Abstract
Neuroblastoma is a primary malignancy mainly occurring in children. We have reported that polymorphisms of several N6-methyladenosine (m6A) RNA modification-related genes contributed to neuroblastoma risk in previous studies. YTHDF2, a “reader” of RNA m6A modification, is involved in cancer progression. Here, we estimated the association between a YTHDF2 gene rs3738067 A>G polymorphism and neuroblastoma susceptibility in 898 neuroblastoma patients and 1,734 healthy individuals from China. We found that the rs3738067 A>G could decrease neuroblastoma risk [AG vs. AA: adjusted odds ratio (OR) = 0.76, 95% confidence interval (CI) = 0.64–0.90, P = 0.002; AG/GG vs. AA: adjusted OR = 0.81, 95% CI = 0.69–0.95, P = 0.011). Besides, the rs3738067 AG/GG genotype was related to reduced neuroblastoma risk in the following subgroups: children aged 18 months and under, boys, patients with tumors originating from retroperitoneal, patients at clinical stage IV, and cases at clinical stages III plus IV. Importantly, false-positive report probability analysis proved our significant results worthy of close attention of. The expression quantitative trait locus analysis results revealed that the rs3738067 was associated with the expression of YTHDF2.
Keywords: neuroblastoma, YTHDF2, rs3738067, polymorphism, susceptibility
Introduction
Neuroblastoma is a prevalent malignancy originating from precursor cells of the sympathetic nervous system, and it mainly affects infants and children under 5 years of age (1). Neuroblastoma with high aggressiveness often progresses quickly, leading to a disappointing prognosis and high recurrence rate. Although some patients experience mild or no treatment exhibit spontaneous regression (2), more than half of patients with high-risk neuroblastoma die even with multimodality treatment (3). Due to the complex nature of the disease, the pathogenesis of neuroblastoma is still far from clear. Increasing evidence suggests that the gradual accumulation of adverse genetic alterations leads to the transformation of normal cells to cancer cells (4). Therefore, it is essential to uncover the detrimental genetic changes in neuroblastoma to screen for high-risk individuals and explore potentially effective treatment.
In recent years, researchers have achieved dramatic advancements in the genetic etiology of neuroblastoma (5). Of note, genome-wide association studies (GWASs) have emerged as a powerful tool for exploring the causal genetic mechanisms of human diseases, including tumors (6). Chromosome instability was considered as one of the major causes in neuroblastoma oncogenesis (7). Two studies demonstrated that neuroblastoma shares common DNA variants with malignant cutaneous melanoma (8) and congenital heart disease (9). Currently, some single-nucleotide polymorphisms (SNPs) related to neuroblastoma susceptibility have been identified by GWASs and studies with candidate gene strategy, including LMO1 (10, 11), METTL14 (12), PARP1 (13), MTHFR and VDR (14). Nevertheless, the genetic variations known presently are not sufficient to fulfill the genetic landscape in neuroblastoma.
N6-methyladenosine (m6A) is the most popular post-transcriptional modification of RNAs in eukaryotes, particularly in messenger RNAs (mRNAs) (15). RNA m6A modification is a dynamic and reversible process regulated by methyltransferases (known as writers) and demethylases (known as erasers). RNAs with m6A modifications can be recognized by some RNA binding proteins (named readers), which decide the different destinies of the modified RNA (16). As a member of the YTH domain family, YTHDF2 functions as an m6A reader to modulate the translation, location, and stability of targeted mRNA (17). Emerging evidence has suggested that dysregulated m6A modifications are tightly implicated in various diseases, especially cancers (18). Many studies have demonstrated the involvement of YTHDF2 in the regulation of m6A modified targets in cancer development (19). However, there are few reports about SNPs in the YTHDF2 gene and tumor risk.
We carried out a multi-center epidemiology study among Chinese children to analyze the association between the SNPs in the key m6A modification modulator gene YTHDF2 and neuroblastoma susceptibility.
Materials and Methods
Sample Selection
This work was conducted with the approval of the Institutional Review Board of Guangzhou Women and Children's Medical Center, with 898 neuroblastoma patients registered in eight hospitals (Guangzhou, Zhengzhou, Wenzhou, Xi'an, Taiyuan, Kunming, Changsha, Shenyang) in China and 1,734 age- and gender-matched healthy controls involved in previous studies (Supplementary Table S1) (20, 21). All participants have signed informed consent.
Polymorphism Selection and Genotyping
Only one potential functional SNP in the YTHDF2 gene (rs3738067 A>G) was chosen and genotyped in this study. Selection criteria and genotyping by TaqMan methodology were described previously (22, 23). The YTHDF2 gene rs3738067 A>G is located in transcription factor binding sites (TFBS) and might affect transcription activity as predicted by SNPinfo (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html).
Statistical Analysis
The Chi-square test was applied to measure the compliance of alleles at individual loci in controls with the Hardy-Weinberg equilibrium (HWE) and the differences of selected demographic variables between patients and controls. Logistic regression analyses determined crude or adjusted odds ratios (ORs, adjusted for age and gender) with respective 95% confidence intervals (CIs) to analyze the association of YTHDF2 gene polymorphism with neuroblastoma risk. False-positive report probability (FPRP) analysis was applied to estimate the deserving attention of YTHDF2 gene polymorphism in neuroblastoma as described before (12). In brief, three parameters were used to determine FPRP values, including statistical power, P-value, and prior probability representing a real association between the SNP and disease. We set 0.2 as an FPRP threshold and assigned a prior probability of 0.1 to detect an OR of 1.5 (for risk effects) or 0.67 (for protective effects) for the association of genotypes with neuroblastoma susceptibility. The association of the rs3738067 A>G with YTHDF2 expression was determined in the GTEx portal (https://www.gtexportal.org/home/) via eQTLs analysis. P < 0.05 was taken as statistically significant. Analyses were processed with SAS 9.1 (SAS Institute).
Results
Association of YTHDF2 rs3738067 A>G With Neuroblastoma Risk
The genotyping of YTHDF2 was successfully screened in 896 neuroblastoma patients and 1,733 controls. The genotype distribution of YTHDF2 rs3738067 A>G polymorphism and its relation to neuroblastoma susceptibility was indicated in Table 1. The frequency of the YTHDF2 rs3738067 A>G genotype coincided with HWE among the controls (HWE=0.359). The minor allele frequency (MAF) of YTHDF2 rs3738067 A>G polymorphism, was 0.2591 for the controls and 0.2410 for the cases. Based on the results of the single-locus analysis, we found that the G carriers of the rs3738067 were associated with decreased neuroblastoma risk (AG vs. AA: adjusted OR = 0.76, 95% CI = 0.64–0.90, P = 0.002; AG/GG vs. AA: adjusted OR = 0.81, 95% CI = 0.69–0.95, P = 0.011).
Table 1.
YTHDF2 rs3738067 A>G polymorphism and neuroblastoma susceptibility.
| Genotype | Cases (%) (N = 896) | Controls (%) (N = 1,733) | P a | Crude OR (95% CI) | P | Adjusted OR (95% CI)b | P b |
|---|---|---|---|---|---|---|---|
| rs3738067 (HWE=0.359) | |||||||
| AA | 535 (59.71) | 944 (54.47) | 1.00 | 1.00 | |||
| AG | 292 (32.59) | 680 (39.24) | 0.76 (0.64–0.90) | 0.002 | 0.76 (0.64–0.90) | 0.002 | |
| GG | 69 (7.70) | 109 (6.29) | 1.12 (0.81–1.54) | 0.498 | 1.12 (0.82–1.55) | 0.478 | |
| Additive | 0.134 | 0.91 (0.79–1.03) | 0.134 | 0.91 (0.80–1.03) | 0.143 | ||
| Dominant | 361 (40.29) | 789 (45.53) | 0.010 | 0.81 (0.69–0.95) | 0.010 | 0.81 (0.69–0.95) | 0.011 |
| Recessive | 827 (92.30) | 1,624 (93.71) | 0.172 | 1.24 (0.91–1.70) | 0.173 | 1.25 (0.91–1.71) | 0.165 |
OR, odds ratio; CI, confidence interval; HWE, Hardy-Weinberg equilibrium.
Values were in bold if the P-values <0.05 or the 95% CIs excluded 1.
χ2 test for genotype distributions between neuroblastoma cases and cancer-free controls.
Adjusted for age and gender.
Stratification Analysis
After that, we assessed the relation between YTHDF2 gene polymorphism and neuroblastoma susceptibility in subgroups classified via age, gender, sites of origins as well as clinical stages. As presented in Table 2, we detected that the rs3738067 AG/GG genotype carriers were linked to reduced neuroblastoma risk in subgroups of children with the age of 18 months and under (adjusted OR = 0.76, 95% CI = 0.58–0.98, P = 0.036), males (adjusted OR = 0.75, 95% CI = 0.60–0.93, P = 0.009), patients with retroperitoneal tumors (adjusted OR = 0.69, 95% CI = 0.54–0.88, P = 0.003), patients at clinical stage IV (adjusted OR = 0.68, 95% CI = 0.51–0.91, P = 0.009) and those at clinical stages III+IV (adjusted OR = 0.74, 95% CI = 0.59–0.93, P = 0.009).
Table 2.
Stratify analysis for YTHDF2 rs3738067 A>G polymorphism and neuroblastoma susceptibility.
| Variables | rs3738067 (cases/controls) | OR (95% CI) | P | AOR (95% CI)a | P a | |
|---|---|---|---|---|---|---|
| AA | AG/GG | |||||
| Age, month | ||||||
| ≤ 18 | 204/373 | 140/340 | 0.75 (0.58–0.98) | 0.033 | 0.76 (0.58–0.98) | 0.036 |
| >18 | 331/571 | 221/449 | 0.85 (0.69–1.05) | 0.128 | 0.86 (0.70–1.06) | 0.158 |
| Gender | ||||||
| Females | 238/415 | 168/329 | 0.89 (0.70–1.14) | 0.353 | 0.90 (0.71–1.15) | 0.411 |
| Males | 297/529 | 193/460 | 0.75 (0.60–0.93) | 0.010 | 0.75 (0.60–0.93) | 0.009 |
| Sites of origin | ||||||
| Adrenal gland | 141/944 | 107/789 | 0.91 (0.69–1.19) | 0.481 | 0.91 (0.69–1.19) | 0.477 |
| Retroperitoneal | 202/944 | 116/789 | 0.69 (0.54–0.88) | 0.003 | 0.69 (0.54–0.88) | 0.003 |
| Mediastinum | 121/944 | 92/789 | 0.91 (0.68–1.21) | 0.518 | 0.92 (0.69–1.22) | 0.552 |
| Others | 67/944 | 38/789 | 0.68 (0.45–1.02) | 0.063 | 0.68 (0.45–1.03) | 0.066 |
| Clinical stages | ||||||
| I | 179/944 | 130/789 | 0.87 (0.68–1.11) | 0.261 | 0.87 (0.68–1.12) | 0.275 |
| II | 91/944 | 69/789 | 0.91 (0.65–1.26) | 0.559 | 0.92 (0.66–1.27) | 0.601 |
| III | 98/944 | 65/789 | 0.79 (0.57–1.10) | 0.166 | 0.80 (0.58–1.11) | 0.186 |
| IV | 147/944 | 84/789 | 0.68 (0.52–0.91) | 0.009 | 0.68 (0.51–0.91) | 0.009 |
| 4 s | 14/944 | 4/789 | 0.34 (0.11–1.04) | 0.059 | 0.37 (0.12–1.15) | 0.085 |
| I+II+4 s | 270/944 | 199/789 | 0.88 (0.72–1.08) | 0.232 | 0.88 (0.72–1.09) | 0.239 |
| III+IV | 245/944 | 149/789 | 0.73 (0.58–0.91) | 0.006 | 0.74 (0.59–0.93) | 0.009 |
OR, odds ratio; CI, confidence interval; AOR, adjusted odds ratio.
Values were in bold if the P-values <0.05 or the 95% CIs excluded 1.
Adjusted for age and gender, omitting the corresponding stratify factor.
FPRP Analysis
An FPRP analysis was implemented to verify whether our significant findings deserve attentions. As shown in Table 3, the significant association for rs3738067 A>G (AG vs. AA, AG/GG vs. AA, males, retroperitoneal, clinical stage IV, and III+IV) remained noteworthy at the prior probability level of 0.1.
Table 3.
False-positive report probability analysis for significant findings for the association between YTHDF2 rs3738067 A>G polymorphism and neuroblastoma susceptibility.
| Genotype | Crude OR(95% CI) | P a | Statistical powerb | Prior probability | ||||
|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||||
| AG vs. AA | 0.76 (0.64–0.90) | 0.002 | 0.929 | 0.005 | 0.016 | 0.153 | 0.646 | 0.948 |
| AG/GG vs. AA | 0.81 (0.69–0.95) | 0.010 | 0.986 | 0.030 | 0.086 | 0.508 | 0.913 | 0.991 |
| ≤ 18 months | 0.75 (0.58–0.98) | 0.033 | 0.808 | 0.108 | 0.267 | 0.800 | 0.976 | 0.998 |
| Males | 0.75 (0.60–0.93) | 0.010 | 0.833 | 0.033 | 0.093 | 0.530 | 0.919 | 0.991 |
| Retroperitoneal | 0.69 (0.54–0.88) | 0.003 | 0.582 | 0.015 | 0.043 | 0.330 | 0.833 | 0.980 |
| Stage IV | 0.68 (0.52–0.91) | 0.009 | 0.561 | 0.045 | 0.124 | 0.608 | 0.940 | 0.994 |
| Stage III+IV | 0.73 (0.58–0.91) | 0.006 | 0.765 | 0.021 | 0.061 | 0.416 | 0.878 | 0.986 |
OR, odds ratio; CI, confidence interval.
The results were in bold if the false-positive report probability <0.200.
Chi-square test was used to calculate the genotype frequency distributions.
Statistical power was calculated using the number of observations in the subgroup and the OR and P-values in this table.
Effect of rs3738067 A>G on the Expression of YTHDF2
To confirm the functional relevance of rs3738067 A>G to the mRNA expression of YTHDF2, Cis-expression quantitative trait loci (eQTLs) analysis of the rs3738067 A>G and YTHDF2 expression was estimated using GTEx data. Results manifested that the rs3738067 A allele was related to increased YTHDF2 expression levels in the whole blood [Figure 1, P=1.9*10−5, normalized effect size (NES)=0.084].
Figure 1.
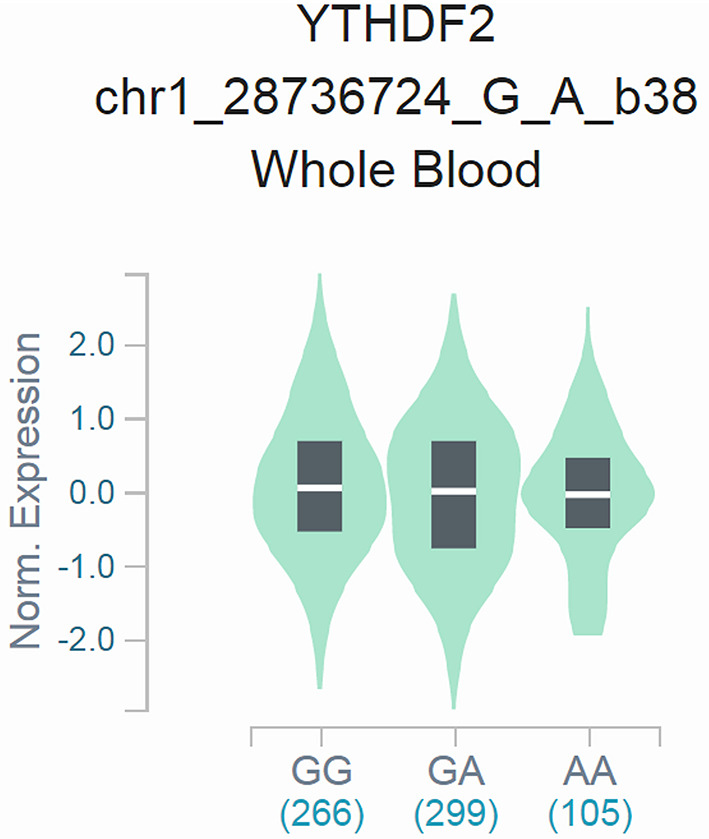
Functional relevance of rs3738067 A>G to YTHDF2 expression in GTEx database. The rs3738067 A allele had a significant association with YTHDF2 level alteration in the whole blood [P=1.9*10−5, normalized effect size (NES) = 0.084].
Discussion
Though many genetic variants linking to neuroblastoma susceptibility have been recognized, further efforts are needed to fully understand this disease's genetic landscape. The present work verified the YTHDF2 rs3738067 A>G could reduce neuroblastoma risk for the first time. YTHDF2 is an m6A modification “reader” which recognizes m6A-modified mRNAs to modulate the translation and stability of targeted mRNA (24). The roles of YTHDF2 in tumors are critical but controversial. For instance, As shown by Zhong et al., YTHDF2 restrained tumor cell growth in hepatocellular carcinoma (25). Shen et al. reported that YTHDF2 repressed cell growth in gastric cancer through modulating FOXC2 expression (26). In contrast, Li and colleagues disclosed that YTHDF2 promoted cell proliferation and migration in ovarian cancer (27). However, the role of YTHDF2 in neuroblastoma remains largely unknown.
Increasing evidence has indicated that genetic variations, including SNPs in m6A modification modulators, correlate closely with cancer progression (28). Also, a report has pointed that YTHDF2 rs3738067 A>G polymorphism exhibits a significant inverse association with glioma risk (29). Our study evaluated the association of the YTHDF2 gene SNP (rs3738067 A>G) with neuroblastoma susceptibility. The results showed that rs3738067 AG/GG genotype was related to reduced neuroblastoma risk in several subgroups, including children aged 18 months and under, males, patients with tumors originating from retroperitoneal, patients in clinical stages IV, and patients in clinical stages III+IV. We also performed FPRP tests to confirm if the obtained associations were noteworthy or not to provide further evidence of the reliability of our results. The association of the rs3738067 A>G with YTHDF2 expression was determined in the GTEx portal via eQTLs analysis. The integrative analyses of eQTL and SNP information may provide more understanding about the complex disease-modulating network (30). However, further studies are needed to substantiate the association between the rs3738067 A>G polymorphism and mRNA expression levels of YTHDF2.
There are several limitations to this study. First, only one SNP (rs3738067 A>G) in the YTHDF2 gene was evaluated. More studies will be performed to find other potential functional SNPs in the YTHDF2 gene. Second, only Chinese children were involved in this study. Thus, the results of this study may not be applicable to other ethnic groups. Moreover, as neuroblastoma is a multifactorial tumor, only genetic analysis is not enough to estimate neuroblastoma risk, and this study failed to incorporate environmental and genetic-environmental factors.
Our work for the first time verified the significant correlation of YTHDF2 gene rs3738067 A>G polymorphism with neuroblastoma risk, and this polymorphism is an intriguing locus for in-depth researches. However, the underlying biological mechanisms remain to be explored.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Board of Guangzhou Women and Children's Medical Center. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author Contributions
HZe, TY, and JH contributed to conception and design of the study. ML and JL organized the original data. JZhu, JC, YL, JZha, ZY, LL, HZh, and SL provided the clinical tissue and blood samples for the study. HX and YZ provided some technical guidance. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 82173593), Natural Science Foundation of Guangdong Province (No. 2019A1515010360), and the Major Science and Technology Special Project of Wenzhou (No. ZY2020021). The Natural science foundation of Guangdong Province (No. 2020A1515011569). Guangzhou Science and Technology Innovation Commission (No. 201607010395). Guangzhou Health science and Technology Project (No. 20201A010018). Guangzhou Health science and Technology Project (No. 20211A011033).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.797195/full#supplementary-material
References
- 1.Nakagawara A, Li Y, Izumi H, Muramori K, Inada H, Nishi M. Neuroblastoma. Jpn J Clin Oncol. (2018) 48:214–41. 10.1093/jjco/hyx176 [DOI] [PubMed] [Google Scholar]
- 2.Swift CC, Eklund MJ, Kraveka JM, Alazraki AL. Updates in diagnosis, management, and treatment of neuroblastoma. Radiographics. (2018) 38:566–80. 10.1148/rg.2018170132 [DOI] [PubMed] [Google Scholar]
- 3.Newman EA, Abdessalam S, Aldrink JH, Austin M, Heaton TE, Bruny J, et al. Update on neuroblastoma. J Pediatr Surg. (2019) 54:383–9. 10.1016/j.jpedsurg.2018.09.004 [DOI] [PubMed] [Google Scholar]
- 4.Ritenour LE, Randall MP, Bosse KR, Diskin SJ. Genetic susceptibility to neuroblastoma: current knowledge and future directions. Cell Tissue Res. (2018) 372:287–307. 10.1007/s00441-018-2820-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aygun N. Biological and genetic features of neuroblastoma and their clinical importance. Curr Pediatr Rev. (2018) 14:73–90. 10.2174/1573396314666180129101627 [DOI] [PubMed] [Google Scholar]
- 6.Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: current insights and future perspectives. Nat Rev Cancer. (2017) 17:692–704. 10.1038/nrc.2017.82 [DOI] [PubMed] [Google Scholar]
- 7.Tonini GP, Capasso M. Genetic predisposition and chromosome instability in neuroblastoma. Cancer Metastasis Rev. (2020) 39:275–85. 10.1007/s10555-020-09843-4 [DOI] [PubMed] [Google Scholar]
- 8.Avitabile M, Succoio M, Testori A, Cardinale A, Vaksman Z, Lasorsa VA, et al. Neural crest-derived tumor neuroblastoma and melanoma share 1p13.2 as susceptibility locus that shows a long-range interaction with the SLC16A1 gene. Carcinogenesis. (2020) 41:284–95. 10.1093/carcin/bgz153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Testori A, Lasorsa VA, Cimmino F, Cantalupo S, Cardinale A, Avitabile M, et al. Exploring shared susceptibility between two neural crest cells originating conditions: neuroblastoma and congenital heart disease. Genes. (2019) 10:663. 10.3390/genes10090663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oldridge DA, Wood AC, Weichert-Leahey N, Crimmins I, Sussman R, Winter C, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. (2015) 528:418–21. 10.1038/nature15540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He J, Zou Y, Wang T, Zhang R, Yang T, Zhu J, et al. Genetic variations of GWAS-identified genes and neuroblastoma susceptibility: a replication study in Southern Chinese children. Transl Oncol. (2017) 10:936–41. 10.1016/j.tranon.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuo Z, Lu H, Zhu J, Hua RX, Li Y, Yang Z, et al. METTL14 gene polymorphisms confer neuroblastoma susceptibility: an eight-center case-control study. Mol Ther Nucleic Acids. (2020) 22:17–26. 10.1016/j.omtn.2020.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avitabile M, Lasorsa VA, Cantalupo S, Cardinale A, Cimmino F, Montella A, et al. Association of PARP1 polymorphisms with response to chemotherapy in patients with high-risk neuroblastoma. J Cell Mol Med. (2020) 24:4072–81. 10.1111/jcmm.15058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olivera GG, Yanez Y, Gargallo P, Sendra L, Alino SF, Segura V, et al. MTHFR and VDR polymorphisms improve the prognostic value of MYCN status on overall survival in neuroblastoma patients. Int J Mol Sci. (2020) 21:2714. 10.3390/ijms21082714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. (2019) 20:608–24. 10.1038/s41580-019-0168-5 [DOI] [PubMed] [Google Scholar]
- 16.Sun W, Zhang B, Bie Q, Ma N, Liu N, Shao Z. The role of RNA methylation in regulating stem cell fate and function-focus on m(6)A. Stem Cells Int. (2021) 2021:8874360. 10.1155/2021/8874360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. (2019) 25:137–48.e6. 10.1016/j.stem.2019.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Zhang M, Ge S, Huang W, Lin X, Gao J, et al. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. (2019) 8:4766–81. 10.1002/cam4.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. (2018) 67:2254–70. 10.1002/hep.29683 [DOI] [PubMed] [Google Scholar]
- 20.Tang J, Lu H, Yang Z, Li L, Li L, Zhang J, et al. Associations between WTAP gene polymorphisms and neuroblastoma susceptibility in Chinese children. Transl Pediatr. (2021) 10:146–52. 10.21037/tp-20-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Cheng J, Li L, Li Y, Zhou H, Zhang J, et al. YTHDF1 gene polymorphisms and neuroblastoma susceptibility in Chinese children: an eight-center case-control study. J Cancer. (2021) 12:2465–71. 10.7150/jca.54496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Zhuo Z, Li L, Hua RX, Li L, Zhang J, et al. The contribution of YTHDF2 gene rs3738067 A>G to the Wilms tumor susceptibility. J Cancer. (2021) 12:6165–9. 10.7150/jca.62154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He J, Wang F, Zhu J, Zhang R, Yang T, Zou Y, et al. association of potentially functional variants in the XPG gene with neuroblastoma risk in a Chinese population. J Cell Mol Med. (2016) 20:1481–90. 10.1111/jcmm.12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, et al. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer. (2020) 19:91. 10.1186/s12943-020-01158-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. (2019) 442:252–61. 10.1016/j.canlet.2018.11.006 [DOI] [PubMed] [Google Scholar]
- 26.Shen X, Zhao K, Xu L, Cheng G, Zhu J, Gan L, et al. YTHDF2 inhibits gastric cancer cell growth by regulating FOXC2 signaling pathway. Front Genet. (2020) 11:592042. 10.3389/fgene.2020.592042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Wu L, Pei M, Zhang Y. YTHDF2, a protein repressed by miR-145, regulates proliferation, apoptosis, and migration in ovarian cancer cells. J Ovarian Res. (2020) 13:111. 10.1186/s13048-020-00717-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng Y, Li S, Gu D, Xu K, Du M, Zhu L, et al. Genetic variants in m6A modification genes are associated with colorectal cancer risk. Carcinogenesis. (2020) 41:8–17. 10.1093/carcin/bgz165 [DOI] [PubMed] [Google Scholar]
- 29.He J, Yuan L, Lin H, Lin A, Chen H, Luo A, et al. Genetic variants in m(6)A modification core genes are associated with glioma risk in Chinese children. Mol Ther Oncolytics. (2021) 20:199–208. 10.1016/j.omto.2020.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Cotarelo P, Gonzalez-Jimenez A, Agudo-Jimenez T, Abarca-Zabalia J, Aladro Y, Pilo B, et al. Genetic variation in NDFIP1 modifies the metabolic patterns in immune cells of multiple sclerosis patients. Sci Rep. (2021) 11:21371. 10.1038/s41598-021-00528-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.


