Abstract
The QUANTIPLEX HIV-1 RNA assay, version 3.0 (a branched DNA, version 3.0, assay [bDNA 3.0 assay]), was evaluated by analyzing spiked and clinical plasma samples and was compared with the AMPLICOR HIV-1 MONITOR Ultrasensitive (ultrasensitive reverse transcription-PCR [US-RT-PCR]) method. A panel of spiked plasma samples that contained 0 to 750,000 copies of human immunodeficiency virus type 1 (HIV-1) RNA per ml was tested four times in each of four laboratories (1,344 assays). Negative results (<50 copies/ml) were obtained in 30 of 32 (94%) assays with seronegative samples, 66 of 128 (52%) assays with HIV-1 RNA at 50 copies/ml, and 5 of 128 (4%) assays with HIV-1 RNA at 100 copies/ml. The assay was linear from 100 to 500,000 copies/ml. The within-run standard deviation (SD) of the log10 estimated HIV-1 RNA concentration was 0.08 at 1,000 to 500,000 copies/ml, increased below 1,000 copies/ml, and was 0.17 at 100 copies/ml. Between-run reproducibility at 100 to 500 copies/ml was <0.10 log10 in most comparisons. Interlaboratory differences across runs were ≤0.10 log10 at all concentrations examined. A subset of the panel (25 to 500 copies/ml) was also analyzed by the US-RT-PCR assay. The within-run SD varied inversely with the log10 HIV-1 RNA concentration but was higher than the SD for the bDNA 3.0 assay at all concentrations. Log-log regression analysis indicated that the two methods produced very similar estimates at 100 to 500 copies/ml. In parallel testing of clinical specimens with low HIV-1 RNA levels, 80 plasma samples with <50 copies/ml by the US-RT-PCR assay had <50 copies/ml when they were retested by the bDNA 3.0 assay. In contrast, 11 of 78 (14%) plasma samples with <50 copies/ml by the bDNA 3.0 assay had ≥50 copies/ml when they were retested by the US-RT-PCR assay (median, 86 copies/ml; range, 50 to 217 copies/ml). Estimation of bDNA 3.0 values of <50 copies/ml by extending the standard curve of the assay showed that these samples with discrepant results had higher HIV-1 RNA levels than the samples with concordant results (median, 34 versus 17 copies/ml; P = 0.0051 by the Wilcoxon two-sample test). The excellent reproducibility, broad linear range, and good sensitivity of the bDNA 3.0 assay make it a very attractive method for quantitation of HIV-1 RNA levels in plasma.
The objective of antiretroviral therapy is to maximally suppress human immunodeficiency virus (HIV) type 1 (HIV-1) replication (U.S. Department of Health and Human Services and Henry J. Kaiser Family Foundation, guidelines for the use of antiretroviral agents in HIV-infected adults and adolescents [http://www.atis.org]). In clinical practice, suppression of HIV-1 replication is considered equivalent to achieving plasma HIV-1 RNA levels below the limit of detection of current assays (∼50 HIV-1 RNA copies/ml). However, recent studies suggest that there is ongoing HIV-1 replication in the blood and lymphatic tissue compartments of individuals with undetectable HIV-1 RNA in their plasma (4, 6, 23). Therefore, monitoring of the response to antiretroviral therapy and HIV-1 pathogenesis studies require the use of sensitive methods for quantitation of HIV-1 RNA in plasma.
A variety of commercial assays are available for quantitation of HIV-1 RNA in plasma. These assays are based on different amplification and detection methods, including reverse transcription (RT) followed by PCR and colorimetric detection (RT-PCR assay; Roche Molecular Systems, Inc., Branchburg, N.J.) (14), nucleic acid sequence-based amplification and colorimetric detection (Organon Teknika Corporation, Durham, N.C.) (21), DNA hybridization and colorimetric detection (Digene assay; Digene Diagnostics, Inc., Silver Spring, Md.), and nucleic acid hybridization and branched DNA (bDNA) signal amplification (bDNA assay; Bayer Nucleic Acid Diagnostics) (15). Of these, the RT-PCR assay is the only one approved by the Food and Drug Administration and the most widely used method for quantitation of HIV-1 RNA in plasma. The most recent version of the RT-PCR assay (AMPLICOR HIV-1 MONITOR Ultrasensitive assay [US-RT-PCR assay]) has a limit of detection of 50 HIV-1 RNA copies/ml of plasma (20).
The bDNA assay has been used in pivotal studies that have established the importance of plasma HIV-1 RNA levels in predicting clinical progression and survival in HIV-infected individuals (11, 12) and defined the dynamics of HIV-1 replication in vivo (16). The limit of quantitation of the version of the bDNA assay used in those studies was 500 HIV-1 RNA copies/ml (11, 12, 16). Recently, a more sensitive version of the bDNA assay (bDNA, version 3.0, assay [bDNA 3.0 assay]) has been developed to detect HIV-1 RNA present at below 100 copies/ml. The bDNA 3.0 assay uses isoC and isoG nucleotides in the amplification molecules to reduce background signals and allow stronger amplification (3). The detection limit of this new bDNA assay is 50 copies/ml (3).
Because of the importance of assessing the performance characteristics of new assays for HIV-1 RNA detection in plasma and of comparing them with other quantitative methods in use, we conducted a multilaboratory study to evaluate the bDNA 3.0 assay by testing a panel of coded replicate samples of negative plasma spiked with HIV-1 RNA. The characteristics of the bDNA 3.0 assay analyzed included the limit of detection, the linear range, and the reproducibility of estimates of HIV-1 RNA concentrations within the linear range of the assay. In addition, the bDNA 3.0 assay was compared to the US-RT-PCR assay by testing a subset of the panel with low HIV-1 RNA concentrations (25 to 500 copies/ml). The aspects compared included the HIV-1 RNA concentrations estimated by the two assays for the same specimens, the number of estimates of each HIV-1 RNA level above or below the limit of detection of each assay, and assay reproducibility. Finally, the bDNA 3.0 and the US-RT-PCR assays were compared by testing clinical plasma samples that contained very low HIV-1 RNA levels.
MATERIALS AND METHODS
HIV-1 RNA panels.
Coded panels of uninfected human plasma to which HIV-1 RNA was added were prepared by the Virology Quality Assurance laboratory (Rush-Presbyterian-St. Luke's Medical Center, Chicago, Ill.) as described elsewhere (22). The nominal HIV-1 RNA concentrations in the panels ranged from levels that were below the limit of detection of the bDNA 3.0 assay specified by the manufacturer (50 HIV-1 RNA copies/ml) to 750,000 HIV-1 RNA copies/ml (Table 1). The number of replicates in the panel varied inversely with the nominal HIV-1 RNA concentration to ensure that the reproducibility at low HIV-1 RNA levels and the limit of detection of the bDNA 3.0 assay could be adequately assessed.
TABLE 1.
Composition of the quantitative HIV RNA panels
| Nominal concn (no. of HIV RNA copies/ml) | No. of replicates
|
|
|---|---|---|
| bDNA 3.0 assay | US-RT-PCR assay | |
| 750,000 | 2 | |
| 500,000 | 2 | |
| 250,000 | 2 | |
| 100,000 | 2 | |
| 50,000 | 3 | |
| 25,000 | 3 | |
| 10,000 | 3 | |
| 5,000 | 3 | |
| 2,500 | 3 | |
| 1,000 | 3 | |
| 500 | 8 | 3 |
| 200 | 8 | 3 |
| 100 | 8 | 3 |
| 75 | 8 | 3 |
| 50 | 8 | 4 |
| 25 | 8 | 4 |
| 12.5 | 8 | |
| 0 | 2 | |
A total of six laboratories participated in the study. All the laboratories had prior experience with molecular biology-based techniques for the detection of HIV-1 RNA in plasma and participated in the Virology Quality Assurance proficiency testing program for HIV-1 RNA quantitation for either or both assays (22). Panels were shipped to four laboratories (laboratories A, B, C, and D) for analysis by the bDNA 3.0 assay. A subset of the panel (25 to 500 HIV-1 RNA copies/ml) was analyzed by the US-RT-PCR assay in four laboratories (laboratories A, D, E, and F), including two that participated in the analysis of the panel of specimens by the bDNA 3.0 assay plus two additional laboratories (Table 1). Both panels were assayed four times on separate days in each laboratory.
Clinical specimens.
Blood samples from HIV-1-infected individuals receiving antiretroviral therapy were collected by one of the investigators (R.W.C.). Samples were obtained from two different groups of individuals: those with <50 HIV-1 RNA copies/ml of plasma, as measured by the bDNA 3.0 assay, and those with <50 HIV-1 RNA copies/ml of plasma, as measured by the US-RT-PCR assay. Plasma was separated from whole blood by centrifugation within 6 h of collection and was stored frozen at −70°C until tested.
Quantitation of HIV-1 RNA. (i) bDNA 3.0 assay.
The QUANTIPLEX HIV-1 RNA assay, version 3.0 (bDNA 3.0 assay), was performed according to the manufacturer's instructions provided with the assay kit. The limit of detection of the assay indicated by the manufacturer was 50 HIV-1 RNA copies/ml (3).
(ii) US-RT-PCR assay.
The US-RT-PCR assay was performed according to the manufacturer's instructions provided with the assay kit. A result was considered below the limit of detection of the assay when all six optical densities for an amplified sample were <0.20.
Statistical analysis.
Prior to data analysis, estimates of HIV-1 RNA concentrations were transformed to a log10 scale to stabilize variances and improve the fit to a normal distribution.
(i) Limit of detection and specificity.
The limit of detection and specificity of both assays were examined by determining the number and percentage of positive results for each expected or nominal HIV-1 RNA concentration of the panel after pooling of the data across runs and laboratories.
(ii) Linear range.
The linear range of the bDNA 3.0 assay was determined by plotting the median difference between the log10 estimated HIV-1 RNA concentration and the log10 nominal HIV-1 RNA concentrations against the log10 nominal HIV-1 RNA concentration. Under the assumption that the difference between the log10-transformed estimated and nominal RNA concentrations did not vary with the nominal concentration in the linear range, this plot would form a horizontal band within the linear range of the assay. Censored observations were set at the censoring limits before differences were calculated (50 or 500,000 copies/ml). This approach would affect the plot only if more than half of the values at a given nominal concentration were censored.
(iii) Within-run variation.
The within-run standard deviation (SD) of log10-transformed estimates at each nominal HIV-1 RNA concentration was estimated from the mean square error of a nested analysis of variance in which laboratory and run within laboratory were the predictors. For the bDNA 3.0 assay, censored observations (<50 or >500,000 copies/ml) were recalculated as uncensored by extending the standard curve of the assay. Observations at 12.5 HIV-1 RNA copies/ml were excluded from the analysis. The within-run SD was also calculated for the US-RT-PCR assay. Observations that were below the limit of detection of this assay (i.e., all six HIV-1 optical densities were <0.20) were treated as left censored at the calculated limits of detection. Model fitting took place in PROC LIFEREG of SAS to accommodate censored data. Because PROC LIFEREG does not allow nested predictors, a one-way linear model was used in which the predictor variable was a set of indicators for run and laboratory. Within-run variation for the two assays was compared by F tests. Separate tests were performed at each nominal concentration from 25 through 500 HIV-1 RNA copies/ml.
(iv) Between-run, interlaboratory, and interassay variations.
Linear regressions of the log10 estimated HIV-1 RNA concentration on the log10 nominal HIV-1 RNA concentration were used to assess variation between runs for each assay in each laboratory, followed by analysis of interlaboratory variation for each assay and by analysis of interassay variation. This was a logical progression in building a linear model to compare the two assays. For example, if no statistically significant differences were found among the four runs within each combination of assay and laboratory, then the data for the four runs could be combined into a single regression per method and laboratory in the second and third stages of the analysis. A statistically significant between-run variation would indicate that a more complicated model, with runs nested within laboratory, would be required. Similar reasoning was used for the analysis of interlaboratory differences.
The initial model for variation among runs within a laboratory allowed variation in both the slopes and intercepts of the regressions. Variation in slopes was tested first because this variation would imply that differences in estimated HIV-1 RNA concentrations among runs varied with nominal concentration. If no significant differences among slopes were identified, the intercepts were compared by using a model under which it was assumed that the regression lines were parallel (i.e., differences among runs were the same at all nominal concentrations). In these models, the difference between two intercepts is an estimate of the average difference in the estimated HIV-1 RNA concentrations between two runs. Intercepts were not compared when differences in slopes were identified. The analysis of interlaboratory variation and between-assay variation also involved initial comparisons of slopes followed by comparisons of intercepts where slopes did not differ significantly. The initial set of predictors for each stage in this process depended on the results obtained at earlier steps. For example, the analysis of interlaboratory variation took into account any differences identified among runs within laboratories. Further information is provided in the Results section.
RESULTS
Samples and assays.
The analysis of the complete panel (0 to 750,000 HIV-1 RNA copies/ml) represented a total of 1,344 bDNA 3.0 assays. The analysis of the subset of the panel (25 to 500 HIV-1 RNA copies/ml) represented a total of 320 US-RT-PCR assays. In addition, a total of 158 clinical samples were analyzed. These included a group of 78 samples that had <50 HIV-1 RNA copies/ml by the bDNA 3.0 assay and that were retested by the US-RT-PCR assay and a group of 80 samples that had <50 HIV-1 RNA copies/ml by the US-RT-PCR assay and that were subsequently retested by the bDNA 3.0 assay.
Limit of detection and specificity.
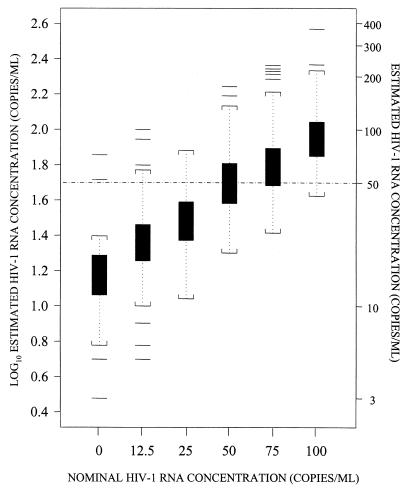
The number of samples that gave HIV-1 RNA values above or below the limit of detection for each assay is summarized in Table 2. The percentage of results below the limit of detection specified by the manufacturer was greater for the bDNA 3.0 assay than for the US-RT-PCR assay at all nominal HIV-1 RNA concentrations that were used to compare the two assays. However, this could be attributed in part to the different algorithms used by each assay to distinguish positive results from negative results. By the bDNA 3.0 assay, estimates that are <50 HIV-1 RNA copies/ml are considered de facto below the limit of detection and are censored by the software provided with the assay. By the US-RT-PCR assay, a result is considered below the limit of detection when all six optical densities from the serial dilutions of the amplified sample are <0.20. Thus, it is possible to obtain a positive result for a sample with <50 HIV-1 RNA copies/ml by the US-RT-PCR assay but not the bDNA 3.0 assay. For instance, at a nominal concentration of 50 HIV-1 RNA copies/ml, 95% of the samples had positive US-RT-PCR results and 48% of the samples had positive bDNA 3.0 assay results (Table 2). However, 21 (34%) of the 61 samples with positive results by the US-RT-PCR assay at a nominal concentration of 50 HIV-1 RNA copies/ml actually had <50 copies/ml. If the US-RT-PCR results are censored at 50 HIV-1 RNA copies/ml (as in the bDNA 3.0 assay), then the proportion of positive results in the US-RT-PCR assay at a nominal concentration of 50 HIV-1 RNA copies/ml would have been 63%; this is closer to the proportion of positive results obtained by the bDNA 3.0 assay for this HIV-1 RNA concentration (Table 2). Therefore, to investigate the effect of the censoring of bDNA 3.0 results at 50 HIV-1 RNA copies/ml, we determined HIV-1 RNA concentrations in samples with censored values by extrapolating the standard curve below the assigned limit of detection of the assay. Although this results in very imprecise results, as evidenced from the wide confidence intervals of the estimated HIV-1 RNA concentrations (Fig. 1), this extrapolation was necessary for comparison of the abilities of the two assays to detect and quantitate HIV-1 RNA in clinical plasma samples, as described below.
TABLE 2.
HIV-1 RNA measurements above or below the limit of detection of each assay
| Nominal concn (no. of HIV-1 RNA copies/ml) | No. (%) of samples
|
||||
|---|---|---|---|---|---|
| bDNA 3.0 assay with the following no. of HIV-1 RNA copies/ml:
|
US-RT-PCR assay
|
||||
| <50a | ≥50, ≤500,000 | >500,000b | <LODc | ≥LOD | |
| 0 | 30 (94) | 2 | 0 | ||
| 12.5 | 122 (95) | 6 | 0 | ||
| 25 | 120 (94) | 8 | 0 | 22 (34) | 42 |
| 50 | 66 (52) | 62 | 0 | 3 (5) | 61d |
| 75 | 38 (30) | 90 | 0 | 1 (2) | 47 |
| 100 | 5 (4) | 123 | 0 | 0 | 48 |
| 200–500 | 0 | 256 | 0 | 0 | 95e |
| 1,000–250,000 | 0 | 352 | 0 | ||
| 500,000 | 0 | 24 | 8 (25) | ||
| 750,000 | 0 | 4 | 28 (88) | ||
Results of <50 HIV-1 RNA copies/ml are reported as “<50” by the bDNA 3.0 assay software; no estimate of HIV-1 RNA concentration is provided.
Results of >500,000 HIV-1 RNA copies/ml are reported as “>500,000” by the bDNA 3.0 assay software; no estimate of RNA concentration is provided.
LOD, limit of detection of the assay: all six optical densities for an amplified sample are <0.2.
One sample (34%) had a measurement that was greater than the limit of detection but had <50 HIV-1 copies/ml.
One sample with an invalid measurement was excluded.
FIG. 1.
Box and whiskers plots of log10 estimated HIV-1 RNA concentration at nominal HIV-1 RNA concentrations up to 100 HIV-1 RNA copies/ml after recalculation of censored observations by using the standard curve for the bDNA 3.0 assay. Boxes extend from the 25th percentile (lower edge) to the 75th percentile (upper edge) HIV-1 RNA levels. The horizontal lines in the middle of the boxes indicate median HIV-1 RNA values. Whiskers are the vertical lines that end in brackets above and below the boxes. The horizontal lines beyond the brackets represent outliers (more than 1.5 times the interquartile range below the 25th percentile or above the 75th percentile). The brackets represent the largest and smallest observed values that are not outliers.
To assess the specificity of the bDNA 3.0 assay, HIV-1-negative plasma not spiked with HIV-1 RNA was included in the panel. Two (6.2%) of the 32 HIV-1-negative replicates were positive by the bDNA 3.0 assay (i.e., 51 and 71 HIV-1 RNA copies/ml, respectively). HIV-1-negative replicates were not included in the subset of the panel that was analyzed by the US-RT-PCR assay.
The limit of detection was defined as the concentration at which 95% of results are positive. Under the existing assay algorithms, the limits of detection were approximately 100 HIV-1 RNA copies/ml for the bDNA 3.0 assay and approximately 50 HIV-1 RNA copies/ml for the US-RT-PCR assay (Table 2). This difference could be attributed in part to the algorithms used by the two assays to distinguish positive results from negative results.
Linear range of bDNA 3.0 assay.
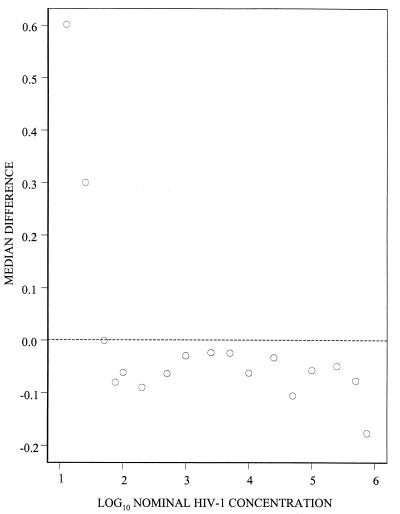
To determine the linear range of the bDNA 3.0 assay, the median difference between log10-transformed estimated and nominal HIV-1 RNA concentrations was plotted against the log10 nominal HIV-1 RNA concentration (Fig. 2). The plot forms the expected horizontal band over nominal HIV-1 RNA concentrations of 75 to 500,000 copies/ml. Setting the censored estimates at the censoring points before calculation of the differences biased the median differences upward at 12.5, 25, and 50 HIV-1 RNA copies/ml and downward at 750,000 HIV-1 RNA copies/ml.
FIG. 2.
Median difference between log10 estimated HIV-1 RNA concentrations and log10 nominal HIV-1 concentrations from the bDNA 3.0 assay plotted against log10 nominal HIV-1 RNA concentrations. Censored observations were set at the censoring point of the assay (50 or 500,000 HIV-1 RNA copies/ml).
Within-run variation for bDNA 3.0 and US-RT-PCR assays.
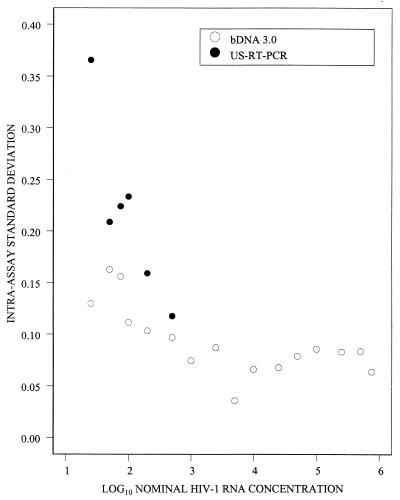
The within-run SD of the log10 estimated HIV-1 RNA concentration for the bDNA 3.0 assay varied inversely with log10 nominal HIV-1 RNA concentrations below 1,000 copies/ml but did not vary systematically at higher nominal concentrations (Fig. 3). Specifically, the within-run SD was 0.17 at 50 copies/ml and fluctuated between 0.05 and 0.09 at 1,000 to 500,000 copies/ml. The within-run SD for the US-RT-PCR assay also varied inversely with the log10 HIV-1 RNA nominal concentration and was higher than the SD for the bDNA 3.0 assay at all concentrations that were used to compare the two assays (25 to 500 HIV-1 RNA copies/ml). For example, the within-run SDs at 500 copies/ml were 0.12 for the US-RT-PCR assay and 0.10 for the bDNA 3.0 assay; at 100 copies/ml, the SDs were 0.22 and 0.11, respectively (Fig. 3). These differences were statistically significant at all nominal HIV-1 RNA concentrations except 500 copies/ml (P = 0.075).
FIG. 3.
Intra-assay standard deviation of log10 estimated HIV-1 RNA concentrations versus log10 nominal HIV-1 RNA concentrations.
Between-run variation.
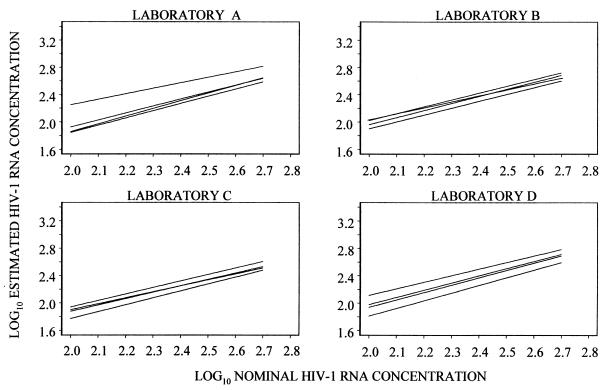
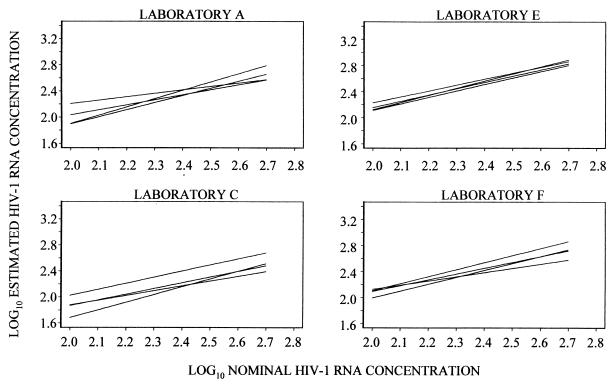
Given the results of the analysis of the limits of detection of the bDNA 3.0 assay and our goal of comparing it to the US-RT-PCR assay, the analysis of between-run variation was limited to results obtained for nominal HIV-1 RNA concentrations between 100 and 500 copies/ml. Fitted regression lines over this HIV-1 RNA copy range are plotted in Fig. 4 and 5. No significant differences among slopes were identified in any laboratory for either assay, indicating that there was no detectable variation with nominal HIV-1 RNA concentration in the differences among runs in any of the laboratories.
FIG. 4.
Regressions of log10 estimated HIV-1 RNA concentrations on log10 nominal HIV-1 RNA concentrations for the bDNA 3.0 assay, using nominal concentrations of 100 to 500 HIV-1 RNA copies/ml.
FIG. 5.
Regressions of log10 estimated HIV-1 RNA concentrations on log10 nominal HIV-1 RNA concentrations for the US-RT-PCR assay, using nominal concentrations of 100 to 500 HIV-1 RNA copies/ml.
Differences among intercepts were found at each of the four laboratories in which the bDNA 3.0 assay was used. The range of the intercepts within each laboratory (maximum intercept minus minimum intercept) provides a measure of the spread of results in each set of four runs. The largest spread was 0.39 log10 in laboratory A. However, three of the four intercepts in this laboratory differed by only 0.068 log10 or less, implying that the average difference between the results from any two of the three runs was <17%. In laboratory D, the range of the four intercepts was 0.25 log10, but three of the four differed by no more than 0.15 log10; i.e., the average difference between estimates from any of the three runs was <41%. Differences among the four regressions at each of the other two laboratories were <0.15 log10 (Fig. 4).
Statistically significant differences among the intercepts for the four regressions for the US-RT-PCR assay were found in only one laboratory (P = 0.0356). The maximum spread of intercepts in this laboratory was 0.258 log10. However, three of four regressions differed by ≤0.08 log10; i.e., the average difference between estimates was <20%. Ranges of <0.125 log10 (<33%) were obtained in the other three laboratories (Fig. 5).
Interlaboratory variation.
The analysis of interlaboratory variation was limited to nominal HIV-1 RNA concentrations of 100 to 500 copies/ml. As noted earlier, the selection of predictors for the analysis of interlaboratory variation was based on the results of the analysis of between-run variation. Thus, under the initial models for interlaboratory variation, it was assumed that the regressions for each laboratory formed four parallel lines with different intercepts. The model also included terms to test for differences among the slopes and the elevations of the regressions in the four laboratories. However, no differences in regression slopes among laboratories were identified (for the concentration-run interaction, bDNA 3.0 assay, P = 0.20; US-RT-PCR assay, P = 0.20; for the concentration-laboratory interaction, bDNA 3.0 assay, P = 0.24; US-RT-PCR assay, P = 0.24). Therefore, the model for the results from each assay was reduced to a set of 16 parallel regression lines. Terms for variation among intercepts within laboratories and for variation in average intercepts among laboratories were still included.
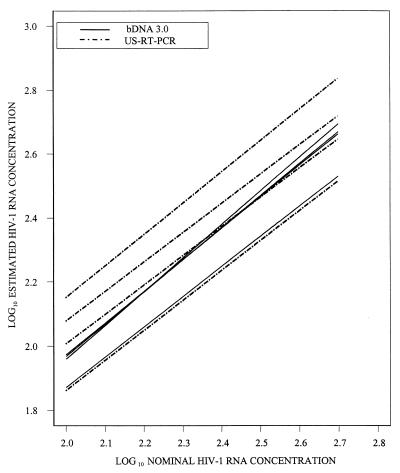
No statistically significant differences in intercepts among laboratories were identified for the bDNA 3.0 assay (P = 0.264). The small magnitude of the interlaboratory differences for this assay can be illustrated by comparing regressions among laboratories after combining data across runs into a single regression line for each laboratory (Fig. 6). Three of these regression lines are nearly superimposable, and the fourth differs from the other three by less than 0.10 log10; i.e., the average difference between estimates from the fourth laboratory and the other three was <26%. In contrast, statistically significant differences among laboratories were found for the US-RT-PCR assay (P < 0.0001). The four regression lines that were formed by combining data across runs within each laboratory were roughly evenly spaced (Fig. 6). The intercepts for the uppermost and lowermost lines differed by approximately 0.30 log10, implying a twofold average difference between estimates in these two laboratories. This was considerably larger than the differences observed by the bDNA 3.0 assay.
FIG. 6.
Regressions of log10 estimated HIV-1 RNA concentrations on log10 nominal HIV-1 RNA concentrations for data combined across runs within each laboratory.
Interassay variation.
The analysis of interassay variation was also limited to nominal HIV-1 RNA concentrations of 100 to 500 copies/ml. Given the results presented above, the regression lines for the 16 runs by each assay were assumed to be parallel for the comparison of data between assays. The initial model did allow for differences in slopes and average intercepts between assays. However, the slopes of regressions from the two assays did not differ significantly (P = 0.16). Thus, there was no detectable variation with nominal HIV-1 RNA concentration in the estimates from the two assays, so the model was reduced to a set of 32 parallel lines. The intercepts for the regressions from the bDNA 3.0 assay were, on average, 0.28 log10 lower than for the regressions for the US-RT-PCR assay (P <0.0001). However, two of the four regression lines for the US-RT-PCR assay were very similar to the regression lines for the bDNA 3.0 assay (Fig. 6). Thus, the estimates from two of the laboratories in which the US-RT-PCR assay was used were very close to the estimates for the bDNA 3.0 assay.
Clinical specimens.
Eleven (14%) of the 78 clinical samples that contained <50 HIV-1 RNA copies/ml by the bDNA 3.0 assay had >50 HIV-1 RNA copies/ml when they were subsequently tested by the US-RT-PCR assay (median, 86 HIV-1 RNA copies/ml; 25th to 75th percentiles, 74 to 120 HIV-1 RNA copies/ml; range, 50 to 217 HIV-1 RNA copies/ml). In contrast, none of the 80 plasma samples with <50 HIV-1 RNA copies/ml by the US-RT-PCR assay had >50 HIV-1 RNA copies/ml when they were retested by the bDNA 3.0 assay (Table 3). To investigate this further, we ignored the specified censoring value of 50 HIV-1 RNA copies/ml for the bDNA 3.0 assay and used the luminescence values to determine the HIV-1 RNA concentrations for all specimens (Table 3). The HIV-1 RNA levels by the bDNA 3.0 assay were significantly lower for samples that contained <50 HIV-1 RNA copies/ml when they were retested by the US-RT-PCR assay compared to those for samples that contained >50 HIV-1 RNA copies/ml when they were retested by the US-RT-PCR assay (P = 0.0051 by the Wilcoxon two-sample test). Similarly, the 80 plasma samples with <50 HIV-1 RNA copies/ml when they were initially analyzed by the US-RT-PCR assay and retested by the bDNA 3.0 assay had low HIV-1 RNA levels when their luminescence values were extrapolated below the censoring value of the bDNA 3.0 assay (Table 3). Overall, the HIV-1 RNA concentrations measured by the bDNA 3.0 assay were higher in the 78 samples that were selected for retesting on the basis of having <50 HIV-1 RNA copies/ml by the bDNA 3.0 assay than in the 80 plasma samples that were selected for retesting on the basis of having <50 HIV-1 RNA copies/ml by the US-RT-PCR assay. However, as indicated earlier, quantitation of bDNA 3.0 assay results by extrapolation of the standard curve for the bDNA 3.0 assay to below 50 HIV-1 RNA copies/ml is very imprecise (Fig. 1).
TABLE 3.
HIV-1 RNA results obtained by the bDNA 3.0 assay for clinical samples after recalculating censored observations
| Initial assaya and subsequent assay, result | No. of specimens | bDNA 3.0 assay result (no. of HIV-1 RNA copies/ml)
|
||
|---|---|---|---|---|
| Median | 25th to 75th percentiles | Range | ||
| bDNA 3.0 assay and US-RT-PCR assay | ||||
| ≥50 copies/mlb | 11 | 34 | 18–36 | 15–49 |
| <50 copies/mlc | 67 | 17 | 11–25 | 2–50 |
| US-RT-PCR assay and bDNA 3.0 assay | ||||
| ≥50 copies/ml | 0 | |||
| <50 copies/mld | 80 | 3 | 1–9 | <1–40 |
All results were <50 HIV-1 RNA copies/ml.
Median, 86 HIV-1 RNA copies/ml; 25th to 75th percentiles, 74 to 120 HIV-1 RNA copies/ml; range, 50 to 217 HIV-1 RNA copies/ml.
Six (8.9%) of the 67 samples were positive by the US-RT-PCR assay (median, 31.5 HIV-1 RNA copies/ml).
Twelve (15%) of the 80 samples were positive by the US-RT-PCR assay (median, 32 HIV-1 RNA copies/ml).
DISCUSSION
The limit of detection of the bDNA 3.0 assay for plasma HIV-1 RNA in this study was higher than the limit of detection of the US-RT-PCR assay found in this and other studies (20). For each of the two assays, we defined the detection limit as the HIV-1 RNA concentration that gave positive results in at least 95% of replicate reactions. As such, the limit of detection for the bDNA 3.0 assay was 100 HIV-1 RNA copies/ml and that for the US-RT-PCR assay was 50 HIV-1 RNA copies/ml. This twofold difference in sensitivity arises, in part, from the different algorithms used by each assay to distinguish positive results from negative results. By the bDNA 3.0 assay, results of <50 HIV-1 RNA copies/ml are always considered negative. In contrast, results of <50 HIV-1 RNA copies/ml could be positive by the US-RT-PCR assay. If the US-RT-PCR assay is censored like the bDNA 3.0 assay at 50 HIV-1 RNA copies/ml, then the proportion of positive results at this concentration in our study would be 62%, moving the limit of detection of the US-RT-PCR assay to between 50 and 75 HIV-1 RNA copies/ml (Table 2). However, the assignment of meaningful detection limits must consider the within-assay precision (usually represented by the SD for replicate samples) in addition to the assay design (i.e., internal standard versus external standards) that are used to estimate the viral RNA copy number.
The clinical importance of using assays for the detection of HIV-1 RNA in plasma capable of measuring HIV-1 RNA levels below 100 copies/ml is not clear at present. Current guidelines for the treatment of patients with HIV-1 infection recommend the use of combination antiretroviral therapy to decrease plasma HIV-1 RNA levels to below 50 copies/ml (2). In clinical trials, suppression of plasma HIV-1 RNA levels below 20 copies/ml has been associated with a longer response to antiretroviral therapy compared with that achieved when viral suppression is below 500 HIV-1 RNA copies/ml (17). In addition, recent studies have shown that there is ongoing HIV-1 replication in the blood and lymphoid tissues of patients with undetectable plasma HIV-1 RNA levels during potent antiretroviral therapy (4, 6, 23). Therefore, the ability to measure very low concentrations of HIV-1 RNA in plasma is of interest. However, the clinical benefit of achieving plasma HIV-1 RNA levels of <50 copies/ml with antiretroviral therapy and the clinical consequences of ongoing viral replication in patients with suppressed plasma viremia are not known at present. In some studies, potent antiretroviral therapy has been associated with clinical and immunologic responses even when suppression of plasma HIV-1 RNA levels has not been achieved (8, 9, 13). Whether a clinical difference exists between suppression of plasma HIV-1 RNA levels below 100 or below 50 copies/ml requires carefully designed prospective clinical trials. Results from those trials would determine the advantage of using assays capable of detecting and quantitating with good precision HIV-1 RNA levels in plasma below 100 copies/ml.
In our study, 6.2% of HIV-1-negative replicates in the panel had ≥50 HIV-1 RNA copies/ml when they were analyzed by the bDNA 3.0 assay. False-positive results by this version of the assay have been reported in a study in which the bDNA 3.0 assay was used to detect HIV-1 RNA in plasma samples from individuals with sexual or parenteral exposure to HIV-1 (M. E. Roland, J. N. Martin, D. Chernoff, B. McGovern, J. Bamberger, M. M. Katz, T. J. Coates, and J. O. Kahn, 6th Conf. Retrovirus Opportunistic Infections, 1999). Among those who did not acquire HIV-1 infection, the false-positivity rate of the bDNA 3.0 assay ranged from 9.1% (4 weeks after exposure) to 27.6% (26 weeks after exposure) (Roland et al., 6th Conf. Retrovirus Opportunistic Infections). Another study reported two cases of false-positive results when the bDNA assay was used to diagnose HIV-1 infection (18). However, the version of the bDNA assay used in that study was not specified. Of note is that the HIV-1 RNA levels in samples with false-positive results in that study were low (1,254 and 1,574 HIV-1 RNA copies/ml, respectively) (18). False-positive results have also been reported by the US-RT-PCR assay when it was used for the diagnosis of HIV-1 infection (1, 18). When interpreting these results, it is important to consider that assays for the quantitation of HIV-1 RNA in plasma have not been developed for the diagnosis of HIV-1 infection and that their specificities have not been established when the assays are applied to individuals who are negative for HIV-1 antibodies.
In our study, the linear range for the bDNA 3.0 assay was 100 to 500,000 HIV-1 RNA copies/ml. This represents a 10-fold increase compared to the linear range of the US-RT-PCR assay (50 to 75,000 HIV-1 RNA copies/ml) (5, 20). This larger linear range would likely reduce the need to dilute and retest plasma samples that contain high HIV-1 RNA levels when these samples are analyzed by the bDNA 3.0 assay. This is of practical importance for diagnostic and research laboratories that use assays to quantitate HIV-1 RNA in plasma, particularly the HIV-1 RNA levels in the plasma of pediatric patients with HIV-1 infection, in whom HIV-1 RNA levels are generally much greater than those in HIV-1-infected adults (10, 19).
We found that the within-run variations for both assays varied inversely with the concentration of HIV-1 RNA, although the magnitude of this variation was smaller for the bDNA 3.0 assay than for the US-RT-PCR assay. The within-run variation that we found in our study for the US-RT-PCR assay was higher than that reported by others. For instance, the mean within-run SD of the log10 number of HIV-1 RNA copies per milliliter in one study of plasma samples with low HIV-1 RNA concentrations (30 to 500 HIV-1 RNA copies/ml) was 0.09 (5). In contrast, the SD of the log10 number of HIV-1 RNA copies per milliliter in our study for similar concentrations among four different laboratories ranged from 0.12 to 0.22.
The between-run variations differed significantly among all four laboratories that used the bDNA 3.0 assay but in only one of the four laboratories that used the US-RT-PCR assay in our study. However, plots of the fitted regression lines indicate that the differences among runs within laboratories were roughly the same for the two assays (Fig. 4 and 5). Because the within-run variation of the bDNA 3.0 assay was lower, the statistical power to detect differences among runs was greater for this assay than for the US-RT-PCR assay. In a previous study that analyzed the precision of the US-RT-PCR assay, the largest source of variation was due to within-run differences between replicates; the between-run variation of the US-RT-PCR assay in that study was smaller than the within-run variation (20). In our study, the interlaboratory variation was greater for the US-RT-PCR assay than for the bDNA 3.0 assay.
The correlation and agreement between the bDNA 3.0 assay and the US-RT-PCR assay have been analyzed in a recent study of 318 plasma samples from 59 randomly selected HIV-1-infected individuals (7). The HIV-1 RNA concentrations in the samples studied ranged from 2.0 to 5.5 log10 copies/ml. That study showed that the bDNA 3.0 and the US-RT-PCR assays were highly correlated (r = 0.98) and had good agreement (mean difference in log10 copies per milliliter ± 2 SD = 0.041 ± 0.176) and that the log10 values obtained by the US-RT-PCR assay were, on average, 1.05-fold higher than the log10 values obtained by the bDNA 3.0 assay (7). Similarly, the HIV-1 RNA levels obtained by the US-RT-PCR assay in our study were on average twofold higher than the levels obtained by the bDNA 3.0 assay for the same sample.
Results of our comparison of the two HIV-1 RNA assays by parallel testing of clinical plasma samples have practical importance for clinical trials and the management of patients on the basis of the HIV-1 RNA levels in plasma. For instance, if management decisions incorporate the categorical detection of HIV-1 RNA at the limit of detection of the assay, then the management decisions will depend on the assay used. For example, we found that 14% of clinical specimens that had <50 HIV-1 RNA copies when they were analyzed by the bDNA 3.0 assay had detectable levels when they were retested by the US-RT-PCR assay. However, this apparent discordance was explained by the difference between the two assays' censoring levels and precision, revealing the low HIV-1 RNA copy number in the specimens once the bDNA 3.0 assay detection parameters were extrapolated below the cutoff of 50 HIV-1 RNA copies/ml. Insufficient specimen volume was available for retesting of these clinical specimens with discordant results by using the bDNA 3.0 assay; however, on the basis of the 95% confidence interval of the assay around the cutoff of 50 HIV-1 RNA copies/ml (Fig. 1), some of these specimens would be expected to give a value of ≥50 HIV-1 RNA copies/ml when retested by the bDNA 3.0 assay. In contrast, all of the 80 specimens with <50 HIV-1 RNA copies/ml when they were initially analyzed by the US-RT-PCR assay also had <50 HIV-1 RNA copies/ml when they were retested by the bDNA 3.0 assay. Taken together, these data suggest that clinical specimens that repeatedly have <50 HIV-1 RNA copies/ml by either assay are probably comparable with regard to the 50-copy/ml detection limit. Nevertheless, our study confirms previous recommendations that the same assay used for the detection of HIV-1 RNA levels in plasma be used to monitor infected individuals (Guidelines for the use of antiretroviral agents in HIV-infected adults and adolescents, U.S. Department of Health and Human Services and Henry J. Kaiser Family Foundation [http://www.atis.org]).
In summary, the bDNA 3.0 assay has excellent reproducibility, a broad linear range, and good sensitivity for the quantitation of HIV-1 RNA in plasma down to 100 copies/ml. Plasma specimens with HIV-1 RNA concentrations that are below a defined detection limit of 50 copies/ml by both the bDNA 3.0 and the US-RT-PCR assays have comparable low levels of HIV-1 RNA for the purpose of using 50 copies/ml as a cutoff value for assessment of viral suppression in clinical trials and in clinical practice. The characteristics of the bDNA 3.0 assay make it a very attractive method for the quantitation of HIV-1 RNA levels in the plasma of HIV-1-infected persons. Because of the potential for false-positive results when plasma samples from non-HIV-infected individuals are tested, assays for HIV-1 RNA detection should not be used for the diagnosis of HIV-1 infection without additional confirmatory clinical and laboratory data.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI 27664 (to R.W.C.) and AI 30731 (to R.W.C.), AACTG Virology Advanced Laboratory contract 96VC010 (to J.B.J.), and the Virology Advanced Technology Laboratory subcontract of AI-2776 (to A.E.). The HIV-1 RNA detection kits used in this study were provided by Bayer Nucleic Acid Diagnostics.
We are indebted to the following individuals for their technical assistance and expertise: at the University of Minnesota; Wuyi Li; at the University of Washington, Joan Dragavon, Reggie Sampoleo, and Michelle Jack; at the Cleveland Clinic Foundation, Colleen Starkey; at Johns Hopkins University, Estelle Piwowar-Manning, Antoine Simmons, and Andy Korpela; and at the AIDS Clinical Trials Group Operations Center, Holly Schneider.
REFERENCES
- 1.Brown A E, Jackson B, Fuller S A, Sheffield J, Cannon M A, Lane J R. Viral RNA in the resolution of human immunodeficiency virus type 1 diagnostic serology. Transfusion. 1997;37:926–929. doi: 10.1046/j.1537-2995.1997.37997454019.x. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter C C J, Cooper D A, Fischl M A, Gatell J M, Gazzard B G, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S G, Richman D D, Saag M S, Schechter M, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A. Antiretroviral therapy in adults. Updated recommendations of the International AIDS Society-USA panel. JAMA. 2000;283:381–390. doi: 10.1001/jama.283.3.381. [DOI] [PubMed] [Google Scholar]
- 3.Collins M L, Irvine B, Tyner D, Fine E, Zayati C, Chang C-A, Horn T, Ahle D, Detmer J, Shen L-P, Kolberg J, Bushnell S, Urdea M S, Ho D D. A branched DNA signal amplification assay for quantitation of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dornadula G, Zhang H, VanUitert B, Stern J, Livornese L, Ingerman M J, Witek J, Kedanis R J, Natkin J, DeSimone J, Pomerantz R J. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA. 1999;282:1627–1632. doi: 10.1001/jama.282.17.1627. [DOI] [PubMed] [Google Scholar]
- 5.Erali M, Hillyard D R. Evaluation of the ultrasensitive Roche Amplicor HIV-1 Monitor assay for quantitation of human immunodeficiency virus type 1 RNA. J Clin Microbiol. 1999;37:792–795. doi: 10.1128/jcm.37.3.792-795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furtado M R, Callaway D S, Phair J P, Kunstman K J, Stanton J L, Macken C A, Perelson A S, Wolinsky S M. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N Engl J Med. 1999;340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 7.Highbarger H C, Alvord W G, Jiang M K, Shah A S, Metcalf J A, Lane H C, Dewar R L. Comparison of the Quantiplex version 3.0 assay and a sensitized Amplicor Monitor assay for measurement of human immunodeficiency virus type 1 RNA levels in plasma samples. J Clin Microbiol. 1999;37:3612–3614. doi: 10.1128/jcm.37.11.3612-3614.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaufmann D, Pantaleo G, Sudre P, Telenti A. CD4-cell count in HIV-1-infected individuals remaining viraemic with highly active antiretroviral therapy (HAART). Swiss HIV Cohort Study. Lancet. 1998;351:723–724. doi: 10.1016/s0140-6736(98)24010-4. [DOI] [PubMed] [Google Scholar]
- 9.Lederberger B, Egger M, Opravil M, Telenti A, Hirschel B, Battegay M, Vernazza P, Sudre P, Flepp M, Furrer H, Francioli P, Weber R. Clinical progression and virologic failure on highly active antiretroviral therapy in HIV-1 patients: a prospective cohort study. Swiss HIV Cohort Study. Lancet. 1999;353:863–868. doi: 10.1016/s0140-6736(99)01122-8. [DOI] [PubMed] [Google Scholar]
- 10.McSherry G D, Shapiro D E, Coombs R W, McGrath N, Frenkel L M, Britto P, Culnane M, Sperling R S. The effects of zidovudine in the subset of infants infected with human immunodeficiency virus type-1 (Pediatric AIDS Clinical Trials Group Protocol 076) J Pediatr. 1999;134:717–724. doi: 10.1016/s0022-3476(99)70287-8. [DOI] [PubMed] [Google Scholar]
- 11.Mellors J W, Muñoz A, Giorgi J V, Margolick J B, Tassoni C J, Gupta P, Kingsley L A, Todd J A, Saah A J, Detels R, Phair J P, Rinaldo C R. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 12.Mellors J W, Rinaldo C R, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 13.Mezzaroma I, Carlesimo M, Pinter E, Muratori D S, Di Sora F, Chiarotti F, Cunsolo M G, Sacco G, Aiuti F. Clinical and immunologic response without decrease in virus load in patients with AIDS after 24 months of highly active antiretroviral therapy. Clin Infect Dis. 1999;29:1423–1430. doi: 10.1086/313520. [DOI] [PubMed] [Google Scholar]
- 14.Mulder J, Resnick R, Saget B, Sceibel S, Herman S, Payne H, Harrigan R, Kwok S. A rapid and simple method for extracting human immunodeficiency virus type 1 RNA from plasma: enhanced sensitivity. J Clin Microbiol. 1997;35:1278–1280. doi: 10.1128/jcm.35.5.1278-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pachl C, Todd J A, Kern D G, Sheridan P J, Fong S F, Stempien M, Hoo B, Besemer D, Yeghiazarian T, Irvine B, Kolberg J, Kokka R, Neuwald P, Urdea M S. Rapid and precise quantification of HIV-1 RNA in plasma using a branched DNA (bDNA) signal amplification assay. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;8:446–454. doi: 10.1097/00042560-199504120-00003. [DOI] [PubMed] [Google Scholar]
- 16.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 17.Raboud J M, Montaner J S G, Conway B, Rae S, Reiss P, Vella S, Cooper D, Lange J, Harris M, Wainberg M, Robinson P, Myers M, Hall D. Suppression of plasma viral load below 20 copies/ml is required to achieve long-term response to therapy. AIDS. 1998;12:1619–1624. doi: 10.1097/00002030-199813000-00008. [DOI] [PubMed] [Google Scholar]
- 18.Rich J D, Merriman N A, Mylonakis E, Greenough T C, Flanifan T, Mady B J, Carpenter C C J. Misdiagnosis of HIV infection by HIV-1 plasma viral load testing: a case series. Ann Intern Med. 1999;130:37–39. doi: 10.7326/0003-4819-130-1-199901050-00007. [DOI] [PubMed] [Google Scholar]
- 19.Shearer W T, Quinn T C, LaRussa P, Lew J F, Mofenson L, Almy S, Rich K, Handelsman E, Diaz C, Pagano M, Smeriglio V, Kalish L A the Women and Infants Transmission Study Group. Viral load and disease progression in infants infected with human immunodeficiency virus type 1. N Engl J Med. 1997;336:1337–1342. doi: 10.1056/NEJM199705083361901. [DOI] [PubMed] [Google Scholar]
- 20.Sun R, Ku J, Jayakar H, Kuo J-C, Brambilla D, Herman S, Rosenstraus M, Spadoro J. Ultrasensitive reverse transcription-PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:2964–2969. doi: 10.1128/jcm.36.10.2964-2969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Gemen B, Kievits T, Schukkink R, van Strijp D, Malek L T, Sooknanan R, Huisman H G, Lens P. Quantification of HIV-1 RNA in plasma using NASBA during HIV-1 primary infection. J Virol Methods. 1993;43:177–188. doi: 10.1016/0166-0934(93)90075-3. [DOI] [PubMed] [Google Scholar]
- 22.Yen-Lieberman B, Brambilla D, Jackson B, Bremer J, Coombs R, Cronin M, Herman S, Katzestein D, Leung S, Ju Lin H, Palumbo P, Rasheed S, Todd J, Vahey M, Reichelderfer P. Evaluation of a quality assurance program for quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trials Group virology laboratories. J Clin Microbiol. 1996;34:2695–2701. doi: 10.1128/jcm.34.11.2695-2701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Ramratnam B, Tenner-Racz C, He Y, Vesanen M, Lewin S, Talal A, Racz P, Perelson A S, Korber B T, Markowitz M, Ho D D. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N Engl J Med. 1999;340:1605–1613. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]