Summary
Background
Nosocomially acquired severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) infection has become the most significant pandemic of our lifetime. Though its transmission was essentially attributed to droplets from an infected person, with recent advancements in knowledge, aerosol transmission seems to be a viable pathway, as well. Because of the lower biological load in ambient aerosol, detection of SARS-CoV-2 is challenging. A few recent attempts of sampling large aerosol volumes and using next-generation sequencing (NGS) to detect the presence of SARS-CoV-2 in the air at very low levels gave positive results. These results suggest the potential of using this technique to detect the presence of SARS-CoV-2 and use it as an early warning signal for possible outbreak or recurrence of coronavirus disease 2019 (COVID-19).
Aim
To assess efficacy of comprehensive respiratory viral panel (CRVP) sequencing and RT-PCR for low-level identification of SARS-CoV-2 and other respiratory viruses in indoor air.
Methods
A large volume of indoor aerosol samples from three major hospitals involved in COVID-19 care in Kuwait was collected. Viral RNA was isolated and subjected to comprehensive respiratory viral panel sequencing (CRVP) as per the standard protocol to detect the SARS-CoV-2 and other respiratory viruses in the hospital aerosol and monitor variations within the sequences. RT-PCR was also employed to estimate the viral load of SARS-CoV-2.
Findings
13 of 15 (86.7%) samples exhibited SARS-CoV-2 with a relative abundance of 0.2–33.3%. The co-occurrence of human adenoviruses (type C1, C2, C5, C4), respiratory syncytial virus (RSV), influenza B, and non-SARS-CoV-229E were also recorded. Alignment of SARS-CoV-2 sequences against the reference strain of Wuhan China revealed variations in the form of single nucleotide polymorphisms (SNPs-17), insertions and deletions (indels-1). These variations were predicted to create missense (16), synonymous (15), frameshift (1) and stop-gained (1) mutations with a high (2), low (15), and moderate (16) impact.
Conclusions
Our results suggest that using CRVP on a large volume aerosol sample was a valuable tool for detecting SARS-CoV-2 in indoor aerosols of health care settings. Owing to its higher sensitivity, it can be employed as a surveillance strategy in the post COVID times to act as an early warning system to possibly control future outbreaks.
Keywords: SARS-CoV-2, RT-PCR, Hospital infection, Surveillance, Mutations, Next-generation sequencing
Introduction
Pathogenic microbes are common within hospital aerosols despite infection control procedures strictly being followed [1]. The presence of severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) in hospital environments has also been reported [2,3]. The airborne route is the primary means of spread [[4], [5], [6], [7]]. Intra-hospital transmission has led to additional outbreaks [2,8,9] and makes health care workers susceptible to further infection [3,10,11]. The biggest challenge in identifying SARS-CoV-2 in aerosols is the low viral load rendering it challenging to be detected by routine procedures, like reverse transcription-polymerase chain reaction (RT-PCR) [12].
Health care staff act as a source of pathogens that can be transmitted to individuals they come into contact with [[13], [14], [15]]. Risk assessment studies established a risk ratio of 2.5 to attain a viral or bacterial infection when in frequent contact with a health care staff, or a regular visitor to a healthcare facility [16,17]. Hospital acquired infections often complicate the case of COVID patients especially those admitted in the intensive care unit or have a compromised immunity [18].
The declining costs of next-generation sequencing (NGS) have enabled researchers to test the presence of pathogens with higher precision and accuracy [19]. NGS in field settings proved critical in developing countermeasures for the Ebola virus epidemic during 2014–2016 in West Africa [20,21]. This approach has also been used as a surveillance strategy for SARS-CoV-2 in Cambodia [22] and Akershus University Hospital, Norway [9]. More recently, targeted virome capture sequening has been used to detect SARS-CoV-2 [[23], [24], [25]]. Comprehensive respiratory panel sequencing (CRVP) uses a similar approach, offering simultaneous amplification of innumerable genetic loci, avoidance of multiplexing, and optimal performance even with low quantity samples [26].
RNA viruses exhibit high mutation rates. Mutations are a natural part of the virus life cycle owing to the lack of proofreading activity of their polymerases especially the RNA viruses [27]. SARS-CoV-2 is also known to mutate to creat new variants [28]. Understanding rates of mutations is needed to gain knowledge about viral spread and prevention [29]. Variant alignment led towards contact tracing of SARS-CoV-2 in the middle eastern region [30]. At the same time, the identification of viral quasispecies mutations sheds light on their general prevalence [28]. NGS data will also provide information on the change in sequence and can shed information on tracing mutations.
Given the above, we collected a large volume of aerosol, employed the RT-PCR, NGS and Comprehensive Respiratory Viral Panel (CRVP) [26] to detect the presence of the SARS-CoV-2 and other respiratory viruses in indoor hospital aerosols in Kuwait. The study also aimed to look at the feasibility of identifying single nucleotide polymorphisms (SNPs), insertions and deletions (indels), and corresponding mutations in the sequences of the SARS-CoV-2 genome.
Materials and methods
Sample collection
Aerosol samples were collected between August and October 2020 from hospitals (Mubarak Al-Kabeer, Sheikh Jaber and Amiri) in Kuwait (Table I) after obtaining permission from the Ministry of Health, Kuwait. The samples were collected using a custom-made sampling device [31], where the ambient air was extracted at the rate of 30 L min−1 for 2 hours, resulting in a 3.6 m3 sample. In addition to hospital samples, two samples were collected from the Kuwait Institute for Scientific Research (KISR) indoor areas as it was dealing with SARS-CoV-2 sample analyses. Since the viral load was lyzed concurrent to the collection of samples were processed for RNA isolation in a BSL2 cabinet.
Table I.
Sampling details for three major hospitals and the KISR site in Kuwait
| Hospital Name/GPS location | Sampling point | Code | Remarks |
|---|---|---|---|
|
Mubarak Al Kabeer Hospital 29.3260° N, 48.0350° E |
Waiting Area near Pharmacy | MKHP | Open space outside the main pharmacy delivering medications |
| Main Gate Entrance | MKHE | Common area in close proximity with outdoor air | |
| Pediatric Casualty | MKHPC | The area receiving paediatric patients | |
| Laboratory 1 | MKHL1 | Central laboratory receiving patient samples COVID-19 for testing | |
| Laboratory 2 | MKHL2 | Interior area of central lab exclusively dealing with COVID-19 samples | |
| COVID-19 Isolation Area | MKHCO | Common ward dealing with COVID-19 suspects | |
| COVID-19 Ward | MKHCW | Common ward taking care of symptomatic COVID-19 patients | |
|
Sheikh Jaber Hospital 29.2768oN, 48.0063oE |
Cytology Laboratory | SJHCL | Laboratory processing samples for cytological testing |
| COVID-19 Observation Area | SJHCO | Common ward dealing with COVID-19 suspects | |
| COVID-19 Ward | SJHCW | Common ward for symptomatic COVID-19 patients | |
| Virology Laboratory | SJHVL | Laboratory processing samples for viral testing | |
|
Amiri Hospital 29.3878° N, 47.9875° E |
Laboratory Reception Area | AMHLR | Reception of the virology laboratory receiving samples and dispatching results regularly |
| Virology Laboratory | AMHVL | Laboratory processing samples for COVID-19 testing | |
|
KISR 29.3369° N, 47.9064° E |
The pavement on the First Floor | KFF | The area near the staircase with continual human traffic |
| Main Reception Area | KGF | Main entrance observing regular staff in & out |
RNA isolation
The standard procedure of Trizol™ reagent (APB, Biosciences, Rockville, MD) was followed to purify total RNA [32]. In brief, the lysed aerosols (in Trizol) were left at room temperature for 10 mins and the crude RNA was separated by adding 0.2 volumes of chloroform (Sigma Aldrich, WGK, Germany). The aqueous chloroform layer was transferred to 0.5 volumes of isopropanol (Merck, Darmstadt, Germany) to precipitate the RNA and washed twice subsequently washed with 70% Ethanol (Merck, MO, US). The pellet was air-dried and dissolved in RNase-free water (Ambion, Austin, Texas). The high sensitivity Qubit HS ssRNA kit was used for fluorometric estimation of isolated RNA on a Qubit 4 fluorometer (Thermo Scientific, Paisley, UK).
RT-PCR for SARS-CoV-2 detection
The VIASURE SARS-CoV- Real-Time PCR Detection Kit-CE-IVD (Certest-Biotec) was employed for quantitative estimation of SARS-CoV-2. The PCR reaction was assembled as per the protocol described by Habibi et al. [5]. In brief 5 μl of isolated RNA was added to the master mix provided in the kit. Serial dilutions of the positive control were prepared and amplified simultaneously. The PCR reaction mix was carried out on the QuantStudio 5 Real-Time PCR System (Applied Biosystems, CA). The amplification profile was set for reverse transcription (15 min at 45°C), initial denaturation (2 min at 95°C) and then 45 cycles of denaturation (10 s at 95°C), and annealing/extension (50 s at 60°C).
Library preparation and sequencing
The protocol described in Thorburn et al. was used for library preparation and sequencing [26]. The extracted total RNA from the above step was converted to complementary deoxy-ribo nucleic acid (cDNA). The Celemics cDNA (Celemics, Korea) synthesis kit was used for this purpose. Firstly, the RNA was fragmented for 1 min at 85°C, then reverse-transcribed to first-strand cDNA synthesis by adding random hexamer primers annealing at 50°C for 10 mins. This was followed by enzyme inactivation at 80 °C for 10 mins. The cDNA:RNA hybrid was finally converted to double-stranded cDNA (dscDNA). Clean-up of dscDNA was done using the Celemag Clean-up beads [26]. The cDNA was amplified after an adapter ligation step followed by end repair. The Celemics target enrichment kit was used for hybridization capture of multiple (n=39) viruses (Fig. S1). The completed paired-end libraries were sequenced on a NextSeq 500 (Illumina, San Diego, CA) using the Mid/High output v2.5 kit for 300 cycles (2 x 150bp).
Data analysis
Raw reads with an N sequence ratio over 10% and low mean base quality (bq< 20, ratio >5%) were filtered out employing the fastx_toolkit 0.0.14 [33]. Reads with adapter sequence were trimmed by AdapterRemoval 2.2.2 [34]. The mpileup pipeline in Samtools 1.1 was used to generate ∗.sam files [35]. Also, the duplicate reads were removed through the MarkDuplicates tool [36]. The Burrows Wheeler Alignment 0.7.10 (BWA) tool was used to align the sequences to the viral genomes (Celemics Respiratory Virus Panel Reference fa) [37]. Before this, the BWA-ALN was done against the host genome (GRCh37 0.6.0 genome fa). The Genome Analysis Tool Kit (GATK) Haplotype caller 4.0.4.0 performed the variant calling and generated the variant caller format (∗.vcf) card [38]. Variations in the SARS-CoV-2 genome were visualized on the Integrated genome viewer (IGV) [39]. The SnpEff tool was used to annotate the variants observed in the SARS-CoV-2 sequences [40].
Data availability
All data generated or analyzed during this study are included with this article or provided as supplementary files. The raw DNA sequencing data were deposited at the public repository of the National Centre for Biotechnology Information (NCBI) under the accession number PRJNA728972 (SRR145227557-SRR14527571).
Results
SARS-CoV-2 detection by RT-PCR
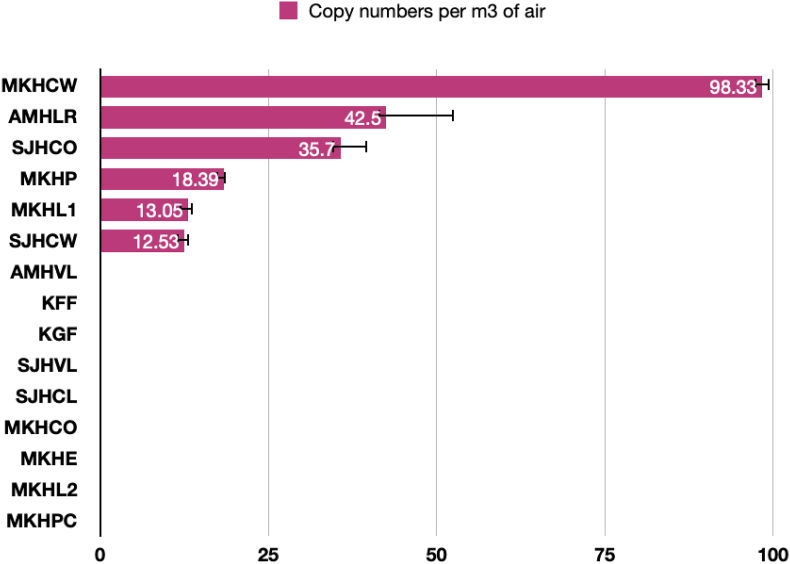
RT-PCR revealed the presence of SARS-CoV-2 at six out of fifteen (40%) locations. The cycle threshold (Ct) ranged from 35.11 to 38.18 (Table S1). The magnitude of the SARS-CoV-2 concentration at different locations is given in Figure 1. The highest copy numbers were 98.33 ± 1.18 copies per m3 of air at MKHCW (main ward admitting COVID patients in MKH hospital). This was followed by 42.50 ± 10.01 copies per m3 of air at AMHLR (reception area outside the virology lab receiving samples and dispatching results at AMH hospital), 35.70 ± 3.94 copies per m3 of air at SJHCO (Receiving area for COVID-19 suspects at SJH hospital), 18.39 ± 0.23 copies per m3 of air at MKHP (waiting area near the pharmacy near the main laboratory entrance at SJH hospital), 13.05 ± 0.70 copies per m3 of air at MKHL1 (central lab receiving patient samples at MKH hospital), and 12.53 ± 0.73 copies per m3 of air at SJHCW (main ward treating COVID patients at the SJH hospital).
Figure 1.
Viral copy numbers of SARS-CoV-2 as revealed by RT-PCR.
SARS-CoV-2 and Other Viruses as Detected by CRVP Sequencing.
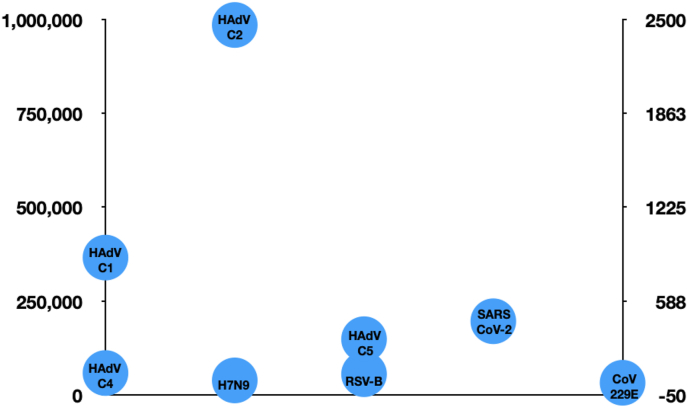
The NGS libraries of 15 samples generated 63828214 paired-end reads, of which 22499296 were used for mapping against the viral genomes after demultiplexing and quality control (Average-4255215; Maximum- 29905906; Minimum-1350006) (Table S2). The average Phred score was Q > 30 as revealed by the Fastxtoolkit analysis. Among the viral reads maximum mapped against the human adenoviruses type 1(HAdV-C1), at MKHL2 (2423460) and minimum (41536) at KGF. The highest reads of HAdV-C2 (6302519) was also found at MKHL2 whereas the lowest was 41536 at KGF. The read counts of HAdV-C5 were maximum (964157) at MKHL2 and minimum (17050) at KGF (Table S3). The HAdV-C1, HAdV-C2 and HAdV-C5 were found at all the locations. SARS-CoV-2 followed human adenoviruses in distribution. It was found at 13 out of 15 locations. It was maximum at SJHCO (1944) and minimum at SJHVL (34). Respiratory syncytial virus was maximum at MKHCW (165) and minimum at MKHP (25) found at 5 out of 15 locations. The non-SARS CoV-229E was present in three locations with the highest reads of 60 at SJHCW and a minimum of 20 at MKHL2 and MKHPC. The average read counts of each viral type detected by CRVP are shown in Figure 2. In general, the genome coverage of these viruses was low. This might be due to their low copy numbers in the aerosols.
Figure 2.
Average reads of viruses present in the indoor aerosols collected from hospitalized and non-hospitalized settings of Kuwait as captured by the CRVP.
Relative abundance of respiratory viruses and SARS-CoV-2 in indoor aerosols
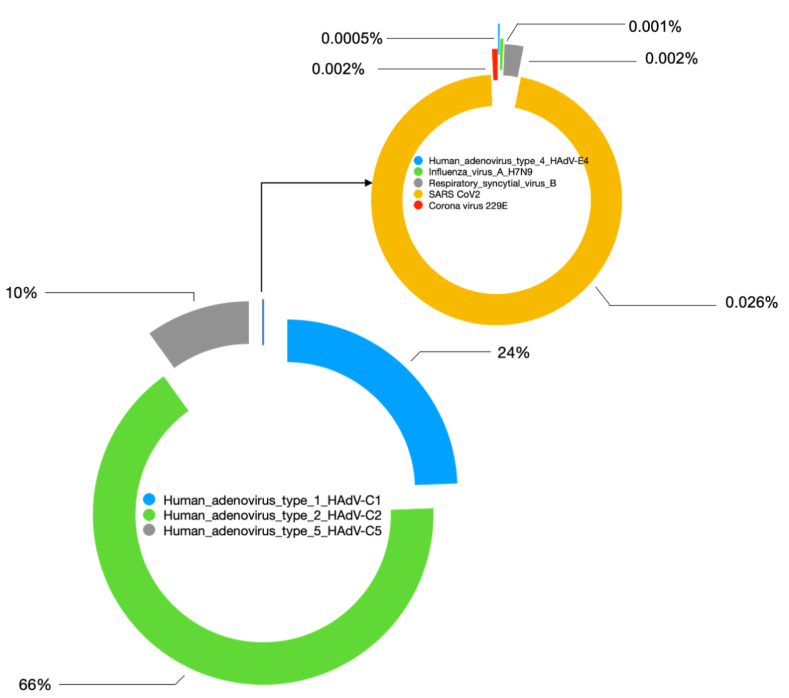
The relative abundance (RA) of the respiratory viruses and SARS-CoV-2 was calculated based on read distribution at a particular location. Human adenoviruses were the dominant viral population in the indoor hospital aerosols. The RA of the viruses was highest for Human adenoviruses (HAdV) type C2 (66%) followed by C1 (24%), C5 (10%), SARS-CoV-2 (0.026%), RSV (0.002%), non-SARS Coronavirus 229E (0.002%), H7N9 (0.001%) and HAdV-C4 (0.0005%) (Figure 3).
Figure 3.
Relative abundance/Ratio of respiratory viruses found in the aerosols of hospital environments in Kuwait.
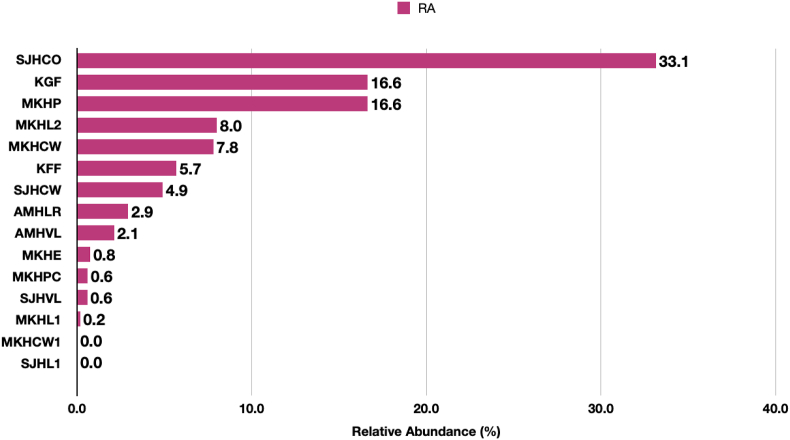
Among SARS-CoV-2, the maximum abundance was recorded at SJHCO (33.1%) followed by KGF and MKHP (16.6%). At MKHL2, MKHCW, KFF, SJHCW, AMHLR and AMHVL the RA was 8.0%, 7.8%, 5.7%, 4.9%, 2.9% and 2.1 % respectively (Figure 4). The RA was less than 1% at MKHE, MKHPC, SJHVL and MKHL1. The SARS-CoV-2 was found in both locations of non-hospitalized settings (KGF-16.6%; KFF-5.7%). The reason for presence of SARS-CoV-2 can be individuals exhalation from non-symptomatic patients and the recirculation of indoor air without any sterilization or passing through HEPA filters at KISR.
Figure 4.
Relative Abundance of SARS-CoV-2 in the indoor aerosols detected by the CRVP sequencing. Values at the end of each bar are the corresponding RA at each site. The read counts were used to calculate the RA and plotted on the X-axis. The Y-axis presents the location.
Variant calling and SNP annotation
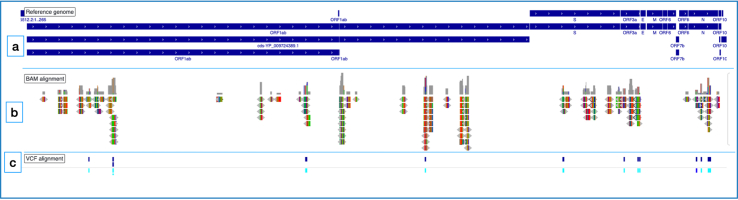
The genome sequences of SARS-CoV-2 was also used to look for variants. Alignment of the ∗.bam files and∗.vcf card file on the IGV browser against the SARS-CoV-2 (NC_045512.2-Wuhan China strain) revealed variations (single nucleotide polymorphisms, insertions and deletions) within the mapped regions of SARS-CoV-2 (Figure 5).
Figure 5.
Graphical view of captured SARS-CoV-2 reads. (a) Blue tracks representing the reference strain (NC_045512.2) of the SARS-CoV-2; (b) Bam files aligned against the reference genome (c) VCF files aligned against the reference genome.
Variant annotation by SNPEff informed all the variations as single nucleotide polymorphisms (SNPs) except for one insertion (C/CTTACAAAGGCACG) recorded at 26212nt at SJHCO. The corresponding nucleotide change and the position in the sequence are presented in Table II. Most of the SNPs occurred in the ORF1ab, ORF 3a, and N genes. One SNP was associated with the S gene in the SJHVL sample.
Table II.
Variations within the mapped SARS-CoV-2 genome
| Sample ID | Number of variants | SNPs | INS | Del | Nucleotide change | Position | Genes |
|---|---|---|---|---|---|---|---|
| SJHVL | 1 | 1 | 0 | 0 | C/T | 28814 | N |
| SJHCW | 2 | 2 | 0 | 0 | G/T; C/A | 12070; 12114 | ORF1ab |
| SJHCO | 7 | 6 | 1 | 0 | C/A; A/G; G/T; C/CTTACAAAGGCACG; T/A; T/C; A/G | 2880; 3916; 26144; 26212; 28611; 29107; 29125 | ORF1ab; ORF3a, N |
| MKHL2 | 2 | 2 | 0 | 0 | A/G | 3916 | S |
| MKHE | 1 | 1 | 0 | 0 | G/T | 25563 | ORF3a |
| MKHCW | 2 | 2 | 0 | 0 | G/T; T/C | 29179; 29213 | N |
| KGF | 1 | 1 | 0 | 0 | T/C | 17137 | ORF1ab |
| MKHL1 | 1 | 1 | 0 | 0 | A/G | 3916 | ORF1ab |
| Total | 17 | 16 | 1 | 0 |
SNPs- Single Nucleotide polymorphism; INS-Insertion; Del-Deletion.
The corresponding transcripts and the impact created by these variations are presented in Table III. Mainly the annotated SNPs caused a low (15) to moderate impact (16). One SNP in the N-gene and the ORF3a gene created a high impact. The effect produced by these mutations was either missense (16; 48.4%) or synonymous (15; 45.5%). However, one frameshift (3.03%) and stop gained (3.03%) mutations were also recorded. However, more complete genomes would provide insights into the overall rate of mutations, which was not covered in the present study.
Table III.
Annotation of mutations and their impact in the SARS-CoV-2
| Sample id | GeneName | Gene Id | Transcript Id | BioType | Variants Impact |
Variants Effect |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HIGH | LOW | MOD | FS | MS | SYN | SG | |||||
| SJHCO | N | GU280_gp10 | GU280_gp10 | Protein Coding | 2 | 1 | 1 | 2 | |||
| ORF1ab | GU280_gp01 | GU280_gp01 | Protein Coding | 1 | 1 | 1 | 1 | ||||
| ORF1ab | GU280_gp01 | GU280_gp01.2 | Protein Coding | 1 | 1 | 1 | 1 | ||||
| ORF1ab | GU280_gp01 | YP_009725299.1 | Protein Coding | 1 | 1 | 1 | 1 | ||||
| ORF1ab | GU280_gp01 | YP_009742610.1 | Protein Coding | 1 | 1 | 1 | 1 | ||||
| ORF3a | GU280_gp03 | GU280_gp03 | Protein Coding | 1 | 1 | 1 | 1 | ||||
| SJHCW | ORF1ab | GU280_gp01 | GU280_gp01 | Protein Coding | 1 | 1 | 1 | 1 | |||
| ORF1ab | GU280_gp01 | GU280_gp01.2 | Protein Coding | 1 | 1 | 1 | 1 | ||||
| ORF1ab | GU280_gp01 | YP_009725303.1 | Protein Coding | 1 | 1 | ||||||
| ORF1ab | GU280_gp01 | YP_009725304.1 | Protein Coding | 1 | 1 | ||||||
| ORF1ab | GU280_gp01 | YP_009742614.1 | Protein Coding | 1 | 1 | ||||||
| ORF1ab | GU280_gp01 | YP_009742615.1 | Protein Coding | 1 | 1 | ||||||
| SJHVL | N | GU280_gp10 | GU280_gp10 | Protein Coding | 1 | 1 | |||||
| KGF | ORF1ab | GU280_gp01 | GU280_gp01 | Protein Coding | 1 | 1 | |||||
| ORF1ab | GU280_gp01 | YP_009725308.1 | Protein Coding | 1 | 1 | ||||||
| MKHCW | N | GU280_gp10 | GU280_gp10 | Protein Coding | 1 | 1 | 1 | 1 | |||
| MKHE | ORF3a | GU280_gp03 | GU280_gp03 | Protein Coding | 1 | 1 | |||||
| MKHL2 | S | GU280_gp02 | GU280_gp02 | Protein Coding | 2 | 2 | |||||
| MKHL1 | ORF1ab | GU280_gp01 | GU280_gp01 | Protein Coding | 1 | 1 | |||||
| ORF1ab | GU280_gp01 | GU280_gp01.2 | Protein Coding | 1 | 1 | ||||||
| ORF1ab | GU280_gp01 | YP_009725299.1 | Protein Coding | 1 | 1 | ||||||
| ORF1ab | GU280_gp01 | YP_009742610.1 | Protein Coding | 1 | 1 | ||||||
| Total | 2 | 15 | 16 | 1 | 16 | 15 | 1 | ||||
Mod-Moderate; FS-Frameshift; MS-Missense; SG-Stop Gained; SYN-Synonymous.
Discussion
Hospital-acquired SARS-CoV-2 infection has not only impacted the patients and the health care workers but has spread beyond to other places via secondary transmission [3,9]. To ensure the safety of patients and health care workers during these challenging times, it is imperative to monitor the viral communities in the hospital aerosol regularly. NGS has been known to better understand nosocomial occurrence [3,9,41,42]. Within NGS, targeted virome capture has gained popularity and ise routinely being used to identify viruses in clinical specimens [[23], [24], [25], [26],43]. Very few studies have demonstrated its use for monitoring SARS-CoV-2 in environmental samples [4,26].
We found targetted CRVP sequencing on viral identification from aerosol samples collected from indoor air of three major hospitals involved in COVID-19 care in Kuwait to be very effective. RT-PCR detected SARS-CoV-2 in only 6 out of 15 locations (40%), whereas the CRVP captured reads at 13 of 15 sites (86%) [4,5]. Lower coverage of respiratory viruses was expected owing to the low biological content especially in the current environmental milieu (where indoor hospital air is filtered using HEPA filters). In environmental samples collected from hospital surfaces in the University of California - Davis, the majority of the samples depicted a coverage below 15% (2.19–14.78%). Our results were comparable with this study [44]. Low viral load, matrix interference and integrity damage are known and pose a significant challenge for the detection of viruses in environmental samples [45,46].
Our observation through RT-PCR detected very low copy numbers of SARS-CoV-2, i.e. at Ct-35-38 in 13–99 copies m3 of air. RT-PCR although powerful in terms of rapid turnaround time and quantification, is limited in sensitivity when the viral loads are less than 10 copies m3 of air [5,47]. The CRVP was, however, sensitive enough to pick SARS-CoV-2 from locations that tested negative by RT-PCR. Our results were congruent with a study conducted on hospital surface samples demonstrating that NGS protocol could detect SARS-CoV-2 in samples with undetermined Ct scores by RT-PCR [48]. The relative abundance of SARS-CoV-2 ranged from 0.2 to 33.1% as demonstrated by CRVP across indoor aerosols collected from 15 sites. Among these 16.6% and 5.7% belonged to the non-hospitalized locations of KGF and KFF, respectively. The spread of SARS-CoV-2 from hospitals to other places have been known [8,9,49,50]. Thus, CRVP would be a useful tool for monitoring the presence of SARS-CoV-2 in both hospital and non-hospital settings. This approach can be used as early warning to assess if there are viral cells in aerosol in low concentration; the information can also be used to adjust indoor - outdoor air exchange. The information becomes more important in both the hot and cold countries where people stay indoors and there is limited air exchanges. We believe such low concentrations in indoor air can be from exhaled air of non-symptomatic patients.
The infective dose of SARS-CoV-2 in humans is still unknown, as such studies are not feasible currently and need further research. However, the virus transmits rapidly and appears highly contagious, suggesting that the minimum infective dose is low [51]. The human infective dose for the closely related non-SARS-CoV-229E was reported as 13 tissue culture infectivity dose (TCID50) [52]. The TCID50 of rhinovirus and adenoviruses through aerosols was 0.68 and 0.5, respectively [53]. Clinical trials on African green monkeys revealed the infectivity dose of SARS-CoV-2 to be less via the aerosol route as compared to other modes of inoculation [54,55]. Thus the low viral loads found in the present study cannot be ignored and call for additional investigations on its infectivity.
With the added value of characterizing multiple viral genomes, the CRVP also picked viral sequences of HAdV-C2, C1, C5. RSV, non-SARS CoV 229E, H7N9 and HAdV- C4. We have previously demonstrated the presence of multiple viruses in similar surroundings through RT-PCR [4,5,56]. Differences in viral types are attributed to the difference in sensitivity of RT-PCR and NGS techniques depending on the viral load, and sequencing depth [26,57]. The CRVP approach was less sensitive to bocavirus, enterovirus and rhinoviruses identification when the viral loads were less [57]; however, the proposed approach of sampling a large aerosol volume is likely to address this concern. CRVP being better than RT-PCR in sensitivity for detection of SARS-CoV-2 [3,5,8,9,48] and the ability of RT-PCR to provide an estimation of viral counts suggests that both these approaches can complement each other for the monitoring of SARS-CoV-2 and other respiratory viruses from aerosol of hospitals and even non-hospital settings. The presence of a considerable portion of non-viral reads is speculated from bacterial and fungal species observed as a part of aerosol communities [56,[58], [59], [60]]. Characterization of these respiratory pathogens is warranted through 16s rRNA and ITS amplicon sequencing.
Although, SARS-CoV-2 has been classified as a slow mutating virus, researchers at the time of writing had detected 12,706 mutations in its genome [61], the majority arising from SNPs. In the present study, we have observed variations in the form of SNPs [56]. In yet another study, 298 frameshift variants, 789 missense variants, 1 start lost, 13 start gained, 1 stop lost, 249 synonymous variants and 720 upstream gene variants from 2200 genome variants of SARS-CoV-2 were reported [30]. Our results were in partial agreement with the above, where the maximum effect was missense followed by synonymous variants. Whether these variations could be the starting point for hospital-mediated outbreaks in the region remains a question worthy of further investigation.
Conclusions
This study demonstrates the superiority of CRVP in detection of SARS-CoV-2 in trace concentrations in the aerosols. The approach also provides meaningful information on co-existing respiratory viruses that can have significant epidemiological implications. The detection of a low level of SARS-CoV-2 can also be an early warning that may warrant increasing air exchange, and sanitization to minimize the transmission risk of COVID and provide surveillance with higher efficiency of virus detection in hospital environments.
Acknowledgements
The authors are thankful to the Kuwait Institute for Scientific Research and the Kuwait Foundation for the Advancement of Sciences for funding and supporting.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.infpip.2021.100199.
Conflict of interest statement
The authors have none to declare.
Funding
The study was funded by Kuwait Institute for Scientific Research (KISR grant no. FB157C) and the Kuwait Foundation for the Advancement of Sciences (KFAS grant no. PR18-14SE-01 and PN20-43BO-01).
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Stockwell R.E., Ballard E.L., O'Rourke P., Knibbs L.D., Morawska L., Bell S.C. Indoor hospital air and the impact of ventilation on bioaerosols: a systematic review. Journal of Hospital Infection. 2019;103(2):175–184. doi: 10.1016/j.jhin.2019.06.016. [DOI] [PubMed] [Google Scholar]
- 2.Duverger C., Souyri V., Monteil C., Fournier S., Espinasse F., Gramer M., et al. Controlling healthcare-associated transmission of SARS-CoV-2 Variant of Concern 202012/01 in a large hospital network. Journal of Hospital Infection. 2021 doi: 10.1016/j.jhin.2021.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paltansing S., Sikkema R., de Man S., Koopmans M., Munnink B.O., de Man P. Transmission of SARS-CoV-2 among healthcare workers and patients in a teaching hospital in the Netherlands confirmed by whole-genome sequencing. Journal of Hospital Infection. 2021;110:178–183. doi: 10.1016/j.jhin.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Habibi N., Uddin S., Al Salameen F., Al Amad S., Kumar V., Al Otaibi M. 2021. Identification and characterization of novel corona and associated respiratory viruses in aerosols. [Google Scholar]
- 5.Habibi N., Uddin S., Al Salameen F., Al Amad S., Kumar V., Al Otaibi M., et al. SARS-CoV - 2, Other Respiratory Viruses and Bacteria in Aerosols: Report from Kuwait’s Hospitals. Indoor Air. 2021 doi: 10.1111/ina.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., et al. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582(7813):557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- 7.Lednicky J.A., Lauzard M., Fan Z.H., Jutla A., Tilly T.B., Gangwar M., et al. Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. International Journal of Infectious Diseases. 2020;100:476–482. doi: 10.1016/j.ijid.2020.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbas M., Zhu N.J., Mookerjee S., Bolt F., Otter J.A., Holmes A.H., et al. Hospital-onset COVID-19 infection surveillance systems: A systematic review. Journal of Hospital Infection. 2021 doi: 10.1016/j.jhin.2021.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Løvestad A.H., Jørgensen S.B., Handal N., Ambur O.H., Aamot H.V. Investigation of intra-hospital SARS-CoV-2 transmission using nanopore whole-genome sequencing. Journal of Hospital Infection. 2021;111:107–116. doi: 10.1016/j.jhin.2021.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suárez-García I., de Aramayona López M.M., Vicente A.S., Abascal P.L. SARS-CoV-2 infection among healthcare workers in a hospital in Madrid, Spain. Journal of Hospital Infection. 2020;106(2):357–363. doi: 10.1016/j.jhin.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davido B., Gautier S., Riom I., Landowski S., Lawrence C., Thiebaut A., et al. The first wave of COVID-19 in hospital staff members of a tertiary care hospital in the greater Paris area: A surveillance and risk factors study. International Journal of Infectious Diseases. 2021;105:172–179. doi: 10.1016/j.ijid.2021.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nasir J.A., Kozak R.A., Aftanas P., Raphenya A.R., Smith K.M., Maguire F., et al. A Comparison of Whole Genome Sequencing of SARS-CoV-2 Using Amplicon-Based Sequencing, Random Hexamers, and Bait Capture. Viruses. 2020;12(8):895. doi: 10.3390/v12080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar P.S., Subramanian K. Demystifying the mist: Sources of microbial bioload in dental aerosols. Journal of Periodontology. 2020;91(9):1113–1122. doi: 10.1002/JPER.20-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunwar A., Tamrakar S., Poudel S., Sharma S., Parajuli P. Bacteriological assessment of the indoor air of different hospitals of Kathmandu District. International Journal of Microbiology. 2019;2019 doi: 10.1155/2019/5320807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habibi N., Mustafa A.S., Khan M.W. Composition of nasal bacterial community and its seasonal variation in health care workers stationed in a clinical research laboratory. PLoS One. 2021;16(11) doi: 10.1371/journal.pone.0260314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacIntyre R., Dwyer D., Seale H., Quanyi W., Yi Z., Yang P., et al. High risk procedures and respiratory infections in hospital health care workers–quantifying the risk. International Journal of Infectious Diseases. 2012;16:e379. [Google Scholar]
- 17.Macintyre C., Seale H., Yang P., Zhang Y., Shi W., Almatroudi A., et al. Quantifying the risk of respiratory infection in healthcare workers performing high-risk procedures. Epidemiology & Infection. 2014;142(9):1802–1808. doi: 10.1017/S095026881300304X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu G., Li X., Hu L., Jiang G. ACS Publications; 2020. An imperative need for research on the role of environmental factors in transmission of novel coronavirus (COVID-19) [DOI] [PubMed] [Google Scholar]
- 19.Hsih W.-H., Cheng M.-Y., Ho M.-W., Chou C.-H., Lin P.-C., Chi C.-Y., et al. Featuring COVID-19 cases via screening symptomatic patients with epidemiologic link during flu season in a medical center of central Taiwan. Journal of Microbiology, Immunology and Infection. 2020;53(3):459–466. doi: 10.1016/j.jmii.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gire S.K., Goba A., Andersen K.G., Sealfon R.S., Park D.J., Kanneh L., et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345(6202):1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Organization W.H. World Health Organization; 2020. Laboratory testing for coronavirus disease ( COVID-19) in suspected human cases: interim guidance, 19 March 2020. [Google Scholar]
- 22.Hoenen T., Groseth A., Rosenke K., Fischer R.J., Hoenen A., Judson S.D., et al. Nanopore sequencing as a rapidly deployable Ebola outbreak tool. Emerging Infectious Diseases. 2016;22(2):331. doi: 10.3201/eid2202.151796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K.W., Deveson I.W., Pang C.N.I., Yeang M., Naing Z., Adikari T., et al. Respiratory viral co-infections among SARS-CoV-2 cases confirmed by virome capture sequencing. Scientific Reports. 2021;11(1):1–9. doi: 10.1038/s41598-021-83642-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briese T., Kapoor A., Mishra N., Jain K., Kumar A., Jabado O.J., et al. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. MBio. 2015;6(5) doi: 10.1128/mBio.01491-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan S.H., Alamouti S.M., Kwok B.S., Lee M.-H., Khattra J., Daneshpajouh H., et al. Target Capture Sequencing of SARS-CoV-2 Genomes Using the ONETest Coronaviruses Plus. Diagnostic Microbiology and Infectious Disease. 2021;101(3):1–6. doi: 10.1016/j.diagmicrobio.2021.115508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorburn F., Bennett S., Modha S., Murdoch D., Gunson R., Murcia P.R. The use of next generation sequencing in the diagnosis and typing of respiratory infections. Journal of Clinical Virology. 2015;69:96–100. doi: 10.1016/j.jcv.2015.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grubaugh N.D., Petrone M.E., Holmes E.C. We shouldn’t worry when a virus mutates during disease outbreaks. Nature Microbiology. 2020;5(4):529–530. doi: 10.1038/s41564-020-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau B.T., Pavlichin D., Hooker A.C., Almeda A., Shin G., Chen J., et al. Profiling SARS-CoV-2 mutation fingerprints that range from the viral pangenome to individual infection quasispecies. Genome Medicine. 2021;13(1):1–23. doi: 10.1186/s13073-021-00882-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanjuán R., Nebot M.R., Chirico N., Mansky L.M., Belshaw R. Viral mutation rates. Journal of Virology. 2010;84(19):9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bindayna K.M., Crinion S. Variant analysis of SARS-CoV-2 genomes in the Middle East. Microbial Pathogenesis. 2021;153:104741. doi: 10.1016/j.micpath.2021.104741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habibi N., Behbehani M., Uddin S., AL Salamin F., Shajan A., Zakir F. In: Environmental resilience and transformation in times of COVID-19. Ramanathan A.L., Chidambaram S., Jonathan M.P., Munoz-Arriola F., Prasanna M.V., Kumar P., editors. Elsevier; 2021. A safe and effective sample collection method for assessment of SARS-CoV-2 in aerosol samples; pp. 194–199. [DOI] [Google Scholar]
- 32.Rio D.C., Ares M., Hannon G.J., Nilsen T.W. Purification of RNA using TRIzol (TRI reagent) Cold Spring Harbor Protocols. 2010;2010(6) doi: 10.1101/pdb.prot5439. pdb. prot5439. [DOI] [PubMed] [Google Scholar]
- 33.Anonymous. 2021. Fastx-Toolkit 0.0.14. [Google Scholar]
- 34.Lindgreen S. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Research Notes. 2012;5(1):1–7. doi: 10.1186/1756-0500-5-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tischler G., Leonard S. biobambam: tools for read pair collation based algorithms on BAM files. Source Code for Biology and Medicine. 2014;9(1):1–18. [Google Scholar]
- 37.Li H. Broad Institute; 2010. Aligning new-sequencing reads by BWA. [Google Scholar]
- 38.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pillay S., Giandhari J., Tegally H., Wilkinson E., Chimukangara B., Lessells R., et al. Whole genome sequencing of SARS-CoV-2: adapting Illumina protocols for quick and accurate outbreak investigation during a pandemic. Genes. 2020;11(8):949. doi: 10.3390/genes11080949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parcell B.J., Gillespie S.H., Pettigrew K.A., Holden M.T. Clinical perspectives in integrating whole-genome sequencing into the investigation of healthcare and public health outbreaks–hype or help? Journal of Hospital Infection. 2021;109:1–9. doi: 10.1016/j.jhin.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klempt P., Brož P., Kašný M., Novotný A., Kvapilová K., Kvapil P. Performance of targeted library preparation solutions for SARS-CoV-2 whole genome analysis. Diagnostics. 2020;10(10):769. doi: 10.3390/diagnostics10100769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coil D.A., Albertson T., Banerjee S., Brennan G., Campbell A.J., Cohen S.H., et al. SARS-CoV-2 detection and genomic sequencing from hospital surface samples collected at UC Davis. PLoS One. 2021;16(6) doi: 10.1371/journal.pone.0253578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fennelly K.P., Tribby M.D., Wu C.-Y., Heil G.L., Radonovich L.J., Loeb J.C., et al. Collection and measurement of aerosols of viable influenza virus in liquid media in an Andersen cascade impactor. Virus Adapt Treat. 2015;7:1–9. [Google Scholar]
- 46.Yao L., Zhu W., Shi J., Xu T., Qu G., Zhou W., et al. Detection of coronavirus in environmental surveillance and risk monitoring for pandemic control. Chemical Society Reviews. 2021;50(6):3656–3676. doi: 10.1039/d0cs00595a. [DOI] [PubMed] [Google Scholar]
- 47.García-Arroyo L., Prim N., Martí N., Roig M.C., Navarro F., Rabella N. Benefits and drawbacks of molecular techniques for diagnosis of viral respiratory infections. Experience with two multiplex PCR assays. Journal of Medical Virology. 2016;88(1):45–50. doi: 10.1002/jmv.24298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coil D.A., Albertson T., Banerjee S., Brennan G., Campbell A.J., Cohen S.H., et al. SARS-CoV-2 detection and genomic sequencing from hospital surface samples collected at UC Davis. medRxiv. 2021 doi: 10.1371/journal.pone.0253578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jerry J., O'Regan E., O'Sullivan L., Lynch M., Brady D. Do established infection prevention and control measures prevent spread of SARS-CoV-2 to the hospital environment beyond the patient room? Journal of Hospital Infection. 2020;105(4):589–592. doi: 10.1016/j.jhin.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kampf G., Brüggemann Y., Kaba H.E., Steinmann J., Pfaender S., Scheithauer S., et al. Potential sources, modes of transmission and effectiveness of prevention measures against SARS-CoV-2. Journal of Hospital Infection. 2020 doi: 10.1016/j.jhin.2020.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karimzadeh S., Bhopal R., Nguyen Tien H. Review of infective dose, routes of transmission, and outcome of COVID-19 caused by the SARS-CoV-2 virus: Comparison with other respiratory viruses. Preprints. 2020;2020070613:20944. [Google Scholar]
- 52.Watanabe T., Bartrand T.A., Weir M.H., Omura T., Haas C.N. Development of a dose-response model for SARS coronavirus. Risk Analysis. An International Journal. 2010;30(7):1129–1138. doi: 10.1111/j.1539-6924.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Couch R.B., Cate T.R., Douglas R.G., Jr., Gerone P.J., Knight V. Effect of route of inoculation on experimental respiratory viral disease in volunteers and evidence for airborne transmission. Bacteriological Reviews. 1966;30(3):517–529. doi: 10.1128/br.30.3.517-529.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnston S.C., Ricks K.M., Jay A., Raymond J.L., Rossi F., Zeng X., et al. Development of a coronavirus disease 2019 nonhuman primate model using airborne exposure. PLoS One. 2021;16(2) doi: 10.1371/journal.pone.0246366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blair R.V., Vaccari M., Doyle-Meyers L.A., Roy C.J., Russell-Lodrigue K., Fahlberg M., et al. ARDS and cytokine storm in SARS-CoV-2 infected caribbean vervets. bioRxiv. 2020 [Google Scholar]
- 56.Behbehani M., Uddin S., Habibi N., Al Salameen F., Sajid S., Abdulrazzack N., et al. Kuwait Institute for Scientific Research; Kuwait: 2021. 210Po in Ultrafine aerosol particles and its likelihood to mutate the microbial community. FB160C. [Google Scholar]
- 57.Prachayangprecha S., Schapendonk C.M., Koopmans M.P., Osterhaus A.D., Schürch A.C., Pas S.D., et al. Exploring the potential of next-generation sequencing in detection of respiratory viruses. Journal of Clinical Microbiology. 2014;52(10):3722–3730. doi: 10.1128/JCM.01641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Al Salameen F., Habibi N., Uddin S., Al Mataqi K., Kumar V., Al Doaij B., et al. Spatio-temporal variations in bacterial and fungal community associated with dust aerosol in Kuwait. PLoS One. 2020;15(11) doi: 10.1371/journal.pone.0241283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salameen F.A., Habibi N., Uddin S., Mataqi K.A., Doaij B.A., Amad S.A., et al. Figshare; 2021. Characterization and identification of microorganisms associated with airborne dust in Kuwait. [Google Scholar]
- 60.Habibi N., Uddin S., Salameen F.A., Behbehani M., Shirshikhar F., Razzack N.A., et al. Collection of Bacterial Community Associated with Size Fractionated Aerosols from Kuwait. Data. 2021;6(12):123. [Google Scholar]
- 61.Akkiz H. Implications of the Novel Mutations in the SARS-CoV-2 Genome for Transmission, Disease Severity, and the Vaccine Development. Frontiers in Medicine. 2021;8 doi: 10.3389/fmed.2021.636532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included with this article or provided as supplementary files. The raw DNA sequencing data were deposited at the public repository of the National Centre for Biotechnology Information (NCBI) under the accession number PRJNA728972 (SRR145227557-SRR14527571).