Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) protein determines virus entry and the palmitoylation of S protein affects virus infection. An acyltransferase complex ZDHHC5/GOGAL7 that interacts with S protein was detected by affinity purification mass spectrometry (AP-MS). However, the palmitoylated cysteine residues of S protein, the effects of ZDHHC5 or GOLGA7 knockout on S protein’s subcellular localization, palmitoylation, pseudovirus entry and the enzyme for depalmitoylation of S protein are not clear.
Methods
The palmitoylated cysteine residues of S protein were identified by acyl-biotin exchange (ABE) assays. The interactions between S protein and host proteins were analyzed by co-immunoprecipitation (co-IP) assays. Subcellular localizations of S protein and host proteins were analyzed by fluorescence microscopy. ZDHHC5 or GOGAL7 gene was edited by CRISPR-Cas9. The entry efficiencies of SARS-CoV-2 pseudovirus into A549 and Hela cells were analyzed by measuring the activity of Renilla luciferase.
Results
In this investigation, all ten cysteine residues in the endodomain of S protein were palmitoylated. The interaction of S protein with ZDHHC5 or GOLGA7 was confirmed. The interaction and colocalization of S protein with ZDHHC5 or GOLGA7 were independent of the ten cysteine residues in the endodomain of S protein. The interaction between S protein and ZDHHC5 was independent of the enzymatic activity and the PDZ-binding domain of ZDHHC5. Three cell lines HEK293T, A549 and Hela lacking ZDHHC5 or GOLGA7 were constructed. Furthermore, S proteins still interacted with one host protein in HEK293T cells lacking the other. ZDHHC5 or GOLGA7 knockout had no significant effect on S protein’s subcellular localization or palmitoylation, but significantly decreased the entry efficiencies of SARS-CoV-2 pseudovirus into A549 and Hela cells, while varying degrees of entry efficiencies may be linked to the cell types. Additionally, the S protein interacted with the depalmitoylase APT2.
Conclusions
ZDHHC5 and GOLGA7 played important roles in SARS-CoV-2 pseudovirus entry, but the reason why the two host proteins affected pseudovirus entry remains to be further explored. This study extends the knowledge about the interactions between SARS-CoV-2 S protein and host proteins and probably provides a reference for the corresponding antiviral methods.
Keywords: SARS-CoV-2, Spike protein, ZDHHC5/GOLGA7, APT2, Virus-host interaction
Introduction
SARS-CoV-2 is a highly transmissible and pathogenic beta-coronavirus and causes the coronavirus disease 19 (COVID-19) pandemic, threatening human health and public safety. SARS-CoV-2 contains a single-stranded positive-sense RNA genome that encodes at least 29 proteins, including four typical structural proteins, spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins [1–3]. However, the pathogenic mechanism of SARS-CoV-2 including the interaction between virus and host remains to be further explored.
SARS-CoV-2 S protein is the major target of virus-neutralizing antibodies, forming a trimer on the surface of the virus particle and binding to the receptor to initiate the viral replication cycle [4]. Each S protomer comprises the S1 and S2 subunits and activation of S protein requires cleavage of S1/S2 by furin-like protease and undergoes a conformational change from prefusion to postfusion [5]. S protein undergoes palmitoylation and alteration of the ten cysteine residues in the endodomain (cytoplasmic tail) of S protein decreased the efficiency of syncytium formation, cell–cell fusion and pseudotyped SARS-CoV-2 infectivity [6]. The palmitoylations of S proteins from other coronaviruses have also been reported. For example, palmitoylation of the cysteine-rich endodomain of S protein from SARS-CoV (severe acute respiratory syndrome coronavirus) is important for spike-mediated cell fusion [7]. The palmitoylation of S protein from murine coronavirus is essential for virion assembly and infectivity [8].
Protein palmitoylation is a dynamic and reversible post-translational modification that is catalyzed by 23 mammalian palmitoyltransferases ZDHHCs and is reversed by several acyl protein thioesterases (such as APT1 and APT2) [9, 10]. Palmitoylation dynamically regulates different aspects in the life of a protein, including stability, localization, interactome and function, playing critical roles in cellular physiology. Each of ZDHHCs intracellular domains contains a conserved Asp-His-His-Cys (DHHC) motif as the catalytic center [11]. Other accessory proteins such as GOLGA7 (or GCP16) are essential components of cell palmitoylation system and can regulate specific ZDHHC enzyme activity, stability and transport [12]. Virus protein palmitoylation is usually very important for viral protein function and viral replication. Therefore, ZDHHCs enzyme necessary for palmitoylation of viral proteins can be used as a potential drug target [13].
Two high confidence human proteins ZDHHC5 and GOLGA7 that interacted with SARS-CoV-2 S protein were identified by AP-MS [14]. ZDHHC5 and accessory protein GOLGA7 form a mutually stable acyltransferase complex [15]. ZDHHC5 can palmitoylate many protein substrates and participate in some important physiological processes, such as fatty acid uptake and immune response [16, 17]. ZDHHC5 usually interacts with substrate proteins through its own PDZ (PSD-95/Discs-large/ZO-1 homology) binding domain and substitution of the cysteine residue in DHHC motif by a serine residue leads to enzyme inactivation [18]. However, the palmitoylated cysteine residues of S protein, the effects of ZDHHC5 or GOLGA7 knockout on S protein’s subcellular localization, palmitoylation, pseudovirus entry, and the enzyme for depalmitoylation of S protein are not clear. In this study, we mainly identified the palmitoylated cysteine residues of S protein, investigated the effects of ZDHHC5 or GOLGA7 knockout on S protein’s subcellular localization, palmitoylation, pseudovirus entry and analyzed the interactions of S protein with two depalmitoylases APT1/APT2.
Methods
Cell lines
Human embryonic kidney (HEK) 293 T cells provided by Prof. Hai-Yan Ren, human lung cancer A549 cells purchased from National Collection of Authenticated Cell Cultures and human cervical carcinoma Hela cells provided by Prof. Feng Shao were grown at 37 °C in 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS). HEK293T cells were used for the ABE and co-IP assays, since the transfection efficiency of HEK293T cells is very high. A549 and Hela cells were used for the fluorescence microscopy and SARS-CoV-2 spike-mediated pseudovirus entry assays, since the two kinds of cells adhered well to the surface of coverslips placed in 6-well plates and on the bottom of 96-well white plates.
Plasmid construction
Total RNA was extracted from HEK293T cells with Trizol reagent (Invitrogen) and was then used for the synthesis of single-stranded cDNA by reverse transcription. The plasmid pcDNA3.1-S expressing SARS-CoV-2 isolate Wuhan-Hu-1 (GenBank accession number NC_045512.2) S protein was provided by Prof. Ai-Ping Tong. Different kinds of nucleic-acid fragments were amplified with the corresponding primers (Table 1). The plasmids used for ABE, co-IP and fluorescence microscopy were constructed. All constructs used in this study were confirmed by DNA sequencing. The operations are as follows.
Table 1.
Primers used in this study
| Name | Sequence(5'to 3')a | Usage |
|---|---|---|
| S-F2 | CTAGCGTTTAAACTTAAGCTTATGTTTGTTTTTCTTGTTTTATTGCCACTAGTCTCTAGTCAGTGCGTGAACCTGACCACA (HindIII) | pcDNA3.1-S-HA, pcDNA3.1-SC10A-HA/Co-IP, ABE |
| S-HA-R | TGCTGGATATCTGCAGAATTCTTAAGCGTAATCTGGAACATCGTATGGGTACATGGTGTAGTGCAGCTTCACGCC (EcoRI) | |
| SA1235C-F | TATCGTGATGGTGACCATCATGCTGTGCGCTATGACATCC | pcDNA3.1-SA1235C-HA/ABE |
| SA1235C-R | GGATGTCATAGCGCACAGCATGATGGTCACCATCACGATA | |
| SA1236C-F | TATCGTGATGGTGACCATCATGCTGGCCTGTATGACATCC | pcDNA3.1-SA1236C-HA/ABE |
| SA1236C-R | GGATGTCATACAGGCCAGCATGATGGTCACCATCACGATA | |
| SA1240C-F | CATCATGCTGGCCGCTATGACATCCTGCGCTTCTGCCCTG | pcDNA3.1-SA1240C-HA/ABE |
| SA1240C-R | CAGGGCAGAAGCGCAGGATGTCATAGCGGCCAGCATGATG | |
| SA1241C-F | CATCATGCTGGCCGCTATGACATCCGCCTGTTCTGCCCTG | pcDNA3.1-SA1241C-HA/ABE |
| SA1241C-R | CAGGGCAGAACAGGCGGATGTCATAGCGGCCAGCATGATG | |
| SA1243C-F | GCTGGCCGCTATGACATCCGCCGCTTCTTGCCTGAAGGGC | pcDNA3.1-SA1243C-HA/ABE |
| SA1243C-R | GCCCTTCAGGCAAGAAGCGGCGGATGTCATAGCGGCCAGC | |
| SA1247C-F | GACATCCGCCGCTTCTGCCCTGAAGGGCTGCGCTAGCGCT | pcDNA3.1-SA1247C-HA/ABE |
| SA1247C-R | AGCGCTAGCGCAGCCCTTCAGGGCAGAAGCGGCGGATGTC | |
| SA1248C-F | GACATCCGCCGCTTCTGCCCTGAAGGGCGCCTGTAGCGCT | pcDNA3.1-SA1248C-HA/ABE |
| SA1248C-R | AGCGCTACAGGCGCCCTTCAGGGCAGAAGCGGCGGATGTC | |
| SA1250C-F | TCTGCCCTGAAGGGCGCCGCTAGCTGTGGCTCCGCCGCTA | pcDNA3.1-SA1250C-HA/ABE |
| SA1250C-R | TAGCGGCGGAGCCACAGCTAGCGGCGCCCTTCAGGGCAGA | |
| SA1253C-F | TCTGCCCTGAAGGGCGCCGCTAGCGCTGGCTCCTGCGCTA | pcDNA3.1-SA1253C-HA/ABE |
| SA1253C-R | TAGCGCAGGAGCCAGCGCTAGCGGCGCCCTTCAGGGCAGA | |
| SA1254C-F | CTGAAGGGCGCCGCTAGCGCTGGCTCCGCCTGTAAGTTTG | pcDNA3.1-SA1254C-HA/ABE |
| SA1254C-R | CAAACTTACAGGCGGAGCCAGCGCTAGCGGCGCCCTTCAG | |
| ZDHHC5-F | CTTGGTACCGAGCTCGGATCCATGCCCGCAGAGTCTGGAAAG (BamHI) | pcDNA3.1-ZDHHC5-3Flag/co-IP |
| ZDHHC5-3Flag-R | TGCTGGATATCTGCAGAATTCTCACTTATCGTCGTCATCCTTGTAATCGATCTTATCGTCGTCATCCTTGTAATCTCCCTTATCGTCGTCATCCTTGTAATCCACCGAAATCTCATAGGTGG (EcoRI) | |
| GOLGA7-F | CTTGGTACCGAGCTCGGATCCATGAGGCCGCAGCAGGCGCC (BamHI) | pcDNA3.1-GOLGA7-3Flag/co-IP |
| GOLGA7-3Flag-R | TGCTGGATATCTGCAGAATTCTTACTTATCGTCGTCATCCTTGTAATCGATCTTATCGTCGTCATCCTTGTAATCTCCCTTATCGTCGTCATCCTTGTAATCTCTTCCACTGCTCATGCCTC (EcoRI) | |
| ZDHHC5-C134S-F | TGTGGAGGAATTTGATCATCACTCCCCCTGGGTGAATAAC | pcDNA3.1-ZDHHC5-C134S-3Flag/co-IP |
| ZDHHC5-C134S-R | GTTATTCACCCAGGGGGAGTGATGATCAAATTCCTCCACA | |
| ZDHHC5△PDZ-3Flag-R | TGCTGGATATCTGCAGAATTCTCACTTATCGTCGTCATCCTTGTAATCGATCTTATCGTCGTCATCCTTGTAATCTCCCTTATCGTCGTCATCCTTGTAATCGGTGGTACCACCAACCCCTG (EcoRI) | pcDNA3.1-ZDHHC5△PDZ-3Flag/co-IP |
| ZDHHC5△PDZ-R | TTATCTAGATCCGGTGGATCCTCAGGTGGTACCACCAACCCCTG (BamHI) | pDsRed2-ZDHHC5△PDZ/localization |
| S-F | TCAGATCTCGAGCTCAAGCTTATGTTTGTTTTTCTTGTTTTATTGCCACTAGTCTCTAGTCAGTGCGTGAACCTGACCACA (HindIII) | pEGFP-S/localization |
| S-R | CACCATGGTGGCGATGGATCCGGTGTAGTGCAGCTTCACGCC (BamHI) | |
| SC10A-R | CACCATGGTGGCGATGGATCCGGTGTAGTGCAGCTTCACGCCCTTCAGCACTGGCTCGGAATCGTCCTCATCAAACTTAGCGGCGGAGCCAGCGCTAGCGGCGCCCTTCAGGGCAGAAGCGGCGGATGTCATAGCGGCCAG (BamHI) | pEGFP-SC10A/localization |
| ZDHHC5-F2 | TCGAGCTCAAGCTTCGAATTCTATGCCCGCAGAGTCTGGAAAG (EcoRI) | pDsRed2-ZDHHC5/localization |
| ZDHHC5-R2 | TTATCTAGATCCGGTGGATCCTCACACCGAAATCTCATAGG (BamHI) | |
| GOLGA7-F2 | TCGAGCTCAAGCTTCGAATTCTATGAGGCCGCAGCAGGCGCC (EcoRI) | pDsRed2-GOLGA7/colocalization |
| GOLGA7-R2 | TTATCTAGATCCGGTGGATCCTTATCTTCCACTGCTCATGC (BamHI) | |
| ZDHHC5-sgRNA-F | CACCGGAACCACGTGAAATCCCGTG | lentiCRISPRv2-ZDHHC5 sgRNA/gene editing |
| ZDHHC5-sgRNA-R | AAACCACGGGATTTCACGTGGTTCC | |
| ZDHHC5-F3 | TGTATTGGTCGCCGGAACTA | PCR |
| ZDHHC5-R3 | CAAGATCACGCCACTGGATG | |
| ZDHHC5-seq-F | CACGTATCCTTCAAGGCC | Sequencing |
| GOLGA7-sgRNA-F | CACCGGGCGGCCAGTCATATCTCGA | lentiCRISPRv2-GOLGA7 sgRNA/gene editing |
| GOLGA7-sgRNA-R | AAACTCGAGATATGACTGGCCGCCC | |
| GOLGA7-F3 | CATTGCCTGTTCTGCTTGCA | PCR |
| GOLGA7-R3 | CCCTCATGCCCAAAGATGGT | |
| GOLGA7-seq-F | GTTTTGGTAACTTAGGCCCAG | Sequencing |
| APT1-F | CTTGGTACCGAGCTCGGATCCATGTGCGGCAATAACATGTC (BamHI) | pcDNA3.1-APT1-3Flag/co-IP |
| APT1-3Flag-R | TGCTGGATATCTGCAGAATTCTCACTTATCGTCGTCATCCTTGTAATCGATCTTATCGTCGTCATCCTTGTAATCTCCCTTATCGTCGTCATCCTTGTAATCATCAATTGGAGGTAGGAGTT (EcoRI) | |
| APT2-F | CTTGGTACCGAGCTCGGATCCATGTGTGGTAACACCATGTC (BamHI) | pcDNA3.1-APT2-3Flag/co-IP |
| APT2-3Flag-R | TGCTGGATATCTGCAGAATTCTCACTTATCGTCGTCATCCTTGTAATCGATCTTATCGTCGTCATCCTTGTAATCTCCCTTATCGTCGTCATCCTTGTAATCGACAGGAGGCAGCAGCTTCT (EcoRI) |
aSequences of restriction sites are underline
To generate the plasmids for analysis of palmitoylation levels of SARS-CoV-2 S protein and its mutant SC10A which represents that all ten cysteine residues in the endodomain (1234–1273 aa) were changed to alanine residues (Fig. 1a), the open reading frame (ORF) of SARS-CoV-2 S gene was amplified using pcDNA3.1-S plasmid as template with primers S-F2/S-HA-R and cloned into pcDNA3.1(+) to produce pcDNA3.1-S-HA. The mutant SC10A was amplified using the plasmid pEGFP-SC10A described below as a template with primers S-F2/S-HA-R and cloned into pcDNA3.1(+) to produce pcDNA3.1-SC10A-HA.
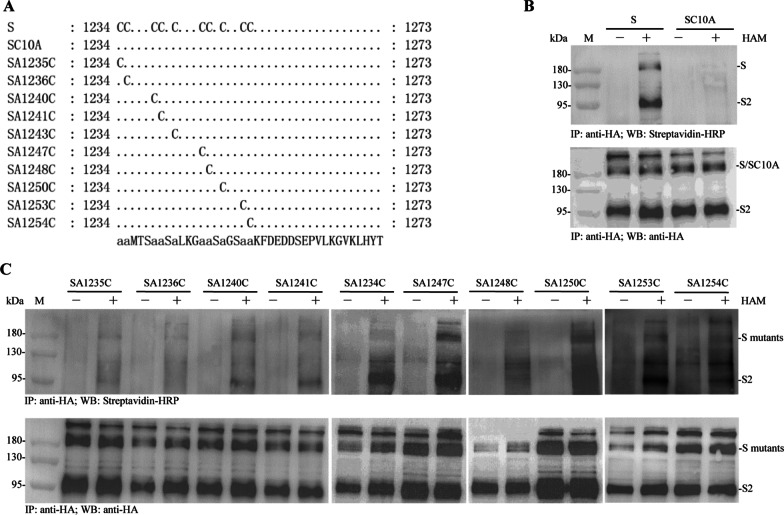
Fig. 1.
The palmitoylation levels of S protein and its mutants measured by ABE assays. a Multiple amino acid sequence alignments of the endodomains (1234–1273 aa) of SARS-CoV-2 S protein and its eleven mutants. The dark spots indicate amino acids on the last line. The small letter “a” indicates that the cysteine residue was replaced with an alanine residue. SC10A represents that all ten cysteine residues in the endodomain were changed to alanine residues. SA1235C ~ SA1254C represent that only one cysteine residue in the endodomain was reserved and the other nine cysteine residues were changed to alanine residues. b The palmitoylation levels of S protein and its mutant SC10A. HEK293T cells were transfected with plasmid expressing S-HA or SC10A-HA for 48 h. The palmitoylation levels of S and SC10A were measured by ABE assays and the cells were then immunoblotted with streptavidin-HRP and anti-HA antibody. M: protein molecular mass marker. C The palmitoylation levels of ten mutants SA1235C ~ SA1254C. HEK293T cells were transfected with ten mutant plasmids expressing SA1235C ~ SA1254C for 48 h. The palmitoylation levels of the ten mutants were measured by ABE assays
To generate plasmids for identification of the palmitoylated cysteine residues of SARS-CoV-2 S protein, ten mutants SA1235C ~ SA1254C (in which only one cysteine residue in the endodomain was reserved and the other nine cysteine residues were changed to alanine residues) were constructed (Fig. 1a). For example, two fragments SA1235C-N and SA1235C-C were amplified from pcDNA3.1-SC10A-HA plasmid with primers S-F2/SA1235C-R and SA1235C-F/S-HA-R, respectively. The fragment SA1235C was amplified via overlap PCR using SA1235C-N and SA1235C-C as templates and cloned into plasmid pcDNA3.1(+) to produce pcDNA3.1-SA1235C-HA. As described above, primers S-F2/SA1236C-R and SA1236C-F/S-HA-R were used to construct the plasmid pcDNA3.1-SA1236C-HA. Similarly, eight other plasmids expressing S protein mutants were constructed.
To generate the plasmids for co-IP assays, the ORFs of the ZDHHC5 and GOLGA7 genes were amplified using HEK293T cDNA as templates with primers ZDHHC5-F/ZDHHC5-3Flag-R and GOLGA7-F/GOLGA7-3Flag-R, respectively. To generate a plasmid expressing the mutant ZDHHC5-C134S (cysteine residue 134 in the DHHC motif of ZDHHC5 was changed to a serine residue). Two fragments ZDHHC5-C134S-N and ZDHHC5-C134S-C were amplified with primers ZDHHC5-F/ZDHHC5-C134S-R and ZDHHC5-C134S-F/ZDHHC5-3Flag-R, respectively. The fragment ZDHHC5-C134S was amplified via overlap PCR using ZDHHC5-C134S-N and ZDHHC5-C134S-C as templates. ZDHHC5△PDZ representing ZDHHC5 lacking a PDZ-binding domain (aa 711 ~ 715, YEISV) was amplified with primers ZDHHC5-F/ZDHHC5△PDZ-3Flag-R. The ORFs of APT1/APT2 genes were amplified using HEK293T cDNA as templates with primers APT1-F/APT1-3Flag-R and APT2-F/APT2-3Flag-R, respectively. All six fragments obtained above were separately cloned into pcDNA3.1(+) to construct the corresponding plasmids.
Two plasmids expressing S protein fused with enhanced green fluorescent protein (S-EGFP) or SC10A fused with EGFP (SC10A-EGFP) were constructed to analyze their subcellular localizations. The ORFs of SARS-CoV-2 S gene and the mutant SC10A were amplified with primers S-F/R and S-F/SC10A-R and then cloned into pEGFP-N3 to produce pEGFP-S and pEGFP-SC10A, respectively. Four plasmids expressing ZDHHC5, ZDHHC5-C134S, ZDHHC5△PDZ and GOLGA7 separately fused with red fluorescent protein (RFP) were constructed to analyze their subcellular localizations. The ORF of the ZDHHC5 or GOLGA7 gene was separately amplified with primers ZDHHC5-F2/R2 or GOLGA7-F2/R2. The fragment ZDHHC5-C134S was amplified with primers ZDHHC5-F2/R2 using pcDNA3.1-ZDHHC5-C134S as template. The fragment ZDHHC5△PDZ was amplified with primers ZDHHC5-F2/ZDHHC5△PDZ-R using pcDNA3.1-ZDHHC5-3Flag as template. The four fragments obtained above were separately cloned into pDsRed2-C1 to produce the corresponding plasmids.
Two pairs of oligos ZDHHC5-sgRNA-F/R and GOLGA7-sgRNA-F/R were designed according to a previous report [15]. The fragments ZDHHC5-sgRNA and GOLGA7-sgRNA were obtained by annealing and separately cloned into plasmid lentiCRISPRv2. The plasmids pMD2-G and psPAX2 were purchased from Addgene (www.addgene.org).
ABE assays
The ABE assays were performed as previously described with minor modifications [17, 19]. In brief, HEK293T, HEK293T-ZDHHC5KO or HEK293T-GOLGA7KO cells transiently expressing HA-tagged S or its mutants SC10A and SA1235C ~ SA1254C were harvested at 48 h post transfection (hpt) and washed with cold phosphate-buffered saline (PBS). Prior to cell lysis, N-ethylmaleimide (NEM) was dissolved in 100% EtOH and added to the Lysis Buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% IGEPAL CA-630, 10% glycerol) with 1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitor cocktail (Sigma) to a final concentration of 50 mM. Cells were then suspended in NEM containing Lysis buffer for 1 h at 4 °C and the supernatants were incubated with Red Anti-HA Affinity Gel (Sigma) overnight at 4 °C. After incubation, the beads were washed one time with Lysis Buffer with 10 mM NEM, one time with Stringent Buffer (Lysis Buffer with 10 mM NEM and 0.1% SDS), and then three times with Lysis Buffer of pH 7.2. Then, beads were incubated with a freshly prepared hydroxylamine (HAM)-containing Lysis Buffer of pH 7.2 with 1 M HAM at room temperature for 1 h and washed four times with Lysis Buffer of pH 7.2 and three times with Lysis Buffer of pH 6.2. Subsequently, beads were treated with Biotin-BMCC (5 μM) in Lysis Buffer of pH 6.2 at 4 °C for 1 h. The immunoprecipitated samples were analyzed by Western blot analysis using anti-HA mouse monoclonal antibody (1:1000, Cell Signaling) and streptavidin-HRP (1:1250).
Co-IP assays
Co-IP assays were performed as previously described with minor modifications [20]. Briefly, HEK293T cells seeded in 10 cm dishes were cotransfected with 7.5 μg of pcDNA3.1-S-HA and 7.5 μg of pcDNA3.1-ZDHHC5-3Flag or other indicated plasmids. As a control, 7.5 μg of empty vector pcDNA3.1(+) was transfected in parallel. At 24 hpt, cells were lysed with 1 mL of a radio immunoprecipitation assay (RIPA) buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate) containing protease inhibitor cocktail (Sigma) and 1 mM PMSF for 1 h. The cell lysates were centrifuged to remove cell debris and the lysate supernatants were collected and divided into two parts. Five hundred microliters of lysate supernatant was used to analyze the co-expression of S protein and other proteins by Western blot analysis using anti-HA and anti-Flag antibodies. Other 500 μL of lysate supernatant was incubated with Red Anti-HA Affinity Gel (Sigma) overnight at 4 °C. The precipitates were collected by centrifugation, washed with ice-cold PBS and RIPA buffer, eluted with 40 μL PBS, and finally subjected to Western blot analysis. Since the band of S protein in cell supernatants was usually very weak by Western blot analysis using anti-HA antibody as shown in Fig. 2a, 4b, 440 μL of lysate supernatants were used to enrich S protein with 25 μL of Red Anti-HA Affinity Gel and other 60 μL of lysate supernatants were used to analyze the expression of other proteins when the interaction between S protein and ZDHHC5, ZDHHC5-C134S, ZDHHC5△PDZ, APT1 or APT2 was detected (Fig. 2c, 4a, 7).
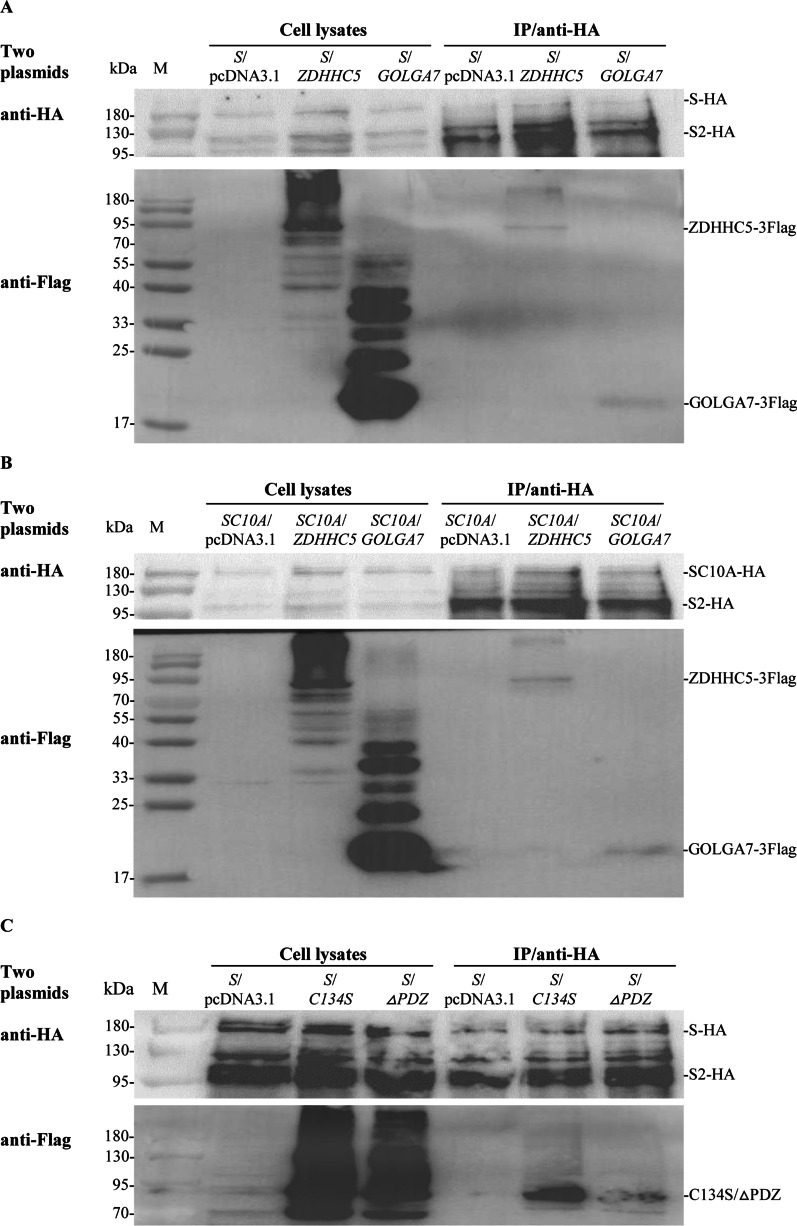
Fig. 2.
Analyzing interactions between SARS-CoV-2 S, SC10A and ZDHHC5, its mutants or GOLGA7 by co-IP. a Interaction between SARS-CoV-2 S protein and ZDHHC5 or GOLGA7. Cell lysates and immunoprecipitated protein complexes (IP) from HEK293T cells cotransfected with indicated plasmids S/pcDNA3.1, S/ZDHHC5 and S/GOLGA7 were subjected to Western blot analysis using anti-HA and anti-Flag antibodies. Cells lysates and IP showed the bands of S-HA, S2-HA, ZDHHC5-3Flag (81 kDa) and GOLGA7-3Flag (19 kDa). b Interaction between SC10A and ZDHHC5 or GOLGA7. Cell lysates and IP from HEK293T cells cotransfected with the indicated plasmids (SC10A/pcDNA3.1, SC10A/ZDHHC5 and SC10A/GOLGA7) were subjected to Western blot analysis. c Interaction between S protein and ZDHHC5-C134S (C134S) or ZDHHC5△PDZ (△PDZ) by co-IP. Cell lysates and IP from HEK293T cells cotransfected with the indicated plasmids (S/pcDNA3.1, S/C134S and S/△PDZ) were subjected to Western blot analysis
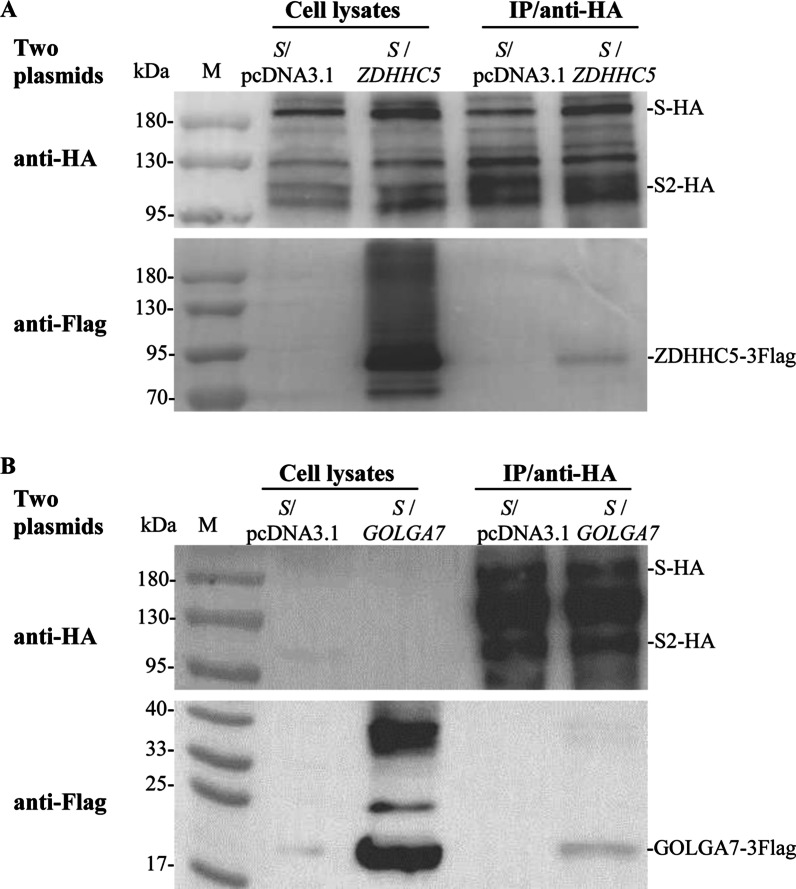
Fig. 4.
Analyzing interactions between SARS-CoV-2 S and ZDHHC5/GOLGA7 in HEK293T cells lacking one protein by co-IP. a Interaction between S protein and ZDHHC5 in HEK293T-GOLGA7KO cells. b Interaction between S protein and GOLGA7 in HEK293T-ZDHHC5KO cells
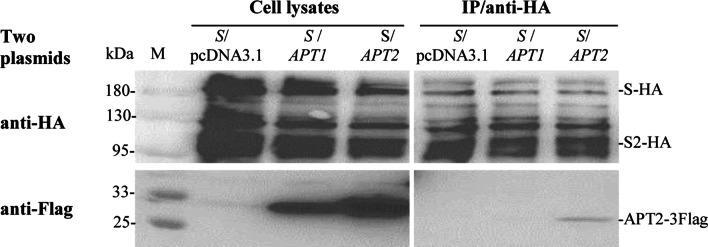
Fig. 7.
Analyzing interactions between SARS-CoV-2 S protein and APT1/APT2 by co-IP. Cell lysates and IP from HEK293T cells cotransfected with indicated plasmids S/pcDNA3.1, S/APT1 and S/APT2 were subjected to Western blot analysis using anti-HA and anti-Flag antibodies
Western blot analysis
Western blot analysis was performed as described previously [20]. Protein samples were resolved by SDS-PAGE, followed by electroblotting to polyvinylidene difluoride (PVDF) membranes. The blots were probed with anti-HA mouse monoclonal antibody (1:1000, Cell Signaling), anti-Flag rabbit monoclonal antibody (1:1000, Cell Signaling), ZDHHC5-specific rabbit polyclonal antiserum (1:100, Sigma), and GOLGA7-specific rabbit polyclonal antiserum (1:1000, Abclonal). Peroxidase-conjugated goat anti-mouse or anti-rabbit IgG (H + L) antibody was used as the secondary antibody. The signals were detected with a chemiluminescent horseradish peroxidase (HRP) substrate (Millipore).
CRISPR/Cas9 gene editing
CRISPR/Cas9 gene editing was performed as previously described with minor modifications [15]. HEK293T, A549 and Hela cells lacking ZDHHC5 or GOLGA7 were generated as follows. Approximately 8 × 105 HEK293T cells in six-well plates were cotransfected with 0.51 μg of pMD2-G, 0.78 μg of psPAX2 and 1.20 μg of lentiCRISPRv2-ZDHHC5 sgRNA or lentiCRISPRv2-GOLGA7 sgRNA. At 48 hpt and 72 hpt, the supernatants were collected and mixed. HEK293T, A549 or Hela cells in six-well plates were inoculated with 500 μL of harvested supernatant for 24 h and then selected with DMEM supplemented with 10% FBS containing 0.9 μg/mL (for HEK293T cells), 2 μg/mL (for A549 cells) or 1 μg/mL (for Hela cells) puromycin until clones formed. All clones were sorted into 96-well plates by flow cytometer (FACSAria SORP) and each well contained only one cell. Individual clones formed in 96-well plates were further transferred into 6-well plates for culture and then identified by DNA sequencing and Western blot analysis. For DNA sequencing, the genomes of cells in 6-well plates were extracted and amplified by PCR using ZDHHC5-F3/R3 or GOGLA7-F3/R3 primers. The PCR products were sequenced using ZDHHC5-seq-F or GOLGA7-seq-F primer.
Fluorescence microscopy
Hela or A549 cells seeded on the surface of coverslips placed in 6-well plates were alone transfected with plasmid pEGFP-N3, pEGFP-S, pEGFP-SC10A, pDsRed2-C1, pDsRed2-ZDHHC5, pDsRed2-ZDHHC5-C134S, pDsRed2-ZDHHC5△PDZ, pDsRed2-GOLGA7 or cotransfected with plasmids pEGFP-S + pDsRed2-ZDHHC5/GOLGA7, pEGFP-SC10A + pDsRed2-ZDHHC5/GOLGA7. At 24 hpt, cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, stained by Hoechst 33,342 and observed under a Leica DMi8 fluorescence microscope (objective 64 ×) or a Leica Stellaris laser confocal microscope (objective 100 ×), as described previously [21].
Immunofluorescence assay (IFA)
To further confirm the subcellular localization of SARS-CoV-2 S protein, an immunofluorescence assay was performed as previously described with minor modifications [22]. Hela cells transfected with plasmid pcDNA3.1-S-HA for 24 h were fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.2% Triton X-100 and blocked in 10% bovine serum albumin (BSA) at room temperature for 1 h. The cells were incubated with anti-HA mouse monoclonal antibody (1:100) in 1% BSA for 2 h, rinsed three times for 10 min each with PBS containing 1% BSA, and then incubated with Alexa-488-conjugated goat anti-mouse IgG (1:500, Invitrogen). The cells were stained by Hoechst 33,342 and examined under a Leica Stellaris laser confocal microscope (objective 100 ×).
SARS-CoV-2 spike-mediated pseudovirus entry assay
To analyze the effects of ZDHHC5 or GOLGA7 knockout on SARS-CoV-2 pseudovirus entry, 5 × 104 A549, A549-ZDHHC5KO, A549-GOLGA7KO, Hela, Hela-ZDHHC5KO or Hela-GOLGA7KO cells were seeded in 96-well white plates and grown overnight. The culture medium was replaced with fresh medium containing 8 μg/mL polybrene for 1 h and then the cells were inoculated with 4E + 6 RLU (relative light units) VSV-SARS-2-S-luc pseudovirus purchased from Delivectory Biosciences Inc. At 16 h post infection (hpi), the culture medium was replaced with fresh medium. Entry efficiency was quantified at 48 hpi by measuring the activity of Renilla luciferase in cell lysates using the ONE-Glo™ Luciferase Assay (E6120, Promega, USA) according to the manufacturer’s instructions (PekinElmer Envision). The infection experiments were performed under biosafety level 2 (BSL2) laboratory conditions.
Results
Identification of palmitoylated cysteine residues of SARS-CoV-2 S protein
The palmitoylation levels of SARS-CoV-2 S protein and its eleven mutants SC10A, SA1235C ~ SA1254C (Fig. 1a) were analyzed by ABE assays. Ectopically expressed S protein and S2 subunit were obviously palmitoylated, but SC10A was not palmitoylated. Loss of signal upon omission of HAM treatment demonstrated that S protein incorporates palmitate through a thioester linkage (Fig. 1b). All ten mutants SA1235C ~ SA1254C were palmitoylated, although the palmitoylation levels of the two mutants SA1236C and SA1248C or their S2 subunits were very weak (Fig. 1c).
Confirmation of interaction between SARS-CoV-2 S protein and ZDHHC5 or GOLGA7 by co-IP
The interaction between SARS-CoV-2 S protein and ZDHHC5 or GOLGA7 was analyzed by co-IP assays. S protein and ZDHHC5 or GOLGA7 were coexpressed in HEK293T cells by cotransfection with indicated plasmids (S/ZDHHC5 and S/GOLGA7). The cell lysates and immunoprecipitated protein complexes (IP) were detected by Western blot analysis using anti-HA and anti-Flag antibodies. As the results, the bands of S protein and S2 subunit were detected in both cell lysates and IP by anti-HA, which were weak in cell lysates and strong in IP (Fig. 2a, anti-HA). This suggested that the fusion protein S-HA and S2 subunit were effectively immunoprecipitated by anti-HA affinity agarose gel. The bands of ZDHHC5-3Flag (81 kDa) and GOLGA7-3Flag (19 kDa) were detected in both cell lysates and IP by anti-Flag. In addition, a band of more than 180 kDa was observed when ZDHHC5-Flag was detected in both cell lysates and IP (Fig. 2a, anti-Flag). As a negative control (S/pcDNA3.1), only S-HA and S2 subunit were detected in cell lysates and IP. These further confirmed that SARS-CoV-2 S protein interacted with ZDHHC5 or GOLGA7.
The effects of the ten cysteine residues in the endodomain of S protein on the interaction between S protein and ZDHHC5 or GOLGA7 were analyzed by co-IP. SC10A and ZDHHC5 or GOLGA7 were coexpressed in HEK293T cells by cotransfection with indicated plasmids (SC10A/ZDHHC5 and SC10A/GOLGA7). The bands of SC10A and its S2 subunit were detected in both cell lysates and IP by anti-HA (Fig. 2b, anti-HA). The bands of ZDHHC5-3Flag and GOLGA7-3Flag were detected in both cell lysates and IP by anti-Flag (Fig. 2b, anti-Flag). These showed that SARS-CoV-2 SC10A interacted with ZDHHC5 or GOLGA7.
The interactions between S protein and the two mutants of ZDHHC5 including ZDHHC5-C134S or ZDHHC5△PDZ were further analyzed by co-IP. The results showed that S protein still interacted with the two mutants (Fig. 2c).
Construction of three cell lines lacking ZDHHC5 or GOLGA7 by CRISPR/Cas9
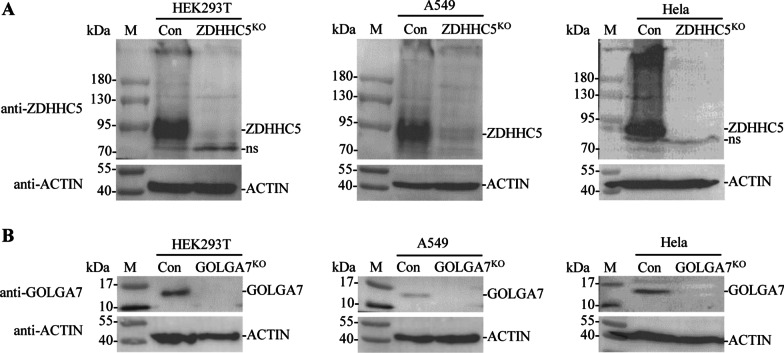
The ZDHHC5 or GOLGA7 gene in three cell lines HEK293T, A549 and Hela was edited by CRISPR/Cas9 to investigate the effects of ZDHHC5 or GOLGA7 knockout on S protein’s subcellular localization, palmitoylation levels and pseudovirus entry. ZDHHC5 or GOLGA7 gene editing was first verified by PCR amplification and DNA sequencing. The DNA fragments were amplified from the genomes of edited cells with primers ZDHHC5-F3/R3 or GOLGA7-F3/R3 and then sequenced, indicating that ZDHHC5 or GOLGA7 gene was edited in all three cell lines. Western blot analysis showed that a specific band of about 78 kDa or 16 kDa was detected in the control cells, but not in the edited cells (Fig. 3a, b), confirming that the expression of ZDHHC5 or GOLGA7 was completely interrupted in the three cell lines.
Fig. 3.
Validation of ZDHHC5 or GOLGA7 gene editing by CRISPR-Cas9. a ZDHHC5 or b GOLGA7 expression in gene-disrupted HEK293T, A549 and Hela cells was determined by Western blot analysis using antibody anti-ZDHHC5 or anti-GOLGA7. ns indicates non-specific band
The interaction between S protein and ZDHHC5 or GOLGA7 in HEK293T cells lacking the other
The interaction between S protein and ZDHHC5 in HEK293T-GOLGA7KO cells and the interaction between S protein and GOLGA7 in HEK293T-ZDHHC5KO cells were further analyzed by co-IP, respectively. The results demonstrated that S protein still interacted with ZDHHC5 in HEK293T-GOLGA7KO cells (Fig. 4a) and with GOLGA7 in HEK293T-ZDHHC5KO cells (Fig. 4b).
Localizations of SARS-CoV-2 S protein, ZDHHC5 and GOLGA7
Fluorescence microscopy showed that S-EGFP distributed in the cytoplasm of Hela and A549 cells, although a weak fluorescence signal was observed in the plasma membrane of some cells. An immunofluorescence assay (IFA) showed that S-HA distributed in the cytoplasm of Hela cells. SC10A-EGFP also distributed in the cytoplasm of Hela. S-EGFP also distributed in the cytoplasm of Hela-ZDHHC5KO and Hela-GOLGA7KO cells (Fig. 5a). As a control, the EGFP distributed in both the cytoplasm and the nucleus of Hela cells.
Fig. 5.
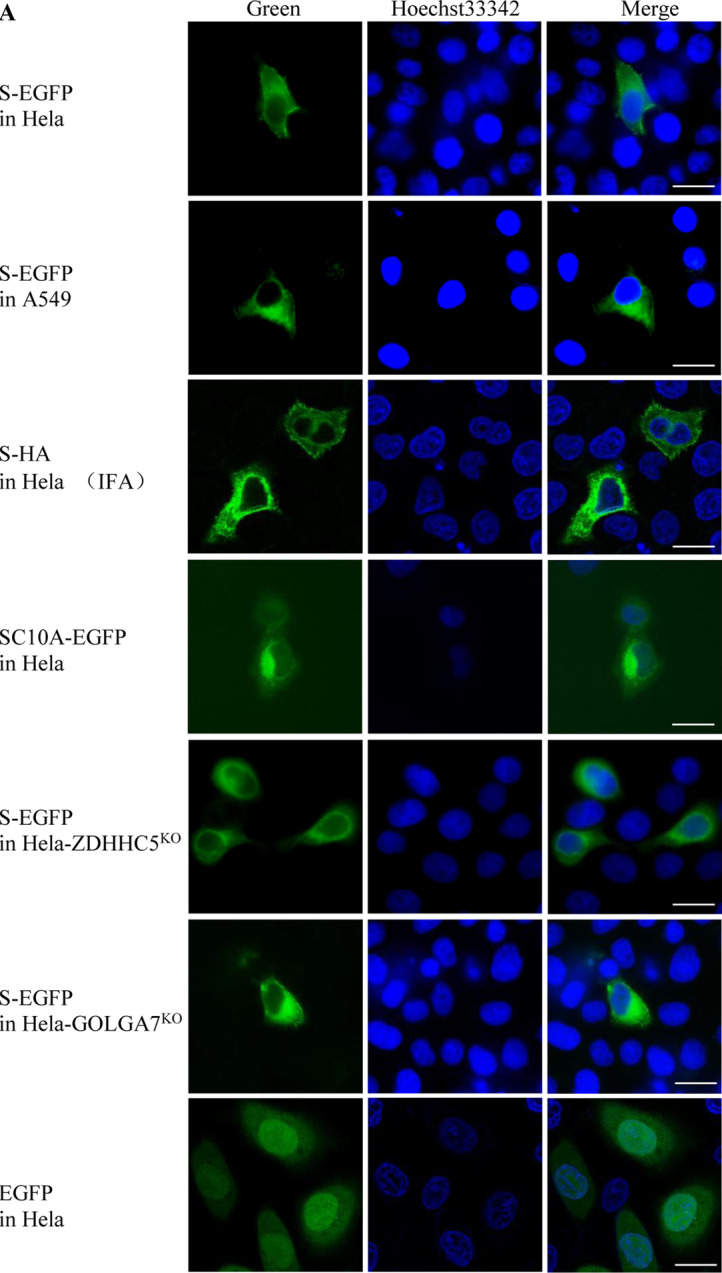
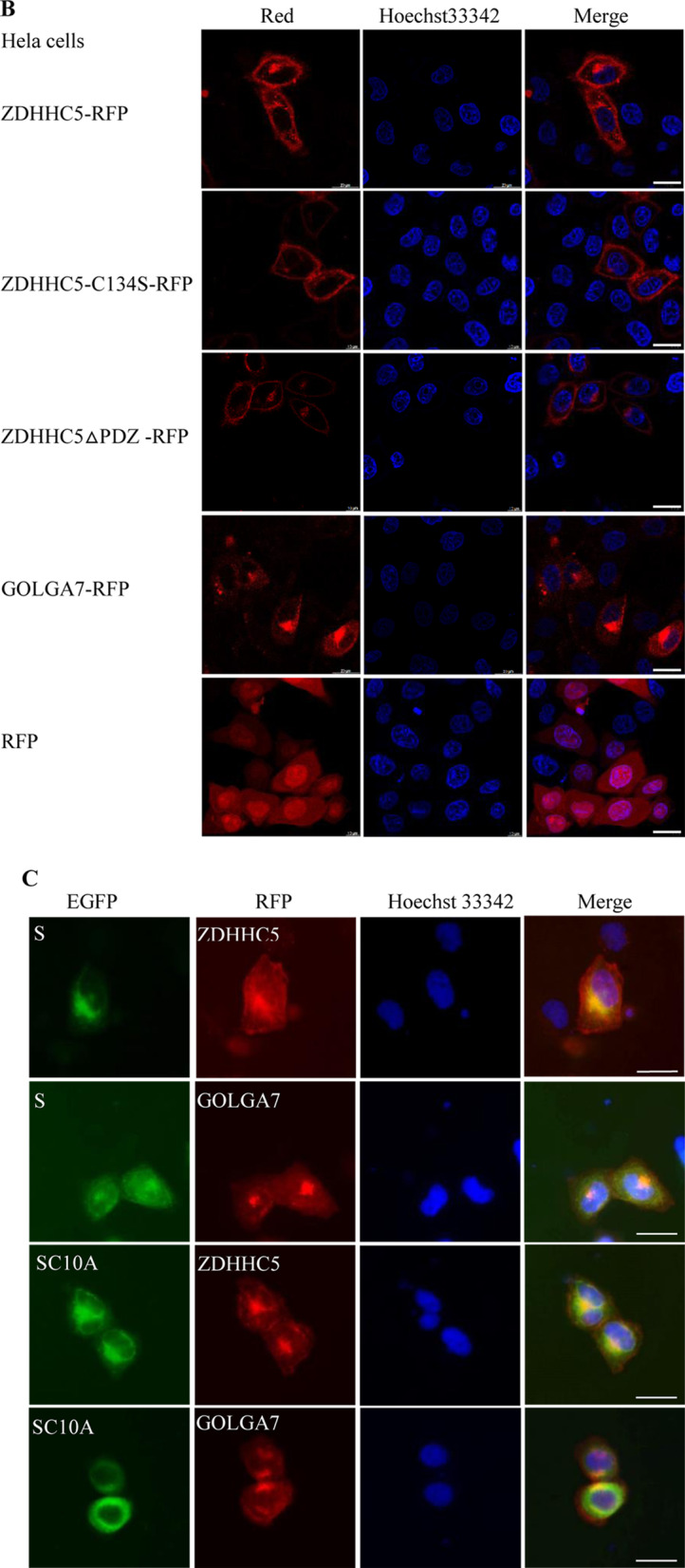
Fluorescence micrographs of cells expressing a single protein or co-expressing two proteins. a Expressing protein S-EGFP, S-HA or SC10A-EGFP alone in Hela, A549, Hela-ZDHHC5KO or Hela-GOLGA7KO cells. Scale bar: 20 μm. b Expressing protein ZDHHC5-RFP, ZDHHC5-C134S-RFP, ZDHHC5△PDZ-RFP or GOLGA7-RFP alone in Hela cells. Scale bar: 20 μm. c Co-expressing two proteins, S-EGFP + ZDHHC5-RFP/GOLGA7-RFP and SC10A-EGFP + ZDHHC5-RFP/GOLGA7-RFP in Hela cells, respectively. S-EGFP (green), SC10A-EGFP (green), ZDHHC5-RFP (red), GOLGA7-RFP (red), nucleus (blue), and colocalization (yellow). Scale bar: 20 μm
ZDHHC5-RFP, ZDHHC5-C134S-RFP, ZDHHC5△PDZ-RFP all distributed in the cytoplasm and plasma membrane of Hela cells. GOLGA7 mainly distributed in the cytoplasm and little distributed in the plasma membrane (Fig. 5b). As a control, the RFP distributed in both the cytoplasm and the nucleus of Hela cells.
When Hela cells were cotransfected, S-EGFP/SC10A-EGFP colocalized with ZDHHC5-RFP/GOLGA7-RFP in the cytoplasm, respectively (Fig. 5c). These not only further confirmed the interaction of S protein with ZDHHC5 or GOLGA7, but also indicated that the palmitoylation of S protein had no significant effect on their colocalization.
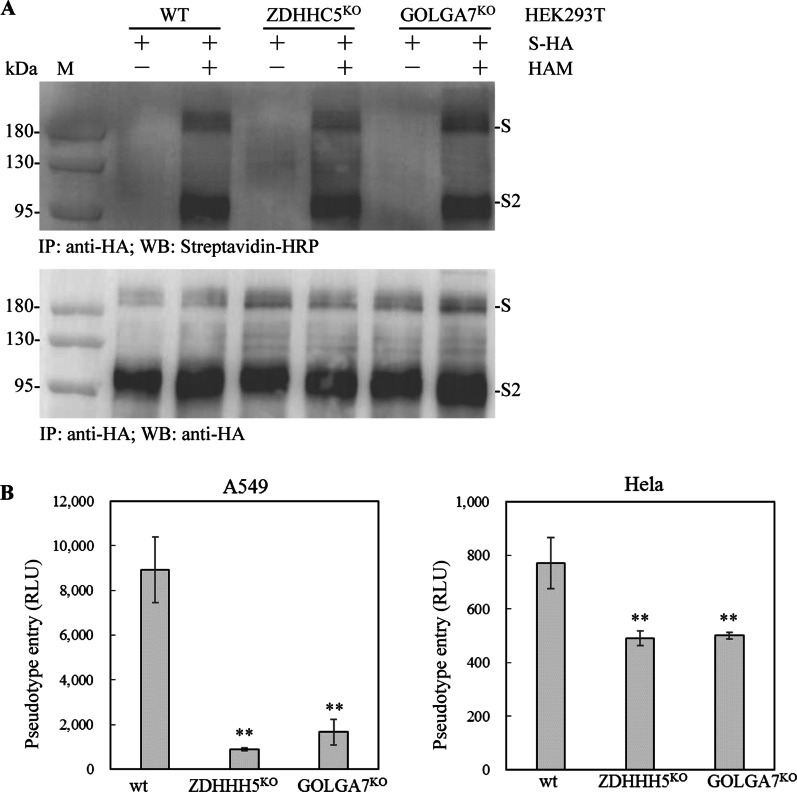
The effects of ZDHHC5 or GOLGA7 knockout on the palmitoylation levels of SARS-CoV-2 S protein and pseudovirus entry
The effect of ZDHHC5 or GOLGA7 knockout on the palmitoylation levels of S protein was analyzed by ABE assays. Ectopically expressed S protein and S2 subunit were palmitoylated in HEK293T, HEK293T-ZDHHC5KO and HEK293T-GOLGA7KO cells and the palmitoylation levels were comparable (Fig. 6a).
Fig. 6.
The effects of ZDHHC5 or GOLGA7 knockout on S protein palmitoylation and pseudovirus entry. a HEK293T, HEK293T-GOLGA7KO and HEK293T-ZDHHC5KO cells were transfected with plasmid expressing S-HA for 48 h. The palmitoylation levels of S protein were measured by ABE assays. b Cells were infected with an equal number of SARS-CoV-2 pseudotyped virions and the entry efficiencies were quantified at 48 hpi by measuring luciferase activity (in relative light units, RLU) (n = 3). ZDHHC5 or GOLGA7 knockout in A549 and Hela cells significantly reduced pseudovirus infection. Error bars indicate standard deviations. ** represents P < 0.01
The effects of ZDHHC5 or GOLGA7 knockout on SARS-CoV-2 pseudovirus entry into Hela and A549 cells were further analyzed. The entry efficiency of SARS-CoV-2 pseudovirus into A549 cells was significantly higher than that into A549-ZDHHC5KO (P = 0.0007) and A549-GOLGA7KO (P = 0.0013) cells. Similarly, the entry efficiency of SARS-CoV-2 pseudovirus into Hela cells was significantly higher than that into Hela-ZDHHC5KO (P = 0.0084) and Hela-GOLGA7KO (P = 0.0086) cells (Fig. 6b).
Furthermore, the entry efficiency of SARS-CoV-2 pseudovirus into A549 cells was significantly higher than that into Hela cells by approximately 11.5-fold (P = 0.0007).
Interaction between SARS-CoV-2 S protein and APT2 determined by co-IP
The interactions between S protein and two depalmitoylases APT1/APT2 were also analyzed by co-IP assays. The results demonstrated that S protein interacted with APT2, but not APT1 (Fig. 7).
Discussion
The aims of this study were to identify the palmitoylated cysteine residues of S protein, investigate the effects of ZDHHC5 or GOLGA7 knockout on S protein’s subcellular localization, palmitoylation, pseudovirus entry and identify the enzyme for depalmitoylation of S protein. The main results showed that ten cysteine residues in the endodomain of SARS-CoV-2 S protein were palmitoylated. The interaction and colocalization of S protein with ZDHHC5 or GOLGA7 were independent of the ten cysteine residues in the endodomain of S protein. ZDHHC5 or GOLGA7 knockout had no significant effect on S protein’s subcellular localization or palmitoylation, but significantly decreased the entry efficiencies of SARS-CoV-2 pseudovirus into A549 and Hela cells. Moreover, the S protein interacted with the depalmitoylase APT2.
S proteins of several coronaviruses have been shown to be palmitoylated. For example, palmitoylation of SARS-CoV S protein is important for spike-mediated cell fusion and is necessary for partitioning into detergent-resistant membranes [7, 23]. The palmitoylation of S protein from murine coronavirus is essential for virion assembly and infectivity and is important for interaction with the M protein [8]. SARS-CoV-2 S protein undergoes palmitoylation and alteration of the ten cysteine residues in the endodomain of S protein decreases the efficiency of syncytium formation, cell–cell fusion and pseudotyped SARS-CoV-2 infectivity [6], but the exact palmitoylated cysteine residues are not yet known. Here, ABE assays showed that ten cysteine residues in the endodomain of S protein were palmitoylated, although the palmitoylation levels of the two mutants SA1236C and SA1248C or their S2 subunits were very weak. It has been reported that the replacement of each of four cysteine clusters in the endodomain of SARS-CoV-2 S protein by two or three alanine residues decreased the palmitoylation levels to different degrees [7]. It is possible that different cysteine residues in the endodomain of SARS-CoV-2 S protein have differential rates of palmitate turnover.
The complex ZDHHC5/GOGAL7 that interacts with S protein was first detected by AP-MS [14]. Here, the interaction of S protein with ZDHHC5 or GOLGA7 was confirmed by co-IP. The mutant SC10A interacted and colocalized with ZDHHC5 or GOLGA7, suggesting that the interaction and colocalization of S protein with ZDHHC5 or GOLGA7 were independent of the ten cysteine residues in the endodomain of S protein. S protein still interacted with the two mutants of ZDHHC5 including ZDHHC5-C134S or ZDHHC5△PDZ, indicating that the interaction between S protein and ZDHHC5 was independent of the enzymatic activity and the PDZ-binding domain of ZDHHC5. ZDHHC5 usually interacts with substrate proteins through its own PDZ binding domain and its DHHC motif also affects its interactions with substrate proteins [17, 18]. These implied that ZDHHC5 might interact with different substrate proteins via different binding sites. A band of more than 180 kDa was observed when ZDHHC5 was detected by co-IP. The band should be the trimer of ZDHHC5, since it can form monomer and aggregation form of higher molecular mass [16].
Fluorescence microscopy showed that S-EGFP distributed in the cytoplasm of Hela and A549 cells. S-HA also distributed in the cytoplasm of Hela cells. Obviously, the localizations of S protein in the two kinds of cells from different tissues were consistent. SARS-CoV-2 S protein was distributed in the cytoplasm of Human Epithelial-2 (HEp-2) cells in a previous report [24]. SC10A-EGFP also distributed in the cytoplasm of Hela, indicating that alteration of the ten cysteine residues in the endodomain had no significant effect on the subcellular localization of S protein. S-EGFP also distributed in the cytoplasm of Hela-ZDHHC5KO and Hela-GOLGA7KO cells, revealing that ZDHHC5 or GOLGA7 knockout had no significant effect on the subcellular localization of S protein. ZDHHC5-RFP, ZDHHC5-C134S-RFP, ZDHHC5△PDZ-RFP all distributed in the cytoplasm and plasma membrane of Hela cells, indicating that the enzymatic activity and PDZ-binding domain of ZDHHC5 had no significant effect on its subcellular localization.
In this study, ZDHHC5 knockout had no significant effects on the subcellular localization or palmitoylation levels of S protein, but significantly decreased the entry efficiencies of SARS-CoV-2 pseudovirus into A549 and Hela cells. These implied that one or more other palmitoyltransferases might be involved in the palmitoylation of S protein. During the preparation of this manuscript, Mesquita et al. reported that ZDHHC5 knockdown in Hela cells or ZDHHC5 knockout in HAP-1 (human astrocyte precursor) cells had no significant effect on the palmitoylation of ectopically expressed S protein, but ZDHHC5 knockdown in Vero E6 (African green monkey kidney cells) significantly decreased S protein palmitoylation during SARS-CoV-2 infection [25]. Moreover, S protein palmitoylation was mainly mediated by ZDHHC8,9,20 via analyzing spike-incorporated radioactivity in Hela cells cotransfected with individual siRNAs targeting all human ZDHHCs [25], while Puthenveetil et al. reported that ZDHHC2,3,6,11,20,21,24 were as putative palmitoylation enzymes for SARS-CoV-2 S protein modification by click-chemistry-based analyses of coexpression of S protein with individual ZDHHC [26]. Our results are consistent with the report of Mesquita et al. However, in other reports, the interactions between SARS-CoV-2 S protein and ZDHHC5/GOLGA7 were confirmed by co-IP and the two host protein overexpressions in HEK293T cells enhanced S protein palmitoylation synergistically and ZDHHC5 knockdown decreased S protein palmitoylation and pseudovirus infection [27, 28]. All these distinct results were likely caused by different analytic systems, such as cell lines and detection methods. Therefore, how ZDHHC5 impacts the palmitoylation of the S protein for virus entry needs more investigation.
Why did ZDHHC5 knockout generate the two different outcomes in pseudovirus entry? Probably because ZDHHC5 knockout could affect the palmitoylation of S protein at earlier infected stage, which led to the decrease of the pseudovirus entry efficiency, suggesting that the palmitoylation of S protein at later infected stages is catalyzed by other palmitoyltransferase members. Moreover, the ZDHHC5 knockout likely affects the functions of other cognate substrates, such as nucleotide oligomerization domain (NOD)-like receptors 1 and 2 (NOD1/2) which are involved in infecting of SARS-CoV-2, to decrease the entry efficiency of pseudovirus [17, 18]. As previously reported, NOD1 is required for recognition of SARS-CoV-2 in lung epithelial cells [29]. A drug that targets NOD2 was shown to have potent broad-spectrum antiviral activity against several viruses, including SARS-CoV-2 [30]. However, more investigation are required to address above mentioned questions.
In addition to ZDHHC5, the role of the accessory protein GOLGA7 was also investigated. S protein interacted with ZDHHC5 in HEK293T-GOLGA7KO cells and with GOLGA7 in HEK293T-ZDHHC5KO cells. GOLGA7 knockout had no significant effects on the subcellular localization or palmitoylation levels of S protein, but significantly decreased the entry efficiencies of SARS-CoV-2 pseudovirus into A549 and Hela cells. It has been reported that GOLGA7 interacts with ZDHHC8,9, and regulates the enzyme activity and stability of ZDHHC9 [12, 31]. ZDHHC8,9 were involved in the palmitoylation of S protein in a recent report [25]. These implied that GOLGA7 knockout might impair the enzymatic activity and disrupt the stability of interacted proteins ZDHHC5,8,9, resulting in the decrease of the entry efficiencies of SARS-CoV-2 pseudovirus into A549 and Hela cells.
An interesting phenomenon is that the entry efficiencies of SARS-CoV-2 spike-mediated pseudovirus into the two different cell lines A549 and Hela are significantly different. The entry efficiency of SARS-CoV-2 spike-mediated pseudovirus into A549 cells was significantly higher than that into Hela cells, indicating that A549 cells were more susceptible to SARS-CoV-2 pseudovirus infection than Hela cells. It has been reported that the lung is the primary tropism of SARS-CoV-2 [32]. However, whether this phenomenon was linked to the pathogenesis of SARS-CoV-2 in different tissues needs more evidence.
Protein palmitoylation is a dynamic and reversible post-translational modification [10]. The depalmitoylation of ectopically expressed S protein in Hela cells was detected [25], but the enzyme responsible for the depalmitoylation of S protein is unclear. APT1 and APT2 are two main depalmitoylases. It has been reported that APT1 can depalmitoylate the glycoproteins of several enveloped viruses in vitro [33]. APT1 is predominantly localized in mitochondria [34] and APT2 is a cytosolic protein [35]. In this study, the depalmitoylase APT2 that interacted with S protein was detected by co-IP. This strongly implied that APT2 might participate in the depalmitoylation and function of S protein. The mechanism and biological significance of interaction between APT2 and S protein need to more evidence.
Conclusions
This report confirmed and elucidated the interactions of three host proteins including ZDHHC5, GOLGA7, and APT2 with SARS-CoV-2 S protein, revealing that ZDHHC5 and GOLGA7 played important roles in SARS-CoV-2 pseudovirus entry. The reason why ZDHHC5 or GOLGA7 knockout decreased pseudovirus entry remains unclear. In the current study, the interactions between proteins of pathogen and host provide potential drug targets for antiviral therapy, extending the road to the treatment of infectious diseases, though it has long way to go.
Acknowledgements
We thank Hai-Yan Ren and Feng Shao for providing HEK293T and Hela cells. We thank Ai-Ping Tong for providing plasmid pcDNA3.1-S. The authors would like to express their heartfelt gratitude to Hui-Fang Li and Yan Wang (Core Facility of West China Hospital) for their help with the flow cytometry.
Abbreviations
- ABE
Acyl-biotin exchange
- BSA
Bovine serum albumin
- BSL2
Biosafety level 2
- co-IP
Co-immunoprecipitation
- COVID-19
Coronavirus disease 19
- EGFP
Enhanced green fluorescent protein
- FBS
Fetal bovine serum
- HAM
Hydroxylamine
- hpi
H post infection
- hpt
H post transfection
- IFA
Immunofluorescence assay.
- IP
Immunoprecipitated protein complexes
- NEM
N-ethylmaleimide
- NOD1/2
Nucleotide oligomerization domain (NOD)-like receptors 1 and 2
- ORF
Open reading frame
- PBS
Phosphate-buffered saline
- PDZ
PSD-95/Discs-large/ZO-1 homology
- PMSF
Phenylmethylsulfonyl fluoride
- PVDF
Polyvinylidene difluoride
- RFP
Red fluorescent protein
- RIPA
Radio immunoprecipitation assay
- RLU
Relative light units
- SARS-CoV
Severe acute respiratory syndrome coronavirus
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
Authors' contributions
WC and XTZ conceived and designed the study. XTZ performed experiments. XXY carried out cell culture. WC and XTZ analyzed the data and wrote the manuscript. All authors have read and approved the final manuscript.
Funding
This work is supported by grants from the National Natural Science Foundation of China (32071236), Key Science and Technology Research Projects in Key Areas of the Corps (Grant No. 2018AB019), the 1.3.5 Project for Disciplines Excellence of West China Hospital, Sichuan University (ZYYC20005), Project funded by China Postdoctoral Science Foundation (2020M683304), Sichuan Science and Technology Support Project (2021YJ0502) and Post-Doctor Research Project, West China Hospital, Sichuan University (2020HXBH082).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent to publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
The original version of this article was revised: We have corrected the second author name as Xiao-Xi Yu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/3/2022
A Correction to this paper has been published: 10.1186/s12985-022-01750-0
Contributor Information
Xiao-Tao Zeng, Email: zengxt@scu.edu.cn.
Wei Cheng, Email: chengweilab@163.com.
References
- 1.Chen B, Tian EK, He B, Tian LJ, Han RY, Wang SW, Xiang QR, Zhang S, El Arnaout T, Cheng W. Overview of lethal human coronaviruses. Signal Transduct Target Ther. 2020;5:89. doi: 10.1038/s41392-020-0190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J, Wang W, Chen Z, Lu S, Yang F, Bi Z, Bao L, Mo F, Li X, Huang Y, et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 5.Yao HP, Song YT, Chen Y, Wu NP, Xu JL, Sun CJ, Zhang JX, Weng TH, Zhang ZY, Wu ZG, et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen HT, Zhang SJ, Wang Q, Anang S, Wang J, Ding HT, Kappes JC, Sodroski J. Spike glycoprotein and host cell determinants of SARS-CoV-2 entry and cytopathic effects. J Virol. 2020;95:e02304–e2320. doi: 10.1128/JVI.02304-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petit CM, Chouljenko VN, Iyer A, Colgrove R, Farzan M, Knipe DM, Kousoulas KG. Palmitoylation of the cysteine-rich endodomain of the SARS-coronavirus spike glycoprotein is important for spike-mediated cell fusion. Virology. 2007;360:264–274. doi: 10.1016/j.virol.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorp EB, Boscarino JA, Logan HL, Goletz JT, Gallagher TM. Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. J Virol. 2006;80:1280–1289. doi: 10.1128/JVI.80.3.1280-1289.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Won SJ, Cheung See Kit M, Martin BR. Protein depalmitoylases. Crit Rev Biochem Mol Biol. 2018;53:83–98. doi: 10.1080/10409238.2017.1409191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang MM, Zhou LX, Xu YJ, Yang M, Xu YL, Komaniecki GP, Kosciuk T, Chen X, Lu X, Zou XP, et al. A STAT3 palmitoylation cycle promotes TH17 differentiation and colitis. Nature. 2020;586:434–439. doi: 10.1038/s41586-020-2799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaballa ME, van der Goot FG. The molecular era of protein S-acylation: spotlight on structure, mechanisms, and dynamics. Crit Rev Biochem Mol Biol. 2018;53:420–451. doi: 10.1080/10409238.2018.1488804. [DOI] [PubMed] [Google Scholar]
- 12.Salaun C, Locatelli C, Zmuda F, Cabrera González J, Chamberlain LH. Accessory proteins of the zDHHC family of S-acylation enzymes. J Cell Sci. 2020;133:jcs251819. doi: 10.1242/jcs.251819. [DOI] [PubMed] [Google Scholar]
- 13.Gadalla MR, Veit M. Toward the identification of ZDHHC enzymes required for palmitoylation of viral protein as potential drug targets. Expert Opin Drug Discov. 2020;15:159–177. doi: 10.1080/17460441.2020.1696306. [DOI] [PubMed] [Google Scholar]
- 14.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O'Meara MJ, Rezelj VV, Guo JZ, Swaney DL, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko PJ, Woodrow C, Dubreuil MM, Martin BR, Skouta R, Bassik MC, Dixon SJ. A ZDHHC5-GOLGA7 protein acyltransferase complex promotes nonapoptotic cell death. Cell Chem Biol. 2019;26:1716–1724.e9. doi: 10.1016/j.chembiol.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 16.Kokkola T, Kruse C, Roy-Pogodzik EM, Pekkinen J, Bauch C, Hönck HH, Hennemann H, Kreienkamp HJ. Somatostatin receptor 5 is palmitoylated by the interacting ZDHHC5 palmitoyltransferase. FEBS Lett. 2011;585:2665–2670. doi: 10.1016/j.febslet.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 17.Lu Y, Zheng YP, Coyaud É, Zhang C, Selvabaskaran A, Yu YY, Xu ZZ, Weng XL, Chen JS, Meng Y, et al. Palmitoylation of NOD1 and NOD2 is required for bacterial sensing. Science. 2019;366:460–467. doi: 10.1126/science.aau6391. [DOI] [PubMed] [Google Scholar]
- 18.Woodley KT, Collins MO. Regulation and function of the palmitoyl-acyltransferase ZDHHC5. FEBS J. 2021 doi: 10.1111/febs.15709. [DOI] [PubMed] [Google Scholar]
- 19.Zhang N, Zhao HJ, Zhang LL. Fatty acid synthase promotes the palmitoylation of chikungunya virus nsP1. J Virol. 2019;93:e01747–e1818. doi: 10.1128/JVI.01747-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng XT, Zhang QY. Interaction between two iridovirus core proteins and their effects on ranavirus (RGV) replication in cells from different species. Viruses. 2019;11:416. doi: 10.3390/v11050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng XT, Gao XC, Zhang QY. Rana grylio virus 43R encodes an envelope protein involved in virus entry. Virus Genes. 2018;54:779–791. doi: 10.1007/s11262-018-1606-8. [DOI] [PubMed] [Google Scholar]
- 22.Ke F, He LB, Zhang QY. Nonstructural protein NS80 is crucial in recruiting viral components to form aquareoviral factories. PLoS ONE. 2013;8:e63737. doi: 10.1371/journal.pone.0063737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBride CE, Machamer CE. Palmitoylation of SARS-CoV S protein is necessary for partitioning into detergent-resistant membranes and cell-cell fusion but not interaction with M protein. Virology. 2010;405:139–148. doi: 10.1016/j.virol.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Cruz-Cosme R, Zhuang MW, Liu DX, Liu Y, Teng SL, Wang PH, Tang QY. A systemic and molecular study of subcellular localization of SARS-CoV-2 proteins. Signal Transduct Target Ther. 2020;5:269. doi: 10.1038/s41392-020-00372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mesquita FS, Abrami L, Sergeeva O, Turelli P, Kunz B, Raclot C, Montoya JP, Abriata LA, Peraro MD, Trono D, et al. S-acylation controls SARS-CoV-2 membrane lipid organization and enhances infectivity. Dev Cell. 2021;56:2790–2807. doi: 10.1016/j.devcel.2021.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puthenveetil R, Lun CM, Murphy RE, Healy LB, Vilmen G, Christenson ET, Freed EO, Banerjee A. S-acylation of SARS-CoV-2 spike protein: mechanistic dissection, in vitro reconstitution and role in viral infectivity. J Biol Chem. 2021;297:101112. doi: 10.1016/j.jbc.2021.101112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee M, Mekhail K, Sugiyama M, Mekhail K, Latreille E, Khosraviani N, Wei K, Lee WL. Fatty acid synthase inhibition prevents palmitoylation of SARS-CoV2 spike protein and improves survival of mice infected with murine hepatitis virus. BioRxiv. 2020 doi: 10.1101/2020.12.20.423603. [DOI] [Google Scholar]
- 28.Wu ZC, Zhang ZY, Wang X, Zhang J, Ren CY, Li YM, Gao LF, Liang XH, Wang PH, Ma CH. Palmitoylation of SARS-CoV-2 S protein is essential for viral infectivity. Signal Transduct Target Ther. 2021;6:231. doi: 10.1038/s41392-021-00651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin X, Riva L, Pu Y, Martin-Sancho L, Kanamune J, Yamamoto Y, Sakai K, Gotoh S, Miorin L, De Jesus PD, et al. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep. 2021;34:108628. doi: 10.1016/j.celrep.2020.108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Limonta D, Dyna-Dagman L, Branton W, Mancinelli V, Makio T, Wozniak RW, Power C, Hobman TC. Nodosome inhibition as a novel broad-spectrum antiviral strategy against arboviruses, enteroviruses, and SARS-CoV-2. Antimicrob Agents Chemother. 2021;65:e0049121. doi: 10.1128/AAC.00491-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- 32.Harrison AG, Lin T, Wang PH. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veit M, Schmidt MFG. Enzymatic depalmitoylation of viral glycoproteins with acyl-protein thioesterase 1 in vitro. Virology. 2001;288:89–95. doi: 10.1006/viro.2001.1063. [DOI] [PubMed] [Google Scholar]
- 34.Kathayat RS, Cao Y, Elvira PD, Sandoz PA, Zaballa ME, Springer MZ, Drake LE, Macleod KF, van der Goot FG, Dickinson BC. Active and dynamic mitochondrial S-depalmitoylation revealed by targeted fluorescent probes. Nat Commun. 2018;9:334. doi: 10.1038/s41467-017-02655-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomatis VM, Trenchi A, Gomez GA, Daniotti JL. Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43. PLoS ONE. 2010;5:e15045. doi: 10.1371/journal.pone.0015045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.