Abstract
Bone morphogenetic proteins (BMPs) are growth factors belonging to the TGF-β (Transforming Growth Factor β) superfamily. BMPs were found to regulate multiple cell processes such as proliferation, survival, differentiation and apoptosis. They were originally described to play a pivotal role in inducing bone, cartilage, ligament, and tendon formation at both heterotopic and orthotopic sites, but were found to play significant role in embryogenesis, and development of multiple tissues and organs. Activities of BMPs are regulated by a number of secreted proteins, which modulate their availability to bind cellular receptors. The functions of individual BMPs are highly redundant due to binding the same receptors and inducing overlapping signal transduction pathways. Recently, BMPs were found to regulate cells of the innate and adaptive immune system. BMPs are involved in thymic development of T cells at the early, double negative, as well as later, double positive, stages of thymopoesis. They specifically modulate thymic development of regulatory T cells (Treg). In the periphery, BMPs affect T cell activation, promoting generation of Treg cells. We found that mice deficient for one of the receptors activated by BMPs demonstrated slower growth of transplantable melanoma tumors.
Keywords: regulatory T cells, CD4+ T cells, cancer, BMPR1A
Introduction
Bone Morphogenic Proteins (BMPs) is a protein family acting during vertebrate embryogenesis and organogenesis by regulating tissue patterning. They determine cell fate during lineage determination and in adults regulate self-renewal of tissues of multiple organs including hematopoietic system. The term “BMP” was proposed in late 1960s when it was observed that proteins contained in the demineralized bone segments implanted into muscles were able to induce ectopic bone formation (Urist, 1965). It is now apparent that besides inducing differentiation of bone-forming cells – osteoblasts, BMPs regulate stem cell renewal and epithelial - mesenchymal transition. BMPs are involved in differentiation and maintenance of multiple tissues and organs including bones, teeth, cartilage and non-osseous tissues of lungs, heart, kidneys and vessels (Ferguson and Anderson, 1992; Zou and Niswander, 1996). BMPs’ role in organogenesis is dependent on their potential to control multiple cell functions: proliferation, adhesion, migration, apoptosis and differentiation.
Despite ubiquitous expression and general significance it took the next twenty years for BMPs to be cloned and characterized (Wozney et al., 1988). The role of BMPs in individual organs is subject of active research and their involvement in regulating the development, homeostasis and functions of the immune system started to be appreciated only recently. Since BMPs and their receptors are expressed by cells of innate and adaptive immune system as well as non-immune cells, they are involved in communication between organ stromal cells and infiltrating hematopoietic cells. Defects in BMP production/signaling were found to lead to auto-immunity, inflammation and even cancer. Examining the role of BMPs in the immune system may explain the basis of many pathological conditions and lead to the design of novel therapeutic strategies.
Bone Morphogenic Proteins and their receptors
BMPs represent the largest subgroup of the TGF-β superfamily which also includes activins, nodal proteins, Mullerian Inhibiting Substance, Growth and Differentiation Factors and TGF-β itself (Carreira et al., 2014). BMPs are further subdivided into four subfamilies based on aminoacid sequence homology. Despite structural similarities individual BMPs my exert opposing influences. For example, BMP2, 4 and 7 are known for their osteogenic function, while BMP3 decreases bone density and BMP13 strongly inhibits bone formation (Mueller and Nickel, 2012). To date, 20 BMPs were discovered and characterized in humans. BMPs are formed as large (400-500 amino acids) precursor proteins (Xiao et al., 2007). Their N-terminal fragment is a signal peptide important for the folding of mature protein; this fragment is cleaved during BMP activation (i.e. BMP4 is activated by a protease furin) (Nelsen and Christian, 2009). BMPs are synthesized as large, inactive precursors, which undergo posttranslational processing and act as disulfide-linked homo and heterodimer glycoproteins. Once secreted, BMPs may associate with molecules present in extracellular space which include soluble proteins: noggin, chordin, gremlin or follistatin, membrane/matrix associated proteins like chondroitin sulfate small leucine-rich proteins, co-receptors and pseudoreceptors like BAMBI (BMP and Activin Membrane-Bound Inhibitor) and matrix protein fibrin (Umulis et al., 2009). These interactions regulate BMP availability to bind membrane receptors and control timing of BMP signals. Matrix proteins may also stimulate BMP signaling by increasing activation and release of BMP, increasing movement of BMPs, protecting them from endocytosis and degradation, acting as shuttles to redistribute BMP ligands and increase their concentration far from sites where these proteins are secreted. This complex system of BMP binding proteins regulating their maturation by proteolytic cleavage, degradation, availability, diffusion, ensures that appropriate BMP gradient is established in tissues.
BMPs and other members of TGF-β family bind two types of membrane receptors, which have cytoplasmic domains with activity of serine/threonine kinases. Type I receptors are 50kD proteins while type II receptors are larger (70-80kD). There are seven type I receptors of TGF-β family: ALK1 (ACVRL1), ALK2 (ACVR1), ALK3 (BMPR1A), ALK4 (ACVR1B), ALK5 (TGFβRI), ALK6 (BMPR1B) and ALK-7 (ACVR1C) and four type II receptors, BMPR2, ACTRIIA, ACTRIIB and TGFβRII. Receptor signaling is initiated by binding of a ligand with one of type I receptors (except BMP6 which first binds type II receptor). Ligand binding induces association with type II receptor which leads to a conformational change of a heteromeric receptor γ ligand complex and trans-phosphorylation of type I receptor which induces kinase activity of type I receptors. Activated type I receptors in turn phosphorylate receptor-regulated transcription factors, R-Smads. Binding of TGF-β or activins results in phosphorylation of Smad2 and 3 while binding of a BMP ligand leads to phosphorylation of Smad1, 5 and 8. SARA (Smad anchor for receptor activation) binds Smad2/3 and facilitates their interaction with receptors. Phosphorylated Smads associate with a co-Smad, Smad4, and the whole complex translocates to the nucleus where it acts as a transcription factor by further interaction with multiple co-activators or co-repressors to regulate gene expression (Akhurst and Hata, 2012).
BMPs and other members of TGF-β family, e.g. activins, were found to associate with multiple receptors and individual receptors may bind more than one ligand. Three of type I receptors, BMPR1A (ALK3), BMPR1B (ALK6) and ACVR2A (activin receptor, ALK2), bind BMPs. Of four TGF-β type II receptors, three, BMPR2, ACVR2A and ACVR2B, bind BMP ligands. Type I receptors BMPR1A, BMPR1B and type II receptor BMPR2 are specific for BMPs and ACVR1A, ACVR2A and ACVR2B can bind both BMPs and activins (Kawabata et al., 1998). This promiscuity of ligand receptor interactions contributes to redundant functions of BMPs but also makes it difficult to define roles of individual ligand-receptor complexes.
Signaling pathways
BMP signaling is highly dependent on cellular and environmental context and interactions with multiple proteins modulate availability and functional concentrations of BMPs and ensure that gradients of BMPs that regulate tissue patterning in embryogenesis and organogenesis are established and maintained. Once BMPs bind their receptors they initiate signaling through canonical pathway, involving phosphorylation of Smads, and through multiple non-canonical pathways. In addition to canonical and non-canonical pathways BMP signaling is influenced by cross talk with different signaling pathways, including the PI3K–AKT, WNT, Hedgehog, Notch, interferon (IFN), TNF and RAS pathways. Often signaling modules involving TGFβ/BMPs/activins are studied only in certain cell types or activation states making it difficult to put together a comprehensive picture of signaling circuits and assessing their significance.
Binding of BMPs and activins to BMP receptors induces phosphorylation of Smad1/5/8 or Smad2/3 respectively. Upon phosphorylation both subsets of R-Smads associate with Smad4 and translocate into nucleus to regulate gene expression. DNA motif GTCT/AGAC bound by Smad2/3 (Smad binding element) and GGCGCC/GGAGCC motif bound by Smad1/5/8 (BMP binding element) are present in promoters or enhancers of many genes, including genes regulating lineage commitment of T cell subsets and T cell functions like Runx, Schnurri, C/EBPa, Foxo and Foxp3 (Itoh et al., 2004; Korchynskyi and ten, 2002; Kusanagi et al., 2000; Lin et al., 2006; Morikawa et al., 2011). Id gene (Inhibitors of DNA Binding) family is one of the most studied genes downstream of BMP receptors (Miyazono et al., 2010). The Id proteins inhibit lineage commitment by binding and sequestering basic helix-loop-helix transcription factors and have been implicated in regulating invasion, proliferation, and survival of various cell types. Id3-deficient T cells were found to poorly upregulate Foxp3, what led to defective generation of regulatory CD4+Foxp3+ cells (Treg), accumulation of Th17 cells and autoimmune disease (Maruyama et al., 2011). Another target of BMP induced Smads is Runx family of transcription factors important for regulating T cell functions (Javed et al., 2008). Runx1-3 are Runt-related heteromeric transcription factors consisting of a DNA-binding α-subunits specific for individual proteins and a common non-DNA binding β subunit. In T cells Runx1 and 3 were found to attenuate Th2 cell differentiation and mice with CD4+ T cells deficient in these proteins are prone to Th2 mediated diseases like asthma (Naoe et al., 2007). Transgenic expression of Runx3 augments Th1 cytokine production (Kohu et al., 2009). In Treg cells Runx1 associates with Foxp3 controlling their suppressor function (Ono et al., 2007; Rudra et al., 2012). It is not been extensively studied to what extent Smad proteins responding to BMP signaling bind to Runx family proteins in T cells but such interactions were found in other cell types (Javed et al., 2008). BMP-specific R-Smads1/5/8 are involved in feedback regulation of transcription of inhibitory Smad6, Smad7 and a decoy receptor BAMBI. Signal transduction mediated by activated R-Smads may be modulated by interactions with inhibitory Smad6 or Smad7 (I-Smads), which block their association with common Smad4 and nuclear translocation. Expression of Smad 7 is increased by IFNγ (acting through Jak1/STAT4) and proinflammatory cytokines TNFα and IL-1β (acting through NFκB/RelA).
Another mechanism which antagonizes TGF-γ/BMP signaling involves interactions with other signaling pathways, like phosphorylation of linker region of R-SMADs by CDK8/9, activation of mitogen-activated protein kinases ERK1/2 or GSK-3β (Kretzschmar et al., 1997) (Guo et al., 2008). ERK1/2 may act downstream of Ras, which is activated by TCR signaling (Kretzschmar et al., 1999). Reduction in the expression of Axin/GSK-3β leads to increased Smad3 stability and transcriptional activity. Wnt signaling, which is known to inactivate GSK-3β, reduces Smad ubiquitination and stabilizes the protein. Linker region phosphorylation (different from phosphorylation by BMP receptors) marks R-SMADs for recognition by HECT type E3 ubiquitin ligases SMURF1, SMURF2/NEDD4L, NEDD4-2 which mediate their polyubiquitination and degradation (Sapkota et al., 2007). We have found that Nedd4 is expressed in activated Foxp3+ Treg cells. OTUB1 (ubiquitin thioesterase) protects pSmad2/3 from ubiquitination and proteolytic degradation (Herhaus et al., 2013). In summary, phosphorylation linker region is an important determinant of steady-state stability of Smads which determines cellular sensitivity to TGF-β/BMP and integrates multiple signaling pathways. AKT (Protein Kinase B) may restrict Smad3 activity through a kinase independent mechanism involving the direct binding of AKT to Smad3, which blocks activation of Smad3 by sequestering Smad3 from TGFβRI (Conery et al., 2004). PI3K (Phosphatidylinositide 3-Kinase)/AKT may also inhibit TGF-β/BMP signaling through a kinase-dependent mechanism involving mTOR (Song et al., 2006; Wahdan-Alaswad et al., 2012). AKT is a strong repressor of induction of the Treg phenotype in vitro and in vivo so and inhibiting TGF-β/BMP signaling could complement reported mTOR-dependent inhibition of Foxp3 expression (Haxhinasto et al., 2008) (Sauer et al., 2008). Recently reported association of Smad and Foxo family transcription factors opens one more avenue how AKT could impact BMP signaling (Seoane et al., 2004) (Neurath et al., 2002). Foxo transcription factors control survival and homing of naive T cells by inducing expression of IL-7Rα, adhesion molecule CD62L and cytokine receptor CCR7 (Kerdiles et al., 2009). They also control differentiation and function of Treg cells (Kerdiles et al., 2010) (Ouyang et al., 2010b) (Ouyang et al., 2012). Activity of Foxo proteins is regulated by their subcellular localization, they complex with 14-3-3 proteins and phosphorylation diminishes their translocation into nucleus and binding to DNA. PI3K through PDK1 (Phosphoinositide Dependent Kinase-1) activates AKT and this phosphorylates Foxo proteins barring them, and associated Smads, from entering nucleus and regulating target genes.
In addition to Smad-mediated signaling, BMP receptor binding triggers activation of alternative, non-conventional pathways. BMPR signaling was found to activate MAP-kinase mediated signaling cascades. BMP2/4 binding to receptors led to phosphorylation of MKK4-JNK (and AP-1) and MKK3-p38. TAK1 (TGF-β activated kinase 1) was identified to directly phosphorylate MAP-kinases (Mao et al., 2011; Shim et al., 2009). TAK1 was proposed to act downstream of TRAF6 (TNF receptor associated factor 6) which moderates signaling of TGF-β/BMP receptors and receptors for TNF, IL-1 and Toll-like receptors (Ohkura et al., 2012). TRAF6 is a E3 ubiquitin ligase, which synthesizes polyubiquitin chains, by self-polyubiquitination of Lys63, which do not target protein for degradation but serve as docking sites to assemble signaling complexes (Yamashita et al., 2008). The significance of p38 signaling in peripheral T cells was demonstrated by showing that it is necessary for conversion to induced Treg cells (Huber et al., 2008). This conversion requires TGF-β signaling but is enhanced by BMPs and activin A which is particularly important in inflammatory responses when TGF-β concentration could be low. Interestingly, activin A have minimal effect on Foxp3 induction in thymic-derived Treg cells, highlighting a possibility of different regulation of Foxp3 gene expression in the thymus and peripheral Treg cells (Huber et al., 2009) . The synergistic effect of TGFβ and BMP/activins was dependent on activation of ERK1/2 and JNK and could be blocked by specific small molecule inhibitors (Lu et al., 2010a). Analysis of Smad2/3 deficient mice only partially abrogated generation of induced Treg and Th17 cells. The importance of non-canonical TGF-β/BMP signaling was further revealed by demonstrating that a similar proportion of Smad2/3 deficient and sufficient mice succumbed to experimentally induced EAE suggesting that generation of Th17 cells in vivo is TGF-β dependent but Smad independent (Lu et al., 2010b). While the molecular mechanisms of BMP signaling was not studied extensively in T cells, they are known to utilize signaling pathways acted on by BMPs in other cell types what suggests that they may play more important role than currently appreciated. For example, in colon cancer cells BMPs were found to suppress PTEN expression via RAS/ERK pathway (Beck and Carethers, 2007). In T cells PTEN (Phosphatase and tensin homolog) is regulated in response to TCR signaling and affects AKT/mTOR axis, which has a major effect on effector and Treg cell differentiation in response to antigen (Powell and Delgoffe, 2010). In another example, BMP induced Smad1 interacted with β-catenin in bone marrow stromal cells impeding its transfer into nucleus (Liu et al., 2006). A crosstalk between BMP and Wnt signaling pathways in T cells is known to regulate Th17, Treg and CD8 cells (Ding et al., 2008; Muranski et al., 2011; Zhao et al., 2010). Finally, TGFβ/BMP pathways communicate with Notch signaling pathway and synergistically regulate target genes in many cell types (Guo and Wang, 2009). Activation of Notch is critical at many stages of T cell ontogeny, including original T cell lineage specification (Radtke et al., 1999). While molecular mechanisms of downstream mediators of BMP signaling with Notch are not yet known in T cells, interaction of Notch1 ICD and Smad3 was found to regulate Foxp3 transcription factor in Treg cells (Asano et al., 2008) (Samon et al., 2008). In summary, binding of BMPs to their receptors initiates complex signaling events and biological outcome is highly context dependent and relies on extensive communication with other signaling pathways.
Pathology associated with impaired or abnormal BMP signaling
BMPs are important in virtually all aspects of the organism’s development and function. Therefore, tight regulation of BMP signaling is crucial to maintain integrity and homeostasis of tissues and organs. This is originated in extracellular space by regulation of BMP availability for binding to receptors. Altered concentration of natural BMP antagonists, like noggin, gremlin or chordin during embryogenesis was shown to have detrimental developmental outcomes, including lethality (Bachiller et al., 2000). The most apparent pathology affected skeletal and nervous systems. Deletion of noggin gene caused early mortality as a result of multiple defects (failure to close neural tube, loss of caudal vertebrae) attributed to uncontrolled signaling of BMP2 and BMP4 (McMahon et al., 1998). BMPs were found to sustain stem cell renewal and regulate their differentiation, including tissue specific and cancer stem cells (Ying et al., 2003) (Chen et al., 2008) (He et al., 2004) (Reya et al., 2001). Cancer stem cells were defined as self-renewing cells that can “phenocopy” the tumor they originate from and are considered a cellular component essential for tumor persistence. Recent reports showed that tumors e.g. gliomas, colorectal cancers, developed mechanisms to tune BMP signaling by secreting gremlin. This regulated tumor invasiveness and supported persistence of non-differentiating and hence self-renewing cells (Karagiannis et al., 2013).
Attenuated BMP signaling is also associated with severe developmental and functional abnormalities. Deletion of BMP2 or BMP4 genes in experimental mice leads to embryonic lethality because of defects in mesoderm formation and BMP7-knockout mice die shortly after birth (Zhang and Evans, 1996) (Dudley et al., 1995). Both BMP2, 4 and 7 are ligands of BMPR1A and constitutive knockout of this receptor was found lethal due to defect of mesoderm formation in early gestation (Mishina et al., 1995). Using conditional knockout and mutant mice BMPs and their receptors were found essential in postnatal ontogeny. For example mutations in Smad4 and BMPR1A have been linked to the development of juvenile polyposis leading to formation of polyps in duodenum and stomach (Takaku et al., 1999). Unlike the BMPR1A-knockout mice, animals deficient in BMPR1B or BMPR2 gene functions survive past neonatal period but are prone to develop pulmonary hypertension (Chida et al., 2012; Soubrier et al., 2013).
BMPs regulate development and function of immune system cells
While BMP signaling was thoroughly investigated in multiple systems their role in immune system, particularly in regulating T cell differentiation and function of mature T cells is only emerging. The main features of BMP signaling, acting in concert and in context of local mediators, including other members of TGF-β family, make it difficult to dissect the functions of individual BMPs. Accumulating data (including our own study) suggests that BMPs and their receptors have a significant role in regulating immune responses of CD4+ T cells, particularly in tumor environment. Here we will summarize the current knowledge on the role of BMPs in T cell ontogeny.
BMPs are involved in thymic development of T cells
T cells mature and differentiate in the thymus from bone marrow derived Common Lymphoid Progenitors, a precursor cells capable of differentiating into various subsets of lymphoid cells including T and B lymphocytes and NK cells (Kuo and Leiden, 1999) (Kondo et al., 2001). Earliest T cell precursors do not express markers of mature helper and cytotoxic lymphocytes, CD4 and CD8 respectively, and are called double negative cells (DN). DN cells are further subdivided into four developmental stages based on the surface expression of CD117 (c-kit), CD44, and CD25 (IL-2 receptor α-chain). Development within this population progresses from the CD117+CD44+CD25− cells (DN1) to the CD117−CD44+CD25+ pro-T cells (DN2), and to CD117−CD44−CD25+ pre-T cells (DN3), and finally to the CD117−44−25− DN4 stage. DN2 pro-T cells initiate rearrangement of the β-chain of T-cell antigen receptor (TCR), which is completed at the DN3 stage. Productive rearrangement of the β-chain allows for expression of pre-TCR complexes, proliferation and expansion of DN3 thymocytes which progress through the DN4 stage, upregulate expression of CD4 and CD8 and become double positive (DP) thymocytes (Davis and Littman, 1994) (von Boehmer et al., 1999). DP thymocytes rearrange their TCR α locus, express a mature TCRα/β complex, and undergo processes of positive and negative selection that generate CD4+ and CD8+ single-positive (SP) thymocytes. A fraction of SP CD4+ thymocytes upregulates Foxp3 transcription factor and generates a population of thymic Treg cells, called natural Treg cells. This cell population is critical for maintenance of immune system homeostasis, immune tolerance and control of immune responses to antigens (Sakaguchi et al., 2008). However, dysregulated suppressor function of these cells is attributed to incomplete elimination of pathogens in chronic infectious diseases or compromised immune responses in cancer (Josefowicz et al., 2012) (Zou, 2006).
Interactions of the developing thymocytes with thymic stromal cells are critical for their differentiation to become DP cells and later, to undergo TCR mediated selection. Thymic stromal cells were found to produce BMP2/4 and contribute to the specification of T cell lineage. BMPs promote differentiation of early progenitor cells into thymocyte lineage but arrest DN1 cells at the stage where they are still capable of producing T, B, NK and DC cells (Hager-Theodorides et al., 2002). BMP2/4 signaling is required for T cell progenitor homeostasis and expansion but negatively regulates DN to DP transition. In contrast, BMP4 signaling promotes generation of NK cells by regulating expression of Id3, Nfil3 and CD122 (Nohe et al., 2002). BMP4 added to fetal thymic explants inhibits thymocyte proliferation, enhances early progenitor survival, and arrests thymocyte differentiation at the DN stage. Neutralization of BMP2/4 by treatment with recombinant noggin promotes and accelerates thymocyte differentiation, increasing the expression of CD2 and the proportion of DN4 cells and DP cells. A similar result was obtained by blocking BMP2/4 signaling in epithelium and mesenchymal cells of noggin transgenic mice. Noggin encoding transgene, controlled by FoxN1 promoter, was expressed in epithelial cells of developing thymus and this resulted in marked reduction of thymus volume and its ectopic location in the neck (Bleul and Boehm, 2005). Despite reduced thymus size newborn thymocytes developed normally. Although splenic T cells were present in these mice, their numbers were over 25-fold reduced compared to wild type animals. This inhibitory effect of BMPs is most likely mediated by BMPR1A and BMPR2 (BMPR1B was not detected on any thymocyte population). High levels of both receptors were found in DN1 subset, expression decreased in DN2, and again increased from DN3 to DP cells to reach maximum in SP CD4+ cells. However, thymic stroma was also shown to express BMP receptors as well as BMPs and signaling molecules Smad1/5/8 suggesting that it is able to respond to BMPs. In vitro analysis of thymic reaggregation cultures also showed that BMP treatment arrests thymocyte precursors at DN1 which is due to signaling both to stromal and hematopoietic cells while the effect of BMP2/4 on transition from DN to DP is mediated by stroma alone (Tsai et al., 2003). In contrast to earlier results, the most recent analysis of conditional knockout mice where BMPR1A gene was deleted in hematopoietic cells (by crossing to vav-cre mice) revealed a distinct outcome. Thymic cellularity, proportions of DN, DP and SP CD4+ and CD8+ T cell subsets and expression profiles of CD5, CD3, CD24, CD2, CD69 were normal in adult mice (Hager-Theodorides et al., 2013). Reduced cellularity of DN3 and DN4 populations was observed in day 14.5 of fetal thymi in BMPR1A/vav-cre mice but cell numbers recovered by day 16.5 when DP cells are first observed. More pronounced effect of BMPR1A deletion in fetal thymocytes could be due to higher receptor levels in fetal thymi. The lack of phenotype in BMPR1A/vav-cre mice shows that earlier observed effects of BMP treatment on DN-DP transition might be indirect and involve thymic stroma. Altogether, despite controversial results, BMP signaling seems to regulate early thymopoiesis although interpretation of data is confusing since both BMPs and their receptors are expressed by developing thymocytes and stromal elements. Considering that BMPs act locally and their concentration and gradient were found important for the biological outcome in other experimental systems it might be difficult to dissect the contribution of hematopoietic and stromal elements to T cell generation using in vitro systems.
Analyzes of fetal thymi or BMPR1A/vav-cre mice provided evidence on the role of BMPs in early stages of thymopoiesis. We have investigated development of T cells in mice where BMPR1A signaling is abrogated in late stages of thymocyte differentiation (BMPR1AT− mice)(Kuczma et al., 2014). Mice where the second exon of the BMPR1A gene, encoding kinase domain of the receptor, is flanked by loxP sites were crossed to CD4cre mice (Mishina et al., 2002) (Lee et al., 2001). BMPR1A function in BMPR1AT− mice is eliminated at the DP stage, in SP CD4+ and CD8+ thymocytes and in all T cells in the peripheral organs. Analysis of BMPR1AT− mice showed that thymic cellularity and composition of subsets was normal with the exception of a population of Foxp3+ Treg cells which, was severely decreased (Kuczma et al., 2014). This suggests that signaling through the BMPR1A has a unique role in the process of generating Treg cells. The mechanistic function of BMPR1A in this process is not currently known. BMPR1A may promote renewal of Treg progenitors and initiate instructive process of precursor commitment to Treg lineage. Alternatively, BMPR1A signaling may synergize with TGF-β to expand and protect Treg precursors from apoptosis (Ouyang et al., 2010a). Although the Foxp3+ cell numbers were reduced, the BMPR1AT− mice had normal proportion of CD4+CD25+ cells. This suggests that BMPR1A functions in CD25+ thymocytes to upregulate/sustain Foxp3 expression. Analysis of thymic and peripheral CD4+CD25+Foxp3− cells showed that they do not have suppressor function.
BMP signaling modulates activation of peripheral CD4+ T cells
Despite well-known role of TGF-β in maintaining immune tolerance but also regulating immune responses, contributions of other members of TGF-β family to control functions of peripheral T cells has been addressed only recently (Zhou et al., 2008). TGF-β may induce upregulation of Foxp3 and skew CD4+ T cell lineage commitment towards induced Treg (iTreg) cells when activation occurs in the presence of IL-2 (Davidson et al., 2007). Alternatively, it may promote Th17 cell generation in the presence of IL-6 (Bettelli et al., 2006). The same combination of TGF-β and IL-6 results in upregulation of ACVR2A specifically in Th17 cells, and not in other CD4 lineages, suggesting the role of activins in Th17 function. The role of BMP signaling in regulating CD4+ T cell activation was analyzed by comparing T cells stimulated in vitro in the absence and presence of dorsomorphin (Yoshioka et al., 2012). Dorsomorphin inhibited Smad1/5/8 phosphorylation in Jurkat and mouse T cells, blocked proliferation by suppressing IL-2 production and expression of CD25 and arresting cells in G0/G1 phase. Dorsomorphin treatment inhibited differentiation of Th1, Th17 and iTreg cells but not Th2 by suppressing expression of lineage-specific transcription factors T-bet, RORγt and Foxp3 but not GATA3. BMPs control phosphorylation of Runx1 and promote its association with AML, which is known to regulate IL-2 gene expression in concert with Foxp3 (Ono et al., 2007).
Despite similarities in signaling pathways between TGF-β and BMPs, BMP2/4 acting alone were not able to induce Foxp3 expression, but increased the ability of TGF-β to promote iTreg development (Lu et al., 2010a). This synergistic effect was dependent on activation of ERK1/2 and JNK (effect could be blocked by MAP kinase inhibitors PD98059 and SB203580 respectively). Combining BMP treatment with trichostatin A (HDAC inhibitor which promotes expression of Foxp3 and iTreg induction) resulted in increased proportion of iTreg cells in the gut. Another member of TGF-β family, activin A which may bind BMPR1A (ALK3) was found to synergize with TGF-β to promote iTreg generation through Smad 3 phosphorylation (Huber et al., 2009). Since activin A production is induced in macrophages stimulated by LPS, different members of the TGF-β family may differentially function in inflammation and steady state.
To examine the impact of BMPR1A signaling in peripheral CD4+ T cells we looked at the expression pattern of this receptor and its ligands. In healthy mice low levels of BMPR1A are found on activated CD44+CD62L− helper T cells and Treg cells expressing high level of Foxp3 (Kuczma et al., 2014). BMPR1A is up-regulated upon in vitro activation of conventional CD4+ T-cells (but not cytotoxic cells), especially in converted to become iTreg cells, and in activated Treg cells. This expression pattern suggested that its main function could be to regulate CD4+ T cell lineage commitment. BMPR1A was also expressed in a population of activated human CD4+ T-cells, especially in the presence of TGF-β. To further reveal BMPR1A signaling axis we have examined production of BMPs by T cells. BMPR1A ligands, i.e., BMP2, 4 and 7, were not expressed in naive T cells. BMP2 were produced by Th1 cells and BMP2 and BMP7 were produced by Th2 cells. We failed to detect BMP4 in activated CD4+ T cells. Expression of BMPR1A ligands in activated T cells (especially Th2 cells) suggests that they may act as autocrine or paracrine factors modulating the outcome of T cell activation.
Analysis of BMPR1AT− mice revealed that they have moderately increased proportion of activated CD4+ cells and decreased proportion of Treg cells in lymph nodes. When stimulated with an antigen, CD4+ T cells from mutant mice were highly activated cells and had proliferative advantage over cells isolated from wild type animals. Furthermore, we found that signaling through BMPR1A regulates CD4+ cell lineage commitment. Activation of CD4+Foxp3− cells in the presence of TGF-β resulted in several fold smaller number of Foxp3+ cells when compared with BMPR1A-sufficient littermates despite the same level of activation measured by CD25, CD44 and CD69 expression. In conclusion, signaling through BMPR1A is essential in peripheral helper cells, stimulated by antigens, to promote development and to sustain population of Treg cells. This finding is consistent with the role of BMPs and other members of TGF-β family revealed in previous in vitro studies (Lu et al., 2010a).
BMP signaling in T cells in cancer
Besides impaired generation of Treg cells, absence of BMPR1A signaling shifted the phenotype of CD4 cells towards IFN-γ producing Th1 cells. Taking all the information together – Th1 phenotype of activated CD4+ cells, diminished proportion of thymic derived Treg cells in situ and impaired iTreg formation, we decided to analyze tumor growth in BMPR1AT− mice. We used B16-F1 melanoma, and showed that the tumor growth and composition of T cells infiltrating the tumor mass (like TIL) were different in BMPR1AT− and wild type mice. We consistently observed highly impaired kinetics of tumor growth in mice lacking BMPR1A signaling in CD4 cells. Surprisingly, the tumor mass was devoid of Treg cells that were readily found in tumor-draining lymph nodes in the same mice. The lack of Treg cells in tumor infiltrates is not known, but it might be caused by impaired translocation of Treg cells into the tumor mass as a result of lack of BMPR1AA expression, or by a defect in iTreg generation/renewal in tumors.
In vitro T cell activation data suggest that targeting BMPR1A in T cells should result in generation of tumor-specific effector CD4+ cells. Indeed, helper T cells found in the tumors in BMPR1AT− mice expressed higher amounts of IFN-γ. We also observed significantly increased (2-3x) CD8 cell numbers in tumors growing in BMPR1AT− mice. These CD8 cells expressed much higher level of CD44 activation marker than the same population isolated from BMPR1A-sufficient animals. Expanded population of CD8 cells in tumors may be caused by signaling through other BMP receptors, or lack of Treg cells in the tumor and enhanced expression of IFN-γ by CD4 cells may promote expansion of CD8 cells and increase their anti-tumor response as it was reported (Song et al., 2007).
BMPs were recently implicated in the development and function of myeloid derived suppressor cells (MDSC). These important cells, together with Foxp3+ regulatory T cells (Treg), constitute the main suppressor populations found in multiple types of cancer and their presence in the circulating blood was correlated with poor prognosis for cancer patients. MDSC were characterized as CD11b+Gr1+ or CD11b+CD15+CD33+HLA−DR− in mouse and human, respectively (Gabrilovich and Nagaraj, 2009). MDSC inhibit response of effector helper and cytotoxic T cells using variety of mechanisms including expression of arginase-1, nitric oxide or indoleamine 2,3-dioxygenase (IDO), production of suppressor cytokines, upregulation of surface molecules inhibiting immune responses (Serafini et al., 2006). They were shown to skew immune responses towards generating suppressor T cells. The effect of BMPs on the tumor population of MDSCs is controversial. Some reports suggest that BMPs decreased MDCS population in breast cancer but others claimed that this treatment increased metastatic potential of human breast cancer lines (Cao et al., 2014; Owens et al., 2012). Interaction between MDSCs and infiltrating T cells is likely shaping adaptive immune responses in tumors. Our preliminary data suggest that a subset of myeloid cells could be major source of BMP2/7 in prostate TRAMP tumors. Thus, BMPs produced by myeloid cells (and prostate tumor cells) could complement known immunosuppressive activity of TGF-β and constitute one of suppressor signaling circuits compromising immune responses in tumors.
Concluding remarks
For a long time the importance of BMPs and their receptors in development and differentiation of multiple organs was well known and appreciated. The role of these proteins in controlling immune system functions has started to be uncovered. Reports from several laboratories revealed that BMPs control early stages of thymic development of T cells and regulate functions of mature T cells. The progress to unravel BMP controlled molecular pathways is hampered by highly redundant functions of individual BMP ligands and their receptors as well as by a context dependent biological outcome. This has led us to adopt a strategy of eliminating one of the major BMP receptors in T cells in order to reveal its functions. Our data show that lack of BMPR1A signaling led to impaired generation and function of regulatory T cells. In addition, conventional T cells deprived of BMPR1A signaling are prone to generate Th1 effector cells when activated. While it is likely to require a significant effort to reveal molecular mechanisms of these processes, the inhibition of tumor growth in BMPR1AT− mice suggests that signaling controlled by this receptor plays an important role in adaptive immune response in cancer.
Figure 1.
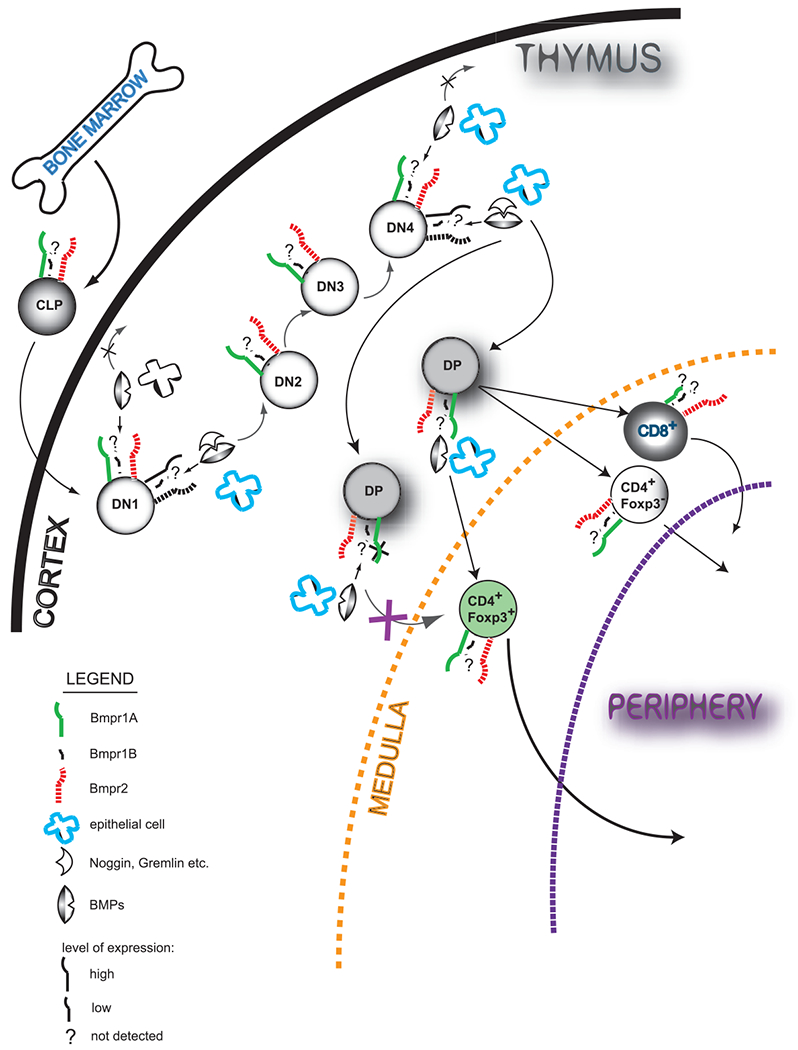
Bone morphogenic proteins (BMP) affect thymopoiesis at multiple stages. Bone marrow derived Common Lymphoid Progenitor (CLP) cell colonize thumus and undergo series of changes to become mature T cell. Upon binding of BMPs (mainly BMP2 and 4) produced by multiple cells in the thymus (i.e. epithelial cells) at Double Negative (DN) stages 1 and 4, thymocyte development in the cortex is arrested. Natural inhibitors of BMP signaling (i.e. Noggin, Chrodin) bind to BMPs sequestering them from their type I (mainly BMPR1A, expression of BMPR1B was not reported in the thymus)and type II (BMPR2) receptors. Regulatory T cells (CD4+Foxp3+) develop only upon signal mediated by BMPs in the context of BMP receptor 1A (BMPR1A) and their generation is largely affected when DP (Double Positive) thymocytes lack its expression. CD8+ T cells express very low level of BMPR1A and their development and function is minimally affected by its deletion.
Figure 2.
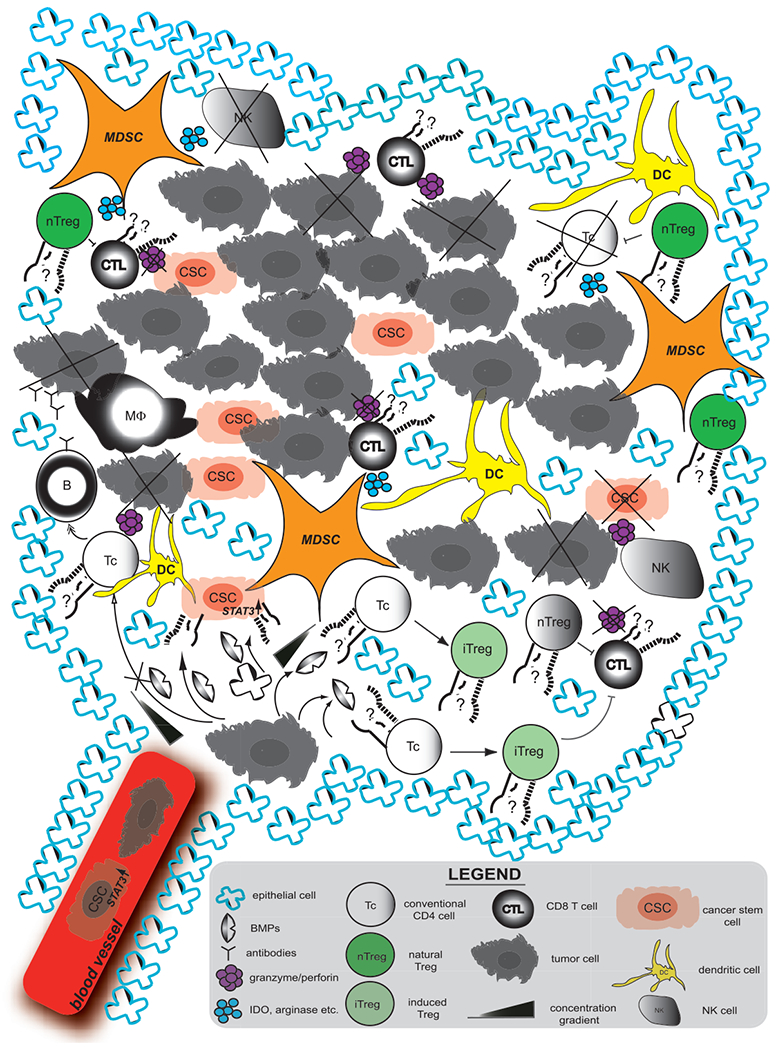
Effect of BMP signaling in the tumor microenvironment. BMPs act locally to enhance self-renewal/stemness of Cancer Stem Cells (CSC) by induction of STAT3 expression. This in turn enhances the metastatic potential of tumor cells. BMPs also induce Foxp3 expression in conventional CD4 cells (CD4+Foxp3−) to become regulatory T cells (induced Treg, iTreg). Treg (both iTreg and thymic-derived natural Treg (nTreg)) and MDSC suppress anti-tumor response targeting granzyme/perforin producing CD8+ T cells, NK cells and CD4+ cells of Th1 lineage by cell-dependent contact and expression of suppressive cytokines or metabolites (TGF-β, IDO, arginase etc.). MDSC-Myeloid Derived Suppressor Cell, NK-Natural Killer, B-B cell, MF-Macrophage, DC-Dendritic Cell
Acknowledgments
This work was supported by the NIAID R21 AI097600 grant to P.K.
REFERENCES
- Akhurst RJ, and Hata A (2012). Targeting the TGFbeta signalling pathway in disease. Nat.Rev.Drug Discov 11, 790–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano N, Watanabe T, Kitani A, Fuss IJ, and Strober W (2008). Notch1 signaling and regulatory T cell function. J.Immunol 180, 2796–2804. [DOI] [PubMed] [Google Scholar]
- Bachiller D, Klingensmith J, Kemp C, Belo JA, Anderson RM, May SR, McMahon JA, McMahon AP, Harland RM, Rossant J, and De Robertis EM (2000). The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature 403, 658–661. [DOI] [PubMed] [Google Scholar]
- Beck SE, and Carethers JM (2007). BMP suppresses PTEN expression via RAS/ERK signaling. Cancer Biol.Ther 6, 1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, and Kuchroo VK (2006). Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238. [DOI] [PubMed] [Google Scholar]
- Bleul CC, and Boehm T (2005). BMP signaling is required for normal thymus development. J.Immunol 175, 5213–5221. [DOI] [PubMed] [Google Scholar]
- Cao Y, Slaney CY, Bidwell BN, Parker BS, Johnstone CN, Rautela J, Eckhardt BL, and Anderson RL (2014). BMP4 inhibits breast cancer metastasis by blocking myeloid-derived suppressor cell activity. Cancer Res 74, 5091–5102. [DOI] [PubMed] [Google Scholar]
- Carreira AC, Alves GG, Zambuzzi WF, Sogayar MC, and Granjeiro JM (2014). Bone Morphogenetic Proteins: structure, biological function and therapeutic applications. Archives of biochemistry and biophysics 561, 64–73. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. (2008). Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117. [DOI] [PubMed] [Google Scholar]
- Chida A, Shintani M, Nakayama T, Furutani Y, Hayama E, Inai K, Saji T, Nonoyama S, and Nakanishi T (2012). Missense mutations of the BMPR1B (ALK6) gene in childhood idiopathic pulmonary arterial hypertension. Circulation journal : official journal of the Japanese Circulation Society 76, 1501–1508. [DOI] [PubMed] [Google Scholar]
- Conery AR, Cao Y, Thompson EA, Townsend CM Jr., Ko TC, and Luo K (2004). Akt interacts directly with Smad3 to regulate the sensitivity to TGF-beta induced apoptosis. Nat.Cell Biol 6, 366–372. [DOI] [PubMed] [Google Scholar]
- Davidson TS, DiPaolo RJ, Andersson J, and Shevach EM (2007). Cutting Edge: IL-2 is essential for TGF-beta-mediated induction of Foxp3+ T regulatory cells. J.Immunol 178, 4022–4026. [DOI] [PubMed] [Google Scholar]
- Davis CB, and Littman DR (1994). Thymocyte lineage commitment: is it instructed or stochastic? Curr Opin Immunol 6, 266–272. [DOI] [PubMed] [Google Scholar]
- Ding Y, Shen S, Lino AC, Curotto de Lafaille MA, and Lafaille JJ (2008). Beta-catenin stabilization extends regulatory T cell survival and induces anergy in nonregulatory T cells. Nat.Med 14, 162–169. [DOI] [PubMed] [Google Scholar]
- Dudley AT, Lyons KM, and Robertson EJ (1995). A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes & development 9, 2795–2807. [DOI] [PubMed] [Google Scholar]
- Ferguson EL, and Anderson KV (1992). Localized enhancement and repression of the activity of the TGF-beta family member, decapentaplegic, is necessary for dorsal-ventral pattern formation in the Drosophila embryo. Development 114, 583–597. [DOI] [PubMed] [Google Scholar]
- Gabrilovich DI, and Nagaraj S (2009). Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 9, 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Ramirez A, Waddell DS, Li Z, Liu X, and Wang XF (2008). Axin and GSK3- control Smad3 protein stability and modulate TGF- signaling. Genes Dev. 22, 106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, and Wang XF (2009). Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 19, 71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager-Theodorides AL, Outram SV, Shah DK, Sacedon R, Shrimpton RE, Vicente A, Varas A, and Crompton T (2002). Bone morphogenetic protein 2/4 signaling regulates early thymocyte differentiation. J.Immunol 169, 5496–5504. [DOI] [PubMed] [Google Scholar]
- Hager-Theodorides AL, Ross SE, Sahni H, Mishina Y, Furmanski AL, and Crompton T (2013). Direct BMP2/4 signaling through BMP receptor IA regulates fetal thymocyte progenitor homeostasis and differentiation to CD4+CD8+ double-positive cell. Cell Cycle 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxhinasto S, Mathis D, and Benoist C (2008). The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J.Exp.Med 205, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, et al. (2004). BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat.Genet 36, 1117–1121. [DOI] [PubMed] [Google Scholar]
- Herhaus L, Al-Salihi M, Macartney T, Weidlich S, and Sapkota GP (2013). OTUB1 enhances TGFbeta signalling by inhibiting the ubiquitylation and degradation of active SMAD2/3. Nat.Commun 4, 2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S, Schrader J, Fritz G, Presser K, Schmitt S, Waisman A, Luth S, Blessing M, Herkel J, and Schramm C (2008). P38 MAP kinase signaling is required for the conversion of CD4+CD25− T cells into iTreg. PLoS.ONE 3, e3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S, Stahl FR, Schrader J, Luth S, Presser K, Carambia A, Flavell RA, Werner S, Blessing M, Herkel J, and Schramm C (2009). Activin a promotes the TGF-beta-induced conversion of CD4+CD25− T cells into Foxp3+ induced regulatory T cells. J.Immunol 182, 4633–4640. [DOI] [PubMed] [Google Scholar]
- Itoh F, Itoh S, Goumans MJ, Valdimarsdottir G, Iso T, Dotto GP, Hamamori Y, Kedes L, Kato M, and ten Dijke Pt P (2004). Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. EMBO J 23, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A, Bae JS, Afzal F, Gutierrez S, Pratap J, Zaidi SK, Lou Y, van Wijnen AJ, Stein JL, Stein GS, and Lian JB (2008). Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J.Biol.Chem 283, 8412–8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu LF, and Rudensky AY (2012). Regulatory T cells: mechanisms of differentiation and function. Annu.Rev.Immunol 30, 531–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis GS, Berk A, Dimitromanolakis A, and Diamandis EP (2013). Enrichment map profiling of the cancer invasion front suggests regulation of colorectal cancer progression by the bone morphogenetic protein antagonist, gremlin-1. Molecular oncology 7, 826–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata M, Imamura T, and Miyazono K (1998). Signal transduction by bone morphogenetic proteins. Cytokine & growth factor reviews 9, 49–61. [DOI] [PubMed] [Google Scholar]
- Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, Depinho RA, and Hedrick SM (2009). Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat.Immunol 10, 176–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerdiles YM, Stone EL, Beisner DR, McGargill MA, Ch’en IL, Stockmann C, Katayama CD, and Hedrick SM (2010). Foxo transcription factors control regulatory T cell development and function. Immunity. 33, 890–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohu K, Ohmori H, Wong WF, Onda D, Wakoh T, Kon S, Yamashita M, Nakayama T, Kubo M, and Satake M (2009). The Runx3 transcription factor augments Th1 and down-modulates Th2 phenotypes by interacting with and attenuating GATA3. J.Immunol 183, 7817–7824. [DOI] [PubMed] [Google Scholar]
- Kondo M, Scherer DC, King AG, Manz MG, and Weissman IL (2001). Lymphocyte development from hematopoietic stem cells. Current opinion in genetics & development 11, 520–526. [DOI] [PubMed] [Google Scholar]
- Korchynskyi O, and ten DP (2002). Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J.Biol.Chem 277, 4883–4891. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, and Massague J (1997). Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature 389, 618–622. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, Timokhina I, and Massague J (1999). A mechanism of repression of TGFbeta/Smad signaling by oncogenic Ras. Genes Dev. 13, 804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczma M, Kurczewska A, and Kraj P (2014). Modulation of bone morphogenic protein signaling in T-cells for cancer immunotherapy. Journal of immunotoxicology 11, 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, and Leiden JM (1999). Transcriptional regulation of T lymphocyte development and function. Annual Review of Immunology 17, 149–187. [DOI] [PubMed] [Google Scholar]
- Kusanagi K, Inoue H, Ishidou Y, Mishima HK, Kawabata M, and Miyazono K (2000). Characterization of a bone morphogenetic protein-responsive Smad-binding element. Molecular biology of the cell 11, 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. (2001). A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15, 763–774. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Lerch TF, Cook RW, Jardetzky TS, and Woodruff TK (2006). The structural basis of TGF-beta, bone morphogenetic protein, and activin ligand binding. Reproduction 132, 179–190. [DOI] [PubMed] [Google Scholar]
- Liu Z, Tang Y, Qiu T, Cao X, and Clemens TL (2006). A dishevelled-1/Smad1 interaction couples WNT and bone morphogenetic protein signaling pathways in uncommitted bone marrow stromal cells. J.Biol.Chem 281, 17156–17163. [DOI] [PubMed] [Google Scholar]
- Lu L, Ma J, Wang X, Wang J, Zhang F, Yu J, He G, Xu B, Brand DD, Horwitz DA, et al. (2010a). Synergistic effect of TGF-beta superfamily members on the induction of Foxp3+ Treg. Eur.J.Immunol 40, 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Wang J, Zhang F, Chai Y, Brand D, Wang X, Horwitz DA, Shi W, and Zheng SG (2010b). Role of SMAD and non-SMAD signals in the development of Th17 and regulatory T cells. J Immunol 184, 4295–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Fan Y, Mou Y, Zhang H, Fu S, and Yang J (2011). TAK1 lysine 158 is required for TGF-beta-induced TRAF6-mediated Smad-independent IKK/NF-kappaB and JNK/AP-1 activation. Cell Signal. 23, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Li J, Vaque JP, Konkel JE, Wang W, Zhang B, Zhang P, Zamarron BF, Yu D, Wu Y, et al. (2011). Control of the differentiation of regulatory T cells and T(H)17 cells by the DNA-binding inhibitor Id3. Nat Immunol 12, 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon JA, Takada S, Zimmerman LB, Fan CM, Harland RM, and McMahon AP (1998). Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes & development 12, 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y, Hanks MC, Miura S, Tallquist MD, and Behringer RR (2002). Generation of Bmpr/Alk3 conditional knockout mice. Genesis. 32, 69–72. [DOI] [PubMed] [Google Scholar]
- Mishina Y, Suzuki A, Ueno N, and Behringer RR (1995). Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes & development 9, 3027–3037. [DOI] [PubMed] [Google Scholar]
- Miyazono K, Kamiya Y, and Morikawa M (2010). Bone morphogenetic protein receptors and signal transduction. Journal of biochemistry 147, 35–51. [DOI] [PubMed] [Google Scholar]
- Morikawa M, Koinuma D, Tsutsumi S, Vasilaki E, Kanki Y, Heldin CH, Aburatani H, and Miyazono K (2011). ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res. 39, 8712–8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller TD, and Nickel J (2012). Promiscuity and specificity in BMP receptor activation. FEBS letters 586, 1846–1859. [DOI] [PubMed] [Google Scholar]
- Muranski P, Borman ZA, Kerkar SP, Klebanoff CA, Ji Y, Sanchez-Perez L, Sukumar M, Reger RN, Yu Z, Kern SJ, et al. (2011). Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity. 35, 972–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, and Taniuchi I (2007). Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J.Exp.Med 204, 1749–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen SM, and Christian JL (2009). Site-specific cleavage of BMP4 by furin, PC6, and PC7. J Biol Chem 284, 27157–27166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurath MF, Weigmann B, Finotto S, Glickman J, Nieuwenhuis E, Iijima H, Mizoguchi A, Mizoguchi E, Mudter J, Galle PR, et al. (2002). The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn’s disease. J.Exp.Med 195, 1129–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohe A, Hassel S, Ehrlich M, Neubauer F, Sebald W, Henis YI, and Knaus P (2002). The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J Biol Chem 277, 5330–5338. [DOI] [PubMed] [Google Scholar]
- Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, Osaki M, Tanaka Y, Yamashita R, Nakano N, et al. (2012). T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 37, 785–799. [DOI] [PubMed] [Google Scholar]
- Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, Miyachi Y, Tsukada T, and Sakaguchi S (2007). Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Beckett O, Ma Q, and Li MO (2010a). Transforming growth factor-beta signaling curbs thymic negative selection promoting regulatory T cell development. Immunity. 32, 642–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Beckett O, Ma Q, Paik JH, Depinho RA, and Li MO (2010b). Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat.Immunol 11, 618–627. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Liao W, Luo CT, Yin N, Huse M, Kim MV, Peng M, Chan P, Ma Q, Mo Y, et al. (2012). Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature 491, 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens P, Pickup MW, Novitskiy SV, Chytil A, Gorska AE, Aakre ME, West J, and Moses HL (2012). Disruption of bone morphogenetic protein receptor 2 (BMPR2) in mammary tumors promotes metastases through cell autonomous and paracrine mediators. Proc Natl Acad Sci U S A 109, 2814–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JD, and Delgoffe GM (2010). The mammalian target of rapamycin: linking T cell differentiation, function, and metabolism. Immunity. 33, 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, and Aguet M (1999). Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10, 547–558. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, and Weissman IL (2001). Stem cells, cancer, and cancer stem cells. Nature 414, 105–111. [DOI] [PubMed] [Google Scholar]
- Rudra D, deRoos P, Chaudhry A, Niec RE, Arvey A, Samstein RM, Leslie C, Shaffer SA, Goodlett DR, and Rudensky AY (2012). Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat.Immunol 13, 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S, Yamaguchi T, Nomura T, and Ono M (2008). Regulatory T cells and immune tolerance. Cell 133, 775–787. [DOI] [PubMed] [Google Scholar]
- Samon JB, Champhekar A, Minter LM, Telfer JC, Miele L, Fauq A, Das P, Golde TE, and Osborne BA (2008). Notch1 and TGFbeta1 cooperatively regulate Foxp3 expression and the maintenance of peripheral regulatory T cells. Blood 112, 1813–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, and Massague J (2007). Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol.Cell 25, 441–454. [DOI] [PubMed] [Google Scholar]
- Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, Knight ZA, Cobb BS, Cantrell D, O’Connor E, et al. (2008). T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc.Natl.Acad.Sci.U.S.A 105, 7797–7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Le HV, Shen L, Anderson SA, and Massague J (2004). Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117, 211–223. [DOI] [PubMed] [Google Scholar]
- Serafim P, Borrello I, and Bronte V (2006). Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression. Semin Cancer Biol 16, 53–65. [DOI] [PubMed] [Google Scholar]
- Shim JH, Greenblatt MB, Xie M, Schneider MD, Zou W, Zhai B, Gygi S, and Glimcher LH (2009). TAK1 is an essential regulator of BMP signalling in cartilage. EMBO J. 28, 2028–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song A, Song J, Tang X, and Croft M (2007). Cooperation between CD4 and CD8 T cells for anti-tumor activity is enhanced by OX40 signals. Eur J Immunol 37, 1224–1232. [DOI] [PubMed] [Google Scholar]
- Song K, Wang H, Krebs TL, and Danielpour D (2006). Novel roles of Akt and mTOR in suppressing TGF-beta/ALK5-mediated Smad3 activation. EMBO J. 25, 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubrier F, Chung WK, Machado R, Grunig E, Aldred M, Geraci M, Loyd JE, Elliott CG, Trembath RC, Newman JH, and Humbert M (2013). Genetics and genomics of pulmonary arterial hypertension. Journal of the American College of Cardiology 62, D13–21. [DOI] [PubMed] [Google Scholar]
- Takaku K, Miyoshi H, Matsunaga A, Oshima M, Sasaki N, and Taketo MM (1999). Gastric and duodenal polyps in Smad4 (Dpc4) knockout mice. Cancer Res 59, 6113–6117. [PubMed] [Google Scholar]
- Tsai PT, Lee RA, and Wu H (2003). BMP4 acts upstream of FGF in modulating thymic stroma and regulating thymopoiesis. Blood 102, 3947–3953. [DOI] [PubMed] [Google Scholar]
- Umulis D, O’Connor MB, and Blair SS (2009). The extracellular regulation of bone morphogenetic protein signaling. Development 136, 3715–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urist MR (1965). Bone: formation by autoinduction. Science 150, 893–899. [DOI] [PubMed] [Google Scholar]
- von Boehmer H, Aifantis I, Feinberg J, Lechner O, Saint-Ruf C, Walter U, Buer J, and Azogui O (1999). Pleiotropic changes controlled by the pre-T-cell receptor. Curr.Opin.Immunol 11, 135–142. [DOI] [PubMed] [Google Scholar]
- Wahdan-Alaswad RS, Bane KL, Song K, Shola DT, Garcia JA, and Danielpour D (2012). Inhibition of mTORC1 kinase activates Smads 1 and 5 but not Smad8 in human prostate cancer cells, mediating cytostatic response to rapamycin. Mol.Cancer Res 10, 821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, and Wang EA (1988). Novel regulators of bone formation: molecular clones and activities. Science 242, 1528–1534. [DOI] [PubMed] [Google Scholar]
- Xiao YT, Xiang LX, and Shao JZ (2007). Bone morphogenetic protein. Biochemical and biophysical research communications 362, 550–553. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, and Zhang YE (2008). TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol.Cell 31, 918–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, and Smith A (2003). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292. [DOI] [PubMed] [Google Scholar]
- Yoshioka Y, Ono M, Osaki M, Konishi I, and Sakaguchi S (2012). Differential effects of inhibition of bone morphogenic protein (BMP) signalling on T-cell activation and differentiation. Eur.J.Immunol 42, 749–759. [DOI] [PubMed] [Google Scholar]
- Zhang C, and Evans T (1996). BMP-like signals are required after the midblastula transition for blood cell development. Developmental genetics 18, 267–278. [DOI] [PubMed] [Google Scholar]
- Zhao DM, Yu S, Zhou X, Haring JS, Held W, Badovinac VP, Harty JT, and Xue HH (2010). Constitutive activation of Wnt signaling favors generation of memory CD8 T cells. J.Immunol 184, 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, et al. (2008). TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature 453, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, and Niswander L (1996). Requirement for BMP signaling in interdigital apoptosis and scale formation. Science 272, 738–741. [DOI] [PubMed] [Google Scholar]
- Zou W (2006). Regulatory T cells, tumour immunity and immunotherapy. Nat.Rev.Immunol 6, 295–307. [DOI] [PubMed] [Google Scholar]


