Abstract
Pneumonia is a life-threatening respiratory lung disease. Children are more prone to be affected by the disease and accurate manual detection is not easy. Generally, chest radiographs are used for the manual detection of pneumonia and expert radiologists are required for the assessment of the X-ray images. An automatic system would be beneficial for the diagnosis of pneumonia based on chest radiographs as manual detection is time-consuming and tedious. Therefore, a method is proposed in this paper for the fast and automatic detection of pneumonia. A deep learning-based architecture ‘MobileNet’ is proposed for the automatic detection of pneumonia based on the chest X-ray images. A benchmark dataset of 5856 chest X-ray images was taken for the training, testing, and evaluation of the proposed deep learning network. The proposed model was trained within 3 Hrs. and achieved a training accuracy of 97.34%, a validation accuracy of 87.5%, and a testing accuracy of 94.23% for automatic detection of pneumonia. However, the combined accuracy was achieved as 97.09% with 0.96 specificity, 0.97 precision, 0.98 recall, and 0.97 F-Score. The proposed method was found faster and computationally lesser expensive as compared to other methods in the literature and achieved a promising accuracy.
Keywords: Pneumonia, Chest X-ray, Deep learning, MobileNet, Automatic detection of pneumonia
Introduction
Pneumonia is an immense respiratory lung disease where millions of children lose their life due to failure of its detection [34]. The chest X-ray is considered the most preferred imaging modality for the diagnosis of pneumonia [20, 29]. The accurate detection of pneumonia based on the chest radiograph is time-consuming [21]. The manual detection of pneumonia requires a radiologist to have a good experience. The learning graph of the radiologist also matters for the correct identification of the disease. As a result, the identification of pneumonia is not easy based on the chest radiograph. Still, there are some cases where it is very difficult to spot the region of interest for pneumonia detection based on chest radiograph. Therefore, it is considered that computer-aided diagnosis [8, 9] can be evolved to assist the radiologist in spotting the region of interest and detect the positive or negative cases of pneumonia. Deep learning neural network methods have demonstrated efficient performance for medical aided diagnostics [4–6]. However, these methods will only be useful for assistance if the accuracy of such methods can be reached to a similar level of human accuracy.
In recent years, researchers have published various articles which were focused on the automatic detection of pneumonia. A brief description of such literature is given in Table 1. The studies by Rajpurkar et al. [31] and Varshni et al. [39] used the dataset ChestX-ray14, which was released by Wang et al. (2017) [40]. The latter used a subset of those images as they were focusing on the detection of pneumonia only, however, the former used the whole dataset as their method was further extended to detect 14 diseases using the dataset. Varshni et al. [39] had proposed DenseNet-169 as a feature extractor and SVM (Support Vector Machine) as a classifier for the detection of pneumonia. Many other studies used a dataset from the Kaggle database [19]. Saraiva et al. [33] presented a convolutional neural network that comprised of seven convolutional layers, three max-pooling layers, and three dense layers. K-fold cross-validation (K = 5) method was also used to validate the model and the accuracy was achieved as 95.30%. Okeke Stephen et al. [36] made a Convolution Neural Network model and trained it from scratch. Hammoudi et al. [13] trained ResNet34, ResNet50, DenseNet169, VGG-16, and Inception ResNet V2 and RNN on the chest X-ray dataset to detect pneumonia [13]. Out of these models, DenseNet169 produced the best results among the mentioned experiments. El Asnaoui, K. [7] used an ensemble learning-based method using on fine-tuned versions of InceptionResNet_V2, ResNet50, and MobileNet_V2. It achieved an F1 score of 94.84% on the task of classifying chest X-rays images among bacterial, viral, COVID-19, and normal cases [7]. Acharya et al. [18] used Siamese CNN (convolutional neural network) architecture to classify the images into three different classes such as bacterial pneumonia, viral pneumonia and normal cases. M.Toğaçar et al. [37] performed an experiment for the detection of pneumonia based on three different deep learning architectures. The study demonstrated three different deep learning pre-trained architectures (AlexNet, VGG-16 and VGG-19) as feature extractors. The minimum redundancy along with the maximum relevance algorithm was used for each model to reduce the number of features to 100 only. The features were then combined and were feeded to the linear discriminant analysis (LDA) for classification. There are several studies are available in the literature for the detection of pneumonia. However, there is a scope of improvement to the available techniques in terms of accuracy and speed of detection of pneumonia.
Table 1.
Literature survey of the methods for the automatic detection of Pneumonia using chest X-ray
| Sr. No | Reference | Objective | Dataset | Method | Result | Remarks |
|---|---|---|---|---|---|---|
| 1 | Hammoudi et al. (2021) [13] | To detect and evaluate pneumonia (normal, bacterial and viral) cases from chest X-rays | 5863 chest X-ray images [19] | CNN based architectures | Accuracy = 95.72% | Along with detecting pneumonia, the type of pneumonia (bacterial or viral) is also identified |
| 2 | El Asnaoui, K. (2021) [7] | To detect pneumonia (bacterial, viral, COVID-19 and normal) |
i) 5856 chest X-ray images [23] ii) COVID chest X-ray (231 images) |
Ensemble deep learning model | F1 score = 94.84% | Used fine-tuned versions of InceptionResNet_V2, ResNet50 and MobileNet_V2) |
| 3 | Acharya et al. (2019) [18] | To automatically diagnose pneumonia from chest radiography images | 5528 chest X-ray images from Kaggle database [19] | Siamese CNN Architecture |
AUC (Area under the curve) for i) Normal vs. Pneumonia = 0.97 ii)Bacterial vs. Viral Pneumonia = 0.95 |
Along with detecting pneumonia, the type of pneumonia (bacterial or viral) is also identified |
| 4 | M.Toğaçar et al. (2019) [37] | To detect pneumonia automatically | 5,849 chest X-ray images from the Kaggle database [19] | Consolidated deep features, mRMR & SGD (Stochastic Gradient Descent) | Accuracy = 99.41% | 3 different architectures AlexNet, VGG-16 and VGG-19 were used as feature extractors. After applying mRMR, features were concatenated and passed to a classifier |
| 5 | Okeke Stephen et al. (2019) [36] | To detect the presence of pneumonia from chest X-ray | 5856 chest X-ray images [23] | CNN model |
Training accuracy = 95.31% Validation accuracy = 93.73% |
Made a CNN model from scratch to extract features |
| 6 |
Aditya Sriram et al. (2019) [35] |
To use radon projections to classify and represent images | 5840 chest X-ray images from Kaggle database [19] | Radon transform | Accuracy = 70.03% | Performed better than two shallow mlp networks |
| 7 |
Ioannis Livieris et al. (2019) [22] |
To classify lung abnormalities from chest X-ray | 5840 chest X-ray images from Kaggle database [19] | Semi-supervised learning | Accuracy = 83.49% | Proposes weighted voting ensemble self-labeled (WvEnSL) algorithm |
| 8 | Karan Jakhar et al. (2018) [16] | To detect pneumonia automatically from chest X-ray | 5863 chest X-ray images from the Kaggle database [19] | Deep CNN | Accuracy = 84% | Used tenfold cross-validation technique to validate the model |
| 9 | Dimpy Varshni et al. (2018) [39] | To develop an automatic system to detect pneumonia |
A subset of 2,862 X-ray images from ChestX-ray14 dataset released by Wang et al. (2017) [40] |
Pretrained model-based feature extraction and classification using a supervised learning algorithm | AUC = 0.8002 | Transfer learning is used to extract the features and a machine learning classifier is used for the classification |
| 10 | Rajpurkar et al. (2017) [31] | To detect pneumonia from X-rays better than practicing radiologist | 112,120 X-ray images from ChestX-ray14 dataset released by Wang et al. (2017) [40] | 121-layer Dense Convolutional Network |
F1 score of 0.435 (95% CI 0.387, 0.481) |
Extended to detect 14 different kinds of diseases such as atelectasis, edema, hernia, pneumonia, etc |
The objective of this study is to detect pneumonia from the chest X-rays images with improved accuracy, high speed, and less computational power requirement. The related articles that focus on detecting pneumonia from the chest X-rays used the models that are very deep and consist of several convolutional layers. Some of them were able to achieve precise accuracy but they require a lot of computational power and time to perform their computations. Thus, to overcome this limitation, we have trained MobileNet architecture which is very lightweight, fast, and computationally less expensive [14]. A comparison between few pre-trained models in terms of their memory requirement and number of parameters is shown in Table 2. There is always a trade-off between accuracy and computational power, however, in this study, we have tried to achieve a precise accuracy using less computational power. Moreover, the proposed method has also been validated on an external dataset to show that the proposed method works well on other datasets too.
Table 2.
Comparison between sizes and number of parameters of various pre-trained models
| Model | Size | Parameters |
|---|---|---|
| VGG16 | 528 MB | 138,357,544 |
| VGG19 | 549 MB | 143,667,240 |
| ResNet50 | 98 MB | 25,636,712 |
| MobileNet | 16 MB | 4,253,864 |
| DenseNet121 | 33 MB | 8,062,504 |
Materials and methods
Materials
We have used two datasets in our study. The first dataset is used to develop a method to detect pneumonia that includes pre-processing, model selection, and hyperparameter tuning. Another dataset is used to validate this method, i.e., to check if the proposed method will work on another dataset or not.
The dataset used in the study is publicly available on Kaggle [19]. It consists of three folders, namely, training, testing, and validation. All these folders are subdivided into two folders, namely, NORMAL and PNEUMONIA. The training, validation, and testing folders contain 5216, 16, and 624 X-ray images, respectively. This was termed as ‘Dataset-1’, using which a model was developed and hyperparameters were tuned.
We also used an external dataset to validate our model [30]. It consists of three folders, namely, train, test, and validation, which are further subdivided into two folders, namely, Normal and Pneumonia. There were 5,500 images in the training folder, 325 images in the validation folder, and 325 images in the test folder. This dataset was termed ‘Dataset-2’.
Methods
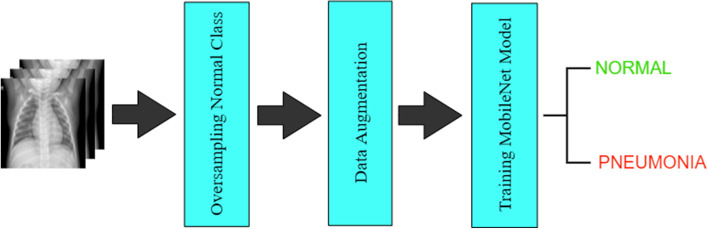
This section describes the applied methodology in detail. This section only uses Dataset-1 to build the model. The proposed pneumonia detection model using the “MobileNet” Architecture is described in Fig. 1. The method for the proposed model includes the following several stages such as the dataset handling stage, the pre-processing and augmentation stage, and the training of the deep learning model. Table 3 demonstrates the list of all layers used in the proposed deep learning architecture.
Fig. 1.
Flow Diagram of the Applied Methodology
Table 3.
Different layers of the proposed Architecture for the automatic detection of pneumonia
| S. No | Layer (type) | Stride | Filter Shape | Output Shape |
|---|---|---|---|---|
| 1 | Input Layer | - | - | (Batch_size, 512, 512, 3) |
| 2 | Conv2D | 2 | (3, 3, 3, 32) | (Batch_size, 256, 256, 32) |
| 3 | DepthwiseConv2D | 1 | (3, 3, 32) | ( Batch_size, 256, 256, 32) |
| 4 | Conv2D | 1 | (1, 1, 32, 64) | ( Batch_size, 256, 256, 64) |
| 5 | DepthwiseConv2D | 2 | (3, 3, 64) | ( Batch_size, 128, 128, 64) |
| 6 | Conv2D | 1 | (1, 1, 64, 128) | ( Batch_size, 128, 128, 128) |
| 7 | DepthwiseConv2D | 1 | (3, 3, 128) | ( Batch_size, 128, 128, 128) |
| 8 | Conv2D | 1 | (1, 1, 128, 128) | ( Batch_size, 128, 128, 128) |
| 9 | DepthwiseConv2D | 2 | (3, 3, 128) | ( Batch_size, 64, 64, 128) |
| 10 | Conv2D | 1 | (1, 1, 128, 256) | ( Batch_size, 64, 64, 256) |
| 11 | DepthwiseConv2D | 1 | (3, 3, 256) | ( Batch_size, 64, 64, 256) |
| 12 | Conv2D | 1 | (1, 1, 256, 256) | ( Batch_size, 64, 64, 256) |
| 13 | DepthwiseConv2D | 2 | (3, 3, 256) | ( Batch_size, 32, 32, 256) |
| 14 | Conv2D | 1 | (1, 1, 256, 512) | ( Batch_size, 32, 32, 512) |
| 15 | DepthwiseConv2D | 1 | (3, 3, 512) | ( Batch_size, 32, 32, 512) |
| 16 | Conv2D | 1 | (1, 1, 512, 512) | ( Batch_size, 32, 32, 512) |
| 17 | DepthwiseConv2D | 1 | (3, 3, 512) | ( Batch_size, 32, 32, 512) |
| 18 | Conv2D | 1 | (1, 1, 512, 512) | ( Batch_size, 32, 32, 512) |
| 19 | DepthwiseConv2D | 1 | (3, 3, 512) | ( Batch_size, 32, 32, 512) |
| 20 | Conv2D | 1 | (1, 1, 512, 512) | ( Batch_size, 32, 32, 512) |
| 21 | DepthwiseConv2D | 1 | (3, 3, 512) | ( Batch_size, 32, 32, 512) |
| 22 | Conv2D | 1 | (1, 1, 512, 512) | ( Batch_size, 32, 32, 512) |
| 23 | DepthwiseConv2D | 1 | (3, 3, 512) | ( Batch_size, 32, 32, 512) |
| 24 | Conv2D | 1 | (1, 1, 512, 512) | ( Batch_size, 32, 32, 512) |
| 25 | DepthwiseConv2D | 2 | (3, 3, 512) | ( Batch_size, 16, 16, 512) |
| 26 | Conv2D | 1 | (1, 1, 512, 1024) | ( Batch_size, 16, 16, 1024) |
| 27 | DepthwiseConv2D | 2 | (3, 3, 1024) | ( Batch_size, 16, 16, 1024) |
| 28 | Conv2D | 1 | (1, 1, 1024, 1024) | ( Batch_size, 16, 16, 1024) |
| 29 | GlobalAveragePooling2D | 1 | Pool (16, 16) | ( Batch_size, 1024) |
| 30 | Sigmoid Classifier | - | Classifier | ( Batch_size, 1) |
|
Total params: 3,229,889 Trainable params: 3,208,001 Non-trainable params: 21,888 | ||||
The Dataset Handling Stage
The training set was highly imbalanced as it contained 3,875 images of the pneumonia class and only 1,341 images of the normal class. To deal with this, the minority (normal) class was oversampled by using data augmentation techniques (rotate and shear) to get 3,876 images of the normal class and a training set of 7,751 images.
The Pre-Processing and Augmentation Stage
The original 3-channel images in the dataset were of different sizes. Since the deep learning models expect all the images to be of the same size, all the images were resized to 512 × 512 pixels. Also, to avoid overfitting and to make the training process easier, training images were augmented and normalized based on their mean and standard deviation. The value of the arguments for the data augmentation is shown in Table 4. Data augmentation was also used to increase the training dataset virtually by including more hypotheses of the object variations in images which will boost the accuracy of the proposed model. In the proposed method, the rotation range was kept as 20 degrees for the inclusion of variations. We did not do any augmentation on the validation and test set images; however, the normalization techniques were used on them.
Table 4.
Parameters used for data augmentation
| Argument | Value |
|---|---|
| Zoom range | 0.1 |
| Rotation range | 20 |
| Sample wise center | True |
| Sample wise std normalization | True |
The boldface entry signifies that MobileNet is the architecture that has been used in this paper
The Training of Deep Learning Model
The MobileNet architecture along with a GlobalAveragePooling2D layer and a dense layer (with sigmoid activation function) was used to train the model. We used MobileNet as its architecture is lightweight and thus, faster to train as compared to other available architectures. This architecture was first proposed by Howard et al. [14]. Instead of performing transfer learning, we trained the model from scratch to get better results.
The architecture of MobileNet is mainly based on depthwise separable convolutions which are faster than the standard convolutions [18]. The convolutional layers consist of many learnable filters and are responsible for performing the convolution operation between the input and the filters. The size of the filters that we have used in our model in the first convolutional layer is 3 × 3. These filters are also called feature detectors. The mathematical convolutional operation between two functions f(t) and g(t) can be defined as in Eq. (1).
| 1 |
The convolution operation is performed between the input and the feature detectors, which results a feature map. The convolutional operation calculates the sum of element-wise multiplication of the input and the filter, and returns a scalar. This operation is continued by sliding the filter over the input with a stride. If the size of the input is , the numbers of filters used are , the stride is s, the padding is p, and the size of the filters is , then the size of the output of the convolutional layer is , where is the floor of . In the proposed model, after the input layer, a convolutional layer is used. The size of the input image is , the number of filters used is 32, the size of the filters is , padding is 1 and the stride value is 2. Consequently, it results in an output of shape . For one convolution operation, the number of multiplications performed is the number of elements in the input, i.e., . Since the filter is slid over the input, the number of multiplications per filter for padding of 0 and a stride of 1 becomes . Total number of multiplications using number of filters in a standard convolution is in Eq. (2).
| 2 |
This standard convolutional operation is computationally expensive and slow. However, this process can speed up by using depth-wise separable convolution.
The depth-wise separable convolution involves two stages: Depth-wise Convolution and Point-wise Convolution. In the depth-wise convolution, the convolution operation is applied to a single input channel at a time. This is not the case with standard convolution as it applies convolution to all the channels. The filters used at this stage have a shape of and since each channel requires a filter, a total of filters are used. For a single filter, the output shape is for a stride of s. When the outputs of all the filters are stacked together, the resulting output shape becomes . Depth-wise convolution is performed in our model using the DepthwiseConv2D layer of the TensorFlow framework. The 1st DepthwiseConv2D layer in our model has an input of shape , a filter size of , padding of 1, and a stride value of 1. Consequently, it results in an output of shape . This is the end of the first stage and from here, the point-wise convolution begins. The output of the depth-wise convolution is the input for point-wise convolution. This stage performs the linear combination of each of the layers. In this stage, the filters of shape are used and hence, convolution is performed over all the layers. If the number of filters used is , the output size is Point-wise convolution is performed in our model using the Conv2D layer of the TensorFlow framework, where the value of f is kept 1 and padding is set to ‘same’ (). In the proposed model, for the 1st convolutional layer in which the point-wise convolution is performed, the input shape is , the filter size is , the number of filters used is 64, the padding value is 0, and the stride value is 1. It results in an output of . The total number of multiplications performed in a depth-wise separable convolution can be calculated by adding together the number of multiplications performed in the depth-wise convolution stage and the point-wise convolution stage. In the depth-wise convolution stage, for a padding of 0 and a stride of 1, the number of multiplications performed is and in the pointwise convolution stage, the number of multiplications performed when using filters is . Thus, the total number of multiplications using number of filters in a depth-wise separable convolution is:
| 3 |
The ratio of the number of multiplications performed in a depth-wise separable convolution to the number of multiplications in a standard convolution is [18]:
| 4 |
Equation (4) confirms that the number of multiplications and hence, computations performed in a depth-wise separable convolution is less than that in a standard convolution. Due to this property of depth-wise separable convolutions, the MobileNet architecture is faster as compared to other deep learning models.
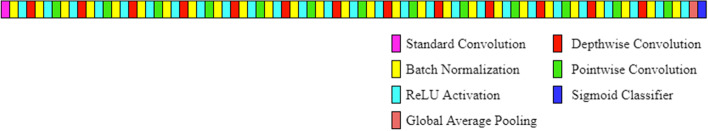
The final model used is defined in Table 3. The first element of the output shape, i.e., the batch size was set to 1. All layers are followed by a batch norm and ReLU (Rectified Linear Unit) nonlinearity except for the final GlobalAveragePooling2D layer which feeds directly into the sigmoid classifier. All the layers of the model are shown in Fig. 2. Batch normalization contributes to speeding up the training of neural networks by normalizing the output of a previous activation layer [15]. It speeds up training by smoothing the optimization landscape in a significant amount. Because of this smoothness, a more predictive and stable behavior of the gradients is induced, which allows faster training [32]. The equations used in batch normalization are shown through Eq. (5)-(8).
| 5 |
| 6 |
| 7 |
| 8 |
Fig. 2.
Proposed Architecture for the Automatic Detection of Pneumonia
Normalization is done by subtracting the batch mean () and dividing by the batch standard deviation (). In Eq. (7), (a small constant) is added in the denominator to avoid division by zero variance. However, after performing this normalization, the weights in the next layer are not optimal [3]. Stochastic gradient descent optimizer undoes this normalization if it’s a way for it to reduce the loss function. So, two trainable parameters are added by batch normalization to each layer, so that the normalized output () is multiplied by a “standard deviation” parameter () and add a “mean” parameter (). Because of these parameters, the stochastic gradient descent optimizer used in the model can do the denormalization by changing only and for each activation. ReLU (Rectified Linear Unit) activation is defined for an input x in Eq. (9) and is zero for a value less than zero. For values greater than or equal to zero, the ReLU activation is the value itself [28].
| 9 |
When an input of shape is passed to a global average pooling layer, it calculates the average of each activation map and returns that average at the output. Since we have number of activation maps, the output shape is . This output is then passed to a sigmoid layer which outputs the probability that the input image belongs to the pneumonia class. If the probability is less than 0.5, the normal class is predicted and if it is greater than or equal to 0.5, the pneumonia class is predicted.
The model was trained with Stochastic Gradient Descent as the optimizer. The objective of the optimizers is to keep updating the weights at every layer until the best learning of parameters in CNN (convolutional neural network) is realized. In the Stochastic Gradient Descent (SGD) method, the weights are updated for every single training set [7]. The formula for SGD optimizer is given in Eq. (10). Here, is the vector of weights to be updated, is the learning rate and is the loss function.
| 10 |
We set the number of epochs to 10 and used ModelCheckpoint callback to save the model’s weights for which the validation accuracy was maximum. The learning rate () was initially set to 0.1 and Exponential Decay was used as the Learning Rate Scheduler. The Learning Rate Scheduler allows reducing the learning rate as the number of epochs increases, which helps train deep learning models. The arguments decay steps, decay rate, and staircase of exponential decay were set to 1,00,000, 0.96, and true respectively. Here, the learning rate is decayed by following a staircase function. Binary Cross Entropy was used as the loss function as given in Eq. (11):
| 11 |
where N is the number of examples, is the actual label, and is the prediction made by the model. The model tries to minimize this loss. The model was trained using GPU (graphics processing unit) on Google Colab for approximately 3 h and we got a training accuracy of 97.34%, a validation accuracy of 87.5%, and a testing accuracy of 94.23%.
Results
To evaluate a model’s performance, we cannot depend on the accuracy only. Various other metrics along with accuracy, were also derived from the confusion matrix to measure the model’s performance, such as specificity, precision, recall, and F1-score. The formulae to compute these metrics are shown through Eq. (12)-(16)
| 12 |
| 13 |
| 14 |
| 15 |
| 16 |
where TP, TN, FP, and FN are represented as true positive, true negative, false positive, and false negative respectively. The specificity answers the question that if the actual label for a chest x-ray is “normal”, how often the model predicts it to be “normal”. Precision tells how often the prediction is correct if the predicted label is “pneumonia”. Recall tells us that if the actual label is “pneumonia”, how often the model is correct. Since there is a trade-off between precision and recall, we have also used the F1-score, which is the harmonic mean of precision and recall. The maximum possible value of specificity, precision, recall, and F1-score is 1. This section displays the results obtained for dataset-1 after training the model, the results obtained for dataset-2 without training the model, and the results obtained for dataset-2 after training the model.
Results on Dataset-1 after training the model on a training set of Dataset-1
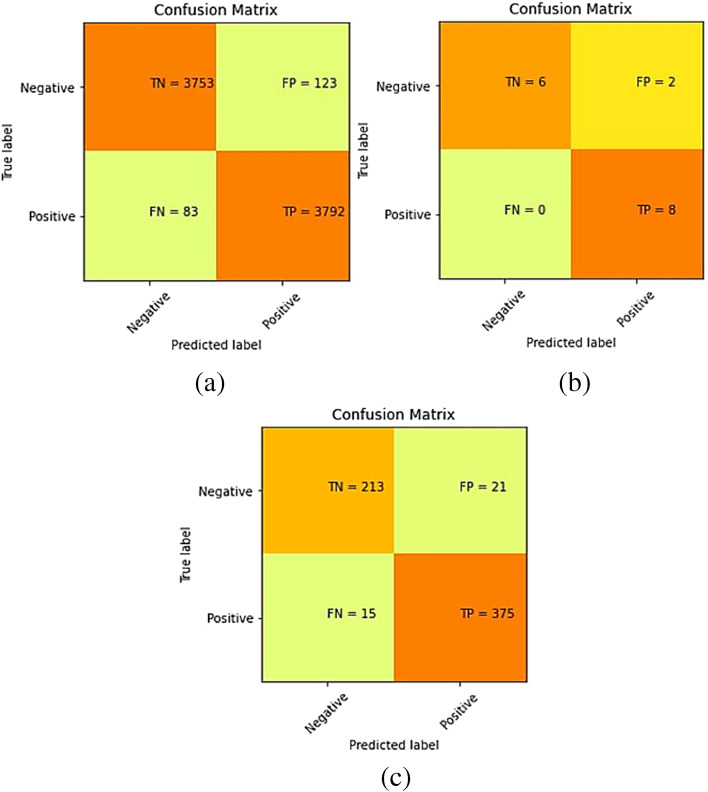
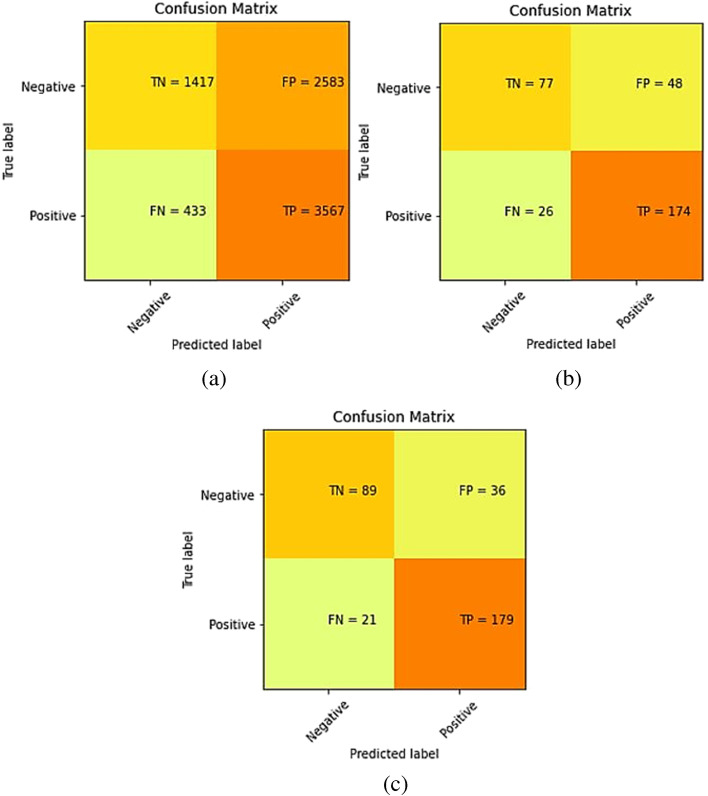
The confusion matrices for the predictions made by our model on training, validation, and test sets of dataset-1 are shown in Fig. 3.
Fig. 3.
Confusion matrix for Dataset-1 after training the model on training set of Dataset-1 (a) Training set predictions, (b) Validation set predictions, and (c) Test set predictions
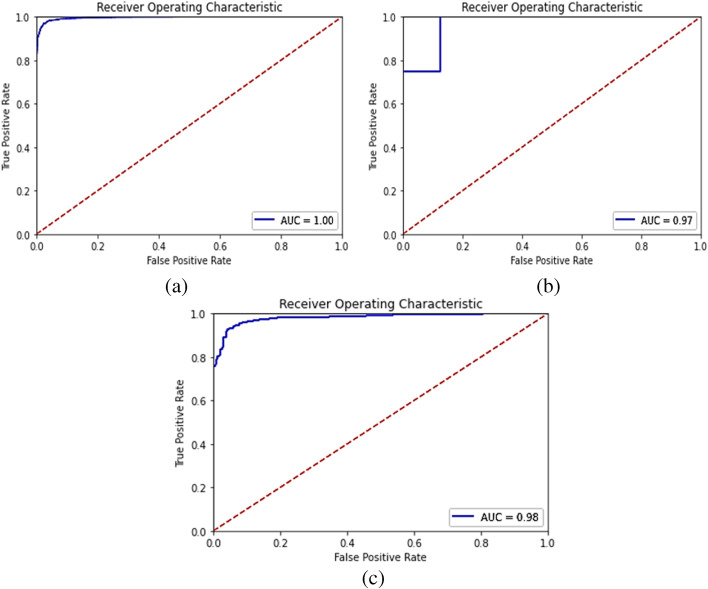
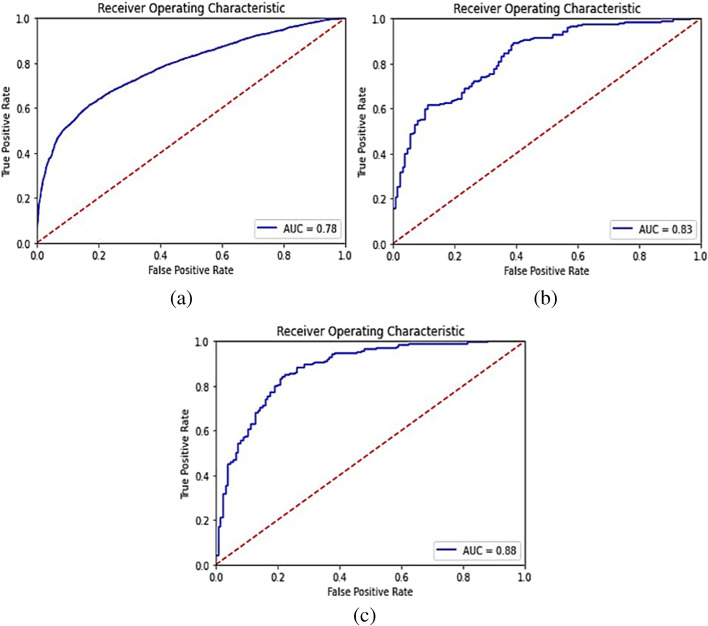
The accuracy, specificity, precision, recall, and F1-score for the proposed model were computed separately for training, validation, and test sets of dataset-1, and shown in Table 5. Also, it demonstrates the combined metrics for the same model. The ROC (receiver operating characteristic) curves is shown in Fig. 4. The model was trained using GPU (graphics processing unit) on Google Colab for approximately 3 h and we got a training accuracy of 97.34%, a validation accuracy of 87.5%, and a testing accuracy of 94.23%. However, the combined accuracy was achieved as 97.09% with 0.96 specificity, 0.97 precision, 0.98 recall, and 0.97 F-Score.
Table 5.
Accuracy, specificity, precision, recall, and F1-score metrics for our model obtained on Dataset-1 after training the model on a training set of Dataset-1
| Metrics | Training | Validation | Test | Combined |
|---|---|---|---|---|
| Accuracy (%) | 97.34 | 87.50 | 94.23 | 97.09 |
| Specificity | 0.97 | 0.75 | 0.91 | 0.96 |
| Precision | 0.97 | 0.80 | 0.95 | 0.97 |
| Recall | 0.98 | 1.00 | 0.96 | 0.98 |
| F1-score | 0.97 | 0.89 | 0.95 | 0.97 |
Fig. 4.
ROC curves for Dataset-1 after training the model on training set of Dataset-1 (a) Training set predictions, (b) Validation set predictions, and (c) Test set predictions
Results on Dataset-2 after training the model on training set of Dataset-1
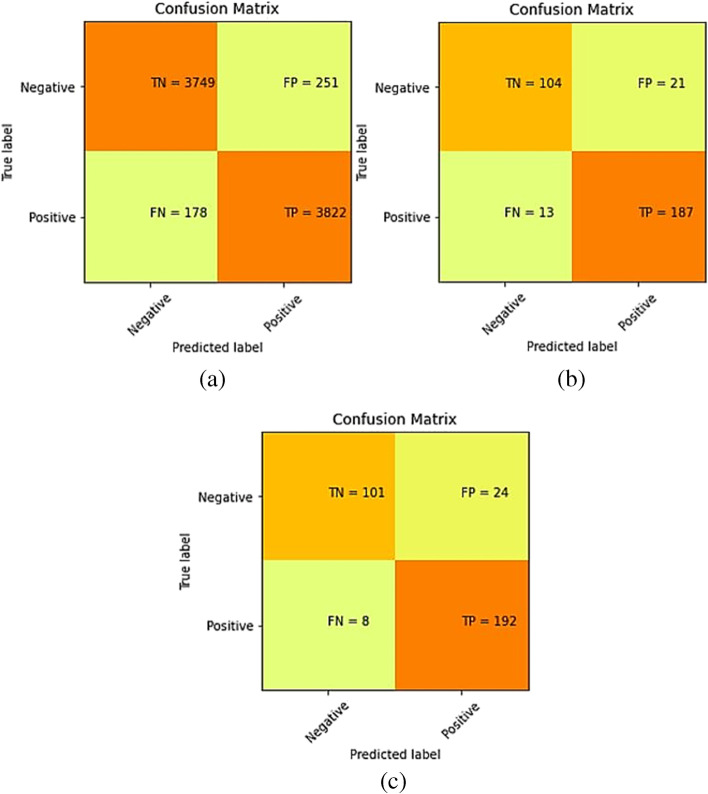
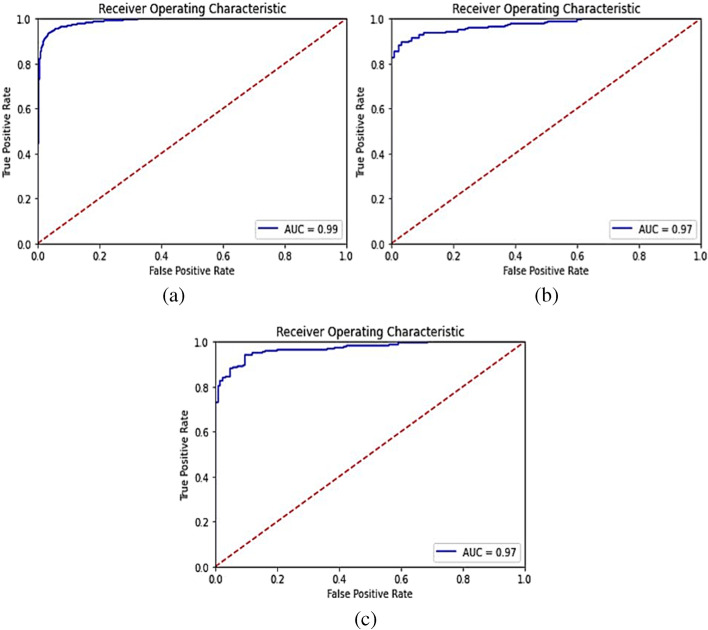
To see that if our proposed model performs well on other chest X-ray datasets too, we used an external dataset (Dataset-2) to validate our model [30]. This dataset has 4,000 images of the pneumonia class and 1,500 images of the normal class in its training set. We applied the same methods (oversampling normal class, data augmentation) on this dataset as we did on Dataset-1, and increased the number of images in the normal class to 4,000. A pre-trained model (as trained on Dataset-1) was used to validate on the training, validation and test image set of Dataset-2. Section 3.2 demonstrates the results when the proposed model was trained only on the training set images of Dataset-1 and validated on all image sets of Dataset-2. The confusion matrices are shown in Fig. 5. The derived metrics from these confusion matrices are shown in Table 6 and the ROC curves are shown in Fig. 6.
Fig. 5.
Confusion matrix for Dataset-2 after training the model on training set of Dataset-1 (a) Training set predictions, (b) Validation set predictions and (c) Test set predictions
Table 6.
Accuracy, specificity, precision, recall and F1-score metrics for our model when validated on Dataset-2 after training the model on training set of Dataset-1
| Metrics | Training | Validation | Test | Combined |
|---|---|---|---|---|
| Accuracy (%) | 62.30 | 77.23 | 82.46 | 63.69 |
| Specificity | 0.35 | 0.62 | 0.71 | 0.37 |
| Precision | 0.58 | 0.78 | 0.83 | 0.59 |
| Recall | 0.89 | 0.87 | 0.89 | 0.89 |
| F1-score | 0.70 | 0.82 | 0.86 | 0.71 |
Fig. 6.
ROC curves for Dataset-2 after training the model on training set of Dataset-1 (a) Training set predictions, (b) Validation set predictions, and (c) Test set predictions
Results on Dataset-2 after training the model on training set of Dataset-2
There was a huge drop in the model’s performance while making predictions on an external dataset using the pre-trained weights. This was because both the datasets (Dataset-1 and Dataset-2) were of entirely different distributions, i.e., taken at different places using different equipment and of people of different age groups. So, we trained the same model (without changing any hyper-parameter) on the external dataset and achieved improved performance. The confusion matrices for the predictions made by our model on the training, validation, and test sets of the external dataset after training are shown in Fig. 7 and other derived metrics from these confusion matrices are shown in Table 7. The ROC curves are shown in Fig. 8.
Fig. 7.
Confusion matrix for Dataset-2 after training the model on training set of Dataset-2 (a) Training set predictions, (b) Validation set predictions, and (c) Test set predictions
Table 7.
Accuracy, specificity, precision, recall, and F1-score metrics for our model when validated on Dataset-2 after training the model on training set of Dataset-2
| Metrics | Training | Validation | Test | Combined |
|---|---|---|---|---|
| Accuracy (%) | 94.63 | 89.53 | 90.15 | 94.31 |
| Specificity | 0.94 | 0.83 | 0.81 | 0.93 |
| Precision | 0.94 | 0.89 | 0.88 | 0.93 |
| Recall | 0.95 | 0.93 | 0.96 | 0.95 |
| F1-score | 0.94 | 0.91 | 0.92 | 0.94 |
Fig. 8.
ROC curves for Dataset-2 after training the model on training set of Dataset-2 (a) Training set predictions, (b) Validation set predictions, and (c) Test set predictions
A comparison of the performance of the model on Dataset-1 (using which its hyper-parameters were tuned) and the external dataset (with and without training) is shown in Table 8. It is evident from Table 8 that our model might not work well on external datasets if they are of different distribution (acquired by different X-ray machines and of people with different age groups and geographical locations) as compared to the dataset using which it was trained. However, when trained on a dataset having similar distribution as the chest x-rays on which the predictions are to be made, our model predicts pneumonia cases with high accuracy. It is recommendable that training should be completed on the similar dataset on which testing is desired. Here, similar dataset means similarity of data acquisition through the X-ray machine, age group of the patients, and geographical location of the patients, etc. However, it is not always mandatory to have the similarity in all these conditions. But, more similarity in these conditions shall improve the accuracy of the detection of pneumonia.
Table 8.
Comparison of performance of the proposed model on the two datasets
| Performance of proposed model on Dataset-1 after training the model on training set of Dataset-1 |
Performance of proposed model on Dataset-2 (external dataset) after training the model on training set of Dataset-1 |
Performance of proposed model on Dataset-2 (external dataset) after training the model on training set of Dataset-2 |
|
|---|---|---|---|
| Test set accuracy (%) | 94.23 | 82.46 | 90.15 |
| Test set specificity | 0.91 | 0.71 | 0.81 |
| Test set precision | 0.95 | 0.83 | 0.88 |
| Test set recall | 0.96 | 0.89 | 0.96 |
| Test set F1-score | 0.95 | 0.86 | 0.92 |
Discussion
Numerous studies in the field of deep learning have been published that focus on the automatic detection of pneumonia. The deep learning techniques are well established and can be used for solving multiple applications in the medical domain [1, 2, 12] as well as in the non-medical domain [38]. Similarly, image processing techniques are also available to solve similar kinds of problems to design an automation system using medical images [10, 11, 27]. There are many advanced schemes to access the relevant data from different sources [17, 24–26].
Livieris et al. [22] proposed a study that introduced a new ensemble semi-supervised learning algorithm based on a new weighted voting scheme to classify abnormalities of lungs from chest X-rays. The pneumonia classification accuracy achieved by their algorithm was 83.49%. However, the same dataset [19] was used in our experiment also, and achieved better accuracy from their experiment. It was observed that Livieris et al.[22] did not mentioned any method to deal with the problem of class imbalance. Therefore, their model did not achieved precise accuracy in detecting pneumonia. Their accuracy may improve if they oversample the minority (normal) class using data augmentation or any other technique to make the number of examples of normal and pneumonia classes equal. However, oversampling is used in our study to avoid the problem of class imbalance.
Jakhar et al. [16] used deep convolutional neural networks (DCNN) for detecting pneumonia from chest X-ray images. They used the dataset from Kaggle [19] which is the same data as we have used. They tried different classifiers with DCNN to perform the classification task but the best results were obtained by using the DCNN model. They used Synthetic Minority Over-sampling Technique (SMOTE) to solve the problem of the imbalanced dataset. They performed K-fold cross-validation (K = 10) and achieved an accuracy of 84%. Our model performs better than their model because they didn’t use data augmentation which prevents overfitting and helps in boosting test accuracy.
Toğaçar et al. [37] had used three pre-trained convolutional neural network models- AlexNet, VGG-16, and VGG-19 for the task of detecting pneumonia from chest X-rays. The features extracted by each model were then reduced to 100 by using the maximum redundancy theorem and concatenated to feed into the classifier. They used the same dataset as ours and achieved a testing accuracy of 98.21%, which is higher than that achieved by our model. This may be because they have used three models to extract features, each of which has a lot more parameters as compared to our model and hence, extract more information. However, their method is computationally very expensive compared to our model. Therefore, our model is recommended in such applications which are already very expensive and need to reduce the significant time. Our model is also helpful in cases where resources are very limited to use.
Stephen et al. [36] made a convolutional neural network from scratch and trained it for the task of pneumonia detection from chest X-ray. They used the same dataset as ours and rearranged it to get 3,722 images in the training set and 2,134 images in the test set. They achieved a training accuracy of 95.31% and a testing accuracy of 93.73%. We achieved a training accuracy of 97.34%, a validation accuracy of 87.5%, and a testing accuracy of 94.23% which is significantly higher than that achieved by the model proposed by Stephen et al. [36]. Stephen et al. [36] had also used data augmentation which helped in achieving higher testing accuracy. The possible reason for a lower training accuracy may be observed that they had used a dropout of 0.5 before the dense_5 layer, it can be interpreted that the drop of the input units of the dense_5 layer is half. However, the dropout also contributed to preventing overfitting and achieving a decent testing accuracy.
The facts that MobileNet architecture is based on depthwise separable convolution and uses batch normalization after every layer contribute significantly to its high speed and less computational power requirement. Other deep learning architectures that are used in other studies are based on standard convolution, which needs to perform a greater number of multiplications as compared to depthwise separable convolution and hence, is computationally more expensive than our proposed model. The model architecture used by Stephen et al. [36] is purely based on standard convolution. Also, it does not include a batch normalization layer and thus, is slower than our model.
We have proposed a model which can detect pneumonia from chest X-rays with high accuracy. Furthermore, our model performs well on other metrics (specificity, precision, recall, and F1-score) too. The metric on which our model performed the lowest, is specificity but there is a trade-off between sensitivity (also called recall) and specificity and for this particular problem of detecting pneumonia, a higher sensitivity is more important than a higher specificity. This is because failure in detecting the presence of pneumonia is more dangerous than incorrectly diagnosed with pneumonia. Moreover, since we have used data augmentation techniques, our model is quite robust. Additionally, as we have used MobileNet architecture, our model is computationally less expensive and faster than those which use architectures like VGG-16, DenseNet-121, Inception, etc. We have also validated our model on an external dataset using pre-trained model’s weights, which showed that our model might not work well on external datasets if they are of different distribution (acquired by different x-ray machines and of people with different age groups and geographical locations) as compared to the dataset using which it was trained. However, when trained on a dataset having similar distribution as the chest x-rays on which the predictions are to be made, our model predicts pneumonia with high accuracy. Thus, the proposed model can be used without any hyperparameter tuning to predict pneumonia in external datasets after training. Furthermore, the proposed model can also be applicable for applications where a number of parameters/variables are very high and want to reduce the training expenses.
Conclusion
We have proposed a fast and computationally less expensive method for automatic detection of pneumonia. The proposed method is based on MobileNet architecture which is a lightweight but achieves a promising accuracy compared to the methods in the literature. This model can be used to predict pneumonia using chest x-rays with high accuracy by training it on a dataset with similar distribution as the data on which the predictions are to be made.
In the future, the proposed model can be trained using a large dataset that contains chest X-ray images of people belonging to different geographical locations and age groups. Training on such a dataset will be useful in using this model for a large-scale purpose. Moreover, since the model is very fast and computationally less expensive, it can also be deployed in a browser or an app.
Data availability
The benchmarked dataset is used and the respective source has been provided.
Code availability
Code can be made available as per request.
Declarations
Ethics approval
Not Applicable.
Consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Conflicts of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Megha Trivedi, Email: meghat525@gmail.com.
Abhishek Gupta, Email: abhishekgupta10@yahoo.co.in.
References
- 1.Ashok M, Gupta A. A Systematic Review of the Techniques for the Automatic Segmentation of Organs-at-Risk in Thoracic Computed Tomography Images. Arch Comput Methods Eng. 2021;28:3245–3267. doi: 10.1007/s11831-020-09497-z. [DOI] [Google Scholar]
- 2.Ashok M, Gupta A (2021) Deep learning-based techniques for the automatic segmentation of organs in thoracic computed tomography images: A Comparative study. in Proceedings - International Conference on Artificial Intelligence and Smart Systems, ICAIS 2021, 198-202
- 3.Batch normalization in Neural Networks. Towards Data Science. Available: https://towardsdatascience.com/batch-normalization-in-neural-networks-1ac91516821c. [Accessed 14th September, 2020].
- 4.Budak Ü, Cömert Z, Çıbuk M, Şengür A. DCCMED-Net: Densely connected and concatenated multi Encoder-Decoder CNNs for retinal vessel extraction from fundus images. Medical Hypotheses. 2020;134:109426. doi: 10.1016/j.mehy.2019.109426. [DOI] [PubMed] [Google Scholar]
- 5.Cömert Z, Kocamaz AF (2019) Fetal Hypoxia Detection Based on Deep Convolutional Neural Network with Transfer Learning Approach, in Software Engineering and Algorithms in Intelligent Systems, Cham, 239–248
- 6.Del Fiol G, Michelson M, Iorio A, Cotoi C, Haynes RB. A Deep Learning Method to Automatically Identify Reports of Scientifically Rigorous Clinical Research from the Biomedical Literature: Comparative Analytic Study,". J Med Internet Res. 2018;20:e10281. doi: 10.2196/10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Asnaoui K. Design ensemble deep learning model for pneumonia disease classification. Int J Multimed Inf Retr. 2021;10:55–68. doi: 10.1007/s13735-021-00204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta A (2019) Current research opportunities of image processing and computer vision. Comput Sci 20(4) 10.7494/csci.2019.20.4.3163
- 9.Gupta A. Challenges for Computer Aided Diagnostics using X-Ray and Tomographic Reconstruction Images in craniofacial applications. Int J Comput Vis Robot. 2020;10:360–371. doi: 10.1504/IJCVR.2020.108153. [DOI] [Google Scholar]
- 10.Gupta A, Kharbanda O, Sardana V, Balachandran R, Sardana H (2015) A knowledge-based algorithm for automatic detection of cephalometric landmarks on CBCT images. Int J Comput Assist Radiol Surg 10:1737–1752 [DOI] [PubMed]
- 11.Gupta A, Kharbanda OP, Sardana V, Balachandran R, Sardana HK. Accuracy of 3D cephalometric measurements based on an automatic knowledge-based landmark detection algorithm. Int J Comput Assist Radiol Surg. 2016;11:1297–1309. doi: 10.1007/s11548-015-1334-7. [DOI] [PubMed] [Google Scholar]
- 12.Gupta RK, Sahu Y, Kunhare N, Gupta A, Prakash D (2021) Deep Learning-based Mathematical Model for Feature Extraction to Detect Corona Virus Disease using Chest X-Ray Images. International Journal of Uncertainty, Fuzziness and Knowledge-Based Systems (in-press)
- 13.Hammoudi K, Benhabiles H, Melkemi M, Dornaika F, Arganda-Carreras I, Collard D, et al. Deep Learning on Chest X-ray Images to Detect and Evaluate Pneumonia Cases at the Era of COVID-19. Journal of medical systems. 2021;45:75–75. doi: 10.1007/s10916-021-01745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T et al (2017) MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv
- 15.Ioffe S, Szegedy C (2015) Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift," presented at the Proceedings of the 32nd International Conference on Machine Learning, Proceedings of Machine Learning Research
- 16.Jakhar K, Hooda N (2018) Big Data Deep Learning Framework using Keras: A Case Study of Pneumonia Prediction. in 2018 4th International Conference on Computing Communication and Automation (ICCCA) 1–5
- 17.Javid T, Gupta MK, Gupta A (2020) A hybrid-security model for privacy-enhanced distributed data mining. Journal of King Saud University - Computer and Information Sciences
- 18.K AA, R S (2020) A Deep Learning Based Approach towards the Automatic Diagnosis of Pneumonia from Chest Radio-Graphs. Biomed Pharmacol J 13
- 19.Kermany D, Zhang K, Goldbaum M (2018) Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification. Mendeley Data V2
- 20.Kolditz M, Ewig S. Community-Acquired Pneumonia in Adults. Dtsch Arztebl International. 2017;114:838–848. doi: 10.3238/arztebl.2017.0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavine M. The Early Clinical X-Ray in the United States: Patient Experiences and Public Perceptions. J Hist Med Allied Sci. 2011;67:587–625. doi: 10.1093/jhmas/jrr047. [DOI] [PubMed] [Google Scholar]
- 22.Livieris IE, Kanavos A, Tampakas V, Pintelas P. A Weighted Voting Ensemble Self-Labeled Algorithm for the Detection of Lung Abnormalities from X-Rays. Algorithms. 2019;12:64. doi: 10.3390/a12030064. [DOI] [Google Scholar]
- 23.Melendez J, Ginneken BV, Maduskar P, Philipsen RHHM, Reither K, Breuninger M, et al. A Novel Multiple-Instance Learning-Based Approach to Computer-Aided Detection of Tuberculosis on Chest X-Rays. IEEE Trans Med Imaging. 2015;34:179–192. doi: 10.1109/TMI.2014.2350539. [DOI] [PubMed] [Google Scholar]
- 24.Namasudra S (2020) Fast and Secure Data Accessing by using DNA Computing for the Cloud Environment. IEEE Transactions on Services Computing, pp. 1–1
- 25.Namasudra S, Chakraborty R, Majumder A, Moparthi NR. Securing Multimedia by Using DNA-Based Encryption in the Cloud Computing Environment. ACM Trans Multimedia Comput Commun Appl. 2020;16:99. [Google Scholar]
- 26.Namasudra S, Roy P (2017) Time saving protocol for data accessing in cloud computing. IET Communications 11(10):1558–1565. https://digital-library.theiet.org/content/journals/10.1049/iet-com.2016.0777
- 27.Neelapu BC, Kharbanda OP, Sardana V, Gupta A, Vasamsetti S, Balachandran R, et al. Automatic localization of three-dimensional cephalometric landmarks on CBCT images by extracting symmetry features of the skull. Dentomaxillofac Radiol. 2018;47:20170054. doi: 10.1259/dmfr.20170054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nwankpa C, Ijomah W, Gachagan A, Marshall S (2018) Activation functions: Comparison of trends in practice and research for deep learning. arXiv preprint arXiv:1811.03378
- 29.Pletz M, Rohde G, Welte T, Kolditz M, Ott S (2016) Advances in the prevention, management, and treatment of community-acquired pneumonia [version 1; peer review: 2 approved]. F1000Research 5 [DOI] [PMC free article] [PubMed]
- 30.Pneumonia Detection (2020) Kaggle. Available: https://www.kaggle.com/mirasel/pneumonia-detection. [Accessed 16th October, 2020]
- 31.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, et al (2017) CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. ArXiv, abs/1711.05225
- 32.Santurkar S, Tsipras D, Ilyas A, Madry A (2018) How Does Batch Normalization Help Optimization?
- 33.Saraiva AA, Ferreira NMF, d. Sousa LL, Costa NJC, Sousa JVM, Santos DBS, et al (2019) Classification of Images of Childhood Pneumonia using Convolutional Neural Networks. BIOIMAGING
- 34.Shen Y, Tian Z, Lu D, Huang J, Zhang Z, Li X, et al. Impact of pneumonia and lung cancer on mortality of women with hypertension. Sci Rep. 2016;6:20. doi: 10.1038/s41598-016-0023-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sriram A, Kalra S, Tizhoosh HR (2019) Projectron -- A Shallow and Interpretable Network for Classifying Medical Images. arXiv
- 36.Stephen O, Sain M, Maduh UJ, Jeong D-U. An Efficient Deep Learning Approach to Pneumonia Classification in Healthcare. J Healthc Eng. 2019;2019:4180949. doi: 10.1155/2019/4180949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toğaçar M, Ergen B, Cömert Z, Özyurt F (2019) A Deep Feature Learning Model for Pneumonia Detection Applying a Combination of mRMR Feature Selection and Machine Learning Models. IRBM
- 38.M. Trivedi and A. Gupta, "Automatic monitoring of the growth of plants using deep learning-based leaf segmentation," International Journal of Applied Science and Engineering, vol. 18, pp. 1–9, 2021/06/01 2021.
- 39.Varshni D, Thakral K, Agarwal L, Nijhawan R, Mittal A (2019) Pneumonia Detection Using CNN based Feature Extraction. in 2019 IEEE International Conference on Electrical, Computer and Communication Technologies (ICECCT), 1–7
- 40.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers R (2017) ChestX-ray14: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The benchmarked dataset is used and the respective source has been provided.
Code can be made available as per request.