Abstract
Polycomb group proteins act through Polycomb group response elements (PREs) to maintain silencing at homeotic loci. The minimal 1.5-kb bithoraxoid (bxd) PRE contains a region required for pairing-sensitive repression and flanking regions required for maintenance of embryonic silencing. Little is known about the identity of specific sequences necessary for function of the flanking regions. Using gel mobility shift analysis, we identify DNA binding activities that interact specifically with a multipartite 70-bp fragment (MHS-70) downstream of the pairing-sensitive sequence. Deletion of MHS-70 in the context of a 5.1-kb bxd Polycomb group response element derepresses maintenance of silencing in embryos. A partially purified binding activity requires multiple, nonoverlapping d(GA)3 repeats for MHS-70 binding in vitro. Mutation of d(GA)3 repeats within MHS-70 in the context of the 5.1-kb bxd PRE destabilizes maintenance of silencing in a subset of cells in vivo but gives weaker derepression than deletion of MHS-70. These results suggest that d(GA)3 repeats are important for silencing but that other sequences within MHS-70 also contribute to silencing. Antibody supershift assays and Western analyses show that distinct isoforms of Polyhomeotic and two proteins that recognize d(GA)3 repeats, the TRL/GAGA factor and Pipsqueak (Psq), are present in the MHS-70 binding activity. Mutations in Trl and psq enhance homeotic phenotypes of ph, indicating that TRL/GAGA factor and Psq are enhancers of Polycomb which have sequence-specific DNA binding activity. These studies demonstrate that site-specific recognition of the bxd PRE by d(GA)n repeat binding activities mediates PcG-dependent silencing.
The developmental fate of parasegments along the antero-posterior axis in Drosophila is determined by the spatially regulated expression of the homeotic genes of the Antennapedia complex and the Bithorax complex (BX-C) (1, 12). Parasegmentally restricted expression of homeotic genes is temporally regulated in two distinct phases: initiation (stages 5 to 11) and maintenance (stages 8 to 17). Initiation of homeotic gene silencing in defined territories occurs because transiently expressed transcription factors encoded by the segmentation genes silence homeotic gene expression in specific parasegments. Later in embryogenesis, maintenance of silencing of homeotic genes requires the action of Polycomb group (PcG) proteins (53). Mutations in PcG genes do not affect initiation of silencing but result in parasegment-selective misexpression of the homeotic genes during the maintenance phase (36, 54, 55, 58). Maintenance of activation of homeotic gene expression requires the action of the trithorax group (trxG) genes (32).
PcG proteins mediate maintenance of silencing at target loci through complex, modular elements termed PcG response elements (PREs). PREs have different modes of silencing, monitored using different assays. PREs were originally defined by their ability to maintain silencing of gene expression regulated by parasegment-specific enhancers in the BX-C (55). In embryos, transgenes regulated by endogenous promoters and parasegment-specific enhancers of homeotic loci show correct initiation of spatial regulation, but during germband extension, transgene expression is derepressed owing to the absence of the PRE sequences required to maintain silencing. Mutations in PcG genes prevent maintenance of silencing mediated by PREs, showing that silencing through PREs is PcG dependent (7–10, 23, 38, 45, 46, 55). Immunohistochemistry experiments on polytene chromosomes (11, 67) and formaldehyde cross-linking experiments show that PcG proteins are associated with known PREs in vivo (40, 60). PREs also silence adjacent reporters used to select transgenic flies (7, 9, 18, 22, 55).
A novel repressive mechanism called pairing-sensitive repression is detected when PREs are in trans. PRE-dependent silencing of the adjacent mini-white reporter gene, contained in the transposon, is stronger from homozygous transgenes than from a heterozygous transgene, presumably because the PREs repress more efficiently when paired. Normally, expression of homozygous transgenes is higher than expression of heterozygous transgene because there are two copies of the transgene rather than one (18, 22, 29, 30). Molecular analysis of PREs shows that the regions responsible for pairing-sensitive repression are separable from the regions required for maintenance of embryonic silencing, as has been shown for the Sex combs reduced PREs (22), the bithoraxoid (bxd) PRE (51), and the infra-abominal-2 (iab-2) PRE (50). In the best-studied case, the shortest fragment from the bxd PRE that exhibits embryonic silencing and pairing-sensitive repression, the 1.5-kb maintenance (M) element, was shown to consist of a central 661-bp fragment required for pairing-sensitive repression flanked by sequences required for embryonic silencing (52).
A central question is how PcG proteins recognize PREs site specifically. No PcG protein binds DNA directly, except for Pleiohomeotic (PHO) (6). PHO is the Drosophila homologue of YY1 and recognizes a short consensus sequence found in many but not all PREs (37). A key observation is that PHO sites are necessary but not sufficient for the pairing sensitivity of an engrailed PRE tested in the white repression assay (6). Mutations of two PHO binding sites in a 160-bp fragment (C3) from the bxd PRE abolished pairing sensitivity of the white gene, supporting the possibility that PHO can recruit other PcG proteins to this fragment (62). PHO sites in the bxd PRE are necessary for silencing of Ultrabithorax (Ubx) in imaginal discs, but no effect was reported on maintenance of embryonic silencing (20). PHO binding sites from the iab-2 regulatory region of abdominal-A are necessary for pairing-sensitive repression of white but are not sufficient to maintain embryonic silencing (50). Recently, it has been shown that if PHO is tethered to a reporter gene, it is unable to silence the reporter, presumably because it is not sufficient to recruit other PcG proteins to this site (47). Together, the data suggest that PHO is necessary for pairing sensitivity and silencing in imaginal discs but that PHO binding sites by themselves are insufficient to confer maintenance of embryonic silencing.
The PHO data strongly imply that sites flanking the pairing-sensitive element in the bxd PRE may recognize other PcG complexes. In vitro binding assays show that subfragments of the 1.5-kb or M element of the bxd PRE that lack PHO binding sites recognize binding activities containing Polycomb (PC) and Posterior Sex Combs (PSC) (26). In the iab-2 regulatory region, sequences from iab-2 that did not contain PHO binding sites could confer embryonic silencing on reporter genes, indicating that PREs are complex, modular elements (50). Many PREs contain d(GA)n repeats, which are recognized by GAGA factor (GAF) (5, 31, 66) and by Pipsqueak (Psq) (35). GAF is an antirepressor for many genes (33, 39) and is encoded by Trithorax-like (Trl), a member of the trxG (17). TRL/GAF colocalizes with PcG proteins at many PREs (59), which could be explained if trxG response elements and PREs are closely situated (62). Trl mutations decrease silencing by the Fab-7 PRE, suggesting that TRL/GAF is required for silencing by this PRE (23). Consistent with this idea, coimmunoprecipitation experiments show that TRL/GAF associates with some PcG complexes that recognize the bxd PRE (26). Psq is a DNA binding protein required in oogenesis and eye development (51, 64), but its role in homeotic gene regulation is unknown.
It is important to identify minimum fragments necessary for PRE function in order to understand the mechanism of site-specific recognition of PREs by PcG-containing DNA binding activities and to determine how PREs are assembled from modular subunits. In this work, we analyze a 485-bp region in the 1.5-kb M element that is downstream of the 661-bp pairing-sensitive region (52), termed DPS (downstream of Pairing-Sensitive region). We use a gel mobility shift assay to screen fragments from DPS for DNA binding activities, using nuclear extract made from Kc cells. One binding site, termed MHS-70, impairs maintenance of embryonic silencing when deleted from a 5.1-kb bxd PRE, demonstrating that this sequence is necessary for PRE function in vivo. Other DNA fragments from within the M element of the bxd PRE compete for binding to MHS-70, suggesting that related sites of recognition are located elsewhere in the bxd PRE. Mutational analysis of MHS-70 shows that in vitro binding depends on d(GA)3 repeat elements. Mutation of the d(GA)3 repeat elements in MHS-70 in the context of the 5.1-kb bxd PRE destabilizes embryonic maintenance of silencing, showing that d(GA)3 elements are required for PRE function. The increased severity of derepression exhibited by the MHS-70 deletion compared to the substitution mutants suggests that other sequences in MHS-70 also contribute to maintenance of silencing. Fractionation of the nuclear extract shows that distinct binding activities recognize MHS-70. One partially purified binding activity is supershifted by antibodies to Polyhomeotic (PH), showing that it contains at least one PcG protein. Western blot and antibody supershift analyses of this activity show that distinct isoforms of PH, and two proteins that recognize d(GA)n repeats, TRL/GAF and Psq, recognize MHS-70 in vitro. Interestingly, the partially purified fraction does not contain PC, Suppressor of zeste [SU(Z)2], or PSC, indicating that we have identified a novel PcG activity which is distinct from the PRC1 complex. Genetic analysis shows that mutations in Trl and psq enhance the homeotic phenotypes of ph. We conclude that TRL/GAF and Psq are enhancers of PC which have sequence-specific DNA binding activity. These results suggest that site-specific recognition of the bxd PRE by d(GA)n repeat binding activities mediate PcG-dependent silencing.
MATERIALS AND METHODS
Plasmid vectors and linker substitution mutants.
The BX-C genomic subclone 3103 containing the 11-kb BamHI bxd fragment of phage L2212 (2) was digested with BamHI and HindIII to isolate a 5.1-kb subfragment of bxd6.5 (Fig. 1A), which was subcloned into pBluescript KS+ [pBS(KS+)] to generate pBS-3101HB. The 1.5-kb StyI-EcoRI fragment or M element (designated 2212StR1.6 in reference 8) was subcloned as a blunt-ended fragment into the EcoRV site of pBS to generate pBS-3103M (Genbank accession number L32205). Restriction fragments generated from the 0.485-kb M fragment (Fig. 1A, DPS) were end repaired, subcloned into the EcoRV site of pBS, and sequenced. The 70-bp MHS-70 fragment was synthesized as blunt-end complementary phosphorylated nucleotides and subcloned into the EcoRV site of pBS. Linker substitutions of MHS-70 (Fig. 5A) and multiple synthetic repeats (MSR) of d(GA)3, d(A)8, and d(A)5 (Fig. 6A) were synthesized as phosphorylated complementary oligonucleotides and subcloned as blunt-ended fragments into the EcoRV site of pBS. Sequences of the upper strands of wild-type, and mutant MHS-70 and MSR of MHS-70 are shown in Table 1.
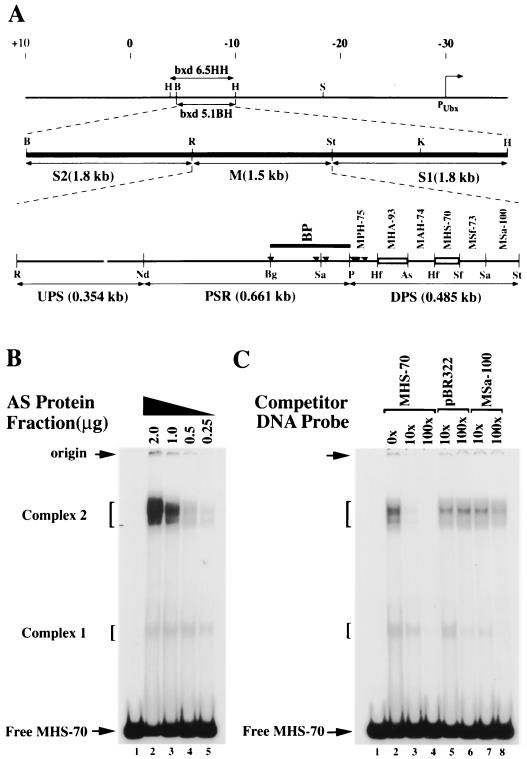
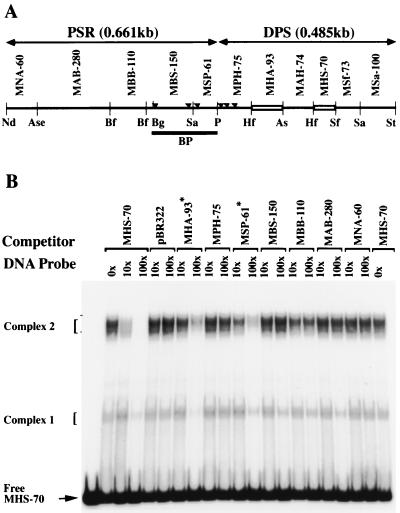
FIG. 1.
Identification of binding activities recognizing MHS-70. (A) Schematic representation of the organization of the bxd PRE. The start site of Ubx transcription is marked by a bent arrow, the coordinates of Bender et al. (2) are marked at the top, and the positions of bxd6.5HH and bxd5.1BH are indicated. The functional regions of bxd5.1, S1, M, and S2 are indicated on the second line and are described in the text. A detailed restriction map of the M element is shown on the third line. Fragments are named by the first letters of the restriction enzymes defining them, followed by their length in base pairs. PSR corresponds to the pairing-sensitive element defined by Sigrist and Pirrotta (52), and the dark line marked BP identifies the fragment described by Horard et al. (26). Small triangles mark the location of PHO binding sites (20). Open boxes represent binding sites identified in fragment DPS. Restriction site abbreviations: As, AseI; B, BamHI; Bg, BglI; H, HindIII; Hf, HinfI; K, KpnI; Nd, NdeI; P, PstI; R, EcoRI; S, SalI; Sa, Sau3A; Sf, SfaNI; St, StyI. (B) Gel mobility shift analyses of complexes formed at MHS-70 with no protein (lane 1) or with decreasing amounts of the AS fraction of KcI cell nuclear extract (lanes 2 to 5). Positions of nucleoprotein complexes formed are shown on the left. (C) Competitive gel mobility shift analysis of complexes formed at MHS-70 (with 0.6 μg of the AS fraction) in the presence of a molar excess of unlabeled, nonspecific (pBR322 BamHI-SalI 276-bp fragment) and specific (MSa-100) competitor DNA fragment (shown on top). Complex 2 formation is sequence specific.
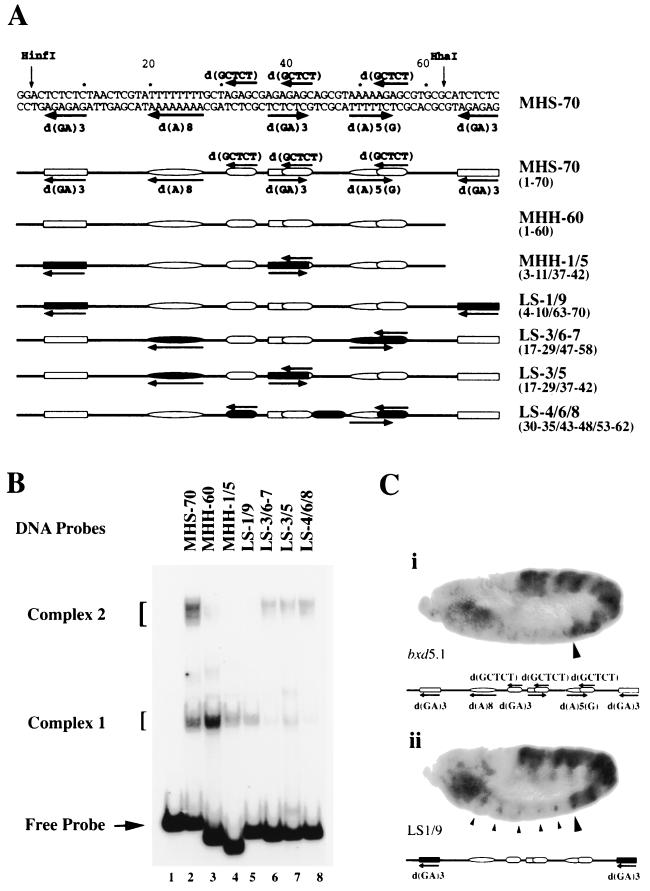
FIG. 5.
Gel shift and in vivo analysis of MHS-70 linker-scanning mutations. (A) Schematic representation of MHS-70 and mutated derivatives. Details of mutations are given in the text. The sequences of interest are marked in the top line and schematically represented in the second line. Sites of mutations are indicated by filled regions in the schematic representations of mutated MHS-70 fragments in lines 3 to 7. Coordinates of the substitution sites are shown on the right in parentheses. (B) Mobility shift analysis of MHS-70 mutants using 0.25 μg of QS0.15. The identities of mutant fragments used in each lane are shown in panel A. Complex 2 formation is abolished by combined substitutions of the d(GA)3 repeats. (C) Derepression caused by d(GA)3 substitution mutations of MHS-70 in bxd5.1 PRE. Conditions and reporters are explained in the legend to Fig. 4. The anterior boundary of PS6 is marked with a large arrowhead. The MHS-70 fragment is schematically represented below each panel. (i) bxd5.1 UbxlacZ from a line exhibiting spotty misexpression in PS1 to PS5 and partial derepression in the head. The PS6 boundary is maintained. (ii) bxd5.1 UbxlacZ mutant for two d(GA)n sites of MHS-70, LS-1/9 (illustrated in Fig. 5A). Note the clear derepression in PS1 to PS5 in a distinct subset of cells (small arrowheads), plus increased derepression in the head.
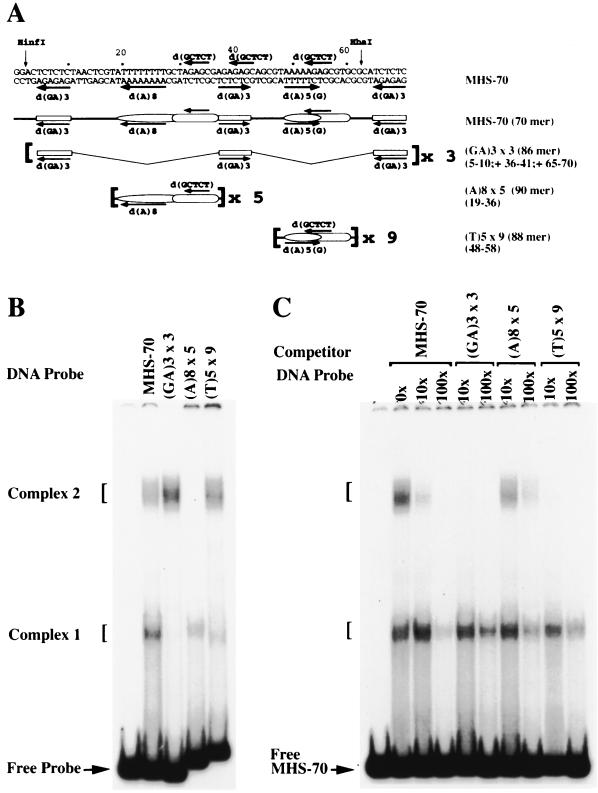
FIG. 6.
Gel shift analysis of MHS-70 MSR elements. (A) Schematic representation of oligomers containing MSR elements used in binding studies. Lines 1 and 2 are the same as in Fig. 5A; lines 3 to 5 show the sequences used to construct oligonucleotides. Sequences of the oligonucleotides are given in Table 1. (B) Mobility shift analysis of oligonucleotides derived from MHS-70 with 0.25 μg of QS0.15. Identities of the oligonucleotides are shown in panel A, and coordinates of the repeat elements are shown on the right in parentheses. Complex 2 is formed at (GA)3 × 3 and (T)5 × 9, and complex 1 is formed at (A)8 × 5 and (T)5 × 9. (C) Competitive mobility shift assay. Molar excesses of the oligonucleotides shown in panel A were used as competitors for the binding of 0.25 μg of QS0.15 to MHS-70. (GA) 3 × 3 and (T)5 × 9 abolished the formation of complex 2. The (A)8 × 5 oligomer did not inhibit the formation of complex 2.
TABLE 1.
Synthetic wild-type and linker mutant oligonucletides used
| Oligonucleotide | Sequencea |
|---|---|
| MHS-70 | 5′-GGACTCTCTCTAACTCGTATTTTTTTTGCTAGAGCGAGAGAGCAGCGTAAAAAGAGCGTGCGCATCTCTC-3′ |
| MHH-60 | 5′-GGACTCTCTCTAACTCGTATTTTTTTTGCTAGAGCGAGAGAGCAGCGTAAAAAGAGCGTG-3′ |
| MHH-1/5 | 5′-GGTGGAACGCCAACTCGTATTTTTTTTGCTAGAGCGCGATCGCAGCGTAAAAAGAGCGTG-3′ |
| LS-1/9 | 5′-GGATGGAAGCTAACTCGTATTTTTTTTGCTAGAGCGAGAGAGCAGCGTAAAAAGAGCGTGCGTGCTAGCG-3′ |
| LS-3/6-7 | 5′-GGACTCTCTCTAACTCGACGCCTAGCCTGAAGAGCGAGAGAGCAGATCGCTAGCCTGCTGCGCATCTCTC-3′ |
| LS-3/5 | 5′-GGACTCTCTCTAACTCGACGCCTAGCCTGAAGAGCGCGATCGCAGCGTAAAAAGAGCGTGCGCATCTCTC-3′ |
| LS-4/6/8 | 5′-GGACTCTCTCTAACTCGTATTTTTTTTGCGGACATGAGAGAGAGTGTGAAAACACTGTAACCCATCTCTC-3′ |
| (GA)3 × 3 | (5′-TCTCTCagcgAGAGAGagcgTCTCTC-3′)3 |
| (A)8 × 5 | (5′-ATTTTTTTTGCTAGAGCG-3′)5 |
| (T)5 × 9 | (5′-TAAAAAGAGCG-3′9 |
Nucleotide substitutions are shown in bold uppercase. Nucleotides shown in bold lowercase in (GA)3 × 3 represent spacer sequences.
Cell culture and fractionation of nuclear extracts.
Drosophila Kc1 cells were grown in spinner cultures with Sf-900 II serum-free medium (Gibco BRL) to a density of 7.5 × 106 cells/ml. Crude nuclear extracts were prepared from approximately 4.5 × 1010 cells by ammonium sulfate (AS) lysis of isolated nuclei as previously described (24, 41). All buffers used for protein extraction and fractionation were freshly supplemented with 1 mM benzamidine-HCl, 1 mM sodium bisulfite, 0.08% bacitracin, 10 μg of pepstatin per ml, 0.15 mM bestatin, 0.02% soybean trypsin inhibitor, 2 trypsin inhibitor units (TIU) of aprotinin, 0.5 mM phenylmethylsulfonyl fluoride, 5 mM KF, 40 μM phenyl arsine oxide, 0.2 mM sodium molybdate, and dithiothreitol (DTT). The AS precipitate of the nuclear lysate was solubilized in one-sixth of the packed cell volume in buffer HEMG0.1 (25 mM HEPES-KOH [pH 7.6], 12.5 mM MgCl2, 1.5 mM DTT, 1 mM EDTA, 1 mM EGTA, 0.01% Brij 36T, 25 μM zinc acetate, 15% [vol/vol] glycerol, (0.1 M KCl) and dialyzed against the same buffer for 3 h at 4°C. The dialysate was stored at −80°C (at a protein concentration of 30 to 40 mg/ml). Approximately 200 mg of the AS fraction was diluted with buffer HEMG0.0 to the conductivity of buffer HEMG0.1 (25 mM HEPES-KOH [pH 7.6], 6.25 mM MgCl2, 1.0 mM DTT, 0.1 mM EDTA, 0.1 mM EGTA, 0.01% Brij 36T, 50 μM zinc acetate, 0.5 mM ATP, 20% [vol/vol] glycerol, (0.1 M KCl) and applied to a Bio-Rex 70 column (200/400 mesh, Na+; Bio-Rad) equilibrated with buffer HEMG0.1 (1.0 by 17.4 cm, 10 mg of protein/ml of resin). The column was washed with 2.5 column volumes (cv) of buffer HEMG0.1 at 2 cv/hr and eluted with step gradients of 2.5 cv of buffer HEMG containing 0.18, 0.3, 0.6, and 0.85 M KCl. The Bio-Rex 70 0.6 M KCl fraction (BR0.6) was diluted with buffer HEMG0.085 to the conductivity of buffer HEMG0.085 (buffer HEMG containing 0.085 M KCl) and applied to a Q Sepharose column (Pharmacia) equilibrated with buffer HEMG0.085 (0.7 by 10 cm, 5 mg of protein/ml of resin). The column was washed with 3 cv of buffer HEMG0.15 at 0.5 ml/min and eluted with step gradients of 3 cv of buffer HEMG containing 0.3, 0.45, and 0.60 M KCl. All chromatographic fractions were stored in their corresponding elution buffers at −80°C. DNA binding activity was tested at each fractionation step using a gel mobility shift assay (21) as described below.
Antibodies and Western blot analysis.
The generation and purification of rabbit anti-Polyhomeotic Proximal (PHP) and -Polyhomeotic Distal (PHD) polyclonal antibody have been previously described (25). Rabbit anti-SU(Z)2 antibody was generated to an Escherichia coli expressed protein of the 213-amino-acid COOH-terminal region of SU(Z)2 (amino acid residues 1153 to 1365) fused to glutathione S-transferase (GST), using previously described procedures (25). Briefly, a BglII restriction fragment from the 3′ end of the Su(z)2 cDNA clone (a kind gift from Paul Adler) was subcloned into the BamHI site of pGEX-3X (Pharmacia) and expressed in E. coli AD202. The fusion protein was affinity purified over a glutathione-agarose column and used to immunize New Zealand White rabbits. Antiserum collected after five booster immunizations was depleted of anti-GST antibodies by using a Sepharose 4B GST affinity column. The depleted antiserum was affinity purified over a Sepharose 4B GST-SU(Z)2 agarose column by elution with 10 mM Na2HPO4-NaOH (pH 12.0)–50% ethylene glycol at 25°C, neutralized with 0.5 M NaH2PO4 (pH 2.5), and dialyzed into phosphate-buffered saline–5% glycerol. Rabbit anti-TRL/GAF P67 polyclonal antibody was a kind gift from Peter Becker and Carl Wu. Rabbit anti-Psq antibody (AS1 [27]) was a kind gift from Celeste Berg. Rabbit anti-PC polyclonal antibodies were obtained from Patrick O'Farrell. For Western blot analysis, 20 μg of protein from the AS, BR0.6, and Q Sepharose 0.15 M KCl (QS0.15) fractions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 9% gel and transferred onto nitrocellulose membranes by using a semidry blotter. The blots were probed with primary antibodies diluted in 5% nonfat dry milk–Tris-buffered saline (TBS; 50 mM Tris-HCl [pH 7.6], (150 mM NaCl) at a titers of 1:3,000 (anti- TRL/GAF P67); 1:1,000 (anti-PHP), 1:5,000 (anti-Psq [AS1]), 1:2,000 (anti-PC), and 1:1,000 [anti-SU(Z)2] for 5 h at room temperature. The blots were subsequently probed with goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (Pierce) diluted in 5% nonfat dry milk–TBS at a titer of 1:200,000 for 90 min at room temperature. The blots were washed with several changes of TBS–0.5% Tween 20 (BDH) for 90 min and once with TBS–0.01% Tween containing 1.0 M NaCl for 60 min. After a final wash with TBS for 15 min, the blot was developed with enhanced chemiluminescence using the Supersignal West Femto substrate (Pierce).
DNA binding assays.
Restriction fragments were excised with EcoRI and HindIII and filled in with α-32P-labeled deoxynucleoside triphosphate in a Klenow reaction. The radiolabeled fragments were desalted in a spin column of Sephadex G-50 and used as probes in gel mobility shift assays. For assays of protein fractions, a 15-μl binding reaction mixture consisting of 3 μl of a 5× binding buffer (125 mM HEPES-KOH) [pH 7.6], 50 μM zinc acetate, 15.625 mM MgCl2, 150 mM KCl, 12.5 mM DTT, 0.05% Brij 36T, 25% glycerol) supplemented with 4% (wt/vol) Ficoll type 400, 1.2 mM ATP, 2.33 to 2.8 mg of bovine insulin (zinc crystals; Gibco BRL) per ml, 1 mM sodium bisulfite, 0.5 mM phenylmethylsulfonyl fluoride, 1.5 μl of poly (dA-dG) · poly (dC-T) (approximately 600 bp; Pharmacia), 1 μl of DNA probe (25,000 cpm/∼0.25 ng of DNA), and various amounts of protein fraction (as indicated in Results) diluted in buffer HEG0.18 (buffer HEMG0.18 lacking MgCl2) was incubated at 25°C for 20 min (incubation step) and placed on ice for an additional 20 min. The reaction mixture was resolved at 110 V (constant voltage) on a 4% polyacrylamide gel (mono/bis ratio of 40/0.5) in 0.5× Tris-borate-EDTA (native PAGE), dried, and exposed to X-ray film at −70°C.
In DNA competition binding assays, the radiolabeled probe and 10- or 100-fold molar excess of unlabeled competitor DNA restriction fragments were added simultaneously to the binding reaction. In gel mobility supershift assays, antibody recognition of protein-DNA complexes was tested by adding antibodies at various titers in TBS (as indicated in Results), and a mixture of 3 μg of biotinylated goat anti-rabbit antibody (Vector Laboratories) and 1.3 μg of streptavidin (Jackson ImmunoResearch) preincubated on ice for 60 min, to the DNA binding reaction after the incubation step. The immune reaction mixture was incubated at 25°C for an additional 15 min, transferred onto wet ice for 40 min, and subsequently resolved by native PAGE.
Fly culture, transgenic lines, and antibody staining of embryos.
Flies were maintained at 22°C on standard cornmeal-sucrose medium containing Tegosept. The wild-type or mutated bxd5.1 BamHI/HindIII (pBS-3103BH) fragments were subcloned after end repair into the filled-in XhoI site of pCaSpeR4 (43). The PCR template used to generate ΔMHS-70 was pBS-3103BH. The primers used were ΔMHS70-p1 (5′-CCGGCTCGAGCCTGTTGCCTTGGCGGCTCT), ΔMHS70-p2 (5′-CCGGGCTAGCCATACGCACGGCTGTTAGAA), ΔMHS70-p3 (5′-CCGGGCTAGCCAAGCGAGAGCTTTTCATAG), and ΔMHS70-p4 (5′-CCGGGCGGCCGCGAAGCCATAACGGCAGAACC). The 200-bp amplified product from primer pair ΔMHS70-p1–ΔMHS70-p2 was digested with XhoI and EcoRI and subcloned into pBS, generating pBS-XR. The 450-bp amplified product from primer pair ΔMHS70-p3–ΔMHS70-p4 was digested with NheI and NotI and subcloned into pBS-XR, generating pBS-XN. A 571-bp BglI fragment in the pBS-3103BH vector was replaced by a 600-bp BglI fragment (containing ΔMHS-70 isolated from pBS-XN) to generate the final construct, pBS-3103BH-ΔMHS70. The 600-bp insert was sequenced to confirm its orientation and recloned into the XhoI site of pCaSpeR4 UbxlacZ transformation vector to generate bxd5.1 ΔMHS70.
The 571-bp BglI fragment from the pBS-3103BH vector was subcloned into pBS, generating pBS-BB. pBS-BB was digested with PstI and AseI to release the MPA-168 fragment, end filled with Klenow enzyme, and religated to generate pBS-BBΔ MPA-168. A 403-bp fragment was excised from vector pBS-BBΔ MPA-168 with BglI and subcloned into a pBS-bxd5.1BH vector in place of the wild-type 571-bp BglI fragment and then recloned into pCaSpeR4 UbxlacZ to generate ΔMPA-168. pBS-XN was digested with PstI and AseI to release MPA-168, end filled with Klenow enzyme, and religated. The double-mutated BglI fragment was subcloned into the pBS-3103BH vector in place of the 571-bp BglI fragment and recloned into pCaSpeR4 UbxlacZ to generate bxd5.1 ΔMHS70 + ΔMPA-168. The mutant MHS70-LS1/9 fragment described above was end filled with Klenow enzyme and ligated into the newly created NheI restriction site of pBS-3103BH-ΔMHS70 to generate bxd5.1-LS1/9. The construct was sequenced to confirm the orientation of the insert.
Staged embryos were collected and fixed by standard methods (36). Murine anti-β-galactosidase antibody (Jackson ImmunoResearch) was incubated with the transgenic embryos overnight at 4°C. A biotinylated goat anti-murine secondary antibody and a Vectastain ABC horseradish peroxidase kit (Vector Laboratories) were used to amplify and detect immune complexes.
RESULTS
Organization of the bxd6.5 PRE.
The minimal Ubx parasegment 6 (PS6) regulatory element shown to mimic the function of the endogenous bxd element in transgenes containing a lacZ reporter fused to the Ubx promoter is bxd14 (55). It contains a 6.5-kb PRE, bxd6.5HH (Fig. 1A), a composite element that silences the UbxlacZ transgene in PS1 to -5 and in odd-numbered PS7 to -13 throughout embryogenesis and enhances the expression of the transgene in even-numbered PS6 to -12 for 12 h of embryogenesis (8, 9). The bxd6.5HH element contains an imaginal enhancer and a shorter 5.1-kb HindIII/BamHI fragment (bxd5.1HB) which can be subdivided into two parasegment-specific enhancers, S1 (1.8 kb) and S2 (1.8 kb), and a silencer M element (1.5 kb) that contains PcG-dependent silencers (Fig. 1A) (8, 9, 44, 52).
The M element contains a 661-bp pairing-sensitive region (PSR) (52) flanked upstream by a 354-bp fragment we term UPS (Upstream of pairing-sensitive region) and downstream by a 485-bp fragment we term DPS (Fig. 1A). Genetic analysis of fragments within the PSR have identified nonoverlapping sites responsive to the PcG genes E(z), Pc, Pcl, pho, Psc, Scm, and Su(z)2 that mediate maintenance of parasegmental silencing of Ubx in embryos as well as pairing-sensitive repression of the white gene (20, 26, 62). Consistent with these findings, binding sites for PHO located within the pairing-sensitive region and DPS (Fig. 1A, marked in sites BP and MPH-75) have been identified (20). Fragment BP is also recognized by PC-containing DNA binding activities (26). We decided to analyze DPS with the goals of identifying minimal fragments recognized by PH-containing DNA binding activities and to permit detailed functional analysis of these fragments.
Identification of DPS DNA binding activities.
To identify DPS DNA binding activities and map their sites of interaction, six restriction fragments spanning DPS (Fig. 1A, MSa-100, MSf-73, MHS-70, MAH-74, MHA-93 and MPH-75) were analyzed in a gel mobility shift assay (21) using AS-fractionated nuclear extract derived from Drosophila Kc1 cells. We reasoned that results might be more easily interpreted if nuclear extracts were made from Kc1 cells, an embryonic mass cell culture that lacks detectable expression of UBX, rather than from embryos that contain mixed populations of cells expressing or repressing UBX. Nucleoprotein complexes formed with fragments within DPS were readily detectable at sites MHS-70 (Fig. 1B, complexes 1 and 2) and MHA-93 (data not shown) but not at sites MSa-100, MSf-73, MAH-74, or MPH-75 (data not shown). As shown in Fig. 1C, nonspecific fragments from pBR322 and the MSa-100 fragment did not inhibit binding to MHS-70, demonstrating that these MHS-70 binding activities identified are sequence specific. As shown in Fig. 1B, MHS-70 is recognized by heterogeneous binding activities that are currently uncharacterized with respect to number and subunit composition. For convenience, we refer to nucleoprotein complexes of similar mobilities in band shift assays as complex 1 and complex 2.
Isolation of a MHS-70 binding activity from nuclear extracts.
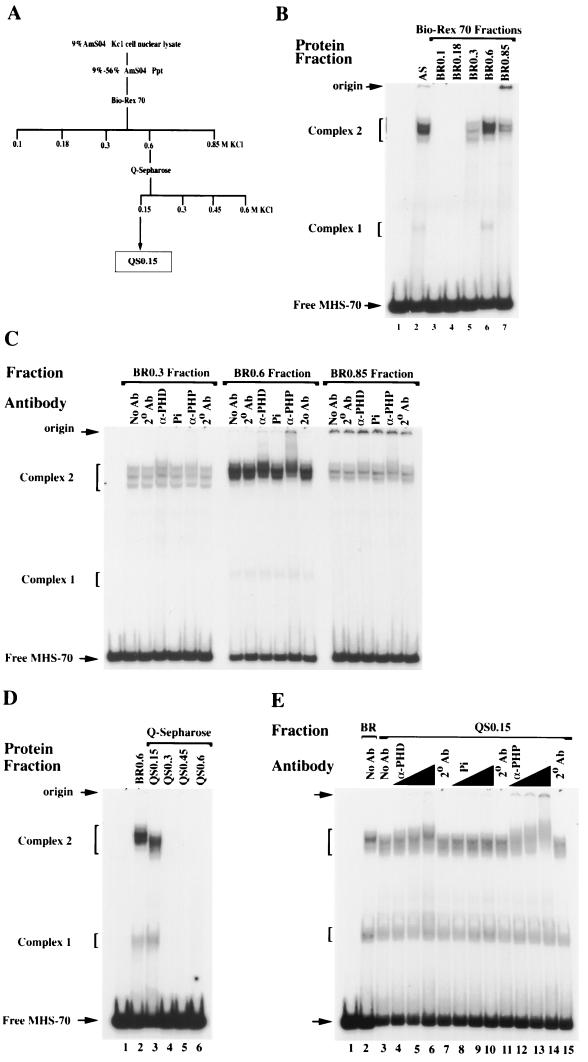
To characterize the MHS-70 binding activities and determine the sequence specificity of DNA binding, the Kc1 AS fraction was fractionated by two chromatographic steps, Bio-Rex 70 and Q Sepharose (Fig. 2A and Materials and Methods). Following chromatography on Bio-Rex 70, MHS-70 complex 2 binding activities were separated into the 0.3, 0.6, and 0.85 M KCl fractions, (BR0.3, BR0.6, and BR0.85), whereas complex 1 binding activities were predominantly in the 0.6 M KCl fraction (Fig. 2B). The apparent differences in the relative mobilities of complex 2 formed by each fraction reflect the heterogeneity of binding activities in the AS fraction.
FIG. 2.
Fractionation and supershift analysis of MHS-70 binding activities. (A) Chromatographic fractionation scheme for MHS-70 binding activities. (B) Gel mobility shift analysis of MHS-70 binding activities present in Bio-Rex 70 fractions, eluted stepwise with buffers containing 0.1 to 0.85 M KCl. Lanes 1 and 2, no protein and 0.6 μg of AS fraction, respectively; lane 6, 0.3 μg of BR0.6; lanes 3, 4, 5, and 7, 1.6 μg of the Bio-Rex 70 fractions shown above the gel. Low mobility complexes migrating close to the origin are detected with fractions eluted at 0.85 M KCl. (C) Supershift analysis of BR0.3, BR0.6, and BR0.85. No antibody (No Ab), antibody for PHD (α-PHD), preimmune serum (Pi), and antibody specific for PHP (α-PHP) at a titer of 1:37.5 were incubated in the binding reactions of the Bio-Rex 70 fractions to MHS-70 in the presence of secondary antibody (2° Ab), and changes in mobility were monitored in the gel shift assay. Partial supershifts of complex 2 are clearly detected in BR 0.6 and are not detected in BR0.3, and are weakly detected in BR0.85. (D) Mobility shift analysis of QS0.15, QS0.3, QS0.45, and QS0.6 with MHS-70. Lanes 1 and 2, no protein and 0.3 μg of BR0.6 respectively; lanes 3 to 6, 0.25 μg of protein from each Q Sepharose fraction. The MHS-70 binding activity is only detectable in QS0.15. (E) Supershift analysis of the QS0.15 binding activity (0.25 μg of protein) with anti-PH antibodies (lanes 4 to 6 and 12 to 14) at titers of 1:75 (lanes 4 and 12), 1:37.5 (lanes 5 and 13), and 1:10 (lanes 6 and 14). Both anti-PH antibodies but not the equivalent titer of preimmune serum (lanes 8 to 10) cause significant supershifts of complex 2.
To determine if the MHS-70 binding activities contained a PcG protein, BR0.3, BR0.6, and BR0.85 were analyzed by supershift assays using antibodies to PH (25). Two different antibodies to PH caused a readily detectable supershift of complex 2 from BR0.6 but not from BR0.3 and weakly from BR0.85 (Fig. 2C). The same activity was tested for the presence of PC and SU(Z)2. Neither PcG protein was detected (data not shown). The data show that the complex 2 binding activities present in BR0.6 contain at least one PcG protein. None of the Bio-Rex fractions analyzed contain detectable PH, PC, or SU(Z)2 in complex 1.
BR0.6 was subsequently fractionated on Q Sepharose, and as shown in Fig. 2D, the MHS-70 binding activities were recovered in the 0.15 M KCl fraction (QS0.15). Complex 2 formed by QS0.15 had a mobility slightly higher than that of the BR0.6 complex 2, whereas complex 1 had a mobility slightly lower than that of BR0.6 complex 1 (Fig. 2D, lanes 2 and 3). To confirm that the partially purified MHS-70 binding activities present in QS0.15 still retained PH, supershift assays were performed in the presence of titered amounts of two PH antibodies. As shown in Fig. 2E, both antibodies clearly supershift complex 2, consistent with the enrichment of PH-containing DNA binding activities in QS0.15. As expected, complex 1 is not supershifted, suggesting that it does not contain detectable PH.
Site specificity of M element recognition by QS0.15 binding activities.
To determine if the interaction of QS0.15 binding activities with the M element was site specific, a competitive gel shift assay was performed using fragments derived from DPS and PSR (Fig. 3A). As expected, the formation of complex 2 is completely inhibited by a 100-fold excess of unlabeled MHS-70, but formation of complex 1 is partially inhibited (Fig. 3B). Two other competing fragments, MHA-93 and MSP-61, partially inhibit complex 2 formation (Fig. 3B), showing that both sites contain sequences related to MHS-70. No other fragment from DPS or PSR of the M element competed for binding. These results indicate that MHS-70 is the predominant binding site of the three distinct sites within the M element recognized by binding activities present in QS0.15.
FIG. 3.
Competitive gel mobility shift analysis of the QS0.15 activity with M element fragments. (A) DNA fragments from the bxd M element, indicated on the top line, were used as competitors in the binding reaction of 0.25 μg of QS0.15 with MHS-70. Abbreviations for restriction enzymes and nomenclature of fragments are given in the legend to Fig. 1A, with the exception of Bf (BfaI). Fragments from the PSR are indicated above the line. (B) Competitive gel mobility shift analysis of complexes formed at MHS-70 in the presence of a molar excess of unlabeled fragments from the bxd M element. As expected, MHS-70 completely inhibits the formation of complex 2. Partial inhibition of complex 2 formation is detected with fragments MHA-93 and MSP-61 (marked with asterisks).
MHS-70 is required for PRE function in embryos.
To test if the MHS-70 fragment identified in the in vitro binding assay was required for PRE function, a germ line transformation assay was conducted. Transgenic lines containing a UbxlacZ reporter fused to a wild-type bxd5.1 PRE or an MHS-70 deletion mutant of bxd5.1 PRE (ΔMHS-70) were tested for the ability to maintain silencing in anterior parasegments of wild-type wiso embryos (Materials and Methods).
The bxd5.1 transgene confers complete maintenance of LacZ expression in PS6 to PS13 and silencing in PS1 to PS5 until late embryogenesis (stage 16) in 2 of 10 lines examined (Fig. 4A) and nearly complete maintenance (with the exception of spotty ectopic expression in the head and thorax) in 8 of 10 lines (Fig. 5C). Because the supershift analysis showed that complex 2 contained PH, it was necessary to test if mutations in ph would cause misexpression of bxd5.1 UbxlacZ. As shown in Fig. 4B, embryos heterozygous for ph2 (a viable deletion that results in a fusion of the phP and phD transcription units to yield one functional but abnormal protein [48]) showed clear misexpression of LacZ in PS1 to PS5. Because the PHP-specific antibody also supershifts complex 2, an inversion that affects phP but not phD (ph409 [13]) was tested for its effect on bxd5.1 UbxlacZ. Misexpression of LacZ was detected in PS1 to PS5, albeit at a much lower level than that seen for ph2, showing that PHP has a role in maintenance of bxd5.1 UbxlacZ silencing (Fig. 4C). Presumably phD can partially compensate for the loss of phP.
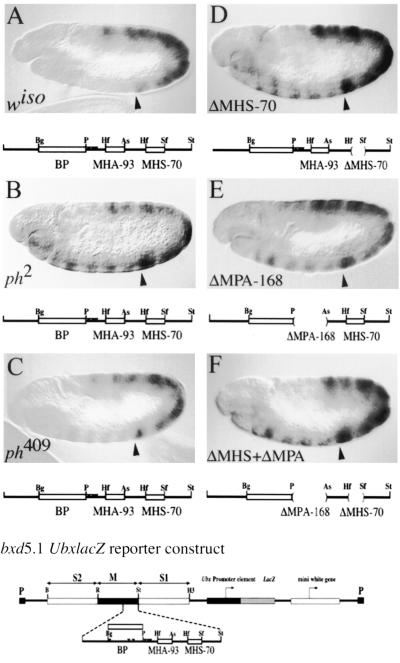
FIG. 4.
Mutational analysis of bxd5.1 UbxlacZ expression in germband-extended embryos. Germband-extended embryos are mounted anterior to the left, dorsal side up. Expression of LacZ was detected immunohistochemically. The anterior boundary of PS6 is marked with an arrowhead. The wild-type bxd5.1 UbxlacZ reporter, consisting of the 5.1-kb bxd PRE, the Ubx promoter, and the lacZ and white reporters flanked by P element long terminal repeats, is shown below the figure, with the DPS fragment expanded to show detail. The DPS fragment is indicated below each panel to show if it is wild type (A to C) or to show the positions and numbers of deletions in mutant constructs (indicated by parentheses in panel D to F). The same bxd5.1 UbxlacZ reporter, which exhibits complete silencing anterior to PS6, is used in panels A to C. (A) Wild-type embryo from a line exhibiting complete silencing of the reporter in PS1 to PS5 and in the head. (B) ph2 mutant embryo, showing partial derepression of bxd5.1 in PS1 to 5 and in the head. (C) ph409 mutant embryo, showing weak derepresion of bxd5.1 in PS 1 to 5. (D) bxd5.1ΔMHS-70 embryo in a wild-type background showing partial derepression of the reporter in PS1 to PS5. (E) bxd5.1ΔMPA-168 embryo exhibiting a derepression pattern similar to that seen in bxd5.1ΔMHS-70. (F) bxd5.1ΔMHS-70 + ΔMPA-168 embryo showing a derepression pattern similar to that seen in either single mutant (compare to panels D and E). Notation is as for Fig. 1A.
In bxd5.1ΔMHS-70 transgenic lines, initial expression of LacZ is identical to that of bxd5.1. By germ band extension, misexpression of LacZ is detected in PS1 to PS5, and this misexpression persists throughout embryogenesis in five of six lines tested, showing that MHS-70 is essential for embryonic maintenance of silencing in the context of bxd5.1 (Fig. 4D).
Because the competition analysis identified other sites related to MHS-70, we tested a deletion of MPA-168 which includes the competing fragment MHA-93 and the adjoining fragment MPH-75 (containing three PHO binding sites). Deletion of MPA-168 caused derepression of LacZ to an extent similar to that seen in ΔMHS-70 in five of six lines tested (Fig. 4E). However, in six of six lines tested, deletion of both MHS-70 and MPA-168 showed the same extent of LacZ misexpression as observed for each individual deletion (Fig. 4F). Therefore, the bxd5.1 PRE function conferred by the MHS-70 and MPA-168 fragments is neither additive nor synergistic, suggesting that there is one functional unit of PRE activity in this region. These results show that MHS-70 and MPA-168 are required for embryonic maintenance of silencing in the context of bxd5.1 PRE.
Sequence specificity of MHS-70 recognition by QS0.15 binding activities.
To characterize the sequence elements in MHS-70 recognized by binding activities present in QS0.15, substitution and deletion mutants of MHS-70 were tested for binding in a gel shift assay. Inspection of the sequence of MHS-70 showed a complex arrangement of three distinct repeat elements (Fig. 5A). These include two direct repeats of d(GA)3 elements flanking a central inverted d(GA)3 element, which constitute binding motifs for TRL/GAF (5) and Psq (35). Interspersed between the d(GA)3 repeats are two d(A) tracts in opposite orientations [Fig. 5A, d(A)8 and d(A)5(G)] as well as three direct repeats of a d(AGAGC) element.
Deletion of one of the terminal d(GA)3 sequences diminished the formation of complex 2 but did not affect the formation of complex 1 (Fig. 5B, lane 3), indicating a requirement of either this repeat element or a full-length fragment for stable binding. Substitution of the two remaining d(GA)3 sequences in the truncated fragment eliminated the formation of complex 2 and caused a twofold reduction in complex 1 formation (Fig. 5B, MHH-1/5), strongly suggesting an essential role for multiple d(GA)3 repeats. As expected, substitution of both terminal d(GA)3 sequences (Fig. 5B, LS-1/9, lane 5) in the full-length fragment completely abolished complex 2. These results show that at least two d(GA)3 repeats are essential for complex 2 but not complex 1 formation and also indicate that MHS-70 might be a multipartite recognition site.
The contribution of the other repeat elements was tested by a dual substitution of the central inverted d(GA)3 sequence plus the d(A)8 sequence (Fig. 5B, LS-3/5, lane 7), which caused a significant reduction in the formation of both complexes 1 and 2. Mutation of both d(A) tracts (Fig. 5B, LS3/6-7, lane 6) caused a similar reduction in formation of both complexes, suggesting that the d(A) tracts alone or in conjunction with a d(GA)3 sequence also contribute to the formation of complexes 1 and 2. Mutation of the three d(AGAGC) direct repeats (Fig. 5B, LS-4/6/8, lane 8) reduced the formation of both nucleoprotein complexes to an extent similar to that of the dual substitution of the d(GA)3 plus d(A)8 sequence (Fig. 5B, compare lane 8 to 7). These results demonstrate that MHS-70 is a multipartite, sequence-specific binding site and define the contribution of the distinct repeat elements, d(GA)3, d(A) tracts, and d(AGAGC) to the formation of complexes 1 and 2.
Mutations of d(GA)3 repeat elements reduce PRE activity.
The foregoing evidence shows that the LS-1/9 mutations which affect the proximal and distal d(GA)3 repeats of MHS-70 abolish complex 2 formation in vitro. To test the contribution of these d(GA)3 repeats for PRE function in vivo, we investigated the effects of the LS-1/9 mutations in the context of the bxd5.1 UbxlacZ transgene. In four of four lines that carried the LS-1/9 mutations, there was partial derepression of the reporter in a small number of cells in the ventral ectoderm in germband-extended embryos in PS1 to PS5 and relatively strong derepression in the head (Fig. 5C). This derepression is more evident in lateral than medial sections. This pattern of derepression is distinct from the bxd5.1ΔMHS-70 mutant phenotype and was not observed in any bxd5.1 UbxlacZ control lines, including those which show spotty misexpression in PS1 to PS5 (an example is shown in Fig. 5C). These results establish a role for d(GA)3 repeats in PRE function in vivo.
Analysis of multiple copies of MHS-70 repeat elements.
To further define the roles of the MHS-70 repeat elements in the recognition of QS0.15 binding activities, the abilities of double-stranded synthetic oligomers containing multiple copies of d(GA)3, d(A) tracts, and d(AGAGC) repeat elements (as shown in Fig. 6A) to bind QS0.15 activities were tested in a gel shift assay. Oligomers containing several copies of the three d(GA)3 repeats [Fig. 6B, (GA)3 × 3] form the low-mobility complex 2 but not the high-mobility complex 1. Conversely, oligomers containing five copies of d(A)8 plus the distal d(AGAGC) repeat [Fig. 6B, (A)8 × 5] form complex 1 but not complex 2. However, oligomers containing nine copies of the d(A)5(G) plus the proximal d(AGAGC) repeat [Fig. 6B, (T)5 × 9] form both complexes 1 and 2. In competitive binding studies carried out with each of these oligomers to determine their ability to compete with MHS-70 (Fig. 6C), both (GA)3 × 3 and (T)5 × 9 oligomers inhibited the formation of complex 2. The (A)8 × 5 oligomers caused a significant reduction in the formation of complex 2. However, all three oligomers caused an approximately twofold reduction in the formation of complex 1 (Fig. 6C).
These experiments define at least two forms of both MHS-70 complex 1 and 2 binding activities in QS0.15. Both d(GA)3 repeats and d(A)5(G)/proximal d(AGAGC) are critical elements for the complex 2 binding activities. They also suggest that the complex 1 binding activities interact with MHS-70 through multiple recognition sequence elements.
Complex 2 binding activities coelute with PH, TRL/GAF, and Psq.
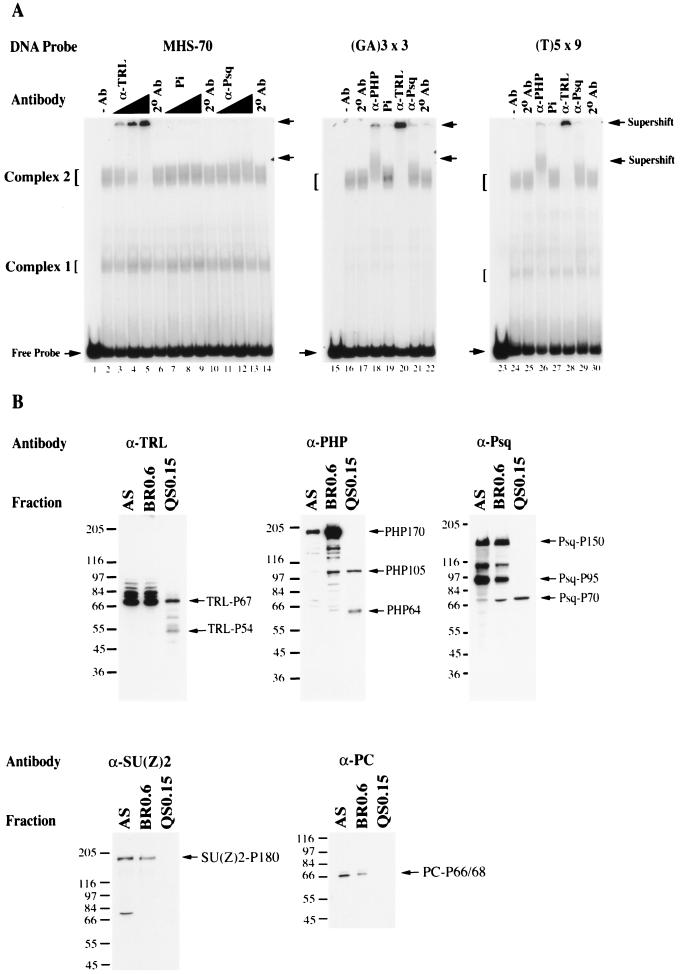
The results of the sequence-specific analysis above suggests that d(GA)n-specific binding factors are present in complex 2. Therefore, antibodies directed against two nuclear factors that bind d(GA)n sequences, TRL/GAF and Psq (5, 35), were tested in binding reactions with MHS-70. Preimmune serum had no effect on the mobility of complexes 1 and 2 (Fig. 7A, lanes 7 to 9). In the presence of increasing amounts of antibody to TRL/GAF, the mobility of complex 2 was significantly retarded, migrating close to the sample well (Fig. 7A, lanes 3 to 5). Antibodies to Psq caused a modest but detectable retardation of complex 2 (Fig. 7A, lanes 11 to 13). Neither of these antibodies altered the mobility of complex 1 (Fig. 7A). These results show that complex 2 contains detectable levels of TRL/GAF and Psq. The significantly reduced mobility of complex 2 in the presence of anti-TRL/GAF antibody presumably results from the ability to induce multimeric aggregates of DNA-TRL/GAF complexes (15, 66). To show that the complexes formed by MHS-70 and the competing oligomers were equivalent, the formation of complex 2 with synthetic oligomers was tested with antibodies to TRL/GAF, PH, and Psq. As shown in Fig. 7B, antibodies to PH, TRL/GAF, and Psq, but not preimmune serum, supershifted complex 2 in both the (GA)3 × 3 and d(T)5 × 9 binding reactions.
FIG. 7.
Supershift and Western analysis of QS0.15. (A) Secondary antibody (2° Ab) in the absence or presence of primary antibodies to PH (α-PHP), TRL/GAF (α-TRL), or Psq (α-Psq) or preimmune serum (Pi) was incubated in binding reactions of 0.25 μg of QS0.15 with MHS-70, (GA)3 × 3, or (T)5 × 9, and the nucleoprotein complexes were examined in a mobility shift assay. Lanes 1, 15, and 23 no protein; lanes 2, 16, and 24, no antibody (−Ab); lanes 6, 10, 14, 17, 22, 25, and 30, only secondary antibody (2° Ab). In the MHS-70 binding reactions, α-TRL titers were 1:240 (lanes 3), 1:120 (lane 4), and 1:60 (lane 5); Pi and α-Psq titers were 1:30 (lanes 7, and 11), 1:10 (lanes 8 and 12), and 1:7.5 (lanes 9 and 13). In the (GA)3 × 3 or (T)5 × 9 binding reactions, α-PHP was used at a titer of 1:10 (lanes 18 and 26), α-TRL was used at 1:60 (lanes 20 and 28), and α-Psq or Pi was used at 1:7.5 (lanes 21 and 29 or lanes 19 and 27). Antibodies to TRL and PHP caused a strong supershift in reactions of all fragments tested, and anti-Psq caused a detectable, partial supershift in reactions of the same fragments. (B) Western analysis of 20 μg of protein derived from AS, BR0.6, and QS0.15 fractions described in Fig. 2A. The fractions were separated by SDS-PAGE on a 9% gel, transferred to nitrocellulose, and probed with the antibodies indicated in panel A as described in Materials and Methods. Specific isoforms of each protein were enriched for and detected in QS0.15 are shown on the right of each gel; positions of protein molecular mass markers (in kilodaltons) (Sigma) are shown on the left.
PHP (25), TRL/GAF (3), and Psq (27) have multiple isoforms. To determine which of these isoforms are potential components of complex 2, Western analysis of the AS, BR0.6 and Q0.15 fractions was undertaken. Q0.15 is enriched for isoforms of TRL/GAF P67 plus TRL/GAF P54, PHP105 plus PHP64, and Psq P70 (Fig. 7B). Taken together with the antibody supershift analysis, these results show that the distinct isoforms of TRL/GAF, PHP and Psq coelute with complex 2 and suggest that these isoforms constitute potential subunits of complex 2 binding activities. It has been previously shown that the full-length isoform of PHP, PHP-170, coimmunoprecipitates with the PcG proteins PC, PSC, SU(Z)2, and SCM (19, 34, 49, 60). Western analyses of the three fractions described above show that there are no detectable levels of PC, SU(Z)2 (Fig. 7B), PSC, or Sex Combs on Midleg (SCM) (data not shown) in Q0.15, indicating that these PcG proteins do not coelute with complex 1 and 2 binding activities. These results suggest that the complex 2 activity is a novel PcG activity containing distinct isoforms of PHP, TRL/GAF, and Psq.
Genetic tests of Trl and psq function in PRE-mediated silencing.
Trl null or hypomorphic alleles (Trl13C, Trl62, and Trl85) do not affect maintenance of silencing in vivo by bxd5.1 UbxlacZ (data not shown), in agreement with results previously obtained (26). Similarly, embryos mutant for psqRF13 (deletion of psq) and Df(2R)psq-lolaΔ18 (deletion of psq, termed psqlola hereafter) (27) show wild-type bxd5.1 UbxlacZ silencing (data not shown). One potential reason for these results is that maternally deposited Trl or psq protein or mRNA rescue the effects of absence of zygotic proteins on embryonic silencing (4, 27).
Genetic interactions between PcG genes are monitored by the enhancement of PcG mutations, providing a sensitive genetic assay for genes required in PcG-mediated silencing (52). Therefore, we tested the ability of Trl and Psq mutations to enhance the extra sex combs phenotype of ph. It has previously been shown that Trl enhances the extra sex combs phenotype of Pc (59). Similarly, we observed that Trl62 enhances the extra sex combs phenotype of ph2 and ph409 (Table 2; Fig 8). We also tested the effects of psqlola-Δ18 and psq2403 on enhancement of ph2 and ph409. As shown in Table 2, there is strong enhancement of the expressivity of the extra sex combs phenotype. These results are consistent with a role for TRL/GAF and Psq in PcG-mediated silencing of homeotic loci and indicate that TRL/GAF and Psq are enhancers of PC which have sequence-specific DNA binding activity.
TABLE 2.
Trl and Psq enhance homeotic phenotypes of PcG mutationsa
| Cross | Genotype | n | Avg no. of legs with sex combs |
|---|---|---|---|
| ph2/ph2; +/+ × +/Y; Trl62/TM3 | ph2/Y; TM3/+ | 89 | 2.15 |
| ph2/Y; Trl62/+ | 76 | 4.33 | |
| ph409/FM7c; +/+ × +/Y; Trl62/TM3 | ph409/Y; TM3/+ | 46 | 4.51 |
| ph409/Y; Trl62/+ | 55 | 5.43 | |
| ph2/ph2; +/+ x +/Y; psqlola/CyO | ph2/Y;/CyO/+ | 53 | 2.46 |
| ph2/Y; psqlola/+ | 44 | 4.02 | |
| ph2/ph2; +/+ x +/Y; psq2403/CyO | ph2/Y; CyO/+ | 84 | 2.48 |
| ph2/Y; psq2403/+ | 68 | 5.03 | |
| psqlola/CyO; +/+ × +/+; Pc4/Tm6B | CyO/+; Pc4/+ | 48 | 2.23 |
| psqlola/+; TM6B4/+ | 52 | 2.0 | |
| psqlola/+; Pc4/+ | 50 | 4.04 | |
| psq2403/CyO; +/+ × +/+; Pc4/TM6B | CyO/+; Pc4/+ | 58 | 3.77 |
| psq2403/+; TM6B/+ | 57 | 2.0 | |
| psq2403/+; Pc4/+ | 58 | 5.22 |
Male progeny of the indicated crossed were scored for the number of legs with extra sex comb teeth. Wild-type males have sex combs on the prothoracic legs only, and hence have 2.0 legs with sex combs. A fly with sex combs on prothoracic, mesothoracic, and metathoracic legs would have 6.0 legs with sex combs. The numbers of sex combs present on individual flies were determined by observation under a dissecting microscope, and the results are expressed as the average number of legs with sex combs found in the files scored. All values are statistically significant at P < 0.05, using a chi-squared test.
FIG. 8.
Trl and psq mutations enhance the extra sex combs phenotype of ph mutants. Wild-type Drosophila males have two sex combs (marked by arrowheads): one on each first leg (prothoracic leg) (A) and absent on the second (mesothoracic) (B) and third (metathoracic) (C) legs. Males hemizygous for ph mutations exhibit posterior homeotic transformations where the second and third legs are frequently transformed into the first, thus increasing the total number of sex combs per male fly. Male flies transheterozygous for trl or psq and ph mutations exhibit an enhancement of the extra sex combs phenotype of ph. For example, ph2/Y; Trl62/+ males always exhibit sex combs on the first legs (D), frequently on the second legs (E), and sometimes on the third legs (F).
DISCUSSION
Our results show that specific DNA binding activities recognize distinct fragments of the bxd1.5 M element in vitro and that two of these fragments, MHS-70 and MPS-168, are necessary for PRE function in vivo. Detailed analysis of the binding requirements of MHS-70 show that this binding site is multipartite and involves repeats of d(GA)3, d(A) tracts, and d(AGAGC) sequences. A partially purified activity recognizing MHS-70 contains specific isoforms of two d(GA)n recognition factors, TRL/GAF and Psq, and the PcG protein PH which have not been identified in any previously characterized PcG protein complex. The d(GA)n sequences are required for in vivo function of the bxd5.1 PRE, but other sequences also contribute to function of the MHS-70 element. We show that TRL/GAF are enhancers of PcG mutations with sequence-specific binding activity, raising the possibility that these proteins recruit PcG proteins to PREs. Below, we discuss the implications of these results for PRE structure and function.
Multipartite structure of the MHS-70 binding site.
Inspection of the MHS-70 sequence revealed a complex arrangement of three distinct repeat elements likely to be important for QS0.15 binding activities: d(GA)3, d(A) tracts, and d(AGAGC). In a competitive inhibition gel shift assay, the contribution of each repeat element was tested by linker-scanning mutagenesis. No single mutation inhibited binding (J. W. Hodgson and H. W. Brock, unpublished results). As shown in Results, combined substitution of the repeat elements in MHS-70 greatly reduced or abolished binding of the activities present in QS0.15. These observations could be explained if multiple copies of the repeat elements were required for MHS-70 recognition. In this case, mutations of individual sites might lower the affinity of binding but not abolish it. Alternatively, protein-protein interactions between factors bound to multiple copies of these repeat elements might stabilize weak protein-DNA interactions at single repeats. Another possibility is that binding to the MHS-70 fragment depends on conformation of the DNA, and single mutations have little effect on conformation adopted by the nucleoprotein complexes. In this context, it should be noted that TRL/GAF binds DNA as multimers, that binding to repeated sites is strongly favored over binding to single sites, and the interaction involves DNA distortion (31, 66).
Experiments in which multiple sites were mutated support the idea that MHS-70 is a multipartite binding site. Mutation of the two terminal d(GA)3 repeats in LS-1/9 abolishes binding by QS0.15 complex 2 activity (Fig. 5B, lane 5), whereas mutation of either repeated copies of the d(A) tracts or d(AGAGC) significantly reduced binding. These results are consistent with previous results obtained in an immunoprecipitation assay with the BP fragment (illustrated in Fig. 1) that showed it was necessary to mutate two d(GA)n sequences in order to abolish binding by activities containing PC (26).
The results of the binding assays with oligomers containing multiple copies of unique arrangement of d(GA)3 repeats and the d(A)8/d(AGAGC) element indicated that QS0.15 contains distinct binding activities. These results show that the d(GA)n repeats are sufficient for complex 2 formation (Fig. 6B) and that the (A)8 × 5 construct is sufficient for complex 1 formation (Fig. 6B), strongly supporting independence of binding. Surprisingly, the d(T)5 × 9 oligomer can form complexes 1 and 2, suggesting additional independent modes of DNA recognition by activities present in QS0.15. The ability of antibodies to TRL/GAF and Psq to supershift d(T)5 × 9 complex 2 suggests that DNA recognition may involve either (i) the multivalent recognition of multiple copies of the minimum d(GAG) trinucleotide in the repeat unit, d(AAAAAGAGCG), by TRL/GAF (15, 65) or DNA conformation (27) or (ii) DNA bending by d(A) tracts in the d(T)5 × 9 repeat unit, d(AAAAAGAGCG) (56). Therefore, it is possible that complex 1 and complex 2 represent two mutually exclusive nucleoprotein conformations. The precise relationships between these three potential binding activities are currently being analyzed with more highly purified protein fractions (Hodgson and Brock, unpublished).
In vivo analysis of MHS-70 function.
We chose to examine the effect of the MHS-70 deletion in the context of the bxd5.1 PRE because this element shows more stable and longer-lasting embryonic silencing and less line-to-line variation than the M element alone. Therefore, results of small mutations in the context of bxd5.1 are easier to interpret, the main complications being conveyed by the presence of redundant recognition sequences in bxd5.1. However, it is important to note that competition studies revealed that the only binding fragments that competed for MHS-70 binding activity are found in the M element (Fig. 3 and results not shown).
Mutation of the terminal d(GA)3 sequences within MHS-70 abolished QS0.15 complex 2 activity in vitro, but the effect of these mutations on derepression of the reporter in germband-extended embryos was less severe than that of the deletion of MHS-70. The simplest explanation for this result is that other sequences in MHS-70 contribute to silencing in vivo, including the poly(A) tracts that bind complex 1 activity and the d(AGAGC) repeats that bind complex 1 and 2 activities. It will be important to test these possibilities directly. We cannot rule out the possibilities that the MHS-70 deletion changes the spacing, or changes the rotational position or flexibility of the DNA, whereas the point mutations in the d(GA)3 repeats do not. Our data show a requirement for d(GA)3 sites in conjunction with other MHS-70 repeat elements in vivo.
These in vivo experiments demonstrate a direct role for the d(GA)3 repeats in maintenance of silencing by a PRE. Recognition of MHA-93 may be involved in the maintenance of silencing, because MHA-93 contains d(GA)n repeats and competes with MHS-70 in binding reactions of activities present QS0.15 (Fig. 3B). If so, mutation of the d(GA)n repeats in MHS-70 may be compensated for by the presence of neighboring d(GA)n sites in MHA-93, thus lessening the effects of the d(GA)3 mutations in MHS-70. Our in vivo data suggest that only one unit of PRE function exists in the region containing MHS-70 and MPA-168, because deletions of these fragments by themselves or in combination have similar rather than additive or synergistic effects on derepression of the bxd5.1 UbxlacZ (Fig. 4). The MHS-70 and MPA-168 fragments have only d(GA)n repeats in common, which suggests an interaction between activities recognizing d(GA)n sequences in MHS-70 and MPA-168 that is required for embryonic silencing mediated by DPS.
The role of TRL/GAF and Psq in PcG-mediated silencing.
The two proteins known that bind d(GA)n repeats are TRL/GAF and Psq. Our in vitro binding studies assay the ability of preformed complexes that have been extracted from chromatin and the nuclear matrix to bind naked DNA and therefore do not directly address how TRL/GAF and Psq function in vivo. An attractive model is that in vivo, one or both of these proteins bind the d(GA)n repeats and recruit PcG proteins to the MHS-70 fragment to assemble a functional repressive complex. Alternatively, TRL/GAF or Psq could bind as part of a preassembled factor.
Because TRL/GAF antagonizes repressors, it is surprising that TRL/GAF might have a role in repression by PREs. Chromatin immunoprecipitation experiments show that PcG and TRL/GAF proteins colocalize in vivo (59), but this result could be explained if trxG and PcG response elements were intermingled (62). TRL/GAF coimmunoprecipitates with PC, providing independent evidence for the existence of complexes containing PcG proteins and the TRL/GAF (26). There are several reports that Trl mutations permit derepression of reporters mediated by PREs in transgenic flies, but these effects are often line specific and allele specific, affect repression in imaginal discs, or are PRE specific (23, 26, 59). We do not see an effect of Trl mutations on misexpression of the bxd5.1 PRE. However, interpretation of these experiments is complicated by perdurance of maternally deposited TRL/GAF, which makes it difficult to detect a zygotic effect of Trl mutants on PRE-mediated silencing. Despite ambiguities in the published evidence, Trl is an enhancer of the extra sex combs phenotype of ph (Table 2) and of Pc (59), supporting a role for Trl in both silencing and activation.
In vivo the TRL/GAF may have an architectural role in chromatin structure that allows access to both PcG and trxG proteins, explaining its dual role. Alternatively, TRL/GAF may be a subunit of activating and repressing transcription complexes involved in sequence-specific targeting of both types of complexes. It has been shown that TRL/GAF interacts with dSAP18 (Sin3-associated polypeptide) and mediates repression through iab-6, perhaps by recruiting the Sin3-histone deacetylase complex (16). These two hypotheses are not mutually exclusive. Further characterization of complex 2 activity may provide support for either or both hypotheses.
We are not aware of any previous suggestion that Psq has a role in PcG-mediated silencing. As noted in the results, two psq mutations had no effect on silencing of the bxd5.1 PRE, but this may be due to maternally deposited protein or mRNA. Nevertheless, psq mutations show strong enhancement of the extra sex combs phenotype of two PcG mutations, ph and Pc, one of the defining characteristics of PcG mutations (53). We conclude that Psq is a novel enhancer of PC with sequence-specific binding activity.
The complex 2 binding activity of QS0.15 is novel.
Previous studies have identified two PcG protein complexes in Drosophila: the PRC1 complex containing the PHP-170 isoform, PC, PSC, and SCM and lacking Enhancer of Zeste [E(Z)] (49), and a complex containing E(Z) and Extra Sex Combs (ESC) but not PH (61). Using in vitro protein binding assays, we have also shown direct interaction between the SAM domains of PH and SCM (42). Consequently, it might be expected that a binding activity containing PHP should contain PC, PSC, or SCM. However, a number of observations support the conclusion that the complex 2 binding activities present in QS0.15 represent novel PcG activities. First, PC, PSC, and SCM are not present in the QS0.15 fraction that contains the MHS-70 binding activities. Second, the PH isoform, PHP-170, that coimmunoprecitates with PC and is reported in PRC1 (49) is not present in QS0.15. Third, PRC1 does not contain TRL/GAF (49), which is present in complex 2, as shown by supershift analysis, and in QS0.15, as shown by Western analysis. Fourth, the complex 2 binding activities present in QS0.15 are sequence specific, whereas the DNA binding activities of PRC1 are unknown. Fifth, the complex 2 activity is chromatographically distinct from PRC1 which elutes in BR0.85 (data not shown). The complex 2 activity is also distinct from the BP binding activity (Fig. 1A), previously identified in coimmunoprecipitation experiments which contains PC and TRL/GAGA (26), as QS0.15 does not contain PC. The MHS-70 complex 2 activity therefore represents a novel PcG sequence-specific activity containing distinct isoforms of PHP, TRL/GAF, and Psq, which may be involved in site-specifically targeting repressive complexes to the bxd PRE. Experiments are currently in progress to determine the subunit composition of more highly fractionated complex 2 activity (Hodgson and Brock, unpublished).
Complex organization of the M element.
Previous studies on the structural organization of the bxd1.5 PRE (M element) revealed three regions: a 661-bp pairing-sensitive fragment which contained distinct PHO binding sites, and PC-containing DNA binding activities flanked upstream by a 354-bp fragment and downstream by a 485-bp fragment (DPS) which contained PHO sites (marked in sites BP and MPH-75 in Fig. 1) (20, 26, 53). Our results identify two additional sites (MHS-70 and MPA-168) located within the DPS fragment required for maintenance of silencing (Fig. 1). These observations are in keeping with the idea that the M element is built up from sites with differing functions (26).
A limited region of the M element is required for pairing-sensitive repression that is separable from regions required for embryonic silencing (26). This conclusion is supported by similar observations in the iab-2 regulatory region of abdominal-A. The sequences required for pairing-sensitive repression by the iab-2 PRE are not sufficient for embryonic silencing, which requires additional sequences in the iab-2 enhancer (50). Consistent with this observation, deletion of MHS-70 and MPA-168, which lie outside the pairing-sensitive region of the bxd5.1 PRE, have no effect on pairing-sensitive repression by bxd5.1 (B. Argiropoulos and H. W. Brock, unpublished results).
Modular organization of the bxd PRE.
Genetic evidence shows that mutations in many PcG genes causes derepression of the bxd PRE (55). It has been shown that distinct fragments of the bxd PRE respond differentially to different PcG mutations (26, 62), supporting the idea that this PRE is recognized by multiple PcG binding activities. This idea is strongly supported by observations that antibodies to PC and PSC preferentially precipitate different fragments of the M element in vitro (26). As we argue above, we show that even within the MHS-70 element, there are recognition sequences for at least two distinct binding activities, neither of which contains PC.
Our results are inconsistent with a model which posits that PHO is the primary recognition and PcG recruitment factor for the bxd PRE. The MHS-70 fragment, which does not contain PHO sites, is required for maintenance of embryonic silencing, ruling out a role for PHO on this fragment. The results of the MHS-70 competition experiments also rule out a model in which PHO binding recruits PcG proteins which cause a change in chromatin structure that spreads from PHO binding sites, independent of the flanking DNA sequences. Instead, these results support a model in which PRE function is built up from sequences with different functions, that recognize specific binding activities which contribute to the overall function of the PRE. This organization may be analogous to the modular organization of viral and cellular enhancers (14).
ACKNOWLEDGMENTS
We thank D. Hogness and J. Mueller for providing clones from the Ubx regulatory region; P. Becker and C. Wu for the gift of anti-TRL/GAF antibody; C. Berg for anti-Psq antibody and mutant fly strains; P. O'Farrell, J. Simon, and J.-M. Dura for antibodies to PC, SCM, and PSC, respectively; P. Adler for the Su(Z)2 cDNA construct; and M. Biggin for mutant Trl fly strains. We thank L. Matsuuchi for discussions on the precipitin reaction and members of the Brock lab for critical comments on the manuscript. We thank V. Pirrotta and members of his lab for sharing information prior to publication and for stimulating discussion.
This work was supported by a grant from the Medical Research council of Canada to H.W.B.
REFERENCES
- 1.Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987;101:1–22. [PubMed] [Google Scholar]
- 2.Bender W, Akam M, Karch F, Beachy P A, Peifer M, Spierer P, Lewis E B, Hogness D S. Molecular genetics of the Bithorax complex in Drosophila melanogaster. Science. 1983;221:23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- 3.Benyajati C, Mueller L, Xu N, Pappano M, Gao J, Mosammaparast M, Conklin D, Granok H, Craig C, Elgin S. Multiple isoforms of GAGA factor, a critical component of chromatin structure. Nucleic Acids Res. 1997;25:3345–3353. doi: 10.1093/nar/25.16.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhat K M, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development. 1996;122:1513–1522. doi: 10.1242/dev.122.4.1113. [DOI] [PubMed] [Google Scholar]
- 5.Biggin M D, Tjian R. Transcription factors that activate the Ultrabithorax promotor in developmentally staged extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- 6.Brown J L, Mucci D, Whitely M, Dirksen M-L, Kassis J A. The Drosophila Polycomb Group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 7.Busturia A, Bienz M. Silencers in Abdominal-B, a homeotic Drosophila gene. EMBO J. 1993;12:1415–1425. doi: 10.1002/j.1460-2075.1993.tb05785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan C-S, Rastelli L, Pirotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang A, O'Connor M B, Paro R, Simon J, Bender W. Discrete Polycomb-binding sites in each parasegmental domain of the bithorax complex. Development. 1995;121:1681–1689. doi: 10.1242/dev.121.6.1681. [DOI] [PubMed] [Google Scholar]
- 10.Christen B, Bienz M. Imaginal disc silencers from Ultrabithorax: evidence for Polycomb response elements. Mech Dev. 1994;48:255–266. doi: 10.1016/0925-4773(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 11.DeCamillis M A, Cheng N, Pierre D, Brock H W. The polyhomeotic gene of Drosophila encodes a chromatin protein that shares polytene chromosome binding sites with Polycomb. Genes Dev. 1992;6:223–232. doi: 10.1101/gad.6.2.223. [DOI] [PubMed] [Google Scholar]
- 12.Duncan I. The bithorax complex. Annu Rev Genet. 1987;21:285–319. doi: 10.1146/annurev.ge.21.120187.001441. [DOI] [PubMed] [Google Scholar]
- 13.Dura J-M, Randsholt N B, Deatrick J, Erk I, Santamaria P, Freeman D J, Freeman S F, Weddell D, Brock H W. Maternal and zygotic requirement for polyhomeotic, a genetically complex locus required for normal expression of segmental identity and cuticular development. Cell. 1987;51:829–839. doi: 10.1016/0092-8674(87)90106-1. [DOI] [PubMed] [Google Scholar]
- 14.Dynan W S. Modularity in promoters and enhancers. Cell. 1989;58:1–4. doi: 10.1016/0092-8674(89)90393-0. [DOI] [PubMed] [Google Scholar]
- 15.Espinas M L, Jimenez-Garcia E, Vaquero A, Canudas S, Bernues J, Azorin F. The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J Biol Chem. 1999;274:16461–16469. doi: 10.1074/jbc.274.23.16461. [DOI] [PubMed] [Google Scholar]
- 16.Espinas M L, Canudas S, Fanti L, Pimpinelli S, Casanova J, Azorin F. The GAGA factor of Drosophila interacts with SAP18, a Sin3-associated polypeptide. EMBO Rep. 2000;1:253–259. doi: 10.1093/embo-reports/kvd046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farkas G, Guasz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 18.Fauvarque M-O, Dura J-M. polyhomeotic regulatory sequences induce development regulator-dependent variegation and targeted P-element insertions in Drosophila. Genes Dev. 1993;7:508–1520. doi: 10.1101/gad.7.8.1508. [DOI] [PubMed] [Google Scholar]
- 19.Franke A, DeCamillis M A, Zink D, Cheng N, Brock H W, Paro R. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 1992;11:2941–2950. doi: 10.1002/j.1460-2075.1992.tb05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fritsch C, Brown J L, Kassis J A, Muller J. The DNA-binding Polycomb group protein Pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development. 1999;126:3905–3913. doi: 10.1242/dev.126.17.3905. [DOI] [PubMed] [Google Scholar]
- 21.Garner M M, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucllic Acids Res. 1981;9:3047–060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gindhart J G, Kaufman T C. Identification of Polycomb and trithorax group responsive elements in the regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics. 1995;139:797–814. doi: 10.1093/genetics/139.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagstrom K, Muller M, Schedl P. A Polycomb and GAGA dependent silencer adjoin the Fab-7 boundary in the Drosophila bithorax complex. Genetics. 1997;146:1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heberlein U, England B, Tjian R. Characterization of Drosophila transcription factors that activate the tandem promoters of the alcohol dehydrogenase gene. Cell. 1985;41:965–977. doi: 10.1016/s0092-8674(85)80077-5. [DOI] [PubMed] [Google Scholar]
- 25.Hodgson J W, Cheng N N, Sinclair D A R, Kyba M, Randsholt N B, Brock H W. The polyhomeotic locus of Drosophila melanogaster is transcriptionally and post-transcriptionally regulated during embryogenesis. Mech Dev. 1997;66:69–81. doi: 10.1016/s0925-4773(97)00091-9. [DOI] [PubMed] [Google Scholar]
- 26.Horard B, Tatout C, Poux S, Pirrotta V. Structure of a Polycomb response element and in vitro binding of Polycomb group complexes containing GAGA factor. Mol Cell Biol. 2000;20:3187–3197. doi: 10.1128/mcb.20.9.3187-3197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horowitz H, Berg C A. The Drosophila pipsqueak gene encodes a nuclear BTB-domain-containing protein required early in oogenesis. Development. 1996;122:1859–1871. doi: 10.1242/dev.122.6.1859. [DOI] [PubMed] [Google Scholar]
- 28.Jimenez-Garcia E, Vaquero A, Espinas M L, Soliva R, Orozco M, Bernues J, Azorin F. The GAGA factor of Drosophila binds triple-stranded DNA. J Biol Chem. 1998;273:24640–24648. doi: 10.1074/jbc.273.38.24640. [DOI] [PubMed] [Google Scholar]
- 29.Kassis J A. Unusual properties of regulatory DNA from the Drosophila engrailed gene: three “pairing-sensitive” sites within a 1.6kb region. Genetics. 1994;136:1025–1038. doi: 10.1093/genetics/136.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kassis J A, VanSickle E P, Sensabaugh S M. A fragment of the engrailed regulatory DNA can mediate transvection of the white gene in Drosophila. Genetics. 1991;128:751–761. doi: 10.1093/genetics/128.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katsani K, Hajibagheri M, Verrijzer C P. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 1999;18:698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennison J A. Transcriptional activation of Drosophila homeotic genes from distant regulatory elements. Trends Genet. 1993;9:75–79. doi: 10.1016/0168-9525(93)90227-9. [DOI] [PubMed] [Google Scholar]
- 33.Kerrigan L A, Croston G E, Lira L M, Kadonaga J T. Sequence-specific transcriptional antirepression of the Drosophila Kruppel gene by the GAGA factor. J Biol Chem. 1991;266:574–582. [PubMed] [Google Scholar]
- 34.Kyba M, Brock H W. The Drosophila Polycomb group protein Psc contacts ph and Pc through specific conserved domains. Mol Cell Biol. 1998;18:2712–2720. doi: 10.1128/mcb.18.5.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehman M, Siegmund T, Lintermann K, Korge G. The Pipsqueak protein of Drosophila melanogaster binds to GAGA sequences through a novel DNA-binding domain. J Biol Chem. 1998;273:28504–28509. doi: 10.1074/jbc.273.43.28504. [DOI] [PubMed] [Google Scholar]
- 36.McKeon J, Brock H W. Interactions of the Polycomb group of genes with homeotic loci of Drosophila. Roux's Arch Dev Biol. 1991;199:387–396. doi: 10.1007/BF01705848. [DOI] [PubMed] [Google Scholar]
- 37.Mihaly J, Mishra R K, Karch F. A conserved sequence motif in Polycomb-response elements. Mol Cell. 1998;1:1065–1066. doi: 10.1016/s1097-2765(00)80107-0. [DOI] [PubMed] [Google Scholar]
- 38.Muller J, Bienz M. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. EMBO J. 1991;10:3147–3156. doi: 10.1002/j.1460-2075.1991.tb04876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okada M, Hirose S. Chromatin remodeling mediated by Drosophila GAGA factor and ISWI activates fushi tarazu gene transcription in vitro. Mol Cell Biol. 1998;18:2455–2461. doi: 10.1128/mcb.18.5.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orlando V, Jane E P, Chinwalla V, Harte P J, Paro R. The binding of Trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 1998;17:5141–5150. doi: 10.1093/emboj/17.17.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker C S, Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell. 1984;36:357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- 42.Peterson A, Kyba M, Bornemann D, Morgan K, Brock H W, Simon J. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol Cell Biol. 1997;17:6683–6692. doi: 10.1128/mcb.17.11.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pirrotta V. Vectors for P-mediated transformation in Drosophila. In: Rodriguez R L, Denhardt D T, editors. Vectors. A survey of molecular cloning vectors and their uses. Boston, Mass: Butterworths; 1988. pp. 437–445. [Google Scholar]
- 44.Pirrotta V, Chan C S, McCabe D, Qian S. Distinct parasegmental and imaginal enhancers and the establishment of the expression pattern of the Ubx gene. Genetics. 1995;141:1439–1450. doi: 10.1093/genetics/141.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pirrota V, Rastelli L. white gene expression, repressive chromatin domains, and homeotic gene regulation in Drosophila. Bioessays. 1994;8:549–556. doi: 10.1002/bies.950160808. [DOI] [PubMed] [Google Scholar]
- 46.Poux S, Kostic C, Pirotta V. Hunchback-independent silencing of late Ubx enhancers by a Polycomb Group Response Element. EMBO J. 1996;15:4713–4722. [PMC free article] [PubMed] [Google Scholar]
- 47.Poux S, McCabe D, Pirrotta V. Recruitment of components of Polycomb Group chromatin complexes in Drosophila. Development. 2001;128:75–85. doi: 10.1242/dev.128.1.75. [DOI] [PubMed] [Google Scholar]
- 48.Saget O, Randsholt N B. Transposon-induced rearrangements in the duplicated locus ph of Drosophila melanogaster can create new chimeric genes functionally identical to wild-type. Gene. 1994;149:227–235. doi: 10.1016/0378-1119(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 49.Shao Z, Raible F, Mollaagabhaba R, Guyon J R, Wu C-T, Bender W, Kingston R E. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 50.Shimell M J, Peterson A J, Burr J, Simon J A, O'Connor M B. Functional analysis of repressor binding sites in the iab-2 regulatory region of the abdominal-A homeotic gene. Dev Biol. 2000;218:38–52. doi: 10.1006/dbio.1999.9576. [DOI] [PubMed] [Google Scholar]
- 51.Siegel V, Jongens T A, Jan L Y, Jan Y N. pipsqueak, an early acting member of the posterior group of genes, affects vasa level and germ cell-somatic cell interaction in the developing egg chamber. Development. 1993;119:1187–1202. doi: 10.1242/dev.119.4.1187. [DOI] [PubMed] [Google Scholar]
- 52.Sigrist C J, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila Polycomb Response Element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 54.Simon J, Chiang A C, Bender W. Ten different Polycomb group genes are required for spatial control of the abd-A and Abd-B homeotic products. Development. 1992;114:493–505. doi: 10.1242/dev.114.2.493. [DOI] [PubMed] [Google Scholar]
- 55.Simon J, Chiang A C, Bender W, Shimell M J, O'Connor M. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev Biol. 1993;158:131–144. doi: 10.1006/dbio.1993.1174. [DOI] [PubMed] [Google Scholar]
- 56.Soto M C, Chou T-B, Bender W. Comparison of germ-line mosaics of genes in the Polycomb group of Drosophila melanogaster. Genetics. 1995;140:231–243. doi: 10.1093/genetics/140.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strahs D, Schlick T. A-tract bending: insights into experimental structures by computational models. J Mol Biol. 2000;301:643–663. doi: 10.1006/jmbi.2000.3863. [DOI] [PubMed] [Google Scholar]
- 58.Struhl G, Akam M E. Altered distribution of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 1985;4:3259–3264. doi: 10.1002/j.1460-2075.1985.tb04075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strutt H, Paro R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strutt H, Paro R. The Polycomb group protein complex of Drosophila melanogaster has different compositions at different target genes. Mol Cell Biol. 1997;17:6773–6783. doi: 10.1128/mcb.17.12.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte P J. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001;128:275–286. doi: 10.1242/dev.128.2.275. [DOI] [PubMed] [Google Scholar]
- 62.Tillib S, Petruk S, Sedkov Y, Kuzin A, Fujioka M, Goto T, Mazo A. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol Cell Biol. 1999;19:5189–5202. doi: 10.1128/mcb.19.7.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsukiyama T, Becker P B, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 64.Weber U, Siegel V, Mlodzik M. pipsqueak encodes a novel nuclear protein required downstream of seven-up for the development of photoreceptors R3 and R4. EMBO J. 1995;14:6247–6257. doi: 10.1002/j.1460-2075.1995.tb00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilkins R C, Lis J. GAGA factor binding of DNA via a single trinucleotide sequence element. Nucleic Acids Res. 1998;26:2672–2678. doi: 10.1093/nar/26.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilkins R C, Lis J. DNA distortion and multimerization: novel functions of the glutamine-rich domain of GAGA factor. J Mol Biol. 1999;285:515–525. doi: 10.1006/jmbi.1998.2356. [DOI] [PubMed] [Google Scholar]
- 67.Zink B, Engstrom Y, Gehring W, Paro R. Direct interaction of the Polycomb protein with Antennapedia regulatory sequences in polytene chromosomes of Drosophila melanogaster. EMBO J. 1991;10:153–162. doi: 10.1002/j.1460-2075.1991.tb07931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]