Abstract
In eukaryotic protein N-glycosylation, a series of glycosyltransferases catalyse the biosynthesis of a dolichylpyrophosphate-linked oligosaccharide before its transfer onto acceptor proteins1. The final seven steps occur in the lumen of the endoplasmic reticulum (ER) and require dolichylphosphate-activated mannose and glucose as donor substrates2. The responsible enzymes—ALG3, ALG9, ALG12, ALG6, ALG8 and ALG10—are glycosyltransferases of the C-superfamily (GT-Cs), which are loosely defined as containing membrane-spanning helices and processing an isoprenoid-linked carbohydrate donor substrate3,4. Here we present the cryo-electron microscopy structure of yeast ALG6 at 3.0 Å resolution, which reveals a previously undescribed transmembrane protein fold. Comparison with reported GT-C structures suggests that GT-C enzymes contain a modular architecture with a conserved module and a variable module, each with distinct functional roles. We used synthetic analogues of dolichylphosphate-linked and dolichylpyrophosphate-linked sugars and enzymatic glycan extension to generate donor and acceptor substrates using purified enzymes of the ALG pathway to recapitulate the activity of ALG6 in vitro. A second cryo-electron microscopy structure of ALG6 bound to an analogue of dolichylphosphate-glucose at 3.9 Å resolution revealed the active site of the enzyme. Functional analysis of ALG6 variants identified a catalytic aspartate residue that probably acts as a general base. This residue is conserved in the GT-C superfamily. Our results define the architecture of ER-luminal GT-C enzymes and provide a structural basis for understanding their catalytic mechanisms.
In the eukaryotic cell, N-glycosylation of secretory proteins is an essential process that involves the transfer of a high-mannose glycan (GlcNAc2Man9Glc3) from a dolichylpyrophosphate (Dol-PP) carrier, catalysed by oligosaccharyltransferase1. The biosynthesis of this donor substrate is a sequential process that is initiated in the cytoplasm, requires the flipping of the GlcNAc2Man5-containing intermediate, and is completed on the luminal side of the ER, where the transfer of the final seven hexoses occurs2 (Fig. 1a). For the cytoplasmic reactions, the donor substrates are soluble, nucleotide-activated sugars. By contrast, the ER-luminal reactions depend on the membrane-embedded donor substrates dolichylphosphate-mannose (Dol-P-Man) and dolichylphosphate-glucose (Dol-P-Glc)2. ALG6 transfers the first of three glucose moieties onto the pre-assembled GlcNAc2Man9 glycan. Following the identification5 and cloning6 of the ALG6 locus, it was shown that deficiencies in ALG6 are a frequent cause of congenital disorders of glycosylation (CDGs)7, in which patients generally have hypo-glycosylated serum glycoproteins8,9. To understand the mechanism of ALG6, we sought to obtain high-resolution structural insight.
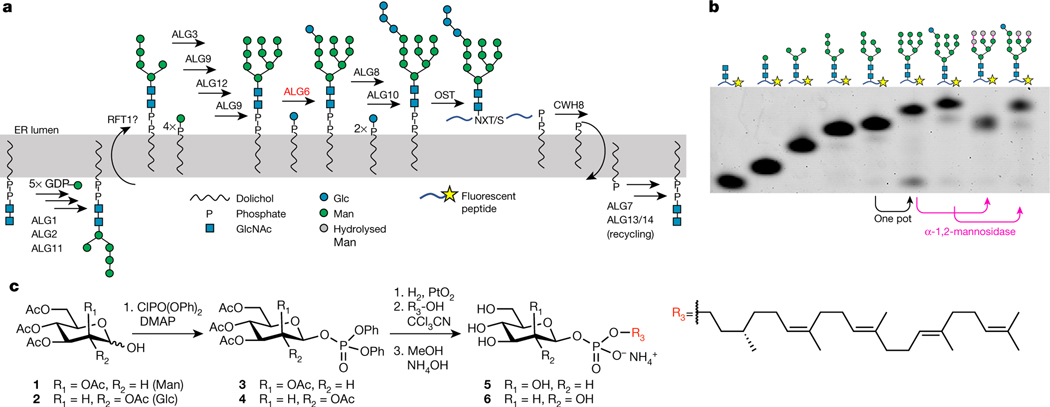
Fig. 1 |. Functional analysis of ALG6.
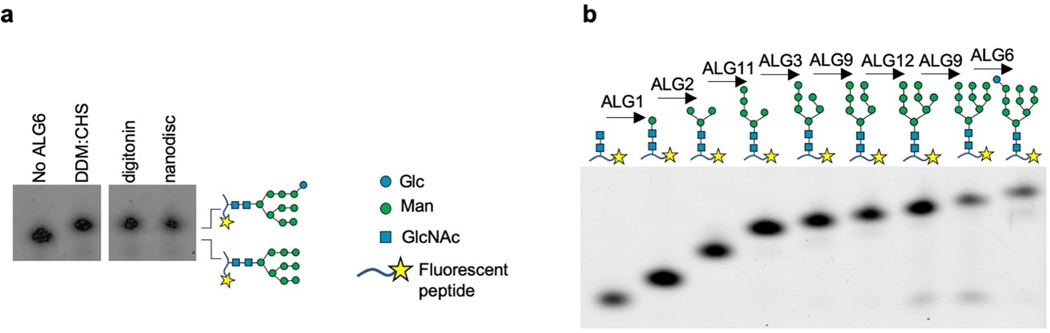
a, Schematic of eukaryotic LLO biosynthesis and glycan transfer by oligosaccharyltransferase (OST)2,36. Enzyme names are indicated above the reaction arrows. The curved arrows across the ER membrane indicate flipping reactions. b, Tricine gel-based analysis of chemo-enzymatic LLO extension and of ALG6-catalysed glucose transfer. Glycans were transferred onto a fluorescent peptide using purified T. brucei STT3A enzyme11. The glycan structures are indicated schematically above the lanes. Note that α−1,2-mannosidase is unable to remove mannoses from the A-branch of GlcNAc2Man9Glc, demonstrating the presence of a glucose moiety. c, Synthesis scheme of Dol-P-hexose analogues including Dol25-P-Man (5) and Dol25-P-Glc (6). Note that the citronellyl-farnesyl moiety of Dol25 faithfully reflects the stereochemistry of native dolichol. DMAP, 4-dimethylaminopyridine.
In vitro activity of purified yeast ALG6
Neither the donor nor the acceptor substrate of ALG6 is commercially available. To generate the acceptor substrate, we started from our previously developed10 method for chemo-enzymatic synthesis and analysis of Dol25-PP-GlcNAc2Man5, where Dol25 refers to a citronellyl-farnesyl moiety. The lipid-linked glycans were analysed by tricine gel analysis following transfer onto a fluorescent acceptor peptide catalysed by the eukaryotic, single-subunit oligosaccharyltransferase STT3A from Trypanosoma brucei10 (Fig. 1b). To append mannose moieties to Dol25-PP-GlcNAc2Man5, we heterologously expressed and purified the membrane-integral enzymes ALG3, ALG9 and ALG12 (Supplementary Fig. 1a) and synthesized the required mannose donor analogue Dol25-P-Man (Fig. 1c). Using a stepwise procedure, we extended Dol25-PP-GlcNAc2Man5 to the ALG6 acceptor substrate Dol25-PP-GlcNAc2Man9 (Fig. 1b). We also synthesized the required donor substrate Dol25-P-Glc de novo (Fig. 1c). Incubation of ALG6 with Dol25-P-Glc and Dol25-PP-GlcNAc2Man9 led to complete conversion to Dol25-PP-GlcNAc2Man9Glc (Fig. 1b). To confirm the physiologically relevant glycan structure, the peptide-attached glycans were incubated with α−1,2-mannosidase, which showed that ALG6-catalysed addition of glucose protected the A-branch of the Man9-containing glycan from mannosidase activity (Fig. 1b). ALG6 was found to be active in different detergents as well as reconstituted in lipid nanodiscs (Extended Data Fig. 1a).
Notably, our chemo-enzymatic approach enabled us to synthesize every lipid-linked oligosaccharide (LLO) intermediate of the biosynthetic pathway (Extended Data Fig. 1b). Our LLO analogues are readily accepted by both the single-subunit OST enzyme from T. brucei11 and the octameric yeast OST enzyme12.
Determination of the structure of ALG6
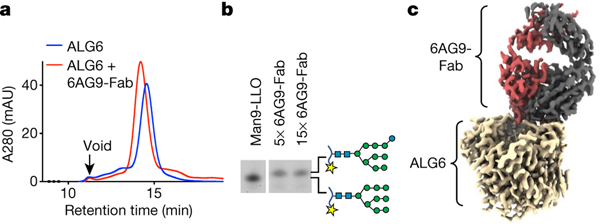
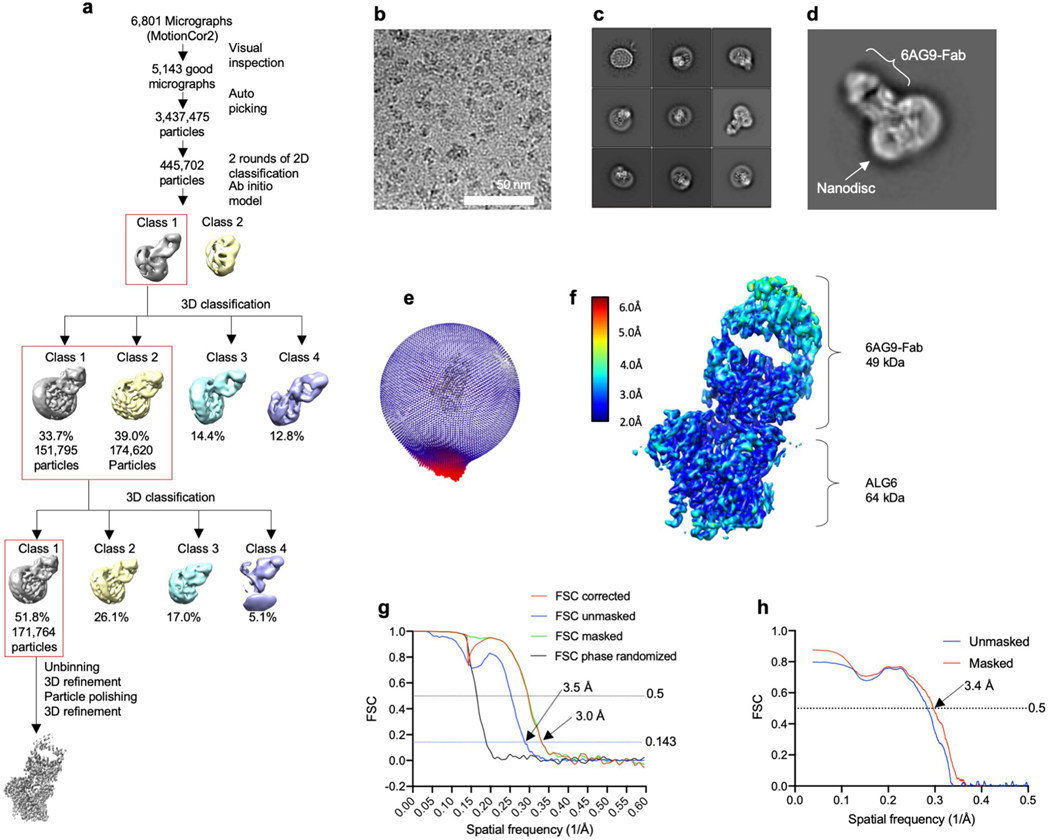
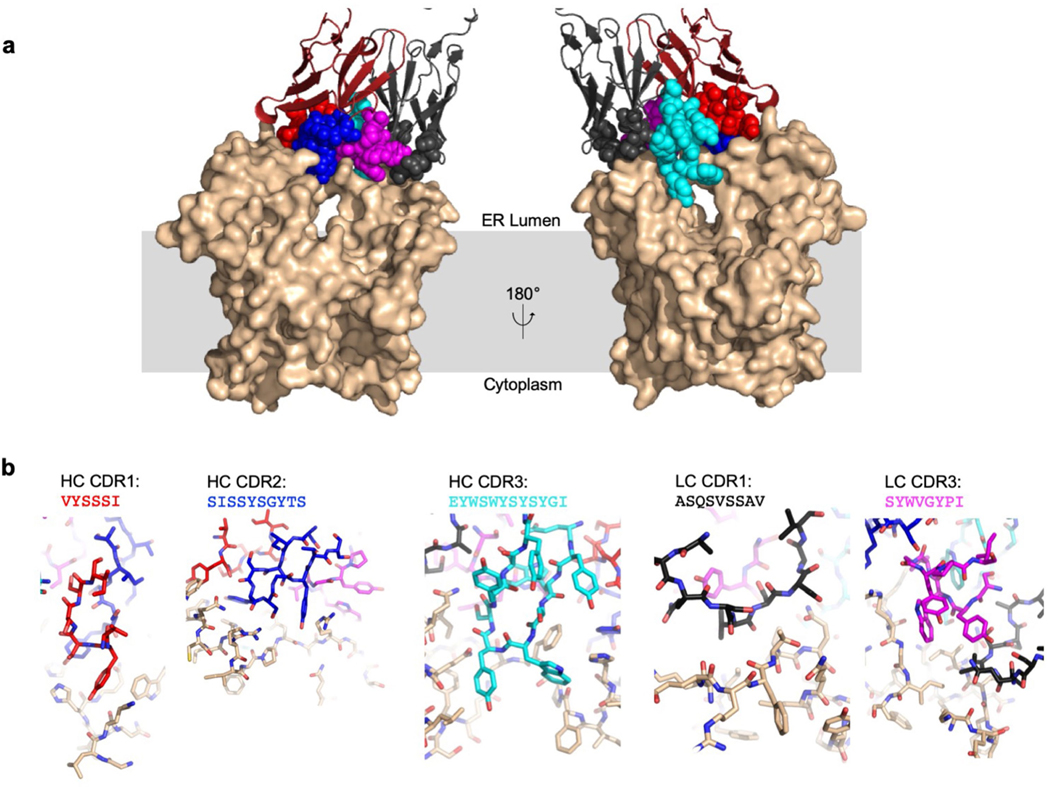
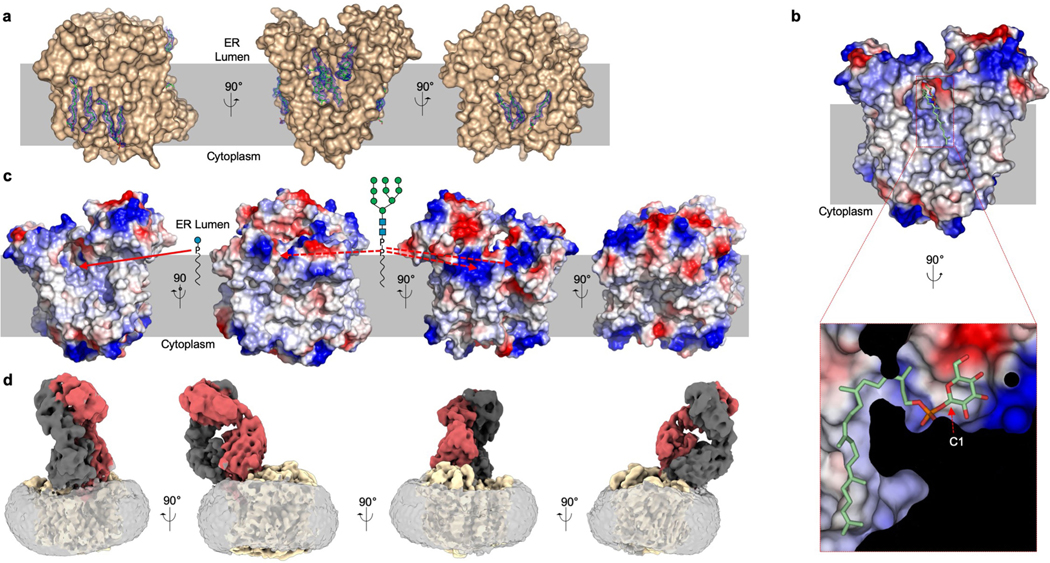
We used phage display and a synthetic library of antigen-binding Fab fragments13 to identify a conformational binder (6AG9-Fab) that increased the size and thermostability of detergent-solubilized ALG6 (Fig. 2a, Supplementary Fig. 2a, b) but did not interfere with its catalytic activity (Fig. 2b). The increased particle mass and size (Extended Data Fig. 2) aided particle alignment, and the electron microscopy (EM) reconstruction of nanodisc-reconstituted apo-ALG6 bound to 6AG9-Fab had an overall resolution of 3.0 Å (Fig. 2c, Extended Data Fig. 2, Extended Data Table 1, Supplementary Fig. 3). This allowed us to build an atomic model of ALG6 that lacked only 37 residues at the presumably flexible N terminus and 28 residues in the cytoplasmic loop that connects transmembrane helix 12 (TM12) and TM13, a segment that contains several short linear motifs including a di-arginine ER-retention motif14. 6AG9-Fab bound to a conformational, ER-luminal epitope of ALG6 and all interacting complementarity-determining region loops of the Fab fragment were well resolved (Extended Data Fig. 3). The EM maps revealed bound lipids and cholesterol hemisuccinate molecules in contact with transmembrane helices of ALG6 (Extended Data Fig. 4a).
Fig. 2 |. ALG6–6AG9-Fab interactions.
a, Size-exclusion chromatography of purified ALG6 in detergent (64 kDa) in the presence and absence of 6AG9-Fab (49 kDa). b, Analysis of ALG6 activity as described in Fig. 1b but in the presence of 6AG9-Fab. 6AG9-Fab was incubated with ALG6 at a 5-fold or 15-fold molar excess to test inhibition. The lane labelled Man9-LLO is a control sample for size comparison. c, Refined and B-factor-sharpened EM map of nanodisc-reconstituted ALG6 bound to 6AG9-Fab coloured by proximity to protein chains of ALG6 (light brown), 6AG9-Fab heavy chain (red) and 6AG9-Fab light chain (grey).
ALG6 topology and GT-C architecture
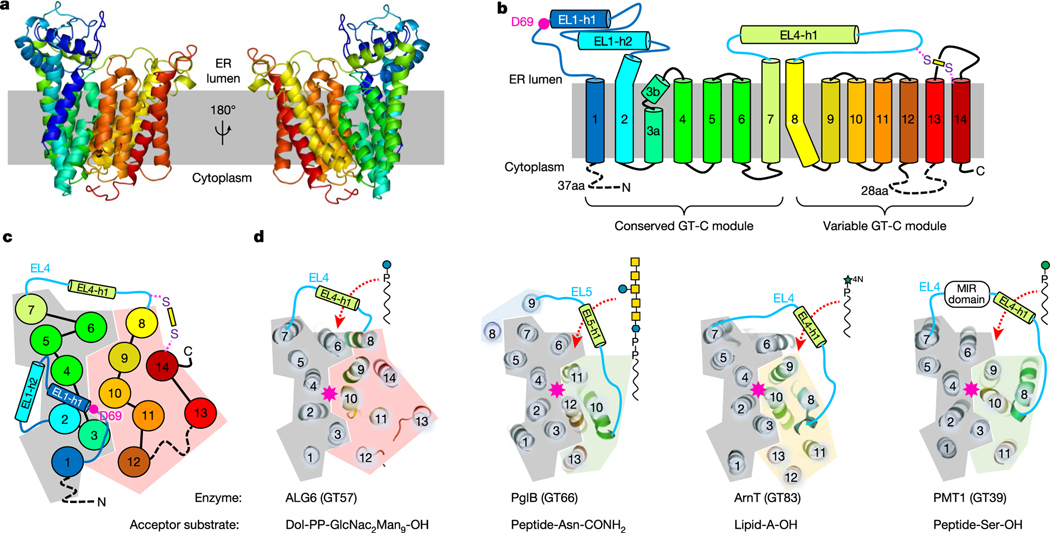
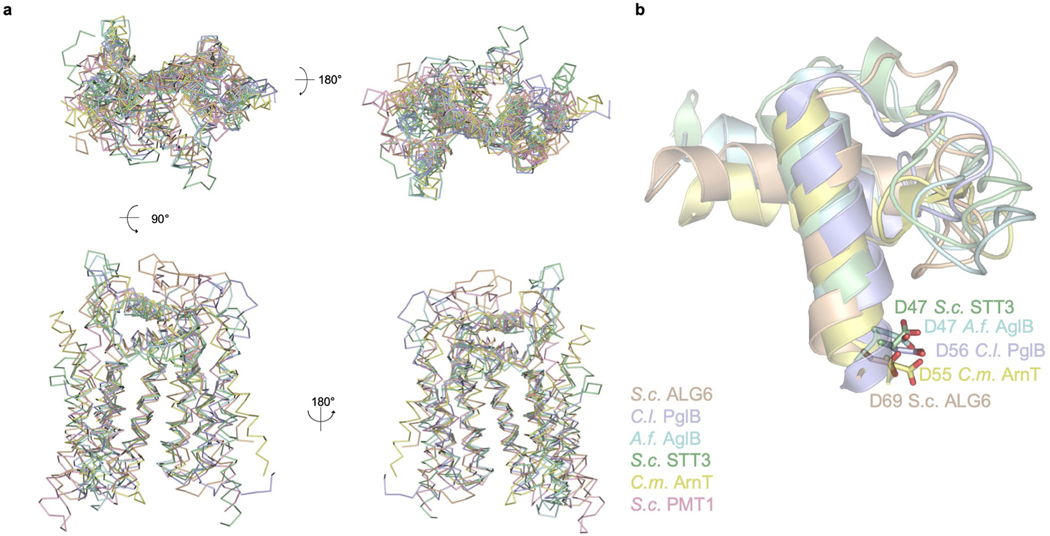
ALG6 has 14 transmembrane helices and two long loops (EL1 and EL4) that form helices in the ER lumen (Fig. 3a–c). A disulfide bridge covalently links the ER-luminal end of TM14 with the external loop EL4 that connects TM7 and TM8 (Fig. 3b, c). While the overall fold and membrane topology of ALG6 are unlike those of previously described structures, we noticed that the transmembrane arrangement and topology of the first half of the protein resemble partial structures of previously reported GT-C enzymes. We superimposed the structure of ALG6, which belongs to the GT57 subfamily according to the CAZy database15, onto those of PglB16 (GT 66), AglB17 (GT66), STT312,18 (GT66), ArnT19 (GT83) and PMT1/PMT220 (GT39). We found that the first seven transmembrane helices (including the luminal helices EL1-h1 and EL1-h2) are structurally similar, whereas the remainder of the proteins differ in fold, topology and number of transmembrane helices (Fig. 3d, Extended Data Fig. 5). This suggests that GT-C enzymes contain a structurally conserved, N-terminal module consisting of TM1–TM7, and a structurally variable C-terminal module (Fig. 3b–d). The structural similarity of the conserved module is not reflected in sequence conservation, as the primary sequences of the compared proteins are dissimilar21. However, the first external loop contains the catalytically essential aspartate residue Asp69 (see below).
Fig. 3 |. ALG6 structure and GT-C architecture.
a, Cartoon representation of ALG6 in rainbow colouring (blue at the N terminus and red at the C terminus). b, Schematic representation of ALG6 topology, with transmembrane and external loop helices numbered and coloured as in a. The pink dot indicates the proposed catalytic residue Asp69. The yellow bar depicts a disulfide bond. aa, amino acids. c, Top view of the schematic representation of ALG6 coloured as in b, with numbered circles depicting transmembrane helices and cylinders depicting helices in the ER-luminal loops. d, Comparison of the topologies of ALG6 and other selected GT-C enzymes of known structure. Ribbon diagrams of the top views of the structures are shown as in c. The constant GT-C modules are shaded grey, whereas the variable GT-C modules are shaded in different colours for each structure. Transmembrane helices are numbered. Note that GT39 and GT66 contain similar variable modules (shaded green). The pink asterisks indicate the locations of the catalytic sites. The red dashed arrows indicate the entry site for the dolichol-linked donor substrates, which are depicted using standard glycobiology symbols.
Our interpretation of a modular architecture is supported by the observation that in all published structures of GT-C enzymes, the active site and the substrate-binding cavities are located at the interface of the conserved and variable modules (pink asterisks and red dashed arrows in Fig. 3d). The architectural similarity further extends to how GT-Cs bind to their dolichyl(pyro)phosphate-linked donor substrates: the dolichol moiety invariably interacts with TM6 of the conserved GT-C module, whereas the distinct attached carbohydrates, which include single sugars or complex glycans, interact with the variable module. The donor substrates reach their respective active sites by ‘diving under’ arch-like structures formed by the external (or ER-luminal) loops that either link the conserved to the variable modules, or are part of the variable modules (Fig. 3d). These loops contain, at the minimum, a helix running parallel to the membrane but may contain entire domains (Fig. 3d). We conclude that the conserved module of GT-Cs probably served as a platform that allowed distinct functionalities to evolve, and that the modularity has allowed GT-C enzymes to accommodate different donor and acceptor substrates and to generate diverse active sites for distinct catalytic mechanisms during glycan transfer reactions.
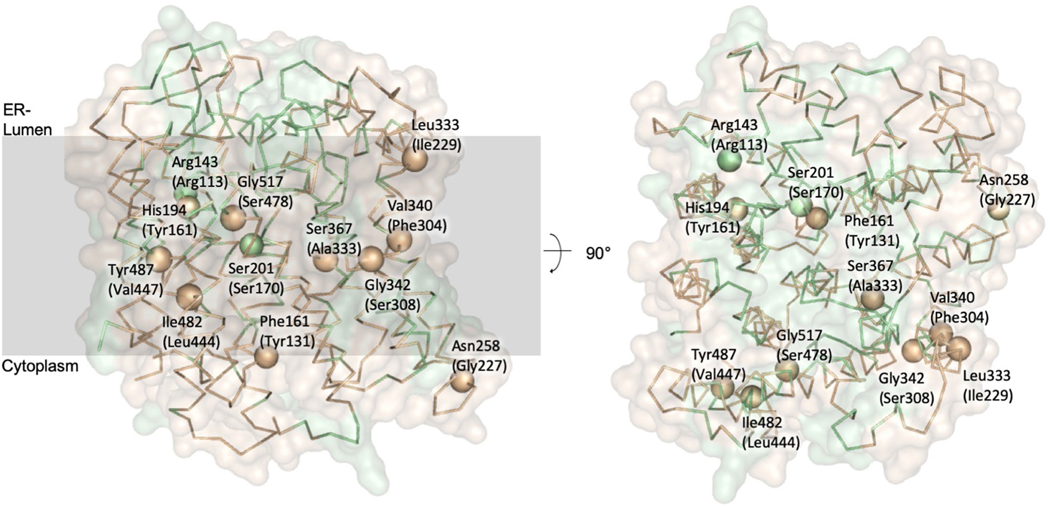
An analysis of residues that have been reported to be implicated in ALG6-associated human CDGs7,22–25 revealed that none of them point directly at the presumed active site. Rather, they are second shell residues with respect to the active site or are located even further away (Extended Data Fig. 6). Such mutations do not fully abolish ALG6 function, but rather reduce the enzymatic activity to a level that is not yet lethal22.
Structure of Dol25-P-Glc-bound ALG6
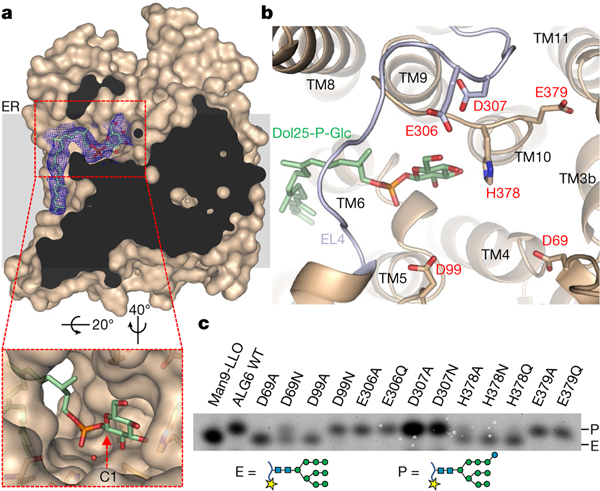
ALG6 forms a large, hydrophilic cavity facing the ER lumen and a groove-shaped cavity facing the lipid bilayer. The residues that line these cavities are not only highly conserved among ALG6 homologues but are also in the functionally related glucosyltrasferase ALG82,26, suggesting that they are involved in substrate binding or form the active site of the enzyme (Extended Data Fig. 7). To investigate the roles of these cavities, we collected a second cryo-EM data set for the ALG6–6AG9-Fab complex (Extended Data Fig. 8, Extended Data Table 1). Here, ALG6 was solubilized in digitonin and pre-incubated with the synthetic donor substrate Dol25-P-Glc. Although the overall resolution of the structure is 3.9 Å and thus lower than that of nanodisc-reconstituted apo-ALG6, there is a well-resolved density for bound donor substrate in a lipid-exposed groove formed by TM6, TM7 and TM8 (Fig. 4a, b, Extended Data Fig. 4b). The shape of the density can clearly accommodate a Dol25-P-Glc molecule. The citronellyl-farnesyl moiety is located in the conserved, lipid-facing groove and interacts mostly with hydrophobic residues from TM6. The loop EL4 forms an arch above the groove on the ER-luminal side, resulting in a funnel-like entrance, and the Dol25 moiety bends under this arch at the level of the membrane boundary. This causes the phosphate group, which represents the leaving group of the glucose transfer reaction, to be lodged in a slightly positively charged surface region of ALG6 (Extended Data Fig. 4b).
Fig. 4 |. Donor substrate binding and catalytic site of ALG6.
a, Surface representation of substrate-bound ALG6 in light brown with an EM density map for Dol25-P-Glc in the blue mesh carved to 3.0 Å, with bound Dol25-P-Glc in stick representation. The front of ALG6 was clipped for clarity. The inset shows a zoomed-in view. The red dashed arrow indicates the anomeric C1 of the glucose moiety. An ordered water molecule close to the C1 carbon is shown as a red sphere. b, Structure of the active site, with ALG6 shown in light brown ribbon representation with EL4 coloured in light blue. Acidic residues in the active site are shown in stick representation and labelled, and bound Dol25-P-Glc is depicted in stick representation, with carbon atoms coloured green. c, Analysis of substrate conversion by ALG6 variants, with mutations indicated above the lanes. WT, wild type.
The EM density revealed a feature large enough to accommodate the glucose moiety of Dol25-P-Glc, but the quality of the map does not allow exact positioning. This is in part due to the fact that this density also covers the adjacent side chain of the conserved histidine residue His378 (Fig. 4a, b). We could nevertheless assign the orientation of the glucose moiety, with one face packing against the surface of ALG6 and the other pointing towards the solvent. We built the model such that the α-anomeric position of the C1 carbon is accessible for a nucleophilic attack by the mannose C3 hydroxyl group of the acceptor glycan. This binding mode can ensure the specificity of ALG6 for Dol-P-Glc over Dol-P-Man, as the axial C2 hydroxyl group of a mannose would cause a steric clash with the enzyme surface (Fig. 4a). The specificity of ALG6 can be demonstrated in vitro, where only glucose, but not mannose, is transferred from a Dol25-P carrier (Extended Data Fig. 9a).
We could not unambiguously identify a second dolichol-binding groove in the ALG6 structure, and multiple smaller grooves are potential candidates for binding the acceptor substrate Dol-PP-GlcNAc2Man9 (Extended Data Fig. 4c). Because several of the transmembrane helices of ALG6 are not perpendicular to the lipid bilayer, the protein appears to induce a distortion of the lipid bilayer of the nanodisc, with the lipid bilayer appearing to thin out at the side of the cavity opposite to the ‘arch’ generated by EL4 (Extended Data Fig. 4d). Such membrane thinning might facilitate the binding of the dolichol-linked donor substrate from the membrane. Deformation of a membrane caused by a protein has been reported previously in the TMEM16 scramblase, where it has been proposed to facilitate or assist in lipid flipping27.
Catalytic mechanism
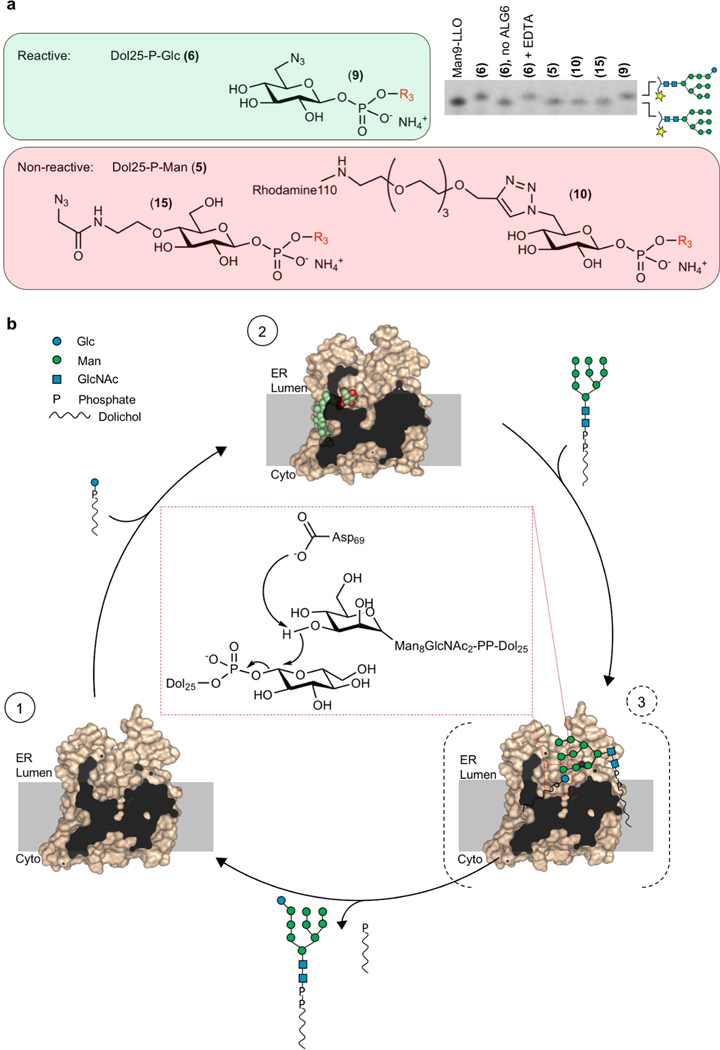
Given that ALG6 is an inverting glycosyltransferase, its reaction is thought to occur via an SN2 mechanism28. This implies that the C3 hydroxyl group of the terminal A-branch mannose of Dol-PP-GlcNAc2Man9 has to be activated for a nucleophilic attack on the anomeric C1 of the glucose moiety to form an α−1–3-glycosidic bond. This is generally achieved by deprotonation of the attacking hydroxyl group by an aspartate or glutamate side chain acting as a general base, either directly or by forming catalytic dyads or triads with suitable groups such as imidazole rings (histidine), hydroxyl groups of protein side chains or ordered water molecules29–31. In our Dol25-P-Glc-bound ALG6 structure, we identified five acidic residues in external loops that could act as a general base (Fig. 4b): Asp69 and Asp99 from EL1, Glu306 and Asp307 from EL4 and Glu379 from EL5. We mutated all five residues to alanine and, in the case of aspartates or glutamates, to asparagine or glutamine. We also mutated His378, which is in direct contact with the glucose moiety, to investigate its potential function in substrate binding. We expressed and purified the resulting ALG6 variants (Supplementary Fig. 1b, c) and tested their in vitro activity using our coupled assay (Fig. 4c). Activity was unaffected by mutation of Glu306, Asp307 and Glu379. Mutating Asp99 to an alanine abolished ALG6 function, whereas mutation to an asparagine did not reduce enzymatic activity. This suggests that Asp99 has a functional role, possibly in acceptor substrate binding, but is not the essential catalytic base. Finally, mutating Asp69 to an alanine abolished ALG6 function, and mutating it to an asparagine strongly reduced activity. We conclude that the structurally conserved Asp69 is the catalytic base in ALG6. The corresponding aspartates (Extended Data Fig. 5b) have been demonstrated to be catalytically essential in oligosaccharyltransferases12,16,17 and the arabinosyltransferase ArnT19 and proposed to be essential in the mannosyltransferase PMT1–PMT220. Hence, an aspartate or glutamate at the N-terminal end of the helix EL-h1 may be catalytically essential in all GT-C enzymes. Mutating His378, which is in the immediate vicinity of the glucose moiety of Dol25-P-Glc, to alanine, asparagine or glutamine, strongly reduced the activity of ALG6 but did not fully abolish it. Given the location of this residue, it is most likely to be involved in the binding and orientation of the donor substrate. Although it has been shown that at least one of the ER-luminal mannose transfer reactions of the LLO biosynthesis pathway is metal-dependent32, our EM map showed no evidence for a bound divalent metal ion (M2+), nor does ALG6 contain a DXD sequence motif similar to those shown to be essential for M2+ binding in oligosaccharyltransferase12,16,20. Purified ALG6 indeed retains activity in the presence of EDTA (Extended Data Fig. 9a), demonstrating that the reaction is metal ion-independent.
To probe the reaction mechanism further, we tested whether ALG6 could process synthetic Dol25-P-Glc analogues in which the glucose moiety carried distinct substituents that were originally designed for labelling studies (Extended Data Fig. 9a). We found that substituting the C4 hydroxyl group with a 2-azido-N-ethylacetamido group abolished glucose transfer, which can be rationalized by the structural data as this would lead to a clash with the side chain of His378 (Fig. 4b). Substituting the C6 hydroxyl group of glucose with an azido group did not impair glucosyl transfer, which is in line with the structural data given that this hydroxyl group is facing the solvent and can probably rotate freely (Fig. 4a). ALG6 activity was abolished when a larger substituent (tetraethylene glycol (PEG4)-linked rhodamine 110) replaced the C6 hydroxyl group (Extended Data Fig. 9a), where it probably hinders access of the acceptor substrate to the active site (Fig. 4a, Extended Data Fig. 4b).
On the basis of our structural and functional data, we propose a three-state mechanism for ALG6 function (Extended Data Fig. 9b). The structure of substrate-bound ALG6 suggests that Dol-P-Glc binds before the Man9-containing acceptor substrate, because the glucose moiety is at the bottom of the active site cavity. We therefore propose that donor and acceptor substrates bind sequentially and that Asp69 acts as a general base that abstracts the proton of the 3-hydroxyl group of the terminal A-branch mannose of the acceptor substrate to activate it for a nucleophilic attack. This may require a conformational change during which the glucose moiety of the donor substrate moves closer to the acceptor substrate and to the catalytic residue Asp69, akin to induced fit. An analogous conformational change has been described in another GT-C enzyme: the bacterial oligosaccharyltransferase PglB33. In ALG6, an induced fit mechanism would allow the enzyme to prevent futile donor substrate hydrolysis. Upon formation of a ternary complex, the glucose moiety may not only become better ordered but might also deviate from the chair conformation, as has been previously observed in other glycosyltransferases and glycosyl hydrolases34,35.
Conclusions
Our ALG6 structure not only defines the architecture of eukaryotic ALG enzymes that are involved in ER-luminal N-glycan synthesis but also suggests an unexpected structural modularity of GT-C enzymes. The conserved GT-C module acts as both a scaffold and a substrate activator by providing a catalytically essential general base residue, whereas the variable module allows different GT-Cs to catalyse diverse glycosyl transfer reactions and to bind to distinct substrates. We identified key residues involved in ALG6 catalysis and provide the basis for future studies of the catalytic mechanism of this and related GT-C enzymes. Our approach of combining synthetic chemistry with chemo-enzymatic glycan extension presents a highly efficient and controlled way to generate LLO analogues that are not only essential for studying ALG6–substrate interactions but may also allow all enzyme-catalysed reactions of the ER-based protein N-glycosylation pathway to be studied in molecular detail.
Methods
Overexpression and purification of ALG3
A synthetic gene (GenScript) of full-length ALG3-PreScission-EYFP-1D4 from Saccharomyces cerevisiae (UniProtKB: P38179) was optimized for expression in Spodoptera frugiperda (GeneArt, Thermo Fisher). The gene was cloned into a pOET1 vector (Oxford Expression Technologies) and was expressed in S. frugiperda (Sf9) cells transfected with baculovirus that was generated with FlashBAC DNA (Oxford Expression Technologies). Cells were cultured in serum-free SF-4 Baculo Express ICM medium (AMIMED) at 27 °C. Cells were transfected at a density of 1 × 106 cells per ml and were harvested after 3 days. For purification, the cells were resuspended in 150 mM NaCl, 50 mM HEPES pH 7.4 with 0.1 mg/ml DNase I, 1:100 protease inhibitor cocktail (Sigma), 0.1 mg/ml PMSF and were lysed by dounce homogenization before solubilization by addition of 1% n-dodecylβ-d-maltopyranosinde (DDM; Antrace), 0.2% cholesteryl hemisuccinate (CHS; Anatrace) and 10% glycerol. After 1 h of solubilization, cell debris was pelleted by centrifugation at 30,000 rpm in a type T45 Ti rotor (Beckmann). The supernatant was added to anti-1D4 antibody-coupled beads and was incubated for 1 h. The beads then were washed with 2 × 20 column volumes (cv.) of washing buffer (150 mM NaCl, 40 mM HEPES pH 7.4, 0.03% DDM and 0.006% CHS). Then the protein was eluted by incubation of washing buffer, supplemented with 0.5 mg/ml 1D4 peptide for 1 h.
Overexpression and purification of ALG6, ALG9 and ALG12
Synthetic genes (GenScript) of Flag-EYFP-PreScission-full length ALG6 (UniProtKB: Q12001), ALG9 (UniProtKB: P53868) or ALG12 (UniProtKB: P53730) from S. cerevisiae were optimized for expression in Homo sapiens (GeneArt, Thermo Fisher). Proteins were expressed, and cells were lysed, solubilized and pelleted as described above for ALG3. The supernatant was added to anti-Flag antibody-coupled resin and was incubated for 1 h. The beads then were washed with 2 × 10 cv. washing buffer (150 mM NaCl, 40 mM HEPES pH 7.4, 0.03% DDM and 0.006% CHS). Then the protein was eluted by incubation of washing buffer, supplemented with 0.2 mg/ml Flag peptide for 1 h. For functional assays, ALG6 was eluted with washing buffer supplemented with 3C-protease. For the substrate-bound structure, the immobilized ALG6 was solubilized, immobilized to Flag resin and washed as described above. Subsequently the immobilized protein was washed with 10 cv. washing buffer supplemented with 0.1% digitonin (Huberlab) and with 2 × 10 cv. digitonin washing buffer (150 mM NaCl, 40 mM HEPES pH 7.4 and 0.1% digitonin). The sample was eluted with 3C-protease and was subsequently purified by size-exclusion chromatography (SEC) in the same buffer.
ALG6-mutant generation
ALG6 mutants were generated by site-directed mutagenesis PCR and were expressed and purified by immobilization to Flag-resin and elution with peptide as describe above for wild-type ALG6. The proteins were further purified by SEC.
Nanodisc reconstitution of ALG6
A mixture of yeast polar lipids (Yeast Extract Polar, Avanti Polar Lipids) and CHS (80:20, w/w) was diluted to a concentration of 10 mM in buffer (150 mM NaCl and 40 mM HEPES pH 7.4). Lipids were solubilized by addition of 10 mM DDM and sonicated for 20 min. Purified EYFP-Pre-Scission-ALG6 in DDM:CHS-supplemented buffer, purified MSP1D1 protein37 and solubilized lipids were mixed in a ratio of 1:6:390. After incubation for 10 min at 4 °C, the mixture was incubated for 20 min at room temperature. Detergent was removed by addition of 0.8 mg activated Biobeads (Bio-Rad) per ml of reconstitution mixture and subsequent incubation at 4 °C overnight. Biobeads were removed and the reconstitution mixture was 10× diluted in washing buffer (150 mM NaCl and 40 mM HEPES pH 7.4). To remove empty nanodiscs, the reconstitution mixture was incubated with anti-Flag antibody-coupled resin for 1 h. After washing the resin with 2 × 10 cv. washing buffer, the sample was eluted by incubation with 3C-protease.
Expression and purification of EYFP
His6-EYFP-1D4 was cloned into a modified pET19b vector (Novagen) and overexpressed in BL21 gold cells (DE3) (Stratagene). Cells were grown in Terrific Broth medium at 37 °C to an OD600 of 3.0 before induction with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). For protein expression, cells were then grown overnight at 25 °C and harvested by centrifugation at 10,000g.
For purification, cells were resuspended in PBS (Gibco) supplemented with 1 mM PMSF and 0.025 mg/ml DNase I. Cells were lysed by three cycles of sonication (50% amplitude and 50% duty cycle). Cell debris was pelleted by ultracentrifugation at 35,000 rpm, in a type T45 Ti rotor, for 30 min. The supernatant was incubated with Ni-NTA resin for 1 h and was subsequently washed with 10 cv. 150 mM NaCl, 50 mM Tris-HCl pH 7.5 and 10 cv. 300 mM NaCl, 40 mM Tris-HCl pH 8.0 and 50 mM imidazole, before elution with 5 cv. 300 mM NaCl, 40 mM Tris-HCl pH 8.0 and 400 mM imidazole. Immediately afterwards, the protein buffer was exchanged into 150 mM NaCl and 50 mM Tris-HCl pH 7.5.
Enzymatic biotinylation of MSP1D1
MSP1D1 fused to a C-terminal Avitag was biotinylated in biotinylation buffer (75 mM NaCl, 50 mM bicine pH 8.3, 10 mM Mg acetate, 10 mM ATP and 0.25 mM biotin) with 2 μM BirA protein, overnight at 4 °C. 3C protease (2.5 μM) was added for 6 h at 4 °C. 3C-protease and BirA were removed by reverse binding to Ni-NTA. Subsequently, the biotinylated MSP1D1 protein was desalted into 150 mM NaCl, 50 mM HEPES pH 7.4 and subsequently used for nanodisc reconstitution as described above.
Synthetic antibody generation
For phage display experiments, enzymatically biotinylated MSP1D1 was used for the nanodisc reconstitution and no reverse binding or cleavage after nanodisc reconstitution was performed. Aggregates were removed by SEC after the nanodisc reconstitution.
To validate the immobilization efficiency of the final sample, pull-down assays of the target proteins immobilized on streptavidin MagneSphere paramagnetic particles (Promega) were performed, followed by biopanning using Fab Library E13,38; DNA was kindly provided by S. Koide (University of Rochester Medical Center, Rochester, NY, USA) and the phage library was prepared by S. Mukherjee (University of Chicago, Chicago, IL, USA). Five subsequent rounds of selection were performed in 25 mM HEPES pH 7.4 and 150 mM NaCl (selection buffer) supplemented with 1% bovine serum albumin (BSA)39,40.
The phage display selection was performed according to published protocols39,41. In brief, in the first round, 400 nM Flag-EYFP-Prescission-ALG6 in nanodiscs was immobilized onto paramagnetic particles for biopanning, followed by three washes and resuspension of the particles directly in log-phase Escherichia coli XL-1 blue cells for infection. Phages were amplified in 2×YT media supplemented with ampicillin (100 μg/ml) and M13-KO7 helper phage (109 p.f.u./ml). From the second to the fifth round, sorting was performed using a Kingfisher magnetic beads handler (Thermo Fisher) with decreasing concentration of target protein: 200 nM for the second round, 100 nM for the third round, 60 nM for the fourth round and 30 nM for the last round. Phage elution was done by disrupting immobilized nanodiscs with 1% Fos-choline 12 in selection buffer. In addition, for rounds 2–5, each phage pool was negatively selected against streptavidin-coated particles and then used as an input for the next round. Throughout the entire selection process, soluble competitors were included in excess: 2 μM non-biotinytaled empty nanodiscs (same reconstitution procedure as for ALG6, without the addition of ALG6 and using non-biotinylated MSP1D1) and 2 μM purified EYFP protein. Phage pools from the fourth and fifth round were used for binder screening.
Synthetic antibody screening
Single-point phage ELISA was performed to screen and validate the binding affinities of selected binders39. Briefly, E. coli XL-1 blue were transfected with selected phage pools and plated on LB/agar plates with ampicillin. Colonies were used to inoculate 2×YT media with 100 μg/ml ampicillin and 109 p.f.u./ml M13-KO7 helper phage. Overnight amplified cultures were pelleted by centrifugation and tenfold diluted supernatants were used for ELISA. For this purpose, 96-well plates coated with 2 μg/ml NeutrAvidin (Thermo Fisher) were blocked with selection buffer with 1% BSA, and Flag-EYFP-Prescission-ALG6 protein reconstituted in biotinylated nanodiscs (50 nM) was immobilized in each well. Subsequent ELISA assays were performed in selection buffer with 2% BSA. Diluted phage particles were assayed against the target protein by using an HRP-conjugated anti-M13 monoclonal antibody (GE Healthcare). Detection was done with TMB substrate (Thermo Fisher) quenched with 1.0 M HCl after 3 min and subsequent absorbance measurement at 450 nm. Biotinylated empty nanodiscs were used as a control and only target-specific binders were selected.
Selected binders were sequenced at the University of Chicago Comprehensive Cancer Center DNA Sequencing facility and cloned into a pRH2.2 vector (kind gift of S. Sidhu (University of Toronto, Toronto, ON, Canada)) using the In-Fusion Cloning kit (Takara). The antigen-binding fragments were expressed in E. coli BL21 gold cells and purified by Protein A chromatography and ion-exchange chromatography as described previously42.
Subsequently, multi-point protein ELISA was performed to estimate the binding affinity. ELISA plate preparation and target immobilization were done similar to single-point ELISA, followed by purified Fabs in threefold dilutions (starting from 3 μM) assayed against target protein by HRP-conjugated mouse anti-human IgG F(ab’)2 monoclonal antibody (Jackson). Detection was done as described above. Data points were plotted assuming sigmoidal dose response and EC50 values were calculated for affinity estimation.
Fab-binding characterization using SEC
For SEC binding analysis, 3C-protease-cleaved ALG6 in DDM:CHS-supplemented buffer was incubated with fourfold molar excess of Fab for 30 min on ice and subsequently analysed by SEC. Peak fractions were subsequently analysed by SDS–PAGE.
Thermostability experiments were performed as described previously43, with 3C-protease-cleaved ALG6 in DDM:CHS-supplemented buffer pre-incubated with threefold molar excess of Fab for 30 min on ice and by measuring A280 instead of fluorescence during SEC.
EM sample preparation
For the apo structure, ALG6 in MSP1D1 lipid nanodiscs was incubated with 6AG9-Fab in 1:2 molar ratio for 1 h at 4 °C and subsequently purified by SEC. Pooled peak fractions were concentrated to 0.33 mg/ml. Quantifoil holey carbon grids, Cu, R 1.2/1.3, 300 mesh, were glow discharged for 45 s, 25 mA using a PELCO easiGLOW glow discharger. Sample (4 μl) was applied to the cryo-EM grids and blotted for 1–2 s before plunge freezing in a liquid ethane–propane mixture with a Vitrobot Mark IV (Thermo Fisher Scientific) operated at 4 °C and 100% humidity.
For the substrate-bound structure, ALG6 in digitonin-supplemented buffer at 7.2 mg/ml was incubated with 1.5-fold excess of 6AG9-Fab for 1 h at 4 °C. Subsequently, the sample was purified by SEC and peak fractions were pooled and were concentrated to 6.3 mg/ml. The sample was supplemented with a final concentration of 150 μM Dol25-P-Glc and incubated for 1.5 h before grid preparation. Grids were prepared the same way as for the nanodisc sample, but with a 3.5-s blotting time.
EM data collection
Data were recorded on a Titan Krios electron microscope (Thermo Fisher Scientific) operated at 300 kV, equipped with a Gatan BioQuantum 1967 filter with a slit width of 20 eV and a Gatan K3 camera. Movies were collected semi-automatically using SerialEM44 at a nominal magnification of 105,000 and a pixel size of 0.42 Å per pixel in super-resolution mode. The defocus range was −0.6 to −2.8 μm. Each movie contained 40 images per stack with a dose per frame of 2.3 electrons/Å2.
EM data processing, model building and refinement
For the apo structure of ALG6 in nanodiscs, 6,801 movies were corrected for beam-induced motion using MotionCor245. The micrographs were visually inspected and 5,143 were used for further processing in RELION 3.046. Contrast transfer function (CTF) was estimated using gCTF47. Particles (n = 3,437,475) were auto-picked and extracted with threefold binning to a pixel size of 2.52 Å/pixel. After two subsequent rounds of 2D classification, 445,702 particles were used to generate an initial model. The particles were subjected to two subsequent rounds of 3D classification and the remaining 171,764 particles were subjected to 3D refinement. Subsequently, the particles were re-extracted at 0.84 Å/pixel and subjected to 3D refinement yielding a map at 3.2 Å resolution. Subsequent particle polishing and 3D refinement lead to a map at 3.1 Å resolution. Application of a tight mask that masked out most of the flexible constant domain yielded a map at 3.0 Å resolution.
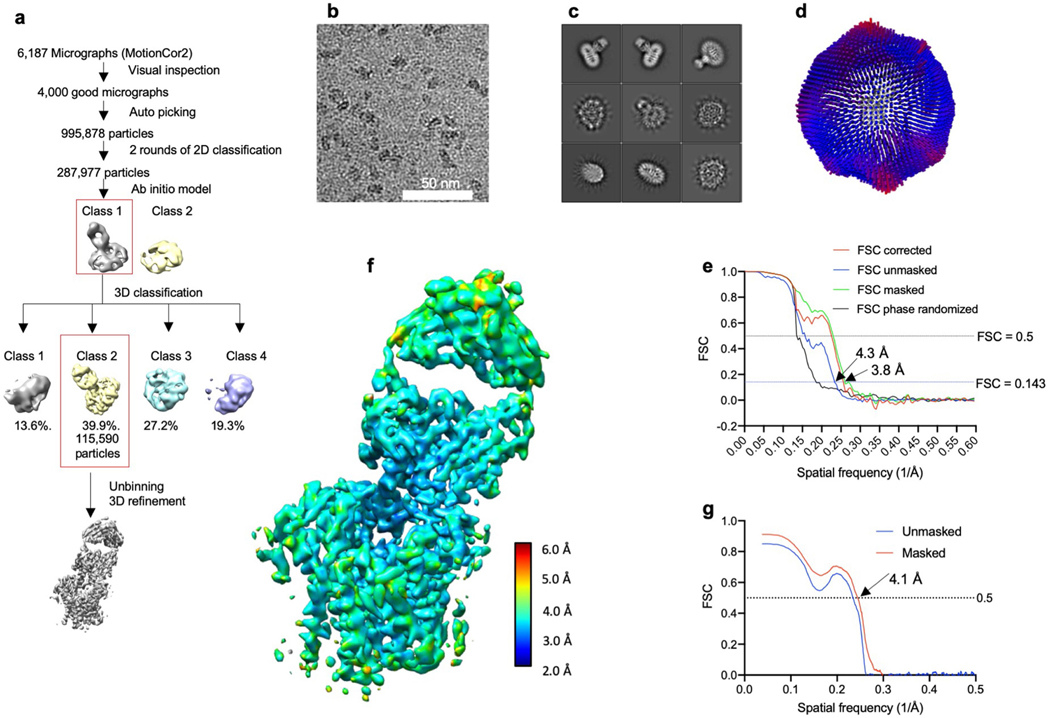
For the substrate-bound structure in digitonin, 6,187 movies were corrected for beam-induced motion using MotionCor245. The micrographs were visually inspected and 4,000 were used for further processing in RELION 3.046. CTF was estimated using gCTF47. Particles (n = 995,878) were auto-picked and extracted with threefold binning to a pixel size of 2.52 Å/pixel. After two subsequent rounds of 2D classification, 287,977 particles were selected and were used to calculate an initial model. After one round of 3D classification, 115,590 particles were selected for one round of 3D refinement and subsequent re-extraction at 0.84 Å/pixel. Subsequent 3D refinement yielded a final map at 3.9 Å.
Model building was performed in Coot48. Most of the Fab, except for the binding interface, was built based on a pre-existing model at high resolution (Protein Data Bank (PDB) ID 5UCB). The structures were refined in PHENIX49 and ligand files were generated in PHENIX eLBOW50. The correlation between model and map was validated using PHENIX mtriage51.
Figure preparation and data analysis
Local resolution estimations were calculated in ResMap52. Figures were prepared in PyMol53 and UCSF Chimera54 and UCSF ChimeraX55. Graphs were generated in GraphPad Prism 8. Protein sequence alignments were performed using CLUSTAL W56, Clustal Omega57 and EMBOSS Needle58. Figure 3d was generated using the following structures: S. cerevisiae ALG6 (this study), Campylobacter lari PglB (PDB 3RCE), Cupriavidus metallidurans ArnT (PDB 5F15) and S. cerevisiae PMT1 (PDB 6P25).
Chemical synthesis of substrate analogues
β-Mannosyl (3) and β-glucosyl (4) triesters were synthesized starting from 2,3,4,6-tetracetylated glycosides with an optimized procedure based on a previously described method59. Diphenyl-protected glycosyl phosphates were readily hydrogenated and coupled to farnesylcitronellol in presence of trichloroacetonitrile. Acetyl deprotection gave β-glycosyl phosphoisoprenoids Dol25-P-Man (5) and Dol25-P-Glc (6). To generate potentially clickable analogues of 6, an azide was introduced in the 6 position following known procedure60, and in the 4 position introducing a short linker via O-alkylation of orthogonal-protected glucopyranoside61 with bromoacetonitrile followed by reduction of the nitrile with BH3•SMe2 (ref. 62) and acylation of the primary amine. β-Phosphorylation of the azide-bearing lactols was performed using diallylphosphoryl chloride63 to avoid hydrogenation steps. The desired configuration was confirmed by ROESY (Supplementary Methods). Diallyl phosphates were deprotected, converted to the phosphoric acid and coupled with farnesylcitronellol11, as described above. More detailed procedures are described in Supplementary Methods.
Synthesis and purification of Dol25-PP-GlcNAc2Man9
Dol25-PP-GlcNAc2Man9 was produced as previously described10. Dol25-PP-GlcNAc2Man5 (20 μM) and Dol25-P-Man (120 μM) were mixed in 150 mM NaCl, 50 mM HEPES pH 7.4, 0.03% DDM and 0.006% CHS. Purified ALG3 (1.5 μM) was added and the mixture was incubated overnight at 4 °C. The reaction was inactivated by heating for 10 min at 95 °C. After cooling down to 4 °C, the reaction was supplemented with 10 mM MgCl2, and 1.5 μM ALG9 and 1.5 μM ALG12 were added and incubated overnight at 4 °C. For purification of Dol25-PP-GlcNAc2Man9, the reaction was lyophilized followed by a first extraction with CHCl3:MeOH (2:1, v/v) to remove contaminants of smaller LLO species. The remaining pellet was then subjected to extraction with CHCl3:MeOH:H2O (10:10:3, v/v/v). Subsequently, the extracted substrate was dried under N2 and was resuspended in 150 mM NaCl and 40 mM HEPES pH 7.4. The yield was quantified as described previously10.
Note that a similar synthetic route for in vitro synthesis of an LLO analogue was previously reported64. However, the lipid carrier in that study was a phytanol moiety instead of dolichol.
In vitro glycosyl transfer assays
Glycosyl transfer reactions for ALG1, ALG2 and ALG11 were performed as previously described10. Glycosyl transfer reactions for ALG3, ALG9 and ALG12 were performed in reaction buffer (150 mM NaCl, 50 mM HEPES pH 7.4, 0.03% DDM and 0.006% CHS) supplemented with 10–20 μM acceptor substrate (starting from purified Dol25-PP-GlcNAc2Man5 as the acceptor substrate with sequential addition and heat inactivation of ALG enzymes) and 50–120 μM donor substrate (Dol25-P-Man). Reactions for ALG9 and ALG12 were supplemented with 10 mM MgCl2. Protein concentrations were ranging from 0.5 to 2 μM. Reactions were carried out overnight at 4 °C. Reactions were stopped by heating at 98 °C for 10 min. This ensured that, during the ALG-mediated glycosyl transfer reactions, no other active ALG enzymes from previous reactions were present and eliminated fluorescence from EYFP fusion proteins, which otherwise would affect subsequent tricine gel analysis.
Reactions for ALG6 were carried out in reaction buffer (150 mM NaCl, 50 mM HEPES pH 7.4, 0.03% DDM and 0.006% CHS) supplemented with 10 μM purified Dol25-PP-GlcNAc2Man9 acceptor substrate, 50 μM of the respective donor substrate and 0.2 μM tag-cleaved ALG6. For reactions in the presence of EDTA or Fab, the entire reaction mixture was preincubated for 1 h on ice, before adding donor and acceptor substrates. The reactions were incubated for 6 h at 10 °C. Subsequently, the reactions were stopped by heating at 98 °C for 10 min. Reactions of ALG6 mutants were carried out as described above, at 10 °C for 24 h. All ALG reaction products were analysed by TbSTT3A-mediated oligosaccharyl-transfer and subsequent tricine gel analysis as previously described10. Owing to lower substrate specificity of TbSTT3A for Dol25-PP-GlcNAc2Man9Glc1, oligosaccharyl-transfer reactions for ALG6 reactions were performed for 12 h at 30 °C. For the oligosaccharyl-transfer reactions for ALG6 reactions that were carried out in the presence of EDTA, 30 μM MnCl2 was added, to avoid inhibition of STT3.
Statistics and reproducibility
Unless otherwise stated, ALG6 assays and tricine gel-based analyses were conducted once as shown in the figures.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
Atomic coordinates of the apo-ALG6 and Dol25-P-Glc-bound ALG6 models were deposited in the RCSB PDB under accession number 6SNI for apo-ALG6 and 6SNH for Dol25-P-Glc-bound ALG6. The 3D cryo-EM maps were deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-10258 for apo-ALG6 and EMD-10257 for Dol25-P-Glc-bound ALG6.
Extended Data
Extended Data Fig. 1 |. ALG6 activity in distinct lipidic environments and chemo-enzymatic synthesis of LLO intermediates.
a, Tricine gel-based analysis of the ALG6-catalysed reaction using Dol25-PP-GlcNAc2Man9 and Dol25-P-Glc substrates. The reactions were carried out in distinct detergents or with ALG6 reconstituted in lipid nanodiscs, as indicated above the gel lanes. The glycans were analysed as described in Fig. 1b. b, Tricine gel-based analysis of ALG reaction cascade intermediates, with reactions analysed as described in Fig. 1b. The structures of glycopeptides are schematically shown above the lanes and respective glycosyltransferase enzymes are indicated above the arrows.
Extended Data Fig. 2 |. Data processing and structure determination of the substrate-free ALG6–6AG9-Fab complex in lipid nanodiscs.
a, Overview of the EM data processing and structure determination pipeline using RELION 3.046. b, Representative cryo-EM micrograph. c, Representative 2D classes. d, A representative 2D class with 6AG9-Fab and the nanodisc belt highlighted. e, Spatial distribution of particles in the final iteration of 3D refinement. f, Refined and B-factor-sharpened EM map, coloured by local resolution estimation as calculated in ResMap52. g, Resolution estimation of the final map via Fourier shell correlation (FSC), as calculated in RELION 3.0. h, The FSC between model and map, as calculated in PHENIX mtriage51.
Extended Data Fig. 3 |. Molecular interactions at the interface of ALG6 and 6AG9-Fab.
a, Surface representation of ALG6 in light brown and cartoon representation of the variable fragment of 6AG9-Fab coloured red (heavy chain) and black (light chain). The five complementarity-determining region (CDR) loops of 6AG9-Fab interacting with ALG6 are shown in sphere representation and make contact mostly with the ER-luminal ‘arch’ formed by EL4 of ALG6. b, Detailed view of the individual CDR:ALG6 interactions, with 6AG9-Fab CDRs and ALG6 in stick representation and coloured as in a. The CDR sequences are indicated above the panels. Note that CDR1 of the light chain (LC) was not randomized. HC, heavy chain.
Extended Data Fig. 4 |. Substrate and lipid interactions of ALG6.
a, Distinct lipid binding sites were identified in the structure of nanodisc-reconstituted ALG6, shown in light brown surface representation. Ordered phospholipids and cholesteryl-hemisuccinate molecules are shown in stick representation (green), and the EM density map was contoured at 5 r.m.s.d. and carved to 1.6 Å. b, Electrostatic surface potential of ALG6, with negative charges coloured in red, neutral charges in white, positive charges in blue and bound Dol25-P-Glc depicted in stick representation with carbons in green. The inset shows a zoomed-in view of the kink induced in the dolichol moiety. c, Electrostatic surface potential of ALG6 shown from four angles to indicate potential binding sites for lipid-linked substrates of ALG6 based on the presence of suitable surface cavities. The red solid arrow indicates the binding site of the observed donor substrate Dol25-P-Glc. The red dashed arrows indicate potential binding sites of the acceptor substrate Dol-PP-GlcNAc2Man9. These sites were chosen based on groove-like features in the transmembrane region that might bind dolichol and positively charged patches at the ER-luminal membrane boundary that could help to bind the pyrophosphate moiety of the acceptor LLO. Binding to these sites would allow the terminal mannose of the A-branch of the acceptor substrate to reach the active site. d, Membrane deformation by ALG6 shown from distinct views. The EM map of the apo-ALG6 complex with the 6AG9-Fab complex in a lipid nanodisc is coloured yellow for ALG6, red (heavy chain) and black (light chain) for 6AG9-Fab, and transparent grey for density indicating the lipid nanodisc. Note that the lipid bilayer of the nanodisc surrounding the ALG6 protein is twisted and thinner in the region opposite the arch formed by EL4.
Extended Data Fig. 5 |. Structural similarity of the conserved module of GT-C enzymes.
a, Cα traces of the first seven transmembrane helices and the first external loop EL1 of selected structures are shown following superposition. These following GT-C family members of known structure were used, with the GT family number indicated: S. cerevisiae (S.c.) PMT1 (GT39, PDB ID 6P25), S.c. ALG6 (GT57, this study), C. lari (C.l.) PglB (GT66, PDB ID 3RCE), A. fulgidus (A.f.) AglB (GT66, PDB ID 3WAJ), S.c. STT3 (GT66, PDB ID 6EZN) and C. metallidurans ArnT (GT83, PDB ID 5F15). b, Close-up view of EL-h1, EL-h2 and the connecting loop of GT-Cs where the catalytic role of the conserved aspartate, shown in stick representation, was experimentally demonstrated.
Extended Data Fig. 6 |. Residues involved in CDGs caused by ALG6 mutations.
The structure of yeast ALG6 is depicted as a Cα trace and as a transparent surface representation coloured in light brown. The surface is coloured green for residues conserved between yeast and human ALG6. Cα atoms of residues that have been shown to cause CDGs in the human homologue of ALG6 are shown in sphere representation. Residue numbers in brackets indicate the corresponding residues of human ALG6 according to pairwise protein sequence alignment with EMBOSS needle58.
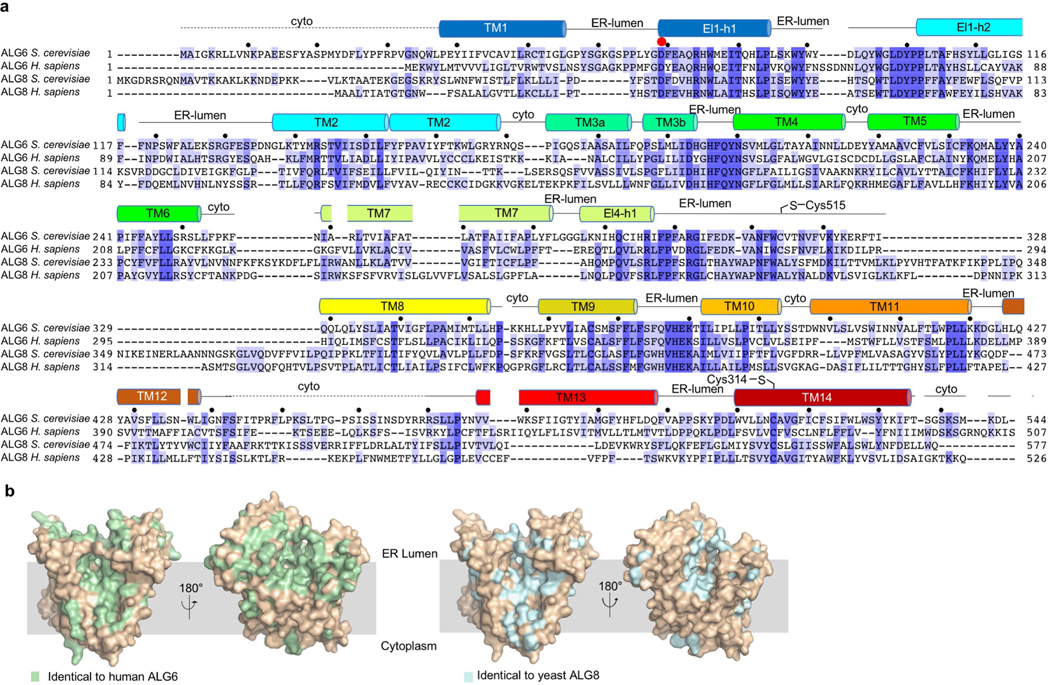
Extended Data Fig. 7 |. Sequence alignment of ALG6 homologues and orthologues.
a, Alignment of amino acid sequences of yeast and human ALG6 and ALG8 proteins, generated with Clustal Omega57 (Uniprot identifiers: Q12001, Q9Y672, P40351 and Q9BVK2). Secondary structure elements of yeast ALG6 are depicted and labelled above the sequence. The dashed lines indicate regions that are disordered in the ALG6 structures. Cytosolic regions are labelled ‘cyto’ and ER-luminal regions are labelled ‘ER-lumen’. The red dot indicates the catalytic base Asp69, which is a structurally conserved aspartate residue that has been observed or proposed to be the catalytic base in all GT-C enzymes of known structures. The black dots indicate every tenth amino acid in the sequence of S. cerevisiae ALG6. b, Surface representation of yeast ALG6 (light brown), with residues identical to human ALG6 highlighted in green (left) and residues identical to yeast ALG8 highlighted in blue (right).
Extended Data Fig. 8 |. Data processing and structure determination of the substrate-bound ALG6–6AG9-Fab complex in detergent solution.
a, Overview of the EM data processing and structure determination pipeline using RELION 3.046. b, A representative cryo-EM micrograph. c, Representative 2D classes. d, Spatial distribution of particles in the final iteration of 3D refinement. e, Resolution estimation of the final map via FSC, as calculated in RELION 3.0. f, The final refined and B-factor-sharpened map, coloured by local resolution estimation, as calculated in ResMap52. g, The FSC between model and map, as calculated in PHENIX mtriage51.
Extended Data Fig. 9 |. Mechanism of ALG6-catalysed glucosyl transfer.
a, Chemical structures of synthetic donor substrate analogues (left) and their functional analysis (right). Compound numbers are indicated in bold and in parentheses. Analysis of ALG6 activity as described in Fig. 1b but in the presence of different substrate analogues as indicated above the lanes. The lane labelled Man9-LLO is a control sample for size comparison. In one of the ALG6 reactions, a pre-incubation of ALG6 with 10 mM EDTA was used to remove any divalent ions from the solution. b, Proposed three-state mechanism of ALG6; intermediate states are numbered and encircled. State 1 represents the apo state of ALG6 based on our EM structure, with ALG6 in surface representation (brown) and clipped for better visualization of the substrate-binding pocket. State 2 represents ALG6 bound to a donor substrate based on our EM structure, with Dol25-P-Glc shown in sphere representation with carbon atoms coloured green and oxygen atoms coloured red. State 3 represents a putative ternary complex based on the apo structure of ALG6, with substrates drawn manually. The inset depicts the proposed reaction mechanism, with Asp69 acting as a general base that deprotonates the C3 hydroxyl group of the attacking mannose moiety.
Extended Data Table 1 |.
Cryo-EM data collection, refinement and validation statisticsa as defined in PHENIX49
| #1 ALG6 apo (EMDB-10258) (PDB 6SNI) | #2 ALG6 Dol25-P-Glc (EMDB-10257) (PDB 6SNH) | |
|---|---|---|
| Data collection and processing | ||
| Magnification | 105.000 | 105,000 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e-/Å2) | 2.3 | 2.3 |
| Defocus range (μm) | −0.6 to 0.8 | −0.6 to 0.8 |
| Pixel size (Å) | 0.42 | 0.42 |
| Symmetry imposed | Cl | Cl |
| Initial particle images (no.) | 3,437,374 | 995,878 |
| Final particle images (no.) | 171,764 | 115,590 |
| Map resolution (Å) | 3.03 | 3.94 |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range (Å) | 25–3.04 | 25–3.89 |
| Refinement | ||
| Initial model used (PDB code) | 5UCB (Fab) | 6SNI |
| Model resolution (Å) | 2.96 | 3.90 |
| FSC threshold | 0.143 | 0.143 |
| Model resolution range (Å)a | 2.96–3.04 | 3.74–3.89 |
| Map sharpening B factor (Å2) | −68.9 | −178.4 |
| Model composition | ||
| Non-hydrogen atoms | 7,510 | 7,315 |
| Protein residues | 918 | 918 |
| Ligands | 8 | 1 |
| B factors (Å2) | ||
| Protein | 19.57 | 48.23 |
| Ligand | 42.76 | 84.99 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.008 |
| Bond angles (°) | 0.848 | 0.912 |
| Validation | ||
| MolProbity score | 3.09 | 2.49 |
| Clashscore | 23.86 | 19.09 |
| Poor rotamers (%) | 8.07 | 1.49 |
| Ramachandran plot | ||
| Favored (%) | 90.55 | 88,90 |
| Allowed (%) | 9.23 | 10.99 |
| Disallowed (%) | 0.22 | 0.11 |
Supplementary Material
Acknowledgements
This research was supported by the Swiss National Science Foundation (SNF) Sinergia programmes TransGlyco (CRSII3_147632) and GlycoStart (CRSII5_173709) to M.A., J.-L.R. and K.P.L, SNF grant 310030B_166672 to K.P.L., as well as by the US National Institutes of Health grant GM117372 to A.A.K. Cryo-EM data were collected at the ScopeM facility at ETH Zürich. We thank the staff of ScopeM for technical support; J. Zürcher for technical support with protein expression and purification; J. Kowal and I. Manolaridis for help with EM data collection; and A. Ramirez and A. Alam for helpful discussions.
Footnotes
Competing interests The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-020-2044-z.
Peer review information Nature thanks Xiaochen Bai and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-020-2044-z.
References
- 1.Kornfeld R. & Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem 54, 631–664 (1985). [DOI] [PubMed] [Google Scholar]
- 2.Burda P. & Aebi M. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426, 239–257 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Liu J. & Mushegian A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 12, 1418–1431 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moremen KW & Haltiwanger RS Emerging structural insights into glycosyltransferase-mediated synthesis of glycans. Nat. Chem. Biol 15, 853–864 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Runge KW, Huffaker TC & Robbins PW Two yeast mutations in glucosylation steps of the asparagine glycosylation pathway. J. Biol. Chem 259, 412–417 (1984). [PubMed] [Google Scholar]
- 6.Reiss G, te Heesen S, Zimmerman J, Robbins PW & Aebi M. Isolation of the ALG6 locus of Saccharomyces cerevisiae required for glucosylation in the N-linked glycosylation pathway. Glycobiology 6, 493–498 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Haeuptle MA & Hennet T. Congenital disorders of glycosylation: an update on defects affecting the biosynthesis of dolichol-linked oligosaccharides. Hum. Mutat 30, 1628–1641 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Jaeken J, Lefeber D. & Matthijs G. Clinical utility gene card for: ALG6 defective congenital disorder of glycosylation. Eur. J. Hum. Genet 23, 1–3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morava E. et al. ALG6-CDG: a recognizable phenotype with epilepsy, proximal muscle weakness, ataxia and behavioral and limb anomalies. J. Inherit. Metab. Dis 39, 713–723 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Ramírez AS et al. Chemo-enzymatic synthesis of lipid-linked GlcNAc2Man5 oligosaccharides using recombinant Alg1, Alg2 and Alg11 proteins. Glycobiology 27, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramírez AS et al. Characterization of the single-subunit oligosaccharyltransferase STT3A from Trypanosoma brucei using synthetic peptides and lipid-linked oligosaccharide analogs. Glycobiology 27, 525–535 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wild R. et al. Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science 359, 545–550 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Fellouse FA et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol 373, 924–940 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Gouw M. et al. The eukaryotic linear motif resource—2018 update. Nucleic Acids Res. 46, D428–D434 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM & Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lizak C, Gerber S, Numao S, Aebi M. & Locher KP X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto S. et al. Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. Proc. Natl Acad. Sci. USA 110, 17868–17873 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai L, Wang T, Zhao G, Kovach A. & Li H. The atomic structure of a eukaryotic oligosaccharyltransferase complex. Nature 555, 328–333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrou VI et al. Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation. Science 351, 608–612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai L, Kovach A, You Q, Kenny A. & Li H. Structure of the eukaryotic protein O-mannosyltransferase Pmt1–Pmt2 complex. Nat. Struct. Mol. Biol 26, 704–711 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albuquerque-Wendt A, Hütte HJ, Buettner FFR, Routier FH & Bakker H. Membrane topological model of glycosyltransferases of the GT-C superfamily. Int. J. Mol. Sci 20, 4842 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imbach T. et al. A mutation in the human ortholog of the Saccharomyces cerevisiae ALG6 gene causes carbohydrate-deficient glycoprotein syndrome type-Ic. Proc. Natl Acad. Sci. USA 96, 6982–6987 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imbach T. et al. Multi-allelic origin of congenital disorder of glycosylation (CDG)-Ic. Hum. Genet 106, 538–545 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Westphal V, Schottstädt C, Marquardt T. & Freeze HH Analysis of multiple mutations in the hALG6 gene in a patient with congenital disorder of glycosylation Ic. Mol. Genet. Metab 70, 219–223 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Dercksen M. et al. ALG6-CDG in South Africa: genotype–phenotype description of five novel patients. JIMD Rep. 8, 17–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Runge KW & Robbins PW A new yeast mutation in the glucosylation steps of the asparagine-linked glycosylation pathway. Formation of a novel asparagine-linked oligosaccharide containing two glucose residues. J. Biol. Chem 261, 15582–15590 (1986). [PubMed] [Google Scholar]
- 27.Lee BC et al. Gating mechanism of the extracellular entry to the lipid pathway in a TMEM16 scramblase. Nat. Commun 9, 3251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lairson LL, Henrissat B, Davies GJ & Withers SG Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem 77, 521–555 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Albesa-Jové D, Cifuente JO, Trastoy B. & Guerin ME Quick-soaking of crystals reveals unprecedented insights into the catalytic mechanism of glycosyltransferases. Methods Enzymol. 621, 261–279 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Notenboom V. et al. Insights into transition state stabilization of the β−1,4-glycosidase Cex by covalent intermediate accumulation in active site mutants. Nat. Struct. Biol 5, 812–818 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Chang A, Singh S, Phillips GN Jr & Thorson JS Glycosyltransferase structural biology and its role in the design of catalysts for glycosylation. Curr. Opin. Biotechnol 22, 800–808 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma CB, Knauer R. & Lehle L. Biosynthesis of lipid-linked oligosaccharides in yeast: the ALG3 gene encodes the Dol-P-Man:Man5GlcNAc2-PP-Dol mannosyltransferase. Biol. Chem 382, 321–328 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Napiórkowska M. et al. Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase. Nat. Struct. Mol. Biol 24, 1100–1106 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Davies GJ, Planas A. & Rovira C. Conformational analyses of the reaction coordinate of glycosidases. Acc. Chem. Res 45, 308–316 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Ardèvol A. & Rovira C. Reaction mechanisms in carbohydrate-active enzymes: glycoside hydrolases and glycosyltransferases. Insights from ab initio quantum mechanics/molecular mechanics dynamic simulations. J. Am. Chem. Soc 137, 7528–7547 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Aebi M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833, 2430–2437 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Denisov IG, Grinkova YV, Lazarides AA & Sligar SG Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc 126, 3477–3487 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Fellouse FA, Wiesmann C. & Sidhu SS Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc. Natl Acad. Sci. USA 101, 12467–12472 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominik PK & Kossiakoff AA Phage display selections for affinity reagents to membrane proteins in nanodiscs. Methods Enzymol. 557, 219–245 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Dominik PK et al. Conformational chaperones for structural studies of membrane proteins using antibody phage display with nanodiscs. Structure 24, 300–309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hornsby M. et al. A high through-put platform for recombinant antibodies to folded proteins. Mol. Cell. Proteomics 14, 2833–2847 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borowska MT, Dominik PK, Anghel SA, Kossiakoff AA & Keenan RJ A YidC-like protein in the archaeal plasma membrane. Structure 23, 1715–1724 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hattori M, Hibbs RE & Gouaux E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure 20, 1293–1299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P. & Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moriarty NW, Grosse-Kunstleve RW & Adams PD Electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Afonine PV et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol 74, 814–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kucukelbir A, Sigworth FJ & Tagare HD Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. [Google Scholar]
- 54.Pettersen EF et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Goddard TD et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson JD, Higgins DG & Gibson TJ CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sievers F. & Higgins DG Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Needleman SB & Wunsch CD A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol 48, 443–453 (1970). [DOI] [PubMed] [Google Scholar]
- 59.Sabesan S. & Neira S. Synthesis of glycosyl phosphates and azides. Carbohydr. Res 223, 169–185 (1992). [Google Scholar]
- 60.Maunier V, Boullanger P, Lafont D. & Chevalier Y. Synthesis and surface-active properties of amphiphilic 6-aminocarbonyl derivatives of d-glucose. Carbohydr. Res 299, 49–57 (1997). [Google Scholar]
- 61.Williams RJ et al. Combined inhibitor free-energy landscape and structural analysis reports on the mannosidase conformational coordinate. Angew. Chem. Int. Edn Engl 53, 1087–1091 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Malet C. & Hindsgaul O. Versatile functionalization of carbohydrate hydroxyl groups through their O-cyanomethyl ethers. J. Org. Chem 61, 4649–4654 (1996). [DOI] [PubMed] [Google Scholar]
- 63.Li T, Tikad A, Pan W. & Vincent SP β-Stereoselective phosphorylations applied to the synthesis of ADP- and polyprenyl-β-mannopyranosides. Org. Lett 16, 5628–5631 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Li ST et al. Reconstitution of the lipid-linked oligosaccharide pathway for assembly of high-mannose N-glycans. Nat. Commun 10, 1813 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates of the apo-ALG6 and Dol25-P-Glc-bound ALG6 models were deposited in the RCSB PDB under accession number 6SNI for apo-ALG6 and 6SNH for Dol25-P-Glc-bound ALG6. The 3D cryo-EM maps were deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-10258 for apo-ALG6 and EMD-10257 for Dol25-P-Glc-bound ALG6.