expression of CD28 Δ30 was even more efficient than for CD28 wt form (Fig. 3B, 17% CD28 wt+ versus up to 90% CD28 Δ30+). Under these conditions, the transcriptional activity of PRRIV/CD28rE was efficiently restored by cotransfection with CD28 full-length cDNA (Fig. 3D). In contrast, cotransfection with the truncated form CD28Δ30 failed to activate PRRIV/CD28rE.
The CD28-deficient JF3 T-cell clone allowed a direct demonstration that CD28 downstream signals alone are sufficient to trigger PRRIV transcriptional activity. Taken together, these data demonstrated that the human PRRIV within IL-2Rα locus corresponds to a new CD28-responsive enhancer.
A CRE/TRE regulatory element within PRRIV is essential for the response to TCR-CD3 and CD28 signaling.
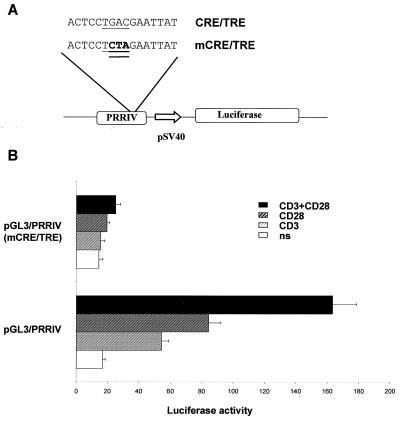
Sequence analysis using the Transcription Element Search Software (TESS [http://www.cbil.upenn.edu/tess]) revealed several putative binding site for known transcription factors which play a major role during T-cell activation, such as NFAT, AP-1, and CREB/ATF family proteins within the DNA sequence of human PRRIV/CD28rE (Fig. 4A). DMS/LM-PCR genomic footprint experiments (2) performed with human primary T cells gave evidence that the CRE/TRE was the only site among the putative regulatory elements significantly modified in response to CD3-CD28 costimulation on the coding strand whereas no clear occupancy was detected on the noncoding strand (Fig. 4B). This site might correspond to a CRE or TRE. Since several reports have shown that CD28 can specifically activate the transcriptional activity of CREB/ATF and AP-1 family proteins (5, 24, 29, 60), we disrupted this CRE/TRE site into the pGL3/PRRIV construct (Fig. 5A) to analyze its involvement in the PRRIV/CD28rE response to CD28 in JH6.2 cells and CD28-deficient Jurkat JF3 T-cell clone. In JH6.2 T cells, disruption of the CRE/TRE motif almost completely abolished the transcriptional activity of PRRIV/CD28rE in response to PMA plus ionomycin (data not shown), CD3, and CD28 stimulation (Fig. 5B). CD28 specificity was further confirmed by cotransfection of wt CRE/TRE and mutated (mCRE/TRE) reporter constructs in JF3 cells. Expression of wt CD28 restored the transcriptional activity of wt PRRIV/CD28rE, whereas the mutated CRE/TRE failed to respond to CD28 signals (data not shown). Hence, the CRE/TRE motif within the PRRIV/CD28rE plays a crucial role in the transcriptional activity of this enhancer in response to TCR-CD3 and CD28 signaling.
FIG. 5.
A CRE/TRE motif is essential for the CD28 response of the human IL-2Rα gene PRRIV/CD28rE. (A) Schematic representation of the wild-type CRE/TRE and mutated (mCRE/TRE) pGL3/PRRIV reporter plasmid. The CRE/TRE motif is underlined, and the nucleotides substituted for its disruption are doubly underlined in boldface. These fragments were inserted upstream of the SV40 minimal promoter (pSV40) in the pGL3 construct. (B) Disruption of the CRE/TRE motif in PRRIV/CD28rE abolished the CD28 response. The wild-type and mCRE/TRE pGL3/PRRIV constructs were transfected into Jurkat JH6.2 T cells. Cells were treated with anti-CD3 289 and/or anti-CD28 248 1 h after transfection. ns, not stimulated.
Identification of transcription factors that bind in vitro to the CRE/TRE motif within the PRRIV/CD28rE in human primary T cells.
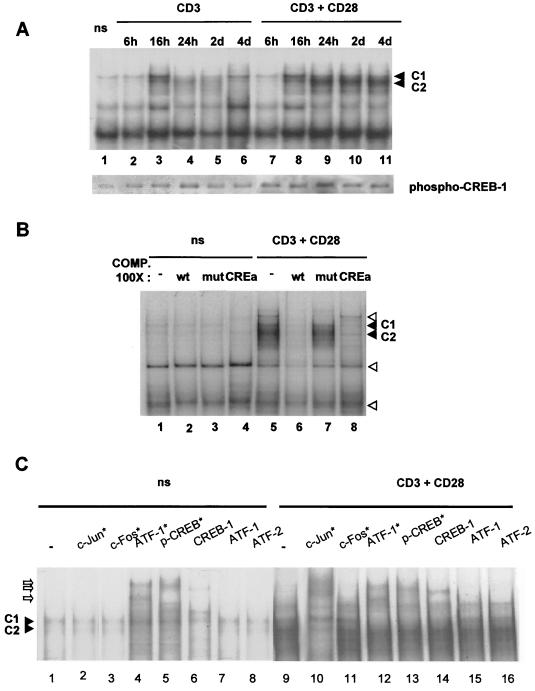
EMSAs performed with a specific radiolabeled double-stranded oligonucleotidic probe identified nuclear factors bound to the functional PRRIV CRE/TRE motif and nuclear extracts from unstimulated and TCR-CD3-and/or CD28-stimulated human primary T cells. Since factors present in the fetal bovine serum used in the cell culture medium might activate the CREB/ATF family proteins (4, 23), we cultured the human primary T cells in serum-free medium. EMSAs performed with the PRRIV CRE/TRE-specific probe revealed several protein-DNA complexes (Fig. 6A). The weak constitutive complex C1 was strongly increased after stimulation by anti-CD3 alone or in combination with anti-CD28. The inducible complexe C2 was present at least 4 days after CD3-CD28 costimulation but was only transiently detected after 16 h in response to anti-CD3 alone (Fig. 6A, compare lanes 3 to 6 with lanes 8 to 11). Western blotting analysis performed with the same nuclear extracts revealed that anti-CD3 alone triggered a more transient phosphorylation of CREB-1 than did the TCR-CD3–CD28 combination, paralleling the detection of complex C2 under both conditions (Fig. 6A, compare the lower and upper panels). Competition assays with unlabeled double-stranded oligonucleotides evidenced that an excess of wt CRE/TRE and CREa abolished the constitutive complex C1 and the inducible complex C2 (Fig. 6B, compare lane 1 with lanes 2 and 4 and lane 5 with lanes 6 and 8). In contrast, the mCRE/TRE competitor did affect complexes C1 and C2 (Fig. 6B, lanes 3 and 7). Taken together, these data demonstrate that the protein-DNA complex C1 and C2 are specific for the PRRIV CRE/TRE motif in human primary T cells.
FIG. 6.
Characterization of transcription factors binding in vitro to CD28 enhancer CRE/TRE motif in human primary T cells. (A) Time course analysis of the protein-DNA complexes revealed by the PRRIV CRE/TRE probe in highly purified human primary T cells stimulated with anti-CD3 and/or anti-CD28 in serum-free medium and harvested at the indicated time points above the upper panel. At the same time points, 5-μg portions of nuclear extracts were analyzed by Western blot analysis using anti-phospho-CREB-1 antibodies (lower panel). (B) Binding specificity of the protein-DNA complexes revealed with the PRRIV CRE/TRE probe. EMSAs were performed with nuclear extracts from resting human primary T cells (lanes 1 to 4) or stimulated with TCR-CD3 plus CD28 for 24 h (lanes 5 to 8) in the presence of the following unlabeled competitors: none (lanes 1 and 5), 100-fold excess of wt CRE/TRE (lanes 2 and 6, wt), mCRE/TRE (lanes 3 and 7, mut), or high-affinity CRE binding site (lanes 4 and 8, CREa). (C) The radiolabeled CRE/TRE probe was incubated with nuclear extracts from unstimulated human primary T cells (lanes 1 to 8) or cells stimulated with anti-CD3 289 plus anti-CD28 248 for 4 days (lanes 9 to 16) without (lanes 1 and 9) or with specific antibodies for the c-Jun/AP-1 family (lanes 2 and 10, c-Jun*); c-Fos/AP-1 family (lanes 3 and 11, c-Fos*); CREB/ATF family (lanes 4 and 12, ATF-1*); phosphorylated CREB-1, CREM, and ATF-1 (lanes 5 and 13, p-CREB*), CREB-1 (lanes 6 and 14); ATF-1 (lanes 7 and 15); or ATF-2 (lanes 8 and 16). Solid arrowheads, positions of the CRE/TRE-specific protein-DNA complexes; open arrowheads, nonspecific protein-DNA complex; open arrows, supershifted protein-DNA complexes. ns, not stimulated.
CD28 activates CREB/ATF family proteins in T lymphocytes (5, 24), and these transcription factors can bind the CRE motif as either homodimers or heterodimers with other CREB/ATF family members or with members of the AP-1 transcription factor family (22, 46). We hypothesized that the protein-DNA complexes C1 and C2 contained some of these proteins, and supershift experiments were performed with a panel of antisera recognizing CREB/ATF and AP-1 family factors. Anti-ATF-1, which recognizes ATF-1 p35, CREB-1 p43, and CREM, efficiently supershifted the constitutive complex C1 (Fig. 6C, compare lanes 1 and 4 with lanes 9 and 12). This complex was also partially supershifted in unstimulated nuclear extracts (compare lane 1 with lanes 5 and 6) and completely abolished in CD3-CD28-stimulated extracts in the presence of anti-phospho-CREB and anti-CREB-1 (compare lane 9 with lanes 13 and 14). Anti-c-Jun, which recognizes c-Jun, Jun B, and Jun D, supershifted the inducible complex C2 (compare lane 9 with lane 10). Anti-c-Fos, which recognizes c-Fos, Fos B, Fra-1, Fra-2, anti-ATF-1, and anti-ATF-2 sera, did not affect the formation of the CRE/TRE-specific protein-DNA complexes C1 and C2 (compare lanes 1, 3, 7, and 8 with lanes 9, 11, 15, and 16). Taken together, these data showed that the constitutive complex C1 bound specifically to the CRE/TRE motif within PRRIV/CD28rE in human primary T cells contained at least CREB-1 p43 and that the TCR/CD3- and/or CD28-inducible complex C2 contained Jun/AP-1 family proteins.
Specific binding of CREB and AP-1 to PRRIV in vivo.
To reconcile the specific binding of CREB and AP-1 to PRRIV in vitro, we used ChIP assays (Fig. 7). The ChIP technique can establish whether a known transcription factor truly binds in the vicinity of a known regulatory element in living cells (45). We prepared resting and CD3-CD28-stimulated human primary T cells and then cross-linked their chromatin before subjecting them to immunoprecipitation with various antisera. The precipitated chromatin DNA was then purified and subjected to PCR analysis with specific DNA primers bracketing PRRIV or PRRIII. ChIP assays using anti-CREB antibody showed that CREB constitutively bound to PRRIV, as shown above in vitro. PRRIV-specific PCR products were generated using phospho-CREB, c-Jun/AP-1, c-Fos, and CBP antibodies only from activated T-cell but not resting T-cell chromatin samples in vivo (Fig. 7A). After T-cell activation, AP-1 apparently cooperated with phosphorylated CREB and CBP to bind to PRRIV. In agreement with EMSAs, ATF-2 was not bound in vivo to PRRIV in primary T cells, like the irrelevant Ets1/2 (Fig. 7A). Conversely, control experiments showed that DNA-protein complexes from CD3-CD28-stimulated primary T cells contained Ets1/2 binding to the PRRIII target sequence, as expected from our previous observations (50), but did not contain CREB (Fig. 7B). Taken together, these results demonstrate that in human resting T cells, CREB constitutively binds to the genomic region containing PRRIV and that T-cell activation triggers the binding of Ser133-phosphorylated CREB with its coactivator CBP and with inducible AP-1 family factors.
Exogenous expression of CREB/ATF and AP-1 family members in COS cells induces synergistic transactivation of PRRIV/CD28rE.
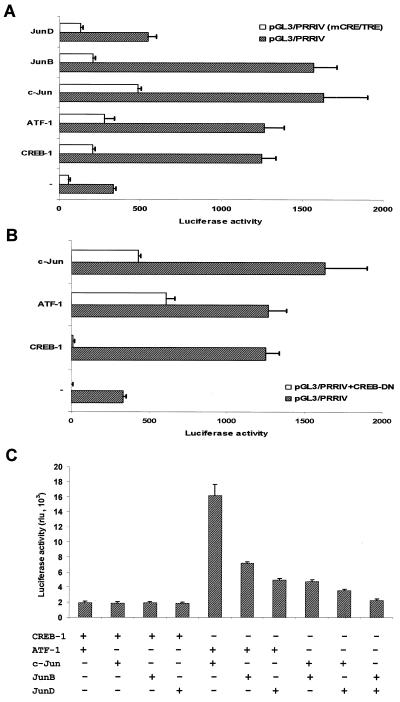
We next investigated the role of CREB-1, ATF-1, and the members of Jun/AP-1 family in transactivating PRRIV/CD28rE, using cotransfection experiments in COS-1 cells. As shown in Fig. 8A, CREB-1, ATF-1, c-Jun, and JunB efficiently transactivated pGL3/PRRIV whereas JunD had a much weaker effect. The disruption of the crucial CRE/TRE motif within PRRIV/CD28rE almost completely abolished these transactivation effects, confirming the DNA specificity of the action on PRRIV/CD28rE.
FIG. 8.
Functional cooperation between CREB/ATF and c-Jun/AP-1 family proteins results in optimal transcriptional activity of PRRIV and depends on the CRE/TRE motif. (A) Transactivation of PRRIV/CD28rE by CREB/ATF and c-Jun/AP-1 family members. COS-1 cells were transfected with expression vectors for CREB-1 (2 μg), ATF-1 (1 μg), c-Jun (1 μg), Jun B (1 μg), or Jun D (1 μg), together with wild-type or mutated pGL3/PRRIV (mCRE/TRE) luciferase reporter constructs (0.5 μg). (B) A CREB dominant negative expression vector inhibited PRRIV/CD28rE transactivation. COS 1 cells were transfected with expression vectors for CREB-1 (2 μg), ATF-1 (1 μg), or c-Jun (1 μg), together with pGL3/PRRIV (0.5 μg) alone or pGL3/PRRIV (0.5 μg) plus CREB dominant negative form (CREB-DN) (1 μg). (C) Functional cooperation between CREB/ATF and c-Jun/AP-1 family members. Cotransfection of COS 1 cells was performed with the same expression vectors and luciferase reporter constructs as those used in the experiment in panel A. rlu, relative light units.
Since phosphorylation of CREB at Ser133 is required for its transcriptional activity but not for its DNA binding ability (37, 55), a CREB dominant negative expression vector containing an alanine substitution at Ser133 (CREB DN) (49) was used to demonstrate the specificity of the transactivation observed with the exogenous expression of CREB-1, ATF-1, and c-Jun in COS-1 cells. Overexpression of CREB DN lead to transcriptional repression through competitive quenching of transactivation. As expected, CREB DN completely abolished the transcriptional activity of pGL3/PRRIV induced by CREB-1 and significantly reduced the effects of ATF-1 and c-Jun (Fig. 8B).
Combined cotransfection of CREB-1, ATF-1, c-Jun, JunB, and JunD expression vectors and the PRRIV/CD28rE luciferase reporter plasmid construct showed a higher activity and synergy between ATF-1 and c-Jun (Fig. 7C). The ATF-1/c-Jun combination induced a nearly eightfold-higher transactivation of PRRIV/CD28rE compared to other tested expression vectors.
Taken together, these data strongly suggest a functional cooperativity between the CREB/ATF family and AP-1 family in the transactivation of PRRIV/CD28rE.
DISCUSSION
In this work, we have identified and characterized an inducible DNase I HS site, DH site 4, located 5′ of the human IL-2Rα gene. Several arguments support the hypothesis that DH site 4 functions as a distal IL-2Rα enhancer in vivo, cooperating with the previously characterized IL-2Rα promoter and regulatory regions to enable optimal TCR-CD3 and CD28 T-cell-specific expression of IL-2Rα. First, the previously characterized promoter and regulatory regions are clearly insufficient to support high-level and CD28-specific transcription of a linked reporter gene (2, 14, 25, 26, 32, 57, 58), and thus distal regulatory regions must be implicated. Second, DH site 4 is contained in a TCR-CD3 and CD28-responsive DNA fragment (PRRIV) of the IL-2Rα gene. Third, the inducible reinforcement of DH site 4 mirrors the features of endogenous IL-2Rα transcription as well as the properties of the PRRIV enhancer in transient-transfection assays. Fourth, the PRRIV enhancer corresponds to a new bona fide CD28-responsive enhancer and contains a functional CRE/TRE motif with several characteristics similar to but distinct from those of the previously reported CD28RE (10, 16, 62).
In the early stage of T-cell activation, CD28 plays a critical role in amplifying the downstream signals of TCR-CD3, but later, absence of the CD28 signal leads to T-cell anergy or triggers apoptosis (52). Interestingly, we observed that anti-CD28 MAbs alone are sufficient to induce the transcriptional activity of PRRIV/CD28rE in transient-transfection assays. This new enhancer might offer useful tool to facilitate the analysis of CD28 signaling in absence of TCR-CD3 engagement.
Efficient production of IL-2 and expression of IL-2Rα by T cells are known to depend on the synergistic activation of T-cell antigen receptor and CD28 molecules on T cells. Several reports have also evidenced that CD28 signal transduction increases the transcriptional activity of the IL-2, IL-4, IL-6, IL-8 (65), IL-3, granulocyte-macrophage colony-stimulating factor, and gamma interferon proximal promoter/enhancers (9, 10, 16, 17, 38, 62). It has been demonstrated that the costimulatory signal acts through κB-like elements called CD28RE within the proximal promoter-enhancer regions of these important immunoregulatory genes. Taken together, these observations suggest that CD28-mediated gene expression is sustained by a synergistic cooperation between several transcriptional factors belonging to unrelated families, such as NF-κB, AP-1, NF-AT, and CREB/ATF. Two different classes of CD28RE exist that differ in their NF-κB binding activity despite sequence similarity (10). The pivotal role played by Rel/NF-κB proteins in CD28 responsiveness has been confirmed for many important immunoregulatory genes (18) and can be in part credited to their presence in the inducible CD28RC. The PRRIV/CD28rE identified in our study differs markedly from the previously characterized CD28 response element since it presents no sequence similarity to known CD28RE. Furthermore, its CD28 responsiveness depends on a crucial CRE/TRE motif, and no members of the Rel/NF-κB family were bound in vitro to this regulatory element. In addition, PRRIV/CD28rE can be activated by CD28 stimulation alone whereas all other CD28RE required additional signals provided by CD3-TCR or mitogenic drugs. This new core enhancer hence defines a distinct class of CD28RE.
In agreement with the identification of CREB/ATF and AP-1 family members bound to the IL-2Rα CRE/TRE site in primary and tumoral T cells, a previous report has established that CD28 responsiveness is conferred by a composite element including the CD28RE and the adjacent NF-IL-2B AP-1 sites in the promoter region of the human IL-2 gene (54). The CD28 costimulatory signals can activate CREB/ATF-1 in primary T lymphocytes (24), and the coordinate transactivation of the IL-2 CD28RE apparently involves cross talk of c-Rel and ATF-1/CREB2 in Jurkat T cells (5). Our results suggest that CREB, ATF-1, c-Jun, Jun B, and Jun D are involved in the transactivation of CRE/TRE on the basis of transient-overexpression experiments. This conclusion is strongly reinforced by the repressor effect of the CREB dominant negative form that binds to CREs but has no transcriptional activity. Furthermore, the ChIP assays have clearly established that T-cell activation triggers the binding of phospho-CREB with its coactivator CREB binding protein (CBP) and with inducible AP-1 family factors to the genomic region containing PRRIV. Therefore, our results clearly point to a pivotal role for the CREB/ATF and AP-1 family proteins in the regulation of PRRIV transcriptional activity in response to TCR and/or CD28 that may require their coordinated interactions.
The importance of Ser133 phosphorylation is demonstrated by the phenotype of transgenic mice expressing the dominant negative CREB (alanine-133-CREB) (3, 59), which are deficient for thymocyte proliferation and IL-2 production (3). In contrast to the canonical model of CREB/ATF activation (15), we observed that phospho-Ser133 CREB and phospho-Ser63 ATF-1 can bind to PRRIV CRE/TRE site in resting primary T cells. Experiments performed with serum-free medium excluded the possibility that serum factors accounted for this discrepancy. However, the constitutive phosphorylation of CREB at Ser133 might be triggered by stress kinases during primary T-cell purification. In line with the constitutive detection of CREB/ATF proteins bound to CRE/TRE, CREB is constitutively targeted to the nucleus via a nuclear localization sequence within the C-terminal basic region (20, 63). Also, DNA binding studies of recombinant CREB indicate that Ser133 phosphorylation has no effect on CREB binding to its cognate binding site CRE (21) but plays a crucial role in activation of CREB transcription potential. CRE/TRE within PRRIV/CD28rE contains a single TGACG motif, and Ser133 phosphorylation increased CREB binding to such low-affinity CRE sites (42). The increase of phospho-CREB expression after stimulation detected by both Western blotting and ChIP assays suggests that additional phosphorylation is required to fully activate CREB transcriptional activity. Although CREB/ATF family proteins constitutively bind to the CRE/TRE site, our observations are in agreement with a critical role in IL-2Rα gene expression in response to CD28 and/or TCR.
We cannot exclude at this stage the involvement of other transcription factors in PRRIV/CD28rE transcriptional regulation since it contains two NF-AT sites and one AP-1 site besides the CRE/TRE motif. However, site-directed mutagenesis and reconstitution experiments in CD28-deficient Jurkat JF3 T cells have firmly established the crucial role played by this regulatory element in CD28 responsiveness. The CD28 signal transduction pathways involve a complex network of several protein kinases and adapters that ultimately results in T-cell proliferation, cytokine secretion, and a sustained T-cell effector response. Thus, the pleiotropic physiological functions of CD28 appear to be sustained by multiple downstream transcription factors that are yet to be fully defined. In this context, the CD28-deficient JF3 T-cell clone provides a novel convenient model to further dissect the CD28-specific functions and signaling pathway.
Recent studies have identified distal elements that mediate long-range cytokine gene regulation and have implicated chromatin reorganization of cytokine gene loci (1). In a previous report (2), we have proposed that preassembled protein-DNA complexes in the proximal-enhancer region of the IL-2Rα gene play a crucial role in the precommitment of T cells to express this gene. In line with this hypothesis, three weak DH sites were already detected within the distal 5′ region in resting T cells. The constitutive CREB/ATF factors bound to the crucial CRE/TRE site of DH site 4 can recruit chromatin-remodeling machineries that locally decondense chromatin. After CD3-CD28 stimulation, activation of the transcriptionally competent locus by binding of inducible phospho-CREB and AP-1 factors probably resulted in the strongly increased sensitivity of the DH4 site, as suggested by the ChIP assays. Further experiments are required to determine whether a long-range cross talk over 9 kb might functionally associate the CREB/ATF and AP-1 transcription factors bound to CRE/TRE within PRRIV/CD28rE and the long-term activated Rel/NF-κB proteins in response to CD28 signal transduction pathway bound to the proximal κB site within PRRI (2, 13). Furthermore, the characterization for the first time of a cluster of four DH sites located between kb −4 to −9 relative to the major transcription initiation site in the human IL-2Rα gene locus opens new perspectives to our understanding of the transcriptional regulation of this important immunoregulatory gene.
ACKNOWLEDGMENTS
We thank V. Coulon for pcDNA3-c-Jun, pcDNA-Jun B, and pcDNA-Jun D; J.-M. Blanchard for pCMV-ATF-1; P. Quinn for pcDNA3-CREB and the CREB dominant negative form, pcDNA3-CREB (S133A); A. Moretta for anti-CD28 248 and anti-CD3 289; and R. Schwinzer for anti-CD28 BW828 (IgG2a). We also thank P. A. Baeuerle, Y. Collette, B. Kahn-Perlès, M.-Z. Lai, P. Rameil, and P. Sassone-Corsi for helpful suggestions.
This work was supported by the Institut National de la Santé et de la Recherche Médicale and by grants from Association pour la Recherche sur le Cancer and Comité des Bouches-du-Rhône de la Ligue Nationale Contre le Cancer. J.-H. Yeh was supported by a fellowship from the French government (BGF).
REFERENCES
- 1.Agarwal S, Rao A. Long-range transcriptional regulation of cytokine gene expression. Curr Opin Immunol. 1998;10:345–352. doi: 10.1016/s0952-7915(98)80174-x. [DOI] [PubMed] [Google Scholar]
- 2.Algarte M, Lecine P, Costello R, Plet A, Olive D, Imbert J. In vivo regulation of interleukin-2 receptor alpha gene transcription by the coordinated binding of constitutive and inducible factors in human primary T-cells. EMBO J. 1995;14:5060–5072. doi: 10.1002/j.1460-2075.1995.tb00188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton K, Muthusamy N, Chanyangam M, Fischer C, Clendenin C, Leiden J M. Defective thymocyte proliferation and IL-2 production in transgenic mice expressing a dominant-negative form of CREB. Nature. 1996;379:81–85. doi: 10.1038/379081a0. [DOI] [PubMed] [Google Scholar]
- 4.Bernal-Mizrachi E, Wice B, Inoue H, Permutt M A. Activation of serum response factor (SRF) in the depolarization induction of Egr-1 transcription in pancreatic islet β-cells. J Biol Chem. 2000;275:25681–25689. doi: 10.1074/jbc.M003424200. [DOI] [PubMed] [Google Scholar]
- 5.Butscher W G, Powers C, Olive M, Vinson C, Gardner K. Coordinate transactivation of the interleukin-2 CD28 response element by c-Rel and ATF-1/CREB2. J Biol Chem. 1998;273:552–560. doi: 10.1074/jbc.273.1.552. [DOI] [PubMed] [Google Scholar]
- 6.Cerdan C, Martin Y, Brailly H, Courcoul M, Flavetta S, Costello R, Mawas C, Birg F, Olive D. IL-1 alpha is produced by T lymphocytes activated via the CD2 plus CD28 pathways. J Immunol. 1991;146:560–564. [PubMed] [Google Scholar]
- 7.Cerdan C, Martin Y, Courcoul M, Brailly H, Mawas C, Birg F, Olive D. Prolonged IL-2 receptor alpha/CD25 expression after T cell activation via the adhesion molecules CD2 and CD28. Demonstration of combined transcriptional and post-transcriptional regulation. J Immunol. 1992;149:2255–2261. [PubMed] [Google Scholar]
- 8.Chambers C A, Allison J P. Costimulatory regulation of T cell function. Curr Opin Cell Biol. 1999;11:203–210. doi: 10.1016/s0955-0674(99)80027-1. [DOI] [PubMed] [Google Scholar]
- 9.Civil A, Geerts M, Aarden L A, Verweij C L. Evidence for a role of CD28RE as a response element for distinct mitogenic T cell activation signals. Eur J Immunol. 1992;22:3041–3043. doi: 10.1002/eji.1830221142. [DOI] [PubMed] [Google Scholar]
- 10.Civil A, Rensink I, Aarden L A, Verweij C L. Functional disparity of distinct CD28 response elements toward mitogenic responses. J Biol Chem. 1999;274:34369–34374. doi: 10.1074/jbc.274.48.34369. [DOI] [PubMed] [Google Scholar]
- 11.Cockerill P N, Shannon M F, Bert A G, Ryan G R, Vadas M A. The granulocyte-macrophage colony-stimulating factor/interleukin 3 locus is regulated by an inducible cyclosporin A-sensitive enhancer. Proc Natl Acad Sci USA. 1993;90:2466–2470. doi: 10.1073/pnas.90.6.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa J J, Keffer J M, Goff J P, Metcalfe D D. Aphidicolin-induced proliferative arrest of murine mast cells: morphological and biochemical changes are not accompanied by alterations in cytokine gene induction. Immunology. 1992;76:413–421. [PMC free article] [PubMed] [Google Scholar]
- 13.Costello R, Lipcey C, Algarte M, Cerdan C, Baeuerle P A, Olive D, Imbert J. Activation of primary human T-lymphocytes through CD2 plus CD28 adhesion molecules induces long-term nuclear expression of NF-κB. Cell Growth Differ. 1993;4:329–339. [PubMed] [Google Scholar]
- 14.Cross S L, Feinberg M, Wolf J B, Holbrook N J, Wong-Staal F, Leonard W J. Regulation of the human interleukin-2 receptor alpha chain promoter: activation of a nonfunctional promoter by the transactivator gene of HTLV-I. Cell. 1987;49:47–56. doi: 10.1016/0092-8674(87)90754-9. [DOI] [PubMed] [Google Scholar]
- 15.De Cesare D, Sassone-Corsi P. Transcriptional regulation by cyclic AMP-responsive factors. Prog Nucleic Acid Res Mol Biol. 2000;64:343–369. doi: 10.1016/s0079-6603(00)64009-6. [DOI] [PubMed] [Google Scholar]
- 16.Fraser J D, Irving B A, Crabtree G R, Weiss A. Regulation of interleukin-2 gene enhancer activity by the T cell accessory molecule CD28. Science. 1991;251:313–316. doi: 10.1126/science.1846244. [DOI] [PubMed] [Google Scholar]
- 17.Fraser J D, Weiss A. Regulation of T-cell lymphokine gene transcription by the accessory molecule CD28. Mol Cell Biol. 1992;12:4357–4363. doi: 10.1128/mcb.12.10.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerondakis S, Grumont R, Rourke I, Grossmann M. The regulation and roles of Rel/NF-κB transcription factors during lymphocyte activation. Curr Opin Immunol. 1998;10:353–359. doi: 10.1016/s0952-7915(98)80175-1. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh P, Tan T H, Rice N R, Sica A, Young H A. The interleukin 2 CD28-responsive complex contains at least three members of the NFκB family: c-Rel, p50, and p65. Proc Natl Acad Sci USA. 1993;90:1696–1700. doi: 10.1073/pnas.90.5.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez G A, Montminy M R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 21.Hagiwara M, Brindle P, Harootunian A, Armstrong R, Rivier J, Vale W, Tsien R, Montminy M R. Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol Cell Biol. 1993;13:4852–4859. doi: 10.1128/mcb.13.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagmeyer B M, Angel P, van Dam H. Modulation of AP-1/ATF transcription factor activity by the adenovirus-ElA oncogene products. Bioessays. 1995;17:621–629. doi: 10.1002/bies.950170708. [DOI] [PubMed] [Google Scholar]
- 23.Herzig R P, Scacco S, Scarpulla R C. Sequential serum-dependent activation of CREB and NRF-1 leads to enhanced mitochondrial respiration through the induction of cytochrome c. J Biol Chem. 2000;275:13134–13141. doi: 10.1074/jbc.275.17.13134. [DOI] [PubMed] [Google Scholar]
- 24.Hsueh Y P, Liang H E, Ng S Y, Lai M Z. CD28-costimulation activates cyclic AMP-responsive element-binding protein in T lymphocytes. J Immunol. 1997;158:85–93. [PubMed] [Google Scholar]
- 25.John S, Reeves R B, Lin J X, Child R, Leiden J M, Thompson C B, Leonard W J. Regulation of cell-type-specific interleukin-2 receptor alpha-chain gene expression: potential role of physical interactions between Elf-1, HMG- I(Y), and NF-κB family proteins. Mol Cell Biol. 1995;15:1786–1796. doi: 10.1128/mcb.15.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.John S, Robbins C M, Leonard W J. An IL-2 response element in the human IL-2 receptor alpha chain promoter is a composite element that binds Stat5, Elf-1, HMG-I(Y) and a GATA family protein. EMBO J. 1996;15:5627–5635. [PMC free article] [PubMed] [Google Scholar]
- 27.June C H, Bluestone J A, Nadler L M, Thompson C B. The B7 and CD28 receptor families. Immunol Today. 1994;15:321–331. doi: 10.1016/0167-5699(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 28.Kanamori H, Suzuki N, Siomi H, Nosaka T, Sato A, Sabe H, Hatanaka M, Honjo T. HTLV-1 p27rex stabilizes human interleukin-2 receptor chain mRNA. EMBO J. 1990;9:4161–4166. doi: 10.1002/j.1460-2075.1990.tb07639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kempiak S J, Hiura T S, Nel A E. The Jun kinase cascade is responsible for activating the CD28 response element of the IL-2 promoter: proof of cross-talk with the I kappa B kinase cascade. J Immunol. 1999;162:3176–3187. [PubMed] [Google Scholar]
- 30.Kuang A A, Novak K D, Kang S M, Bruhn K, Lenardo M J. Interaction between NF-kappa B and serum response factor-binding elements activates an interleukin-2 receptor alpha-chain enhancer specifically in T lymphocytes. Mol Cell Biol. 1993;13:2536–2545. doi: 10.1128/mcb.13.4.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai J H, Horvath G, Subleski J, Bruder J, Ghosh P, Tan T H. RelA is a potent transcriptional activator of the CD28 response element within the interleukin 2 promoter. Mol Cell Biol. 1995;15:4260–4271. doi: 10.1128/mcb.15.8.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lecine P, Algarte M, Rameil P, Beadling C, Bucher P, Nabholz M, Imbert J. Elf-1 and Stat5 bind to a critical element in a new enhancer of the human interleukin-2 receptor alpha gene. Mol Cell Biol. 1996;16:6829–6840. doi: 10.1128/mcb.16.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenschow D J, Walunas T L, Bluestone J A. CD28/B7 system of T cell costimulation. Annu Rev Immunol. 1996;14:233–258. doi: 10.1146/annurev.immunol.14.1.233. [DOI] [PubMed] [Google Scholar]
- 34.Leonard W J, Kronke M, Pfeffer N J, Depper J M, Greene W C. Interleukin 2 receptor gene expression in normal human T lymphocytes. Proc Natl Acad Sci USA. 1985;82:6281–6285. doi: 10.1073/pnas.82.18.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin B B, Cross S L, Halden N F, Roman D G, Toledano M B, Leonard W J. Delineation of an enhancer like positive regulatory element in the interleukin-2 receptor alpha-chain gene. Mol Cell Biol. 1990;10:850–853. doi: 10.1128/mcb.10.2.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linsley P S, Ledbetter J A. The role of the CD28 receptor during T cell responses to antigen. Annu Rev Immunol. 1993;11:191–212. doi: 10.1146/annurev.iy.11.040193.001203. [DOI] [PubMed] [Google Scholar]
- 37.Liu F, Thompson M A, Wagner S, Greenberg M E, Green M R. Activating transcription factor-1 can mediate Ca(2+)− and cAMP-inducible transcriptional activation. J Biol Chem. 1993;268:6714–6720. [PubMed] [Google Scholar]
- 38.Li-Weber M, Giasi M, Krammer P H. Involvement of Jun and Rel proteins in up-regulation of interleukin-4 gene activity by the T cell accessory molecule CD28. J Biol Chem. 1998;273:32460–32466. doi: 10.1074/jbc.273.49.32460. [DOI] [PubMed] [Google Scholar]
- 39.Maggirwar S B, Harhaj E W, Sun S C. Regulation of the interleukin-2 CD28-responsive element by NF-ATp and various NF-κB/Rel transcription factors. Mol Cell Biol. 1997;17:2605–2614. doi: 10.1128/mcb.17.5.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mari B, Imbert V, Belhacene N, Far D F, Peyron J F, Pouyssegur J, Van Obberghen-Schilling E, Rossi B, Auberger P. Thrombin and thrombin receptor agonist peptide induce early events of T cell activation and synergize with TCR cross-linking for CD69 expression and interleukin 2 production. J Biol Chem. 1994;269:8517–8523. [PubMed] [Google Scholar]
- 41.Nelson B H, Willerford D M. Biology of the interleukin-2 receptor. Adv Immunol. 1998;70:1–81. doi: 10.1016/s0065-2776(08)60386-7. [DOI] [PubMed] [Google Scholar]
- 42.Nichols M, Weih F, Schmid W, DeVack C, Kowenz-Leutz E, Luckow B, Boshart M, Schütz G. Phosphorylation of CREB affects its binding to high and low affinity sites: implications of cAMP induced gene transcription. EMBO J. 1992;11:3337–3346. doi: 10.1002/j.1460-2075.1992.tb05412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nunes J, Klasen S, Franco M D, Lipcey C, Mawas C, Bagnasco M, Olive D. Signalling through CD28 T-cell activation pathway involves an inositolphospholipid-specific phospholipase C activity. Biochem J. 1993;293:835–842. doi: 10.1042/bj2930835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pages F, Ragueneau M, Klasen S, Battifora M, Couez D, Sweet R, Truneh A, Ward S G, Olive D. Two distinct intracytoplasmic regions of the T-cell adhesion molecule CD28 participate in phosphatidylinositol 3-kinase association. J Biol Chem. 1996;271:9403–9409. doi: 10.1074/jbc.271.16.9403. [DOI] [PubMed] [Google Scholar]
- 45.Parekh B S, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 46.Pennypacker K R. AP-1 transcription factor complexes in CNS disorders and development. J Fla Med Assoc. 1995;82:551–554. [PubMed] [Google Scholar]
- 47.Pierce J W, Jamieson C A, Ross J L, Sen R. Activation of IL-2 receptor alpha-chain gene by individual members of the rel oncogene family in association with serum response factor. J Immunol. 1995;155:1972–1980. [PubMed] [Google Scholar]
- 48.Plet A, Huet X, Algarte M, Rech J, Imbert J, Philips A, Blanchard J M. Relief of cyclin A gene transcriptional inhibition during activation of human primary T lymphocytes via CD2 and CD28 adhesion molecules. Oncogene. 1997;14:2575–2583. doi: 10.1038/sj.onc.1201103. [DOI] [PubMed] [Google Scholar]
- 49.Quinn P G. Distinct activation domains within cAMP response element-binding protein (CREB) mediate basal and cAMP-stimulated transcription. J Biol Chem. 1993;268:16999–17009. [PubMed] [Google Scholar]
- 50.Rameil P, Lecine P, Ghysdael J, Gouilleux F, Kahn-Perles B, Imbert J. IL-2 and long-term T cell activation induce physical and functional interactions between STAT5 and ETS transcription factors in human T cells. Oncogene. 2000;19:2086–2097. doi: 10.1038/sj.onc.1203542. [DOI] [PubMed] [Google Scholar]
- 51.Roifman C M, Dadi H K. Human interleukin-2 receptor alpha deficiency. Pediatr Res. 2000;48:6–11. doi: 10.1203/00006450-200007000-00004. [DOI] [PubMed] [Google Scholar]
- 52.Rudd C E. Upstream-downstream: CD28 cosignaling pathways and T cell function. Immunity. 1996;4:527–534. doi: 10.1016/s1074-7613(00)80479-3. [DOI] [PubMed] [Google Scholar]
- 53.Schorle H, Holtschke T, Hunig T, Schimpl A, Horak I. Development and function of T cells in mice rendered interleukin-2 deficient by gene targeting. Nature. 1991;352:621–624. doi: 10.1038/352621a0. [DOI] [PubMed] [Google Scholar]
- 54.Shapiro V S, Truitt K E, Imboden J B, Weiss A. CD28 mediates transcriptional upregulation of the interleukin-2 (IL-2) promoter through a composite element containing the CD28RE and NF-IL-2B AP-1 sites. Mol Cell Biol. 1997;17:4051–4058. doi: 10.1128/mcb.17.7.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheng M, Thompson M A, Greenberg M E. CREB: a Ca(2+)-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- 56.Siefken R, Klein-Hessling S, Serfling E, Kurrle R, Schwinzer R. A CD28-associated signaling pathway leading to cytokine gene transcription and T cell proliferation without TCR engagement. J Immunol. 1998;161:1645–1651. [PubMed] [Google Scholar]
- 57.Soldaini E, Pla M, Beermann F, Espel E, Corthesy P, Barange S, Waanders G A, MacDonald H R, Nabholz M. Mouse interleukin-2 receptor alpha gene expression. Delimitation of cis-acting regulatory elements in transgenic mice and by mapping of DNAse-I hypersensitive sites. J Biol Chem. 1995;270:10733–10742. doi: 10.1074/jbc.270.18.10733. [DOI] [PubMed] [Google Scholar]
- 58.Sperisen P, Wang S M, Soldaini E, Pla M, Rusterholz C, Bucher P, Corthesy P, Reichenbach P, Nabholz M. Mouse interleukin-2 receptor alpha gene expression—Interleukin-1 and interleukin-2 control transcription via distinct cis-acting elements. J Biol Chem. 1995;270:10743–10753. doi: 10.1074/jbc.270.18.10743. [DOI] [PubMed] [Google Scholar]
- 59.Struthers R S, Vale W W, Arias C, Sawchenko P E, Montminy M R. Somatotroph hypoplasia and dwarfism in transgenic mice expressing a non-phosphorylatable CREB mutant. Nature. 1991;350:622–624. doi: 10.1038/350622a0. [DOI] [PubMed] [Google Scholar]
- 60.Su B, Jacinto E, Hibi M, Kallunki T, Karin M, Ben-Neriah Y. JNK is involved in signal integration during costimulation of T lymphocytes. Cell. 1994;77:727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 61.Taniguchi T, Minami Y. The IL-2/IL-2 receptor system: a current overview. Cell. 1993;73:5–8. doi: 10.1016/0092-8674(93)90152-g. [DOI] [PubMed] [Google Scholar]
- 62.Verweij C L, Geerts M, Aarden L A. Activation of interleukin-2 gene transcription via the T-cell surface molecule CD28 is mediated through an NF-κB-like response element. J Biol Chem. 1991;266:14179–14182. [PubMed] [Google Scholar]
- 63.Waeber G, Habener J F. Nuclear translocation and DNA recognition signals colocalized within the bZIP domain of cyclic adenosine 3′,5′-monophosphate response element-binding protein CREB. Mol Endocrinol. 1991;5:1431–1438. doi: 10.1210/mend-5-10-1431. [DOI] [PubMed] [Google Scholar]
- 64.Ward S G. CD28: a signalling perspective. Biochem J. 1996;318:361–377. doi: 10.1042/bj3180361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wechsler A S, Gordon M C, Dendorfer U, LeClair K P. Induction of IL-8 expression in T cells uses the CD28 costimulatory pathway. J Immunol. 1994;153:2515–2523. [PubMed] [Google Scholar]
- 66.Willerford D M, Chen J, Ferry J A, Davidson L, Ma A, Alt F W. Interleukin-2 receptor alpha chain regulates the size and content of the peripheral lymphoid compartment. Immunity. 1995;3:521–530. doi: 10.1016/1074-7613(95)90180-9. [DOI] [PubMed] [Google Scholar]
- 67.Wu C. Analysis of hypersensitive sites in chromatin. Methods Enzymol. 1989;170:269–289. doi: 10.1016/0076-6879(89)70052-5. [DOI] [PubMed] [Google Scholar]