Abstract
The vast array of cell types of multicellular organisms must individually fine-tune their internal metabolism. One important metabolic and stress regulatory mechanism is the dynamic attachment/removal of glucose-derived sugar N-acetylglucosamine on proteins (O-GlcNAcylation). The number of proteins modified by O-GlcNAc is bewildering, with 7000+ sites in human cells. The outstanding challenge is determining how key O-GlcNAc sites regulate a target pathway amidst thousands of potential global sites. Innovative solutions are required to address this challenge in cell models and disease therapy. This Perspective shares critical suggestions for the O-GlcNAc field gleaned from the international O-GlcNAc community. Further, we summarize critical tools and tactics to enable newcomers to O-GlcNAc biology to drive innovation at the interface of metabolism and disease. The growing pace of O-GlcNAc research makes this a timely juncture to involve a wide array of scientists and new tool makers to selectively approach regulatory roles of O-GlcNAc in disease.
Graphical Abstract
1. Introduction
Cells continuously use protein modifications to fine-tune signaling and epigenetic activities to maintain homeostasis.1 Human and other eukaryotic cells use an elegant system for nutrient and stress signaling whereby metabolites generated during nutrient flux and stress are directly added to key regulatory proteins as the sugar O-GlcNAc (O-linked N-acetylglucosamine).2 Protein O-GlcNAc modifications are therefore a dynamic sensor of nutrient flux and stress pathways in human cells and tissues (Fig. 1). Diseases ranging from cancer3,4 to diabetes5 and neurodegeneration6,7 affect cellular nutrient flux and result in aberrant O-GlcNAcylation. Cellular stressors like reactive oxygen species, heat shock, DNA damage, or toxins also drive O-GlcNAc.8 An evolving hypothesis in the field is that any disease state with altered nutrient or stress levels has an O-GlcNAc component.9 The complexity of unraveling key sites between diverse diseases is a central challenge for the field. Accordingly, our understanding of central unifying O-GlcNAc features between disease states remains incomplete.10 The goal of this Perspective is to provide actionable recommendations and focal points to expand the O-GlcNAc field and impact our understanding of how metabolism and stress sensing regulates physiology and disease at the molecular level.
Figure 1:
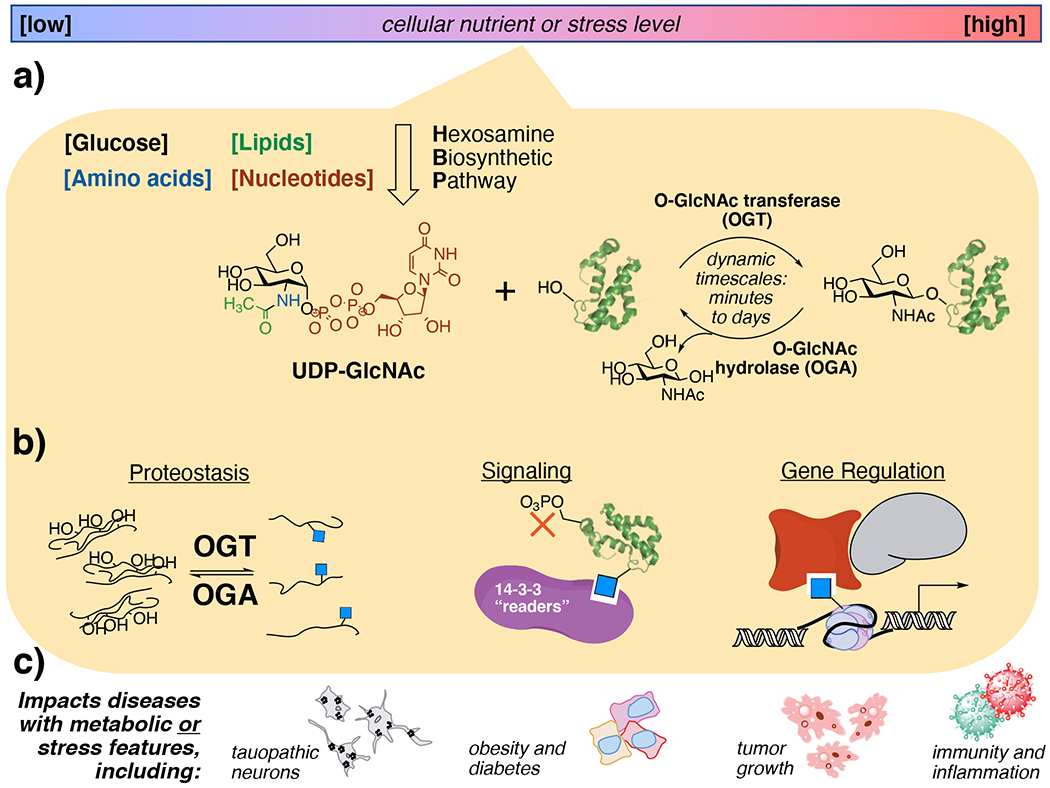
Cells use O-GlcNAc modifications to sense metabolites resulting from altered nutrient or stress levels, a process that alters protein behaviors and can lead to disease. a) Nutrient flux through the through the hexosamine biosynthetic pathway creates UDP-GlcNAc, which is dynamically cycled onto proteins as O-GlcNAcylation by OGT and OGA. b) O-GlcNAc events regulate diverse cellular functions depending on the biological context and the type of protein that is modified. c) Aberrant O-GlcNAcylation is observed in numerous diseases that display altered hexosamine biosynthetic pathway flux.
A critical challenge for determining pathological O-GlcNAc-driven mechanisms is identifying which specific pathways and proteins are affected in each disease state. O-GlcNAc addition events are widespread and modify over 7000 known protein sites.11,12 Just two proteins cycle O-GlcNAc on and off from proteins in humans, O-GlcNAc transferase (OGT)13,14 and O-GlcNAcase/hydrolase (OGA).15 An open question is how these O-GlcNAc processing enzymes differentially recognize key substrates from others. One form of control over global cellular O-GlcNAcylation comes from nutrient flux through the hexosamine biosynthetic pathway (HBP), which incorporates carbohydrate, amino acid, and nucleotide metabolites into the OGT substrate UDP-GlcNAc.16 Differential levels of UDP-GlcNAc from HBP flux allows cellular control over metabolism, signaling, and transcriptional activities through O-GlcNAcylation (Fig 1a).1 Another form of control can come from cellular stressors that increase the overall O-GlcNAc level under harmful stimuli.17 Thus, O-GlcNAc is a functional regulator over nutrient metabolism and stress.
The extreme promiscuities of OGT and OGA, which both accept thousands of substrate O-GlcNAc glycosites on proteins, make detailed functional studies of individual glycosites challenging. The activities of OGT and OGA are dynamic, unevenly distributive rather than systematically processive,18 and can “cycle” O-GlcNAc onto and off of proteins as fast as minutes19 or over the course of days.20 Moreover, each individual O-GlcNAc site may have distinct roles on a protein, for instance affecting its stability, protein interactions, or active conformation (Fig 1b).21 A grand challenge for the field is to determine not only key O-GlcNAc sites for given cellular processes but also spatiotemporal aspects of O-GlcNAc that enable cells to regulate key O-GlcNAc events. Here, we will show how innovative chemical and biological tools have made key steps toward these goals.
Given the regulatory nature of protein O-GlcNAcylation at large, determining the biological functions of a given O-GlcNAc modification on a protein is often the key to unlocking therapeutic insight. It is likely that, in a given disease state, multiple O-GlcNAc sites on multiple proteins synergistically or antagonistically impact pathological processes.9 Indeed, disease states with pathological hyper- or hypoglycemia all display aberrant O-GlcNAc levels (Fig 1c).10 Developing a precise understanding of O-GlcNAc roles is a critical step toward O-GlcNAc-focused disease interventions. We will discuss O-GlcNAc-focused tactics established for mechanistic studies in the model tissues as a platform for moving to detailed studies in disease.
The O-GlcNAc field is rapidly growing, with new labs and collaborations beginning each year. We end the Perspective by channeling input from the global O-GlcNAc community into 5 specific key recommendations that we have heard repeatedly. These are meant to serve as mere guidelines accelerate the potential of O-GlcNAc research for disease impact.
2. Chemical Biology Tools to Track O-GlcNAc
The size of an O-GlcNAc modification on a protein is small, just 212 Daltons, but it can have a major impact on protein conformation, interactions, and stability. Because individual O-GlcNAc modifications are too subtle to be detected without molecular-scale techniques or mass spectrometry, chemical tools and precision analytical strategies are essential.22 There are also very few commercial site-specific antibodies for O-GlcNAc sites—fewer than 10 as of this writing, mostly for histones—making spatiotemporal studies of O-GlcNAc modifications on protein challenging.23 This is in contrast to antibodies for the related phosphorylated serine or threonine sites, which have enabled rigid dissection of phosphorylation events during cellular signaling.24 However, chemical tools are beginning to allow precision for key O-GlcNAc sites. To date, the field of O-GlcNAc-directed chemical tools has centered on pharmacological inhibitors of O-GlcNAc processing proteins, molecular imaging probes, proteomic developments (Fig 2), and, recently, biochemically-directed tools to target specific O-GlcNAc features (Fig 3, below).23
Figure 2:
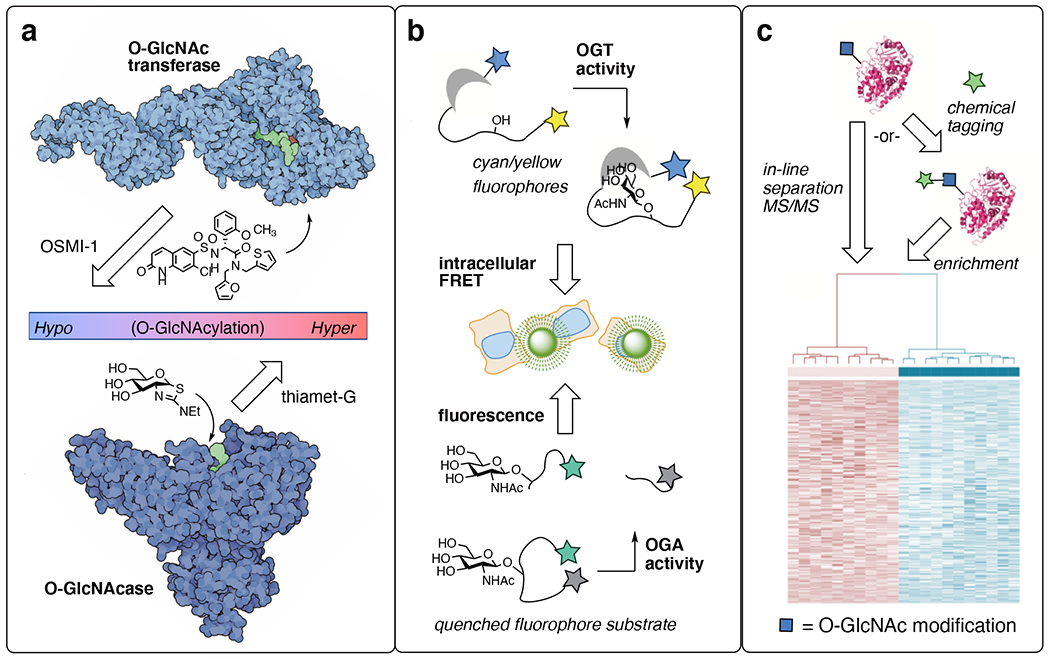
Chemical biology tools reveal a plethora of O-GlcNAc cellular features. a) Selective inhibitors for O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) allow enable O-GlcNAc effects to be implicated and targeted in disease. b) Imaging tools enable global OGT and OGA activity to be observed in living cells. c) Proteomics developments for O-GlcNAc have revealed > 7000 unique human sites. OGT image generated from PDB ID: 3pe4 and 1w3b; OGA image generated from PDB 2cbj.
Figure 3:
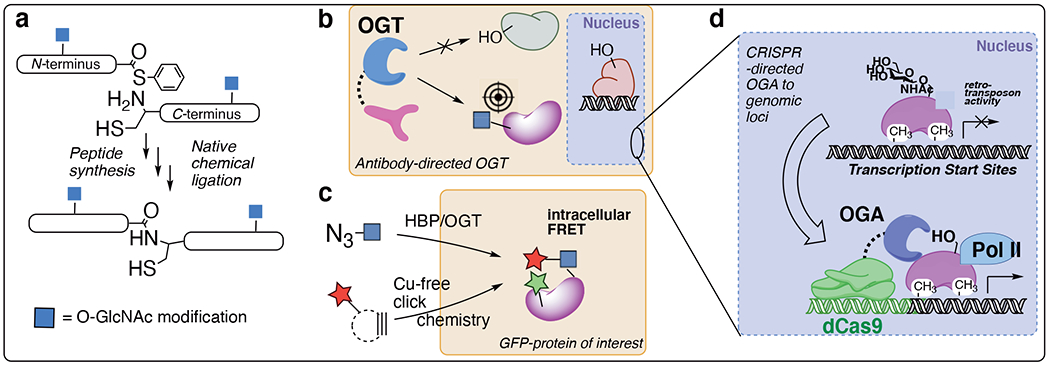
Site-specific chemical tools for O-GlcNAc studies. a) Total protein synthesis of tau and a-synuclein specific O-GlcNAc glycoforms. b) Targeted OGT and OGA tools enable selective GlcNAc glycosylation (or de-glycosylation) of a desired protein. c) Metabolic labeling and intracellular installation of fluorescent sugar tags allows quantification of O-GlcNAcylation of a protein of interest fused to a fluorescent protein. d) O-GlcNAc roles at genomic loci are uncovered using dCas9 nuclease targeting.
Chemical tools to probe and identify global O-GlcNAc functions.
The development of selective pharmacological probes has unlocked the therapeutic potential of targeting O-GlcNAc processing enzymes.25 Depending on whether a disease states is fundamentally hypoglycemic like Alzheimer’s disease or hyperglycemic like cancer, either enhancing or blocking O-GlcNAc levels can be beneficial as a treatment strategy. Cellular O-GlcNAc transferase (OGT) inhibitors like the OSMI series26 (Fig 2a) or the unnatural sulfur heterocycle analog Ac45SGlcNAc27 block O-GlcNAcylation. OGT inhibitors reveal biological mechanisms dependent on O-GlcNAc mechanisms in disease and have been extensively reviewed.25,28 However, none has yet entered the clinic, suggesting challenges with safety and efficacy in vivo. An optimized analog of Ac45SGlcNAc, 5SGlcNHex, exchanges ester groups that are easily cleaved in serum with a cell-permeable lipid tail and has improved pharmacokinetics in mice, delivering a promising OGT inhibitor for in vivo studies.29
O-GlcNAcase (OGA) chemical probes, including thiamet-G (Fig 2a), are active compounds in both cellular as well as animal studies, as reviewed.30 OGA inhibitors have successfully passed through clinic evaluations with federal approval to treat the hypoglycemic neurodegenerative disease progressive supranuclear palsy (PSP).31 PSP is a rare disease, but the first uses of O-GlcNAc cycling inhibitors in humans is a groundbreaking medical advance that will inevitably expand clinical therapeutic uses of O-GlcNAcase inhibitors in disease.
Imaging probes for O-GlcNAc cycling dynamics are another niche in which chemical tools are essential (Fig 2b). The variable timeframes of O-GlcNAcylation installation and persistence, from minutes to hours to days, begs for real-time tools for imaging global O-GlcNAc changes. The Mahal group built fluorescent intracellular O-GlcNAc-binding lectin domains to reveal nutrient-factor-driven changes in OGT activity on the order of minutes.32 Expanding the approach toward spatially-targeted variants highlighted different features and time frames of nutrient sensing between the nucleus, cytosol, and plasma membrane.33 OGA activity has also been imaged by relying on its hydrolase activity to unleash a fluorophore from a quencher.34 Real-time studies with OGA-directed fluorescent probes reveal fast kinetics of OGA in live cells, on the order of seconds to minutes.34 These OGT and OGA imaging tools confirm the dynamic, nutrient/stress-sensing nature of global O-GlcNAcylation events and will be most useful when applied between healthy and disease tissue states, which is an ongoing area of study. PET ligands have been developed, allowing the direct detection of O-GlcNAcase levels in mice, primates and humans.35,36 In addition, enzymatic labeling of O-GlcNAc proteins in fixed histology samples enables detailed spatial resolution.37 Though spatial imaging has not yet be done in live cells, this enzymatic O-GlcNAc labeling technique allows patient samples to be probed for subcellular O-GlcNAc distribution in frozen tissue.37
O-GlcNAc-directed proteomic strategies has also benefitted greatly from chemical tool development (Fig 2c). Improved separation technologies by weak lectin affinity chromatography carefully pick out O-GlcNAc from other glycosylation types, allowing tandem mass spectrometry (MS/MS) experiments to directly detect endogenous O-GlcNAc patterns from tissue samples.38–42 Chemical tagging using bioorthogonal chemical groups enable labeling with affinity enrichment handles. Chemical labeling has also increased the coverage of the “O-GlcNAcome.” Metabolic labeling (also called metabolic engineering or metabolic chemical reporters) hijack biochemical pathways toward UDP-GlcNAc analogs bearing azide or alkyne handles, allowing chemical detection.43 With metabolic labeling, non-enzymatic “off-target” labeling can occur on cysteine residues as S-GlcNAcylation.44 Next-generation metabolic labeling tools can avoid S-GlcNAcylation by avoiding problematic chemical groups.45,46 On the other hand, chemoenzymatic labeling approaches in lysed tissue uses an engineered galactosyltransferase (GalT1 (Y298L)) to selectively label O-GlcNAcylated sites on proteins with click chemistry handles.47 The enzymatic selection also avoids the problem of S-GlcNAc off-target labeling. For both major types of O-GlcNAc chemical labeling, recent advances in isotope-coded bioorthogonal chemistry have improved the sensitivity of both the metabolic and chemoenzymatic labeling approaches, allowing the identification of hundreds or even thousands of O-GlcNAc sites in a single experiment.48,49
O-GlcNAc tools for specific GlcNAc glycosites and proteins.
Most O-GlcNAc chemical tools are “global” tools that study pan-cellular O-GlcNAcylation. A ‘holy grail’ in the field is directed O-GlcNAc systems to target precise proteins, cellular structures, and genomic sites. The role of O-GlcNAc in protein stability has been revealed by global methods, but to unpick the functional role(s) of each O-GlcNAc site discrete glycoforms must by synthesized (Fig 1a).22 The Pratt group has undertaken the comprehensive synthesis of α-synuclein glycoforms,50 including unnatural sugar analogs like glucose and mannose to determine the biophysical mechanisms whereby O-GlcNAc stabilizes proteins.51 A similar approach has been developed by the Hackenberger group for tau.52
An approach to visualizing specific O-GlcNAc protein events is to fuse a fluorescent protein to the protein of interest, which can be recorded in cells using azide metabolic labeling and copper-free click chemistry (Fig 3c).53 The Chen group used this approach to follow β-catenin O-GlcNAcylation in cells.53 Intracellular click chemistry has also been used to determine O-GlcNAc effects on protein maturation.
Cellular studies to determine site-specific O-GlcNAc functions, however, must selectively modify endogenous proteins in living cells. Towards this goal, the Woo lab re-engineered the O-GlcNAc cycling enzymes OGT and OGA with fusions to nanobodies. This approach enables “nano-OGT” to install O-GlcNAc on a precise protein target,54 and “nano-OGA” to remove O-GlcNAc from a chosen target,55 respectively (Fig 3b). In separate type of directed approach, specific roles of O-GlcNAc in chromatin remodeling and transcription at defined genomic loci was probed using an RNA-guided system. The Boulard, Edwards, and Bestor groups fused a dead Cas9 nuclease (dCas9) to OGA, enabling genomic targeting with O-GlcNAc cycling tools using guide-RNA (Fig 3d).56 This dCas9-OGA system reveals that O-GlcNAcylation is a key regulator of viral gene transposon effects at specific genomic loci. This genome-targeted OGA tool, applied at sites of viral transposons, can in principle be targeted to other disease states to carefully pick out O-GlcNAc mechanisms during the epigenetic regulation of any desired genomic loci.
Genetic engineering advances, especially CRISPR (clustered regularly interspaced short palindromic repeats)-guided gene editing allows for rapid mutation of a specific O-GlcNAc serine or threonine site to alanine.57 Ser/Thr to Ala point mutations can selectively eliminate an O-GlcNAc site, allowing “deletion” studies. More recently, the van Aalten group hijacked OGT’s promiscuous thiol glycosylation catalytic activity to install S-linked GlcNAc through Ser/Thr to Cys mutation.58 S-linked GlcNAc is cleaved by OGA approximately 10-fold slower than O-GlcNAc,59 making Ser/Thr->Cys mutations guided by CRISPR gene editing a promising strategy for the “permanent” installation of stabilized GlcNAc residues on precise protein sites. CRISPR-based studies can probe specific glycosites and generate arrays to screen for key functions in phenotypic assays.
Chemical tools have expanded the coverage of O-GlcNAc sites, and now precision tools are catching up for directed O-GlcNAc studies. These tools are set to reveal specific functions of the O-GlcNAcylation of distinct target proteins and await application in real-time functional studies.
3. Biological Tactics to Determine O-GlcNAc Function
Scaling cellular O-GlcNAc studies up to the tissue and organism level—where daily rhythms of feeding, activity, and other hallmarks of physiology add complexity60—requires certain tactical considerations. Foremost, O-GlcNAc glycosylation may be necessary to maintain cellular health. Knockout of OGT or OGA is embryonic or perinatally lethal,13,15 so knockouts may have unintended consequences in developed organs. Viability may vary between the many cell types that comprise an organ. To overcome cellular needs for O-GlcNAc cycling, conditional knockout of OGT or OGA using Cre/Lox-dependent, promoter-specific ablation has been widely applicable.14,61 In general, OGT knockout has a more dramatic phenotypic outcome that knockout of OGA. This OGT effect knockout likely reflects a unique requirement for OGT-catalyzed O-GlcNAc addition, while redundant mechanisms may act in the degradation of O-GlcNAc targets. If knockout is not an option, pharmacological inhibitors can probe O-GlcNAc cycling enzymes in a reversible fashion. Glucose use also varies across tissue types, for instance reaching levels up to 10-fold higher in the brain than in certain tissues in the periphery.62 Owing to this, and the brain’s role in dictating behavior, we discuss tactics in the context of the brain vs. the periphery, but complementary strategies are often employed in each type of tissue.
O-GlcNAc tactics employed in the brain.
O-GlcNAc cycling is important in the brain, which primarily uses glucose for metabolism and has higher OGT expression than peripheral tissues.62 OGT is the most well-conserved glycosyltransferase in humans,63 making inherited mutants that lead to disease exceedingly rare. However, subtle point mutations in OGT’s substrate recognition domain are associated with a form of X-linked intellectual disability, directly demonstrating the importance of O-GlcNAc events in the brain.64,65
Adjusting global levels of OGT and OGA activity controls neurodegeneration6 and neuroprotection66 (Fig 4a). The recent FDA (United States Food and Drug Administration) designation and approval of the OGA inhibitor MK-8719 for tauopathy treatment31 underscores the pharmaceutical potential of neuroprotective O-GlcNAc therapeutics. To overcome the systemic nature of pharmacological OGA inhibition, conditional knockout mice using floxed OGT (OGTf) were crossed with neuron-specific Cre animals to generate neuron-specific OGT cKO models (Fig 4b). Following development, mice bearing forebrain neuron OGTf rapidly underwent neurodegeneration within 7 weeks. Furthermore, the brain tissue isolated from OGTf mice mimicked reduced O-GlcNAc levels in severe human Alzheimer’s disease tissue samples.6
Figure 4:
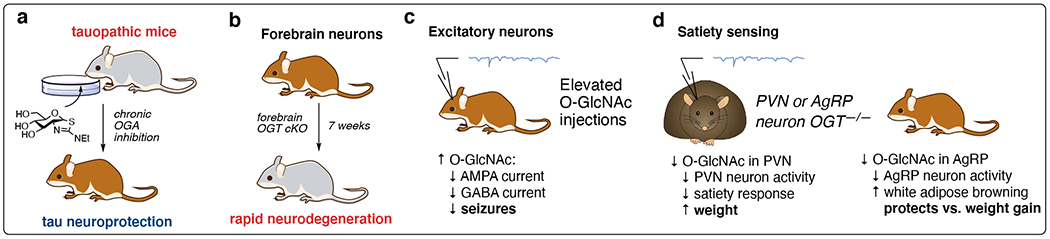
High glucose and GlcNAc utilization in the brain makes neurobiology a key area of study for the O-GlcNAc field. a) Pharmacological elevation of O-GlcNAcylation is protective against neurodegenerative disease. b) Loss of forebrain O-GlcNAcylation leads to neuron death, simulating neurodegeneration. c) O-GlcNAcylation regulates physiology and behavior by modulating neuron activity in specific neuron types. d) O-GlcNAc plays reciprocal roles in feeding behavior regulation between neuron type.
The brain also uses O-GlcNAc to fine-tune neurotransmission through the modification of GABA and glutaminergic receptors, modulating signals in response to local cellular environments (Fig 4c).67 Neuron-specific enhancement of O-GlcNAc regulates satiety in the paraventricular nucleus of the hypothalamus (PVN) and in neurons that express agouti-related protein (AgRP)(Fig 4d). Loss of O-GlcNAc in PVN neurons reduced the homeostatic sensing of nutrients for feedback regulation of satiety and feeding responses, leading to elevated weight.68 Intriguingly, loss of OGT in AgRP neurons protects against diet-induced weight gain. In both cases, loss of OGT is generally deactivating to neurons at the global cellular level. An open question is whether this is due to a loss of a specific O-GlcNAc site or multiple glycosides, which remains difficult to probe with genetic tools. Another question is the impact of hormones in different neurons, because O-GlcNAc regulates multiple, tissue-specific roles in insulin signaling.69
An alternative tactic to study the effects of specific O-GlcNAc sites on neuronal proteins is to use chemical tools. O-GlcNAc regulates proteostasis in axons, with disease mutations leading to neuropathies in a protein- and site-specific manner.70 Specific sites on tau52 in Alzheimer’s disease and α-synuclein50,51 in Parkinson’s disease have been interrogated through protein semi-synthesis (Fig 3a, above). An area that has not yet been met with success is tracking the dynamic nature of O-GlcNAc in specific brain (or tissue) regions, especially during nutrient flux, stress, or signaling.
O-GlcNAc tactics employed in the periphery.
Beyond well-studied metabolic disease roles, current research is expanding into a diverse array of processes including developmental physiology, metabolic regulation, and cancer processes.10 O-GlcNAc research in stem cells reveal roles of OGT/OGA cycling in hemopoiesis71 as well as pancreatic islet cell differentiation.72 Immune cells also respond to nutrient flux, and O-GlcNAc studies suggest that a hyperglycemic diets affect T-cell maturation differently between individuals with obese or non-obese metabolism.73 O-GlcNAc mechanisms do not act in isolation, and current research is also probing cross-talk between other signaling systems including phosphorylation74,75 and lipid signaling76 as emerging areas in O-GlcNAc-linked disease studies. Peripheral O-GlcNAc studies have identified critical metabolic roles for OGT in muscle,77 adipose,78 and the liver.1
An area of intense study is epigenetic regulation via direct transcription factor O-GlcNAcylation including the core pluripotency factors Sox2, Oct4, and Klf4.79,80 Modulation of transcription factors can direct differentiation, and loss of OGT or OGA in developing cells are each perinatally lethal.14,15 Less understood functions of O-GlcNAc include functional activity on chromatin remodeling during embryonic stem cell (ESC) and cancer stem cell (CSC) processes (Fig. 5).81 Current directions in this field focus on DNA methyltransferase (DNMT) proteins56 and demethylase enzymes including Ten-Eleven Translocation proteins (TET), which strongly bind OGT.82 Crosstalk of O-GlcNAcylation and phosphorylation on the RNA polymerase II C-terminal domain (RNA Pol II CTD) is thought to fine-tune transcriptional activity once recruited to a genetic locus.18 After transcription, splicing is regulated by extensive O-GlcNAc/phosphorylation crosstalk, toggling between productive or detained introns for rapid regulation of cellular protein levels that acts in under an hour.19 This altered splicing depending on O-GlcNAc levels is likely another key mechanism for nutrient and stress sensing to complement HBP flux (Fig 1a). Temporal splicing dynamics and epigenetic regulation will be a critical next frontier for O-GlcNAc mechanisms of cellular homeostasis.
Figure 5:
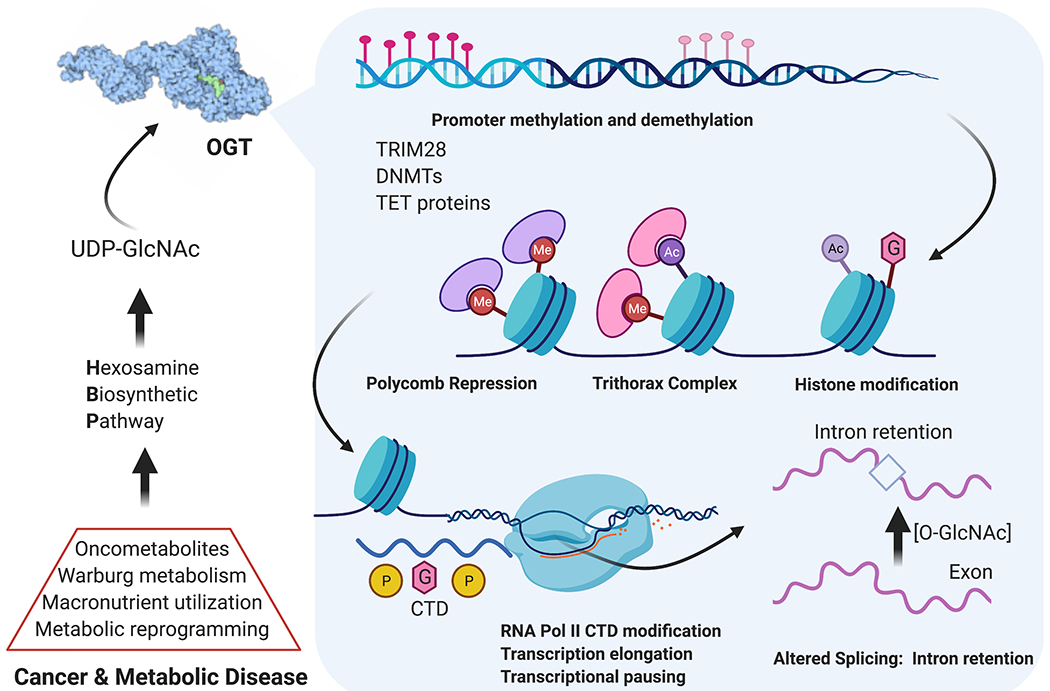
O-GlcNAc dynamics play extensive roles in epigenetic regulation, development, and cancer. The Hexosamine Biosynthetic Pathway acts at the macronutrient level to produce UDP-GlcNAc. Nucleocytoplasmic OGT modifies transcription factors, chromatin remodeling proteins, RNA polymerase II, and splicing factors for extensive nutrient/stress-responsive epigenetic events. In development, stem cells use these programs to differentiate. Numerous cancers harness enhanced HBP flux to activate growth, survival, and metastatic programs through balancing differentiation and de-differentiation to cancer stem cells.
Essential roles for proteostasis, signaling, and epigenetic regulation during development, cancer, and the brain reveal critical functions of O-GlcNAc sites, and we suggest that the surface has only been scratched. OGT and OGA activity is regulated by multiple factors including nutrient levels and cellular stress, enabling individual cells to use O-GlcNAc as a sensor of local environments, including nutrient levels as well as oxidative, cytotoxic, osmotic, or cellular stressors.8 If your team studies a disease associated with stress or metabolic dysfunction, it is almost certain that O-GlcNAc is involved.
4. Recommendations for next-generation O-GlcNAc research
An active discussion in the O-GlcNAc community has led to a community-driven roadmap for rapid growth of O-GlcNAc glycobiology. Harnessing virtual meeting technology, over three hundred global experts including funding agency program officers met and recommended key areas that might fuel future research emerged from this community discussion. We summarize these here. Box 1 distills needs for innovative new technologies and recommendations for continued rapid growth of the O-GlcNAc field, as developed in the international community of O-GlcNAc researchers.
Box 1 |. Call for innovations to advance O-GlcNAc biology.
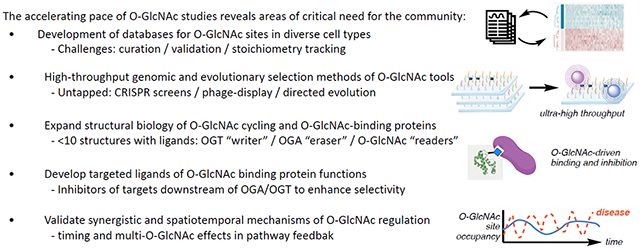
Recommendation 1: Develop a centralized database of curated O-GlcNAc sites linked to physiology/pathophysiology.
Technology has fueled a rapid increase in reported O-GlcNAc sites, now in the thousands.11 However, there lacks a designated and curated repository of O-GlcNAc sites, which is currently a major limitation for informatic and comparative studies of O-GlcNAc pathways. When a site has been defined, there is little information about the abundance of the modification at that site. The highest priority recommendation of this Perspective is to provide support for a comprehensive database accepted by the field that encompasses what is currently known about O-GlcNAc sites in diseases. Several general databases for post-translational modifications include O-GlcNAc, such as PhosphoSitePlus83 and GlyGen.84 Two recent groups have completed comprehensive searches of all known O-GlcNAc sites. As of this writing, the “O-GlcNAcAtlas” database12 contains over 2800 human O-GlcNAc sites and the “O-GlcNAcome” database contains over 7000 human O-GlcNAc sites.11 These literature-spanning databases are a major step toward mapping confirmed O-GlcNAc sites in humans and other species. Their ongoing curation is critical as the pace of O-GlcNAc research increases.
Despite these advances, critical information about O-GlcNAcylation status is lacking. Almost nothing is known about the stoichiometry of most O-GlcNAc sites. Mass shift tools have been used to count the number of O-GlcNAc sites, but these tools have only been applied to a subset of proteins.85 Another challenge is a discrepancy in total numbers of high confidence O-GlcNAc assignments between databases, which underscores that a centralized, accepted depository is still lacking. In O-GlcNAc assignments, care must be taken to validate sites by careful scrutiny of MS/MS data, since metabolic labeling experiments are now suspected to be contaminated with a small but artificial population of S-linked GlcNAc sites.44 S-GlcNAc sites can be readily excluded by careful MS/MS annotation, chemical proteomic validation,86 or OGA overexpression controls. Bioinformatic prediction methods, including the YinOYang server87 and O-GlcNAc-PRED-II88 are limited by lack of access to such a curated database.89 Development of next-generation informatic tools to infer site and regulatory networks is a key corollary future goal.
Recommendation 2: Support the development of genomic and evolutionary selection methods for O-GlcNAc tool development.
The lack of an apparent local consensus sequence for O-GlcNAcylation is a primary hurdle for understanding the diversity of O-GlcNAc-driven effects. The development of genomic tools, such as tiled CRISPR arrays to screen potential O-GlcNAc site effects, would be a tremendous boon to the field. Base editing is limited to a small set of mutations, but prime editing has the potential to expand this strategy by creating or eliminating O-GlcNAc glycosites on genome-wide scales.90
A related challenge is a lack of in vivo biosensors for specific O-GlcNAc sites. Few site-selective O-GlcNAc antibodies have been generated.23 Therefore, the wide variety of O-GlcNAc sites and effects begs for tools developed using highly combinatorial approaches such as directed evolution and phage display. In one potential strategy, though the authors did not develop tools for O-GlcNAc glycans, Cummings et al successfully used yeast surface display to generate site-specific anti-glycan antibodies.91 The use of targeting systems like bionanoparticles or RNA aptamers92 could bring new methods into O-GlcNAc research, including uses as intracellular sensors for specific O-GlcNAc glycosites or UDP-GlcNAc nucleotide levels. This type of O-GlcNAc site-selective detection approach would enable the characterization of critical O-GlcNAc sites as biomarkers to track glycobiological functions in disease.
Recommendation 3: Expand structural biology of O-GlcNAc cycling and reader proteins.
Despite OGT binding and activity on thousands of protein sites, few structures of OGT have been solved with substrates.93 Co-crystal structures of just one protein and several peptides are reported, which limits our knowledge of binding mechanisms beyond those particular glycosites. Biochemical experiments by Vocadlo et al suggest that human OGA binding to O-GlcNAc glycoproteins is primarily driven by sugar interactions with minimal regard to protein sequence, but OGT-substrate affinity is highly variable and suggestive that human OGT is responsible for differentially controlling substrate glycosylation patterns between cell environments.94 Particularly mysterious is the tetratricopeptide repeat (TPR) domain, with up to 13 TPR units that make a superhelix where substrates are thought to bind. Microarray studies over 6000 proteins determined that specific structural elements, including asparagine ladders, contribute to binding, but the dearth of co-crystal structures remains a barrier.95 As an alternative approach to direct binding studies, electrophilic probes were used to determine the effects of key TPR residues via mutation and trapping of a protein substrate.96 However, a “solved” mechanism of OGT substrate recognition remains far from reach as of this writing for thousands of potential OGT substrates. On the flipside, less is known about OGA’s substrate recognition preferences beyond kinetic studies.94
A few O-GlcNAc binding proteins, or “readers,” have been confirmed through biophysical and structural studies.97 Though these O-GlcNAc-driven interactions were observed to be weak (172-324 μM) and likely transient, O-GlcNAc readers represent a promising regulatory motif. We strongly encourage structural biologists with skill in cryo-electron microscopy and protein NMR techniques to intersect with O-GlcNAc research to enable faster, dynamic access to additional O-GlcNAc cycling and reader interaction characterization data. The chemical synthesis of defined O-GlcNAc glycoforms and substrate mimics will facilitate this endeavor.22
Recommendation 4: Develop targeted ligands for O-GlcNAc readers and functions.
The pharmacological landscape of O-GlcNAc targeting remains limited. The OGA inhibitor MK-8719 was granted FDA Orphan drug designation to treat tau-driven neurodegenerative disease.31 However, O-GlcNAc regulatory pathways downstream of this “global” O-GlcNAc cycling enzyme have not been intentionally targeted for therapeutic development. The discovery and structural validation of O-GlcNAc reader proteins is an untapped area for drug design in human disease.97 Targeting O-GlcNAc signaling pathways downstream of OGT and OGA could provide higher specificity and avoid the toxic events associated with OGT loss-of-function.6 A key example is the key stem cell reprogramming factor Sox2, which has a single O-GlcNAc site that determines over 20 protein-protein interactions.98 A challenge is relatively weak binding affinities for O-GlcNAc-driven interactions, reported to be in the high micromolar dissociation constants for 14-3-3, Ebp1, and α-enolase.97 Photocrosslinking is an elegant solution to overcome weak binding,99 and further developments and technologies will no doubt expand our knowledge of O-GlcNAc reader proteins.
Recommendation 5: Validate spatiotemporal and synergistic O-GlcNAc mechanisms.
The diverse and ubiquitous nature of O-GlcNAc protein sites enable cells to fine-tune their response to environment using a wide variety of mechanisms. It is very likely that disease processes are mediated by multiple O-GlcNAc effects over different proteins and pathways and with varied spatiotemporal parameters. The O-GlcNAc community is open to the idea that discrete O-GlcNAc-driven effects may be synergistic or antagonistic to each other in cellular settings, and network-based modeling (or related informatics strategies) might be the key that unlocks complex regulatory relationships involving metabolic and stress pathways. A key example lies in enhancing overall O-GlcNAc levels in the treatment of neurodegenerative disease. Pratt et al have revealed complementary mechanisms to reduce toxic protein aggregation of α-synuclein, both by direct α-synuclein glycosylation50 and by the activities of chaperone proteins that are enhanced by O-GlcNAcylation.100 It is likely that other diseases, like Alzheimer’s, share similar synergistic O-GlcNAc effects on disease progression or treatment. At the global proteomics level, the Walker and Zachara groups revealed distinct protein expression functions of both catalytic and noncatalytic roles of OGT using mutants capable of only one or the other function.63 The resulting data revealed essential roles of for cell proliferation as well as disease pathways. Separating and confirming unique roles poses an active area of study for O-GlcNAc-implicated diseases. Finally, few tools exist for studying dynamic O-GlcNAc modifications on short timescales and within the complex intracellular space. Insulin stimulation causes noted movement of OGT between the plasma membrane, cytosol, and nucleus within minutes,76 so spatiotemporal tools are needed to open a crucial next frontier in O-GlcNAc studies.
Regardless of the potential for multiple O-GlcNAc events, determining the specific sites and mechanisms of individual pathways is crucial for unpicking each arm of complementary or competing pathways. The O-GlcNAc community encourages reviewers and referees to consider both site-specific in addition to multi-site roles for O-GlcNAc regulation as we approach a holistic understanding of this critical sugar for cellular adaptation to environment and disease.
Implications/future directions:
In just the past 10 years, efforts to understand the biological roles for the O-GlcNAc modification have greatly expanded. These advances have been fueled by researchers from diverse fields recognizing the potential importance of O-GlcNAcylation in their area of research. The expansion has been linked to improved methods for perturbing O-GlcNAc addition and removal and the ability to sensitively detect O-GlcNAcylation sites on its diverse protein targets. The increasing pace of O-GlcNAc disease biology and chemical tool development reveals yet more areas that are unexplored. Focused efforts on technology and tactical development to solve challenges the O-GlcNAc glycobiology field currently faces will no doubt further our holistic understanding in mechanisms of cellular physiology and disease.
Acknowledgements:
We would like to thank Dr. Pamela Marino, Program Director at the US National Institute of General Medical Sciences (NIGMS) for her commitment and vision for expanding glycoscience as a national priority. We would also like to acknowledge Dr. Michelle Bond and Dr. Karl Krueger, Program Officers, for their support of glycobiology tools and for discussions and notes. We would further like to thank all of the participants of the international O-GlcNAc workshop held in March, 2020 who contributed to discussion, both during and after the meeting. We gratefully acknowledge the NIGMS grant 1R35GM142637-01 for financial support to CF as well as the National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK) for financial support to JAH.
Footnotes
Competing Interests:
The authors declare no competing interests.
Data Sharing Statement:
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
References
- 1.Hart GW Nutrient regulation of signaling and transcription. J Biol Chem 294, 2211–2231, doi: 10.1074/jbc.AW119.003226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X & Qian K Protein O-GlcNAcylation: emerging mechanisms and functions. Nature Reviews Molecular Cell Biology 18, 452 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrer CM, Sodi VL & Reginato MJ O-GlcNAcylation in Cancer Biology: Linking Metabolism and Signaling. J Mol Biol 428, 3282–3294, doi: 10.1016/j.jmb.2016.05.028 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanover JA, Chen W & Bond MR O-GlcNAc in cancer: An Oncometabolism-fueled vicious cycle. J Bioenerg Biomembr 50, 155–173, doi: 10.1007/s10863-018-9751-2 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Ma J & Hart GW Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev Proteomics 10, 365–380, doi: 10.1586/14789450.2013.820536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang AC, Jensen EH, Rexach JE, Vinters HV & Hsieh-Wilson LC Loss of O-GlcNAc glycosylation in forebrain excitatory neurons induces neurodegeneration. Proceedings of the National Academy of Sciences 113, 15120, doi: 10.1073/pnas.1606899113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper used conditional knockout of OGT to precisely determine hippocampal neuron-specific roles for O-GlcNAc, especially neurodegeneration vs. neuroprotection effects.
- 7.Wheatley EG et al. Neuronal O-GlcNAcylation Improves Cognitive Function in the Aged Mouse Brain. Current Biology 29, 3359–3369.e3354, doi: 10.1016/j.cub.2019.08.003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez MR, Dias TB, Natov PS & Zachara NE Stress-induced O-GlcNAcylation: an adaptive process of injured cells. Biochemical Society Transactions 45, 237–249, doi: 10.1042/bst20160153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang YR & Suh PG O-GlcNAcylation in cellular functions and human diseases. Adv Biol Regul 54, 68–73, doi: 10.1016/j.jbior.2013.09.007 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Slawson C, Copeland RJ & Hart GW O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends in biochemical sciences 35, 547–555, doi: 10.1016/j.tibs.2010.04.005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wulff-Fuentes E et al. The human O-GlcNAcome database and meta-analysis. Scientific Data 8, 25, doi: 10.1038/s41597-021-00810-4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma J, Li Y, Hou C & Wu C O-GlcNAcAtlas: A database of experimentally identified O-GlcNAc sites and proteins. Glycobiology, doi: 10.1093/glycob/cwab003 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shafi R et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proceedings of the National Academy of Sciences 97, 5735, doi: 10.1073/pnas.100471497 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnell N, Zachara NE, Hart GW & Marth JD Ogt-Dependent X-Chromosome-Linked Protein Glycosylation Is a Requisite Modification in Somatic Cell Function and Embryo Viability. Molecular and Cellular Biology 24, 1680, doi: 10.1128/MCB.24.4.1680-1690.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang YR et al. O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell 11, 439–448, doi: 10.1111/j.1474-9726.2012.00801.x (2012). [DOI] [PubMed] [Google Scholar]
- 16.Marshall S, Nadeau O & Yamasaki K Dynamic Actions of Glucose and Glucosamine on Hexosamine Biosynthesis in Isolated Adipocytes. Journal of Biological Chemistry 279, 35313–35319, doi: 10.1074/jbc.M404133200 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Zachara NE, Molina H, Wong KY, Pandey A & Hart GW The dynamic stress-induced “O-GlcNAc-ome” highlights functions for O-GlcNAc in regulating DNA damage/repair and other cellular pathways. Amino Acids 40, 793–808, doi: 10.1007/s00726-010-0695-z (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu L et al. Distributive O-GlcNAcylation on the Highly Repetitive C-Terminal Domain of RNA Polymerase II. Biochemistry 55, 1149–1158, doi: 10.1021/acs.biochem.5b01280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan Z-W et al. O-GlcNAc regulates gene expression by controlling detained intron splicing. Nucleic acids research 48, 5656–5669, doi: 10.1093/nar/gkaa263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper identifies an elegant mechanism for rapid re-balancing of OGT/OGA activity upon O-GlcNAc disruption via O-GlcNAc feedback into alternative splicing regulation.
- 20.Miller MW, Caracciolo MR, Berlin WK & Hanover JA Phosphorylation and Glycosylation of Nucleoporins. Archives of Biochemistry and Biophysics 367, 51–60, doi: 10.1006/abbi.1999.1237 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Eustice M, Bond MR & Hanover JA O-GlcNAc cycling and the regulation of nucleocytoplasmic dynamics. Biochemical Society Transactions 45, 427–436, doi: 10.1042/bst20160171 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Groenevelt JM, Corey DJ & Fehl C Chemical Synthesis and Biological Applications of O-GlcNAcylated Peptides and Proteins. Chembiochem 22, 1854–1870, doi: 10.1002/cbic.202000843 (2021). [DOI] [PubMed] [Google Scholar]; This paper collects known synthetic methods for site-specific O-GlcNAc installation. Also see Gorelik et al (RSC Chem Biol, 2020) and Alteen et al (Curr Opin Struct Biol) for thorough collections of O-GlcNAc detection tools and assays.
- 23.Gorelik A & van Aalten DMF Tools for functional dissection of site-specific O-GlcNAcylation. RSC Chemical Biology 1, 98–109, doi: 10.1039/D0CB00052C (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoevesandt O & Taussig MJ Phospho-specific antibodies by design. Nature Biotechnology 31, 889–891, doi: 10.1038/nbt.2712 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Alteen MG, Tan HY & Vocadlo DJ Monitoring and modulating O-GlcNAcylation: assays and inhibitors of O-GlcNAc processing enzymes. Curr Opin Struct Biol 68, 157–165, doi: 10.1016/j.sbi.2020.12.008 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Martin SES et al. Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors. Journal of the American Chemical Society 140, 13542–13545, doi: 10.1021/jacs.8b07328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gloster TM et al. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nature chemical biology 7, 174–181, doi: 10.1038/nchembio.520 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ju Kim E O-GlcNAc Transferase: Structural Characteristics, Catalytic Mechanism and Small-Molecule Inhibitors. ChemBioChem 21, 3026–3035, doi: 10.1002/cbic.202000194 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Liu TW et al. Metabolic Inhibitors of O-GlcNAc Transferase That Act In Vivo Implicate Decreased O-GlcNAc Levels in Leptin-Mediated Nutrient Sensing. Angewandte Chemie (International ed. in English) 57, 7644–7648, doi: 10.1002/anie.201803254 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elbatrawy AA, Kim EJ & Nam G O-GlcNAcase: Emerging Mechanism, Substrate Recognition and Small-Molecule Inhibitors. Chem Med Chem 15, 1244–1257, doi: 10.1002/cmdc.202000077 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Selnick HG et al. Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies. Journal of Medicinal Chemistry 62, 10062–10097, doi: 10.1021/acs.jmedchem.9b01090 (2019). [DOI] [PubMed] [Google Scholar]; This paper reports the medicinal chemistry efforts that led to the first FDA-sanctioned inhibitor of OGA in human disease.
- 32.Carrillo LD, Krishnamoorthy L & Mahal LK A Cellular FRET-Based Sensor for β-O-GlcNAc, A Dynamic Carbohydrate Modification Involved in Signaling. Journal of the American Chemical Society 128, 14768–14769, doi: 10.1021/ja065835+ (2006). [DOI] [PubMed] [Google Scholar]
- 33.Carrillo LD, Froemming JA & Mahal LK Targeted in vivo O-GlcNAc sensors reveal discrete compartment-specific dynamics during signal transduction. J Biol Chem 286, 6650–6658, doi: 10.1074/jbc.M110.191627 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cecioni S & Vocadlo DJ Carbohydrate Bis-acetal-Based Substrates as Tunable Fluorescence-Quenched Probes for Monitoring exo-Glycosidase Activity. J Am Chem Soc 139, 8392–8395, doi: 10.1021/jacs.7b01948 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Lee J-H et al. PET quantification of brain O-GlcNAcase with [18F]LSN3316612 in healthy human volunteers. EJNMMI Research 10, 20, doi: 10.1186/s13550-020-0616-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul S et al. Evaluation of a PET Radioligand to Image O-GlcNAcase in Brain and Periphery of Rhesus Monkey and Knock-Out Mouse. J Nucl Med 60, 129–134, doi: 10.2967/jnumed.118.213231 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aguilar AL, Hou X, Wen L, Wang PG & Wu P A Chemoenzymatic Histology Method for O-GlcNAc Detection. Chembiochem 18, 2416–2421, doi: 10.1002/cbic.201700515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haynes PA & Aebersold R Simultaneous Detection and Identification of O-GlcNAc-Modified Glycoproteins Using Liquid Chromatography-Tandem Mass Spectrometry. Analytical Chemistry 72, 5402–5410, doi: 10.1021/ac000512w (2000). [DOI] [PubMed] [Google Scholar]
- 39.Vosseller K et al. Quantitative analysis of both protein expression and serine/threonine post-translational modifications through stable isotope labeling with dithiothreitol. PROTEOMICS 5, 388–398, doi: 10.1002/pmic.200401066 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Vosseller K et al. O-Linked N-Acetylglucosamine Proteomics of Postsynaptic Density Preparations Using Lectin Weak Affinity Chromatography and Mass Spectrometry. Molecular & Cellular Proteomics 5, 923–934, doi: 10.1074/mcp.T500040-MCP200 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Chalkley RJ, Thalhammer A, Schoepfer R & Burlingame AL Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proceedings of the National Academy of Sciences 106, 8894, doi: 10.1073/pnas.0900288106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alfaro JF et al. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc Natl Acad Sci U S A 109, 7280–7285, doi: 10.1073/pnas.1200425109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darabedian N & Pratt MR Identifying potentially O-GlcNAcylated proteins using metabolic labeling, bioorthogonal enrichment, and Western blotting. Methods Enzymol 622, 293–307, doi: 10.1016/bs.mie.2019.02.017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin W et al. Artificial Cysteine S-Glycosylation Induced by Per-O-Acetylated Unnatural Monosaccharides during Metabolic Glycan Labeling. Angewandte Chemie International Edition 57, 1817–1820, doi: 10.1002/anie.201711710 (2018). [DOI] [PubMed] [Google Scholar]; This paper reports off-target S-GlcNAcylated “artifacts” in metabolic labeling, suggesting that stringent validation of O-GlcNAc proteomic studies is required.
- 45.Pedowitz NJ et al. Anomeric Fatty Acid Functionalization Prevents Nonenzymatic S-Glycosylation by Monosaccharide Metabolic Chemical Reporters. ACS Chemical Biology, doi: 10.1021/acschembio.1c00470 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hao Y et al. Next-generation unnatural monosaccharides reveal that ESRRB O-GlcNAcylation regulates pluripotency of mouse embryonic stem cells. Nature communications 10, 4065, doi: 10.1038/s41467-019-11942-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khidekel N et al. A Chemoenzymatic Approach toward the Rapid and Sensitive Detection of O-GlcNAc Posttranslational Modifications. Journal of the American Chemical Society 125, 16162–16163, doi: 10.1021/ja038545r (2003). [DOI] [PubMed] [Google Scholar]
- 48.Woo CM et al. Mapping and quantification of over 2000 O-linked glycopeptides in activated human T cells with isotope-targeted glycoproteomics (Isotag). Molecular & Cellular Proteomics 17, 764–775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports exquisitely sensitive O-GlcNAc labeling tools with isotope tagging to enable global characterization of O-GlcNAc sites during T cell activation events.
- 49.Li J et al. An Isotope-Coded Photocleavable Probe for Quantitative Profiling of Protein O-GlcNAcylation. ACS Chemical Biology 14, 4–10, doi: 10.1021/acschembio.8b01052 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Levine PM et al. α-Synuclein O-GlcNAcylation alters aggregation and toxicity, revealing certain residues as potential inhibitors of Parkinson’s disease. Proc Natl Acad Sci U S A 116, 1511–1519, doi: 10.1073/pnas.1808845116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galesic A et al. Comparison of N-Acetyl-Glucosamine to Other Monosaccharides Reveals Structural Differences for the Inhibition of α-Synuclein Aggregation. ACS Chem Biol 16, 14–19, doi: 10.1021/acschembio.0c00716 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwagerus S, Reimann O, Despres C, Smet-Nocca C & Hackenberger CP Semi-synthesis of a tag-free O-GlcNAcylated tau protein by sequential chemoselective ligation. J Pept Sci 22, 327–333, doi: 10.1002/psc.2870 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Lin W, Gao L & Chen X Protein-Specific Imaging of O-GlcNAcylation in Single Cells. ChemBioChem 16, 2571–2575, doi: 10.1002/cbic.201500544 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Ramirez DH et al. Engineering a Proximity-Directed O-GlcNAc Transferase for Selective Protein O-GlcNAcylation in Cells. ACS Chemical Biology, doi: 10.102l/acschembio.0c00074 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ge Y et al. Target protein deglycosylation in living cells by a nanobody-fused split O-GlcNAcase. Nature chemical biology 17, 593–600, doi: 10.1038/s41589-021-00757-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boulard M, Rucli S, Edwards JR & Bestor TH Methylation-directed glycosylation of chromatin factors represses retrotransposon promoters. Proc Natl Acad Sci U S A 117, 14292–14298, doi: 10.1073/pnas.1912074117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adli M The CRISPR tool kit for genome editing and beyond. Nature communications 9, 1911, doi: 10.1038/s41467-018-04252-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gorelik A et al. Genetic recoding to dissect the roles of site-specific protein O-GlcNAcylation. Nature structural & molecular biology 26, 1071–1077, doi: 10.1038/s41594-019-0325-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Macauley MS, Stubbs KA & Vocadlo DJ O-GlcNAcase catalyzes cleavage of thioglycosides without general acid catalysis. Journal of the American Chemical Society 127, 17202–17203, doi: 10.1021/ja0567687 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Kim EY et al. A role for O-GlcNAcylation in setting circadian clock speed. Genes & Development 26, 490–502, doi: 10.1101/gad.182378.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keembiyehetty C et al. Conditional Knock-out Reveals a Requirement for O-Linked N-Acetylglucosaminase (O-GlcNAcase) in Metabolic Homeostasis. Journal of Biological Chemistry 290, 7097–7113, doi: 10.1074/jbc.M114.617779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okuyama R & Marshall S UDP-N-acetylglucosaminyl transferase (OGT) in brain tissue: temperature sensitivity and subcellular distribution of cytosolic and nuclear enzyme. J Neurochem 86, 1271–1280, doi: 10.1046/j.1471-4159.2003.01939.x (2003). [DOI] [PubMed] [Google Scholar]
- 63.Levine ZG et al. Mammalian cell proliferation requires noncatalytic functions of O-GlcNAc transferase. Proceedings of the National Academy of Sciences 118, e2016778118, doi: 10.1073/pnas.2016778118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; OGT has distinct protein regulatory roles through three mechanisms: O-GlcNAc catalytic modification of proteins, O-GlcNAc-driven proteolysis, and non-catalytic functions. Rapid OGT regulatory tools are also developed herein.
- 64.Konzman D et al. O-GlcNAc: Regulator of Signaling and Epigenetics Linked to X-linked Intellectual Disability. Front Genet 11, 605263, doi: 10.3389/fgene.2020.605263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pravata VM et al. An intellectual disability syndrome with single-nucleotide variants in O-GlcNAc transferase. Eur J Hum Genet 28, 706–714, doi: 10.1038/s41431-020-0589-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; X-linked intellectual disability is the first human disease implicated with OGT single nucleotide polymorphisms, indicating a genetic basis. Also see Konzman, et al (2020).
- 66.Yuzwa SA et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nature chemical biology 4, 483–490, doi: 10.1038/nchembio.96 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Stewart LT et al. Acute Increases in Protein O-GlcNAcylation Dampen Epileptiform Activity in Hippocampus. J Neurosci 37, 8207–8215, doi: 10.1523/jneurosci.0173-16.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lagerlöf O et al. The nutrient sensor OGT in PVN neurons regulates feeding. Science 351, 1293–1296, doi: 10.1126/science.aad5494 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reveals that organismal feeding behavior is controlled by O-GlcNAc, which is revealed to influence apetite at the molecular level in PVN neurons.
- 69.Vaidyanathan K & Wells L Multiple tissue-specific roles for the O-GlcNAc post-translational modification in the induction of and complications arising from type II diabetes. J Biol Chem 289, 34466–34471, doi: 10.1074/jbc.R114.591560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen P-H et al. Gigaxonin glycosylation regulates intermediate filament turnover and may impact giant axonal neuropathy etiology or treatment. JCI Insight 5, doi: 10.1172/jci.insight.127751 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abramowitz LK, Harly C, Das A, Bhandoola A & Hanover JA Blocked O-GlcNAc cycling disrupts mouse hematopoeitic stem cell maintenance and early T cell development. Scientific Reports 9, 12569, doi: 10.1038/s41598-019-48991-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baumann D et al. Role of nutrient-driven O-GlcNAc-post-translational modification in pancreatic exocrine and endocrine islet development. Development 147, dev186643, doi: 10.1242/dev.186643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hruby A & Hu FB The Epidemiology of Obesity: A Big Picture. Pharmacoeconomics 33, 673–689, doi: 10.1007/s40273-014-0243-x (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Z et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal 3, ra2–ra2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leney AC, El Atmioui D, Wu W, Ovaa H & Heck AJR Elucidating crosstalk mechanisms between phosphorylation and O-GlcNAcylation. Proceedings of the National Academy of Sciences 114, E7255, doi: 10.1073/pnas.1620529114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang X et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 451, 964–969, doi: 10.1038/nature06668 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Lambert M, Bastide B & Cieniewski-Bernard C Involvement of O-GlcNAcylation in the Skeletal Muscle Physiology and Physiopathology: Focus on Muscle Metabolism. Front Endocrinol (Lausanne) 9, 578, doi: 10.3389/fendo.2018.00578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang Y et al. O-GlcNAc transferase inhibits visceral fat lipolysis and promotes diet-induced obesity. Nature communications 11, 181, doi: 10.1038/s41467-019-13914-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parker MP, Peterson KR & Slawson C O-GlcNAcylation and O-GlcNAc Cycling Regulate Gene Transcription: Emerging Roles in Cancer. Cancers 13, 1666, doi: 10.3390/cancers13071666 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ozcan S, Andrali SS & Cantrell JE Modulation of transcription factor function by O-GlcNAc modification. Biochimica et biophysica acta 1799, 353–364, doi: 10.1016/j.bbagrm.2010.02.005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Akella NM et al. O-GlcNAc Transferase Regulates Cancer Stem-like Potential of Breast Cancer Cells. Molecular Cancer Research 18, 585–598, doi: 10.1158/1541-7786.Mcr-19-0732 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hrit J et al. OGT binds a conserved C-terminal domain of TET1 to regulate TET1 activity and function in development. eLife 7, e34870, doi: 10.7554/eLife.34870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hornbeck PV et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic acids research 43, D512–520, doi: 10.1093/nar/gku1267 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.York WS et al. GlyGen: computational and informatics resources for glycoscience. Glycobiology 30, 72–73 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Darabedian N, Thompson JW, Chuh KN, Hsieh-Wilson LC & Pratt MR Optimization of Chemoenzymatic Mass Tagging by Strain-Promoted Cycloaddition (SPAAC) for the Determination of O-GlcNAc Stoichiometry by Western Blotting. Biochemistry 57, 5769–5774, doi: 10.1021/acs.biochem.8b00648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wells L et al. Mapping Sites of <em>O</em>-GlcNAc Modification Using Affinity Tags for Serine and Threonine Post-translational Modifications. Molecular & Cellular Proteomics 1, 791–804, doi: 10.1074/mcp.M200048-MCP200 (2002). [DOI] [PubMed] [Google Scholar]
- 87.Gupta R & Brunak S Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput, 310–322 (2002). [PubMed] [Google Scholar]
- 88.Jia C, Zuo Y & Zou Q O-GlcNAcPRED-II: an integrated classification algorithm for identifying O-GlcNAcylation sites based on fuzzy undersampling and a K-means PCA oversampling technique. Bioinformatics 34, 2029–2036, doi: 10.1093/bioinformatics/bty039 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Jochmann R, Holz P, Sticht H & Sturzl M Validation of the reliability of computational O-GlcNAc prediction. Biochimica et biophysica acta 1844, 416–421, doi: 10.1016/j.bbapap.2013.12.002 (2014). [DOI] [PubMed] [Google Scholar]
- 90.Anzalone AV et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157, doi: 10.1038/s41586-019-1711-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McKitrick TR et al. Development of smart anti-glycan reagents using immunized lampreys. Communications Biology 3, 91, doi: 10.1038/s42003-020-0819-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zichel R, Chearwae W, Pandey GS, Golding B & Sauna ZE Aptamers as a Sensitive Tool to Detect Subtle Modifications in Therapeutic Proteins. PLOS ONE 7, e31948, doi: 10.1371/journal.pone.0031948 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levine ZG & Walker S The Biochemistry of O-GlcNAc Transferase: Which Functions Make It Essential in Mammalian Cells? Annual review of biochemistry 85, 631–657, doi: 10.1146/annurev-biochem-060713-035344 (2016). [DOI] [PubMed] [Google Scholar]
- 94.Shen DL, Gloster TM, Yuzwa SA & Vocadlo DJ Insights into O-linked N-acetylglucosamine ([0-9]O-GlcNAc) processing and dynamics through kinetic analysis of O-GlcNAc transferase and O-GlcNAcase activity on protein substrates. J Biol Chem 287, 15395–15408, doi: 10.1074/jbc.M111.310664 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; Detailed in vitro kinetic studies of OGT and OGA reveal mechanistic roles in nutrient sensing, adding to a rich body of OGT and OGA mechanistic studies.
- 95.Levine ZG et al. O-GlcNAc Transferase Recognizes Protein Substrates Using an Asparagine Ladder in the Tetratricopeptide Repeat (TPR) Superhelix. Journal of the American Chemical Society 140, 3510–3513, doi: 10.1021/jacs.7b13546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kositzke A et al. Elucidating the protein substrate recognition of O-GlcNAc transferase (OGT) toward O-GlcNAcase (OGA) using a GlcNAc electrophilic probe. Int J Biol Macromol 169, 51–59, doi: 10.1016/j.ijbiomac.2020.12.078 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Toleman CA et al. Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins. Proc Natl Acad Sci U S A 115, 5956–5961, doi: 10.1073/pnas.1722437115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; O-GlcNAcylation was long through to prevent protein-protein interactions, but this report shows O-GlcNAc-driven interactions for the first time, confirmed by structual biology.
- 98.Myers SA et al. SOX2 O-GlcNAcylation alters its protein-protein interactions and genomic occupancy to modulate gene expression in pluripotent cells. eLife 5, e10647, doi: 10.7554/eLife.10647 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu S-H et al. Metabolic labeling enables selective photocrosslinking of O-GlcNAc-modified proteins to their binding partners. Proceedings of the National Academy of Sciences, doi: 10.1073/pnas.1114356109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Balana AT et al. O-GlcNAc modification of small heat shock proteins enhances their anti-amyloid chaperone activity. Nature Chemistry 13, 441–450, doi: 10.1038/s41557-021-00648-8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]


