Abstract
A yeast two-hybrid assay has identified an androgen-dependent interaction of androgen receptor (AR) with amino-terminal enhancer of split (AES), a member of the highly conserved Groucho/TLE family of corepressors. Full-length AR, as well as the N-terminal fragment of AR, showed direct interactions with AES in in vitro protein-protein interaction assays. AES specifically inhibited AR-mediated transcription in a well-defined cell-free transcription system and interacted specifically with the basal transcription factor (TFIIE) in HeLa nuclear extract. These observations implicate AES as a selective repressor of ligand-dependent AR-mediated transcription that acts by directly interacting with AR and by targeting the basal transcription machinery.
Androgen receptor (AR) is a member of the superfamily of ligand-inducible transcription factors and mediates the biological actions of androgens (19). Like other superfamily members, AR contains a central DNA-binding domain, a C-terminal ligand-binding domain with an associated AF-2 activation domain, and a large N-terminal region containing the AF-1 activation domain (4, 26). Nuclear receptors regulate the transcription of their target genes through the agency of various coactivators and corepressors that are recruited to target genes through interactions with promoter-bound receptors (56). Many of the known coactivators for nuclear receptors contain histone acetyltransferase activities and are thought to act mainly through targeted chromatin structural perturbations that facilitate the subsequent recruitment (to the promoter) and function of other transcriptional coactivators and basal transcriptional factors (3). Transcriptional corepressors, by contrast, mediate repression by various nuclear receptors. Some nuclear receptors (including retinoid receptor, thyroid hormone receptor, vitamin D receptor, and certain orphan receptors) that are not associated with heat shock proteins in their unliganded state repress transcription by recruitment of corepressor complexes (15, 35). Corepressor complexes contain histone deacetylase (HDAC) activities that maintain chromatin in a configuration that excludes functional interactions of the general transcriptional machinery with the promoter. In contrast, unliganded steroid receptors (including AR) generally associate with heat shock proteins and, upon ligand binding, dissociate from the heat shock proteins, translocate to the nucleus, and associate with coactivators to activate or repress target genes (30).
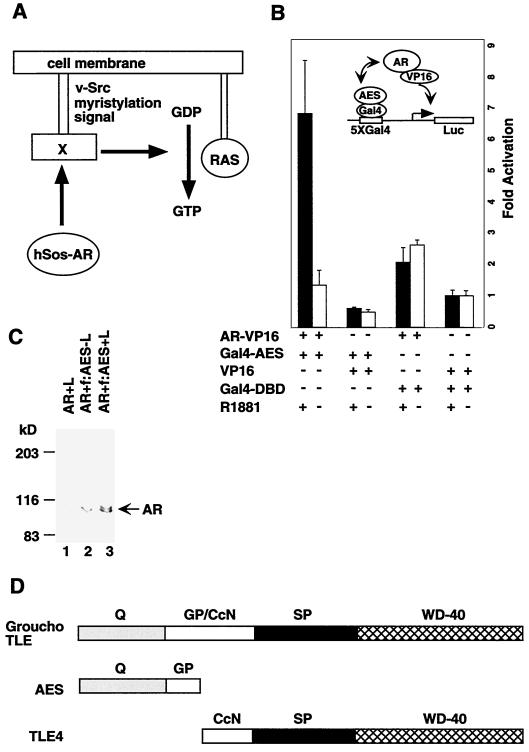
Another type of corepressor, implicated in the function of other types of repressors, is the Groucho/TLE family (see Fig. 1D) (5, 11). The larger family members such as Drosophila Groucho and its mammalian homologues, the TLE proteins (transducin-like enhancer of split [TLE1-3]), share five domains. A carboxyl-terminal WD-40 repeat domain (WD-40) and an amino-terminal glutamine-rich domain (Q) are highly conserved. In the much less well-conserved central region, there is a loosely conserved CcN motif (CcN), consisting of putative cdc2 kinase and casein kinase II phosphorylation sites, and two poorly conserved regions (GP and SP) that are characteristically rich in either glycine and proline (GP) or serine and proline (SP) residues. A shorter family member, human TLE4, is similar except for the absence of the amino-terminal Q and GP domains. The shortest family member, amino-terminal enhancer of split (AES), shares only the first two regions of the amino terminus.
FIG. 1.
AR interacts with AES in vivo. (A) Schematic diagram of the Ras signaling pathway utilized in the yeast two-hybrid system. (B) Mammalian two-hybrid assay with Ga14-AES and AR-VP16 fusion proteins in 293T cells. 293T cells were cotransfected with 1 μg of either AR-VP16, Ga14-AES, VP16, or Ga14-DBD in the presence or absence of R1881 (100 nM), along with 100 ng of pG5-Luc reporter plasmid. A significant interaction was detected only between AR and AES. (C) AR was coimmunoprecipitated with AES. 293T cells were transfected with AR (lane 1) or AR and FLAG-tagged AES (lanes 2 and 3) in the presence (lanes 1 and 3) or absence (lane 2) of R1881 (100 nM). Whole-cell extracts were made from the transfected cells and incubated with M2 agarose beads. The immunoprecipitated proteins were analyzed by Western blotting with an anti-AR antibody. (D) Domain structures of three forms of the Groucho/TLE family proteins.
The Q domain mediates both homo- and hetero-oligomerization between Groucho/TLE family proteins, whereas the WD-40 repeats appear to mediate protein-protein interactions with relevant DNA-binding activators and repressors. Groucho/TLE proteins do not have recognizable DNA-binding domains but can repress transcription directly if tethered to DNA through a Gal4 DNA-binding domain or if recruited to DNA through interactions with other DNA-binding activators and repressors. The function of AES remains controversial. It was suggested that AES might act as an inhibitor of Groucho/TLE corepressors by dominant negative mechanisms (28, 45). On the other hand, AES has been shown to mediate Blimp-1-dependent repression of the beta interferon gene (41) and to repress NF-κB-driven gene expression (51) in vivo.
Here we demonstrate that AES physically interacts with human AR both in vivo and in vitro and that it represses AR-dependent transcription both in transient-transfection assays and in a purified cell-free transcription system. In addition, we find that AES interacts selectively with the basal transcription factor TFIIE. These observations indicate that AES represses AR-driven transcription by directly targeting the basal transcription machinery.
MATERIALS AND METHODS
Yeast screening.
The yeast two-hybrid screening was performed as previously described with minor modifications (57). Briefly, an expression plasmid encoding the Cyto-trap bait was generated by inserting the cDNA sequences of human AR into pSos, a yeast shuttle vector. Saccharomyces cerevisiae strain cdc25H was transformed sequentially with pSos-AR and human prostate cDNA library expression plasmids (Stratagene). The positive clones were those that grow on the plates with galactose and 100 nM R1881 at 37°C but not on galactose plates in the absence of R1881. Plasmids were rescued from each of these positive colonies and identified by nucleotide sequencing.
Mammalian two-hybrid analysis.
Expression vectors that encode hybrid polypeptides were produced by inserting AES cDNA sequences into the pCMV-GAL4 vector or by inserting AR cDNA sequences into the pVP-FLAG7 vector (57). A mammalian two-hybrid assay was conducted in 293T cells as described previously (52), except that when indicated, transfected cells were incubated for 40 h with medium containing 100 nM R1881. The pRL-LUC plasmid was included in each culture of transfected cells as an internal control. The luciferase activity was determined using the Dual-luciferase assay system (Promega).
Transient transfection.
The AR and AES expression vectors for transfection assays were constructed by inserting their corresponding cDNA sequences into pcDNA3.1. The AR-responsive reporter gene ARE4-LUC contains four AR-responsive elements ahead of the E4 basal promoter and the luciferase gene. HeLa Z cells were maintained in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum. Transfections were performed using SuperFect reagent (Qiagen). Briefly, 105 cells were plated onto 24-well plates approximately 24 h before transfection. After the plates were washed with phosphate-buffered saline, cells in each well were transfected with 50 ng of an expression vector (AR, estrogen receptor [ER], or thyroid hormone receptor [TR]), 100 ng of the reporter plasmid, 5 ng of the pRL-LUC internal control plasmid, and the indicated amount of the AES expression vector. The total amount of DNA was adjusted to 1 μg with pcDNA3.1. Transfections were conducted in phenol red-free RPMI 1640 medium, and 2 h later the medium was changed either to phenol red-free RPMI 1640 medium plus 10% charcoal dextran-stripped fetal bovine serum or to regular medium containing 100 nM R1881, 1 μM β-estradiol, or 10 nM T3. The cells were cultured for another 48 h and harvested for luciferase assays (Promega). For trichostatin A (TSA) treatment, 10 ng of TSA per ml was added to transfected cells 24 h before harvest. Three independent experiments were carried out in each case for statistical analysis.
Purification of transcription factors.
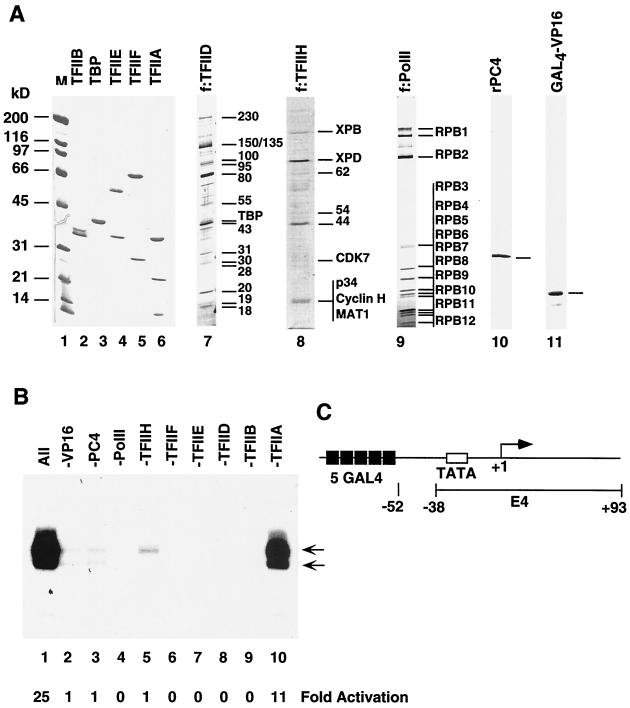
Histidine-tagged TFIIAα and TFIIAγ were expressed in bacteria via the pRSET vector and purified on Ni-nitritotriacetic acid (NTA)-agarose in the presence of 6 M urea (8). TFIIAαγ was reconstituted with a combination of equimolar amounts of purified TFIIAα and TFIIAγ and dialyzed against BC300–0.1% NP-40. The FLAG-tagged TFIIAβ was expressed via vector pET15d and purified on M2 agarose. TFIIA was reconstituted with a combination of equal amounts of TFIIAαγ and TFIIAβ. Bacterially expressed histidine-tagged TFIIB was purified on Ni-NTA-agarose and phosphocellulose. Histidine-tagged TFIIEα and FLAG-tagged TFIIEβ were expressed in bacteria and purified on Ni-NTA-agarose and M2 agarose, respectively, and TFIIE was reconstituted with a combination of two subunits and further purified through M2 agarose. TFIIF was expressed and reconstituted as reported previously (53). Bacterially expressed untagged PC4 was purified through heparin-Sepharose and phosphocellulose (13). Histidine-tagged GAL4-VP16 was expressed in bacteria and purified through Ni-NTA-agarose and S-Sepharose.
Nuclear extract was made from the FLAG-tagged TAF135 cell line (31) and further fractionated by phosphocellulose and DEAE cellulose (DE52) chromatography. FLAG-tagged TFIID (f:TFIID) was isolated from the 0.3 M KCl fraction of a DE52 column by M2 agarose affinity purification. HeLa cell lines stably expressing FLAG-tagged RPB9 and FLAG-tagged XRB1 were established, and FLAG-tagged RNA polymerase II (f:PolII) and TFIIH (f:TFIIH) were purified from these cell lines by described procedures (54). Recombinant human androgen receptor was expressed in Sf9 cells via a baculovirus vector as a FLAG-tagged fusion protein and purified on M2 agarose.
In vitro transcription and primer extension.
To create the template pARE-E4, a DNA fragment containing four copies of the androgen-responsive element (AGAACAGCAAGTGCT) from the PSA promoter was inserted into SphI and XbaI sites of the vector pG5E4. Transcription reactions were carried out in a final volume of 25 μl, and the reaction mixtures contained 90 fmol of supercoiled plasmid DNA template, 20 mM HEPES (pH 7.9), 12% glycerol, 6 mM MgCl2, 70 mM KCl, 5 mM dithiothreitol (DTT), 600 μM each ATP, UTP, CTP, and GTP, 40 U of recombinant RNasin, 0.5 mg of bovine serum albumin per ml, 12 ng of TFIIA, 30 ng of TFIIB, 2 μl of f:TFIID, 0.5 μl of f:TFIIH, 12 ng of TFIIF, 6 ng of TFIIE, 150 ng of PC4, 1 μl of f:PolII, 30 ng of human AR, and various amounts of different cofactors. After a 60-min incubation at 30°C, the transcription reactions were stopped by adding 175 μl of stop solution (1% sodium dodecyl sulfate, 5 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 7.5], 20 μg of glycogen, 40 μg of proteinase K) and incubating the mixture for 20 min at 37°C. RNA was extracted with phenol-chloroform and precipitated with ethanol. It was then hybridized with the kinase 32P-labeled primer CGCCAAGCTATTTAGGTGACACTAT (5′ end labeled; 1 × 106 to 2 × 106 cpm) in 20 μl of hybridization buffer (10 mM Tris-HCl [pH 7.5], 250 mM KCl, 1 mM EDTA) for 90 min at 37°C. The primer extension reaction was started by adding 40 μl of extension reaction solution (75 mM Tris-HCl [pH 8.0], 15 mM DTT, 12 mM MgCl2, 75 μg of actinomycin D per ml, 12 U of recombinant RNasin, 750 μM each dATP, dTTP, dCTP, and dGTP, 100 U of SuperScript RNase H reverse transcriptase), and the reaction mixture was incubated for 90 min at 37°C. The cDNA products were extracted with phenol-chloroform; precipitated with ethanol; dissolved, and denatured (100°C for 3 min) in 10 μl of 95% formamide containing 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol FF; and finally analyzed on a 6% polyacrylamide–7 M urea gel.
Protein-protein interaction assay.
Recombinant glutathione S-transferase (GST) fusion (expressed in bacterial cells) or FLAG-tagged (expressed in insect cells) proteins (1 μg) were immobilized on 10 μl of glutathione or M2 agarose beads, respectively. Then 10 μl of beads was incubated for 2 h at 4°C with 5 μl of rabbit reticulocyte lysate containing [35S]Met-labeled proteins or 100 μl of HeLa nuclear extract (60 μg proteins) in a final volume of 200 μl containing 20 mM HEPES (pH 7.9), 0.2 mM EDTA, 20% glycerol, 2 mM DTT, 150 mM KCl, 0.1% NP-40 and 0.5 mg of BSA per ml. The beads were washed five times (1 ml each) with the incubation buffer, boiled in 10 μl of the 2× SDS gel sample buffer, and analyzed by autoradiography or Western blot analysis. For the coimmunoprecipitation assay, 10 μl of M2 agarose beads was incubated with 250 μl of whole-cell extract from transfected 293T cells in BC150–0.1% NP-40 for 2 h. The beads were washed with the incubation buffer and analyzed by Western blotting.
In situ hybridization.
The archival normal prostate tissues were obtained during radical prostatectomy of prostate cancer patients at New York University Medical Center under an Institutional Review Board-approved protocol. The procedure for in situ hybridization was as described previously (29). Briefly, the sections (4 μm) of prostate tissues were hydrated, postfixed in 4% paraformaldehyde, treated with proteinase K, and deacetylated. The prehybridization and hybridization were performed at 68°C. The 536-bp AR (nucleotides 2224 to 2716) and the 648-bp AES (nucleotides 353 to 957) cDNA fragments containing T7 and T3 promoters at each end was generated by PCR. The 33P-labeled probe RNAs (sense and antisense) were generated by in vitro transcription with T7 and T3 RNA polymerases, respectively, and hybridized to the slides containing prostate tissue specimens. After being washed, the slides were exposed for 2 to 3 weeks and then counterstained with hematoxylin and eosin.
RESULTS
N-terminal domain of AR interacts with the Groucho/TLE family protein AES.
Various coactivators and corepressors have been shown to play a critical role in mediating the functions of nuclear receptors (56). Although a number of AR-interacting coactivators have been identified (reviewed in references 4 and 19), we have used a yeast two-hybrid screening method to search for additional AR-interacting proteins. For this assay, full-length human AR (residues 2 to 919) was fused to human Sos (hSos) as a bait (Fig. 1A). The temperature-sensitive mutant S. cerevisiae strain cdc25H, which contains a point mutation in the yeast homolog (cdc25) of the hSos gene, cannot grow at 37°C but can grow at the permissive temperature (25°C). This yeast strain was used to screen a human prostate cDNA expression library fused to the v-Src myristylation sequence, which anchors the fusion protein to the plasma membrane. If the bait and target proteins physically interact, the hSos protein is recruited to the membrane, thereby activating the Ras signaling pathway and allowing the cdc25H yeast strain to grow at 37°C.
Approximately 2 million transformants from the prostate cDNA library were screened, and 35 positive clones were obtained. Nucleotide sequence determination and comparison with GenBank databases (National Center for Biotechnology Information) revealed seven clones that encoded the human AES (5, 11). To confirm that the interactions between AR and AES are specific, human AES was fused to the Gal4 DNA-binding domain and AR was fused to the VP16 transcriptional activation domain. These constructs were transfected into 293T cells with a reporter containing five Gal4-binding sites and the E1b core promoter fused to the luciferase gene, and activation of luciferase reporter was measured in the absence and presence of ligand (R1881). A sevenfold activation of the reporter gene was observed in the presence of androgen but not in its absence, indicating that AR-AES interactions are hormone-dependent in vivo (Fig. 1B). As negative controls, neither coexpression of AR-VP16 with Gal4-DBD nor coexpression of Gal4-AES with VP16 resulted in significant ligand-dependent activation of the reporter (Fig. 1B). To further confirm the interaction of AR with AES in mammalian cells, we performed a coimmunoprecipitation using immobilized anti-FLAG monoclonal antibody (M2 agarose). AR was coimmunoprecipitated with AES from the whole-cell extract made from cells transfected with AR and FLAG-tagged AES in the presence of 100 nM ligand (R1881) (Fig. 1C, lane 3). In the absence of ligand, only trace amounts of AR were coimmunoprecipitated (lane 2). As a negative control, no AR was immunoprecipitated by M2 agarose when the cell was transfected with AR alone (lane 1).
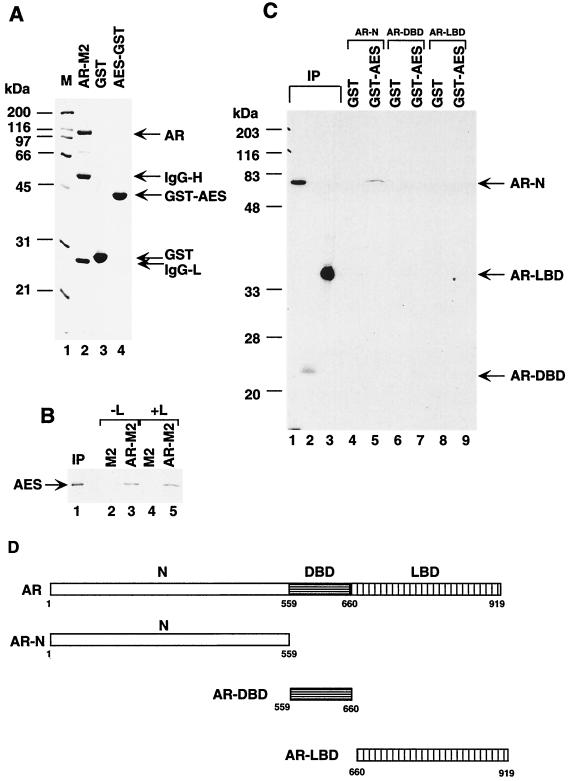
To further investigate the interactions of AES with AR, we performed in vitro protein-protein pull-down assays. In vitro-translated [35S]AES was incubated with FLAG-tagged AR that had been expressed in Sf9 cells and immobilized on M2 agarose beads (Fig. 2A, lane 2). Figure 2B shows that AES bound to AR-M2 (lanes 3 and 5) but not to unliganded M2 agarose beads (lanes 2 and 4). These interactions were found to be ligand independent (Fig. 2B, compare lane 5 with lane 3), a somewhat surprising observation in view of the observed ligand-dependent interactions in vivo (Fig. 1B). This discrepancy is probably because AR associates with heat shock proteins and other chaperones in vivo in the absence of androgen (30), thereby preventing its interactions with AES as well as other cofactors. To identify the AR domain that interacts with AES, the N-terminal, DNA-binding, and ligand-binding domains of AR (Fig. 2D) were expressed as 35S-labeled proteins and incubated with GST and GST-AES fusion protein immobilized on glutathione-agarose beads (Fig. 2A, lanes 3 and 4). As shown in Fig. 2C, the N-terminal part bound to GST-AES (lane 5) but not to GST alone (lane 4) whereas the DNA-binding and ligand-binding domains failed to interact (lanes 7 and 9). This demonstration that AES interacts with the AR N-terminal region is interesting in light of the significant role of this region in target gene activation by liganded AR (26).
FIG. 2.
The N-terminal part of AR directly interacts with AES in vitro. (A) SDS-PAGE (12% polyacrylamide) analysis of the M2 agarose-bound recombinant AR (lane 2), bacterially expressed and purified GST (lane 3), and GST-AES (lane 4) proteins. Standard molecular mass markers (M) (in kilodaltons) are shown in lane 1. IgG light (IgG-L) and heavy (IgG-H) chains of monoclonal antibody (M2) that dissociated from agarose beads by boiling with SDS sample buffer are indicated on the right. (B) AR interacts with AES in vitro independently of the ligand. Radiolabeled AES was incubated with M2 (lanes 2 and 4) or FLAG-tagged AR immobilized on M2 agarose beads (lanes 3 and 5) in the absence (lane 2 and 3) or presence (lanes 4 and 5) of 100 nM R1881. After the beads were washed, bound AES and 5% of the input (IP) (lane 1) were analyzed on by SDS-PAGE (12% polyacrylamide) and visualized by autoradiography. (C) The N-terminal part of AR is sufficient to bind to AES. GST (lanes 4, 6, and 8) or GST-AES (lanes 5, 7, and 9) proteins, immobilized on beads, were mixed with 5 μl of in vitro labeled N-terminal (AR-N) (lanes 4 and 5), DNA-binding (AR-DBD) (lanes 6 and 7), and ligand-binding (AR LBD) (lanes 8 and 9) domains of AR. After the beads were washed, the bound proteins and 5% of the input (IP) (lanes 1 to 3) were analyzed by SDS-PAGE (12% polyacrylamide) and visualized by autoradiography. (D) Diagram of AR, AR DNA-binding domain, and AR ligand-binding domain.
AES represses AR-dependent gene expression.
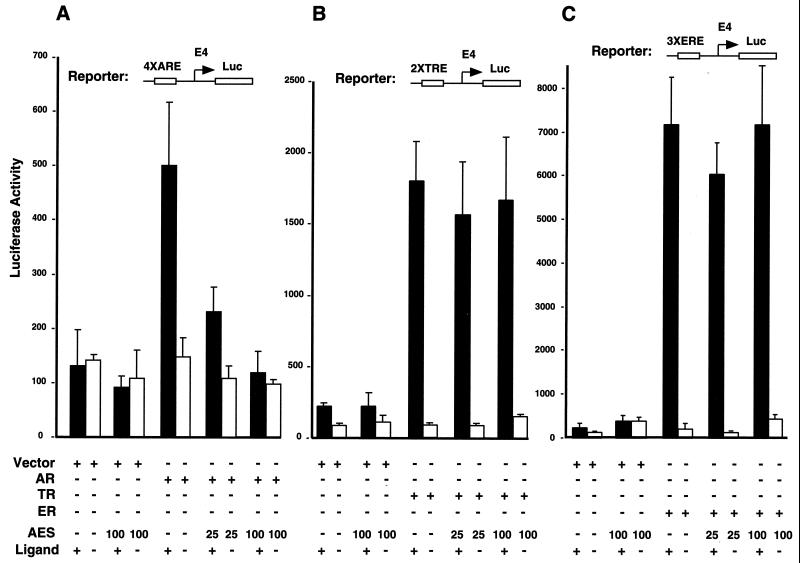
We then investigated the effect of AES on AR-dependent transcription by performing transient-transfection assays. The luciferase reporter plasmid containing four tandem copies of the PSA gene androgen-responsive elements (7) upstream of the minimal adenovirus E4 promoter (see Fig. 5C) was cotransfected with expression vectors for AR and/or AES into HeLa Z cells in the absence or presence of ligand (R1881). As shown in Fig. 3A, AR activated the reporter gene about fourfold in the presence of androgen, and coexpressed AES completely blocked this AR-dependent transactivation in a dose-dependent manner. In the absence of cotransfected AR or ligand (R1881), AES did not influence reporter gene activity, indicating that the inhibitory effect of AES on AR-dependent gene expression was not due to an effect on the E4 promoter. Similar results were obtained with the LNCaP prostate cancer cell line (data not shown).
FIG. 5.
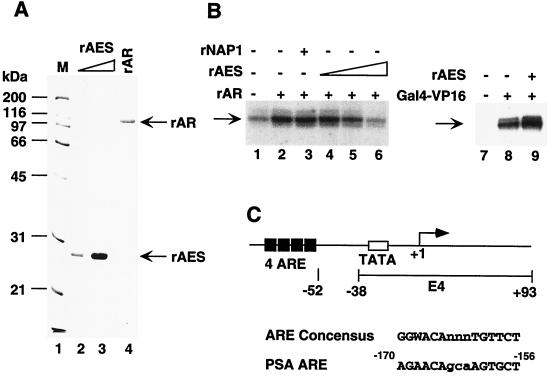
AES represses AR-driven transcription in vitro. (A) SDS-PAGE analysis of recombinant AES and AR proteins. Portions of 50 (lane 2) and 200 (lane 3) ng of purified recombinant 6His-tagged AES expressed in bacteria and 100 ng of purified recombinant human AR (lane 4) expressed in Sf9 cells were subjected to SDS-PAGE with Coomassie blue R250 staining. (B) AES inhibition of AR-dependent transcription. A synthetic template containing four ARE elements (pARE-E4) was transcribed in a system reconstituted with the purified factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, Pol II, and PC4) as shown in Fig. 1, panel A, and 10 ng of affinity-purified (via f:Nut2) TRAP/Mediator complex (27). Other additions included 30 ng of rAR (lanes 2 to 6); 10 (lane 4), 30 (lane 5) or 100 (lanes 6 and 9) ng of rAES; 100 ng of rNAP1 (lane 3); and 30 ng of Gal4-VP16 (lanes 8 and 9). The specifically initiated transcript is indicated by an arrow and was monitored by primer extension. (C) Diagram of the synthetic ARE-containing promoter. The template (pARE-E4) contains four tandem copies of the ARE from the PSA promoter positioned upstream of the adenovirus E4 promoter.
FIG. 3.
AES inhibits AR-mediated transcription in vivo. (A) AES represses AR-dependent luciferase gene expression induced by AR in the presence of R1881. HeLa cells were transfected with 500 ng of 4×ARE-E4-luc reporter plasmid, 30 ng of pCMV-AR, and the indicated amounts of pCMV-AES expression plasmids. Cells were grown in the absence or presence of 100 nM R1881 for 48 h after transfection and then harvested for luciferase activity assays. (B) TR-mediated T3-dependent luciferase gene expression is not suppressed by AES. The reporter construct contains 2×TRE, the E4 core promoter, and the luciferase gene. After transfection, HeLa cells were grown for 48 h in the absence or presence of 10 nM T3 before being harvested for luciferase assays. (C) ER-mediated estradiol-dependent luciferase gene expression is not suppressed by AES. The reporter construct contains 3×ERE, the E4 core promoter, and the luciferase gene. After transfection, HeLa cells were grown for 48 h in the absence or presence of 1 μM estradiol before the luciferase assays were performed.
To further examine whether the inhibitory effect of AES is specific for AR, we compared the effects of AES on the transcription of reporters containing the same E4 promoter under the control of TR and ER. As shown in Fig. 3B and C, TR and ER activated the reporter genes about 17- and 35-fold, respectively, in the presence of their cognate ligands (T3 and estradiol). In contrast to its dramatic effect on AR-mediated transactivation, AES showed no effect on TR- or ER-mediated transcription. Western blot analysis revealed that the expression levels of ER were comparable to those of AR (data not shown). Hence, AES shows nuclear receptor-specific inhibitory effects in vivo.
Establishment of a highly purified in vitro transcription system for activator function.
To study the mechanism of basal and activator-dependent transcription, we established an activator-responsive complementation assay involving homogenous recombinant and FLAG-tagged immunopurified natural general initiation factors (TFIIs) and positive cofactors (PCs) (43, 44). The recombinant factors expressed in and purified from bacteria included TFIIA (three subunits [Fig. 4A, lane 6]), TFIIB (one subunit [lane 2]), TFIIE (two subunits [lane 4]), TFIIF (two subunits [lane 5]), and PC4 (1 subunit [lane 10]). The multisubunit components purified from cell lines expressing FLAG-tagged subunits included f:TFIID (∼15 subunits [Fig. 4A, lane 7]), f:TFIIH (9 subunits [lane 8]), and f:Pol II (12 subunits [lane 9]). Recombinant GAL4-VP16 (lane 11) was used as an activator to establish the functionality of this particular assay system. The GAL4-VP16-responsive template pG5E4 (Fig. 4C) contains five Gal4-binding sites preceding the adenovirus E4 core promoter (from −38 to +93) (36). To determine whether all purified factors are required for transcription in our highly purified transcription system, we first tested a complete mixture of all GTFs, Pol II, Gal4-VP16, and PC4 with supercoiled DNA template (pG5E4) and then omitted individual factors. As shown in Fig. 4B, basal (activator-independent) transcription (lane 2) is completely dependent on TFIID (lane 8), TFIIB (lane 9), TFIIE (lane 7), TFIIF (lane 6), and Pol II (lane 4) whereas activation (up to 25-fold) by GAL4-VP16 (lane 1) absolutely requires Pol II and all initiation factors other than TFIIA.
FIG. 4.
Transcription activation by a model activator in a cell-free system reconstituted with purified factors. (A) SDS-PAGE analysis of purified factors. Coomassie blue R250 staining of purified recombinant activator GAL4-VP16 (lane 11), the general coactivator PC4 (lane 10), and the general initiation factors TFIIA, TFIIB, TFIIE, TFIIF, and TBP (lanes 2 to 6) was performed. Silver staining of the immunopurified FLAG-tagged multisubunit general initiation factors TFIID and TFIIH and RNA polymerase II (lanes 7 to 9) was performed. The subunits identified as integral subunits are indicated by size (in kilodaltons) or by name on the right. Some bands are difficult to visualize because of weak or negative staining. Unmarked bands represent either degradation products or contaminants that can be removed by further purification. Lane 1 shows molecular weight markers (M), (B) Activator-dependent transcription. Transcription was conducted with the purified components shown in panel A and the DNA template indicated in panel C. The two arrows show specifically initiated transcripts assayed by primer extension. A complete reaction with all factors is shown in lane 1, whereas reactions with single-factor omissions (indicated at the top) are shown in lanes 2 to 10. Fold activation above the basal level (-GAL4-VP16, lane 2) is indicated at the bottom. (C) Diagram of the model template. The template contains five tandem GAL4 sites adjacent to the adenovirus E4 core promoter.
AES represses AR-driven transcription in vitro.
For initial tests of AR function, we constructed a synthetic hybrid promoter (pARE-E4) containing four copies of an androgen response element (ARE) from the PSA promoter (7, 42) just upstream of the adenovirus E4 core promoter (Fig. 5C). This template was assayed in the above-described purified system supplemented with an affinity-purified TRAP complex previously shown to facilitate transcription by other nuclear receptors (12, 17, 58). In this system, the purified baculovirus-expressed recombinant AR (Fig. 5A, lane 4) activated transcription threefold (Fig. 4B, compare lane 2 with lane 1). To test its effect on AR-dependent transcription in this system, human AES was expressed in and purified from bacteria (Fig. 5A, lanes 1 and 2). Addition of recombinant AES inhibited AR-dependent transcription in a dose-dependent manner and, at the highest level (100 ng), reduced it to the basal level (compare lane 6 and lane 1). As negative controls, 100 ng of recombinant mouse NAP1 expressed and purified in a manner identical to that for AES had no detectable effect on the AR-driven transcription (compare lane 3 and lane 2) and recombinant AES did not repress but slightly (1.5-fold) enhanced Gal4-VP16-driven transactivation (compare lane 9 and lane 8) in the same reconstituted system. These results suggest that AES specifically and directly represses AR-driven transcription in vitro.
AES interacts with TFIIE.
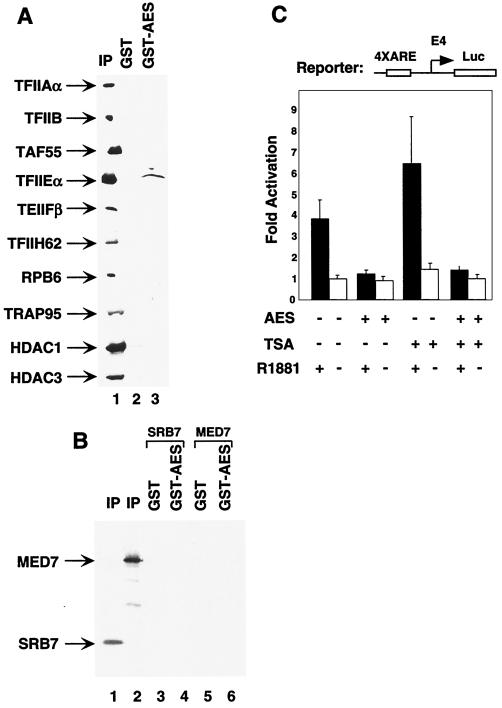
The structural resemblance of Groucho/TLE proteins to Tup1 (21), a general transcription repressor in yeast, suggested that the two proteins may function by a similar mechanism. More recently, Gromoller and Lehming (14) reported TUP1-mediated repression through physical interaction with the SRB7 subunit of the yeast Mediator complex. To test whether human AES uses a similar mechanism to repress transcription, we performed protein-protein pull-down assays. GST-AES failed to bind the human homolog (TRAP-Mediator complex, detected by anti-TRAP95 antibodies) of the yeast Mediator complex (27) in HeLa nuclear extract (Fig. 6A), as well as the independently expressed SRB7 and MED7 subunits of the human TRAP-Mediator complex (Fig. 6B, lanes 4 and 6). These results suggest that human AES, unlike yeast Tup1, may not directly interact with the Mediator complex.
FIG. 6.
AES interacts with TFIIE and represses AR-dependent transcription in the presence of TSA. (A) GST (lane 2) and GST-AES (lane 3), immobilized on agarose beads, were incubated with HeLa nuclear extract. The bound proteins (lanes 2 and 3) and 10% of the input (IP) were analyzed by Western blot assays with the corresponding antibodies indicated on the left. (B) AES does not interact with MED7 and SRB7. GST (lanes 3 and 5) or GST-AES (lanes 4 and 6) proteins, immobilized on beads, were mixed with 5 μl of in vitro-labeled SRB7 (lanes 3 and 4) and MED7 (lanes 5 and 6). After the beads were washed, the bound proteins and 10% of the inputs (IP) (lanes 1 and 2) were analyzed by SDS-PAGE (15% polyacrylamide) and visualized by autoradiography. (C) AES inhibits AR-dependent transcription in the presence of TSA. HeLa cells were transfected with 100 ng of 4×ARE-E4-luc reporter plasmid, 50 ng of pCMV-AR, and 100 ng of pCMV-AES expression plasmid. Cells were grown in the absence or presence of 100 nM R1881 and 10 ng of TSA per ml for 48 h after transfection and then were harvested for luciferase activity assays.
HDAC-containing complexes mediate the function of various corepressors in vivo (15, 35). To investigate whether AES also functions through these complexes, we performed transient-transfection assays in the presence of the general deacetylase inhibitor TSA. Figure 6C shows that AES still actively inhibits AR-dependent transcription in the presence of TSA and at a level similar to that observed in the absence of TSA. These results suggest that AES represses AR-dependent transcription by directly targeting the basal transcriptional machinery rather than through chromatin modifications involving recruitment of HDAC-containing corepressor complexes. This conclusion is further supported by the absence of demonstrable interactions of AES with HDAC1- and HDAC3-containing complexes (16, 25) in HeLa nuclear extract (Fig. 6A). However, since the reporter gene in the transient-transfection assay may not be packaged appropriately into chromatin, we cannot rule out the possibility of the involvement of HDACs in AES function on endogenous genes within chromatin.
To further study the mechanism of action of AES, we performed additional protein-protein pull-down assays to assess possible interactions of AES with components of the basal transcriptional machinery. GST and GST-AES agarose beads were incubated with HeLa nuclear extract, and the bound proteins were analyzed by Western blot assays with polyclonal antibodies against subunits of RNA polymerase II and basal transcription factors. As shown in Fig. 6A, TFIIE (detected by antibodies against TFIIEα) was specifically retained by GST-AES, relative to GST alone, whereas other basal transcription factors (TFIIA, TFIIB, TFIID, TFIIF, and TFIIH) and RNA polymerase II failed to be bound. These observations implicate TFIIE as a possible target for AES.
AES is highly expressed, along with AR, in prostate epithelial cells.
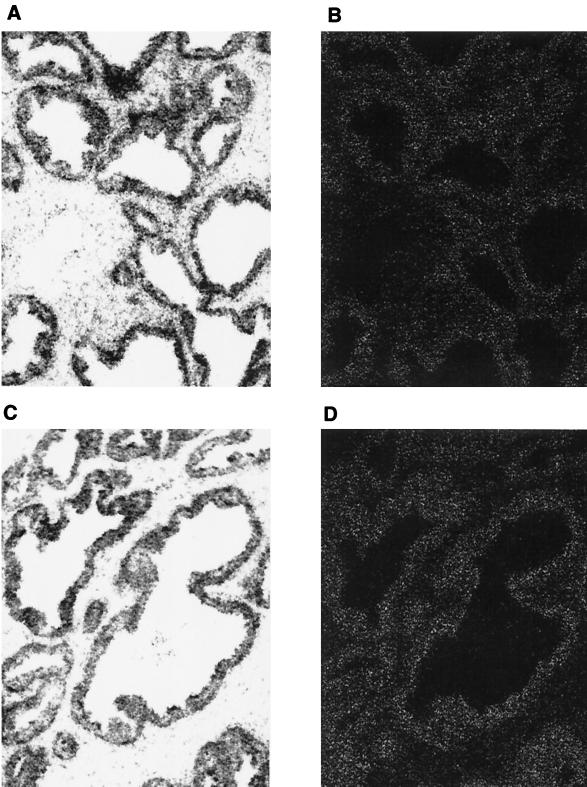
To determine whether AES and AR are expressed in the same cells in humans, we investigated the expression levels of AES and AR in normal prostate tissues by in situ RNA hybridization. Consistent with previous observations (32), the expression levels of AR were high in the epithelial cells of the prostate (Fig. 7A and B). Expression of AES was also evident in the epithelial cells (Fig. 7C and D). As negative controls, no hybridization signals above the background levels were detected with the sense RNA probes, thus indicating that the signals obtained with the antisense probe are specific (data not shown).
FIG. 7.
AES expressed in the epithelial cells of the prostate. The slides containing sections of prostate tissues were hybridized with antisense AR (A and B) or AES (C and D) RNA probes. The emulsion-coated slides were exposed and evaluated under a Nikon microscope with a digital camera interfaced to a computer. The left and right panels show bright-field and dark-field images of the same area of the slides.
DISCUSSION
Various cofactors that have been implicated in the function of AR, as well as a number of other nuclear receptors, include p300/CBP, p160 family proteins, the ARA group (ARA24, ARA45, ARA54, ARA55, ARA70, and ARA160), ARIP3, SNURF, and BAG-1L (18, 19, 46). All of these enhance AR-mediated transcription in vivo, although there is not a clear mechanistic understanding of the function of these factors. The results described here demonstrate (i) that AES is a selective repressor of ligand-dependent AR-mediated transcription and (ii) that AES physically interacts with the N-terminal region of AR and represses AR-driven transcription by targeting the basal transcriptional machinery (possibly TFIIE). These observations thus reveal a new negative regulatory pathway for AR function, as well as new insights into the mechanism of action of mammalian Groucho/TLE proteins.
AES represses AR-dependent transcription.
A number of proteins have been demonstrated to repress AR-dependent transcription in vivo. These include AP-1 (34, 47), NF-κB (39), TR4 (testicular orphan receptor 4) (24) and HBO1 (histone acetyl transferase binding to origin recognition complex 1) (49). AP-1, NF-κB, and TR4 appear to inhibit AR-dependent transcription by mutual transcriptional interference (unexpected interactions of distinct transcription factors). Although the molecular mechanisms that underlie this phenomenon have remained mostly elusive, this may involve competition for a coactivator commonly required by both activators (1).
HBO1 belongs to the MYST family, which is characterized by highly conserved C2HC zinc fingers and a putative histone acetyltransferase domain. HBO1 contains a putative repression domain, interacts with the DBD-LBD of AR, and inhibits AR-dependent transcription in vivo, although the exact mechanism of HBO1 action remains to be determined (49). Based on the results presented here, AES represses AR-driven transcription in a manner more like that of HBO1. Like HBO1, AES physically interacts with AR and specifically represses AR-dependent transcription in transient-transfection assays. Also as reported for HBO1, AES probably does not act broadly as a nuclear hormone receptor corepressor because it represses AR-dependent transcription but not TR- or ER-dependent transcription. These results are consistent with the fact that AES physically interacts with the N-terminal region of AR, which is not conserved in the N-terminal regions of TR and ER (37). Nonetheless, it remains important to determine whether AES might repress other (as yet untested) nuclear receptors and, related, whether other members of the Groucho/TLE family can repress the function of AR or other nuclear receptors.
Like AR, some of the DNA-binding partners for the Groucho/TLE proteins do not always act as transcriptional repressors, and, in fact, some are better characterized as activators (11). For the Groucho-interacting Dorsal and Runt domain proteins (2, 9, 20, 22), the context of the target gene promoter appears to be critical for determining whether activation or repression will occur. These observations suggest that the recruitment of Groucho/TLE proteins and/or their repressor activities might also be dependent on the nature of the target gene promoter. It is also possible that Groucho/TLE proteins might function as coactivators in certain situations. Thus, it will be important to determine whether AES repression of AR-driven transcription is dependent on the target gene promoter context.
Mechanism of AES function.
At present, relatively little is know about the mechanisms by which Groucho/TLE family proteins function as eukaryotic (co)repressors. Various repressors and activators recruit the Groucho/TLE proteins through specific interactions with various regions of Groucho proteins (11). In the well-defined reconstituted transcription system utilized here, we observed repression of AR-dependent transcription from DNA templates by recombinant AES. Consistent with the indication from this result that AES may function through interactions with the basal transcriptional machinery, a specific interaction of AES with the basal transcription factor TFIIE was observed. Similarly, previous studies have shown that the zinc finger protein Kruppel represses transcription through physical interactions with TFIIE (48). Hence, these studies suggest that TFIIE may serve as a more general target for various corepressors and repressors.
TUP1, a general transcriptional corepressor (21, 50), is a yeast analog of the Groucho/TLE proteins. Gromoller and Lehming (14) demonstrated that the essential holoenzyme component SRB7 is a physical and functional target of TUP1. In addition, genetic interactions between Cyc8-Tup1 and a variety of Pol II holoenzyme components (SRB8, SRB10, SRB11, Sin4, Rgr1, Rox3, and Hrs1) have been reported (23). However, we failed to detect direct interactions of AES with human SRB7 or the SRB7-containing TRAP/Mediator complex in protein-protein pull-down assays, indicating that human AES may not directly target human SRB7 or the TRAP/Mediator complex. This observation may reflect the fact that Tup1 and Groucho/TLE proteins show poor sequence conservation (at the amino acid level) in both repression domains and WD-40 repeats. Similar to AES and suggesting a chromatin-independent mechanism, the purified Tup1-containing complex directly represses transcription in a crude yeast extract in vitro (40).
Many corepressor complexes contain HDAC enzymes. The Drosophila HDAC Rpd3 has been identified as a Groucho-interacting protein (6), and, possibly related, Groucho proteins also interact with histone H3 (38). Yeast Tup1 similarly interacts directly and genetically with histones H3 and H4 (10, 38), and mutations in genes encoding the HDACs abolish Tup1-mediated repression (55). These findings have led to a repression model, possibly complementing the more direct mechanisms indicated above, involving Groucho/Tup1 recruitment by promoter-bound factors, HDAC recruitment by Groucho/Tup1, and subsequent function of HDAC to establish and/or maintain a transcriptionally silenced chromatin structure. Our results do not support this model for AES. First, we failed to detect interactions between AES and HDAC1- or HDAC3-containing complexes. Second, the deacetylase inhibitor TSA did not affect AES-mediated inhibition of AR-dependent transcription in transient-transfection assays. Third, we observed a direct inhibition of AR-dependent transcription by recombinant AES in a highly purified reconstituted transcription system on a naked DNA template.
In summary, our results point both to a novel function for AES in mediating repression of AR-dependent transcription and to a mechanism involving direct interactions both with AR and with the basal transcription machinery. AR is an important regulatory factor in the development, differentiation, and maintenance of male reproductive functions, as well as in the regulation of other sexually dimorphic processes ranging from the development of neural tissues to the modulation of immune function (33). Thus, the mammalian Groucho-related protein AES, and possible other family members, may play a pivotal role in these biological processes by modulating the transcriptional activity of AR.
ACKNOWLEDGMENTS
We thank Hua Xiao for the prostate cDNA library and Yun Kyoung Kang for immunopurified TRAP/Mediator complex.
This work was supported partly by a CaP CURE Award to R.G.R. and Z.W. and by an NIH grant to R.G.R. X.Y. is supported by a postdoctoral fellowship of the Cancer Research Institute.
REFERENCES
- 1.Aarnisalo P, Palvimo J J, Janne O A. CREB-binding protein in androgen receptor-mediated signaling. Proc Natl Acad Sci USA. 1998;95:2122–2127. doi: 10.1073/pnas.95.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson B D, Fisher A L, Blechman K, Caudy M, Gergen J P. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger S L. Gene activation by histone and factor acetyltransferases. Curr Opin Cell Biol. 1999;11:336–341. doi: 10.1016/S0955-0674(99)80046-5. [DOI] [PubMed] [Google Scholar]
- 4.Brinkmann A O, Blok L J, de Ruiter P E, Doesburg P, Steketee K, Berrevoets C A, Trapman J. Mechanisms of androgen receptor activation and function. J Steroid Biochem Mol Biol. 1999;69:307–313. doi: 10.1016/s0960-0760(99)00049-7. [DOI] [PubMed] [Google Scholar]
- 5.Chen G, Courey A J. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 6.Chen G, Fernandez J, Mische S, Courey A J. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleutjens K B, van Eekelen C C, van der Korput H A, Brinkmann A O, Trapman J. Two androgen response regions cooperate in steroid hormone regulated activity of the prostate-specific antigen promoter. J Biol Chem. 1996;271:6379–6388. doi: 10.1074/jbc.271.11.6379. [DOI] [PubMed] [Google Scholar]
- 8.DeJong J, Bernstein R, Roeder R G. Human general transcription factor TFIIA: characterization of a cDNA encoding the small subunit and requirement for basal and activated transcription. Proc Natl Acad Sci USA. 1995;92:3313–3317. doi: 10.1073/pnas.92.8.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy J B, Gergen J P. Sex, segments, and the central nervous system: common genetic mechanisms of cell fate determination. Adv Genet. 1994;31:1–28. doi: 10.1016/s0065-2660(08)60394-6. [DOI] [PubMed] [Google Scholar]
- 10.Edmondson D G, Smith M M, Roth S Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 11.Fisher A L, Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 12.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge H, Martinez E, Chiang C M, Roeder R G. Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods Enzymol. 1996;274:57–71. doi: 10.1016/s0076-6879(96)74008-9. [DOI] [PubMed] [Google Scholar]
- 14.Gromoller A, Lehming N. Srb7p is a physiological target of Tup1. EMBO J. 2000;19:6845–6852. doi: 10.1093/emboj/19.24.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 16.Huang E Y, Zhang J, Miska E A, Guenther M G, Kouzarides T, Lazar M A. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 17.Ito M, Yuan C-X, Malik S, Gu W, Fondell J D, Yamamura S, Fu Z-Y, Zhang X, Qin J, Roeder R G. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 18.Janne O A, Moilanen A M, Poukka H, Rouleau N, Karvonen U, Kotaja N, Hakli M, Palvimo J J. Androgen-receptor-interacting nuclear proteins. Biochem Soc Trans. 2000;28:401–405. [PubMed] [Google Scholar]
- 19.Jenster G. The role of the androgen receptor in the development and progression of prostate cancer. Semin Oncol. 1999;26:407–421. [PubMed] [Google Scholar]
- 20.Jiang J, Cai H, Zhou Q, Levine M. Conversion of a dorsal-dependent silencer into an enhancer: evidence for dorsal corepressors. EMBO J. 1993;12:3201–3209. doi: 10.1002/j.1460-2075.1993.tb05989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 22.Kirov N, Zhelnin L, Shah J, Rushlow C. Conversion of a silencer into an enhancer: evidence for a co-repressor in dorsal-mediated repression in Drosophila. EMBO J. 1993;12:3193–3199. doi: 10.1002/j.1460-2075.1993.tb05988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee M, Chatterjee S, Struhl K. Genetic analysis of the role of Pol II holoenzyme components in repression by the Cyc8-Tup1 corepressor in yeast. Genetics. 2000;155:1535–1542. doi: 10.1093/genetics/155.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y F, Shyr C R, Thin T H, Lin W J, Chang C. Convergence of two repressors through heterodimer formation of androgen receptor and testicular orphan receptor-4: a unique signaling pathway in the steroid receptor superfamily. Proc Natl Acad Sci USA. 1999;96:14724–14729. doi: 10.1073/pnas.96.26.14724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Wang J, Nawaz Z, Liu J M, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacLean H E, Warne G L, Zajac J D. Localization of functional domains in the androgen receptor. J Steroid Biochem Mol Biol. 1997;62:233–242. doi: 10.1016/s0960-0760(97)00049-6. [DOI] [PubMed] [Google Scholar]
- 27.Malik S, Roeder R G. Transcriptional regulation through mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 28.Mallo M, Lieberma P M, Gridley T. Possible involvement of the mouse Grg protein in transcription. Cell Mol Biol Res. 1995;41:435–440. [PubMed] [Google Scholar]
- 29.Manova K, Nocka K, Besmer P, Bachvarova R F. Gonadal expression of c-kit encoded at the W locus of the mouse. Development. 1990;110:1057–1069. doi: 10.1242/dev.110.4.1057. [DOI] [PubMed] [Google Scholar]
- 30.Marivoet S, van Dijck P, Verhoeven G, Heyns W. Interaction of the 90-kDa heat shock protein with native and in vitro translated androgen receptor and receptor fragments. Mol Cell Endocrinol. 1992;88:165–174. doi: 10.1016/0303-7207(92)90021-w. [DOI] [PubMed] [Google Scholar]
- 31.Martinez E, Ge H, Tao Y, Yuan C X, Palhan V, Roeder R G. Novel cofactors and TFIIA mediate functional core promoter selectivity by the human TAFII150-containing TFIID complex. Mol Cell Biol. 1998;18:6571–6583. doi: 10.1128/mcb.18.11.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyamoto K K, McSherry S A, Dent G A, Sar M, Wilson E M, French F S, Sharief Y, Mohler J L. Immunohistochemistry of the androgen receptor in human benign and malignant prostate tissue. J Urol. 1993;149:1015–1019. doi: 10.1016/s0022-5347(17)36284-5. [DOI] [PubMed] [Google Scholar]
- 33.Mooradian A D, Morley J E, Korenman S G. Biological actions of androgens. Endocr Rev. 1987;8:1–8. doi: 10.1210/edrv-8-1-1. [DOI] [PubMed] [Google Scholar]
- 34.Murtha P E, Zhu W, Zhang J, Zhang S, Young C Y. Effects of Ca++ mobilization on expression of androgen-regulated genes: interference with androgen receptor-mediated transactivation by AP-I proteins. Prostate. 1997;33:264–270. doi: 10.1002/(sici)1097-0045(19971201)33:4<264::aid-pros7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 35.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 36.Oelgeschlager T, Tao Y, Kang Y K, Roeder R G. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- 37.Owen G I, Zelent A. Origins and evolutionary diversification of the nuclear receptor superfamily. Cell Mol Life Sci. 2000;57:809–827. doi: 10.1007/s000180050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- 39.Palvimo J J, Reinikainen P, Ikonen T, Kallio P J, Moilanen A, Janne O A. Mutual transcriptional interference between RelA and androgen receptor. J Biol Chem. 1996;271:24151–25156. doi: 10.1074/jbc.271.39.24151. [DOI] [PubMed] [Google Scholar]
- 40.Redd M J, Arnaud M B, Johnson A D. A complex composed of tup1 and ssn6 represses transcription in vitro. J Biol Chem. 1997;272:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- 41.Ren B, Chee K J, Kim T H, Maniatis T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 1999;13:125–137. doi: 10.1101/gad.13.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roche P J, Hoare S A, Parker M G. A consensus DNA-binding site for the androgen receptor. Mol Endocrinol. 1992;6:2229–2235. doi: 10.1210/mend.6.12.1491700. [DOI] [PubMed] [Google Scholar]
- 43.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 44.Roeder R G. The role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harbor Symp Quant Biol. 1998;63:201–218. doi: 10.1101/sqb.1998.63.201. [DOI] [PubMed] [Google Scholar]
- 45.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 46.Sadar M D, Hussain M, Bruchovsky N. Prostate cancer: molecular biology of early progression to androgen independence. Endocr Relat Cancer. 1999;6:487–502. doi: 10.1677/erc.0.0060487. [DOI] [PubMed] [Google Scholar]
- 47.Sato M, Sadar M D, Bruchovsky N, Saatcioglu F, Rennie P S, Sato S, Lange P H, Gleave M E. Androgenic induction of prostate-specific antigen gene is repressed by protein-protein interaction between the androgen receptor and AP-1/c-Jun in the human prostate cancer cell line LNCaP. J Biol Chem. 1997;272:17485–17494. doi: 10.1074/jbc.272.28.17485. [DOI] [PubMed] [Google Scholar]
- 48.Sauer F, Fondell J D, Ohkuma Y, Roeder R G, Jackle H. Control of transcription by Kruppel through interactions with TFIIB and TFIIE beta. Nature. 1995;375:162–164. doi: 10.1038/375162a0. [DOI] [PubMed] [Google Scholar]
- 49.Sharma M, Zarnegar M, Li X, Lim B, Sun Z. Androgen receptor interacts with a novel MYST protein, HBO1. J Biol Chem. 2000;275:35200–35208. doi: 10.1074/jbc.M004838200. [DOI] [PubMed] [Google Scholar]
- 50.Smith R L, Johnson A D. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci. 2000;25:325–330. doi: 10.1016/s0968-0004(00)01592-9. [DOI] [PubMed] [Google Scholar]
- 51.Tetsuka T, Uranishi H, Imai H, Ono T, Sonta S, Takahashi N, Asamitsu K, Okamoto T. Inhibition of nuclear factor-kappaB-mediated transcription by association with the amino-terminal enhancer of split, a Groucho-related protein lacking WD40 repeats. J Biol Chem. 2000;275:4383–4390. doi: 10.1074/jbc.275.6.4383. [DOI] [PubMed] [Google Scholar]
- 52.Tsan J T, Wang Z, Jin Y, Hwang L-Y, Bash R O, Baer R. Mammalian cells as hosts for two-hybrid studies of protein-protein interaction. In: Bartal P L, Fields S, editors. The yeast two-hybrid system. Oxford, United Kingdom: Oxford University Press; 1997. pp. 217–232. [Google Scholar]
- 53.Wang B Q, Kostrub C F, Finkelstein A, Burton Z F. Production of human RAP30 and RAP74 in bacterial cells. Protein Expression Purif. 1993;4:207–214. doi: 10.1006/prep.1993.1027. [DOI] [PubMed] [Google Scholar]
- 54.Wang Z, Roeder R G. Three human RNA polymerase III-specific subunits form a subcomplex and participate in specific transcription initiation. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 55.Watson A D, Edmondson D G, Bone J R, Mukai Y, Yu Y, Du W, Stillman D J, Roth S Y. Ssn6-tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 2000;14:2737–2744. doi: 10.1101/gad.829100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu L, Glass C K, Rosenfeld M G. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 57.Yu X, Wu L C, Bowcock A, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 58.Yuan C-X, Ito M, Fondell J D, Fu Z-Y, Roeder R G. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]