Abstract
Candida dubliniensis is an opportunistic fungal pathogen that has been linked to oral candidiasis in AIDS patients, although it has recently been isolated from other body sites. DNA sequence analysis of the internal transcribed spacer 2 (ITS2) region of rRNA genes from reference Candida strains was used to develop molecular beacon probes for rapid, high-fidelity identification of C. dubliniensis as well as C. albicans. Molecular beacons are small nucleic acid hairpin probes that brightly fluoresce when they are bound to their targets and have a significant advantage over conventional nucleic acid probes because they exhibit a higher degree of specificity with better signal-to-noise ratios. When applied to an unknown collection of 23 strains that largely contained C. albicans and a smaller amount of C. dubliniensis, the species-specific probes were 100% accurate in identifying both species following PCR amplification of the ITS2 region. The results obtained with the molecular beacons were independently verified by random amplified polymorphic DNA analysis-based genotyping and by restriction enzyme analysis with enzymes BsmAI and NspBII, which cleave recognition sequences within the ITS2 regions of C. dubliniensis and C. albicans, respectively. Molecular beacons are promising new probes for the rapid detection of Candida species.
Candida dubliniensis is a newly recognized opportunistic pathogen that has been linked to oral candidiasis in human immunodeficiency virus (HIV)-infected patients (16, 25, 45, 47), although it has also been observed in blood isolates from bone marrow transplant patients and oral and vaginal isolates from non-HIV-infected patients (26, 29, 35). C. dubliniensis was initially difficult to distinguish from other Candida species in standard clinical laboratory tests because of its closely shared phenotypic and genotypic characteristics with C. albicans (45, 46). However, more recently, phenotypic characteristics, including carbon assimilation (7, 13, 33, 39), growth temperature (34), immunofluorescence (3), and DNA-based molecular approaches (7–9, 11, 15, 18, 48) have been used to distinguish C. dubliniensis from other Candida species. C. dubliniensis is largely susceptible to existing antifungal agents (30), although it can rapidly develop in vitro fluconazole resistance (29). A small percentage of isolates with fluconazole resistance have been reported (29, 30), with some isolates expressing common classes of multidrug transporters (28). Ultimately, this propensity may present a problem in the oral cavity, where other resistant Candida species from HIV-infected patients are frequently encountered (1, 19, 29, 52). Given the growing recognition of C. dubliniensis as an opportunistic pathogen of immunosuppressed patients, a rapid and reliable method for the identification of this non-C. albicans species is an important clinical goal for proper disease management.
The internal transcribed spacer 2 (ITS2) is a spacer region flanked by the 5.8S and 28S rRNA genes and has been used to identify other clinically important fungi such as Pneumocystis, Aspergillus, and Cryptococcus spp. (11, 20–22, 36, 49). The ITS2 region can be amplified with universal fungal primers ITS3 and ITS4 specific for conserved sequences in the ends of the 5.8 and 28S rRNA genes (22). Use of the ITS2 region for species identification requires either direct sequence analysis, which is highly accurate but time-consuming, or detection with sequence-specific hybridization probes. However, the use of linear probes for detection, whether amplified or not, can pose problems of sensitivity and false-positive results, depending on the probe sequence and hybridization conditions (14). Recently, molecular beacons were introduced to overcome these limitations (51).
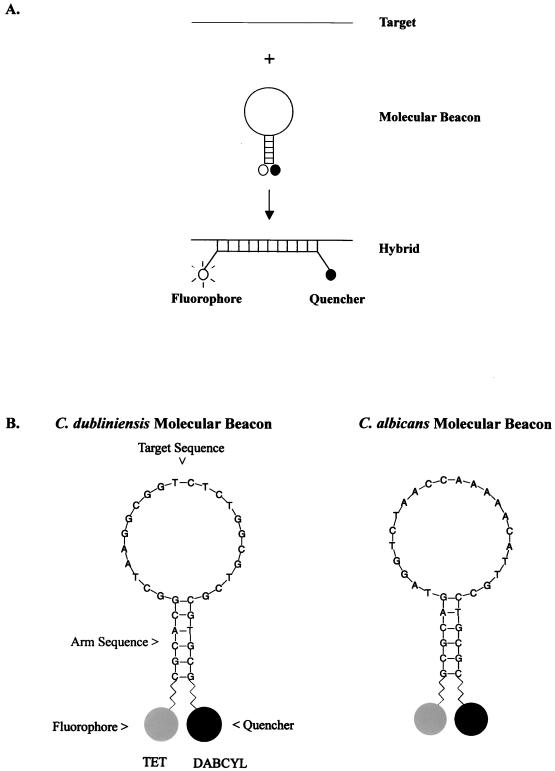
Molecular beacons are small, single-stranded nucleic acid hairpin probes that brightly fluoresce when they are bound to their targets (50, 51). They possess a loop-and-stem structure in which the loop contains the complementary target sequence and the stem forms by the annealing of short complementary nucleotide sequence arms adjacent to the target sequence (Fig. 1A). A fluorophore is covalently linked to the end of the stem sequence, and a quencher is covalently linked to the other end. In free solution, molecular beacons do not fluoresce because the stem structure keeps the fluorophore close to the quencher and the fluorescence energy is absorbed and released as heat. However, in the presence of target DNA, the loop sequence anneals to the target, a probe-target hybrid is formed, forcing the stems that contain the fluorophore and quencher to disassociate, and fluorescence occurs. Molecular beacons have a significant advantage over conventional nucleic acid probes because of their fidelity and ability for allele discrimination (24, 50). This property has recently been exploited for the detection of single-nucleotide base pair changes within the rpoB gene of Mycobacterium tuberculosis, which confers resistance to the antibiotic rifampin (31, 32), and for the detection of allelic differences in the human β-chemokine receptor 5 (CCR5) gene (17). In addition, molecular beacons have been used to rapidly detect and quantitate four retroviruses responsible for AIDS and T-cell lymphoma/leukemia (53).
FIG. 1.
(A) Molecular beacon consists of a stem-loop structure with a fluorophore and a quencher bound to the ends of the probe. In free solution, these probes are nonfluorescent because the stem hybrid keeps the fluorophore close to the quencher. When the probe sequence in the loop hybridizes to its target, forming a rigid double helix, a conformational reorganization occurs that separates the quencher from the fluorophore, restoring fluorescence. The figure is adapted from Tyagi and Kramer (51). (B) Nucleotide sequence of the Candida species-specific molecular beacons. The 22-nucleotide target sequence is complementary to the ITS2 region of each Candida species. The fluorophore tetrachloro-6-fluorescein (TET) was attached to the sulfhydryl group on the 5′ arm sequence and 4-(4′-dimethylaminophenylazo)benzoic acid (DABCYL), a quencher, was attached to an amino group on the 3′ arm sequence to form the stem region of the molecular beacon.
In this report, we describe a method for the rapid identification of C. dubliniensis in which a species-specific molecular beacon probe that recognizes a 22-nucleotide target region in the ITS2 region of this organism is used. The results of application of this probe were compared with those of more conventional molecular biology-based approaches that involve random amplified polymorphic DNA (RAPD) analysis and restriction endonuclease analysis (REA).
MATERIALS AND METHODS
Strains and growth conditions.
Candida reference strains C. albicans ATCC 90028 C. glabrata ATCC 90030, and C. krusei ATCC 6258, were obtained from American Type Culture Collection (Manassas, Va.). C. dubliniensis reference strain NCPF3949 was obtained from the National Collection of Yeast Cultures (Norwich, England). Clinical isolates of C. albicans and C. dubliniensis (isolates M1-23 and CST 23, respectively) were obtained from the Microbiology Laboratory at Memorial Sloan-Kettering Cancer Center. The isolates were obtained from 21 patients over a period of 2 months (6 July 1998 to 13 September 1998) and were not epidemiologically related. Two strains, strains M1 (ATCC 18804) and M3, were laboratory test strains included in the panel of test strains. The yeasts were presumably identified by tests for detection of the formation of germ tubes at 37°C in horse serum (Life Technologies, Grand Island, N.Y.), production of chlamydospores on cornmeal agar with polysorbate 80 (Becton Dickinson Microbiology Systems, Cockeysville, Md.), substrate assimilation with the API 20C AUX and ID 32C systems (bioMérieux Inc., Hazelwood, Mo.), colorimetric growth on CHROMagar Candida plates (DRG International, Mountainside, N.J.), and growth at 45°C. Growth at 45°C was assessed by removing a single colony and streaking it over the surface of a Sabouraud dextrose agar plate (Becton Dickinson), which was incubated at 45°C for 48 h. Colony formation on the last three quadrants of the plate was considered good growth, while growth on the first quadrant was considered poor growth (13, 30). All strains were maintained on Sabouraud dextrose agar (4% [wt/vol] dextrose, 1% [wt/vol] peptone, 1.5% [wt/vol] agar [pH 5.6]) and were grown with shaking (∼250 rpm) at 30°C in YPD medium (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, 2% [wt/vol] dextrose [pH 5.7]).
RAPD analysis of genomic DNA.
Yeast cells were incubated at 37°C for 1 h in a suspension that contained 1 M sorbitol, 0.1 M EDTA, 0.1% (wt/vol) Zymolyase-100T (Seikaguku Corp., Tokyo, Japan), and 1% (vol/vol) 2-mercaptoethanol and that was adjusted to pH 7.5. Chromosomal DNA was purified with the Wizard genomic DNA purification kit (Promega, Madison, Wis.). Standard PCR amplifications were performed in a 50-μl reaction mixture that consisted of 33.5 μl of nuclease-free water (Promega), 5 μl of 10× Buffer A (Promega), 3 μl of 25 mM MgCl2, 4 μl of 2.5 mM deoxynucleoside triphosphates (PE Applied Biosystems, Foster City, Calif.), 2 μl of primer 1 (25 μM; 5′-AACGCGCAAC-3′), 2 μl of primer 2 (25 μM; 5′-GAGGGTGGNGGNTCT-3′) (IDTDNA, Coralville, Iowa), and 2 μl of chromosomal DNA (approximately 100 ng). The reaction was initiated by the addition of 0.5 μl of 5 U of Taq polymerase (Promega) per μl. PCR was performed in a PTC-150 Minicycler (MJ Research, Waltham, Mass.) with 45 cycles of a three-step program that consisted of 94°C for 1 min (step 1), 37°C for 1 min (step 2), and 72°C for 3 min (step 3). The PCR products (4 μl) were separated on 20% polyacrylamide gels in an X-Cell gel apparatus (Novex, Carlsbad, Calif.) for ∼3 h at 175 V.
Statistical analysis.
Similarity coefficients based on the DNA fingerprinting patterns of prominent bands of ≤1,000 bp among all isolates were calculated as the ratio of matches over the total number of bands scored. Similarities were calculated as the arithmetic mean of all pairwise distances between strain fingerprint patterns. Student's t test was used to compare genetic similarities between different groups of isolates. Banding patterns and similarity coefficients were determined with the Molecular Analyst/Fingerprinting Plus v.1.12 (Bio-Rad Laboratories, Hercules, Calif.) and the statistical software GB-STAT 6.5 (Scolari, London, England). P values of <0.05 were considered significant.
ITS2 amplification and REA.
PCR amplification of the ITS2 region was performed with fungus-specific universal primers ITS3 (5′-GCATCGATGAAGAACGCAGC-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) to amplify a conserved portion of the 5.8S ribosomal DNA (rDNA) region, the adjacent ITS2 region, and a small portion of the 28S rDNA region, yielding products of 0.338 kb for C. albicans and 0.343 for C. dubliniensis. Full DNA sequence analysis of the PCR products obtained with universal fungal primers ITS3 and ITS4 specific for rDNA genes was used to confirm the species identification, as described previously (11). The reaction mixture (total volume, 100 μl) consisted of 2 μl of genomic DNA (∼200 ng), 1 μl of 25 mM dNTP (Promega), 1 μl of 5 U of Amplitaq Gold Taq polymerase (PE Applied Biosystems) per μl, 10 μl of 10× buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 15 mM MgCl2, 0.01% [wt/vol] gelatin), 1 μl of universal fungal primers ITS3 and ITS4 (25 μM) (IDTDNA) (14), and 84 μl of nuclease-free water (Promega). The PCR mixture was subjected to PCR with the following cycling conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 58°C for 1 min, and 72°C for 1 min and a final step of 72°C for 5 min. PCR-amplified products were purified with the Qiaquick DNA Purification Kit (Qiagen, Valencia, Calif.). The PCR amplification products (∼500 ng) were digested with 1 μl of restriction enzymes NspBII (C. albicans ITS2 specific) (AP Biotech, Piscataway, N.J.) and BsmAI (C. dubliniensis ITS2 specific) (New England Biolabs, Beverly, Mass.) for 1 h and were analyzed by gel electrophoresis in a 1.5% agarose gel.
Molecular beacon design and analysis.
Molecular beacons specific for the ITS2 region of C. albicans and C. dubliniensis were designed on the basis of published probe sequences (11) that were independently confirmed by DNA sequence analysis at the New York University DNA sequencing facility. Target sequence selection, beacon design, and synthesis were optimized by standard protocols available on the Public Health Research Institute's website for molecular beacons (http://www.molecular-beacons.com), as described by Tyagi and colleagues (50, 51). Each molecular beacon possessed a 6-nucleotide arm sequence and a 22-nucleotide probe target recognition sequence, as follows: 5′-GCTAAGGCGGTCTCTGGCGTCG (C. dubliniensis) and 5′-TAGGTCTAACCAAAACATTGC (C. albicans). The arm sequences 5′-GCGAGG and 3′-CCTCGC were designed to form a stable stem hybrid at the annealing temperature of the PCR to ensure that nonhybridized probes remained in a hairpin conformation (no fluorescence). The fluorophore tetrachloro-6-carboxyfluorescein and the quencher 4-(4′-dimethylaminophenylazo)benzoic acid (Molecular Probes Inc., Eugene, Oreg.) were covalently attached to the 5′ and 3′ ends of the arm sequences, respectively. Real-time PCR amplification was performed with 2 μl of species-specific molecular beacons (100 ng) in a 50-μl reaction volume that contained 2 μl of genomic DNA (200 ng), 0.5 μl of 25 mM nucleotide mix (Promega), 0.5 μl of 5 U of Amplitaq Gold Taq polymerase (PE Applied Biosystems) per μl, 5 μl of 10× buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 15 mM MgCl2, 0.01% [wt/vol] gelatin), 5 μl of 25 mM MgCl2, 1 μl of universal fungal primers ITS3 and ITS4 (1 mg/ml), and 33 μl of nuclease-free water (Promega). The PCR mixture was subjected to PCR with the following cycling conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 58°C for 1 min, and 72°C for 1 min in a Prism 7700 96-well spectrofluorometric thermal cycler (PE Applied Biosystems).
RESULTS
RAPD analysis.
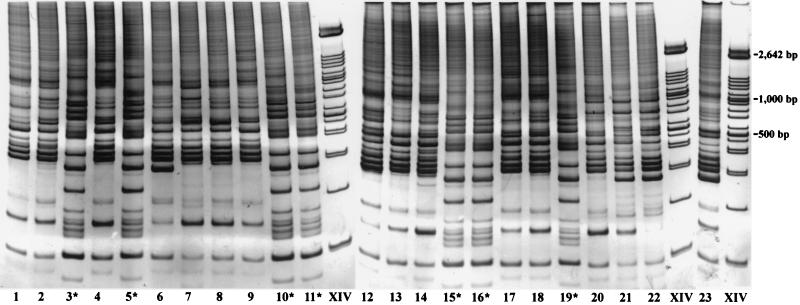
A collection of 23 Candida isolates that mostly contained C. albicans and a smaller number of C. dubliniensis strains was assembled by the Microbiology Laboratory at Memorial Sloan-Kettering Cancer Center from stock cultures. The collection was provided for a blind evaluation of C. dubliniensis. Initially, all 23 strains were subjected to random primed RAPD genotyping analysis to assess the relative genomic relatedness of the strains. The RAPD fragments for all 23 isolates displayed a profile of more than 15 prominent bands, with sizes ranging from 0.1 to 1 kb (Fig. 2), with nearly identical profiles produced in three separate trials. Two distinct banding profiles were identified in which band similarities of greater than 0.8 (band similarity coefficient, 0 to 3 band differences) were found when the band profiles for common strains in the set were compared (P < 0.05), but similarities of less than 0.5 were found when the band profiles between strains of the two sets were compared (P < 0.05). The major group included strains M1, M2, M4, M6 to M9, M12 to M14, M17, M18, and M20 to M23, whose patterns matched that of a reference C. albicans strain, while the minor group consisted of M3, M5, M10, M11, M15, M16, and M19, whose patterns matched the pattern observed from a C. dubliniensis reference strain (not shown).
FIG. 2.
RAPD analysis of 23 Candida isolates. Genomic DNA was extracted and purified from each isolate, and PCR amplification was performed with random primers, as described in Materials and Methods. The PCR-amplified products were run on a 20% Tris-borate-EDTA–polyacrylamide gel and stained with GelStar (FMC Bioproducts). ∗, suspected C. dubliniensis strain.
REA.
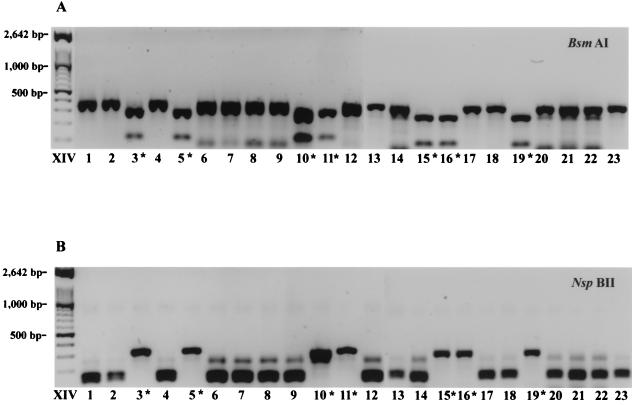
PCR amplification of the ITS2 region with universal fungal primers ITS3 and ITS4 (22) was used to generate ∼0.3-kb fragments from all 23 isolates. The fragments were analyzed with restriction enzyme BsmAI, which has a specific sequence recognition sequence in this region from C. dubliniensis. Figure 3A shows that 7 of the 23 amplified products were cut by this restriction enzyme, yielding fragments of 0.25 and 0.9 kb. These restricted amplicons were derived from the same subset of seven strains identified by RAPD analysis (Fig. 2). The remaining PCR-amplified products could be cut with restriction enzyme NspBII, which specifically recognizes a site in the ITS2 region from C. albicans to yield fragments of ∼0.18 and 0.16 kb (Fig. 3B).
FIG. 3.
The ITS2 region was PCR amplified with universal fungal primers ITS3 and ITS4, and the products were digested with restriction enzymes BsmAI (C. dubliniensis specific) (A) and NspBII (C. albicans specific) (B). The restriction fragments were run on a 1.2% agarose gel and stained with GelStar (FMC Bioproducts). ∗, C. dubliniensis strains.
Specificity and sensitivity of molecular beacon assay.
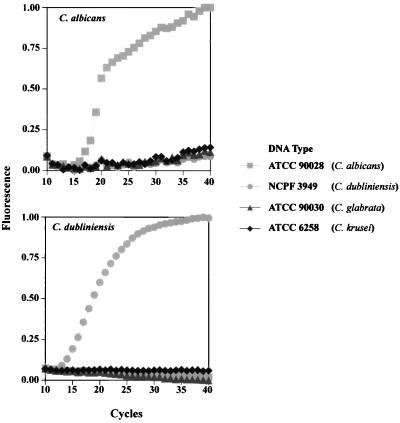
Species-specific molecular beacons were designed to target sequences within the ITS2 regions of C. albicans and C. dubliniensis (Fig. 1B). Real-time PCR was performed with each of the molecular beacons with DNA from reference strains of C. albicans, C. dubliniensis, C. glabrata, and C. krusei. Fluorescence was detected for each of the molecular beacons only in the prescence of its proper DNA target (Fig. 4). There was no effect of excess nontarget amplicons on the signal of the molecular beacon. The sensitivity of the C. dubliniensis-specific molecular beacon was evaluated by serially diluting genomic DNA 105-fold from a starting amount of 100 ng. The molecular beacon was able to detect the target when ∼100 pg of the initial genomic DNA was present (Fig. 5). This result further illustrates that molecular beacons have the ability to quantify target DNA in a real-time PCR assay (6).
FIG. 4.
Selectivities of species-specific molecular beacons for reference Candida strains. In this experiment, species-specific molecular beacons, as indicated in the panels, were used to probe individual DNAs from the four Candida species. Real-time PCR amplification of the ITS2 regions of four reference Candida strains was performed with 100 ng of beacons and ∼200 ng of genomic DNA for 40 cycles in a Prism 7700 96-well spectrofluorometric thermal cycler (PE Applied Biosystems).
FIG. 5.
Real-time PCR amplification for determination of relative sensitivity was performed for 40 cycles with C. dubliniensis genomic DNA (∼100 ng) that was serially diluted 105-fold, as indicated, in the presence of a fixed amount (100 ng) of the molecular beacon. Relative fluorescence was monitored in a PE Applied Biosystems 7700 Prism 96-well spectrofluorometric thermal cycler.
Detection of C. dubliniensis.
The molecular beacon specific for C. dubliniensis was used to probe all 23 Candida isolates, along with reference strains of C. albicans, C. dubliniensis, C. glabrata, and C. krusei. Figure 6 shows that cycle-dependent amplification of the signal from the C. dubliniensis-specific molecular beacon was obtained only for the seven suspected C. dubliniensis strains, strains M3, M5, M10, M11, M15, M16, and M19. All 23 isolates were also probed with the C. albicans-specific molecular beacon, and a fluorescence signal was obtained only for the 16 isolates which the C. dubliniensis probe did not detect (data not shown). Table 1 summarizes the results obtained by the three different DNA-based species identification methods (analysis with molecular beacons, RAPD analysis, and REA) and demonstrates that the results obtained by analysis with molecular beacons showed a 100% correlation with the results obtained by the other approaches.
FIG. 6.
Real-time detection of C. dubliniensis from a blinded panel of 23 Candida isolates was accomplished by PCR amplification of the ITS2 region in the presence of a molecular beacon specific for C. dubliniensis. Relative fluorescence was monitored in a PE Applied Biosystems 7700 Prism 96-well spectrofluorometric thermal cycler. Control DNAs from reference strains of C. albicans, C. krusei, and C. glabrata were also evaluated.
TABLE 1.
DNA-based species identification
| Strain | Isolation date (mo/day/yr) | Isolate source |
Candida detection by:
|
|||||
|---|---|---|---|---|---|---|---|---|
| RAPD analysis
|
REA
|
Analysis with molecular beacons
|
||||||
| C. albicans | C. dubliniensis | C. albicans | C. dubliniensis | C. albicans | C. dubliniensis | |||
| Test strains | ||||||||
| M1 | a | a | • | • | • | |||
| M2 | 09/02/98 | Mouth | • | • | • | |||
| M3 | b | b | • | • | • | |||
| M4 | 09/13/98 | Mouth | • | • | • | |||
| M5 | 09/08/98 | Sputum | • | • | • | |||
| M6 | 07/06/98 | Facial wound | • | • | • | |||
| M7 | 07/08/98 | Throat | • | • | • | |||
| M8 | 07/13/98 | Mouth | • | • | • | |||
| M9 | 09/02/98 | Sputum | • | • | • | |||
| M10 | 07/23/98 | Tongue | • | • | • | |||
| M11 | 08/02/98 | Blood | • | • | • | |||
| M12 | 07/24/98 | Mouth | • | • | • | |||
| M13 | 08/08/98 | Bronchial wash | • | • | • | |||
| M14 | 08/10/98 | Sputum | • | • | • | |||
| M15 | 08/12/98 | Bronchial wash | • | • | • | |||
| M16 | 08/14/98 | Tracheal aspirate | • | |||||
| M17 | 08/25/98 | Pleural fluid | • | • | • | |||
| M18 | 08/11/98 | Sputum | • | • | • | |||
| M19 | 08/26/98 | Stool | • | |||||
| M20 | 08/14/98 | Mouth | • | • | • | |||
| M21 | 08/19/98 | Pleural fluid | • | • | • | |||
| M22 | 08/15/98 | Sputum | • | • | • | |||
| M23 | 08/18/98 | Nasopharnyx | • | • | • | |||
| Control strains | ||||||||
| ATCC 90028 | • | • | • | |||||
| NCPF 3949 | • | • | • | |||||
ATCC 18804.
New York State Health Department proficiency test strain.
DISCUSSION
The increasing prevalence of fungal infections caused by non-C. albicans species that display clinical resistance or reduced susceptibility to common azole-based antifungal drugs has created a need to rapidly differentiate between Candida species early in infection to assist in proper therapeutic management. Conventional morphology and carbon assimilation tests require several days or more for identification (54) and may misidentify some species. C. dubliniensis is an example of such a misidentified organism that is now readily turning up in the stock culture collections of C. albicans from numerous clinical laboratories (45, 47). In the last few years, several new methods have been developed to distinguish the phenotypic properties of this organism from those of other Candida species, including chlamydospore formation (44), carbon assimilation (39), temperature-dependent growth (34), colony coloration on CHROMagar (16), and immunofluorescence (3), as have commercially available systems (API20C AUX, RapID Yeast Plus, VITEK YBC, and VITEK 2 ID-YST) (13, 33). Despite advances in phenotypic detection, species identification by these routes is relatively slow. Genotyping with hybridization probes such as the C. albicans mid-repeat-sequence probes 27A and Ca3 (2, 8, 40) and the C. dubliniensis-specific complex probe Cd25-1 (15) can be used to provide species and subspecies information. These techniques, however, are typically too labor intensive for a clinical laboratory setting and are more suited for epidemiological investigations.
Nucleic acid amplification of species-specific target sequences by PCR or ligase chain reaction, or with RNA-dependent Qβ replicase provides the most rapid alternative to standard testing (14) since few fungal cells are required. This high level of sensitivity is particularly attractive for the early detection of fungemias, which are difficult to detect by conventional procedures like blood culture or those that depend upon a competent immune system (27). The detection of 1 to 2 CFU per ml of blood can generally be achieved, provided the target gene is present in numerous (>100) copies (10). While the sensitivity associated with PCR amplification can be maximized to detect low levels of pathogenic organisms, PCR assays applied in clinical diagnostics have drawbacks due to potential contamination with environmental organisms, coamplification of human DNA, false priming, and/or the necessity for additional hybridization steps (43). Furthermore, PCR products are rarely validated other than by size, and detection schemes that involve linear probes may have limited fidelity and limited overall sensitivity.
Recently, a promising method for rapid identification of Candida spp., including C. dubliniensis, with sequence-specific digoxigenin-labeled hybridization probes in a PCR-enzyme immunoassay format was described (11, 12, 41, 42). This method is robust but requires a posthybridization step to remove the contaminating unhybridized probe that can lead to false-positive results. Typically, any time that linear probes are used for detection, there is a risk that false annealing may occur. Probe-target hybridization is highly temperature dependent, and depending on the nucleotide composition of the probe, random annealing can pose a problem, especially when one is dealing with sequences with high G+C contents, since the temperature profile for annealing is shifted downward (5, 38).
To overcome a number of the inherent problems associated with nucleic acid amplification and detection, we have applied molecular beacon technology to detect PCR-amplified target sequences in the ITS2 regions of C. dubliniensis and C. albicans. Using this technology, we readily detected all seven C. dubliniensis strains from a panel of 23 unknown Candida isolates (Fig. 6). The molecular beacon analysis data were independently validated by less robust molecular approaches involving RAPD analysis-based genotyping and REA of target recognition sequences in the ITS2 regions of C. dubliniensis and C. albicans. The ITS2 region of fungi is a suitable target for species identification because of its variability among species (11, 21, 22). In addition, only a single primer set, ITS3 and ITS4, is required to amplify this region from fungi because of the highly conserved domains of the 5.8S and 28S rDNA genes that flank it. The targeting of high-copy-number RNA genes is also an advantage because it increases the sensitivity of detection of amplified DNA without the need for nested PCR techniques (12). The total time required for sample preparation and analysis of target sequences is typically less than 6 h starting from pregrown colonies of ∼1 mm.
Molecular beacons have a distinct advantage over linear fluorescent probes because of their stem-and-loop structures (4, 6). In this conformation, nonhybridized beacons remain dark because the fluorophore is maintained close to the quencher. When bound to its target, the beacon opens and fluoresces brightly. There is no requirement for isolation of probe-target hybrids to measure fluorescence, which eliminates any possible posthybridization contamination. Thus, molecular beacons can be added prior to amplification and real-time fluorescence can be measured in a single-step assay. Real-time monitoring of the PCR amplification allows a quantitative measure of the starting template based on the fluorescence signal as a function of PCR cycle (23), a feature unavailable to endpoint assays such as PCR-enzyme immunoassay.
The use of hairpin-shaped molecular beacons in PCR assays provides several advantages over the use of linear probes because the stem-loop structure imparts an increased ability to discriminate single-base-pair mismatches compared to that from the use of linear probes such as TaqMan (4, 50). The hairpin shape makes mismatched probe-target hybrids less thermally stable than hybrids between corresponding linear probes. Thus, molecular beacons are more useful for allele discrimination, which adds to their fidelity in monitoring of authentic products in PCR amplifications. To successfully monitor a PCR assay, the molecular beacon should be designed to hybridize to its target at the PCR annealing temperature, whereas the free molecular beacon should stay closed and nonfluorescent at higher temperatures. A probe sequence should be chosen such that the molecular beacon dissociates from its target at a temperature 7 to 10°C higher than the annealing temperature of the PCR amplification (6) (see the Public Health Research Institute website for molecular beacons support). Finally, unlike linear hydrolysis probes, the quenching of molecular beacons has been shown to occur through a collisional mechanism that involves a direct transfer of energy from the fluorophore to the quencher (4). This property enables a common quencher molecule to be used with beacons, which increases the number of possible fluorophores that can be used as reporters. This is especially important for multiplexing of molecular beacons in a PCR assay, which have been reported for the detection of viruses (53) and drug resistance-conferring mutations in M. tuberculosis (31, 37).
In summary, our results indicate that species-specific molecular beacons are a highly reliable tool for molecular biology-based identification of C. dubliniensis that overcomes many of the inherent problems associated with nucleic acid amplification and detection. The results in this study extend previous applications of molecular beacons and demonstrate that they provide a rapid and highly reliable method for detection of sequence-specific amplified DNA. Molecular beacons are ideal tools for clinical diagnostics because of their stability, high signal-noise property, real-time monitoring capability, and high-throughput potential. We are developing a panel of molecular beacons for molecular identification of numerous fungi. Furthermore, since molecular beacons can be used with several different fluorophores, they are amenable for use for detection of multiplex sequences in a single reaction tube (53).
ACKNOWLEDGMENTS
This work was supported by a sole-source contract from the New State Department of Health, Albany, N.Y. (to D.S.P.).
REFERENCES
- 1.Alexander B D, Perfect J R. Antifungal resistance trends towards the year 2000. Implications for therapy and new approaches. Drugs. 1997;54:657–678. doi: 10.2165/00003495-199754050-00002. [DOI] [PubMed] [Google Scholar]
- 2.Anderson J, Srikantha T, Morrow B, Miyasaki S H, White T C, Agabian N, Schmid J, Soll D R. Characterization and partial nucleotide sequence of the DNA fingerprinting probe Ca3 of Candida albicans. J Clin Microbiol. 1993;31:1472–1480. doi: 10.1128/jcm.31.6.1472-1480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bikandi J, Millan R S, Moragues M D, Cebas G, Clarke M, Coleman D C, Sullivan D J, Quindos G, Ponton J. Rapid identification of Candida dubliniensis by indirect immunofluorescence based on differential localization of antigens on C. dubliniensis blastospores and Candida albicans germ tubes. J Clin Microbiol. 1998;36:2428–2433. doi: 10.1128/jcm.36.9.2428-2433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet G, Tyagi S, Libchaber A, Kramer F R. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc Natl Acad Sci USA. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borisova O F, Shchyolkina A K, Chernov B K, Tchurikov N A. Relative stability of AT and GC pairs in parallel DNA duplex formed by a natural sequence. FEBS Lett. 1993;322:304–306. doi: 10.1016/0014-5793(93)81591-m. [DOI] [PubMed] [Google Scholar]
- 6.Cayouette M, Sucharczuk A, Moores J, Tyagi S, Kramer F R. Using molecular beacons to monitor PCR product formation. Strategies Newsl. 1999;12:85–88. [Google Scholar]
- 7.Coleman D, Sullivan D, Harrington B, Haynes K, Henman M, Shanley D, Bennett D, Moran G, McCreary C, O'Neill L. Molecular and phenotypic analysis of Candida dubliniensis: a recently identified species linked with oral candidosis in HIV-infected and AIDS patients. Oral Dis. 1997;3(Suppl. 1):S96–S101. doi: 10.1111/j.1601-0825.1997.tb00384.x. [DOI] [PubMed] [Google Scholar]
- 8.Diaz-Guerra T M, Mellado E, Cuenca Estrella M, Laguna F, Rodriguez-Tudela J L. Molecular characterization by PCR-fingerprinting of Candida dubliniensis strains isolated from two HIV-positive patients in Spain. Diagn Microbiol Infect Dis. 1999;35:113–119. doi: 10.1016/s0732-8893(99)00072-3. [DOI] [PubMed] [Google Scholar]
- 9.Donnelly S M, Sullivan D J, Shanley D B, Coleman D C. Phylogenetic analysis and rapid identification of Candida dubliniensis based on analysis of ACT1 intron and exon sequences. Microbiology. 1999;145:1871–1882. doi: 10.1099/13500872-145-8-1871. [DOI] [PubMed] [Google Scholar]
- 10.Einsele H, Hebart H, Roller G, Loffler J, Rothenhofer I, Muller C A, Bowden R A, van Burik J, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elie C M, Lott T J, Reiss E, Morrison C J. Rapid identification of Candida species with species-specific DNA probes. J Clin Microbiol. 1998;36:3260–3265. doi: 10.1128/jcm.36.11.3260-3265.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita S, Lasker B A, Lott T J, Reiss E, Morrison C J. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J Clin Microbiol. 1995;33:962–967. doi: 10.1128/jcm.33.4.962-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gales A C, Pfaller M A, Houston A K, Joly S, Sullivan D J, Coleman D C, Soll D R. Identification of Candida dubliniensis based on temperature and utilization of xylose and α-methyl-d-glucoside as determined with the API 20C AUX and Vitek YBC systems. J Clin Microbiol. 1999;37:3804–3808. doi: 10.1128/jcm.37.12.3804-3808.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ieven M, Goossens H. Relevance of nucleic acid amplification techniques for diagnosis of respiratory tract infections in the clinical laboratory. Clin Microbiol Rev. 1997;10:242–256. doi: 10.1128/cmr.10.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joly S, Pujol C, Rysz M, Vargas K, Soll D R. Development and characterization of complex DNA fingerprinting probes for the infectious yeast Candida dubliniensis. J Clin Microbiol. 1999;37:1035–1044. doi: 10.1128/jcm.37.4.1035-1044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirkpatrick W R, Revankar S G, McAtee R K, Lopez-Ribot J L, Fothergill A W, McCarthy D I, Sanche S E, Cantu R A, Rinaldi M G, Patterson T F. Detection of Candida dubliniensis in oropharyngeal samples from human immunodeficiency virus-infected patients in North America by primary CHROMagar Candida screening and susceptibility testing of isolates. J Clin Microbiol. 1998;36:3007–3012. doi: 10.1128/jcm.36.10.3007-3012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostrikis L G, Tyagi S, Mhlanga M M, Ho D D, Kramer F R. Spectral genotyping of human alleles. Science. 1998;279:1228–1229. doi: 10.1126/science.279.5354.1228. [DOI] [PubMed] [Google Scholar]
- 18.Kurzai O, Heinz W J, Sullivan D J, Coleman D C, Frosch M, Muhlschlegel F A. Rapid PCR test for discriminating between Candida albicans and Candida dubliniensis isolates using primers derived from the pH-regulated PHR1 and PHR2 genes of C. albicans. J Clin Microbiol. 1999;37:1587–1590. doi: 10.1128/jcm.37.5.1587-1590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Ribot J L, McAtee R K, Lee L N, Kirkpatrick W R, White T C, Sanglard D, Patterson T F. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob Agents Chemother. 1998;42:2932–2937. doi: 10.1128/aac.42.11.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lott T J, Burns B M, Zancope-Oliveira R, Elie C M, Reiss E. Sequence analysis of the internal transcribed spacer 2 (ITS2) from yeast species within the genus Candida. Curr Microbiol. 1998;36:63–69. doi: 10.1007/s002849900280. [DOI] [PubMed] [Google Scholar]
- 21.Lott T J, Holloway B P, Logan D A, Fundyga R, Arnold J. Towards understanding the evolution of the human commensal yeast Candida albicans. Microbiology. 1999;145:1137–1143. doi: 10.1099/13500872-145-5-1137. [DOI] [PubMed] [Google Scholar]
- 22.Lott T J, Kuykendall R J, Reiss E. Nucleotide sequence analysis of the 5.8S rDNA and adjacent ITS2 region of Candida albicans and related species. Yeast. 1993;9:1199–1206. doi: 10.1002/yea.320091106. [DOI] [PubMed] [Google Scholar]
- 23.Manganelli R, Dubnau E, Tyagi S, Kramer F R, Smith I. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol Microbiol. 1999;31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 24.Marras S A, Kramer F R, Tyagi S. Multiplex detection of single-nucleotide variations using molecular beacons. Genet Anal. 1999;14:151–156. doi: 10.1016/s1050-3862(98)00018-7. [DOI] [PubMed] [Google Scholar]
- 25.Meiller T F, Jabra-Rizk M A, Baqui A, Kelley J I, Meeks V I, Merz W G, Falkler W A. Oral Candida dubliniensis as a clinically important species in HIV-seropositive patients in the United States. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1999;88:573–580. doi: 10.1016/s1079-2104(99)70088-0. [DOI] [PubMed] [Google Scholar]
- 26.Meis J F, Ruhnke M, De Pauw B E, Odds F C, Siegert W, Verweij P E. Candida dubliniensis candidemia in patients with chemotherapy-induced neutropenia and bone marrow transplantation. Emerg Infect Dis. 1999;5:150–153. doi: 10.3201/eid0501.990119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morace G, Pagano L, Sanguinetti M, Posteraro B, Mele L, Equitani F, D'Amore G, Leone G, Fadda G. PCR-restriction enzyme analysis for detection of Candida DNA in blood from febrile patients with hematological malignancies. J Clin Microbiol. 1999;37:1871–1875. doi: 10.1128/jcm.37.6.1871-1875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moran G P, Sanglard D, Donnelly S M, Shanley D B, Sullivan D J, Coleman D C. Identification and expression of multidrug transporters responsible for fluconazole resistance in Candida dubliniensis. Antimicrob Agents Chemother. 1998;42:1819–1830. doi: 10.1128/aac.42.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moran G P, Sullivan D J, Henman M C, McCreary C E, Harrington B J, Shanley D B, Coleman D C. Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob Agents Chemother. 1997;41:617–623. doi: 10.1128/aac.41.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaller M A, Messer S A, Gee S, Joly S, Pujol C, Sullivan D J, Coleman D C, Soll D R. In vitro susceptibilities of Candida dubliniensis isolates tested against the new triazole and echinocandin antifungal agents. J Clin Microbiol. 1999;37:870–872. doi: 10.1128/jcm.37.3.870-872.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piatek A S, Telenti A, Murray M R, El-Hajj H, Jacobs W R, Jr, Kramer F R, Alland D. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob Agents Chemother. 2000;44:103–110. doi: 10.1128/aac.44.1.103-110.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piatek A S, Tyagi S, Pol A C, Telenti A, Miller L P, Kramer F R, Alland D. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat Biotechnol. 1998;16:359–363. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 33.Pincus D H, Coleman D C, Pruitt W R, Padhye A A, Salkin I F, Geimer M, Bassel A, Sullivan D J, Clarke M, Hearn V. Rapid identification of Candida dubliniensis with commercial yeast identification systems. J Clin Microbiol. 1999;37:3533–3539. doi: 10.1128/jcm.37.11.3533-3539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinjon E, Sullivan D, Salkin I, Shanley D, Coleman D. Simple, inexpensive, reliable method for differentiation of Candida dubliniensis from Candida albicans. J Clin Microbiol. 1998;36:2093–2095. doi: 10.1128/jcm.36.7.2093-2095.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polacheck I, Strahilevitz J, Sullivan D, Donnelly S, Salkin I F, Coleman D C. Recovery of Candida dubliniensis from non-human immunodeficiency virus-infected patients in Israel. J Clin Microbiol. 2000;38:170–174. doi: 10.1128/jcm.38.1.170-174.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reiss E, Tanaka K, Bruker G, Chazalet V, Coleman D, Debeaupuis J P, Hanazawa R, Latge J P, Lortholary J, Makimura K, Morrison C J, Murayama S Y, Naoe S, Paris S, Sarfati J, Shibuya K, Sullivan D, Uchida K, Yamaguchi H. Molecular diagnosis and epidemiology of fungal infections. Med Mycol. 1998;36(Suppl. 1):249–257. [PubMed] [Google Scholar]
- 37.Rhee J T, Piatek A S, Small P M, Harris L M, Chaparro S V, Kramer F R, Alland D. Molecular epidemiologic evaluation of transmissibility and virulence of Mycobacterium tuberculosis. J Clin Microbiol. 1999;37:1764–1770. doi: 10.1128/jcm.37.6.1764-1770.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rychlik W. Priming efficiency in PCR. BioTechniques. 1995;18:84–86. , 88–90. [PubMed] [Google Scholar]
- 39.Salkin I F, Pruitt W R, Padhye A A, Sullivan D, Coleman D, Pincus D H. Distinctive carbohydrate assimilation profiles used to identify the first clinical isolates of Candida dubliniensis recovered in the United States. J Clin Microbiol. 1998;36:1467. doi: 10.1128/jcm.36.5.1467-1467.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmid J, Voss E, Soll D R. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca3. J Clin Microbiol. 1990;28:1236–1243. doi: 10.1128/jcm.28.6.1236-1243.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin J H, Nolte F S, Holloway B P, Morrison C J. Rapid identification of up to three Candida species in a single reaction tube by a 5′ exonuclease assay using fluorescent DNA probes. J Clin Microbiol. 1999;37:165–170. doi: 10.1128/jcm.37.1.165-170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin J H, Nolte F S, Morrison C J. Rapid identification of Candida species in blood cultures by a clinically useful PCR method. J Clin Microbiol. 1997;35:1454–1459. doi: 10.1128/jcm.35.6.1454-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skladny H, Buchheidt D, Baust C, Krieg-Schneider F, Seifarth W, Leib-Mosch C, Hehlmann R. Specific detection of Aspergillus species in blood and bronchoalveolar lavage samples of immunocompromised patients by two-step PCR. J Clin Microbiol. 1999;37:3865–3871. doi: 10.1128/jcm.37.12.3865-3871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staib P, Morschhauser J. Chlamydospore formation on Staib agar as a species-specific characteristic of Candida dubliniensis. Mycoses. 1999;42:521–524. doi: 10.1046/j.1439-0507.1999.00516.x. [DOI] [PubMed] [Google Scholar]
- 45.Sullivan D, Coleman D. Candida dubliniensis: an emerging opportunistic pathogen. Curr Top Med Mycol. 1997;8:15–25. [PubMed] [Google Scholar]
- 46.Sullivan D, Coleman D. Candida dubliniensis: characteristics and identification. J Clin Microbiol. 1998;36:329–334. doi: 10.1128/jcm.36.2.329-334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan D, Haynes K, Bille J, Boerlin P, Rodero L, Lloyd S, Henman M, Coleman D. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J Clin Microbiol. 1997;35:960–964. doi: 10.1128/jcm.35.4.960-964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan D J, Henman M C, Moran G P, O'Neill L C, Bennett D E, Shanley D B, Coleman D C. Molecular genetic approaches to identification, epidemiology and taxonomy of non-albicans Candida species. J Med Microbiol. 1996;44:399–408. doi: 10.1099/00222615-44-6-399. [DOI] [PubMed] [Google Scholar]
- 49.Turenne C Y, Sanche S E, Hoban D J, Karlowsky J A, Kabani A M. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J Clin Microbiol. 1999;37:1846–1851. doi: 10.1128/jcm.37.6.1846-1851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tyagi S, Bratu D P, Kramer F R. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 51.Tyagi S, Kramer F R. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 52.Vanden Bossche H, Dromer F, Improvisi I, Lozano-Chiu M, Rex J H, Sanglard D. Antifungal drug resistance in pathogenic fungi. Med Mycol. 1998;36(Suppl. 1):119–128. [PubMed] [Google Scholar]
- 53.Vet J A, Majithia A R, Marras S A, Tyagi S, Dube S, Poiesz B J, Kramer F R. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc Natl Acad Sci USA. 1999;96:6394–6399. doi: 10.1073/pnas.96.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warren N G, Hazen K G. Candida, Cryptococcus, and other yeasts of medical importance. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 723–737. [Google Scholar]