Abstract
The MET receptor tyrosine kinase (RTK) and its cognate ligand HGF comprise a signaling axis essential for development, wound healing and tissue homeostasis. Aberrant HGF/MET signaling is a driver of a number of cancers and contributes to drug resistance to several approved therapeutics targeting other RTKs, making MET itself an important drug target. In RTKs, homeostatic receptor signaling is dependent on autoinhibition in the absence of ligand binding and orchestrated set of conformational changes induced by ligand-mediated receptor dimerization that result in activation of the intracellular kinase domains. A fundamental understanding of these mechanisms in the MET receptor remains incomplete, despite decades of research. This is due in part to the complex structure of the HGF ligand, which remains unknown in its full-length form, and a lack of high-resolution structures of the complete MET extracellular portion in an apo or ligand bound state. A current view of HGF-dependent MET activation has evolved from biochemical and structural studies of HGF and MET fragments and here we review what these findings have thus far revealed.
Introduction
MET, also known as Hepatocyte Growth Factor Receptor (HGFR) and its close homolog RON (Recepteur d’Origine Nantais), also known as Macrophage-Stimulating 1 Receptor (MST1R) form a subfamily within the larger receptor tyrosine kinase (RTK) family of membrane receptors (1–5). Like other RTKs, MET consists of an extracellular domain (ECD), a single pass transmembrane domain (TM), and an intracellular region that includes a juxtamembrane domain (JM), kinase domain (KD) and carboxy-terminal tail (C-tail) (5). The ECD of MET includes a sema domain, a Plexin/Semaphorin/Integrin (PSI) domain, and four Integrin/Plexin/Transcription factor (IPT) domains (6, 7). The single pass transmembrane helix of MET connects the ECD module to a long intracellular juxtamembrane region that is followed by a tyrosine kinase domain and a short C-tail (Figure 1A & B). The broad range of physiological outputs downstream from MET includes activating cell motility, altering cellular morphology and driving cell cycle progression, ultimately playing essential roles in development of multiple tissues during embryogenesis and in wound healing and tissue homeostasis in adults (8–10). The growth and motility signals activated by MET make it a potent oncogene when it becomes abnormally activated through gene amplification, enhanced expression or mutation, and this has been observed in a number of solid cancers, including lung, kidney and liver (11–13).
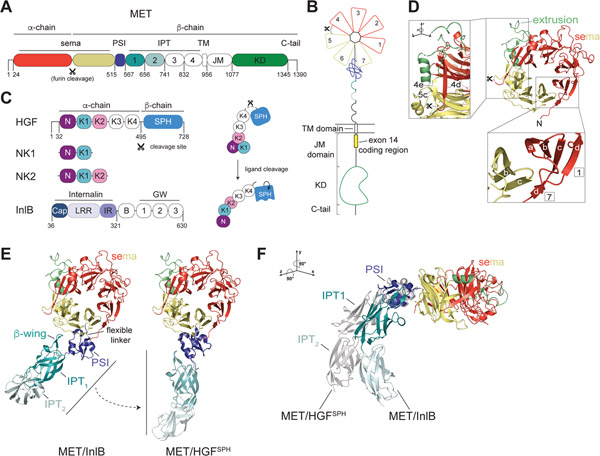
Figure 1: Architecture of the MET extracellular domain.
(A) Domain architecture of the MET receptor which includes the sema domain, PSI (Plexin, Semaphorin, Integrin) domain, IPT (Integrin, Plexin, Transcription factor) domain, TM (transmembrane) domain, JM (juxtamembrane) domain and KD (kinase domain). Residues at domain boundaries are marked. The furin cleavage site in the sema domain is indicated with a scissors sign. (B) Cartoon representation of the domain structure of the full-length MET receptor colored relative to (A). The extracellular domain (ECD) includes the sema domain and the stalk region (PSI-IPT1–4). Numbers in the sema domain denote individual blades of the β-propellor structure. Exon 14 in the membrane proximal region of the JM domain is marked by a yellow box. (C) Domain architecture of MET ligands: HGF, NK1, NK2 and InlB. The HGF cleavage site, which generates the mature α- and β-chains of the ligand, is marked with a scissors sign. Domain abbreviations are: N (N domain), K1-K4 (kringle domains 1–4), SPH (serine protease homology domain), Cap (Cap domain), LRR (leucine rich repeat), IR (inter repeat region), B (B-repeat), GW (Gly-Trp domain). Domains colored in white have no associated high-resolution structural data. Panel on the right illustrates HGF ligand cleavage and proposed associated conformational changes that lead to formation of an active ligand. The N, K1 and K2 domain arrangement is based on crystal structures described in Figure 4A and B. (D) Crystal structure of the MET sema domain is shown in cartoon representation (PDB: 1SHY). PSI and SPH domains are removed for clarity. The extrusion region is colored lime green. (D, lower inset) The 4 anti-parallel β-strands (labelled a-d) that constitute each blade of the sema domain are shown for blades 1 and 7. Strand d of blade 7, which is contributed by the N-terminal region of the sema domain, encloses the structure. (D, high inset) The extrusion region is highlighted by rotation of the sema domain to demonstrate its continuation from blade 5c that continues back to form blade 4e. Two colored dots indicate an unresolved region of the extrusion (Thr 402-Arg 413). (E) Two orientations of the MET ECD in complexes with different ligands illustrate flexibility of the stalk region. Left, structure of MET in the MET/InlB complex (PDB: 6GCU) in which the InlB ligand has been removed for clarity. Right, structure of MET in the MET/HGFSPH complex (PDB: 1SHY), in which the HGFSPH domain has been removed for clarity, is also aligned via the PSI domain on the structure of the PSI-IPT1–2 module (PDB: 5LSP) to highlight the extended conformation of the stalk in this state. (F) The structures of MET/InlB and MET/HGFSPH (shown in grey) were aligned on their sema domains and rotated as noted relative to (E). HGFSPH and InlB ligands were removed for clarity.
The cognate ligand for MET is hepatocyte growth factor (HGF), also known as scatter factor (SF) (14–17). HGF is produced in a pro-form that is proteolytically cleaved to a mature, active α- and β-chain heterodimer held together by a disulfide bond (18) (Figure 1C). Two natural splice variants of HGF, called NK1 and NK2, encompass different fragments of the HGF α-chain (Figure 1C) (19, 20). The prevailing model for MET activation is that the mature HGF ligand can dimerize and bind two MET molecules to form a 2:2 heterocomplex (21–23). While not strictly required for signaling, heparan sulfate can augment MET activation by HGF by driving ligand dimerization or through increasing the local concentration of HGF at the cell surface (24, 25). In addition to HGF, a number of other non-canonical mechanisms of MET activation have been described under pathological and physiological conditions. Internalin B (InlB), an effector protein from Listeria monocytogenes, can directly engage MET receptor forming oligomers that in turn cluster MET, activating it and triggering endocytosis that facilitates entry of the bacterium into cells (26–28). Another protein ligand described to interact with MET is decorin, a leucine rich repeat (LRR) proteoglycan, which upon MET binding induces receptor downregulation through enhanced recruitment of the E3-ligase Cbl (29). In addition, numerous reports have documented co-activation of MET via other transmembrane receptors, including Integrinβ–4 (30), CD44v6 (31), VEGFR (32), ROR1 (33), HER3 (34), HER2 (35), AXL (36), RON (37) and the plexin-B1/SEMA4D complex (38) (Reviewed in (39)). The mechanisms by which these co-receptors activate MET have not been structurally defined, and in many cases, it remains unclear whether they represent direct interactions and how they converge on the canonical receptor activation mechanism that proceeds via MET homodimerization. Signaling crosstalk of MET with other receptors often manifests under pathological conditions and this may present novel therapeutic opportunities for targeting MET (12, 40, 41).
Our knowledge of activation mechanisms and communication across the plasma membrane for many RTKs has been compiled from high-resolution structures of individual domains of the receptors, their biochemical characterizations and cell-based functional studies (5, 42). While several high-resolution structures of fragments of the MET ECD and HGF have been solved (Tables 1 and 2), a number of important structural states are still missing. Consequently, our understanding of how HGF engages with the MET ECD to initiate activation of the intracellular kinase domain remains incomplete. Here we discuss the current state of structural understanding of HGF-dependent MET activation and highlight areas for much needed future investigations.
Table 1.
Structures of the MET ECD
| PDB ID | Technique | MET domains | Other ligands/proteins | |||||
|---|---|---|---|---|---|---|---|---|
| Sema | PSI | IPT1 | IPT2 | IPT3 | IPT4 | |||
| 1SHY | X-ray | + | + | HGF β-chain | ||||
| 2UZX | X-ray | + | + | + | InlB | |||
| 2UZY | X-ray | + | + | + | + | |||
| 4O3T | X-ray | + | + | HGF β-chain + Zymogen Activator Peptide (ZAP.14) | ||||
| 4O3U | X-ray | + | + | HGF β-chain + Zymogen Activator Peptide (ZAP2.3) | ||||
| 6GCU | X-ray | + | + | + | + | InlB and DARPin A3A | ||
| 6I04 | X-ray | + | + | Fab | ||||
| 4K3J | X-ray | + | + | HGF β-chain + Fab (Onartuzumab) | ||||
| 1SSL | NMR | + | - | |||||
| 5LSP | X-ray | + | + | + | Fab (107_A07) | |||
Table 2.
Structures of HGF
| PDB ID | Technique | HGF domains | Other ligands/proteins | |||||
|---|---|---|---|---|---|---|---|---|
| N | K1 | K2 | K3 | K4 | SPH | |||
| 6LZ9 | X-ray | + | Fab (t8E4) | |||||
| 4D3C | X-ray | + | + | monoclonal antibody (SFN68) | ||||
| 5CP9 | X-ray | + | + | MB605 (3-(furan-2-yl)propanoic acid) | ||||
| 5COE | X-ray | + | + | HEPES | ||||
| 5CS5 | X-ray | + | + | PIPES | ||||
| 5CS9 | X-ray | + | + | MES | ||||
| 5CS1 | X-ray | + | + | - | ||||
| 5CS3 | X-ray | + | + | (H)EPPS | ||||
| 5CT1 | X-ray | + | + | CHES | ||||
| 5CT3 | X-ray | + | + | 2FA (3-hydroxypropane-1-sulfonic acid) | ||||
| 5CSQ | X-ray | + | + | MOPS | ||||
| 5CT2 | X-ray | + | + | CAPS | ||||
| 4O3T | X-ray | + | ZAP.14 + MET sema+PSI | |||||
| 4O3U | X-ray | + | ZAP2.3 + MET sema+PSI | |||||
| 4K3J | X-ray | + | Fab (Onartuzumab) + MET sema+PSI | |||||
| 4IUA* | X-ray | + | + | + | *Mouse NK2 | |||
| 3SP8 | X-ray | + | + | + | Heparin DP10 | |||
| 3MKP | X-ray | + | + | Heparin | ||||
| 3HMS | X-ray | + | - | |||||
| 3HN4 | X-ray | + | + | + | HEPES | |||
| 3HMT | X-ray | + | - | |||||
| 2QJ4* | X-ray | + | + | *Mouse NK1 | ||||
| 2QJ2 | X-ray | + | + | - | ||||
| 1SI5 | X-ray | + | - | |||||
| 1SHY | X-ray | + | MET sema + PSI | |||||
| 1GP9 | X-ray | + | + | HEPES | ||||
| 1GMN | X-ray | + | + | Heparin, HEPES | ||||
| 1GMO | X-ray | + | + | Heparin, HEPES | ||||
| 1NK1 | X-ray | + | + | - | ||||
| 1BHT | X-ray | + | + | HEPES | ||||
| 2HGF | NMR | + | - | |||||
Extracellular domain of MET and its relations to the Plexin/Semaphorin system
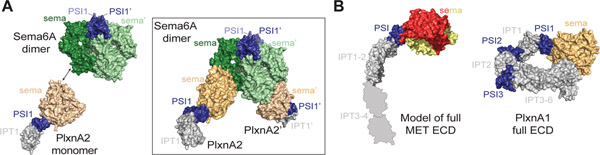
One of the distinct features of MET, and its close homolog RON, compared to other RTKs is the presence of a sema domain within their ECDs. Sema domain is a characteristic feature of another class of membrane receptors, called plexins, and their ligands, semaphorins (43). Instead of the kinase domain, plexins have intracellular GTPase domain as signaling units (44). It is presumed that MET and plexins shared a common ancestor, as early MET-like orthologs identified in both Lophotrochozoa and Echinodermata have extended ECDs like plexins (45), but the ligand systems for MET and plexins have evolved to be quite different. The semaphorins commonly act as sema-domain mediated dimers to activate plexins (46) (Figure 2A). While plexins have their own sema domains, they do not dimerize via these domains but use them to bind semaphorin dimers to form an active tetrameric complex (46) (Figure 2B). The sema domain of MET also provides the predominant binding site for its ligand HGF and like semaphorins, HGF also dimerizes to bring together two MET monomers. However, since HGF does not contain a sema domain the mode of HGF binding to MET is unique.
Figure 2: Architecture of the semaphorin/plexin ECDs.
(A) Surface representation of the Sema6A dimer and a PlxnA2 monomer based on PDB:3OKY. The tetrameric complex of the Sema6A dimer bound to two PlxnA2 monomers is shown boxed on the right-hand side. (B) Model of the complete MET ECD based on low resolution structures (21, 58). The MET sema, PSI and IPT1–2 are shown in surface representation from PDB: 1SHY and 5LSP. The IPT3 and IPT4 domains have not been resolved in high resolution and are shown as IPT domain silhouettes to match low resolution MET EVD models. The crystal structure of the full PlxnA1 ECD (PDB: 5L56) is also shown as a surface representation for comparison to the shorter MET ECD model.
The sema domain of MET forms a canonical β-propeller comprised of seven blades, with each blade containing 4-antiparallel β-strands named a-d moving from the inside to the outside of the propeller structure (Figure 1B & D). Circular arrangement of the 7 blades results in an enclosed structure, with the N-terminus providing the last strand of the seventh blade (Figure 1D, inset). Loops connecting a-b and c-d form the bottom face and b-d and d-a strands form the top face of the propeller (Figure 1D). These loops represent the most divergent sequences in sema domain-containing proteins and are frequently utilized for protein-protein interactions (43, 46). Of note is the extended loop that follows the 5c strand, termed the extrusion (47), which in semaphorins forms part of the dimer interface (47–51). In MET this loop consists of 57 amino acids and extends to form a fifth strand on the fourth blade (4e) before continuing to strand 6a, resulting in only 3 strands in the fifth blade (5a-5c) (52) (Figure 1D). The significance of this unique extrusion in MET is not yet defined but may contribute to its distinct mode of ligand binding from the plexin receptors.
A hallmark feature of MET and RON is cleavage of the sema domain between strands 4d and 5a (Arg 307-Ser 308 in MET) by the endoprotease furin (7, 52, 53). In MET, the resulting α (~50 kDa) and β (~145 kDa) polypeptide chains remain disulfide-linked, forming the mature heterodimeric receptor (Figure 1A & B) (7). Surprisingly, the functional relevance of the furin-mediated cleavage of MET, which is not resolved in existing crystal structures, is unknown. Mutation of this site does not appear to impede receptor maturation, ligand binding, or activation (53, 54). Given its proximity to the putative ligand binding site for the HGF α-chain (discussed below), this site and its proteolytic processing may make currently unappreciated contribution to ligand binding.
With the exception of the viral semaphorin protein A39R, all proteins with a sema domain are followed by a PSI domain (43). The PSI domain is a small cysteine rich knot which in MET is followed by four Ig-like IPT domains that together form the stalk region (PSI-IPT1–4) (7). In MET, two glycine residues present in the sema-PSI linker (Gly 517 and Gly 519) provide flexibility between the sema domain and the stalk, evident in different orientations that the IPT1 domain adopts relative to the sema domain in a number of ligand-bound MET (and RON) ECD structures (27, 52, 55, 56) (Figure 1E & F). The stalk region itself is more rigid in its N-terminal part due to the presence of a loop in the IPT1 domain, termed the β-wing, that provides extended contact with the PSI domain (7, 27, 57) (Figure 1E). This flexible connection between the sema domain and the more rigid stalk in MET could contribute to transitions between inactive and active states of the receptor. The MET ECD stalk forms a curved, hook-like architecture based on low resolution reconstructions of the full MET ECD in early cryo-electron microscopy (EM) tomography and small angle X-ray scattering (SAXS) studies (21, 58) (Figure 2B). The extended stalk region of plexin receptors forms an overall ring-like architecture that has been proposed to mediate receptor autoinhibition by spatially separating two receptors and preventing dimerization (59, 60) (Figure 2B). Further structures of the entire MET ECD are required to define the relationship between its ECD architecture and MET autoinhibition and activation.
Ligand-induced activation
A majority of biochemical evidence and structural models derived from low resolution techniques, such as cryo-EM and SAXS, collectively support the model in which MET activation proceeds via formation of a heterodimeric 2:2 ligand:receptor complex (21, 55, 61–63). In this model HGF dimer bridges two MET ECDs. In contrast to receptors such as EGFR and c-KIT, whose ECDs in solution form dimeric complexes upon ligand binding (64–66), the full MET ECD bound to HGF is monomeric in solution (21). A shorter MET ECD fragment that contains only the sema and PSI domains was shown to form a 2:2 complex with mature HGF (21) or with one of its splice variants, NK1 (62). This suggests that the remaining region of MET ECD (corresponding to the stalk region) exerts an autoinhibitory effect on ligand-dependent ECD dimerization. In the full-length receptor, this autoinhibition may be counteracted upon ligand binding by additional dimer-stabilizing interactions contributed by the transmembrane and intracellular regions.
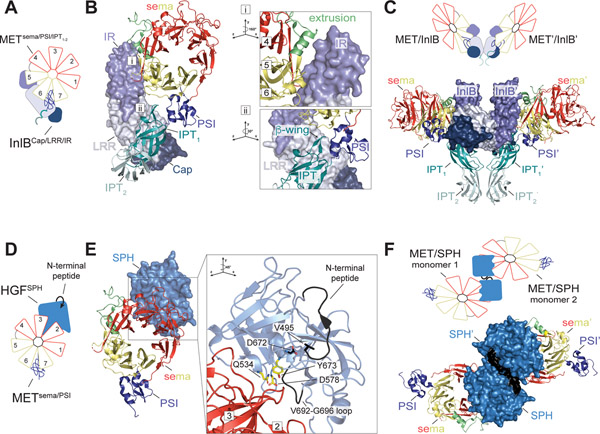
The architecture of the full-length, active HGF-bound MET receptor dimer is unknown. While there have been a number of structures of MET ECD fragments bound to β-chain of HGF, none of these structures yielded definitive dimeric complexes that explain receptor activation. The only known ligand-stabilized dimeric form of MET ECD is a complex between the sema-PSI-IPT1–2 fragment of MET ECD and the InlB toxin (27, 67). InlB contains an Internalin domain, which encompasses a Cap, LRR and an Internalin Repeat (IR), that together comprise the minimal fragment known to efficiently activate MET in cells (68) (Figure 1C). InlB binds MET via two interfaces (27) (Figure 3A & B). As revealed by X-ray crystallography, engagement of these two sites by InlB results in a more bent MET ECD orientation compared to the one seen in the HGF β-chain-bound sema-PSI structures (27) (Figure 1E & F). In the InlB/MET ECD structures, the top face of the sema domain is predicted to tilt towards the plasma membrane, much like a wilted flower (Figure 1F).
Figure 3: Binding modes of MET receptor ligands.
(A) Schematic representation of InlB bound to the MET ECD and (B) crystal structure of the MET ECD (shown as cartoon) bound to InlB (shown in surface mode) based on PDB: 6GCU. Two binding sites of InlB on MET are indicated by ‘i’ and ‘ii’ and are shown at higher magnification in the insets on the right. The first site (i) involves the IR domain, a low affinity binding site, which packs against two α-helices formed by the extrusion region and 5a-b loop as well as 6c-d loop of the MET sema domain. The second site (ii) is a high affinity binding site that involves interaction between the LRR and Cap domains of InlB and the β-wing region of the MET IPT1 domain. Numbers refer to the corresponding blades in the sema domain. (C) InlB-induced dimer of the MET ECD seen in the MET/InlB crystal structure (PDB: 6GCU) is shown (bottom) and cartooned as a schematic model (top). Dimerization interface is formed by the concave face of the LRR repeat of InlB, with a second putative interface formed by the IPT2 domains of the MET ECD. (D) Schematic representation of HGF SPH domain bound to the MET ECD and (E) crystal structure of the MET ECD (shown as cartoon) bound to the SPH domain (shown in surface mode) based on PDB: 1SHY. The inset on the right shows a magnified view of the SPH/MET binding interface seen in the crystal structure. Upon SPH domain cleavage, the liberated SPH N-terminal peptide binds back to the activation pocket via the critical Val 495 residue engaging Asp 672 (colored in black). The pseudo-active site residues Gln 534, Asp 578, and Tyr 673 together with a loop region (Val 692–Gly 696) (colored yellow) of the SPH domain make up the binding interface on HGF which engages the bottom face of blades 2 and 3 on the MET sema domain. (F) HGFSPH-induced dimer of the MET ECD seen in the MET/HGFSPH crystal structure (PDB: 1SHY) is shown (bottom) and cartooned as a schematic model (top).
InlB itself dimerizes via the concave face of its LRR domain and mutation of the LRR dimer interface prevents InlB-induced MET activation (27, 67, 69) (Figure 3C). In MET/InlB dimer, the two MET ECDs also engage via their IPT2 domains revealing a potential dimerization mode for MET in the active dimer (Figure 3C). While the role of the IPT2 interface for InlB or HGF-mediated MET activation has not been characterized, antibodies or engineered protein inhibitors that bind the PSI-IPT1 and IPT1–2 region of the MET stalk abrogate HGF-induced MET activation (57, 70). Thus, the PSI-IPT1–2 regions are likely important for the formation of the ECD domain dimer. High resolution structure of the full-length ECD of MET is needed to unveil the architecture of the functionally relevant active MET ECD dimer.
HGF structure and processing
HGF is a complex growth factor, which is structurally related to the serine protease plasminogen. HGF undergoes analogous processing to plasminogen from its inactive zymogen pro-form to a proteolytically cleaved (between Val 494 – Arg 495) form, which constitutes a biologically active, disulfide-linked α- and β-chain heterodimer (71, 72) (Figure 1C). The α-chain is characterized by an N-terminal (N) finger or PAN domain and four kringle domains (K1–4), and the β-chain encodes a serine protease homology (SPH) domain (Figure 1C). HGF is catalytically inactive as a protease due to mutations in the canonical catalytic triad of the SPH domain (73).
The α- and β-chain of HGF contain independent binding sites for MET, and both binding events are required to activate MET. The α-chain binds MET with high affinity and can engage the receptor in both the un-cleaved and cleaved form of the ligand (71, 72). Conversely, cleavage of HGF is required for the low affinity site located in the β-chain of HGF to bind MET and for receptor activation (71, 72, 74, 75). Consequently, the isolated α- and β-chains of HGF cannot independently activate MET, however, they have can activate MET when combined together from isolated preparations (76). While a crystal structure of the SPH domain bound to the MET sema domain was solved (52), there are no high-resolution structures of MET in complex with the full-length HGF α-chain alone or the NK1 and NK2 variants of HGF that correspond to two different fragments of the α-chain. Hence, major questions remain about collective contributions of the α- and β-chains in full-length HGF to MET activation. Below we discuss key findings that emerged from the structural studies of the individual α- and β-chains of HGF about their binding to MET.
Interactions of the α- and β-chains of HGF with MET
Numerous studies support a model in which the HGF α-chain binds to β-chain fragment of the MET sema domain (blades 5–7). Deletion of the sema domain abolishes interaction of the HGF α-chain with MET and the NK1 ligand selectively cross-links with the β-chain of MET (7, 61, 77). Furthermore, InlB protein and the therapeutic antibody onartuzumab, which bind to the β-chain fragment of MET sema domain, compete with HGF binding (27, 78). Lastly, mutations detected in patients that occur in the α-helix of the extrusion region (V370D and N375L) in the β-chain of MET decrease HGF affinity for MET (79, 80), further substantiating that this region of the sema domain contains the binding site for HGF α-chain. Intriguingly, another primary binding site for HGF α-chain has been proposed by Michieli, Comoglio and colleagues in the IPT3–4 domains of the MET stalk. Monoclonal antibodies targeting this region of MET do indeed reduce HGF-driven signaling (70, 81). Thus, further investigation is required to definitively resolve the α-chain binding site on MET.
The pseudo-plasminogen protease SPH domain of the HGF β-chain is the only fragment of HGF resolved in crystal structures in complex with MET (Figure 3D & E). Like in other plasminogen proteases, the HGF SPH domain is generated via the cleavage that liberates an amino-terminal polypeptide, which then binds back to the SPH domain and allosterically stabilizes the “active site” pocket (52) (Figure 3E). The cleavage results in an approximate 14-fold increase in affinity between the HGF β-chain and MET (75). Short peptides designed to mimic the binding mode of the N-terminal residue of cleaved HGF to the SPH pocket increase HGF activity in the un-cleaved form, underscoring that stabilization of the SPH domain is critical for HGF function as a MET ligand (82). However, the mechanism by which binding of the β-chain to MET sema domain exerts its activating effect on the receptor is unclear. Crystal contacts observed between adjacent SPH domains in the structure of the SPH/MET complex have been proposed as a potential HGF dimerization mechanism (Figure 3F). This observation still awaits experimental validation and as discussed below, it is the α-chain of HGF that likely directly supports MET receptor dimerization.
Insights into HGF α-chain binding and autoinhibition from studies of its fragments: NK1 and NK2
Our understanding of how the HGF α-chain binds to MET and contributes to its dimerization has been predominantly inferred from detailed biochemical and structural analysis of the NK1 and NK2 ligands alone. These two ligands have opposing effects on MET activation. NK1 contains the N domain and the first kringle domain (K1) and is a MET agonist; NK2 contains the N domain and the first two kringle domains (K1 and K2) and is a MET antagonist (83–85) (Figure 1C). Despite the potential mechanistic differences between NK1 and HGF in MET activation discussed below, studies of NK1 and NK2 have provided important insights into how these fragments might behave in the context of the full-length ligand.
The NK1 fragment of the HGF α-chain contains the high affinity binding site for the MET ECD (Kd ~150 nM) (77). Both N and K domains alone can also bind MET, albeit with low affinity (Kd ~1–3 μM) (77). NK1 function as a ligand is dependent on heparan sulfate binding and the N domain contains a high affinity binding site for heparin or heparan sulfate (86–88) (Figure 4A). Heparan sulfate induces NK1 dimerization in solution and increases its affinity for MET (Kd ~3 nM), which is equivalent to the biological activity of the mature HGF molecule in cells (83–85). A direct MET binding site on N domain is not characterized but we know that K1 domain binds to MET via an interface that is commonly used by kringle domains to bind lysine residues (lysine binding site, LBS) (21, 62, 89). The non-canonical LBS of the HGF K1 domain has diverged from other kringle domains and is predicted not to bind lysine side chains (90, 91). While the LBS site is critical for NK1 activity, its precise role in full-length HGF is still contentious. Mutation of LBS site on the K1 domain of HGF abrogates binding to MET and downstream signaling (21, 62) and small molecule binders of the K1 LBS pocket reduce HGF activation of MET (91). However, a recent study demonstrated that while mutation of the K1 binding site residues in full-length HGF decreased binding of the un-cleaved ligand, in its cleaved (mature) form these mutants could still bind the MET ECD and were equally potent as the wild type, mature HGF in activating MET signaling (92).
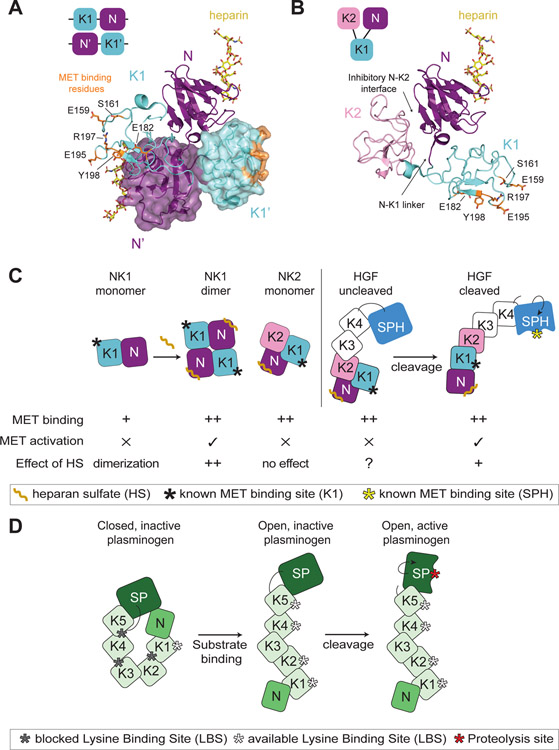
Figure 4: Insights into HGF autoinhibition from the structures of NK1 and NK2 ligands.
(A) Domain organization of the head-to-tail dimer of NK1 is shown schematically in the top left corner. Crystal structure of the NK1 dimer is shown in cartoon representation, with the second monomer shown in a semi-transparent surface mode (PDB: 1GMO). Heparan sulfate is shown in yellow bound to each N domain. Residues important for MET binding in the K1 domain are shown in orange in stick representation. The linker region between the N and K1 domains as well as reciprocal interfaces of the N and K1’ and K1 and N’ domains are shown to contribute to the dimer interface. (B) Domain organization of the autoinhibited NK2 monomer is shown schematically in the top left corner. The NK2 crystal structure is shown in cartoon representation (PDB: 3SP8). The heparan sulfate is modelled in via the alignment of the N domain of the NK2 ligand on the N domain of the NK1 ligand (PDB: 1GMO). The obscured N-K1 linker is indicated by an arrow. (C) Cartoon schematic of NK1, NK2 and HGF (cleaved and uncleaved). The relative binding affinity of these different ligand states for MET is indicated by ‘+’ (low affinity) or ‘++’ (high affinity). The ability of these ligand states to activate MET signaling is denoted by ‘✕’ or ‘✓’. Heparan sulfate (HS) binding to the N domain has different effects on these ligands. NK1: increased binding affinity for MET and ligand dimerization (++), NK2: no effect, HGF-uncleaved: no defined contribution (?), HGF-cleaved: HS promotes HGF dimerization (+) but is not required for signaling. The binding site for MET in the K1 domain and the cleaved SPH domain is indicated by a black and yellow star, respectively. (D) Cartoon schematic showing domain architecture and activation of plasminogen. The inactive molecule is maintained via multiple autoinhibitory intramolecular interactions. Substrate binding by the exposed LBS of K1 domain leads to elongation and accessibility to the additional LBS sites and of the cleavage motif. Maturation of plasminogen by cleavage activates the serine protease domain (SP).
Structural analysis of isolated NK1 and NK2 ligands have elucidated the mechanisms by which they act as an agonist and an antagonist of MET, respectively. Crystals structures of the NK1 ligand consistently show a head to tail dimer, with the dimerization interface formed via the linker region between the N and K1 domains as well as reciprocal binding between the N and K1 domains of opposing ligand monomers (88, 90, 91, 93–95) (Figure 4A). In this orientation, the two LBS motifs in the K1 domains are exposed on opposite sides of the NK1 dimer, providing a hypothesis as to how NK1 brings together two MET receptor molecules for activation (Figure 4A). In NK2, the additional kringle domain (K2) forms interdomain contacts with the N domain, obscuring the dimerization interface thus rendering NK2 monomeric and a MET antagonist (85) (Figure 4B). Mutation of the inhibitory interface between the N and K2 domains turns NK2 into a MET agonist (85). Similar to NK2, the full-length α-chain of HGF (N-K1-K2-K3-K4) is a MET antagonist (96–98), suggesting that it preserves the intermolecular inhibition enforced by the K2 domain. This mechanism to our knowledge has not been investigated in the full-length ligand.
Additional insights into intramolecular inhibition within HGF come from its comparison to plasminogen. In addition to its active SPH domain (β-chain), plasminogen also contains an N domain and five kringle domains (α-chain) (Figure 4D). In crystal structures of its full-length un-cleaved form, plasminogen is in an autoinhibited conformation stabilized in part through intramolecular interactions between lysine residues in the N domain with the LBS region spanning the K4 and K5 domains (99, 100). In this conformation the cleavage site in inaccessible. Binding of plasminogen kringle domains to lysine residues on fibrin is thought to open the molecule and expose the cleavage motif (101) (Figure 4D). While interdomain interactions in plasminogen are not equivalent to those observed in the autoinhibited NK2 structure (23), it is tempting to speculate that HGF binding to MET via its K1 domain serves to initiate a series of similar interdomain rearrangements. Those would lead in turn to accessibility of the HGF cleavage site prefacing HGF cleavage by its binding to MET. Indeed, analysis of the full HGF molecule by low resolution cryo-EM, SAXS and high-speed atomic force microscopy points to a more elongated (open) architecture of cleaved HGF compared to its un-cleaved form (21, 102). The cleaved HGF is also more dynamic, which seems to be essential for its activity as a macrocyclic peptide or a monoclonal antibody that bind to surfaces exposed on the cleaved HGF ligand (between K4-SPH) inhibit HGF by impairing this dynamic motion (102, 103).
Understanding the role of heparan sulfate in MET signaling
Heparan sulfate is an ancillary component of the active HGF/MET complex but the precise role it plays in ligand-induced activation of MET is not entirely clear. Apparent differences between the roles of heparan sulfate in HGF- versus NK1-mediated activation of MET exist. While not strictly required for HGF signaling, heparan sulfate potentiates HGF activation of MET in cells and promotes dimerization of full-length HGF (7, 24, 25). The shorter NK1 ligand is strictly dependent on heparan sulfate for dimerization and activation of the receptor (83, 84) (Figure 4C). Heparan sulfate also enhances the affinity of NK1 for MET, a function which is independent of its ability to dimerize the ligand (95) (Figure 4C). Surprisingly, the mechanisms by which heparan sulfate facilitates ligand dimerization (HGF and NK1) or enhances affinity (NK1) are not known. Crystal structures of apo NK1 and of NK1 bound to heparan sulfate contain identical NK1 dimer forms, and thus did not provide mechanistic insight into how heparan sulfate alters the ligand (88).
Similarly to full-length HGF, the NK2 variant binds MET with high affinity independently of heparan sulfate (85) (Figure 4C). This suggests that the K2 domain might stabilize the MET binding interface in the NK2 and full-length HGF, and in the NK1 variant, which misses the K2 domain, heparan sulfate plays this stabilizing role. It is likely that the answer to these mechanistic distinctions will only be provided by the structural characterization of the HGF α-chain bound to MET. Collectively studies of NK1, NK2 and HGF suggest that HGF engages MET via multiple interfaces and that receptor activation likely follows a stepwise model in which HGF cleavage, ligand binding to heparan sulfate, MET binding by the HGF α-chain and β-chain, and receptor dimerization are all in concert to activate the receptor.
Activation and regulation of MET at the membrane and inside the cell
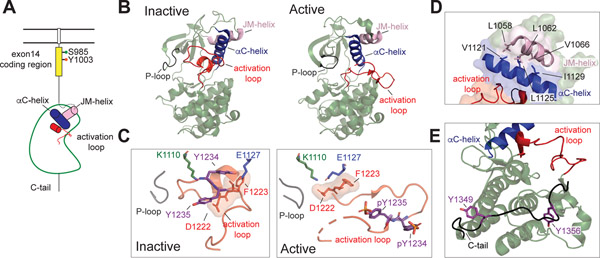
In contrast to substantial structural characterization of the MET ECD and HGF fragments, the intracellular portion of MET remains largely uncharacterized except for the kinase domain. More than seventy deposited crystal structures of the MET kinase domain have revealed that MET contains a canonical kinase domain (Figure 5A & B) (104). A majority of MET kinase structures include an additional N-terminal α-helix (JM helix, Asn 1058–Val 1070) that packs on top of the kinase N-lobe and is likely required for stability of the kinase fold (Figure 5B & D). In the inactive state, MET kinase adopts a Src/CDK-like inactive conformation in which the activation loop forms a partial helix and packs against the catalytic cleft, displacing helix αC away from the active site and preventing the helix αC Glu 1127 from engaging the catalytic lysine (Lys 1110) (Figure 5B). The activation loop is tethered to the kinase through multiple interactions, including Tyr 1234 engaging the catalytic Glu 1127, Tyr 1230 and Lys 1232 binding the P-loop, and the active site is partially blocked by Met 1229 (105). Activation of the MET kinase domain requires sequential phosphorylation of tandem tyrosine residues in the activation loop (Tyr 1235 and Tyr 1234). Tyr 1235 is first phosphorylated due to greater solvent exposure, thereby promoting destabilization of activation loop interactions, followed by phosphorylation of Tyr 1234, which together stabilize the active state of the kinase (105, 106) (Figure 5C). Consequently, mutation of Tyr 1234 to a phenylalanine results in reduced kinase activity through stabilization of the autoinhibited state (106, 107).
Figure 5: MET kinase domain states.
(A) Cartoon representation of the MET receptor intracellular domains. The region encoded by exon 14 is colored yellow. Two residues (Ser 985, Tyr 1003) that are important for MET regulation upon being phosphorylated are marked. The αC helix and JM helix in the kinase domain are indicated in blue and pink, respectively. The activation loop is shown in red. (B) Crystal structures of an inactive (PDB: 2G15) and active (PDB: 3R7O) MET kinase domain are shown in cartoon representation. (C, left panel) The activation loop of the inactive MET structure forms a short helix that obscures the αC-helix from swinging into the active site and from forming the salt bridge required for catalysis (Lys 1110 and Glu 1127). In this Src/CDK-like inactive conformation, the DFG motif is in the DFG-in conformation, as indicated by the Asp 1222 pointing towards the active site. The JM-helix is shown in light pink. (C, right panel) In the active conformation, the activation loop tyrosines (Tyr 1234 and Tyr 1235) are phosphorylated (denoted as ‘pY’), releasing the activation loop from the active site. (D) Zoomed-in view of the hydrophobic residues that form the interface between the JM helix and the αC helix in the MET kinase domain (PDB: 2G15), including Leu 1058, Leu 1062, Val 1066 on the JM-helix and Val 1121, Leu 1125, Iso 1129 on the αC-helix. (E) The kinase proximal region of the C-terminal tail (Gly 1346-Lys 1360) is shown in black bound to the back of the kinase C-lobe (PDB: 1R0P). Tyr 1349 and Tyr 1356 are shown in purple.
MET cancer mutations, described in papillary renal cell carcinoma (PRCC) and other solid cancers including hepatocellular carcinomas and advanced head and neck squamous cell carcinoma (HNSCC) (12, 108, 109), act predominantly through destabilization of the autoinhibited kinase structure (104–107, 110). For example, mutation of the activation loop Tyr 1235 to a negatively charged aspartate, found in PRCC, HNSCC and lung adenocarcinoma, destabilizes the autoinhibited conformation of the activation loop. Likewise, Asp 1228 mutations (His/Val/Asn), detected in PRCC, break an active site salt bridge with Lys 1240. Mutations of Tyr 1230 (His/Asp/Cys) that also occur in PRCC, interrupt interactions observed between the hydrophobic P-loop and the activation loop needed to stabilize ATP binding (104–107, 110, 111).
Much less is known about the structure of other intracellular domains of MET. The juxtamembrane (JM) region in MET is substantially longer (107 residues) in comparison to most other RTKs and is predicted to be largely unstructured. It has two important regulatory motifs located within the membrane proximal region (defined here as amino acids Asp 963-Asp 1010, encoded by exon 14) (Figure 5A). One of them is centered around Tyr 1003, which when phosphorylated recruits the E3 ligase Cbl leading to MET endocytosis and lysosomal degradation (112, 113). The second site involves Ser 985, which is phosphorylated by PKC and also results in enhanced MET internalization and downregulation (114–116). The JM segment encoded by human exon 14 is skipped in approximately 4% of non-small-cell lung cancer patients due to mutations in the intron/exon boundary of exon 14 (117–119). One of the functional consequences of exon 14 skipping is loss of Cbl-mediated ubiquitination and receptor internalization, leading to prolonged HGF-induced signaling (120, 121). Single point mutation of Tyr 1003 to Phe is also transformative in cell-based studies (112, 119). However, there is evidence for other regulatory elements in the exon 14 besides Cbl binding. Addition of the exon 14 coding sequence to the cytoplasmic TPR-MET, in which exon 14 coding region is lost from the MET ICD during the gene fusion event, reduces transforming capacity even when the Cbl-binding site is mutated (121–123). This suggests that the inhibitory role of the membrane proximal half of the JM domain might extend beyond the control of receptor ubiquitination-dependent downregulation and could potentially involve direct interactions with the kinase domain.
On the opposite site of the kinase domain, the C-terminal tail has been directly implicated in regulation of MET kinase activity. Addition of peptides mimicking the C-terminal tail to cells inhibited MET autophosphorylation and MET-dependent signaling (124). Consequently, truncation of the entire tail increased receptor activation (125). A similar observation has been made for the RON receptor, where C-terminal tail truncations also activated the kinase (126). Structurally, the resolved kinase proximal region of the C-terminal tail wraps around the back side of the C-lobe towards the activation loop (104) (Figure 5E). Further extension of the C-terminal tail may partially block the substrate binding region or stabilize the activation loop in an inactive conformation, before Tyr 1349 and Tyr 1356 become phosphorylated and disengage from the kinase C-lobe (Figure 5E). Interestingly, in the case of MET, removal of the most terminal 26 amino acids, which correspond to approximately half of the entire C-terminal tail length, results in decreased receptor activation rather than activation (125). These early studies highlight a complex role that the C-terminal tail plays in regulation of receptor activity, where in addition to providing the docking sites for downstream effectors, the tail may also play direct positive and negative regulatory roles in regulating kinase activity.
The transmembrane domains (TM) of the receptor might also play an important role since it contains a GxxxG motif (Gly 933-Leu-Iso-Ala-Gly 937) that facilitates homo-dimerization between membrane-embedded helices (127–129). Peptides that correspond to the MET TM can form homodimers in an E. coli-based membrane dimerization assay (130). How all the intracellular domains come together upon ligand binding to form an active MET receptor complex is largely unknown.
Conclusions
Given the complexity of the HGF and MET structures and their cleavage-dependent rearrangements, it is not surprising that studies of individual domains of these proteins have not yet revealed all the features of receptor activation. While we know that HGF needs to be cleaved to promote formation of the active MET receptor complex, we do not know all its binding interfaces on MET and how it cooperates with heparin sulfate to promote formation of active MET dimers. The structure of the full-length HGF that encompasses both α- and β-chains is still missing, and the structure of full-length MET ECD and the importance of its cleavage remains unknown. Consequently, HGF-dependent structural rearrangements within the ECD remain uncharacterized. Lastly, we have no knowledge on how the conformational transitions in the MET ECD translate to the intracellular juxtamembrane and kinase domains. The apparent promiscuity of MET in forming co-complexes with other ligands and membrane receptors adds another dimension of mystery to understanding how MET cellular signaling is regulated at the level of receptor activation. While revealing any of these long-standing MET secrets will not be trivial, it is highly anticipated and has potential to inform new and more effective strategies for disrupting misregulated MET signaling in disease.
Perspectives.
The MET receptor tyrosine kinase and its cognate ligand HGF comprise a signaling axis essential for development, wound healing and tissue homeostasis. Aberrant HGF/MET signaling has been a target of therapies in a number of human cancers.
Understanding the mechanisms of HGF-mediated MET activation and signaling is dependent on the characterization of its basal and activated states. Studies on the fragments of HGF and MET proteins support a model in which HGF is activated by cleavage to form a dimer, which then engages two MET monomers to form an active complex. However, currently there are no high-resolution structures of the active HGF/MET dimer complex that directly visualize these ligand/receptor interactions.
Defining the ligand binding site of the HGF α-chain on MET, along with complete structures of the inactive and active, full-length HGF ligand is needed to fully understand how basal activity of HGF and its binding to MET is regulated.
Acknowledgements
We thank Raphael Trenker and Devan Diwanji of the Jura lab for ongoing and insightful discussions regarding regulation of MET and HGF. The authors sincerely apologize to all colleagues whose work was omitted in this review owing to space constraints. This work was supported in part by grants from the National Cancer Institute to N.J. and E.A.C (R01 CA230263) and E.A.C (R01 CA239604, R01 CA222862, R01 CA227807, U24 CA210974).
Footnotes
Declaration of interests
E.A. Collisson is consultant at Takeda, Merck, Loxo and Pear Diagnostics, reports receiving commercial research grants from Astra Zeneca, Ferro Therapeutics, Senti Biosciences, Merck KgA and Bayer and stock ownership of Tatara Therapeutics, Clara Health, BloodQ, Guardant Health. All other authors declare no competing interests.
References
- 1.Cooper CS, Park M, Blair DG, Tainsky MA, Huebner K, Croce CM, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311(5981):29–33. [DOI] [PubMed] [Google Scholar]
- 2.Park M, Dean M, Kaul K, Braun MJ, Gonda MA, Vande Woude G. Sequence of MET protooncogene cDNA has features characteristic of the tyrosine kinase family of growth-factor receptors. Proc Natl Acad Sci U S A. 1987;84(18):6379–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. 1991;251(4995):802–4. [DOI] [PubMed] [Google Scholar]
- 4.Ronsin C, Muscatelli F, Mattei MG, Breathnach R. A novel putative receptor protein tyrosine kinase of the met family. Oncogene. 1993;8(5):1195–202. [PubMed] [Google Scholar]
- 5.Lemmon MA, Schlessinger J. Cell Signaling by Receptor Tyrosine Kinases. Cell. 2010;141(7):1117–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bork P, Doerks T, Springer TA, Snel B. Domains in plexins: links to integrins and transcription factors. Trends Biochem Sci. 1999;24(7):261–3. [DOI] [PubMed] [Google Scholar]
- 7.Gherardi E, Youles ME, Miguel RN, Blundell TL, Iamele L, Gough J, et al. Functional map and domain structure of MET, the product of the c-met protooncogene and receptor for hepatocyte growth factor/scatter factor. Proc Natl Acad Sci U S A. 2003;100(21):12039–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nature Publishing Group. 2010;11(12):834–48. [DOI] [PubMed] [Google Scholar]
- 9.Petrini I Biology of MET: a double life between normal tissue repair and tumor progression. Ann Transl Med. 2015;3(6):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato T Biological roles of hepatocyte growth factor-Met signaling from genetically modified animals (Review). Biomedical reports. 2017:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Silva DM, Roy A, Kato T, Cecchi F, Lee YH, Matsumoto K, et al. Targeting the hepatocyte growth factor/Met pathway in cancer. Biochemical Society Transactions. 2017;45(4):855–70. [DOI] [PubMed] [Google Scholar]
- 12.Comoglio PM, Trusolino L, Boccaccio C. Known and novel roles of the MET oncogene in cancer: a coherent approach to targeted therapy. Nature Publishing Group. 2018;18(6):341–58. [DOI] [PubMed] [Google Scholar]
- 13.Koch JP, Aebersold DM, Zimmer Y, Medová M. MET targeting: time for a rematch. Oncogene. 2020:1–18. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura T, Nawa K, Ichihara A, Kaise N, Nishino T. Purification and subunit structure of hepatocyte growth factor from rat platelets. FEBS Lett. 1987;224(2):311–6. [DOI] [PubMed] [Google Scholar]
- 15.Stoker M, Gherardi E, Perryman M, Gray J. Scatter factor is a fibroblast-derived modulator of epithelial cell mobility. Nature. 1987;327(6119):239–42. [DOI] [PubMed] [Google Scholar]
- 16.Gherardi E, Stoker M. Hepatocytes and scatter factor. Nature. 1990;346(6281):228. [DOI] [PubMed] [Google Scholar]
- 17.Weidner KM, Arakaki N, Hartmann G, Vandekerckhove J, Weingart S, Rieder H, et al. Evidence for the identity of human scatter factor and human hepatocyte growth factor. Proc Natl Acad Sci U S A. 1991;88(16):7001–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura T, Nishizawa T, Hagiya M, Seki T, Shimonishi M, Sugimura A, et al. Molecular cloning and expression of human hepatocyte growth factor. Nature. 1989;342(6248):440–3. [DOI] [PubMed] [Google Scholar]
- 19.Cioce V, Csaky KG, Chan AM, Bottaro DP, Taylor WG, Jensen R, et al. Hepatocyte growth factor (HGF)/NK1 is a naturally occurring HGF/scatter factor variant with partial agonist/antagonist activity. J Biol Chem. 1996;271(22):13110–5. [DOI] [PubMed] [Google Scholar]
- 20.Chan AM, Rubin JS, Bottaro DP, Hirschfield DW, Chedid M, Aaronson SA. Identification of a competitive HGF antagonist encoded by an alternative transcript. Science. 1991;254(5036):1382–5. [DOI] [PubMed] [Google Scholar]
- 21.Gherardi E, Sandin S, Petoukhov MV, Finch J, Youles ME, Ofverstedt LG, et al. Structural basis of hepatocyte growth factor/scatter factor and MET signalling. Proc Natl Acad Sci U S A. 2006;103(11):4046–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12(2):89–103. [DOI] [PubMed] [Google Scholar]
- 23.Niemann HH. Structural basis of MET receptor dimerization by the bacterial invasion protein InlB and the HGF/SF splice variant NK1. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2013;1834(10):2195–204. [DOI] [PubMed] [Google Scholar]
- 24.Zioncheck TF, Richardson L, Liu J, Chang L, King KL, Bennett GL, et al. Sulfated oligosaccharides promote hepatocyte growth factor association and govern its mitogenic activity. J Biol Chem. 1995;270(28):16871–8. [DOI] [PubMed] [Google Scholar]
- 25.Kemp LE, Mulloy B, Gherardi E. Signalling by HGF/SF and Met: the role of heparan sulphate co-receptors. Biochem Soc Trans. 2006;34(Pt 3):414–7. [DOI] [PubMed] [Google Scholar]
- 26.Shen Y, Naujokas M, Park M, Ireton K. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell. 2000;103(3):501–10. [DOI] [PubMed] [Google Scholar]
- 27.Niemann HH, Jager V, Butler PJ, van den Heuvel J, Schmidt S, Ferraris D, et al. Structure of the human receptor tyrosine kinase met in complex with the Listeria invasion protein InlB. Cell. 2007;130(2):235–46. [DOI] [PubMed] [Google Scholar]
- 28.Radoshevich L, Cossart P. Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat Rev Microbiol. 2018;16(1):32–46. [DOI] [PubMed] [Google Scholar]
- 29.Goldoni S, Humphries A, Nyström A, Sattar S, Owens RT, McQuillan DJ, et al. Decorin is a novel antagonistic ligand of the Met receptor. The Journal of cell biology. 2009;185(4):743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trusolino L, Bertotti A, Comoglio PM. A signaling adapter function for alpha6beta4 integrin in the control of HGF-dependent invasive growth. Cell. 2001;107(5):643–54. [DOI] [PubMed] [Google Scholar]
- 31.Orian-Rousseau V, Chen L, Sleeman JP, Herrlich P, Ponta H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes & development. 2002;16(23):3074–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu KV, Chang JP, Parachoniak CA, Pandika MM, Aghi MK, Meyronet D, et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell. 2012;22(1):21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gentile A, Lazzari L, Benvenuti S, Trusolino L, Comoglio PM. The ROR1 pseudokinase diversifies signaling outputs in MET-addicted cancer cells. Int J Cancer. 2014;135(10):2305–16. [DOI] [PubMed] [Google Scholar]
- 34.Frazier NM, Brand T, Gordan JD, Grandis J, Jura N. Overexpression-mediated activation of MET in the Golgi promotes HER3/ERBB3 phosphorylation. Oncogene. 2019;38(11):1936–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong LR, Mohamed Salleh NAB, Ong RW, Tan TZ, Syn NL, Goh RM, et al. A common MET polymorphism harnesses HER2 signaling to drive aggressive squamous cell carcinoma. Nat Commun. 2020;11(1):1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W, Xiong X, Abdalla A, Alejo S, Zhu L, Lu F, et al. HGF-induced formation of the MET-AXL-ELMO2-DOCK180 complex promotes RAC1 activation, receptor clustering, and cancer cell migration and invasion. J Biol Chem. 2018;293(40):15397–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benvenuti S, Lazzari L, Arnesano A, Li Chiavi G, Gentile A, Comoglio PM. Ron kinase transphosphorylation sustains MET oncogene addiction. Cancer Res. 2011;71(5):1945–55. [DOI] [PubMed] [Google Scholar]
- 38.Giordano S, Corso S, Conrotto P, Artigiani S, Gilestro G, Barberis D, et al. The semaphorin 4D receptor controls invasive growth by coupling with Met. Nat Cell Biol. 2002;4(9):720–4. [DOI] [PubMed] [Google Scholar]
- 39.Lai AZ, Abella JV, Park M. Crosstalk in Met receptor oncogenesis. Trends in Cell Biology. 2009;19(10):542–51. [DOI] [PubMed] [Google Scholar]
- 40.Viticchie G, Muller PAJ. c-Met and Other Cell Surface Molecules: Interaction, Activation and Functional Consequences. Biomedicines. 2015;3(1):46–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013;3(6):658–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diwanji D, Thaker T, Jura N. More than the sum of the parts: Toward full-length receptor tyrosine kinase structures. IUBMB Life. 2019;71(6):706–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gherardi E, Love CA, Esnouf RM, Jones EY. The sema domain. Curr Opin Struct Biol. 2004;14(6):669–78. [DOI] [PubMed] [Google Scholar]
- 44.Pascoe HG, Wang Y, Zhang X. Structural mechanisms of plexin signaling. Prog Biophys Mol Biol. 2015;118(3):161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alves JM, Martins AH, Lameu C, Glaser T, Boukli NM, Bassaneze V, et al. Kinin-B2 Receptor Activity in Skeletal Muscle Regeneration and Myoblast Differentiation. Stem Cell Rev Rep. 2019;15(1):48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siebold C, Jones EY. Structural insights into semaphorins and their receptors. Semin Cell Dev Biol. 2013;24(3):139–45. [DOI] [PubMed] [Google Scholar]
- 47.Love CA, Harlos K, Mavaddat N, Davis SJ, Stuart DI, Jones EY, et al. The ligand-binding face of the semaphorins revealed by the high-resolution crystal structure of SEMA4D. Nat Struct Biol. 2003;10(10):843–8. [DOI] [PubMed] [Google Scholar]
- 48.Antipenko A, Himanen JP, van Leyen K, Nardi-Dei V, Lesniak J, Barton WA, et al. Structure of the semaphorin-3A receptor binding module. Neuron. 2003;39(4):589–98. [DOI] [PubMed] [Google Scholar]
- 49.Janssen BJ, Robinson RA, Perez-Branguli F, Bell CH, Mitchell KJ, Siebold C, et al. Structural basis of semaphorin-plexin signalling. Nature. 2010;467(7319):1118–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H, Juo ZS, Shim AH, Focia PJ, Chen X, Garcia KC, et al. Structural basis of semaphorin-plexin recognition and viral mimicry from Sema7A and A39R complexes with PlexinC1. Cell. 2010;142(5):749–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nogi T, Yasui N, Mihara E, Matsunaga Y, Noda M, Yamashita N, et al. Structural basis for semaphorin signalling through the plexin receptor. Nature. 2010;467(7319):1123–7. [DOI] [PubMed] [Google Scholar]
- 52.Stamos J, Lazarus RA, Yao X, Kirchhofer D, Wiesmann C. Crystal structure of the HGF beta-chain in complex with the Sema domain of the Met receptor. The EMBO journal. 2004;23(12):2325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Komada M, Hatsuzawa K, Shibamoto S, Ito F, Nakayama K, Kitamura N. Proteolytic processing of the hepatocyte growth factor/scatter factor receptor by furin. FEBS Lett. 1993;328(1–2):25–9. [DOI] [PubMed] [Google Scholar]
- 54.Mark MR, Lokker NA, Zioncheck TF, Luis EA, Godowski PJ. Expression and characterization of hepatocyte growth factor receptor-IgG fusion proteins. Effects of mutations in the potential proteolytic cleavage site on processing and ligand binding. J Biol Chem. 1992;267(36):26166–71. [PubMed] [Google Scholar]
- 55.Niemann HH. Structural insights into Met receptor activation. Eur J Cell Biol. 2011;90(11):972–81. [DOI] [PubMed] [Google Scholar]
- 56.Chao KL, Tsai IW, Chen C, Herzberg O. Crystal structure of the Sema-PSI extracellular domain of human RON receptor tyrosine kinase. PLoS One. 2012;7(7):e41912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DiCara DM, Chirgadze DY, Pope AR, Karatt-Vellatt A, Winter A, Slavny P, et al. Characterization and structural determination of a new anti-MET function-blocking antibody with binding epitope distinct from the ligand binding domain. Scientific reports. 2017;7(1):9000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Niemann HH, Petoukhov MV, Hartlein M, Moulin M, Gherardi E, Timmins P, et al. X-ray and neutron small-angle scattering analysis of the complex formed by the Met receptor and the Listeria monocytogenes invasion protein InlB. J Mol Biol. 2008;377(2):489–500. [DOI] [PubMed] [Google Scholar]
- 59.Kong Y, Janssen BJ, Malinauskas T, Vangoor VR, Coles CH, Kaufmann R, et al. Structural Basis for Plexin Activation and Regulation. Neuron. 2016;91(3):548–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suzuki K, Tsunoda H, Omiya R, Matoba K, Baba T, Suzuki S, et al. Structure of the Plexin Ectodomain Bound by Semaphorin-Mimicking Antibodies. PLoS One. 2016;11(6):e0156719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kong-Beltran M, Stamos J, Wickramasinghe D. The Sema domain of Met is necessary for receptor dimerization and activation. Cancer cell. 2004;6(1):75–84. [DOI] [PubMed] [Google Scholar]
- 62.Youles M, Holmes O, Petoukhov MV, Nessen MA, Stivala S, Svergun DI, et al. Engineering the NK1 fragment of hepatocyte growth factor/scatter factor as a MET receptor antagonist. J Mol Biol. 2008;377(3):616–22. [DOI] [PubMed] [Google Scholar]
- 63.Koschut D, Richert L, Pace G, Niemann HH, Mély Y, Orian-Rousseau V. Live cell imaging shows hepatocyte growth factor-induced Met dimerization. BBA - Molecular Cell Research. 2016;1863(Part A):1552–8. [DOI] [PubMed] [Google Scholar]
- 64.Lemmon MA, Pinchasi D, Zhou M, Lax I, Schlessinger J. Kit receptor dimerization is driven by bivalent binding of stem cell factor. J Biol Chem. 1997;272(10):6311–7. [DOI] [PubMed] [Google Scholar]
- 65.Reshetnyak AV, Opatowsky Y, Boggon TJ, Folta-Stogniew E, Tome F, Lax I, et al. The strength and cooperativity of KIT ectodomain contacts determine normal ligand-dependent stimulation or oncogenic activation in cancer. Mol Cell. 2015;57(1):191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dawson JP, Bu Z, Lemmon MA. Ligand-induced structural transitions in ErbB receptor extracellular domains. Structure. 2007;15(8):942–54. [DOI] [PubMed] [Google Scholar]
- 67.Andres F, Iamele L, Meyer T, Stüber JC, Kast F, Gherardi E, et al. Inhibition of the MET Kinase Activity and Cell Growth in MET-Addicted Cancer Cells by Bi-Paratopic Linking. Journal of molecular biology. 2019;431(10):2020–39. [DOI] [PubMed] [Google Scholar]
- 68.Banerjee M, Copp J, Vuga D, Marino M, Chapman T, van der Geer P, et al. GW domains of the Listeria monocytogenes invasion protein InlB are required for potentiation of Met activation. Mol Microbiol. 2004;52(1):257–71. [DOI] [PubMed] [Google Scholar]
- 69.Ferraris DM, Gherardi E, Di Y, Heinz DW, Niemann HH. Ligand-mediated dimerization of the Met receptor tyrosine kinase by the bacterial invasion protein InlB. J Mol Biol. 2010;395(3):522–32. [DOI] [PubMed] [Google Scholar]
- 70.Basilico C, Hultberg A, Blanchetot C, de Jonge N, Festjens E, Hanssens V, et al. Four individually druggable MET hotspots mediate HGF-driven tumor progression. J Clin Invest. 2014;124(7):3172–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hartmann G, Naldini L, Weidner KM, Sachs M, Vigna E, Comoglio PM, et al. A functional domain in the heavy chain of scatter factor/hepatocyte growth factor binds the c-Met receptor and induces cell dissociation but not mitogenesis. Proc Natl Acad Sci U S A. 1992;89(23):11574–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lokker NA, Mark MR, Luis EA, Bennett GL, Robbins KA, Baker JB, et al. Structure-function analysis of hepatocyte growth factor: identification of variants that lack mitogenic activity yet retain high affinity receptor binding. EMBO J. 1992;11(7):2503–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maun HR, Kirchhofer D, Lazarus RA. Pseudo-active sites of protease domains: HGF/Met and Sonic hedgehog signaling in cancer. Biol Chem. 2010;391(8):881–92. [DOI] [PubMed] [Google Scholar]
- 74.Naldini L, Tamagnone L, Vigna E, Sachs M, Hartmann G, Birchmeier W, et al. Extracellular proteolytic cleavage by urokinase is required for activation of hepatocyte growth factor/scatter factor. EMBO J. 1992;11(13):4825–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kirchhofer D, Yao X, Peek M, Eigenbrot C, Lipari MT, Billeci KL, et al. Structural and Functional Basis of the Serine Protease-like Hepatocyte Growth Factor β-Chain in Met Binding and Signaling. The Journal of biological chemistry. 2004;279(38):39915–24. [DOI] [PubMed] [Google Scholar]
- 76.Matsumoto K, Kataoka H, Date K, Nakamura T. Cooperative interaction between alpha- and beta-chains of hepatocyte growth factor on c-Met receptor confers ligand-induced receptor tyrosine phosphorylation and multiple biological responses. J Biol Chem. 1998;273(36):22913–20. [DOI] [PubMed] [Google Scholar]
- 77.Holmes O, Pillozzi S, Deakin JA, Carafoli F, Kemp L, Butler PJG, et al. Insights into the structure/function of hepatocyte growth factor/scatter factor from studies with individual domains. Journal of molecular biology. 2007;367(2):395–408. [DOI] [PubMed] [Google Scholar]
- 78.Merchant M, Ma X, Maun HR, Zheng Z, Peng J, Romero M, et al. Monovalent antibody design and mechanism of action of onartuzumab, a MET antagonist with anti-tumor activity as a therapeutic agent. Proc Natl Acad Sci U S A. 2013;110(32):E2987–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miao W, Sakai K, Sato H, Imamura R, Jangphattananont N, Takagi J, et al. Impaired ligand-dependent MET activation caused by an extracellular SEMA domain missense mutation in lung cancer. Cancer Sci. 2019;110(10):3340–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tode N, Kikuchi T, Sakakibara T, Hirano T, Inoue A, Ohkouchi S, et al. Exome sequencing deciphers a germline MET mutation in familial epidermal growth factor receptor-mutant lung cancer. Cancer Sci. 2017;108(6):1263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Basilico C, Arnesano A, Galluzzo M, Comoglio PM, Michieli P. A high affinity hepatocyte growth factor-binding site in the immunoglobulin-like region of Met. The Journal of biological chemistry. 2008;283(30):21267–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Landgraf KE, Steffek M, Quan C, Tom J, Yu C, Santell L, et al. An allosteric switch for pro-HGF/Met signaling using zymogen activator peptides. Nature Chemical Biology. 2014;10(7):567–73. [DOI] [PubMed] [Google Scholar]
- 83.Schwall RH, Chang LY, Godowski PJ, Kahn DW, Hillan KJ, Bauer KD, et al. Heparin induces dimerization and confers proliferative activity onto the hepatocyte growth factor antagonists NK1 and NK2. J Cell Biol. 1996;133(3):709–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sakata H, Stahl SJ, Taylor WG, Rosenberg JM, Sakaguchi K, Wingfield PT, et al. Heparin binding and oligomerization of hepatocyte growth factor/scatter factor isoforms. Heparan sulfate glycosaminoglycan requirement for Met binding and signaling. J Biol Chem. 1997;272(14):9457–63. [DOI] [PubMed] [Google Scholar]
- 85.Tolbert WD, Daugherty-Holtrop J, Gherardi E, Vande Woude G, Xu HE. Structural basis for agonism and antagonism of hepatocyte growth factor. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(30):13264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sakata H, Stahl SJ, Taylor WG, Rosenberg JM, Sakaguchi K, Wingfield PT, et al. Heparin binding and oligomerization of hepatocyte growth factor/scatter factor isoforms. Heparan sulfate glycosaminoglycan requirement for Met binding and signaling. The Journal of biological chemistry. 1997;272(14):9457–63. [DOI] [PubMed] [Google Scholar]
- 87.Zhou H, Mazzulla MJ, Kaufman JD, Stahl SJ, Wingfield PT, Rubin JS, et al. The solution structure of the N-terminal domain of hepatocyte growth factor reveals a potential heparin-binding site. Structure. 1998;6(1):109–16. [DOI] [PubMed] [Google Scholar]
- 88.Lietha D, Chirgadze DY, Mulloy B, Blundell TL, Gherardi E. Crystal structures of NK1-heparin complexes reveal the basis for NK1 activity and enable engineering of potent agonists of the MET receptor. EMBO J. 2001;20(20):5543–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lokker NA, Presta LG, Godowski PJ. Mutational analysis and molecular modeling of the N-terminal kringle-containing domain of hepatocyte growth factor identifies amino acid side chains important for interaction with the c-Met receptor. Protein Eng. 1994;7(7):895–903. [DOI] [PubMed] [Google Scholar]
- 90.Ultsch M, Lokker NA, Godowski PJ, de Vos AM. Crystal structure of the NK1 fragment of human hepatocyte growth factor at 2.0 A resolution. Structure. 1998;6(11):1383–93. [DOI] [PubMed] [Google Scholar]
- 91.Sigurdardottir AG, Winter A, Sobkowicz A, Fragai M, Chirgadze D, Ascher DB, et al. Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding. Chem Sci. 2015;6(11):6147–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Umitsu M, Sakai K, Tamura-Kawakami K, Matsumoto K, Takagi J. The constitutive high-affinity Met-binding site in the kringle domain is dispensable for the signalling activity of hepatocyte growth factor. J Biochem. 2020;167(6):577–86. [DOI] [PubMed] [Google Scholar]
- 93.Chirgadze DY, Hepple JP, Zhou H, Byrd RA, Blundell TL, Gherardi E. Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding. Nat Struct Biol. 1999;6(1):72–9. [DOI] [PubMed] [Google Scholar]
- 94.Watanabe K, Chirgadze DY, Lietha D, de Jonge H, Blundell TL, Gherardi E. A new crystal form of the NK1 splice variant of HGF/SF demonstrates extensive hinge movement and suggests that the NK1 dimer originates by domain swapping. J Mol Biol. 2002;319(2):283–8. [DOI] [PubMed] [Google Scholar]
- 95.Tolbert WD, Daugherty J, Gao C, Xie Q, Miranti C, Gherardi E, et al. A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist. Proc Natl Acad Sci U S A. 2007;104(37):14592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Date K, Matsumoto K, Shimura H, Tanaka M, Nakamura T. HGF/NK4 is a specific antagonist for pleiotrophic actions of hepatocyte growth factor. FEBS Lett. 1997;420(1):1–6. [DOI] [PubMed] [Google Scholar]
- 97.Date K, Matsumoto K, Kuba K, Shimura H, Tanaka M, Nakamura T. Inhibition of tumor growth and invasion by a four-kringle antagonist (HGF/NK4) for hepatocyte growth factor. Oncogene. 1998;17(23):3045–54. [DOI] [PubMed] [Google Scholar]
- 98.Matsumoto K, Nakamura T. Mechanisms and significance of bifunctional NK4 in cancer treatment. Biochem Biophys Res Commun. 2005;333(2):316–27. [DOI] [PubMed] [Google Scholar]
- 99.Law RH, Caradoc-Davies T, Cowieson N, Horvath AJ, Quek AJ, Encarnacao JA, et al. The X-ray crystal structure of full-length human plasminogen. Cell Rep. 2012;1(3):185–90. [DOI] [PubMed] [Google Scholar]
- 100.Xue Y, Bodin C, Olsson K. Crystal structure of the native plasminogen reveals an activation-resistant compact conformation. J Thromb Haemost. 2012;10(7):1385–96. [DOI] [PubMed] [Google Scholar]
- 101.Wu G, Quek AJ, Caradoc-Davies TT, Ekkel SM, Mazzitelli B, Whisstock JC, et al. Structural studies of plasmin inhibition. Biochem Soc Trans. 2019;47(2):541–57. [DOI] [PubMed] [Google Scholar]
- 102.Sakai K, Passioura T, Sato H, Ito K, Furuhashi H, Umitsu M, et al. Macrocyclic peptide-based inhibition and imaging of hepatocyte growth factor. Nature Chemical Biology. 2019;15(6):598–606. [DOI] [PubMed] [Google Scholar]
- 103.Umitsu M, Sakai K, Ogasawara S, Kaneko MK, Asaki R, Tamura-Kawakami K, et al. Probing conformational and functional states of human hepatocyte growth factor by a panel of monoclonal antibodies. Sci Rep. 2016;6:33149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, et al. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003;100(22):12654–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang W, Marimuthu A, Tsai J, Kumar A, Krupka HI, Zhang C, et al. Structural characterization of autoinhibited c-Met kinase produced by coexpression in bacteria with phosphatase. Proc Natl Acad Sci U S A. 2006;103(10):3563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chiara F, Michieli P, Pugliese L, Comoglio PM. Mutations in the met oncogene unveil a “dual switch” mechanism controlling tyrosine kinase activity. J Biol Chem. 2003;278(31):29352–8. [DOI] [PubMed] [Google Scholar]
- 107.Cristiani C, Rusconi L, Perego R, Schiering N, Kalisz HM, Knapp S, et al. Regulation of the wild-type and Y1235D mutant Met kinase activation. Biochemistry. 2005;44(43):14110–9. [DOI] [PubMed] [Google Scholar]
- 108.Duplaquet L, Kherrouche Z, Baldacci S, Jamme P, Cortot AB, Copin MC, et al. The multiple paths towards MET receptor addiction in cancer. Oncogene. 2018;37(24):3200–15. [DOI] [PubMed] [Google Scholar]
- 109.Recondo G, Che J, Janne PA, Awad MM. Targeting MET Dysregulation in Cancer. Cancer Discov. 2020;10(7):922–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Timofeevski SL, McTigue MA, Ryan K, Cui J, Zou HY, Zhu JX, et al. Enzymatic characterization of c-Met receptor tyrosine kinase oncogenic mutants and kinetic studies with aminopyridine and triazolopyrazine inhibitors. Biochemistry. 2009;48(23):5339–49. [DOI] [PubMed] [Google Scholar]
- 111.Schmidt L, Duh FM, Chen F, Kishida T, Glenn G, Choyke P, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16(1):68–73. [DOI] [PubMed] [Google Scholar]
- 112.Peschard P, Fournier TM, Lamorte L, Naujokas MA, Band H, Langdon WY, et al. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol Cell. 2001;8(5):995–1004. [DOI] [PubMed] [Google Scholar]
- 113.Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S. The endophilin-CIN85-Cbl complex mediates ligand-dependent downregulation of c-Met. Nature. 2002;416(6877):187–90. [DOI] [PubMed] [Google Scholar]
- 114.Kermorgant S, Zicha D, Parker PJ. Protein kinase C controls microtubule-based traffic but not proteasomal degradation of c-Met. J Biol Chem. 2003;278(31):28921–9. [DOI] [PubMed] [Google Scholar]
- 115.Hashigasako A, Machide M, Nakamura T, Matsumoto K, Nakamura T. Bi-directional regulation of Ser-985 phosphorylation of c-met via protein kinase C and protein phosphatase 2A involves c-Met activation and cellular responsiveness to hepatocyte growth factor. J Biol Chem. 2004;279(25):26445–52. [DOI] [PubMed] [Google Scholar]
- 116.Nakayama M, Sakai K, Yamashita A, Nakamura T, Suzuki Y, Matsumoto K. Met/HGF receptor activation is regulated by juxtamembrane Ser985 phosphorylation in hepatocytes. Cytokine. 2013;62(3):446–52. [DOI] [PubMed] [Google Scholar]
- 117.Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5(8):850–9. [DOI] [PubMed] [Google Scholar]
- 118.Paik PK, Drilon A, Fan PD, Yu H, Rekhtman N, Ginsberg MS, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015;5(8):842–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, Mendoza N, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66(1):283–9. [DOI] [PubMed] [Google Scholar]
- 120.Abella JV, Peschard P, Naujokas MA, Lin T, Saucier C, Urbe S, et al. Met/Hepatocyte growth factor receptor ubiquitination suppresses transformation and is required for Hrs phosphorylation. Mol Cell Biol. 2005;25(21):9632–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lu X, Peled N, Greer J, Wu W, Choi P, Berger AH, et al. MET Exon 14 Mutation Encodes an Actionable Therapeutic Target in Lung Adenocarcinoma. Cancer Res. 2017;77(16):4498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vigna E, Gramaglia D, Longati P, Bardelli A, Comoglio PM. Loss of the exon encoding the juxtamembrane domain is essential for the oncogenic activation of TPR-MET. Oncogene. 1999;18(29):4275–81. [DOI] [PubMed] [Google Scholar]
- 123.Mak HHL, Peschard P, Lin T, Naujokas MA, Zuo D, Park M. Oncogenic activation of the Met receptor tyrosine kinase fusion protein, Tpr–Met, involves exclusion from the endocytic degradative pathway. Oncogene. 2007;26(51):7213–21. [DOI] [PubMed] [Google Scholar]
- 124.Bardelli A, Longati P, Williams TA, Benvenuti S, Comoglio PM. A peptide representing the carboxyl-terminal tail of the met receptor inhibits kinase activity and invasive growth. J Biol Chem. 1999;274(41):29274–81. [DOI] [PubMed] [Google Scholar]
- 125.Gual P, Giordano S, Anguissola S, Comoglio PM. Differential requirement of the last C-terminal tail of Met receptor for cell transformation and invasiveness. Oncogene. 2001;20(39):5493–502. [DOI] [PubMed] [Google Scholar]
- 126.Yokoyama N, Ischenko I, Hayman MJ, Miller WT. The C terminus of RON tyrosine kinase plays an autoinhibitory role. J Biol Chem. 2005;280(10):8893–900. [DOI] [PubMed] [Google Scholar]
- 127.Sternberg MJ, Gullick WJ. A sequence motif in the transmembrane region of growth factor receptors with tyrosine kinase activity mediates dimerization. Protein Eng. 1990;3(4):245–8. [DOI] [PubMed] [Google Scholar]
- 128.Teese MG, Langosch D. Role of GxxxG Motifs in Transmembrane Domain Interactions. Biochemistry. 2015;54(33):5125–35. [DOI] [PubMed] [Google Scholar]
- 129.Lemmon MA, Treutlein HR, Adams PD, Brunger AT, Engelman DM. A dimerization motif for transmembrane alpha-helices. Nat Struct Biol. 1994;1(3):157–63. [DOI] [PubMed] [Google Scholar]
- 130.Finger C, Escher C, Schneider D. The single transmembrane domains of human receptor tyrosine kinases encode self-interactions. Sci Signal. 2009;2(89):ra56. [DOI] [PubMed] [Google Scholar]