Abstract
The utility of the urinary proteome in infectious diseases remains unclear. Here, we analyzed the proteome and metabolome of urine and serum samples from patients with COVID-19 and healthy controls. Our data show that urinary proteins effectively classify COVID-19 by severity. We detect 197 cytokines and their receptors in urine, but only 124 in serum using TMT-based proteomics. The decrease in urinary ESCRT complex proteins correlates with active SARS-CoV-2 replication. The downregulation of urinary CXCL14 in severe COVID-19 cases positively correlates with blood lymphocyte counts. Integrative multiomics analysis suggests that innate immune activation and inflammation triggered renal injuries in patients with COVID-19. COVID-19-associated modulation of the urinary proteome offers unique insights into the pathogenesis of this disease. This study demonstrates the added value of including the urinary proteome in a suite of multiomics analytes in evaluating the immune pathobiology and clinical course of COVID-19 and, potentially, other infectious diseases.
Keywords: COVID-19, proteomics, metabolomics, serum, urine, CXCL14, ESCRT super-complex, renal injury
Graphical abstract
The value of the urine proteome is largely underestimated compared to the blood proteome. Bi et al. systematically compare proteins and metabolites in COVID-19 urine and sera and show the urine proteome to be a valuable source of biomarkers, offering unique insights into the pathogenesis of infectious diseases.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic threatens hundreds of millions of people in the world. Over 137 million people have been infected by severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), with over 5.5 million of deaths globally (Worldometer, 2021) as of January 9th, 2022. Approximately 80% of patients with COVID-19 are not severely ill, displaying mild symptoms with a good prognosis. The remaining 20% of patients develop severe illness requiring intensive care, including oxygen therapy and/or assisted ventilation (WHO, 2020b). The ability to identify early infected patients who will or will not progress to severe disease relieves the overall burden and improves the effectiveness of clinical care but demands fundamental understanding of the molecular pathogenesis of COVID-19.
Multiple serological alterations exhibit specific changes in severe COVID-19, such as interleukin-6 (IL-6), d-dimer (d-D), glucose, thrombin time, fibrinogen, C-reactive protein (CRP), and blood lymphocyte count (Gao et al., 2020; Qin et al., 2020; Tan et al., 2020). Systematic screening of proteins, metabolites, and lipids has uncovered aberrant regulation of physiological processes, including the complement system, macrophage functions, and platelet degranulation in the sera of severe cases (Shen et al., 2020; Wu et al., 2020). Combinations of these blood analytes could be used to classify the severity of COVID-19 (Messner et al., 2020; Shen et al., 2020).
Urine is derived from the peripheral circulation and is a more accessible source for diagnosing several diseases (Adachi et al., 2006; Barratt and Topham, 2007). It has been reported that multiple urinary biochemical analytes, including glucose (GLU-U), proteinuria (Rui et al., 2020), urine β2-microglobulin, and liver-type fatty acid-binding proteins (Katagiri et al., 2020), correlated with COVID-19 severity. Proteomic studies of urine have been used to discover novel disease biomarkers, such as recurrent urinary tract infections (Muntel et al., 2015; Vitko et al., 2020) and familial Parkinson’s disease (Virreira Winter et al., 2021). Proteomic analysis of the urine of 6 patients with COVID-19 and 32 healthy controls identified 214 uniquely altered proteins in COVID-19 urine (Li et al., 2020). Tian et al. (2020) reported the downregulation of immune-related proteins such as tyrosine phosphatase receptor type C, leptin, and tartrate-resistant acid phosphatase type 5 by analyzing the urine proteome of 14 patients with COVID-19 and 23 controls. These studies suggest the potential value of urinary proteins in understanding host responses in COVID-19. However, the sample sizes of these studies were relatively small. What remains unclear are the association of blood and urinary proteins and the interplay between proteins and metabolites. While several metabolomic studies of COVID-19 serum have been reported (Heer et al., 2020; Shen et al., 2020; Thomas et al., 2020; Wu et al., 2020), whether and how urinary metabolites are modulated in COVID-19 is unknown.
In this study, we systematically investigated the proteome and metabolome of COVID-19 urine and matched serum specimens. Our data show the modulation of proteins and metabolites in COVID-19 urine and sera, which uncover immune responses to SARS-CoV-2. We uncovered intriguing disparities between urine and serum proteomes. Integrative analysis of the proteome and metabolome revealed evidence of renal injuries induced by immune dysregulation. This study presents proof-of-principle evidence for the feasibility of using urine as an additional and informative biospecimen for understanding the pathogenesis of COVID-19 and other infectious diseases.
Results
Proteomic and metabolomic profiling of COVID-19 urine and sera
A cohort of 71 patients with COVID-19 comprising 23 severe cases and 48 non-severe cases were recruited for this study. Another 17 non-COVID-19 cases with flu-like symptoms such as cough and fever and 27 healthy controls were enrolled as controls (Figure 1 A; Table 1 ; Table S1). Age and gender were matched between cases and controls. Proteomic analyses were performed on matched serum and urine samples from 50 patients with COVID-19 (39 non-severe and 11 severe), 17 non-COVID-19 cases, and 23 healthy controls (Figures S1A–S1C; Table S1). In addition, 106 urine samples (27 healthy controls, 15 non-COVID-19, 44 non-severe, and 20 severe) and 75 serum samples (24 healthy controls, 15 non-COVID-19, 30 non-severe, and 6 severe) from 106 individuals were obtained for metabolomic analysis (Figure S1C; Table S1).
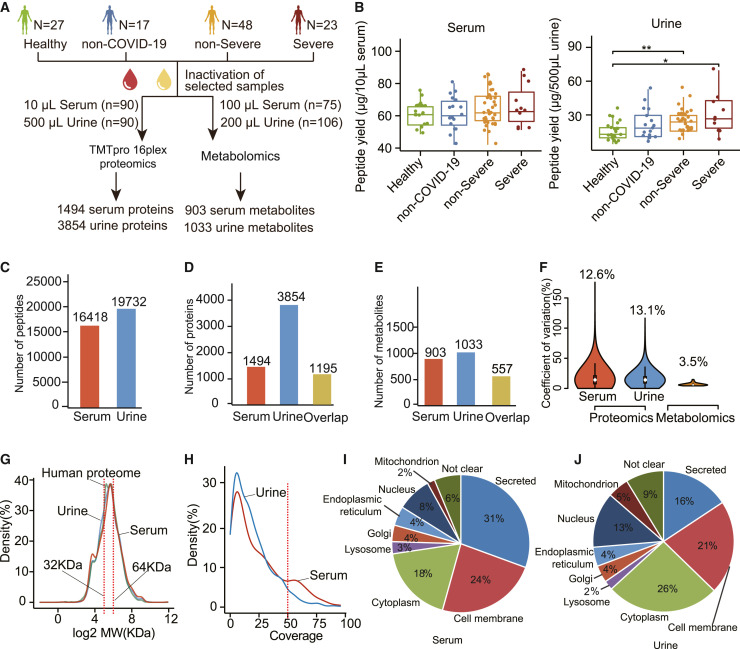
Figure 1.
Overview of the serum and urine proteomics and metabolomics data
(A) Study design. Four groups—healthy control (n = 27), non-COVID-19 control (n = 17), patients with non-severe COVID-19 (n = 48), and patients with severe COVID-19 (n = 23)—were included in this study.
(B) Peptide yields of the 4 groups in serum and urine samples.
(C–E) Number of characterized and overlapped peptides (C), proteins (D), and metabolites (E) in serum and urine.
(F) Coefficients of variation (CVs) of the protein abundance from control samples by proteomics and metabolomics.
(G) Molecular weight (MW) distributions of quantified proteins in the serum, the urine, and the entire human proteome.
(H) Sequence coverage distribution of each quantified protein in serum and urine.
(I and J) Subcellular localization composition of proteins identified in the (I) serum and (J) urine.
p value between two groups were calculated by two-sided unpaired Student’s t test and adjusted by the Benjamini and Hochberg correction. Adjusted p values: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. H, healthy; n-S, non-severe COVID-19; S, severe COVID-19.
Table 1.
Demographics and baseline characteristics of COVID-19 patients
| Variables | Healthy control (n = 27) | Non-COVID-19 (n = 17) | COVID-19 |
||
|---|---|---|---|---|---|
| Total (n = 71) | Non-severe (n = 48) | Severe (n = 23) | |||
| Sex, no.(%) | |||||
| Male | 18 (66.7) | 9 (52.9) | 46 (64.8) | 32 (66.7) | 14 (60.9) |
| Female | 9 (33.3) | 8 (47.1) | 25 (35.2) | 16 (33.3) | 9 (39.1) |
| Age, year | |||||
| Mean ± SD | 47.1 ± 9.8 | 54.5 ± 19.6 | 48.6 ± 14.5 | 44.6 ± 13.7 | 56.8 ± 12.7 |
| Median (IQR) | 48.0 (40.0–53.0) | 59.0 (40.0–69.0) | 51.0 (36.0–59.0) | 44.0 (36.0–54.8) | 56.0 (47.0–65.0) |
| Range | 30.0–72.0 | 15.0–85.0 | 4.0–77.0 | 4.0–70.0 | 33.0–77.0 |
| BMI, kg/m2 | |||||
| Mean ± SD | 24.4 ± 2.7 | 21.7 ± 2.3 | 24.9 ± 3.0 | 24.4 ± 3.2 | 25.9 ± 2.3 |
| Median (IQR) | 24.3 (22.2–26.9) | 22.0 (19.2–23.7) | 24.9 (22.8–27.0) | 24.4 (22.0–26.8) | 25.8 (24.6–27.2) |
| Range | 20.5–29.1 | 17.7–25.1 | 16.0–31.3 | 16.0–30.7 | 22.2–31.3 |
| Symptoms, no. (%) | |||||
| Fever | 10 (58.8) | 46 (64.8) | 28 (58.3) | 18 (78.3) | |
| Cough | 7 (41.2) | 28 (39.4) | 17 (35.4) | 11 (47.8) | |
| Headache | 3 (17.6) | 5 (7.0) | 3 (6.3) | 2 (8.7) | |
| Fatigue | 2 (11.8) | 5 (7.0) | 2 (4.2) | 3 (13.0) | |
| Pharyngalgia | 3 (17.6) | 3 (4.2) | 3 (6.3) | 0 (0.0) | |
| Expectoration | 5 (29.4) | 6 (8.5) | 4 (8.3) | 2 (8.7) | |
| Diarrhea | 0 (0.0) | 3 (4.2) | 2 (4.2) | 1 (4.3) | |
| Chest tightness | 4 (23.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Chest CT, no. (%) | |||||
| Involvement of chest radiographs | 3 (17.6) | 67 (94.4) | 44 (91.7) | 23 (100.0) | |
| Comorbidity, no. (%) | |||||
| Hypertension | 3 (17.6) | 9 (12.7) | 6 (12.5) | 3 (13.0) | |
| Diabetes | 1 (5.9) | 7 (9.9) | 5 (10.4) | 2 (8.7) | |
| Respiratory system | 2 (11.8) | 3 (4.2) | 0 (0.0) | 3 (13.0) | |
| Other endocrine system | 0 (0.0) | 4 (5.6) | 1 (2.1) | 3 (13.0) | |
| Chronic kidney disease | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Digestive system | 0 (0.0) | 3 (4.2) | 2 (4.2) | 1 (4.3) | |
| Oxygenation index, mmHg | |||||
| Mean ± SD | 458.4 ± 123.4 | 483.9 ± 126.7 | 391.9 ± 85.0 | ||
| Median (IQR) | 440.5 (380.9–502.4) | 447.6 (319.1–528.6) | 361.1 (323.8–469.0) | ||
| Range | 295.2–890.5 | 319.1–890.5 | 295.2–490.0 | ||
| Treatment, no. (%) | |||||
| Oxygen inhalation | 3 (17.6) | 62 (87.3) | 39 (81.3) | 23 (100) | |
| Antibiotics | 7 (41.2) | 7 (9.9) | 5 (10.4) | 2 (8.7) | |
| Antiviral drug | 3 (17.6) | 71 (100.0) | 48 (100.0) | 23 (100.0) | |
| Immunoglobulin | 0 (0.0) | 14 (19.7) | 1 (2.1) | 13 (56.5) | |
| Methylprednisolone | 2 (11.8) | 20 (28.2) | 7 (14.6) | 13 (56.5) | |
| Chinese medicine | 0 (0.0) | 71 (100.0) | 48 (100.0) | 23 (100.0) | |
CT, computed tomography; IQR, interquartile range; SD, standard deviation.
Peptide yields from serum samples were not significantly different among the four groups (healthy, non-COVID-19, non-severe, and severe), indicating the reproducibility of our sample preparation method (Figure 1B). However, peptide yields from urine specimens were significantly higher in severe and non-severe cases than from healthy controls (Figure 1B). This observation confirms a report of proteinuria in patients infected with SARS-CoV-2 (Su et al., 2020).
We identified and quantified 16,148 peptides and 1,494 proteins from sera using tandem mass tag (TMT)-based proteomics, while 19,732 peptides and 3,854 proteins were identified from urine using similar methodology (Figures 1C and 1D; Table S2; STAR Methods). The proteomic depth of urine specimens achieved in this study exceeds most other published studies (Table S3) and is reasonably deep to encompass most urinary proteins. Moreover, we characterized 1,033 urine metabolites and 903 serum metabolites in patients with COVID-19 (Figure 1E; Table S2). The serum and urine proteome dataset showed minimal batch effects (Figures S1D and S1E). The median coefficients of variance (CVs) of the quality control (QC) samples were 13% for proteomic data and 3.5% for metabolomic data, indicating the robust quality of our data (Figure 1F).
80% of detectable serum proteins were detected in urine
We compared the proteins thus quantified from matched pairs of serum and urine specimens. Overall, the number of proteins identified in urine was ∼2.5 times greater than that in sera. Eighty percent of proteins identified in sera (i.e., 1,195 proteins) were also detected in urine (Figure 1D), indicating that a majority of serum proteins are detectable in urine. In contrast, our data showed that the numbers of quantified metabolites in sera and urine are similar (Figure 1E; 903 versus 1,033). Unlike proteins, however, 62% of serum metabolites (i.e., 557 metabolites) were detectable in urine (Figure 1E). The discrepancy in protein and metabolite detection is probably due to differences in their abundance and stability in sera and urine.
It is generally assumed that the molecular weight (MW) cutoff for glomerular filtration is 30–50 kDa (Haraldsson et al., 2008), but whether other proteins beyond that weight range can be detected in urine remains unclear. The MW distribution analysis of matched urine and serum proteomes in our data showed the MW ranges of proteins in serum and urine were approximately identical to that in the human proteome (Figure 1G), indicating that urinary proteins are not limited by low MW. More proteins in the urinary proteome had relatively low sequence coverage (Figure 1H), suggesting that low-abundance proteins are more readily detectable in the urine.
Analysis of the subcellular localization of proteins identified in serum and urine showed that secreted proteins constituted the largest proportion of the serum proteome (31%), followed by membrane proteins (24%) and cytoplasmic proteins (18%) (Figure 1I). In contrast, cytoplasmic proteins (26%) and membrane proteins (21%) were the most abundant protein groups in the urinary proteome, while the proportion of secreted proteins was only 16% (Figure 1J). Of interest was the higher proportion of nuclear proteins in urine than in serum (13% versus 8%) (Figures 1I and 1J). This suggests that the urinary proteome thus measured contained more intracellular compartment proteins released from tissues, compared to the serum proteome at similar limits of detection.
Machine learning model using urinary proteins identified severe COVID-19 cases
Proteins circulating in the blood have been used to build machine learning models to classify COVID-19 severity (Messner et al., 2020; Shen et al., 2020). However, the invasive nature of blood sampling limits the wide application of blood-based tests. Here, we investigated whether urinary proteins could be used in machine learning modeling for classifying COVID-19 severity. Based on the rank of the mean decrease in accuracy, we selected the top 20 proteins in the serum and urine data, respectively (Figures 2A and 2C). The top 20 serum proteins were related to platelet degranulation, acute phase response, and immune cell proliferation, consistent with published reports (Chen and John Wherry, 2020; Messner et al., 2020; Shen et al., 2020) (Figure 2B). The top 20 urinary proteins were involved in cell adhesion, cell development, secretion, digestion, and extracellular matrix or structure organization related to biological processes (Figure 2D). We further explored the power of these top 20 urinary proteins for classifying the severity compared to that of sera. By increasing the number of proteins in the model, we optimized 20 random forest models for each specimen type and evaluated the performance of the respective classifier in terms of accuracy and area under the curve (AUC) after internal cross-validation, as detailed in the STAR Methods. Our data showed that urine-derived models performed as well as those from sera (Figure 2E). As the number of features in the model exceeded the top 4, the accuracy of the models for both sample types rose beyond 0.9, and the AUC was higher than 0.95 (Figure 2E).
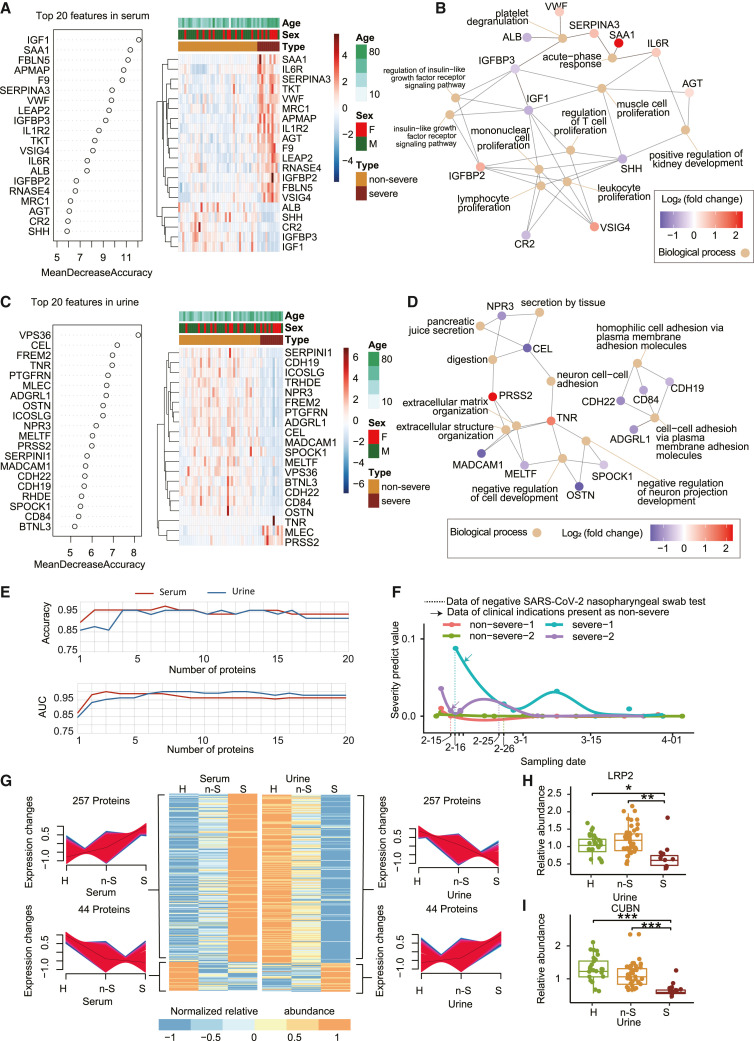
Figure 2.
Identification of severe and non-severe COVID-19 cases at the proteomics level
(A and C) The top 20 feature proteins in serum (A) or urine (C) proteomics data selected by random forest analysis and ranked by the mean decrease in accuracy.
(B and D) The biological process involved in the top 20 urine (B) or serum (D) proteins were annotated by Gene Ontology (GO) database and visualized by the clusterProfiler R package.
(E) Line chart shows the accuracy and AUC values of the 20 serum or urine models. The features in each model were selected from top n (number of feature) important variables in the serum and urine data.
(F) Severity prediction value of 4 patients with COVID-19 at different urine sampling times.
(G) Heatmap shows 301 proteins identified in both serum and urine with opposite expression patterns in different patient groups. The 301 proteins are a union of 257 proteins that are upregulated in serum but downregulated in urine and 44 proteins that are downregulated in serum but upregulated in urine. The relative intensity values of proteins were Z score normalized.
(H and I) The relative abundance of LRP2(H) and CUBN (I) in urine. The y axis means the protein expression ratio by TMT-based quantitative proteomics.
To further evaluate the performance of such urinary proteins for classifying COVID-19 severity, we trained a model using the 20 urinary proteins above and tested it on an independent TMT-labeled urinary proteomic dataset of 13 patients with COVID-19 (Table S2) and a label-free data-independent acquisition (DIA) urinary proteomics dataset (Tian et al., 2020) of 14 patients with COVID-19. The AUC values of the model were 0.89 and 0.80 in the 2 datasets, and the accuracy values were 0.69 and 0.71, respectively (Figures S1F and S1G). We also trained a logistic regression model using the 20 urinary proteins described above and tested it on an independent dataset of 4 patients with COVID-19 whose urine samples were collected at different time points (Figure 2F). For severe COVID-19 cases, the severity prediction value trended lower when samples were collected at later time points after hospital admission (Figure 2F). These data further support the utility of our urinary protein model for predicting progression to clinical severity in early infection.
Our data showed that urinary proteomics can be as informative as that of sera in terms of classifying and predicting COVID-19 severity. Considering its non-invasive nature and easy accessibility, urine could be a widely used sample source for COVID-19 management. Nevertheless, more independent validation is required before this could become the clinical standard of care.
301 proteins showed opposite expression patterns in urine and sera
We examined the correlation between serum and urine proteomic data in COVID-19 cases. A total of 24 proteins showed negative correlation (Pearson’s correlation coefficient <−0.3, p < 0.05) and 60 proteins showed positive correlation (Pearson’s correlation coefficient >0.3, p < 0.05) (Figure S1H).
Interestingly, we found that 301 proteins (i.e., 25% of the 1,195 proteins) identified in both urine and matched sera, showed opposite expression patterns in urine and serum in mean relative protein abundance levels among healthy, non-severe, and severe groups (Figure 2G). Blood proteins are filtered by the glomerulus and reabsorbed by the renal tubules before urine is formed. In addition, proteins may be released into urine from the urinary tract. Levels of most proteins vary greatly within the nephron during glomerular filtration and tubular reabsorption. Two important regulators involved in tubular reabsorption identified in our urine proteome, megalin (LRP2) (Figure 2H) and cubilin (CUBN) (Figure 2I), were both downregulated in the urine, indicating potential dysregulation of reabsorption. Decreased concentrations of LRP2 and CUBN in urine were further confirmed by parallel reaction monitoring (PRM)-based targeted mass spectrometry (MS) assay and enzyme-linked immunosorbent assay (ELISA), as shown in Figure S1I. In addition, the ligand proteins transported by LRP2 and CUBN, such as selenoprotein P (SELENOP), plasminogen activator, urokinase (PLAU), epidermal growth factor (EGF), galactosidase alpha (GLA), and apolipoprotein-H (APOH), were also downregulated in urine (Norden et al., 2002) (Figure S1J). Thus, the tubular reabsorption process appears dysregulated in the patients with COVID-19, resulting in a downregulation pattern of certain urinary proteins. From these collective findings, we hypothesize that the intricate process of protein transport from blood to urine and disordered tubular reabsorption in patients with severe COVID-19 may account for the divergent presence of these 301 proteins in serum and urine. This discrepancy of serum-urine protein expression, as discovered here in patients with COVID-19, may also be present in other disorders, which awaits further investigation.
197 cytokines and their receptors identified in urine, while 124 identified in sera
Uncontrolled inflammatory innate responses have caused cytokine storm in patients with COVID-19, contributing to high mortality (Cao, 2020). In this study, we identified 124 cytokines and their receptors in serum and 197 in urine, totaling 234 cytokines and receptors. They were grouped into 6 types, namely chemokines, interferons, ILs, transforming growth factor-β (TGF-β) family, tumor necrosis factor (TNF) family, and other cytokines (Figures 3 A and S2A; STAR Methods). Eighty-seven cytokines were present in both biofluids (Figures S2B and S2D).
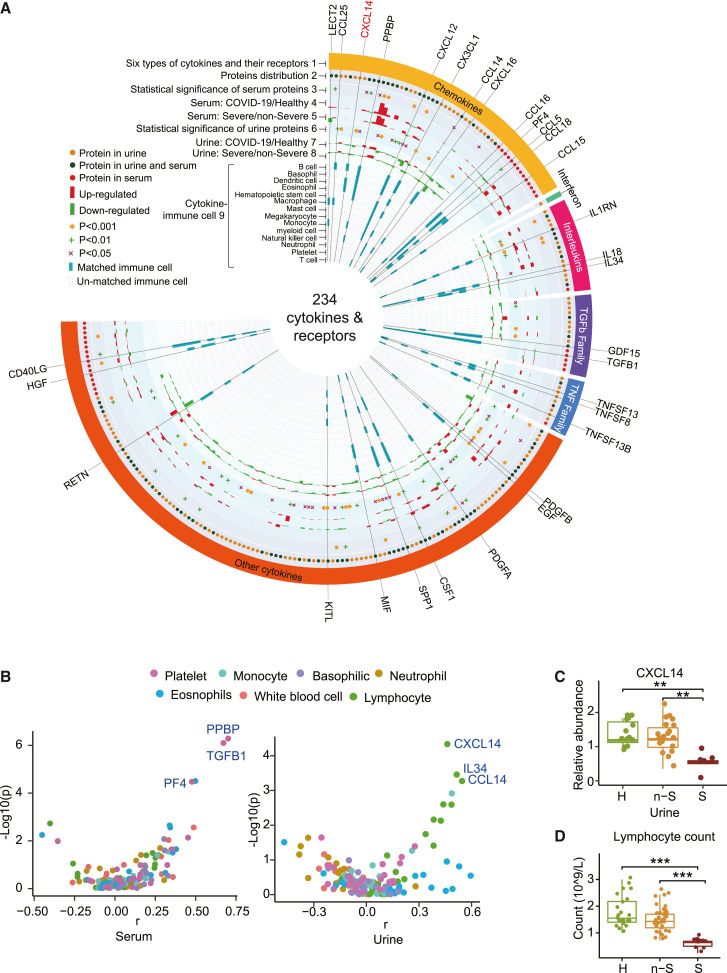
Figure 3.
Cytokines characterized in the urine and serum
(A) Circos plot integrating the relative expression and cytokine-immune cell relationship of 234 cytokines and their receptors. Track 1, the outermost layer, represents 234 cytokines and their receptors, which are grouped into six classes. Track 2 shows the cytokines detected from our urine and/or serum proteomics data, as indicated by different colored dots. Tracks 3 and 6, cytokines from the urine or serum, with a cutoff of p < 0.05 when comparing healthy donors and non-severe and severe cases using one-way ANOVA, were regarded as statistically significant. Tracks 4 and 7 represent serum or urine cytokine abundance distribution in COVID-19 (includes non-severe and severe) group and healthy group. Tracks 5 and 8 represent serum or urine cytokine abundance distribution in severe and non-severe groups. Track 9, the inner circle, shows the immune cells related to each cytokine inferred by immuneXpresso.
(B) Spearman’s rank correlation coefficients between serum or urine cytokines and immune cells.
(C) Expression pattern of CXCL14 in the urine.
(D) Lymphocyte count in healthy donors and COVID-19 cases.
We identified 33 significantly dysregulated cytokines and receptors from COVID-19 serum (Figure 3A, track 3), and 68 cytokines and receptors from COVID-19 urine (Figure 3A, track 6). These modulated cytokines and receptors were enriched for the STAT3 pathway and hepatic fibrosis (Figure S2C). Most cytokines and receptors in urine (i.e., 136 of 197, 69%) were downregulated in patients with COVID-19 compared to healthy controls (Figure 3A, track 7), while 77 of 124 cytokines (62%) were upregulated in the serum of patients with COVID-19 (Figure 3A, track 4).
Cytokines produced by immune cells mediate diverse immune processes. In our data, 31 cytokines were involved in the functions of multiple immune cell types (Figure 3A, track 9), as described in the STAR Methods. Serum PPBP, TGFB1, and PF4 showed the highest Spearman’s rank correlation coefficient (r) with platelet counts, but were not significantly perturbed in COVID-19 cases (Figures 3B and S2G). Interestingly, the concentration of CXCL14 in urine, confirmed by PRM and ELISA (Figure S2H), showed the most significant correlation with lymphocyte counts of COVID-19 cases (Figure 3B). CXCL14 was detected only in urine and was significantly downregulated in severe cases (Figure 3C), consistent with the reduction in lymphocyte counts (Figure 3D). CXCL14 has been reported to enhance T cell activation and proliferation (Chen et al., 2010). As lymphopenia is characteristic of severe COVID-19 (Tan et al., 2020), urinary CXCL14 may be a biomarker of COVID-19 severity. Moreover, urinary IL34 and CCL14 also showed significant correlation with lymphocyte counts and were downregulated in severe cases (Figures 3B and S2I); both are worth investigating further as additional biomarkers of disease severity.
In summary, more dysregulated cytokines and receptors were discovered in COVID-19 urine than in serum. Urinary CXCL14, together with IL-34 and CCL14, are potential biomarkers reflecting the lymphocyte counts of patients with COVID-19 and may be used to monitor the severity of COVID-19 disease.
Dysregulated ESCRT super-complex suggests virus replication
From the 1,195 proteins identified in both COVID-19 urine and sera (Figure 1D), we found 330 proteins that were differentially expressed in either serum or urine compared to healthy controls (Table S4). Defining criteria of differentially expressed proteins (DEPs) are outlined in the STAR Methods. Eighteen virus budding-related DEPs were dysregulated in urine but not in sera. Of note, all 18 proteins were downregulated in patients with COVID-19. Sixteen of the 18 proteins were selected for targeted proteomic analysis using PRM on 73 unfractionated urine specimens (Table S2; Figure S5A). Twelve PRM-detected proteins showed a strong correlation (p < 0.01) with TMT data (Figure S5B), confirming the downregulation of these proteins in severe cases (Figure 4 A). Thirteen of the 18 proteins belong to the endosomal sorting complexes required for transport (ESCRT) super-complex (Figures 4A and 4B). Our data showed suppression of the major components of ESCRT-I (TSG101, VPS28, and VPS37D), ESCRT-II (VPS36, SNF8, and VPS25) (Hurley and Hanson, 2010), and the ESCRT-III CHMP protein family including CHMP1B, CHMP2A, CHMP3, CHMP4A, CHMP4B, CHMP4C, and CHMP5 (Adell and Teis, 2011) (Figure 4A). The intriguing significant decrease in ESCRT super-complex proteins was observed only in urine, plausibly suggesting intense consumption of the ESCRT super-complex during active replication of SARS-CoV-2 viruses in severe cases since the budding of enveloped viruses depends on the function of the host cell ESCRT complex. We further explored the correlation of these 18 DEPs with the cycle threshold (CT) of SARS-CoV-2 reverse transcriptase-polymerase chain reaction (RT-PCR) tests. Figure S5C shows positive correlation of the virus budding-related proteins VPS28, CHMP4B, CHMP2A, CHMP4A, VPS4A, VPS4B, TSG101, and PDCD6IP with CT values (r > 0.4, p < 0.05), which supports the potential association of urinary virus budding-associated proteins with COVID-19 viral load in vivo. The ESCRT super-complex proteins were not detected in the serum proteomic data, probably due to their relatively low abundance or instability in blood. The urine PRM assays established in this study could be further developed into potential clinical assays for COVID-19 monitoring.
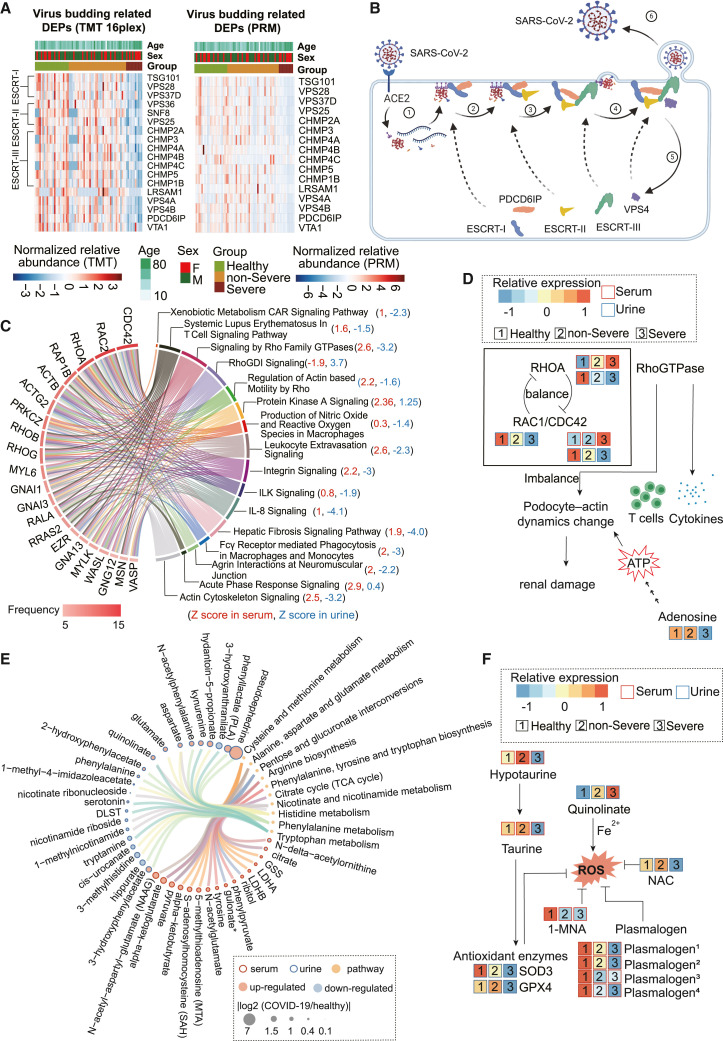
Figure 4.
Dysregulated proteins and metabolites in the serum and urine of patients with COVID-19
(A) Virus budding-related DEPs uniquely regulated in the urine were identified by untargeted TMT 16plex proteomics and confirmed by PRM.
(B) Schematic diagram of the virus budding process.
(C) The top 21 regulated proteins are ranked by the frequency with which they are enrolled in the overlapped 16 out of 20 pathways between the serum and the urine by ingenuity pathway analysis (IPA).
(D) Schematic diagram of the dynamic balance of Rho GTPases. The imbalance affects the functional integrity of glomerular podocytes and results in renal damage.
(E) DEPs and differentially expressed microRNAs (DEMs) were involved in the 10 KEGG pathways.
(F) Schematic diagram of metabolites participating in the oxidative stress in COVID-19.
Imbalanced CDC42-RHOA-RAC1 molecular switches indicate disrupted immune system and renal injury
Our data showed consistent upregulation or downregulation of 148 serum proteins, 580 urinary proteins, 161 serum metabolites, and 138 urinary metabolites of healthy controls compared with patients with non-severe COVID-19 who progressed to severe COVID-19 (Figures S4B–S4E; Table S4). The dysregulated proteins in serum were enriched in 23 pathways and those in urine in 108 pathways. Interestingly, 20 pathways from the serum data also appeared in the urine data (Figure 4C; Table S5), suggesting that the most dysregulated pathways detected in serum could also be detected in urine. Twenty-one proteins frequently occurred in 16 of the 20 shared pathways (Figure 4D). Of note, CDC42, RAC1/RAC2, and RHOA from the Rho GTPase family stood out as the most dominantly dysmodulated proteins in urine and/or serum (Figures 4C and S6A). In our proteomic data, RAC1 was detected only in urine and RAC2 was detected only in serum (Figure S6A). Concentrations of CDC42, RAC1, and RHOA in urine were further confirmed by PRM and ELISA (Figure S6B). RhoGTPase also functions as a key regulator of lymphocytes in adaptive immunity. It participates in the development and migration of T cells (Saoudi et al., 2014) (Figure 4D) and also regulates signal transduction evoked in the presentation of pathogen-derived antigens and in cytokine and chemokine receptor signaling (Bros et al., 2019; Reif and Cantrell, 1998) (Figure 4D). CDC42, RHOA and RAC1 act as molecular switches in the dynamic regulation of actin cytoskeleton in podocytes (Figure 4D), which are essential in maintaining glomerular permselectivity (Perico et al., 2016). RHOA enhances the contraction of actin fibers, while RAC1/CDC42 are antagonistic, promoting cell movement by lamellipodia/filopodia formation (Saleem and Welsh, 2019). Fine modulation of the GTPases is required for the motility of podocytes, and disruption of this process may lead to glomerular sclerosis and renal damage (Saleem and Welsh, 2019). In addition, podocyte-actin dynamics requires substantial ATP consumption (Imasawa and Rossignol, 2013). Our data showed that adenosine, a product of ATP metabolism, was significantly reduced in the urine of severe cases (Figures 4D and S6C), suggesting the possible impairment of podocyte motility, dysfunction of immune system, and renal injury.
Standard clinical laboratory tests showed that urine creatinine, specific gravity, and the estimated glomerular filtration rate (eGFR) decreased, while urine pH increased significantly in the severe cases (Figure S6C), suggesting some degree of renal dysfunction (Ronco et al., 2019). Significant reduction of cyclic AMP (cAMP) in patients with renal injuries has been reported, probably due to impaired glomerular filtration (Mocan et al., 1998). Urinary cAMP is a sensitive biomarker for the onset of acute renal failure and subsequent recovery (Vitek et al., 1977). In our study, both eGFR and urinary cAMP of severe cases were significantly reduced (Figures S6D and S6E), consistent with renal impairment in severe COVID-19, and which may partly account for the discrepancy of protein dysregulation patterns in urine and serum.
Activation of reactive oxygen species (ROS) and impaired immune cells in COVID-19
The dysregulated metabolites in COVID-19 urine and serum were enriched in 10pathways based on Kyoto Encyclopedia of Genes and Genomes (KEGG) (Table S5), including tryptophan biosynthesis and metabolism (Figure 4E; Table S5). There are three metabolic pathways for tryptophan. The first results in tryptamine through the action of aromatic-l-amino acid decarboxylase. The second pathway forms serotonin through the action of tryptophan hydroxylase. The third pathway converts >95% of free tryptophan to N-formylkynurenine (NFK), which is further metabolized into kynurenine and 3- hydroxyanthranilate by kynureninase. Activation of the kynurenine pathway could prevent hyperinflammation and induce long-term immune tolerance through the generation of T regulatory (Treg) cells and modulation of immune phenotypes of dendritic cells (Sorgdrager et al., 2019). In our data, tryptamine and serotonin were downregulated and 3-hydroxyanthranilate and kynurenine were upregulated in the urine samples of patients with COVID-19 (Figures S6F and S6G). These results indicated that serotonin and tryptamine metabolic pathways were suppressed, while NFK production was enhanced to trigger the activation of anti-inflammatory mechanisms in patients with COVID-19.
Like other viral infections, SARS-COV-2 infection has been reported to trigger oxidative stress by generating an imbalance between the oxidant and antioxidant systems in vivo (Cecchini and Cecchini, 2020; Ntyonga-Pono, 2020). Taurine, hypotaurine, and 1-methylnicotinamide (1-MNA) were significantly downregulated in COVID-19 serum (Figures 4F and S6H). Taurine and hypotaurine have antioxidant effects that can protect immune cells from oxidative stress damage (Learn et al., 1990; Marcinkiewicz and Kontny, 2014). 1-MNA inhibits ROS generation and has anti-inflammatory actions on vascular endothelium (Biedroń et al., 2008). Against this background suggestive of oxidative stress, multiple antioxidant enzymes such as SOD3 and GPX4 were also found to be downregulated in the urine of severe COVID-19 cases in the proteomic data (Figures 4F and S6H). Plasmalogen, which regulates inflammation (Wallner and Schmitz, 2011) and neutralizes reactive oxygen molecules (Broniec et al., 2011), was downregulated in the COVID-19 serum (Figure 4F and S6I).
In the COVID-19 urine, some significantly changed metabolites related to ROS were also identified, such as downregulated N-acetylcysteine (NAC) and upregulated quinolinate (Figures 4E, 4F, and S6J). NAC functions in the nicotinate and nicotinamide metabolism pathway and is a precursor of the antioxidant glutathione, which can improve cell-mediated immunity against influenza virus (Shi and Puyo, 2020). Quinolinate mediates ROS generation by complexing with Fe2+ (Lugo-Huitrón et al., 2013). Quinolinate can induce inflammation by increasing TNF-α (Block and Schwarz, 1994) and IL-6 expression (Schiefer et al., 1998). Activated macrophages are known to produce more quinolinate after an inflammatory response (Heyes, 1993).
Taken as a whole, the metabolomic data point to widely activated ROS production, which could lead to a variety of immune-mediated tissue injuries in patients with COVID-19.
Inflammation-induced renal injuries as revealed by multiomics data
The 20 pathways prominent in both serum and urine were related mainly to immunity (Table S6). We found that most immunity-related pathways were downregulated in urine but upregulated in serum, except for protein kinase A signaling, coagulation system, acute phase response signaling, and liver X receptor (LXR)/retinoid X receptor (RXR) activation, which were upregulated in both serum and urine (Table S6). Protein kinase A signaling was reported to be involved in the innate immunity of activated macrophage (Wan et al., 2007) and autophagy (Stephan et al., 2009). Inhibition of LXR/RXR has proatherogenic effects of arsenic in macrophages (Padovani et al., 2010). The interplay between inflammation and coagulation has been studied extensively (Levi and van der Poll, 2010).
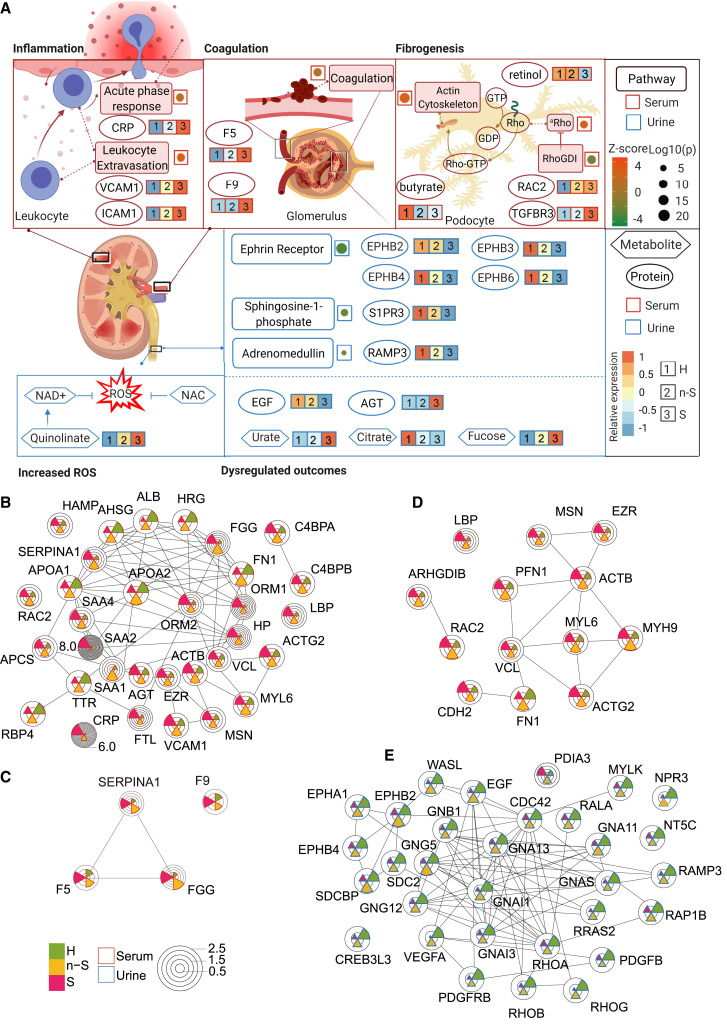
We then analyzed all of the urine and serum proteomic and metabolomic data to explore whether COVID-19-induced inflammation could have led to immune-related renal injuries (Figure 5 A). We identified multiple dysregulated pathways involved in inflammation in agreement with the literature (Schulte-Schrepping et al., 2020; Shen et al., 2020) (Table S5). Our dataset enabled the discovery of more enriched pathways that were missed in other studies with relatively fewer protein identifications (Messner et al., 2020; Shen et al., 2020). In the 23 enriched serum pathways found in this study (Table S5), the leukocyte extravasation signaling pathway stood out for its activation level (Z score 2.6) (Figure 5A; Table S5). Vascular cell adhesion protein 1 (VCAM-1) and intercellular adhesion molecule 1 (ICAM-1), which participate in leukocyte extravasation (Muller, 2013), were upregulated in COVID-19 sera (Figures 5A, 5B, and S7A). Inflammation may initiate coagulation, an innate immune process (Esmon, 2005). F5 and F9 are key proteins in coagulation (Franchini and Mannucci, 2011; Segers et al., 2007), and both were upregulated in the sera of patients with COVID-19 (Figures 5A, 5C, and S7A). Coagulation may also activate downstream fibrosis (Suárez-Álvarez et al., 2016). Renal fibrosis is the main cause of chronic renal disease, and Rho guanosine triphosphatase (GTPase) is highly related to this fibrotic process. The Rho pathway is essential for the function of glomerular podocytes by modulating the actin cytoskeleton (Perico et al., 2016). The Rho pathway-related proteins, namely RAC1, RAC2, and TGFBR3, were dysregulated in both serum and urine (Figures 5A, 5D, S6A, and S7A), suggesting their potential for inducing renal fibrogenesis. The decreased levels of serum metabolites, retinol, and butyrate support this hypothesis (Figures 5A and S7A). Retinol derivatives (retinoids) can protect damaged podocytes through anti-inflammatory and anti-fibrotic effects, thereby repairing renal injuries (Mallipattu and He, 2015). Butyrate, which is mainly produced by gut microbes, has been proposed as a potential therapeutic agent for decreasing systemic inflammation and ameliorating renal damage (Felizardo et al., 2019). Reduced serum retinol and butyrate levels of patients with COVID-19 suggests immune-related renal damage.
Figure 5.
The hypothetic model of immune dysregulation and increased ROS that induces renal injuries in patients with severe COVID-19
(A) Pathways are displayed in square boxes, proteins are displayed in circles, while metabolites are displayed in hexagons. The Z score of the activity of a pathway is displayed as dots beside the respective pathway in a red (for serum) or blue (for urine) box, with its size representing the log10(p value) of each pathway and its color representing the Z score value. Relative protein or metabolite expression is labeled beside the respective molecule. aRho, regulation of actin-based motility by Rho.
(B) Serum DEPs involved in the acute phase response and leukocyte extravasation signaling.
(C) Serum DEPs involved in the coagulation system.
(D) Serum DEPs involved in the actin cytoskeleton and Rho signaling.
(E) Urine DEPs involved in the ephrin receptor signaling, sphingosine-1-phosphate signaling, and adrenomedullin signaling. The relative expression values of proteins are shown in the pie chart.
In urine, multiple pathways are also enriched based on DEPs (Figures 5A and 5E). These pathways include ephrins and Eph receptor signaling (Coulthard et al., 2012; Wu et al., 2019), sphingosine-1-phosphate signaling (Lee et al., 2011), and adrenomedullin signaling pathways (Kubo et al., 1998), all of which are involved in the renal injury process. Ephrins signal through Eph receptors EphB2, EphB3, EphB4, and EphB6, and affect renal reabsorption (Ogawa et al., 2006), and they were all significantly downregulated in COVID-19 urine (Figures 5A, 5E, and S7B). Sphingosine-1-phosphate receptor 3 (S1PR3) mediates sphingosine-1-phosphate signaling and is downregulated in the urine of patients with severe COVID-19. Another evidence for possible renal damage in patients with COVID-19 comes from the downregulated receptor activity-modifying protein 3 (RAMP3) (Figures 5A, 5E, and S7B), a critical protein for the activation of adrenomedullin receptors (Kuwasako et al., 2001).
In addition, several known protein or metabolite biomarkers for renal injuries are present in the current proteomic and metabolomic datasets. Our data showed a decline in urinary EGF (Figures 5A and S7B), suggesting renal damage in patients with COVID-19 (Li et al., 2018). NAC and quinolinate, as described above, are related to ROS and renal damage. They were dysregulated in COVID-19 urine (Figures 5A and S6G). Increased core fucose levels may contribute to the pathogenesis of renal fibrosis (Shen et al., 2013), and the fucose level was upregulated in COVID-19 urine. We also observed increased urate and decreased citrate in urine (Figures 5A and S7C), consistent with previous findings in chronic kidney disease (Kalantari and Nafar, 2019). These findings suggest that patients with COVID-19 may sustain renal impairment related to immune dysfunctions and are manifested in many altered proteins and metabolites in urine.
Discussion
In this study, we have shown urine to be a useful specimen for understanding the pathogenesis of COVID-19 and monitoring its clinical progression. The datasets from this study are a resource characterizing changes of 3,854 proteins and 1,033 metabolites in the COVID-19 urine, as well as the changes of 1,494 proteins and 903 metabolites in the COVID-19 serum. Interestingly, while 80% of detectable serum proteins were measured in urine, only 31% of urinary proteins were detected in serum. The range of protein MWs identified in urine and sera was similar, indicating that the excretion and secretion of urinary proteins were not restricted to those of low MW.
We found 301 overlapping serum and urinary proteins that showed opposite expression patterns. Despite the discrepancy, urinary proteins can be used to effectively classify COVID-19 severity, with an accuracy comparable to that when using serum proteins.
Urinary proteins offer unique insights into the pathogenesis of COVID-19 compared with blood proteome. The downregulation of key reabsorption regulators in renal tubules, such as LRP2 and CUBN, and their ligands, including SELENOP, PLAU, EGF, GLA, and APOH, point to disrupted renal reabsorption in patients with COVID-19. Urine contains more detectable cytokines than serum. Dysregulation of urinary CXCL14, as well as IL-34 and CCL14, correlated with blood lymphocyte counts.
ESCRT, a super-protein complex that facilitates virus budding, was downregulated in COVID-19 urine, suggesting active consumption of ESCRT for virus replication. Due to its relatively low abundance, this protein complex was absent in our serum proteomic data.
CDC42, RAC1, and RHOA proteins from the Rho GTPase family are the most dominantly modulated proteins in COVID-19, which could contribute to impaired adaptive immunity and renal injuries. Activated ROS was found in COVID-19, which may result in damage to immune cells. Furthermore, integrative analysis of proteomic and metabolomic data suggests that renal injuries may be induced by abnormal inflammatory responses.
Targeted MS-based protein quantification, such as multiple reaction monitoring (MRM)-MS and parallel reaction monitoring (PRM)-MS, is the commonly used validation method in proteomics. In this study, PRM-MS results showed that four proteins, namely CUBN, CXCL14, RHOA, and RAC1, were significantly downregulated in severe cases (Figures S1I, S2H, and S6B). LRP2 and CDC42 also showed a declining trend, although they were not statistically significant. ELISA showed similar results, but did not achieve statistical significance (Figure S1I). This may be because ELISA is an antibody-based chromogenic reaction whose performance is crucially dependent on antibody quality (sensitivity and specificity), while PRM-MS is antibody independent and provides more precise quantification of proteins. Nevertheless, ELISA remains the main method for semi-quantitative protein analysis in clinical laboratories due to its ease of use.
Overall, this study presents a comprehensive proteomic and metabolomic analysis of paired serum and urine samples from patients with COVID-19 and demonstrates that selected urinary proteins may be used for the classification of COVID-19 severity. Evidence for dysregulated immune responses and renal injuries in patients with COVID-19 uncovered in this study should be further investigated to advance COVID-19 diagnosis and therapy. Our approach more generally supports the utility of urine as an informative biospecimen to understand disease pathogenesis and develop new therapeutic strategies for infectious diseases.
Limitations of the study
In this study, ∼35% non-COVID-19 cases and 37% patients with COVID-19 had comorbidities such as hypertension and diabetes (Table 1). We cannot completely exclude the effects of comorbidities on changes in the proteomic or metabolomic data. However, we took care to ensure that COVID-19 and non-COVID-19 patient groups had equivalent burdens of comorbidities. The opposite protein expression patterns observed between urine and serum (Figure 2G) may be a partial result of disrupted renal reabsorption. However, the present study did not directly confirm this with independent evidence. Due to the limited independent cohort size, the predictive nature of the 20-protein signature awaits further verification.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Serum samples | Taizhou Hospital | This paper (Table S1_Patient ID) |
| Urine samples | Taizhou Hospital | This paper (Table S1_Patient ID) |
| Chemicals, peptides, and recombinant proteins | ||
| Triethylammonium bicarbonate buffer (TEAB) | Sigma-Aldrich | Cat # T7408 |
| Urea | Sigma-Aldrich | Cat # U1250 |
| Tris (2-carboxyethyl) phosphine (TCEP) | Adamas-beta | Cat # 61820E |
| Iodoacetamide (IAA) | Sigma-Aldrich | Cat # I6125 |
| Trypsin | Hualishi Tech | Cat # HLS TRY001C |
| Lys-C | Hualishi Tech | Cat # HLS LYS001C |
| Trifluoroacetic acid (TFA) | Thermo Fisher Scientific | Cat # 85183 |
| Water | Thermo Fisher Scientific | Cat # W6-4 |
| Acetonitrile | Thermo Fisher Scientific | Cat # A955-4 |
| Formic acid (FA) | Thermo Fisher Scientific | Cat # A117-50 |
| Ammonium hydroxide solution | Sigma-Aldrich | Cat # 221228 |
| Methanol | Sigma-Aldrich | Cat # 3486 |
| Critical commercial assays | ||
| TMTpro 16plex reagents | Thermo Fisher Scientific | Cat # A44520 |
| Creatinine | Beckman | Cat # OSR6178 |
| Glucose | Beckman | Cat # GL7210 |
| SARS-CoV-2 nucleic acid detection kit-1 | Shanghai Zhijiang | Cat # Z-RR-0479-02025 |
| SARS-CoV-2 nucleic acid detection kit-2 | Shanghai Zhijiang | Cat # Z-RR-0479-02-50 |
| SARS-CoV-2 nucleic acid detection kit-3 | Shanghai Zhijiang | Cat # Z-RR-0479-02AT-5002025 |
| Urine creatinine | Beckman Coulter | Cat # OSR6178 |
| Urine pH value | Guilin URIT | Cat #56208349 |
| Urine specific gravity | Guilin URIT | Cat #56208349 |
| High Select™ Top-14 Abundant Protein Depletion Mini Spin Columns | Thermo Fisher Scientific | Cat #A36370 |
| SOLAμ | Thermo Fisher Scientific | Cat # 62209-001 |
| Human Low Density Lipoprotein Receptor Related Protein 2 (LRP2)ELISA Kit | Bioswamp Co., Ltd | Cat # HM12145 |
| Human Cubilin (CUBN) ELISA Kit | Bioswamp Co., Ltd | Cat # HM12367 |
| Human Breast And Kidney Expressed Chemokine (BRAK)ELISA Kit | Bioswamp Co., Ltd | Cat # HM10258 |
| Human Ras Homolog Gene Family, Member A (RHOA)ELISA Kit | Bioswamp Co., Ltd | Cat # HM11399 |
| Human Cell Division Cycle Protein 42 (CDC42) ELISA Kit | Bioswamp Co., Ltd | Cat # HM12354 |
| Human Ras Related C3 Botulinum Toxin Substrate 1 (Rac1) ELISA Kit | Bioswamp Co., Ltd | Cat # HM11359 |
| Human Low Density Lipoprotein Receptor Related Protein 2 (LRP2)ELISA Kit | Bioswamp Co., Ltd | Cat # HM12145 |
| Human Cubilin (CUBN) ELISA Kit | Bioswamp Co., Ltd | Cat # HM12367 |
| Deposited data | ||
| Mass spectrometry data | https://www.iprox.org | PXD030662 |
| Data analysis codes | https://zenodo.org/ | https://doi.org/10.5281/zenodo.5642579 |
| Software and algorithms | ||
| Xcalibur | Thermo Fisher Scientific | Cat # OPTON-30965 |
| Proteome Discoverer Version 2.4.1.15 | Thermo Fisher Scientific | N/A |
| R version 4.0.2 | R Project | https://www.r-project.org |
| Ingenuine Pathway Analysis | QIAGEN Digital Insights | N/A |
| IMMPORT | Northrop Grumman Information Technology Health Solutions | https://www.immport.org/ |
| UniProt | UniProt Consortium | https://www.uniprot.org/ |
Resource availability
Lead contact
Further information should be directed to and will be fulfilled by the Lead Contact Tiannan Guo (guotiannan@westlake.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Patient information
This study included 71 patients with COVID-19 who were hospitalized in Taizhou Public Health Center from February to April 2020. The samples from this study are from a clinical trial that our team initiated and registered in the Chinese Clinical Trial Registry with an ID of ChiCTR2000031365. Samples were collected from residual samples after medical test. This study and waiver of Informed Consent have been approved by the Ethical/Institutional Review Board of Taizhou Hospital and Westlake University. Consents from patients were waived by the boards.
All COVID-19 patients were classified into four categories (scores 3-6) according to World Health Organization ordinal scale (WOS) (WHO, 2020a). In this study, patients scored as 3 or 4 were defined as non-severe patients, and patients scored as score 5 or 6 were defined as severe patients (Table S1). We also included 17 non-COVID-19 cases, who had clinical symptoms similar to COVID-19, including fever and/or cough, but the nucleic acid test results were negative, and 27 healthy examinees as the control group (Figure 1A; Table S1). Among the patients with COVID-19, 64.8% showed fever, 39.4% had cough symptoms, 94.4% had lung imaging changes. Hypertension and diabetes were the main underlying diseases, accounting for 12.7% and 9.9% of all patients with COVID-19, respectively. All patients took antiviral drugs and traditional Chinese medicine (TCM) treatment. Besides, 81.3% of patients received oxygen treatment, and 10.4% of patients were treated with antibiotics (Table 1).
The fasting venous blood and the first-morning midstream urine of all subjects were collected in the morning. The venous blood samples were centrifuged at 1500 g for 10 min to separate the serum. The urine samples were centrifuged at 400 g for 5 min. The serum and urine supernatants were collected into fresh tubes and were frozen at -80°C for further analysis.
For proteomics analysis, paired serum and urine samples from 90 subjects including 23 healthy donors, 17 non-COVID-19 cases, 39 patients with non-severe COVID-19 and 11 patients with severe COVID-19 were collected. We also collected 13 urine samples from COVID-19 patients as a test cohort. For metabolomics analysis, 106 urine samples from 27 healthy donors, 15 non-COVID-19 cases, 44 patients with non-severe COVID-19 and 20 patients with non-severe COVID-19 were collected.
Method details
Paired serum and urine proteome analysis
Serum samples were inactivated and sterilized at 56°C for 30 min and processed as reported previously with some modifications (Shen et al., 2020). Ten μL of serum for each sample was depleted using High Select Top-14 Abundant Protein Depletion Mini Spin Columns according to the manufacturer’s instructions. The eluates were denatured in 50 μL buffer (8 M urea in 100 mM triethylammonium bicarbonate, TEAB) and were enriched using a 3 kDa Millipore super filtration membrane column. The protein lysates were reduced and alkylated with 10 mM tris (2-carboxyethyl) phosphine (TCEP) and 40 mM iodoacetamide (IAA) in darkness for 30 min at 30°C. The solution was further diluted with 200 μL of 100 mM TEAB. Then the protein was digested firstly with 2 μg of trypsin at 32°C for 4 hours, followed by the second digestion using 2 μg of trypsin at 32°C for overnight. The reaction was stopped by adding 30 μL 10% trifluoroacetic acid (TFA).
Urine samples were inactivated and sterilized at 56°C for 30 min and 500 μL urine sample was then precipitated overnight with cold acetone (urine: acetone = 1:4, v/v, -20°C). The precipitated urine samples were centrifuged at 3000 g for 5 min. The pellet was resuspended in 200 μL 8M urea in 100 mM TEAB. The protein lysates were reduced and alkylated with 10 mM TCEP and 40 mM IAA in darkness for 30 min at 30°C. The solution was further diluted with 200 μL of 100 mM TEAB. The digestion was completed by using an enzyme mixture of 5 μg of trypsin and 1 μg of Lys-C at 32°C for 12 h and the reaction was stopped by adding 110 μL 10% trifluoroacetic acid (TFA).
The serum and urine peptides were labeled by TMTpro 16 plex reagents. Before TMTpro 16plex labeling, a batch design procedure was performed to minimize the batch effect. We randomly divided four different groups of samples into 6 batches for TMTpro 16plex labeling, with each batch containing the identical number of samples. A pooled sample was labeled with TMT channel 126 and included in each group as the quality control. In each batch, the peptides were fractionated and analyzed as described before (Shen et al., 2020) with minor modifications. In brief, the TMT samples were fractionated by the DIONEX UltiMate 3000 RSLCnano System (Thermo Fisher Scientific, San Jose, USA) coupled with an XBridge Peptide BEH C18 column (300 Å, 5 μm, 4.6 mm ∗250 mm) (Waters, Milford, MA, USA). The samples were separated using a gradient from 5% to 35% acetonitrile (ACN) in 10 mM ammonia (pH = 10.0) at a flow rate of 1 mL/min. Peptides were separated into 60 fractions and combined into 30 fractions. The fractions were then dried and redissolved in 2% ACN/0.1% formic acid (FA) of MS grade. The re-dissolved peptides were analyzed with the same U3000 HPLC system coupled to a Q Exactive HF hybrid Quadrupole-Orbitrap (Thermo Fisher Scientific, San Jose, USA) in data dependent acquisition (DDA) mode. For each fraction, peptides were loaded onto a pre-column (3 μm, 100 Å, 20 mm∗75 mm i.d.) and then analyzed with a 60 min LC gradient at a flow rate of 300 nL/min (analytical column: 1.9 μm, 120 Å, 150 mm∗75 mm i.d.; Buffer A: 2% ACN and 0.1% FA; Buffer B: 98% ACN and 0.1% FA). The gradient was uniformly changed from 5% to 28% buffer B.
For MS acquisition, the m/z range of MS1 was 350-1,800 Da with the resolution at 60,000, AGC target was set at 3e6, and maximum ion injection time (max IT) is 50 ms. Top 15 precursors were selected for MS/MS experiment, with the resolution of 45,000, AGC at 2e5, as well as the max IT of 120 ms. The isolation window of selected precursor was 0.7 m/z.
Metabolome analysis
Ethanol was added to the urine and serum samples and shaken vigorously to inactivate any potential viruses, then dried in a biosafety hood. Each 100 μL of serum or urine was extracted by adding 300 μL of methanol extraction solution. And the protein precipitate was removed by centrifugation and the supernatant contained metabolites of different chemical nature. To ensure the quantity and reliability of metabolite detection, non-targeted metabolomics assays were performed on four platforms as described previously with the same LC-MS/MS data acquisition parameters (Shen et al., 2020).
All UPLC-MS/MS methods were performed on ACQUITY 2D UPLC system (Waters, Milford, MA, USA) and Q Exactive HF hybrid Quadrupole-Orbitrap (Thermo Fisher Scientific, San Jose, USA) with HESI-II heated ESI source and Orbitrap mass analyzer.
ELISA analysis
All urine samples were centrifuged at 400 g for 5 min. LRP2, CUBN, CXCL14, RHOA, CDC42, RAC1 were detected by HUMAN ELISA kits (Wuhan Bioswamp Co., Ltd., Wuhan, China) according to the manufacturer’s instructions. The diluted standards and samples were all added to appropriate wells and then the HRP-conjugate reagent was added. The mixtures were then incubated for 30 min at 37°C. The wells were aspirated and washed for 5 times. The chromogen solution A and B were added to each well and incubated for 10 min. The stop solution was added to each well. The absorbance was read on a Thermo Fisher Scientific Multiskan FC microplate reader at 450 nm (Thermo Fisher Scientific, USA). The final concentration of protein was corrected by the concentration of the total protein respectively in each sample for statistical analysis. For validation of DEPs by ELISA, 29 urine samples were chosen from all 71 patients with COVID-19 including 21 non-severe and 8 severe. We recollected urine samples from 18 healthy donors due to the shortage of previous samples.
PRM analysis
The expression of 16 virus budding related proteins in Figure 4A was verified by PRM. Unfractionated urine peptides were separated at 300 nL/min along with a 30 min gradient and scheduled acquisition for 50 peptides (including 11 iRT peptides) by Q Exactive HF Hybrid Quadrupole-Orbitrap Mass Spectrometer as descript previously(Zheng et al., 2020).
Quantification and statistical analysis
Database search for quantification
For proteomics data analysis, MS raw data were searched using Proteome Discoverer (Version 2.4.1.15, Thermo Fisher Scientific) against a manually annotated and reviewed Homo sapiens protein FASTA database (SwissProt, 27 April 2020). The enzyme digestion was set to full-specific trypsin with two missed cleavages. Static modifications were set to TMTpro of lysine residues and N-terminus peptides, and carbamidomethylation of cysteine. Variable modifications were set to oxidation of methionine and acetylation of N-terminus peptides. Precursor ion mass tolerance was set to 10 ppm, and product ion was 0.02 Da. The peptide-spectrum-match allowed 1% target false discovery rate (FDR).
The protein abundance ratios of target samples to the pooled sample within each batch were used as the relative protein abundance for data analysis.
For metabolomics data analysis, metabolites were identified by searching an in-house library of more than 3,300 standards whose data entries are generated by running purified compound standards through the experimental platform. Metabolite’s abundance in serum or urine sample were quantified by in-house software. All the metabolites were identified within three strict criteria as described previously (Shen et al., 2020): narrow window retention index (RI), mass deviation less than 10 ppm and MS/MS spectra with high forward and reverse scores. For serum metabolomics data, the metabolite abundance was used to data mining. For urine metabolomics data mining, to reduce the effects caused by urine sample heterogeneity, the metabolite abundance was normalized by creatinine abundance.
Quality control analysis
The quality control analysis for proteomics and metabolomics data was as described previously (Shen et al., 2020) with minor modifications. For proteomics analysis, serum or urine samples from four groups were randomly distributed in six batches. A pooled peptide sample was the control sample labeled by TMT pro-126 in each batch. To ensure proteomics data stability, we first analyzed the CV values of protein abundance of six pooled urine samples and six pooled serum samples (Figure 1D). Then we checked the distribution of 90 urine and serum samples in six batches using UMAP with default parameters (Figures S1D and S1E) (McInnes et al., 2018). Z-Score normalization of relative protein abundance ratio for each sample was used as the input features. Proteins that are detected in over 80% of all samples were analyzed. For metabolomics analysis, as described previously, we also analyzed the CV of internal standards intensity in each sample to ensure the reliability of metabolomics data (Figure 1D).
Differential expression analysis and cluster analysis of proteins and metabolites
For more strict analysis, we narrowed the original 1,494 serum proteins, 3,854 urine proteins, 903 serum metabolites and 1,033 urine metabolites to a refined list of molecules that were detected in over 80% samples. The missing values of the protein matrix were imputed as zero. Protein or metabolites fold change (FC) between two groups was calculated using the mean relative protein abundance ratio. The p value was calculated by two-sided unpaired Welch’s t-test using relative protein abundance ratio between groups, followed by Benjamini & Hochberg correction. Proteins or metabolites with adjusted p value less than 0.05 and absolute log2 (FC) over 0.25 were regards as DEPs or DEMs.
All DEPs or DEMs were obtained from comparing COVID-19 groups (including severe COVID-19 and non-severe COVID-19 groups) with the healthy group, comparing the non-severe COVID-19 group with the severe COVID-19 group, and comparing the non-COVID-19 group with the healthy group (Figures S3B–S3E). To exclude the false positive influence of the non-COVID-19 groups, we filtered out the DEPs or DEMs between non-COVID-19 and healthy groups (Figure S3A). After filtering, 171 serum proteins, 805 urine proteins, 269 serum metabolites, and 338 urine metabolites were identified. Then the expression pattern in healthy, non-severe COVID-19, severe COVID-19 groups of these DEPs and DEMs was analyzed by Mfuzz (Kumar and Futschik, 2007). Mfuzz analysis was performed with default parameters to divide DEPs or DEMs into eight clusters (Figures S4B–S4E). For serum Mfuzz analysis, we selected 148 DEPs in clusters 1, 2, 3 and 4 (Figure S4B) and 161 DEMs in cluster 1, 3, 6 and 7 (Figure S4D) for further pathway enrichment analysis. For urine, we selected 580 DEPs in clusters 1, 3, 5, 6 and 7 (Figure S4C) and 138 DEMs in clusters 2, 6 and 8 for further pathway enrichment analysis (Figure S4E).
Machine learning
An R package RandomForest (version 4.6.14) was used to select important variables in the serum and urine data and build a machine learning model to distinguish patients with severe COVID-19 from non-severe ones. For feature selection, 1384 serum proteins and 3737 urine proteins in 39 non-severe and 11 severe COVID-19 cases were selected as input features. Finally, the 20 proteins, whose mean decrease accuracy ranked top 20, were screened out to build the classification model, and 4-fold cross validation were performed in each model. The AUC of the receiver operating characteristic curve and diagnostic accuracy was used to evaluate metrics for calculating the performance of the model.
After selecting 20 proteins, we adopt the Logistic Regression (LR) algorithm, within a Python package scikit-learn (version 0.24.2), to classify non-severe and severe. In LR algorithm, the C and penalty are basic parameters in LR. In this paper, we set the parameter C =1.0 and penalty = ‘l2’. We built a computational model to predict severe and non-severe and the probability of each sample was finally obtained.
Cytokine analysis
We classified the 234 cytokines into six types based on IMMPORT database(Updated: July 2020) (ImmPort, 2020). The one-way analysis of variance (ANOVA) was used to determine whether the cytokines show statistically significant differences among healthy, severe, and non-severe groups in serum and urine. According to an online database called immuneXpresso (Kveler et al., 2018), we matched the association between 234 cytokines and immune cells. 31 cytokines from our data were involved in the function of multiple immune cells and highlighted in Figure 3A. The correlation of cytokine expression and immune cells count in COVID-19 cases was calculated by the Spearman’s correlation coefficient. The shinyCircos (Yu et al., 2018) was used to visualize the proteomics data of Figure 3A.
Pathway enrichment analysis
For subcellular localization of each protein, the online UniProt database (https://www.uniprot.org/) was applied. The DEMs pathway analysis was performed by MetaboAnalyst (Pang et al., 2020). The Ingenuine Pathway Analysis (IPA) (Krämer et al., 2013) software was used to enrich DEPs or COVID-19 associated cytokines to signaling pathways. Log2(FC) of DEPs were used as the observation value for IPA analysis. The p value of IPA analysis was calculated with the right-tailed Fisher's exact test and was considered significant if less than 0.05.
Additional resources
This research is part of the work of a clinical trial named “To explore the pathogenesis and course prediction of novel coronavirus pneumonia (COVID-19) severe patients”. This research explored urine biomarkers for severe COVID-19 identification. The clinical trial was registered in the Chinese Clinical Trial Registry with an ID of ChiCTR2000031365 (https://www.chictr.org.cn/hvshowproject.aspx?id=25407).
Acknowledgments
This work is supported by grants from the National Key R&D Program of China (no. 2020YFE0202200), the National Natural Science Foundation of China (nos. 81972492, 21904107, and 81672086), the Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholars (no. LR19C050001), the Hangzhou Agriculture and Society Advancement Program (no. 20190101A04), the China Postdoctoral Science Foundation (no. 2020T130106ZX), and the Tencent Foundation (2020). We thank the Westlake University Supercomputer Center for assistance in data generation and storage, and the Mass Spectrometry & Metabolomics Core Facility at the Center for Biomedical Research Core Facilities of Westlake University for sample analysis.
Author contributions
T.G., B.S., J.X., H. Liu, and Y. Zhu designed and supervised the project. B.S., X.B., Y. Zheng, X. Zhu, J.D., H. Lyu, D.Y., Z.X., S.Z., Y.L., P.X., G.Z., D.W., H. Zhu, S.C., J.L., and H. Zhao collected the samples and clinical data. W.L., X.D., S.L., X.Y., N.X., L.X., S.Q., C.Z., W.G., X. Zhan., and J.H. conducted proteomics and metabolomics analysis. The data were interpreted and presented by all of the co-authors. X.B., W.L., X.D., S.L., Y. Zhu, and T.G. wrote the manuscript, with input from all of the other authors.
Declaration of interest
The research group of T.G. is partly supported by Pressure Biosciences. T.G. and Y. Zhu are shareholders of Westlake Omics. W.L., X.Y., N.X., W.G., and X. Zhan are currently employees of Westlake Omics. S.Q., C.Z., and H.L. are employees of Calibra Lab at DIAN Diagnostics. The remaining authors declare no competing interests.
Published: January 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110271.
Supplemental information
The protein matrix for 13 urine samples (13 patients) or 30 urine samples (4 patients) with COVID-19 for testing model, related to Figure 2, and the virus budding-related protein matrix for 69 urine samples, related to Figure 4.
Data and code availability
-
•
The proteomics raw data have been deposited at ProteomeXchange Consortium and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
All original codes have been deposited at Zenodo and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adachi J., Kumar C., Zhang Y., Olsen J.V., Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adell M.A., Teis D. Assembly and disassembly of the ESCRT-III membrane scission complex. FEBS Lett. 2011;585:3191–3196. doi: 10.1016/j.febslet.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt J., Topham P. Urine proteomics: the present and future of measuring urinary protein components in disease. CMAJ. 2007;177:361–368. doi: 10.1503/cmaj.061590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedroń R., Ciszek M., Tokarczyk M., Bobek M., Kurnyta M., Słominska E.M., Smoleński R.T., Marcinkiewicz J. 1-Methylnicotinamide and nicotinamide: two related anti-inflammatory agents that differentially affect the functions of activated macrophages. Arch. Immunol. Ther. Exp. (Warsz) 2008;56:127–134. doi: 10.1007/s00005-008-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block F., Schwarz M. Expression of GFAP in the striatum and its projection areas in response to striatal quinolinic acid lesion in rats. Neuroreport. 1994;5:2237–2240. doi: 10.1097/00001756-199411000-00008. [DOI] [PubMed] [Google Scholar]
- Broniec A., Klosinski R., Pawlak A., Wrona-Krol M., Thompson D., Sarna T. Interactions of plasmalogens and their diacyl analogs with singlet oxygen in selected model systems. Free Radic. Biol. Med. 2011;50:892–898. doi: 10.1016/j.freeradbiomed.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bros M., Haas K., Moll L., Grabbe S. RhoA as a key regulator of innate and adaptive immunity. Cells. 2019;8 doi: 10.3390/cells8070733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini R., Cecchini A.L. SARS-CoV-2 infection pathogenesis is related to oxidative stress as a response to aggression. Med. Hypotheses. 2020;143:110102. doi: 10.1016/j.mehy.2020.110102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Guo L., Tian J., He H., Marinova E., Zhang P., Zheng B., Han S. Overexpression of CXC chemokine ligand 14 exacerbates collagen-induced arthritis. J. Immunol. 2010;184:4455–4459. doi: 10.4049/jimmunol.0900525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., John Wherry E. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020;20:529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthard M.G., Morgan M., Woodruff T.M., Arumugam T.V., Taylor S.M., Carpenter T.C., Lackmann M., Boyd A.W. Eph/Ephrin signaling in injury and inflammation. Am. J. Pathol. 2012;181:1493–1503. doi: 10.1016/j.ajpath.2012.06.043. [DOI] [PubMed] [Google Scholar]
- Esmon C.T. The interactions between inflammation and coagulation. Br. J. Haematol. 2005;131:417–430. doi: 10.1111/j.1365-2141.2005.05753.x. [DOI] [PubMed] [Google Scholar]
- Felizardo R.J.F., de Almeida D.C., Pereira R.L., Watanabe I.K.M., Doimo N.T.S., Ribeiro W.R., Cenedeze M.A., Hiyane M.I., Amano M.T., Braga T.T., et al. Gut microbial metabolite butyrate protects against proteinuric kidney disease through epigenetic- and GPR109a-mediated mechanisms. FASEB J. 2019;33:11894–11908. doi: 10.1096/fj.201901080R. [DOI] [PubMed] [Google Scholar]
- Franchini M., Mannucci P.M. Inhibitors of propagation of coagulation (factors VIII, IX and XI): a review of current therapeutic practice. Br. J. Clin. Pharmacol. 2011;72:553–562. doi: 10.1111/j.1365-2125.2010.03899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Li T., Han M., Li X., Wu D., Xu Y., Zhu Y., Liu Y., Wang X., Wang L. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J. Med. Virol. 2020;92:791–796. doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraldsson B., Nyström J., Deen W.M. Properties of the glomerular barrier and mechanisms of proteinuria. Physiol. Rev. 2008;88:451–487. doi: 10.1152/physrev.00055.2006. [DOI] [PubMed] [Google Scholar]
- Heer C.D., Sanderson D.J., Alhammad Y.M.O., Schmidt M.S., Trammell S.A.J., Perlman S., Cohen M.S., Fehr A.R., Brenner C. Coronavirus and PARP expression dysregulate the NAD Metabolome: a potentially actionable component of innate immunity. bioRxiv. 2020 doi: 10.1101/2020.04.17.047480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyes M.P. Quinolinic acid and inflammation. Ann. N. Y. Acad. Sci. 1993;679:211–216. doi: 10.1111/j.1749-6632.1993.tb18300.x. [DOI] [PubMed] [Google Scholar]
- Hurley J.H., Hanson P.I. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imasawa T., Rossignol R. Podocyte energy metabolism and glomerular diseases. Int. J. Biochem. Cell Biol. 2013;45:2109–2118. doi: 10.1016/j.biocel.2013.06.013. [DOI] [PubMed] [Google Scholar]
- ImmPort . ImmPort; 2020. Gene Summary. [Google Scholar]
- Kalantari S., Nafar M. An update of urine and blood metabolomics in chronic kidney disease. Biomark. Med. 2019;13:577–597. doi: 10.2217/bmm-2019-0008. [DOI] [PubMed] [Google Scholar]
- Katagiri D., Ishikane M., Asai Y., Kinoshita N., Ota M., Moriyama Y., Ide S., Nakamura K., Nakamoto T., Nomoto H., et al. Evaluation of coronavirus disease 2019 severity using urine biomarkers. Crit. Care Explor. 2020;2:e0170. doi: 10.1097/CCE.0000000000000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2013;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Kurioka H., Minamino N., Nishitani Y., Sato H., Nishino T., Iwano M., Shiiki H., Kangawa K., Matsuo H., Dohi K. Plasma and urinary levels of adrenomedullin in chronic glomerulonephritis patients with proteinuria. Nephron. 1998;80:227–230. doi: 10.1159/000045172. [DOI] [PubMed] [Google Scholar]
- Kumar L., Futschik M.E. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwasako K., Kitamura K., Ito K., Uemura T., Yanagita Y., Kato J., Sakata T., Eto T. The seven amino acids of human RAMP2 (86) and RAMP3 (59) are critical for agonist binding to human adrenomedullin receptors. J. Biol. Chem. 2001;276:49459–49465. doi: 10.1074/jbc.M108369200. [DOI] [PubMed] [Google Scholar]
- Kveler K., Starosvetsky E., Ziv-Kenet A., Kalugny Y., Gorelik Y., Shalev-Malul G., Aizenbud-Reshef N., Dubovik T., Briller M., Campbell J., et al. Immune-centric network of cytokines and cells in disease context identified by computational mining of PubMed. Nat. Biotechnol. 2018;36:651–659. doi: 10.1038/nbt.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learn D.B., Fried V.A., Thomas E.L. Taurine and hypotaurine content of human leukocytes. J. Leukoc. Biol. 1990;48:174–182. [PubMed] [Google Scholar]
- Lee S.Y., Kim D.H., Sung S.A., Kim M.G., Cho W.Y., Kim H.K., Jo S.K. Sphingosine-1-phosphate reduces hepatic ischaemia/reperfusion-induced acute kidney injury through attenuation of endothelial injury in mice. Nephrology (Carlton) 2011;16:163–173. doi: 10.1111/j.1440-1797.2010.01386.x. [DOI] [PubMed] [Google Scholar]
- Levi M., van der Poll T. Inflammation and coagulation. Crit. Care Med. 2010;38:S26–S34. doi: 10.1097/CCM.0b013e3181c98d21. [DOI] [PubMed] [Google Scholar]
- Li B., Zhang Y., Wang F., Nair V., Ding F., Xiao H., Yao Y., Kretzler M., Ju W., Ding J. Urinary epidermal growth factor as a prognostic marker for the progression of Alport syndrome in children. Pediatr. Nephrol. 2018;33:1731–1739. doi: 10.1007/s00467-018-3988-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang Y., Liu H., Sun W., Ding B., Zhao Y., Chen P., Zhu L., Li Z., Li N., et al. Urine proteome of COVID-19 patients. Urine. 2020;2:1–8. doi: 10.1016/j.urine.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugo-Huitrón R., Ugalde Muñiz P., Pineda B., Pedraza-Chaverrí J., Ríos C., Pérez-de la Cruz V. Quinolinic acid: an endogenous neurotoxin with multiple targets. Oxid. Med. Cell Longev. 2013;2013:104024. doi: 10.1155/2013/104024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallipattu S.K., He J.C. The beneficial role of retinoids in glomerular disease. Front. Med. (Lausanne) 2015;2:16. doi: 10.3389/fmed.2015.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinkiewicz J., Kontny E. Taurine and inflammatory diseases. Amino Acids. 2014;46:7–20. doi: 10.1007/s00726-012-1361-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L., Healy J., Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. arXiv. 2018 1802.03426. [Google Scholar]
- Messner C.B., Demichev V., Wendisch D., Michalick L., White M., Freiwald A., Textoris-Taube K., Vernardis S.I., Egger A.-S., Kreidl M., et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020;11:11–24.e14. doi: 10.1016/j.cels.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocan M.Z., Erem C., Ulusoy S. Urinary cAMP activity in chronic renal failure. Int. Urol. Nephrol. 1998;30:215–222. doi: 10.1007/bf02550580. [DOI] [PubMed] [Google Scholar]
- Muller W.A. Getting leukocytes to the site of inflammation. Vet. Pathol. 2013;50:7–22. doi: 10.1177/0300985812469883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntel J., Xuan Y., Berger S.T., Reiter L., Bachur R., Kentsis A., Steen H. Advancing urinary protein biomarker discovery by data-independent acquisition on a quadrupole-orbitrap mass spectrometer. J. Proteome Res. 2015;14:4752–4762. doi: 10.1021/acs.jproteome.5b00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden A., Lapsley M., Igarashi T., Kelleher C., Lee P., Matsuyama T., Scheinman S., Shiraga H., Sundin D., Thakker R., et al. Urinary megalin deficiency implicates abnormal tubular endocytic function in Fanconi syndrome. J. Am. Soc. Nephrol. 2002;13:125–133. doi: 10.1681/ASN.V131125. [DOI] [PubMed] [Google Scholar]
- Ntyonga-Pono M.P. COVID-19 infection and oxidative stress: an under-explored approach for prevention and treatment? Pan Afr. Med. J. 2020;35:12. doi: 10.11604/pamj.2020.35.2.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K., Wada H., Okada N., Harada I., Nakajima T., Pasquale E.B., Tsuyama S. EphB2 and ephrin-B1 expressed in the adult kidney regulate the cytoarchitecture of medullary tubule cells through Rho family GTPases. J. Cell Sci. 2006;119:559–570. doi: 10.1242/jcs.02777. [DOI] [PubMed] [Google Scholar]
- Padovani A.M., Molina M.F., Mann K.K. Inhibition of liver x receptor/retinoid X receptor-mediated transcription contributes to the proatherogenic effects of arsenic in macrophages in vitro. Arteriosc. Thromb. Vasc. Biol. 2010;30:1228–1236. doi: 10.1161/atvbaha.110.205500. [DOI] [PubMed] [Google Scholar]
- Pang Z., Chong J., Li S., Xia J. MetaboAnalystR 3.0: toward an optimized workflow for global metabolomics. Metabolites. 2020;10 doi: 10.3390/metabo10050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perico L., Conti S., Benigni A., Remuzzi G. Podocyte-actin dynamics in health and disease. Nat. Rev. Nephrol. 2016;12:692–710. doi: 10.1038/nrneph.2016.127. [DOI] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif K., Cantrell D.A. Networking Rho family GTPases in lymphocytes. Immunity. 1998;8:395–401. doi: 10.1016/s1074-7613(00)80545-2. [DOI] [PubMed] [Google Scholar]
- Ronco C., Bellomo R., Kellum J.A. Acute kidney injury. Lancet. 2019;394:1949–1964. doi: 10.1016/s0140-6736(19)32563-2. [DOI] [PubMed] [Google Scholar]
- Rui L., Qingfeng M., Huan H., Hanwen S., Fang L., Kailang W., Wei W., Chengliang Z. The value of urine biochemical parameters in the prediction of the severity of coronavirus disease 2019. Clin. Chem. Lab. Med. 2020;58:1121–1124. doi: 10.1515/cclm-2020-0220. [DOI] [PubMed] [Google Scholar]
- Saleem M.A., Welsh G.I. Podocyte RhoGTPases: new therapeutic targets for nephrotic syndrome? F1000Res. 2019;8 doi: 10.12688/f1000research.20105.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saoudi A., Kassem S., Dejean A., Gaud G. Rho-GTPases as key regulators of T lymphocyte biology. Small GTPases. 2014;5 doi: 10.4161/sgtp.28208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefer J., Töpper R., Schmidt W., Block F., Heinrich P., Noth J., Schwarz M. Expression of interleukin 6 in the rat striatum following stereotaxic injection of quinolinic acid. J. Neuroimmunol. 1998;89:168–176. doi: 10.1016/s0165-5728(98)00133-7. [DOI] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers K., Dahlbäck B., Nicolaes G.A. Coagulation factor V and thrombophilia: background and mechanisms. Thromb. Haemost. 2007;98:530–542. [PubMed] [Google Scholar]
- Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020 doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N., Lin H., Wu T., Wang D., Wang W., Xie H., Zhang J., Feng Z. Inhibition of TGF-β1-receptor posttranslational core fucosylation attenuates rat renal interstitial fibrosis. Kidney Int. 2013;84:64–77. doi: 10.1038/ki.2013.82. [DOI] [PubMed] [Google Scholar]
- Shi Z., Puyo C.A. N-acetylcysteine to combat COVID-19: an evidence review. Ther. Clin. Risk Manag. 2020;16:1047–1055. doi: 10.2147/TCRM.S273700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgdrager F.J.H., Naudé P.J.W., Kema I.P., Nollen E.A., Deyn P.P. Tryptophan metabolism in inflammaging: from biomarker to therapeutic target. Front. Immunol. 2019;10:2565. doi: 10.3389/fimmu.2019.02565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan J.S., Yeh Y.Y., Ramachandran V., Deminoff S.J., Herman P.K. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. U S A. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H., Yang M., Wan C., Yi L.X., Tang F., Zhu H.Y., Yi F., Yang H.C., Fogo A.B., Nie X., Zhang C. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Álvarez B., Liapis H., Anders H.J. Links between coagulation, inflammation, regeneration, and fibrosis in kidney pathology. Lab. Invest. 2016;96:378–390. doi: 10.1038/labinvest.2015.164. [DOI] [PubMed] [Google Scholar]
- Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.Q., Wang Q., Miao H. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct. Target Ther. 2020;5:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T., Stefanoni D., Reisz J.A., Nemkov T., Bertolone L., Francis R.O., Hudson K.E., Zimring J.C., Hansen K.C., Hod E.A., et al. COVID-19 infection results in alterations of the kynurenine pathway and fatty acid metabolism that correlate with IL-6 levels and renal status. medRxiv. 2020 doi: 10.1101/2020.05.14.20102491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W., Zhang N., Jin R., Feng Y., Wang S., Gao S., Gao R., Wu G., Tian D., Tan W., et al. Immune suppression in the early stage of COVID-19 disease. Nat. Commun. 2020;11:5859. doi: 10.1038/s41467-020-19706-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virreira Winter S., Karayel O., Strauss M.T., Padmanabhan S., Surface M., Merchant K., Alcalay R.N., Mann M. Urinary proteome profiling for stratifying patients with familial Parkinson's disease. EMBO Mol. Med. 2021:e13257. doi: 10.15252/emmm.202013257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitek V., Gold P.H., Gill W., Lang D.J., Conn A., Cowley R.A. Urinary cyclic AMP and post-traumatic acute renal failure. Clin. Chim. Acta. 1977;75:401–414. doi: 10.1016/0009-8981(77)90359-x. [DOI] [PubMed] [Google Scholar]
- Vitko D., Cho P.S., Kostel S.A., DiMartino S.E., Cabour L.D., Migliozzi M.A., Logvinenko T., Warren P.G., Froehlich J.W., Lee R.S. Characterizing patients with recurrent urinary tract infections in vesicoureteral reflux: a pilot study of the urinary proteome. Mol. Cell Proteomics. 2020;19:456–466. doi: 10.1074/mcp.RA119.001873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner Stefan, Schmitz Gerd. Plasmalogens the neglected regulatory and scavenging lipid species. Chem Phys Lipids. 2011;164(6):573–589. doi: 10.1016/j.chemphyslip.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Wan H., Versnel M.A., Cheung W.Y., Leenen P.J., Khan N.A., Benner R., Kiekens R.C. Chorionic gonadotropin can enhance innate immunity by stimulating macrophage function. J. Leukoc. Biol. 2007;82:926–933. doi: 10.1189/jlb.0207092. [DOI] [PubMed] [Google Scholar]
- WHO . WHO; 2020. COVID-19 Therapeutic Trial Synopsis. [Google Scholar]
- WHO Report of the WHO-China joint mission on coronavirus disease 2019 (COVID-19) 2020. https://www.who.int/publications-detail/report-of-the-who-china-joint-mission-on-coronavirus-disease-2019-(covid-19
- Worldometer Coronavirus updates. 2021. https://www.worldometers.info/
- Wu B., Rockel J.S., Lagares D., Kapoor M. Ephrins and Eph receptor signaling in tissue repair and fibrosis. Curr. Rheumatol. Rep. 2019;21:23. doi: 10.1007/s11926-019-0825-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Shu T., Yang X., Song J.-X., Zhang M., Yao C., Liu W., Huang M., Yu Y., Yang Q., et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020;7:1157–1168. doi: 10.1093/nsr/nwaa086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Ouyang Y., Yao W. shinyCircos: an R/Shiny application for interactive creation of Circos plot. Bioinformatics. 2018;34:1229–1231. doi: 10.1093/bioinformatics/btx763. [DOI] [PubMed] [Google Scholar]
- Zheng X., Xu K., Zhou B., Chen T., Huang Y., Li Q., Wen F., Ge W., Wang J., Yu S., et al. A circulating extracellular vesicles-based novel screening tool for colorectal cancer revealed by shotgun and data-independent acquisition mass spectrometry. J. Extracell. Vesicles. 2020;9:1750202. doi: 10.1080/20013078.2020.1750202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The protein matrix for 13 urine samples (13 patients) or 30 urine samples (4 patients) with COVID-19 for testing model, related to Figure 2, and the virus budding-related protein matrix for 69 urine samples, related to Figure 4.
Data Availability Statement
-
•
The proteomics raw data have been deposited at ProteomeXchange Consortium and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
All original codes have been deposited at Zenodo and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.