Abstract
In modern clinical practice, digital pathology plays a crucial role and is increasingly a technological requirement in the laboratory environment. The advent of whole slide imaging, the availability of faster networks, and cheaper storage solutions make it easier for pathologists to manage digital slide images and share them for clinical use. In parallel, unprecedented advances in machine learning have enabled synergy of artificial intelligence and digital pathology, which offers image-based diagnosis possibilities that were only limited to radiology and cardiology in the past. With integration of digital slides into the pathology workflow, advanced algorithms and computer-aided diagnostic techniques extend the frontiers of the pathologist’s view beyond a microscopic slide and enable true utilization and integration of knowledge beyond human limits and boundaries. There is clear potential for artificial intelligence breakthroughs in digital pathology. In this review article, we discuss recent advancements in digital slide based image diagnosis along with some challenges and opportunities for artificial intelligence in digital pathology.
Keywords: artificial Intelligence, deep learning, digital pathology, digital slides, whole slide imaging
INTRODUCTION
Digital pathology plays a critical role in modern clinical practice and is increasingly a technology requirement within the laboratory environment [1]. Advances in computing power, faster networks, and cheaper storage have enabled pathologists to manage digital slide images with more ease and flexibility than in the past decade and to share images for telepathology and clinical use. In the last two decades, digital imaging in pathology has seen the inception and evolution of whole slide imaging (WSI) which allows entire slides to be imaged and permanently stored at high resolution.
In particular, whole slide imaging (WSI) serves as an enabling platform for the application of artificial intelligence (AI) in digital pathology. Until now, AI has been mostly used for image-based diagnosis in radiology and cardiology. Its application to pathology is an expanding field of active research with several research groups and dedicated companies. Images produced by WSI are the source of rich set of information – complexity is higher than many other imaging modalities because of their large size (100k x 100k size is not uncommon), presence of color information (H&E and immunohistochemistry), no apparent anatomical orientation as in radiology, availability of information at multiple scales (e.g. 4x, 20x), multiple z-stack levels (each slice contain a finite thickness and depending on the plane of focus, it will generate different images). Clearly, it is not humanly possible to extract all visual information by a human reader.
With integration of digital slides into the pathology workflow, advanced algorithms and computer-aided diagnostic techniques extend the frontiers of the pathologist’s view beyond a microscopic slide and enable true utilization and integration of knowledge beyond human limits and boundaries [1]. AI already enables pathologists to identify unique imaging markers associated with disease processes with the goal of improving early detection, determining prognosis, and selecting treatments most likely to be effective. This allows pathologists to serve more patients while maintaining diagnostic and prognostic accuracy. This is especially important considering the ever-increasing number of patients in an aging population and the less than 2% of the medical graduates going into pathology [2].
While AI is slated to benefit many areas of clinical health sciences (oncology, drug development, etc.), the focus of this review is to highlight the use in digital pathology and whole slide imaging, including education (e.g. digital slide teaching sets), quality assurance (e.g. second opinions, proficiency testing, archiving), and clinical diagnosis (i.e. telepathology). Further, we explore how AI has advanced these areas of digital pathology as well as specific use cases and applications of AI in research, image analysis, CAD, and with a discussion of the techniques used, challenges, and barriers [3]. Finally, we discuss the ultimate goal of AI and WSI – integration of pathological image information with clinical data – and its reservations. With WSI as an enabling platform for AI, digital pathology will have meaningful and measurable impact on both clinical and research components of pathology workflow [1, 2].
AI AND EDUCATION
WSI is already used for teaching at conferences, virtual workshops, presentations, and tumor boards [1]. Equipped with WSI, AI tools can help further training of the next generation of pathologists by providing on demand, standardized, interactive digital slides that can be shared with multiple users anywhere at any time [1, 2]. Additionally, AI tools can provide automated annotations as quiz mode for trainees. With the help of these interactive tools, trainees can view, pan, and zoom enhanced digital slides which can provide a real-time, dynamic tutoring environment. Our group has developed some of these approaches – specifically generation of synthetic digital slides, which will be discussed here.
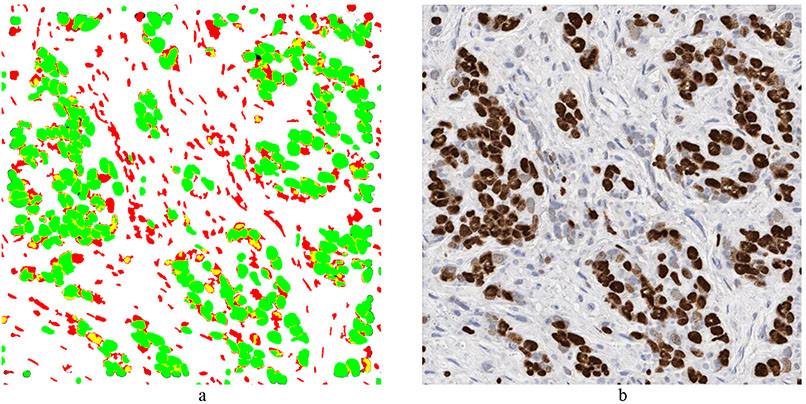
In our first attempts at generating synthetic images, we extracted individual as well as clusters of both positively and negatively stained nuclei from real WSI images, and systematically placed the extracted nuclei clumps on an image canvas – a cut-and-paste approach [4]. These images were evaluated by four board certified pathologists in the task of estimating the ratio of positive to total number of nuclei. The resulting concordance correlation coefficients (CCC) between the pathologist and the true ratio range from 0.86 to 0.95 (point estimates). In our follow-up study, we employed an approach known as conditional Generative Adversarial Networks (cGANs) [5]. cGAN method included two main components: generator, G, and discriminator, D. While the generator tries to create “fake” stained images, discriminator tries to “catch” these fake images, each getting better in an iterative manner. The main idea is to force G to learn the underlying distribution of the images from the training data. The accuracy of five experts (3 pathologists and 2 image analysts) in distinguishing between 15 real and 15 synthetic images was only 47.3% (±8.5%). Generation of synthetic histopathology images could be useful for educational purposes as they will give countless number of different combinations for pathology trainees to test their skills. In addition, these approaches can be very useful for quality control and understanding the perceptual and cognitive challenges that pathologists face. Figure 1 shows an example of synthetic breast cancer image generation.
Figure 1:
Example of synthetic breast cancer image generation. a) Input image with desired Ki67 positive (green) and negative nuclei (red). b) Synthetic image generated by cGAN. The nuclei location and size in the synthetic image are the same as in the input image. c) Randomly drawn lines. d) Synthetic bladder cancer image generated based on the randomly drawn lines in (c).
AI AND QUALITY ASSURANCE (QA)
The development of automated, high-speed, high-resolution WSI has had a significant impact on quality assurance. Digitized slides readily available to pathologists in the laboratory information system (LIS) or an intranet can be used for several QA tasks, including teleconsultation, gauging both inter- and intra-observer variance, proficiency testing, and archiving of slides. For example, the College of American Pathologists now optionally sends WSIs in addition to glass slides of certain proficiency testing cases.
AI can serve an important role in quality assurance. It is very difficult for pathologist and radiologist alike to stay current in all organ systems and cancer types. As with all disciplines, frequency of interactions builds confidence and skills, and helps keep practitioners current with evolving diagnostic tools. In addition, providing feedback either manually or with intelligent deep learning and AI tools, pathologist performance has the potential to keep improving. AI can be used as a supplement to these manual digital reviews both in routine diagnostic workflow or as a complement to the more formal quality reviews that are part of pathology laboratory quality management process. AI can also provide a quality check on the diagnosis rendered by a pathologist by applying automated diagnostic algorithms prospectively or retrospectively. These methods can continue to serve as patient safety mechanism to improve the quality of diagnosis and error prevention.
AI FOR CLINICAL DIAGNOSIS
Rendering routine pathologic diagnoses using WSI is feasible. Several studies have been published comparing the diagnostic interpretation made using digital slides to diagnoses rendered using glass slides and a conventional light microscope. Today, there are some pathology laboratories that have gone slide-less, such as the general pathology laboratory at Kalmar County Hospital in Kalmar, Sweden. Others such as The Ohio State University have made significant advances towards conversion to a digital pathology workflow. Essential requirements for the integration of digital pathology into LIS include accurate digital reproduction of the scanned glass slide, running the slide scanner continually with limited use of lab personnel, and saving, archiving, and later retrieved of the image without degradation. AI offers to improve on current solutions to the first of these essentials through detection of out-of-focus areas and color standardization.
The quality of images produced by whole slide imaging (WSI) scanners has a direct influence on the readers’ performance and their reliability of diagnosis [6]. Most modern WSI scanners come equipped with autofocus (AF) optics system to select focal planes to accurately capture the three-dimensional tissue morphology as a two-dimensional digital image [7]. To account for varying thickness of tissue sections, AF optics systems determine a set of focus points at different focal planes. From these focal planes, scanners capture images to produce sharp tissue representation. However, WSI scanners may still produce digital images with out-of-focus/blurry areas if their AF optics system erroneously selects focus points that lie in a different plane than the proper height of the tissue [8]. A naïve solution would be to add several extra focus points. This is impractical, as it would long delays to slide scanning. AI provides a better alternative by automatically identifying out-of-focus regions and allowing the WSI scanners to add a few extra focal points in those regions. AI achieves this by either “feature engineering” or via “representation learning” approach. In [8], the authors adopted a feature engineering approach by handcrafting texture features from gray level co-occurrence matrices and gradient information. These features were used in conjunction with decision trees to classify 200x200 pixel-sized regions as focused or blurred. Unfortunately, the method is only sensitive to high level of blurriness, and it requires modifying program parameters to adopt it to images acquired at different resolutions. Another approach, DeepFocus [7], based on representation learning, discovers features automatically from the images to identify blurry regions. Because DeepFocus automatically learns features at different levels of abstraction, it can generalize to different types of tissues and even to color variations due to different types of staining, H&E and IHC.
Standardization of the color displayed by digital slides is important for the accuracy of AI. Color variations in digital slides are often produced due to different lots or manufacturers of staining reagents, variations in thickness of tissue sections, difference in staining protocols, and disparity in scanning characteristics. One such example of color variation is shown in Figure 2. These variations often impose obstacles to clinical diagnosis/prognosis carried out by humans as well as machines [9, 10]. Moreover, these variations are one of the main hurdles in generalization of the machine learning algorithms to multiple sites. For this reason, lack of color normalization (standardization) in AI pipeline could negatively affect the performance of machine learning algorithms [10]. For a long time, collecting color statistics to perform color matching across images has remained the main source of color normalization. However, recent progress in generative models have presented novel ways of color normalization. In [11], the authors adopted a generative adversarial (GAN) [12] network for stain normalization. Unlike conventional GAN, which uses noise as an input, this network requires the grey-scale image as an input. This enables the network to preserve the image structure while manipulating the color information. It also requires a color system matrix (consisting of stain vectors, similar to color de-convolutional matrix) as an input for the generator in the network. This network can simultaneously learn the chromatic space of H&E images and can normalize it to a template image using the color system matrix. The non-parametric nature of the network makes it applicable to a wide variety of histopathological images. In a similar effort [13], the authors adopted the concept of style transfer [14] for color normalization. Their objective was to transfer the staining appearance of tissue images across different datasets to avoid color variations caused by batch effects. When histopathological images are acquired in different experimental setups and tested on pretrained diagnostic models, the prediction performance can suffer due to batch effects, i.e., nonbiological experimental variations such as age of sample, method of slide preparation, specifications of the imaging device, and type of postprocessing software [15]. Style transfer provides one plausible solution by finding image representations that independently model variations in the semantic image content and the style in which it is presented [13]. However, the method lacks a mechanism to standardize a dataset to a given slide without retraining the whole network. In [9], the authors used the concept of Cycle-Consistent Adversarial Networks [16] to perform color normalization. CycleGAN, a variant of GAN, permits the unpaired Image-to-Image translation through cycle-consistency. CycleGAN allows the images to be mapped to a specified color-model and preserves the same tissue structure.
Figure 2:
Example of color normalization. a) Reference image: T1 bladder cancer image cropped from a whole slide image. b) Image to be normalized to Reference Image. The objective is to change the colors of (b) so that it has a similar color appearance as the Reference image. c) Color normalized image produced by cGAN.
AI AND IMAGE ANALYSIS
Image analysis tools can automate and quantify with greater consistency and accuracy than light microscopy. Computer aided diagnosis (CAD) is widely used for ER, PR, and HER2/neu assessments in breast cancer, Ki67 assessment in carcinoid tumors, as well as multiple other clinical and research stains. The reliability of these CAD methods in general requires the standardization of the image acquisition step, which has been discussed previously. The development of WSI has facilitated large growth in the number of researchers and companies seeking to utilize CAD to analyze WSI and to develop new software tools to assist pathologists. Prior to WSI, the field of image analysis was limited by the requirement of pathologists having to select regions of interest to be analyzed. Since WSI allows the entire slide to be available for analysis, field selection can be automated. The following section summarizes various AI methods to enable this region of interest selection. First, the task of nuclei detection and nuclear segmentation is discussed, followed by a brief discussion of region identification.
Nuclear segmentation in WSI enables extraction of high-quality features for nuclear morphometrics and other analysis in computational pathology [17]. For this reason, automatic nuclei segmentation is among the most studied problems in AI [18]. AI enables efficient a range of nuclear segmentation tasks including segmenting of all nuclei from a WSI to identifying a subset of nuclei within specific anatomical regions.
Like other areas of pathology, deep learning algorithms are well known for their state-of-the-art performance on nuclei segmentation task [18]. In general, these algorithms estimate a probability map of the nuclear and non-nuclear (two-class) regions based on the learned nuclear appearances and rely on complex post-processing methods to obtain the final nuclear shapes and separation between touching nuclei [17]. For instance, [19] used a multiscale convolutional network to generate a nuclear probability map. This map was subjected to graph-partitioning to segment individual nuclei from the image. Similarly, [20] exploited the topology of the probability map using region-growing [21] to segment the individual nuclei. Unfortunately, these methods require retraining for their generalization to unseen datasets. Moreover, these methods also fail to generalize if the training and test images belongs to different organs. To overcome these issues, there is growing trend to train the nuclei segmentation methods on images taken from different organs.
In [17], the authors created a well annotated database consisting of 30 WSI of digitized tissue samples from several organs. The WSI were taken from the publically available database, The Cancer Genomic Atlas (TCGA) [22]. The images were generated at 18 different hospitals, which adds to the diversity of this dataset in terms of variation in slide preparation protocols among labs. Over 21,000 nuclei were manually annotated to train a deep learning algorithm. Unlike former methods, the authors formulated the nuclei segmentation as a three-class problem. They considered the nuclei edges as a third class while generating the ternary probability map. This map was subjected to region-growing to segment the individual nuclei. In another attempt [23], a generative model was adapted to perform nuclei segmentation in images taken from different organs. They trained a generative model using images from four different organs to synthetically generate pathology images. These synthetically generated images were combined with real images to train a CNN to perform nuclei segmentation.
During most pathological analysis, pathologists are interested in identifying a subset of nuclei in a certain anatomical regions [own]. For instance, in T1 bladder cancer, the pathologists are interested in identifying the tumor nuclei within lamina propria [24]. Similarly, in breast and pancreatic neuroendocrine tumors, pathologists are only interested in the ratio of tumor positive to total tumor cells within hotspots [25, 26]. In neuroblastoma, the analysis is only limited to the presence of centroblasts within follicles [27, 28]. For these reasons, there is an increasing interest in developing AI algorithms that can identify a subset of cells within a certain anatomical region. In [26], a transfer learning method [29] was adapted to identify tumor positive and negative nuclei from images of Ki67 breast cancer tissues. Similarly, a dual cGAN was designed along a dictionary-learning framework [30] to identify tumor regions from non-tumor regions in colorectal patients [31].
Although far less established and routine than their use in clinical workflows in radiology, CAD is an active research area for tumor biopsies. Currently, manual interpretation of these images often involves extremely laborious tasks such as cell counting. Moreover, these quantitative measures are far from exhaustive and typically consider only specific portions of biopsies (i.e. hotspots [32]) and specific anatomical regions. CAD offers increased efficiency, accurate quantification, and potentially novel, subsensory features for analysis and interpretation of histopathological images, thus mitigating pathologist workload and inter/intra variability.
The majority of research in the automated analysis of digital tumor biopsies rides the recent wave of deep learning [33]. In the context of images, deep learning allows computers to mimic the process of human visual perception through a cascade of layered, interconnected computational units which vaguely resemble biological neurons. Due to the sheer size (i.e. file size) of digital biopsies and the computational demands and complexities of deep learning, research has mostly focused on physically smaller tasks which look at small portions of the image like mitosis detection [34], anatomical region identification [35], and cancer identification [36]. However, recent research attempts to bypass these computational barriers through specialized deep learning methods which take advantage of whole-slide information. These methods are whole-slide in two senses – utilizing 1) the entire tissue section and 2) anatomical regions, which are not typically considered for diagnosis and decision making. Ehteshami et al. developed a fully automatic method to detect ductal carcinoma in situ using whole-slide H&E stained biopsies [37, 38]. Their method initially partitions a whole-slide into “superpixels” based on similarity at some magnification. Superpixels are grouped into anatomical regions (specifically epithelium) based on graph clustering [39]. Finally, each cluster is classified as DCIS or benign/normal based on features extracted by deep learning [38]. Niazi et al. utilized deep learning to identify tumor regions in pancreatic neuroendocrine tumors [26]. Their method employed transfer learning, whereby the features of a pretrained deep learning algorithm (i.e. trained on some other classification task) are fined-tuned and retrained to classify tumor and stroma regions. Both Ki67 positive and negative tumor cells were utilized. Cruz-Roa et al. developed an adaptive, automated sampling method for whole-slide images [40]. Their method approached deep learning computational barriers by carefully picking regions of whole slides using quasi- Monte Carlo sampling [41]. Their method was able to detect invasive breast cancer with DICE coefficient of 76% across multiple institutions, scanners, and preparation protocols. Finally, Niazi et al. developed a novel ROI selection method for hotspot detection in breast cancer to minimize magnitude of data transfer [25]. Clearly, whole-slide image based decisions are within reach and are preferred to methods utilizing only portions of slides.
INTEGRATION OF AI WITH OTHER CLINICAL DATA
Histopathological image analysis is not only limited to visual analysis, several other sources of data need to be included coming from -omics, clinical records, patient demographic information [42, 43]. Clinical data (e.g. demographic information, medical history, lab and clinical results) are mostly in unstructured free-text reports. Natural language processing (NLP) technologies can be used to extract relevant information and tie those to the information in histopathological slides [44]. NLP has also started to benefit from recent deep learning-based AI technologies. AI will be essential to comb through these disparate sources of information and help pathologists arrive at the best clinical decisions for patients. AI will be able to discover complicated or subtle connections than a human would. AI treatment needs to include not only images but also clinical and outcome data, enabling high dimensional analysis that is beyond what human brain alone can accomplish. Rich sources of data will lead to transform pathology from a clinical science to informatics science where the tissue would be only one of the sources of data.
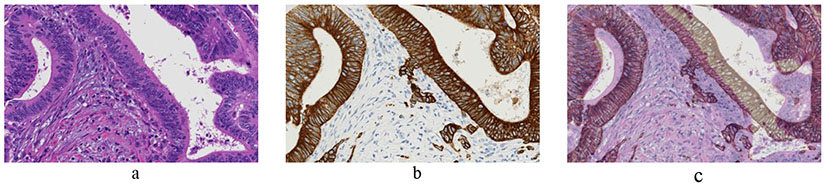
In anatomical pathology, immunohistochemical (IHC) staining plays a profound role in diagnosis (determining the biological characteristics of wide variety of tumors) [45], prognosis [46] and selecting appropriate systemic therapy for cancer patients [4, 47]. As genomic technologies are increasingly utilized, DNA-level and transcriptional-level features obtained from homogenized tissues will be evaluated for their utility in creating new sub-classifications of malignancies to predict future disease behavior and treatment response. Understanding the relationships between genomic features and quantitative IHC features will be critical for further illuminating protein-genomic associations, and for creating improved molecular classifications that combine nucleotide-resolution information generated at exome scales with spatial and subcellular protein-level information. As a first towards this process, AI is helping us to perform a cell level registration among adjacent sections of the tumor tissue [48]. Image registration is required to compensate for the non-linear geometric deformation induced by the staining process. Figure 4 shows an example of such non-linear deformation.
Figure 4:
Adjacent tissue sections of colorectal cancer patient. a) H&E image. b) Adjacent tissue section of (a) stained for pan-cytokeratin to assist in the identification of tumor buds. c) H&E and pan-cytokeratin images overlaid on each other to depict the non-linear deformation between the tissue sections.
Image registration is enabling us to study the behavior of different biomarkers within the same cell [49, 50]. It is also paving the path towards better understanding of tumor microenvironment. Moreover, image registration is enabling us to combine information from different modalities [51]. Having said that, image registration is still relatively unexplored frontiers of digital pathology. The dream of combing pathology with -omics or other modalities hinges on the development of reliable image registration methods. With the advent of deep learning, we expect to see progress in this area in the near future.
PERCEPTION AND LIMITATIONS OF AI
Many of the AI approaches, particularly deep-learning based systems, are criticized for not being able to explain how they arrive at their decisions, hence called “black boxes.” While these algorithms will still offer benefits, clinical, legal and regulatory issues need to be sorted out going forward. At the same time, there’s active research to make the algorithms easier to interpret by humans and provide insight on how they work, e.g. by providing some of the features that the algorithm is focusing on or by dividing the AI algorithm execution into steps, each of which make logical sense to a human expert. These will provide some transparency to AI algorithms but will often come at the expense of performance hit as a trade-off. On the regulatory side, there could be restrictions, e.g. the General Data Protection Regulation of Europe stipulates that “The data subject shall have the right not to be subject to a decision based solely on automated processing…” whose medical implications should be carefully reviewed.
Is AI going to replace pathologists? The notion that AI will replace pathologists is just a speculation at this point while the AI can be extremely helpful. Expert-AI combination will yield results that are more accurate, consistent and useful than what an expert can do alone. While the AI will continue to make decisions in narrow fields, humans can take several factors into account and are better at synthesizing information to arrive at decisions. AI will be trained to extract information and connect it with other complementary sources of information. For example, we evaluated multiple regions of single slides as well as multiple sections from different patients’ tumors using computational histologic analysis and semi-quantitative proteomic profiling of neuroblastoma slides. We found that both approaches determined that inter-tumor heterogeneity was greater than intratumor heterogeneity and both techniques can supplement pathologist review of neuroblastoma for refined risk stratification.
Some exciting developments in AI haven’t been applied to medicine yet. For example, one shot learning is learning from only a small number of training samples as opposed to a large number of samples. It’s done typically by means of transferring knowledge from other domains or models or extracting from the context. This could be particularly useful in pathology where deep truthing is challenging due to the complexity of the images and their sheer size. In reinforcement learning, algorithms are trained to reach complex goals by comparing the immediate actions with long term outcomes. As in human learning, the algorithm has to wait (e.g. until the end of the game) to find out whether particular actions will lead to success or failure. These could be particularly useful in training some algorithms to make complex decisions in pathology where the outcome may be known yet what particular biological factors or treatment options led to that outcome may not be completely apparent.
While pathology is rapidly moving towards digital and with new developments in AI, bringing these together are expected to bring exciting changes to healthcare while a large number of technical, ethical and legal questions need to be answered first.
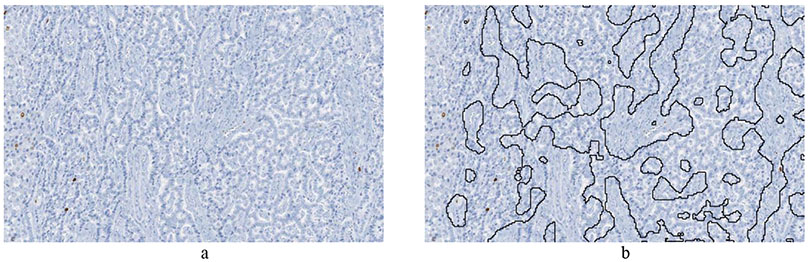
Figure 3:
Example of tumor identification from Ki67 stained slides of pancreatic neuroendocrine tumor. a) Image cropped from Ki67 slide of pancreatic neuroendocrine tumor patient. b) The non-tumor regions are automatically outlined by a deep learning algorithm.
Source of Funding:
The project described was supported in part by Awards Number R01CA134451 (PI: Gurcan), U24CA199374 (PI: Gurcan) from the National Cancer Institute, and OSU CCC Intramural Research (Pelotonia) Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute, or the National Institutes of Health.
Footnotes
Conflicts of Interest: We have no conflicts of interest to declare.
Search Strategy and Selection Criteria section: As this is a relatively new area of research, we performed a direct search that involved reviewing recent articles in this domain. Apart from it, we also used Google Scholar to identify recent development in this area.
References
- [1].Farahani N, Parwani AV, and Pantanowitz L, "Whole slide imaging in pathology: advantages, limitations, and emerging perspectives," Pathol Lab Med Int, vol. 7, pp. 23–33, 2015. [Google Scholar]
- [2].Zarella MD, Bowman D, Aeffner F, Farahani N, Xthona A, Absar SF, Parwani A, Bui M, and Hartman DJ, "A Practical Guide to Whole Slide Imaging: A White Paper From the Digital Pathology Association," Archives of pathology & laboratory medicine, 2018. [DOI] [PubMed] [Google Scholar]
- [3].Tizhoosh HR and Pantanowitz L, "Artificial intelligence and digital pathology: Challenges and opportunities," Journal of pathology informatics, vol. 9, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Niazi MKK, Abas FS, Senaras C, Pennell M, Sahiner B, Chen W, Opfer J, Hasserjian R, Louissaint A Jr, and Shana'ah A, "Nuclear IHC enumeration: A digital phantom to evaluate the performance of automated algorithms in digital pathology," PloS one, vol. 13, p. e0196547, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mirza M and Osindero S, "Conditional generative adversarial nets," arXiv preprint arXiv: 1411.1784, 2014. [Google Scholar]
- [6].Shrestha P, Kneepkens R, Vrijnsen J, Vossen D, Abels E, and Hulsken B, "A quantitative approach to evaluate image quality of whole slide imaging scanners," Journal of pathology informatics, vol. 7, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Senaras C, Niazi MKK, Lozanski G, and Gurcan MN, "DeepFocus: Detection of out-of-focus regions in whole slide digital images using deep learning," PloS one, vol. 13, p. e0205387, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lopez XM, D'Andrea E, Barbot P, Bridoux A-S, Rorive S, Salmon I, Debeir O, and Decaestecker C, "An automated blur detection method for histological whole slide imaging," PloS one, vol. 8, p. e82710, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shaban MT, Baur C, Navab N, and Albarqouni S, "StainGAN: Stain Style Transfer for Digital Histological Images," arXiv preprint arXiv: 1804.01601, 2018. [Google Scholar]
- [10].Komura D and Ishikawa S, "Machine learning methods for histopathological image analysis," Computational and Structural Biotechnology Journal, vol. 16, pp. 34–42, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zanjani FG, Zinger S, Bejnordi BE, van der Laak JA, and de With PH, "Stain normalization of histopathology images using generative adversarial networks," in 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), 2018, pp. 573–577. [Google Scholar]
- [12].Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, and Bengio Y, "Generative adversarial nets," in Advances in neural information processing systems, 2014, pp. 2672–2680. [Google Scholar]
- [13].Bentaieb A and Hamarneh G, "Adversarial stain transfer for histopathology image analysis," IEEE transactions on medical imaging, vol. 37, pp. 792–802, 2018. [DOI] [PubMed] [Google Scholar]
- [14].Gatys LA, Ecker AS, and Bethge M, "Image style transfer using convolutional neural networks," in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 2414–2423. [Google Scholar]
- [15].Kothari S, Phan JH, Stokes TH, Osunkoya AO, Young AN, and Wang MD, "Removing batch effects from histopathological images for enhanced cancer diagnosis," IEEE Journal of Biomedical and Health Informatics, vol. 18, pp. 765–772, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhu J-Y, Park T, Isola P, and Efros AA, "Unpaired image-to-image translation using cycle-consistent adversarial networks," arXiv preprint, 2017. [Google Scholar]
- [17].Kumar N, Verma R, Sharma S, Bhargava S, Vahadane A, and Sethi A, "A dataset and a technique for generalized nuclear segmentation for computational pathology," IEEE transactions on medical imaging, vol. 36, pp. 1550–1560, 2017. [DOI] [PubMed] [Google Scholar]
- [18].Xing F and Yang L, "Robust nucleus/cell detection and segmentation in digital pathology and microscopy images: a comprehensive review," IEEE reviews in biomedical engineering, vol. 9, pp. 234–263, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Song Y, Zhang L, Chen S, Ni D, Lei B, and Wang T, "Accurate segmentation of cervical cytoplasm and nuclei based on multiscale convolutional network and graph partitioning," IEEE Transactions on Biomedical Engineering, vol. 62, pp. 2421–2433, 2015. [DOI] [PubMed] [Google Scholar]
- [20].Xing F, Xie Y, and Yang L, "An automatic learning-based framework for robust nucleus segmentation," IEEE transactions on medical imaging, vol. 35, pp. 550–566, 2016. [DOI] [PubMed] [Google Scholar]
- [21].Tremeau A and Borel N, "A region growing and merging algorithm to color segmentation," Pattern Recognition, vol. 30, pp. 1191–1203, 1997. [Google Scholar]
- [22].Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, and Network CGAR, "The cancer genome atlas pancancer analysis project," Nature genetics, vol. 45, p. 1113, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mahmood F, Borders D, Chen R, McKay GN, Salimian KJ, Baras A, and Durr NJ, "Deep Adversarial Training for Multi-Organ Nuclei Segmentation in Histopathology Images," arXiv preprint arXiv: 1810.00236, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Niazi MKK, Tavolara T, Arole V, Parwani A, Lee C, and Gurcan M, "MP58-06 automated staging of T1 bladder cancer using digital pathologic h&e images: a deep learning approach," The Journal of urology, vol. 199, p. e775, 2018. [Google Scholar]
- [25].Niazi MKK, Lin Y, Liu F, Ashok A, Marcellin MW, Tozbikian G, Gurcan M, and Bilgin A, "Pathological image compression for big data image analysis: Application to hotspot detection in breast cancer," Artificial intelligence in medicine, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Niazi MKK, Tavolara TE, Arole V, Hartman DJ, Pantanowitz L, and Gurcan MN, "Identifying tumor in pancreatic neuroendocrine neoplasms from Ki67 images using transfer learning," PloS one, vol. 13, p. e0195621, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Komaropoulos EN, Niazi M, Lozanski G, and Gurcan MN, "Histopathological image analysis for centroblasts classification through dimensionality reduction approaches," Cytometry Part A, vol. 85, pp. 242–255, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sertel O, Lozanski G, Shana’ah A, and Gurcan MN, "Computer-aided detection of centroblasts for follicular lymphoma grading using adaptive likelihood-based cell segmentation," IEEE Transactions on Biomedical Engineering, vol. 57, pp. 2613–2616, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Szegedy C, Vanhoucke V, Ioffe S, Shlens J, and Wojna Z, "Rethinking the inception architecture for computer vision," in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 2818–2826. [Google Scholar]
- [30].Mairal J, Ponce J, Sapiro G, Zisserman A, and Bach FR, "Supervised dictionary learning," in Advances in neural information processing systems, 2009, pp. 1033–1040. [Google Scholar]
- [31].Tavolara T, Niazi MKK, Chen W, Frakel W, and Gurcan MN, "Colorectal Tumor Identification by Tranferring Knowledge from Pan-cytokeratin to H&E," SPIE Medical Imaging, February, 2019. 2019. [Google Scholar]
- [32].Niazi MKK, Hartman DJ, Pantanowitz L, and Gurcan MN, "Hotspot detection in pancreatic neuroendocrine tumors: Density approximation by α-shape maps," in SPIE Medical Imaging, 2016, pp. 97910B–97910B. [Google Scholar]
- [33].LeCun Y, Bengio Y, and Hinton G, "Deep learning," Nature, vol. 521, p. 436, 2015. [DOI] [PubMed] [Google Scholar]
- [34].Albarqouni S, Baur C, Achilles F, Belagiannis V, Demirci S, and Navab N, "Aggnet: deep learning from crowds for mitosis detection in breast cancer histology images," IEEE transactions on medical imaging, vol. 35, pp. 1313–1321, 2016. [DOI] [PubMed] [Google Scholar]
- [35].Khalid Khan TETM, Arole Vidya, and Parwani b. Anil V., Lee Cheryl, Gurcan Metin N., "Automated T1 bladder risk stratification based on depth of lamina propria invasion from H&E tissue biopsies: A deep learning approach " SPIE Medical Imaging, 2018. [Google Scholar]
- [36].Litjens G, Sanchez CI, Timofeeva N, Hermsen M, Nagtegaal I, Kovacs I, Hulsbergen-Van De Kaa C, Bult P, Van Ginneken B, and Van Der Laak J, "Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis," Scientific reports, vol. 6, p. 26286, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bejnordi BE, Zuidhof G, Balkenhol M, Hermsen M, Bult P, van Ginneken B, Karssemeijer N, Litjens G, and van der Laak J, "Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images," Journal of Medical Imaging, vol. 4, p. 044504, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bejnordi BE, Balkenhol M, Litjens G, Holland R, Bult P, Karssemeijer N, and van der Laak JA, "Automated detection of DCIS in whole-slide H&E stained breast histopathology images," IEEE transactions on medical imaging, vol. 35, pp. 2141–2150, 2016. [DOI] [PubMed] [Google Scholar]
- [39].Schaeffer SE, "Graph clustering," Computer science review, vol. 1, pp. 27–64, 2007. [Google Scholar]
- [40].Cruz-Roa A, Gilmore H, Basavanhally A, Leldman M, Ganesan S, Shih N, Tomaszewski J, Madabhushi A, and Gonzalez L, "High-throughput adaptive sampling for whole-slide histopathology image analysis (HASHI) via convolutional neural networks: Application to invasive breast cancer detection," PloS one, vol. 13, p. e0196828, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Caflisch RE, "Monte carlo and quasi-monte carlo methods," Acta numerica, vol. 7, pp. 1–49, 1998. [Google Scholar]
- [42].Natrajan R, Sailem H, Mardakheh FK, Garcia MA, Tape CJ, Dowsett M, Bakal C, and Yuan Y, "Microenvironmental heterogeneity parallels breast cancer progression: a histology–genomic integration analysis," PLoS medicine, vol. 13, p. e1001961, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Heindl A, Nawaz S, and Yuan Y, "Mapping spatial heterogeneity in the tumor microenvironment: a new era for digital pathology," Laboratory investigation, vol. 95, p. 377, 2015. [DOI] [PubMed] [Google Scholar]
- [44].Louis DN, Gerber GK, Baron JM, Bry L, Dighe AS, Getz G, Higgins JM, Kuo FC, Lane WJ, and Michaelson JS, "Computational pathology: an emerging definition," Archives of pathology & laboratory medicine, vol. 138, pp. 1133–1138, 2014. [DOI] [PubMed] [Google Scholar]
- [45].Zaha DC, "Significance of immunohistochemistry in breast cancer," World journal of clinical oncology, vol. 5, p. 382, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Niazi MKK, Downs-Kelly E, and Gurcan MN, "Hot spot detection for breast cancer in Ki-67 stained slides: image dependent filtering approach," in SPIE Medical Imaging, 2014, pp. 904106–904106-8. [Google Scholar]
- [47].Das H, Wang Z, Niazi MKK, Aggarwal R, Lu J, Kanji S, Das M, Joseph M, Gurcan M, and Cristini V, "Impact of diffusion barriers to small cytotoxic molecules on the efficacy of immunotherapy in breast cancer," PloS one, vol. 8, p. e61398, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sertel O, Dogdas B, Chiu CS, and Gurcan MN, "Muscle histology image analysis for sarcopenia: Registration of successive sections with distinct atpase activity," in Biomedical Imaging: From Nano to Macro, 2010 IEEE International Symposium on, 2010, pp. 1423–1426. [Google Scholar]
- [49].Johnson S, Brandwein M, and Doyle S, "Registration parameter optimization for 3D tissue modeling from resected tumors cut into serial H&E slides," in Medical Imaging 2018: Digital Pathology, 2018, p. 105810T. [Google Scholar]
- [50].Yigitsoy M and Schmidt G, "Hierarchical patch-based co-registration of differently stained histopathology slides," in Medical Imaging 2017: Digital Pathology, 2017, p. 1014009. [Google Scholar]
- [51].Chappelow J, Bloch BN, Rofsky N, Genega E, Lenkinski R, DeWolf W, and Madabhushi A, "Elastic registration of multimodal prostate MRI and histology via multiattribute combined mutual information," Medical physics, vol. 38, pp. 2005–2018, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]