Abstract
Coronavirus disease 2019 (COVID-19), which started out as an outbreak of pneumonia, has now turned into a pandemic due to its rapid transmission. Besides developing a vaccine, rapid, accurate, and cost-effective diagnosis is essential for monitoring and combating the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its related variants on time with precision and accuracy. Currently, the gold standard for detection of SARS-CoV-2 is Reverse Transcription Polymerase Chain Reaction (RT-PCR), but it lacks accuracy, is time-consuming and cumbersome, and fails to detect multi-variant forms of the virus. Herein, we have summarized conventional diagnostic methods such as Chest-CT (Computed Tomography), RT-PCR, Loop Mediated Isothermal Amplification (LAMP), Reverse Transcription-LAMP (RT-LAMP), as well new modern diagnostics such as CRISPR–Cas-based assays, Surface Enhanced Raman Spectroscopy (SERS), Lateral Flow Assays (LFA), Graphene-Field Effect Transistor (GraFET), electrochemical sensors, immunosensors, antisense oligonucleotides (ASOs)-based assays, and microarrays for SARS-CoV-2 detection. This review will also provide an insight into an ongoing research and the possibility of developing more economical tools to tackle the COVID-19 pandemic.
Keywords: SARS-CoV-2, diagnostics, RT-PCR, LAMP, biosensors, point of care, CRISPR-Cas
Graphical Abstract.
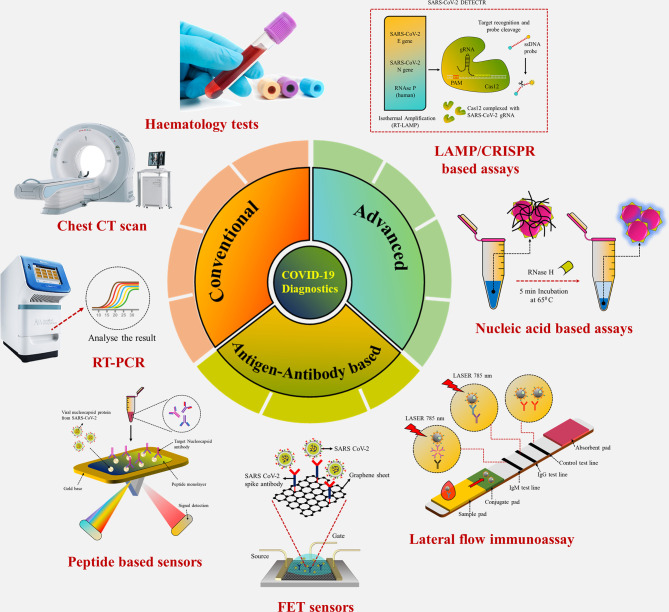
Conventional laboratory-based techniques for point-of-care detection of SARS-CoV-2.
Introduction
Coronavirus was discovered in Wuhan, China due to an outbreak of pneumonia in December 2019 (1–5) and the World Health Organization (WHO) gave it the nomenclature 2019 novel coronavirus (2019‐nCoV). Based on a study on coronavirus, the International Committee on Taxonomy of Viruses, on comparing the phylogeny and taxonomy, declared this virus similar to severe acute respiratory syndrome coronavirus (SARS‐CoV) and named it SARS‐CoV‐2 (6). SARS-CoV was first reported as early as 2002 in Guangdong, China (1), while Saudi Arabia reported cases of MERS-CoV in June 2012. As of June 2021, a total of 176 million COVID-19 confirmed cases have been reported around the world. Transmission of SARS-CoV was primarily through direct contact with an infected person and had overall a low transmittable rate. Therefore, in June 2003, the outbreak was primarily contained within households, with only a few cases of infection and mortality being recorded. However, in stark contrast, SARS-CoV-2, which surfaced in December 2019, has turned into a pandemic.
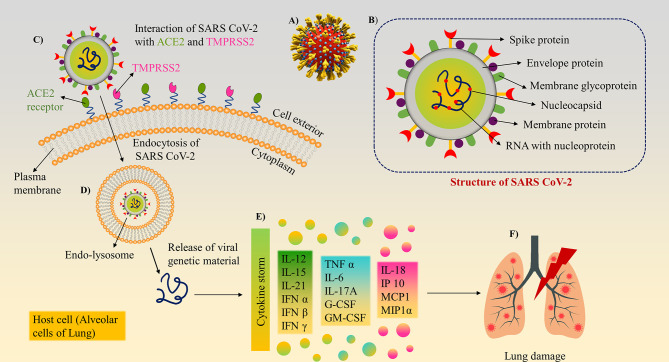
SARS CoV-2 is a beta-coronavirus with a crown-like appearance due to the presence of spike S1 glycoprotein on its surface (7). Members of this family cause hepatic, enteric, respiratory, and neurological diseases in humans as well as different animal species, including bats, cattle, cats, and camels (8). The complete description of human Coronaviridae family and Orthocoronavirinae subfamily have been listed in Table 1 . Coronavirus is a positive, single-stranded ribonucleic acid (RNA) (+ssRNA) virus (30 kb in length) that contains the following major structural proteins, i.e., spike (S), nucleocapsid (N), envelope (E), and membrane (M) protein as shown in Figures 1A, B (15).
Table 1.
Different genus, subgenus, species, and genome size of human Coronaviridae family and Orthocoronavirinae subfamily.
| Genus | Subgenus | Species (Abbreviations) | Genome Size | Reference |
|---|---|---|---|---|
| Alphacoronavirus | Duvinaco virus | Human coronavirus (HCoV-229E) | 27,317 | (9) |
| Setraco virus | Human coronavirus (HCoV-NL63) | 27,553 | (10) | |
| Betacorona virus | Embeco virus | Beta coronavirus (HCoV-OC43) | 30,738 | (11) |
| Human coronavirus (HCoV-HKU1) | 29,926 | (12) | ||
| Merbeco virus | Middle East respiratory syndrome-related coronavirus (MERSr-CoV) | 30,119 | (13) | |
| Sarbeco virus | Severe acute respiratory syndrome-related coronavirus (SARSr-CoV) | 29,751 | (14) |
Figure 1.
Structure and mechanism of action/pathogenesis of SARS-CoV-2: (A) 3D model of SARS-CoV-2 with surface spike glycoprotein. (B) Different structural and non-structural proteins of SARS-CoV-2, as a target biomarker for the development of diagnostics. (C) SARS-CoV-2 binds to ACE2 and Transmembrane protease serine 2 (TMPRSS2) receptors present on lung cells using its surface spike glycoprotein and internalised. (D) The virion further releases the viral genetic material into the cytoplasm. (E) Cytokine storm induction inside the cells. (F) Inflammatory response cause tissue/organ damage in the lungs.
Among the structural proteins, spike protein comprises two subunits, S1 and S2, to guide viral attachment, fusion, and entry. S1 corresponds to the large receptor binding domain (RBD), while S2 is helpful in the formation of the stalk of the spike protein. RBD present on S1 binds to the Angiotensin-converting enzyme 2 (ACE-2) receptor enabling entry of the virus and Transmembrane protease serine 2 (TMPRSS2) receptor enabling S protein priming (16), which in turn is essential for viral transmission and pathogenesis. Other than the spike protein, the most abundant M protein has three transmembrane domains, which help shape the virion. E proteins are primarily found in small quantities and facilitate the assembly and release of the virus. N protein is composed of two domains capable of binding to the RNA of the virus. Hemagglutinin-esterase (HE) is the fifth structural protein present in β-coronaviruses, which has been recently discovered. It acts as a hemagglutinin and binds to sialic acid, thus enhancing the S-protein-mediated cellular entry of the virus. SARS-CoV-2 predominantly affects the airways and the epithelial cells of the lungs (17) and causes massive apoptosis of the epithelial and endothelial cells (18). Viral infection and replication in the body causes aggressive inflammation, and acute lung injury, along with the secretion of a cytokine storm of interleukins (IL-1β, IL-10, and IL-4), interferon (IFN-γ), IFN-γ produced protein (IP-10), and monocyte chemoattractant protein (MCP-1), as shown in Figures 1C – F .
Currently, there are multiple vaccines that are under various stages of development and trials, with mass vaccination drives underway in most countries. However, the transmission and evolution rate of the virus is much faster than the rate of vaccinating the entire world’s population, and hence, quick diagnostics is essential to manage the ongoing pandemic. Currently, there are mainly three types of diagnostic techniques available, Chest Computed Tomography (CT), RT-PCR (gold standard), and lateral flow-based chromatographic strip (19). CT scan and RT-PCR are limited to larger hospitals, are significantly time-consuming, and call for the requirement of skilled professionals (20). Moreover, CT scan can only detect the symptoms of the disease, but it cannot identify the specific virus strain present on the basis of these symptoms unlike in the way artificial intelligence (AI) can (21). However, antibody-based detection is currently unreliable for detecting early and asymptomatic cases (22). Hence, antibody-based tests, along with RT-PCR are being used in a joint effort to gain quick, accurate, specific, and highly sensitive COVID-19 diagnostic techniques. Herein, sensitivity is the ability of an assay to identify individuals with the disease (true-positive rate) on the basis of the lowest quantity of the given analyte that the assay can detect, whereas specificity is the ability of the assay to correctly identify those without the disease (true-negative rate) by reducing cross-reactive results. Accuracy is the umbrella term for both sensitivity and specificity that relates to the ability of the assay to correctly discriminate between the diseased and healthy individual.
To overcome certain limitations posed by conventional techniques, the development of different types of biosensors/biochips have begun to replace the conventional methods (23). Various bio-recognition elements such as aptamers, nucleic acids, peptides, antigens (Ags), and antibodies (Abs), can be immobilized on the sensors for highly specific detection of various biomarkers (24). Deployment of nanomaterial, such as gold and silver nanoparticles, graphene, quantum dots, carbon nanotubes, and molecularly imprinted polymers can further enhance the sensitivity of the diagnostic assay (25–30). This review article focuses on giving a brief overview of the current detection methods and provides a base for further developing highly efficient portable sensors for SARS-CoV-2 detection.
Conventional Diagnostic Techniques
Currently, RT-PCR, CT scan, and hematology assays are the basic diagnostic tests for SARS-CoV-2 and are deemed as the gold standards. However, multiple serological assays are under development for quick diagnosis of COVID-19 but have the disadvantage of sensitivity as compared to the conventional techniques currently being used. However, it was observed that, when serological tests were combined with molecular tests, both sensitivity and specificity increased by several folds.
Chest CT Scan
CT scan involves taking serial x-ray images from various angles and combining them, following which a computer program creates a cross-section image of the lungs. It is used to detect pneumonia caused by COVID-19 infection by demonstrating typical radiographic features including consolidated multifocal patchy, ground-glass opacity, and/or interstitial differences distributed peripherally (31). Patients displaying clinical symptoms but negative RT-PCR results may be diagnosed by typical CT features. A positive correlation was found between C-reactive protein, lactate dehydrogenase, and sedimentation rate of erythrocytes with the severity of pneumonia as seen via CT (32). Deep learning (DL) extracts CT images and divides the lesion area into segments, splitting COVID-19 into 6 stages: healthy, ultra-early, early, rapid progression, consolidation, and dissipation (33). CT is preferred over other techniques due to its high resolution, efficacy, sensitivity, and its reliability in evaluating the severity of respiratory infection. Bilateral thickening and irregular pleural lines are some of the patterns observed in the case of COVID-19. A more advanced version being used to detect low levels of lung infection in COVID-19 patients is high-resolution computed tomography (HRCT). In contrast to helical CT, HRCT uses narrow beam collimation to capture thin slice images of the lung parenchyma and provides higher definition images of lung alveoli, airways, interstitium, and pulmonary vasculature. However, it is a costly technique, and available only in central hospitals. This technique also has other limitations—it fails to distinguish between different viruses, and is of no use in asymptomatic cases.
RT-PCR
The detection of trace amounts of gene transcripts via RT-PCR has made it the ideal choice for various contagious disease diagnostics (34, 35). Several laboratories have been developing diagnostic assays, but RT-PCR remains the most widely used detection technique for SARS-CoV-2 as it directly measures parts of the viral genome or viral transcripts instead of secondary biomarkers. The genes such as N, E, and RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 are used for targeted detection via RT-PCR (36). RT-PCR fundamentally involves 2 steps: (a) reverse transcription of genome RNA or individual viral transcripts to form complementary DNA (cDNA) using an enzyme; (b) cDNA amplification by PCR using specific primers and labeled with fluorescent hydrolysis probes ( Figure 2 ).
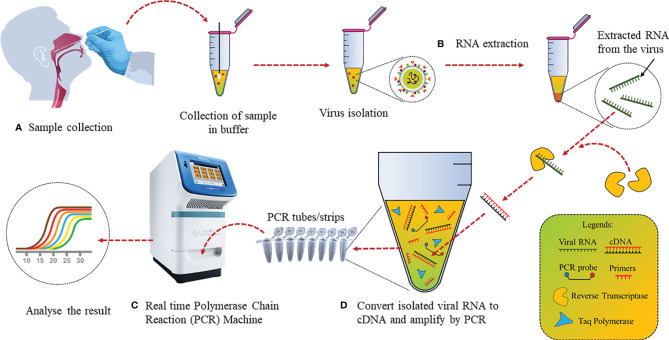
Figure 2.
Detection of SARS-CoV-2 by conventional RT-PCR: (A) Nasopharyngeal swab samples collection in media and virus isolation. (B) RNA extraction from isolated virus using an RNA extraction kit. (C) Conversion of RNA to cDNA using reverse transcriptase. (D) cDNA PCR amplification with specific primers and fluorescent hydrolysis probes followed by analysis.
cDNA templates are produced in Step 1 and the yield is increased by repeating the thermal cycles. This template is then used for Step 2. Gene-specific primers amplify the selected region of the cDNA template, whereas the probes give fluorescent signals every time specific gene regions are amplified, resulting in quantifiable data (37). On January 11, 2020, as soon as the entire genome sequence of SARS-CoV2 was uploaded, Malaysian Institute for Medical Research (IMR) developed RT-PCR primers and probes for the same, while several other countries developed RT-PCR kits. A list of primers used by different research groups have been listed in Table 2 .
Table 2.
Primers available for different target genes of SARS-CoV-2 for RT-PCR.
| Detection Method | Target Gene | Forward Primer | Reverse Primer | Reference |
|---|---|---|---|---|
| TaqMan RT-PCR | Nucleocapsid | TAATCAGACAAGGAACTGATTA | CGAAGGTGTGACTTCCATG | (38) |
| TaqMan RT-PCR | RdRp/Helicase | CGCATACAGTCTTRCAGGCT | GTGTGATGTTGAWATGACATGGTC | (39) |
| TaqMan RT-PCR | Spike | CCTACTAAATTAAATGATCTCTGCTTTACT | CAAGCTATAACGCAGCCTGTA | (39) |
| TaqMan RT-PCR | Nucleocapsid | GCGTTCTTCGGAATGTCG | TTGGATCTTTGTCATCCAATTTG | (39) |
| TaqMan RT-PCR | RdRp gene | GTGARATGGTCATGTGTGGCGG | CARATGTTAAASACACTATTAGCATA | (40) |
| TaqMan RT-PCR | Envelope | ACAGGTACGTTAATAGTTAATAGCGT | ATATTGCAGCAGTACGCACACA | (40) |
| TaqMan RT-PCR | Nucleocapsid | CACATTGGCACCCGCAATC | GAGGAACGAGAAGAGGCTTG | (40) |
| TaqMan RT-PCR | ORF1b | TGGGGYTTTACRGGTAACCT | AACRCGCTTAACAAAGCACTC | (40) |
| TaqMan RT-PCR | NSP2 | ATGCATTTGCATCAGAGGCT | TTGTTATAGCGGCCTTCTGT | (41) |
Recently, due to the increasing need for bulk sample testing, pooled sample testing for RT-PCR has also been approved. In 1943, this method of mixing several samples together in a “batch” or pooled sample was introduced by Dorfman and proved helpful in correctly identifying all infected individuals using fewer resources. RT-PCR being a robust technology with high sensitivity and specificity has already been used to test sample pools for human immunodeficiency virus and hepatitis B and C and is also now being employed for SARS-CoV-2. This is an effective method for large-scale screening of the population without wastage of resources and time. Even though RT-PCR is the most widely used SARS-CoV-2 diagnostic technique so far, it is time-consuming, and hence, the number of tests that can be carried out in a 24-h time period is low. This is a limiting factor that prevents reliable data of viral transmission to be collected in the entire population so as to gain an understanding of the world scenario. Due to low viral titer in the early days of the infection, RT-PCR may not be sensitive enough to detect the small amounts of RNA sample, leading to false-negative results. Therefore, multiple tests of different body fluids may be required for a confirmatory test, and a combination of RT-PCR with serological tests for confirmation.
Hematology Tests
Blood tests have played a vital role in disease diagnosis over the years, and hematology tests, which include the study of blood samples, have also been introduced for the diagnosis of SARS-CoV-2. In case of SARS-CoV-2, blood profiling of COVID-19-positive cases showed lymphopenia and increased lactate dehydrogenase (LDH), and these patients were critical in the intensive care unit (ICU). Patients admitted to the ICU had a higher nadir absolute lymphocyte count (ALC), nadir absolute monocyte count (AMC), nadir hemoglobin (Hb), higher peak absolute neutrophil count (ANC), and high LDH levels as compared to less critically ill patients who did not require ICU admission (42). Other hemagglutination tests (HAT) have also been performed to detect SARS-CoV-2 antibodies to study seroconversion in the population. These simple, inexpensive and rapid tests rely on the hemagglutination activity of the virus and the ability of specific antibodies to inhibit the virus from agglutinating the erythrocytes (43). Townsend and group have reported a HAT that shows 90% sensitivity and 99% specificity in the detection of Abs after RT-PCR. This is comparable to commercially available kits for receptor-binding domain (RBD) Ab detection, and can be applied as point-of-care testing (POCT) (44). Kruse et al. tested the possibility of using the red blood cell agglutination assay for SARS-CoV-2 Ab detection. They created a fusion protein consisting of RBD from spike protein and scFv targeting H antigen on red blood cells. The agglutination test was performed by mixing the fusion protein with red blood cells and the serum of a patient recovered from COVID-19 (specimen collected at least 28 days post symptoms and a negative PCR test upon discharge). Optimal agglutination was achieved at the highest fusion protein concentration, i.e., 100% within 5 min of incubation (45).
Novel Approaches for SARS-CoV-2 Diagnostics
Nucleic Acid Based Assays for SARS-CoV-2
RT-PCR is a cumbersome and time-consuming process, requiring both expensive equipment and trained personnel. Moreover, it does not have the screening capacity to keep pace with the rapidly growing pandemic. Hence, an alternative technique is the Loop-Mediated Isothermal Amplification (LAMP) method, which is highly similar to RT-PCR in terms of sensitivity and specificity, but with one exception, the nucleic acid amplification occurs at a single temperature (isothermal amplification), and hence, there is no need for a thermocycler, which, in turn, makes the process faster (46). In the LAMP process, amplification of the gene occurs via two types of loop region elongation reactions (i.e., self-elongation of template strand from the stem loop at 3’ terminals followed by binding and elongation of new primers to the loop region), which keep on repeating. Loopamp kit, based on the loop-mediated isothermal amplification (LAMP), was compared with RT-PCR for SARS-CoV-2 detection. A total of 76 nasopharyngeal samples showed 100% sensitivity, 97.6% specificity, and limit of detection (LOD) up to 1.0 × 101 copies/μl. Using four primers to detect six various regions in the RNA increased the sensitivity of this test (47). Reverse Transcription Loop-mediated isothermal amplification (RT-LAMP) method for SARS-CoV detection employed four primers to increase the assay sensitivity, and a fluorescent dye was used to determine the amplified product without any sample preparation (48). However, it needs a laboratory setup, and only one sample can be run at a time. RT-LAMP was done in a single 30-min step at 63°C and optical density (OD) was measured at 400 nm (with color changing to green from orange). The assay could identify N, E, and Open Reading Frame-1ab (ORF1ab) genes simultaneously with 92.3%, 98.5%, and 99% accuracy, respectively. All three genes were combined and amplified, and the results were correlated with RT-PCR test results on 208 clinical samples (20-fold dilutions) with LOD up to 1000 copies/ml. Another RT-LAMP assay could detect SARS-CoV-2 in 30 min (49). The GenBank MN908947 strain which showed high similarity to other SARS-CoV-2 strains but not with SARS-like bat coronavirus, was used to design six primers for RT-LAMP. To avoid tetraplex structure formation, which may interfere with RT-LAMP, the primers were made such that no four guanines came in a row. According to the gel electrophoresis data, 63°C for 30 min was the optimum condition for all six primers. The results matched with RT-PCR and LOD was approximately 1.02 fg. Various types of spiked samples, including urine, saliva, serum, nasopharyngeal, and oropharyngeal swabs, were used together with clinical urine, and plasma without any treatment. Urine samples showed more significant signals, and no cross-reactivity. When three colorimetric isothermal amplification methods [LAMP, cross-priming amplification (CPA), and polymerase spiral reaction (PSR)] were compared, LAMP showed the best results in naso- and oropharyngeal swabs with the highest sensitivity (43 copies) (50). LAMP or RT-LAMP based techniques are highly successful in SARS-CoV-2 detection. Since these techniques have certain practical limitations, these can be overcome using novel diagnostic methods such as CRISPR or antigen/antibody-based rapid detection kits.
The most widely applied diagnostic test for SARS-CoV-2, i.e., RT-PCR, is based on nucleic acid. Research is being carried out to develop rapid diagnostic sensor assays based on a similar principle of detecting viral nucleic acid. The fundamental basis of nucleic acid-based assays is the process of hybridization of a pair of nucleic acid, i.e., DNA/DNA or cDNA/RNA, based on their dependence as complementary sequences (51). They primarily make use of DNA, RNA, peptide nucleic acid (PNA), and aptamers (both DNA and RNA) as oligonucleotide probes. The basic principle of DNA, RNA, and PNA is based on sequence complementarity as per Chargaff’s rules of base pairing. However, in the case of aptamers, the principle is based more on receptor–ligand interactions. Table 3 compares different nucleic acid-based point-of-care diagnostic kits for SARS-CoV-2, which follow different principles such as RT-PCR, LAMP, and RT-LAMP.
Table 3.
Commercially available PoCT kits for SARS-CoV-2 detection with nucleic acid target analytes.
| Detection Method | Target Analyte | Sensitivity (%) | Specificity (%) | Detection Time | Reference |
|---|---|---|---|---|---|
| RCA with magnetic nanoparticles | Synthetic complementary DNA (RdRp) | N/A | N/A | <2 h | (52) |
| CRISPR-based LAMP with lateral flow assay | RNA (E, N genes) | 95 | 100 | <1 h | (53) |
| Real-time qRT-PCR | RNA (RdRp, E, N genes) | 100 | 100 | >4 h | (40) |
| Reverse transcription-LAMP | RNA | N/A | N/A | <1 h | (49) |
| LAMP with colorimetric readout | RNA | N/A | N/A | <1 h | (54) |
| Digital PCR | RNA | N/A | N/A | <1 h | (55) |
| Reverse transcription-LAMP | RNA (ORF1ab, S genes) | 100 | 100 | <1 h | (56) |
| Reverse transcription-LAMP | RNA | N/A | N/A | <1 h | (57) |
N/A, not available.
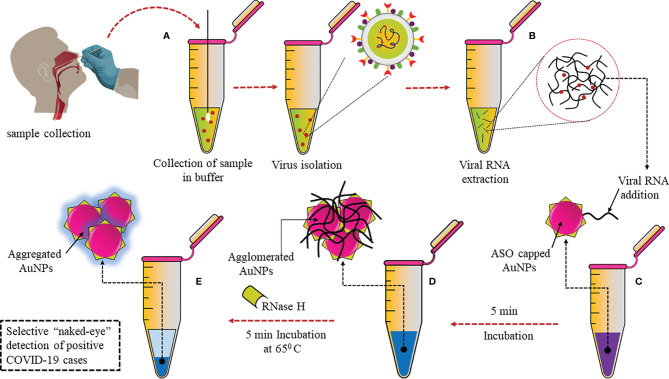
A plasmonic sensor with dual functions has been developed, combining localized surface plasmon resonance (LSPR) with plasmonic photothermal (PPT) effect. In this sensor, cDNA bioreceptors were functionalized onto two-dimensional gold nanoislands (AuNIs) to detect specific sequences of the SARS-CoV-2 genome through hybridization of nucleic acids. At its plasmonic resonance frequency, the AuNIs chip was illuminated, and thermoplasmonic heat was generated for better performance. This heat enhanced the temperature of in situ hybridization and ensured sensitive and specific differentiation between two gene sequences that may be similar. This dual-functional sensor had a very low LOD of 0.22 pM and was highly sensitive towards SARS-CoV-2, which allowed precise detection in a mixture of similar genes (58). Moitra et al. have developed a colorimetric assay that the naked eye can detect, as can be seen in Figure 3 . Thiolated antisense oligonucleotides (ASOs) specific for N-gene are functionalized onto gold nanoparticles (AuNPs). The thiolated ASO-AuNPs aggregate when the target RNA sequence (N-gene) is present and a change in the surface plasmon resonance (SPR) can be detected within 10 min in the RNA extracted from clinical samples. Furthermore, RNA was cleaved from the RNA–DNA conjugate by adding RNaseH, which resulted in precipitation that could be visually detected due to the aggregation of AuNPs. This developed assay had negligible cross-reactivity with MERS-CoV and LOD at 0.18 ng/μl (59).
Figure 3.
Selective naked eye SARS-CoV-2 detection: (A) Nasopharyngeal swab sample collected and virus isolated. (B) Viral RNA extraction. (C) Addition of AuNPs conjugated with thiolated ASOs specific for N gene. (D) Aggregation of ASO-AuNPs when target N-gene is present. (E) Addition of RNaseH to cleave RNA from RNA–DNA conjugate resulted in aggregation.
Peptide-Based Assays for SARS-CoV-2
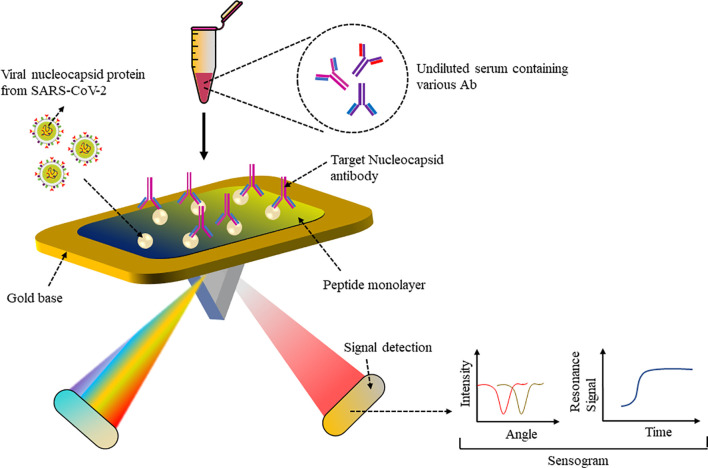
A biomimetic biosensor has an artificial bioreceptor component that can mimic natural bioreceptor activity and can be developed based on peptide bioreceptors (60). Numerous living organisms produce anti-microbial peptides (AMPs) to neutralize invading pathogens (61). For example, the AMP Cecropin-A from the Hyalophora cecropia moth, the first AMP to be isolated and characterized, shows inhibitory activity against herpes simplex virus 1 and 2 as well as human immunodeficiency virus 1. These short (mainly < 40 amino acids), amphipathic (contain both positively charged as well as hydrophobic residues), cationic proteins can have different secondary structures that can bind to receptors. Since they have a common building block, peptides with a specific sequence may be substituted for proteins in biological analysis. The advantage of using peptides is their smaller protein structure, making them more stable when coated on a biosensor and less prone to degradation rather than the entire large protein. Moreover, artificial peptides have various desirable properties such as chemical and structural versatility, high affinity to proteins or any analyte, standardized synthesis protocol and easy modification and hence make good bioreceptors for fabrication of biosensors. The surface plasmon resonance (SPR) sensor was able to detect nucleocapsid antibodies (Abs), which are specific for SARS-CoV-2. Abs expressed against SARS-CoV-2 N protein upon infection were detected in a number of patients immunized against SARS-CoV-2, which can be used to support vaccine research (62). As shown in Figure 4 , a peptide monolayer functionalized with recombinant N Ag was coated onto the SPR sensor, to detect anti-SARS-CoV-2 Abs in nM concentration. The assay was conducted on undiluted human serum and a portable SPR machine was used to detect the assay results, which were obtained within 15 min. This strategy enables further research on label-free rapid point-of-care testing for Abs.
Figure 4.
SPR-based sensor: Peptide monolayer functionalized with N protein coated onto gold base of SPR sensor. Addition of the clinical serum samples led to Ag–Ab interaction and change in SPR signal was recorded to detect SARS-CoV-2 Ab.
CRISPR-Based Diagnostics for SARS-CoV-2
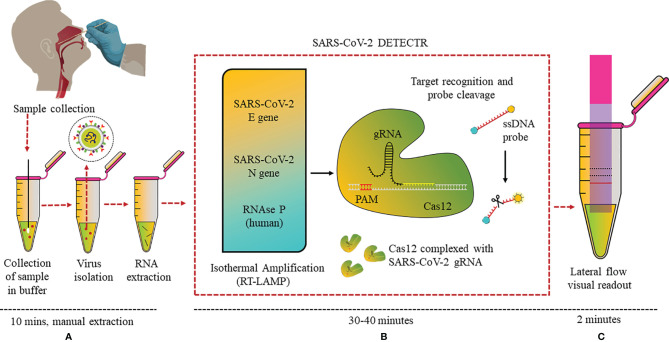
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technology has not only revolutionized genome editing, but also helped in the development of advanced diagnostic assays. CRISPR systems are an essential part of the adaptive immune system of microbes and has the ability to recognize foreign nucleic acids based on their sequence and remove them via endonuclease activity of the CRISPR-associated (Cas) enzyme, and this property has been made use of in the diagnostic field. CRISPR-Cas-based biosensors target a specific sequence in the genome, making use of CRISPR-Cas protein–RNA complex as the bioreceptor for specific recognition of the sequence (63). Combining RT-LAMP, CRISPR, and LFA in one assay, a detection kit was reported from the California Department of Public Health, University of California, Mammoth Biosciences Inc., and Abbott Viral Diagnostics and Discovery Inc. (53). This “SARS-CoV-2 DETECTR” assay extracted RNA from samples and, using RT-PCR (10 min), increased the copy numbers of RNAse P, N, and E genes (62°C for 20–30 min). CRISPR-Cas12 enzyme detected copies of the specific sequences of the genome and produced a fluorescence signal when the reporter dye was cleaved (37°C for 10 min) ( Figure 5 ).
Figure 5.
DETECTR LFA for SARS-CoV-2: (A) Nasopharyngeal swab viral RNA extracted from isolated virus to be used as the input sample or the sample matrix in the DETECTR assay. (B) RT-LAMP pre-amplification of E, N, and RNase P gene using ssDNA probes followed by Cas12-based detection. (C) Visual readout of the results on lateral flow strip by a fluorescence reader.
Eleven COVID-19 and 12 influenza nasopharyngeal swab samples were tested on this optimized assay. SARS-CoV-2 DETECTR proved to be 90% sensitive and 100% specific, and the results matched with that of the Centers for Disease Control and Prevention (CDC) RT-PCR standard assay. Moreover, this assay could specifically differentiate between SARS-CoV-2, SARS-CoV, and MERS-CoV with a single-nucleotide difference due to the specific primers and probe designed for N gene. The group is now focused on clinical validation of the kit for Emergency Use Authorisation (EUA) from the FDA. Similarly, Ding et al. have made an all-in-one dual CRISPR-Cas12a assay (AIOD-CRISPR) targeting the N protein gene and can detect even a few copies of the nucleic acids (DNA or RNA) (64). A PoC lateral flow diagnostic kit named FnCas9 Editor Linked Uniform Detection Assay (FELUDA) was developed using a Cas9 ortholog from Francisella novicida, for detection of single-nucleotide variants (SNV) in the nucleotide sequence of SARS-CoV-2 by the naked eye, which may be modified for diagnostics of various other diseases as well (65).
Antibody Based Detection for SARS-CoV-2
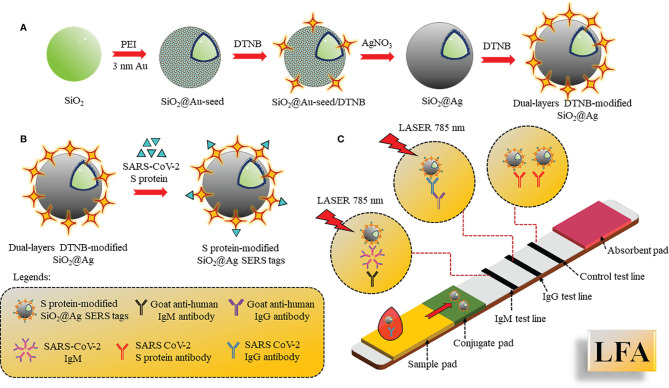
B lymphocytes of the adaptive immune system produce and secrete antibodies during the invasion of foreign pathogens. Since these antibodies are specifically produced against a specific antigen, the antibody–antigen reaction is highly specific and hence can be used as specific biomarkers for the diagnosis of diseases. Immune response against SARS-CoV-2 is triggered after a week that enhances production of immunoglobulins IgG/IgM in the blood to fight against the virus, and detection of these antibodies can help diagnose the infection. IgM is the first antibody to respond that helps eliminate the pathogens, while IgG provides the second, more robust, antibody-based immunity. As shown by Zhang et al., IgM and IgG can be detected after 5 days of infection by enzyme-linked immunosorbent assay (ELISA) (66). Pan and co-workers showed that the sensitivity of colloidal gold nanoparticle LFA for detection of IgG and IgM were 11.1%, 92.9%, and 96.8% within 7 days of infection, 8–14 days of infection, and after 14 days of infection, respectively (67). Corresponding to the above work, both IgG and IgM first appeared on the fourth day in confirmed cases, and positive detection of IgG and IgM were 3.6% and 11.1% respectively, in early stage. It was also observed that concentration of IgM stayed positive in approximately 75% intermediate and late-stage cases, whereas the IgG rate increased in the intermediate phase and rose to 96.8% in late-stage cases, leading to precise detection of IgG. Hence, detection of IgM and IgG in combination resulted in maximal testing efficacy, mainly during the intermediate stage. Furthermore, whole blood and plasma samples showed consistent results of 100% and 97.1% with excellent sensitivity in IgM and IgG assays (68, 69). A quick approach was investigated for the detection and targeting of viral IgG or IgM antibodies using colloidal gold-based immunochromatographic (ICG) assay and compared with RT-PCR (70). Label-free nanophotonic biosensor was developed, which can be integrated into small portable devices that could be given to different clinics (71). The LOD was approximately 0.2 pM, corresponding to 2 × 104 copies of SARS-CoV-2. A multiplexed grafting-coupled fluorescent plasmonics (GC-FP) biosensor was designed for sensitive and rapid detection of Abs in dried blood spots and serum of humans (72). This GC-FP technique could measure Ag–Ab interactions at multiple targets simultaneously in one sample with 100% sensitivity and selectivity. Upon measuring IgG levels in the serum against three antigens of COVID-19, N protein, spike S1S2, and spike S1 yielded linear and quantitative response with dilution as low as 1:1600, and the results produced corresponded with a Luminex-based microsphere immunoassay and ELISA. The efficacy of dried blood samples had 100% selectivity and 86.7% sensitivity in COVID-19 diagnostics. This test was further conducted to detect multiple Ig isotypes with sensitive detection of IgG, IgM, and IgA. Another widely used method is lateral flow immunoassay (LFA). LFA is a simple cellulose-based device and has been considered as one of the most popular PoCT devices to detect the Ab present in the blood. It consists of a sample pad and conjugate pad, embedded onto a nitrocellulose membrane. Sample (blood, cells, and antibodies) is loaded onto the sample pad and allowed to move to the conjugate pad, where the antibody binds to pre-immobilized AuNPs-conjugated antigen (SARS-CoV-2 spike), and continues to move to the test and control lines due to capillary action. Molecules such as fluorophores, latex nanoparticles, and gold nanoparticles are used for the colorimetric visualization. Surface Enhanced Raman Spectroscopy (SERS)-based lateral flow immunoassay (LFA) using dual-layered 5,5’-dithiobis-(2-nitrobenzoic acid) DTNB-modified SiO2@Ag NPs, and SARS-CoV-2 S protein-modified SiO2@Ag SERS tags ( Figure 6 ) were also reported to show ultra-sensitive and quick detection of IgM/IgG in clinical samples for diagnosis of COVID-19.
Figure 6.
SERS-based LFA: (A) Preparation of double-layered DTNB-SiO2@AgNPs by PEI-mediated absorption method followed by addition of DNTB. (B) S protein conjugation with SiO2@Ag SERS tags. (C) LFA assembled with S protein-SiO2@Ag SERS tags drop casted onto the conjugate pad and 2 test lines coated with IgM, IgG, and control line coated with S protein Ab. The sample was applied on the sample pad and SERS signal intensities were recorded.
The dual-layered DTNB-modified SiO2@AgNPs was synthesized by the absorption of 3 nm Au seed NPs on SiO2 mediated by polyethyleneimine (PEI) and the addition of 5,5’-dithio-bis-(2-nitrobenzoic acid) (DNTB) to fabricate SiO2@Au-seed/DTNB NPs, from which SiO2@Ag core-shell NPs synthesis was carried out using seed growth technique by AgNO3 reduction using formaldehyde. The second layer of DTNB was modified in such a way onto the SiO2@AgNPs surface that it formed dual-layered DTNB-loaded nanotags. S protein was further conjugated with SiO2@Ag SERS tags. The LFA was assembled with S protein-SiO2@Ag SERS tags drop cast onto the conjugate pad and two test lines coated with anti-human IgM, IgG, and the control line coated with S protein Ab. Different concentrations of IgM/IgG were analyzed simultaneously by analyzing the intensity of SERS signal on the test/control lines (73).
Antigen-Based Diagnostics for SARS-CoV-2
Targeting a specific antigen produced by a foreign pathogen either secreted in the circulatory system or on the surface of the pathogen is a highly specific method for detection of that pathogen. Since the results obtained are rapid, they are often used for a first laboratory investigation/screening in a suspected case. SARS-CoV-2 surface or secretory Ag can be detected in clinical samples within 2 days of infection, even when the viral load is low before the manifestation of symptoms, and hence it can help in early diagnosis, especially when compared to Ab-based detection, as antibodies develop later. For example, IgM can be detected in 5 days to 1 week and IgG after a week. A comparative Table 4 has been listed for Ag and Ab-based diagnostic tests for SARS-CoV-2.
Table 4.
Various antigen- and antibody-based diagnostic kits available for the detection of SARS-CoV-2.
| Type of POCT Kit | Target Analyte | Sensitivity (%) | Specificity (%) | Detection Time | Reference |
|---|---|---|---|---|---|
| Graphene field effect transistor (Gra-FET) | Antibodies | N/A | N/A | N/A | (74) |
| LFIA | IgM and IgG | 88.8 | 90.5 | 15 min | (22) |
| LFIA | IgM and IgG | 97.7 | N/A | N/A | (74) |
| Colloidal gold-based immune-chromatographic (ICG) strip | IgM or IgG | 96.7 | N/A | 15 min | (67) |
| Chemi-luminescence immunoassay | IgM and IgG (recombinant nucleocapsid) | 82.3 | 97.4 | 23 min | (75) |
| Immune-precipitation and parallel DNA sequencing | Antibody | 90–97 | ~98 | N/A | (76) |
| Colloidal Gold Immuno chromatographic assay (GICA) | Nucleoprotein | 57.8 | 99.6 | 15 min | (77) |
| GICA | Nucleoprotein | 30.3 | 100 | 15 min | (78) |
| Fluorescence immuno chromatographic assay (FICA) | SARS-CoV-2 antigen | 93.8 | 100 | <15 min | (79) |
| GICA | Nucleoprotein | 50.5 | 100 | N/A | (80) |
| Microfluidic FICA | SARS-CoV-2 antigen/IgG/IgM | N/A | N/A | <15min | (81) |
| Chemiluminescence Immunoassay (CLIA) | Nucleocapsid protein | 55.3 | 99.7 | 30 min | (82) |
| FICA | Nucleocapsid protein | 11.6 | N/A | 30 min | (83) |
| FICA | Nucleocapsid protein | 67.8 | 100 | 10 min | (84) |
| GICA | IgG and IgM | 86.88 | 99.40 | 5–10 min | (85) |
| GICA | IgG and IgM | 95.10 | 91.3 | 15 min | (86) |
| CLIA | IgG and IgM | 100 | N/A | N/A | (87) |
| CLIA | IgG and IgM for nucleocapsid protein | 81.54 | 96.62 | N/A | (88) |
| ELISA | IgG and IgM | 81.33 | N/A | N/A | (89) |
| ELISA | IgG and IgM for nucleocapsid and spike protein | 82.3 | N/A | N/A | (90) |
| ELISA | IgA, IgM, and IgG | 85.5 | N/A | N/A | (91) |
| ELISA | IgG, IgA against spike protein | N/A | N/A | N/A | (92) |
N/A, not available.
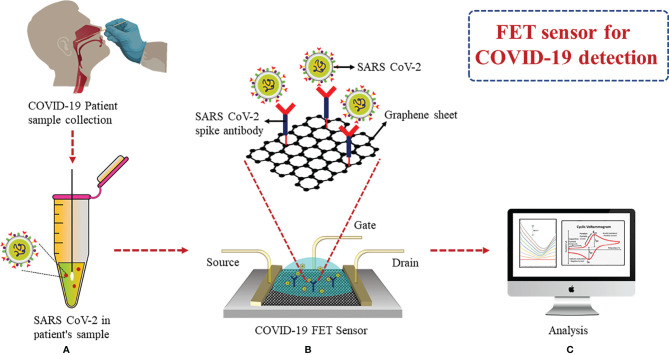
A FET biosensor using a graphene base was able to detect S protein and the complete virus in clinical samples. Spike Ab was immobilized onto the FET sensor via 1-pyrenebutyric acid N-hydroxysuccinimide ester for spike Ag detection ( Figure 7 ). According to the results, LOD was found to be 1 fg/ml when prepared in phosphate buffer saline (PBS) and 100 fg/ml in clinical transport medium. It was demonstrated that SARS-CoV-2 showed an LOD of 1.6 pfu/ml when in culture and 2.42 copies/ml in clinical samples. It was possible to differentiate between SARS-CoV-2 spike protein from MERS-CoV in nasopharyngeal swabs (in the transport medium) using this FET sensor (93). A similar graphene-based FET sensor was developed by Zhang et al., which detected the target biomarker Spike protein S1 containing the RBD within 2 min in real time without any labeling and showed a highly sensitive LOD of 0.2 pM (94).
Figure 7.
FET-based graphene sensor: (A) Collection of nasopharyngeal swab. (B) Immobilization of spike Ab via 1-pyrenebutyric acid N-hydroxysuccinimide ester on graphene FET sensor. (C) Analysis of binding events of Ag–Ab through electrical signals obtained for detection of spike Ag.
A nanostar and graphene-based electrochemical detection system was developed for SARS-CoV-2 in an aqueous solution, composed of Screen-Printed Carbon Electrode (SPCE). The SPCE working electrode was activated using graphene oxide (GO) and gold nanostars (AuNS), which can detect viruses in biological media (blood, saliva, and oropharyngeal/nasopharyngeal swab). The nanosensor demonstrated a superior LOD of 1.68 × 10−22 μg/ml in biological media, while blind testing of 100 clinical samples further proved the sensitivity and specificity (95). Mertens et al. developed a colloidal gold immunochromatographic assay where nucleoprotein MAbs-gold conjugate was coated on LFA. The Respi-strip was accurate by 82.6% and had overall sensitivity and specificity of 57.6% and 99.5%, respectively, and the results were obtained within 15 min on nasopharyngeal swab samples (77). A fluorescence immunochromatographic assay was developed for N protein detection in nasopharyngeal swab and urine samples in 10 min. The bioreceptor used on the test line was nucleocapsid MAb and carboxylate-modified polystyrene Europium (III) chelate microparticles conjugated with anti-N protein MAbs on the conjugate pad. In 73.6% of confirmed positive cases, the N Ag was detected in the urine on the same day (84). Another Fluorine Doped Tin Oxide (FTO)-based diagnostic electrode was fabricated to detect spike Ag in saliva samples. Spike Ab was immobilized on the electrode and FTO results were measured on a potentiostat. Under optimum conditions, the sensor showed a LOD of 0.63 fM and 120 fM in buffer and spiked saliva, respectively (96). Another low-cost electrochemical advanced diagnostic (LEAD) technology biosensor used inexpensive graphite leads modified with human angiotensin-converting enzyme 2 (ACE2) antigen as the receptor for on-site detection of SARS-CoV-2 Spike protein in clinical saliva and nasopharyngeal samples. The developed technology provided results within 6.5 min and showed an LOD of 229 fg/ml (97).
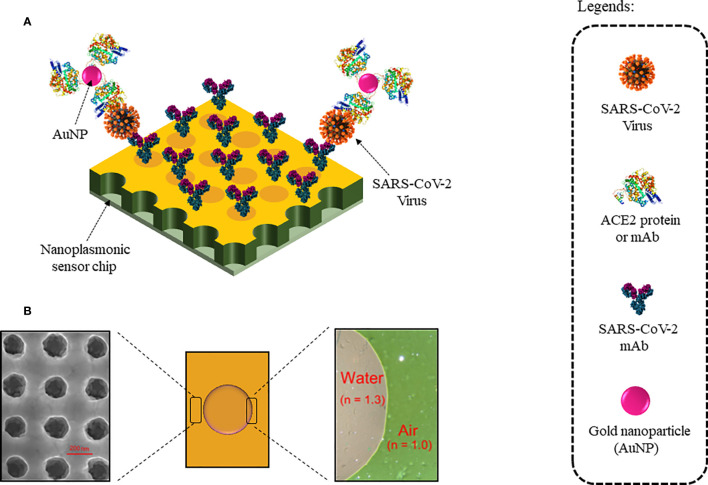
Nanoplasmonic sensors use nanoscale light below the diffraction limit (which typically is half the width of the wavelength of light being used to view the specimen) by converting free photons into localized charge-density oscillations (so-called surface plasmons) on noble-metal nanostructures, which serve as nanoscale analogs of radio antennas and are typically designed by using antenna theory concepts. One such nanoplasmonic resonance sensor coated with SARS-CoV-2 monoclonal antibodies (mAbs) was developed for a one-step direct optical rapid detection of SARS-CoV-2 virus particles, i.e., 15 min without any sample preparation with a LOD of 370 vp/ml as shown in Figure 8 . Measurements could be taken on both a classic microplate reader and a handheld smartphone connected to the device, making it an excellent POCT (98).
Figure 8.
Nanoplasmonic Sensor for label-free detection of COVID-19: (A) Sensor chip coated with SARS-CoV-2 mAbs. Colorimetric detection with AuNPs conjugated with ACE2 protein or mAb. (B) Au nanocup array chip with a drop of water on top. (Left) Zoomed SEM image of the nanocup array. (Right) TEM image showed air and water interfaces exhibiting green and far red pink colors, respectively.
As discussed above, conventional techniques, such as RT-PCR and a few commercial antibody detection assays, are the most prominently used diagnostic tests for the quick identification and immediate isolation of COVID-19 patients to prevent further transmission of COVID-19 infection. However, these assays have certain limitations, such as low specificity and sensitivity, elaborate sample preparation requirements that, in turn, demands skilled personnel, high response time, cumbersome protocols as most of them require a laboratory setup and equipment, and often expensive. The low viral load (less RNA) in asymptomatic patients and low levels of other potential biomarkers (e.g., Spike, Nucleocapsid, Envelope, and RdRp proteins) in body fluids for early screening of COVID-19 patients make point-of-care detection challenging. In comparison to the existing conventional techniques, recent detection methods such as antigen-based assays and peptide- and aptamer-based biosensors showed higher sensitivity, more specificity, and rapid detection time, as can be seen from the assays listed in Tables 3 and 4 , and hence, can potentially overcome the limitations. These assays can be used to develop PoCT devices for large-scale sensitive screening of populations for SARS-CoV-2 especially when the titer is low such as in the case of asymptomatic patients or early stages of the disease.
Conclusion
The knowledge gained from similar viral outbreaks of SARS-CoV and MERS-CoV in the past have led to the development of rapid diagnostics during the onset of SARS-CoV-2. Genome sequence identification of SARS-CoV-2 paved the way towards nucleic acid-based assays, which helped control the spread of COVID-19. The antibody and antigen-based methods provide further understanding, if compared with nucleic acid-based assays for COVID-19-infected patients. We have discussed the recent methods developed for SARS-CoV-2 diagnostics including its drawbacks. Currently, not much research exists in the development of peptide- and aptamer-based biosensors, which could lead to much higher sensitivity and specificity in SARS-CoV-2 detection. The diagnostic market has an immense potential to deal with the challenges associated with the existing methods for rapid and accurate detection at early stage of infection, which indeed is and must be the primary criteria. Keeping this view in our minds, we have emphasized on potential methods that are being used to control the transmission of SARS-CoV-2. However, these methods can further be employed for pandemic outbreaks in the future for swift and immediate control in hospitals, clinics, healthcare centers, and rural areas.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
The authors would like to acknowledge the support provided by Science and Engineering Research Board-Intensification of Research in High Priority Area (SERB-IRHPA) (Grant Number-IPA/2020/000069). RC and MH gratefully acknowledge the Slovenian Research Agency (ARRS) through the funding of programme P1-0143. AR would like to acknowledge DST-INSPIRE fellowship IF180729 sponsored by DST, New Delhi.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A Novel Coronavirus From Patients With Pneumonia in China. N Engl J Med (2020) 382:727–33. doi: 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A New Coronavirus Associated With Human Respiratory Disease in China. Nature (2020) 579:265–9. doi: 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen L, Liu W, Zhang Q, Xu K, Ye G, Wu W, et al. RNA Based mNGS Approach Identifies a Novel Human Coronavirus From Two Individual Pneumonia Cases in 2019 Wuhan Outbreak. Emerg Microbes Infect (2020) 9:313–9. doi: 10.1080/22221751.2020.1725399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ren L-L, Wang Y-M, Wu Z-Q, Xiang Z-C, Guo L, Xu T, et al. Identification of a Novel Coronavirus Causing Severe Pneumonia in Human. Chin Med J (Engl) (2020) 133:1015–24. doi: 10.1097/cm9.0000000000000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan JFW, Yuan S, Kok KH, To KKW, Chu H, Yang J, et al. A Familial Cluster of Pneumonia Associated With the 2019 Novel Coronavirus Indicating Person-to-Person Transmission: A Study of a Family Cluster. Lancet (2020) 395:514–23. doi: 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, et al. Severe Acute Respiratory Syndrome-Related Coronavirus: The Species and Its Viruses-a Statement of the Coronavirus Study Group. BioRxiv (2020). doi: 10.1101/2020.02.07.937862 [DOI] [Google Scholar]
- 7. Li F, Li W, Farzan M, Harrison SC. Structural Biology: Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed With Receptor. Science (2005) 309:1864–8. doi: 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- 8. Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, et al. Molecular Basis of Binding Between Novel Human Coronavirus MERS-CoV and Its Receptor CD26. Nature (2013) 500:227–31. doi: 10.1038/nature12328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thiel V, Herold J, Schelle B, Siddell SG. Infectious RNA Transcribed In Vitro From a cDNA Copy of the Human Coronavirus Genome Cloned in Vaccinia Virus. J Gen Virol (2001) 82:1273–81. doi: 10.1099/0022-1317-82-6-1273 [DOI] [PubMed] [Google Scholar]
- 10. Abdul-Rasool S, Fielding BC. Understanding Human Coronavirus HCoV-Nl63. Open Virol J (2010) 4:76–84. doi: 10.2174/1874357901004010076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. St-Jean JR, Jacomy H, Desforges M, Vabret A, Freymuth F, Talbot PJ. Human Respiratory Coronavirus OC43: Genetic Stability and Neuroinvasion. J Virol (2004) 78:8824–34. doi: 10.1128/jvi.78.16.8824-8834.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lau SKP, Woo PCY, Yip CCY, Tse H, Tsoi HW, Cheng VCC, et al. Coronavirus HKU1 and Other Coronavirus Infections in Hong Kong. J Clin Microbiol (2006) 44:2063–71. doi: 10.1128/JCM.02614-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cui J, Li F, Shi ZL. Origin and Evolution of Pathogenic Coronaviruses. Nat Rev Microbiol (2019) 17:181–92. doi: 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tratner I. SRAS : 1. Le Virus. Médecine/Sciences. Paris: EDP Sciences; Vol. 19. (2003). pp. 885–91. doi: 10.1051/medsci/20031989885. [DOI] [PubMed] [Google Scholar]
- 15. Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, et al. Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine. Cell Mol Immunol (2020) 17:613–20. doi: 10.1038/s41423-020-0400-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The Spike Protein of SARS-CoV - A Target for Vaccine and Therapeutic Development. Nat Rev Microbiol (2009) 7:226–36. doi: 10.1038/nrmicro2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen IY, Moriyama M, Chang MF, Ichinohe T. Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Front Microbiol (2019) 10:50. doi: 10.3389/fmicb.2019.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fu Y, Cheng Y, Wu Y. Understanding SARS-CoV-2-Mediated Inflammatory Responses: From Mechanisms to Potential Therapeutic Tools. Virol Sin (2020) 35:266–71. doi: 10.1007/s12250-020-00207-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cui F, Zhou HS. Diagnostic Methods and Potential Portable Biosensors for Coronavirus Disease 2019. Biosens Bioelectron (2020) 165:112349. doi: 10.1016/j.bios.2020.112349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaushik A, Khan R, Solanki P, Gandhi S, Gohel H, Mishra YK. From Nanosystems to a Biosensing Prototype for an Efficient Diagnostic: A Special Issue in Honor of Professor Bansi D. Malhotra Biosens (2021) 11:359–62. doi: 10.3390/bios11100359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al. Using Artificial Intelligence to Detect COVID-19 and Community-Acquired Pneumonia Based on Pulmonary CT: Evaluation of the Diagnostic Accuracy. Radiology (2020) 296:E65–71. doi: 10.1148/radiol.2020200905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, et al. Development and Clinical Application of a Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J Med Virol (2020) 92:1518–24. doi: 10.1002/jmv.25727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ahirwar R, Gandhi S, Komal K, Tripathi PP, Vyas MS, Kumar K, et al. Biochemical Composition, Transmission and Diagnosis of SARS-CoV-2. Biosci Rep (2021) 41:BSR20211238. doi: 10.1042/BSR20211238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roberts A, Chauhan N, Islam S, Mahari S, Ghawri B, Gandham RK, et al. Graphene Functionalized Field-Effect Transistors for Ultrasensitive Detection of Japanese Encephalitis and Avian Influenza Virus. Sci Rep (2020) 10:14546. doi: 10.1038/s41598-020-71591-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tey JN, Gandhi S, Wijaya IPM, Palaniappan A, Wei J, Rodriguez I, et al. Direct Detection of Heroin Metabolites Using a Competitive Immunoassay Based on a Carbon-Nanotube Liquid-Gated Field-Effect Transistor. Small (2010) 6:993–8. doi: 10.1002/smll.200902139 [DOI] [PubMed] [Google Scholar]
- 26. Gandhi S, Caplash N, Sharma P, Raman Suri C. Strip-Based Immunochromatographic Assay Using Specific Egg Yolk Antibodies for Rapid Detection of Morphine in Urine Samples. Biosens Bioelectron (2009) 25:502–5. doi: 10.1016/j.bios.2009.07.018 [DOI] [PubMed] [Google Scholar]
- 27. Pingarrón JM, Yáñez-Sedeño P, González-Cortés A. Gold Nanoparticle-Based Electrochemical Biosensors. Electrochim Acta (2008) 53:5848–66. doi: 10.1016/j.electacta.2008.03.005 [DOI] [Google Scholar]
- 28. Islam S, Shukla S, Bajpai VK, Han Y-K, Huh YS, Kumar A, et al. A Smart Nanosensor for the Detection of Human Immunodeficiency Virus and Associated Cardiovascular and Arthritis Diseases Using Functionalized Graphene-Based Transistors. Biosens Bioelectron (2019) 126:792–9. doi: 10.1016/j.bios.2018.11.041 [DOI] [PubMed] [Google Scholar]
- 29. Roberts A, Tripathi PP, Gandhi S. Graphene Nanosheets as an Electric Mediator for Ultrafast Sensing of Urokinase Plasminogen Activator Receptor-A Biomarker of Cancer. Biosens Bioelectron (2019) 141:111398. doi: 10.1016/j.bios.2019.111398 [DOI] [PubMed] [Google Scholar]
- 30. Shahdeo D, Roberts A, Abbineni N, Gandhi S. Graphene Based Sensors. In: Comprehensive Analytical Chemistry. Amsterdam: Elsevier B.V; (2020). p. 175–99. doi: 10.1016/bs.coac.2020.08.007 [DOI] [Google Scholar]
- 31. Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, et al. CT Imaging Features of 2019 Novel Coronavirus (2019-NCoV). Radiology (2020) 295:202–7. doi: 10.1148/radiol.2020200230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, et al. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology (2020) 296:32–40. doi: 10.1148/radiol.2020200642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y, Liu G, Zhang Q. Deep Learning and Medical Diagnosis. Lancet (2019) 394:1709–10. doi: 10.1016/S0140-6736(19)32501-2 [DOI] [PubMed] [Google Scholar]
- 34. Pabbaraju K, Wong S, Wong AA, Appleyard GD, Chui L, Pang XL, et al. Design and Validation of Real-Time Reverse Transcription-PCR Assays for Detection of Pandemic (H1N1) 2009 Virus. J Clin Microbiol (2009) 47:3454–60. doi: 10.1128/JCM.01103-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee MS, Chang PC, Shien JH, Cheng MC, Shieh HK. Identification and Subtyping of Avian Influenza Viruses by Reverse Transcription-PCR. J Virol Methods (2001) 97:13–22. doi: 10.1016/S0166-0934(01)00301-9 [DOI] [PubMed] [Google Scholar]
- 36. Xu L, Li D, Ramadan S, Li Y, Klein N. Facile Biosensors for Rapid Detection of COVID-19. Biosens Bioelectron (2020) 170:112673. doi: 10.1016/j.bios.2020.112673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nolan T, Hands RE, Bustin SA. Quantification of mRNA Using Real-Time RT-PCR. Nat Protoc (2006) 1:1559–82. doi: 10.1038/nprot.2006.236 [DOI] [PubMed] [Google Scholar]
- 38. Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, et al. Molecular Diagnosis of a Novel Coronavirus (2019-Ncov) Causing an Outbreak of Pneumonia. Clin Chem (2020) 66:549–55. doi: 10.1093/clinchem/hvaa029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chan JFW, Yip CCY, To KKW, Tang THC, Wong SCY, Leung KH, et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and With Clinical Specimens. J Clin Microbiol (2020) 58:e00310–20. doi: 10.1128/JCM.00310-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 Novel Coronavirus (2019-Ncov) by Real-Time RT-PCR. Euro Surveill (2020) 25:2000045–53. doi: 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yip CCY, Chan WM, Ip JD, Seng CWM, Leung KH, Poon RWS, et al. Nanopore Sequencing Reveals Novel Targets for Detection and Surveillance of Human and Avian Influenza a Viruses. J Clin Microbiol (2020) 58:1–10. doi: 10.1128/JCM.02127-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fan BE, Chong VCL, Chan SSW, Lim GH, Lim KGE, Tan GB, et al. Hematologic Parameters in Patients With COVID-19 Infection. Am J Hematol (2020) 95:131–4. doi: 10.1002/ajh.25774 [DOI] [PubMed] [Google Scholar]
- 43. Landolt GA, Townsend HGG, Lunn DP. Equine Influenza Infection. In: Equine Infectious Diseases, 2nd ed. Amsterdam: Elsevier Inc; (2013). p. 141–151.e7. doi: 10.1016/B978-1-4557-0891-8.00013-0 [DOI] [Google Scholar]
- 44. Townsend A, Rijal P, Xiao J, Kit Tan T, Huang K-YA, Huo J, et al. A Haemagglutination Test for Rapid Detection of Antibodies to SARS-CoV-2 1 2. medRxiv (2020). doi: 10.1101/2020.10.02.20205831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kruse R, Huang Y, Smetana H, Gehrie E, Amukele T, Tobian A, et al. A Rapid, Point of Care Red Blood Cell Agglutination Assay for Detecting Antibodies Against SARS-CoV-2. bioRxiv (2020). doi: 10.1101/2020.05.13.094490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thompson D, Lei Y. Mini Review: Recent Progress in RT-LAMP Enabled COVID-19 Detection. Sens Actuators Rep (2020) 2:100017. doi: 10.1016/j.snr.2020.100017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kitagawa Y, Orihara Y, Kawamura R, Imai K, Sakai J, Tarumoto N, et al. Evaluation of Rapid Diagnosis of Novel Coronavirus Disease (COVID-19) Using Loop-Mediated Isothermal Amplification. J Clin Virol (2020) 129:104446–9. doi: 10.1016/j.jcv.2020.104446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Park GS, Ku K, Baek SH, Kim SJ, Kim S, BT K, et al. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J Mol Diagn (2020) 22:729–35. doi: 10.1016/j.jmoldx.2020.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lamb LE, Bartolone SN, Ward E, Chancellor MB. Rapid Detection of Novel Coronavirus (COVID-19) by Reverse Transcription-Loop-Mediated Isothermal Amplification. medRxiv (2020). doi: 10.1101/2020.02.19.20025155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tran DH, Cuong HQ, Tran HT, Le UP, Do HDK, Bui LM, et al. A Comparative Study of Isothermal Nucleic Acid Amplification Methods for SARS-CoV-2 Detection at Point of Care. BioRxiv (2020). doi: 10.1101/2020.05.24.113423 [DOI] [Google Scholar]
- 51. Hahn S, Mergenthaler S, Zimmermann B, Holzgreve W. Nucleic Acid Based Biosensors: The Desires of the User. Bioelectrochem (2005) 67:151–4. doi: 10.1016/j.bioelechem.2004.07.006 [DOI] [PubMed] [Google Scholar]
- 52. Tian B, Gao F, Fock J, Dufva M, Hansen MF. Homogeneous Circle-to-Circle Amplification for Real-Time Optomagnetic Detection of SARS-CoV-2 RdRp Coding Sequence. Biosens Bioelectron (2020) 165:112356. doi: 10.1016/j.bios.2020.112356 [DOI] [PubMed] [Google Scholar]
- 53. Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR–Cas12-Based Detection of SARS-CoV-2. Nat Biotechnol (2020) 38:870–4. doi: 10.1038/s41587-020-0513-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang Y, Odiwuor N, Xiong J, Sun L, Nyaruaba RO, Wei H, et al. Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP. medRxiv (2020). doi: 10.1101/2020.02.26.20028373 [DOI] [Google Scholar]
- 55. Li Y, Xie Z, Lin W, Cai W, Wen C, Guan Y, et al. An Exploratory Randomized, Controlled Study on the Efficacy and Safety of Lopinavir/Ritonavir or Arbidol Treating Adult Patients Hospitalized With Mild/Moderate COVID-19 (ELACOI). medRxiv (2020). doi: 10.1101/2020.03.19.20038984 [DOI] [Google Scholar]
- 56. Eikenberry SE, Mancuso M, Iboi E, Phan T, Eikenberry K, Kuang Y, et al. To Mask or Not to Mask: Modeling the Potential for Face Mask Use by the General Public to Curtail the COVID-19 Pandemic. Infect Dis Model (2020) 5:293–308. doi: 10.1016/j.idm.2020.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kilic T, Weissleder R, Lee H. Molecular and Immunological Diagnostic Tests of COVID-19: Current Status and Challenges. iScience (2020) 23:101406. doi: 10.1016/j.isci.2020.101406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano (2020) 14:5268–77. doi: 10.1021/acsnano.0c02439 [DOI] [PubMed] [Google Scholar]
- 59. Moitra P, Alafeef M, Alafeef M, Alafeef M, Dighe K, Frieman MB, et al. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano (2020) 14:7617–27. doi: 10.1021/acsnano.0c03822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kesarwani V, Gupta R, Vetukuri RR, Kushwaha S, Gandhi S. Identification of Unique Peptides for SARS-CoV-2 Diagnostics and Vaccine Development by an In Silico Proteomics Approach. Front Immunol (2021) 12:725240. doi: 10.3389/fimmu.2021.725240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hoyos-Nogués M, Gil FJ, Mas-Moruno C. Antimicrobial Peptides: Powerful Biorecognition Elements to Detect Bacteria in Biosensing Technologies. Molecules (2018) 23:1683–707. doi: 10.3390/molecules23071683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Djaileb A, Charron B, Jodaylami MH, Thibault V, Coutu J, Stevenson K, et al. A Rapid and Quantitative Serum Test for SARS-CoV-2 Antibodies With Portable Surface Plasmon Resonance Sensing. ChemRxiv (2020). doi: 10.26434/CHEMRXIV.12118914.V1 [DOI] [Google Scholar]
- 63. Uygun ZO, Yeniay L, Gi̇rgi̇n Sağın F. CRISPR-Dcas9 Powered Impedimetric Biosensor for Label-Free Detection of Circulating Tumor DNAs. Anal Chim Acta (2020) 1121:35–41. doi: 10.1016/j.aca.2020.04.009 [DOI] [PubMed] [Google Scholar]
- 64. Ding X, Yin K, Li Z, Lalla RV, Ballesteros E, Sfeir MM, et al. Ultrasensitive and Visual Detection of SARS-CoV-2 Using All-in-One Dual CRISPR-Cas12a Assay. Nat Commun (2020) 11:4711–21. doi: 10.1038/s41467-020-18575-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Azhar M, Phutela R, Ansari AH, Sinha D, Sharma N, Kumar M, et al. Rapid, Field-Deployable Nucleobase Detection and Identification Using Fncas9. BioRxiv (2020). doi: 10.1101/2020.04.07.028167 [DOI] [Google Scholar]
- 66. Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B, et al. Molecular and Serological Investigation of 2019-Ncov Infected Patients: Implication of Multiple Shedding Routes. Emerg Microbes Infect (2020) 9:386–9. doi: 10.1080/22221751.2020.1729071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pan Y, Li X, Yang G, Fan J, Tang Y, Zhao J, et al. Serological Immunochromatographic Approach in Diagnosis With SARS-CoV-2 Infected COVID-19 Patients. J Infect (2020) 81:e28–32. doi: 10.1016/j.jinf.2020.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee HK, Lee BH, Seok SH, Baek MW, Lee HY, Kim DJ, et al. Production of Specific Antibodies Against SARS-Coronavirus Nucleocapsid Protein Without Cross Reactivity With Human Coronaviruses 229E and OC43. J Vet Sci (2010) 11:165–7. doi: 10.4142/jvs.2010.11.2.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhuoyue W, Xin Z, Xinge Y. IFA in Testing Specific Antibody of SARS Coronavirus. South China J Prev Med (2003) 29:36–7. [Google Scholar]
- 70. Huang L, Tian S, Zhao W, Liu K, Ma X, Guo J. Multiplexed Detection of Biomarkers in Lateral-Flow Immunoassays. Analyst (2020) 145:2828–40. doi: 10.1039/c9an02485a [DOI] [PubMed] [Google Scholar]
- 71. Soler M, Estevez MC, Cardenosa-Rubio M, Astua A, Lechuga LM. How Nanophotonic Label-Free Biosensors Can Contribute to Rapid and Massive Diagnostics of Respiratory Virus Infections: COVID-19 Case. ACS Sens (2020) 5:2663–78. doi: 10.1021/acssensors.0c01180 [DOI] [PubMed] [Google Scholar]
- 72. Cady NC, Tokranova N, Minor A, Nikvand N, Strle K, Lee WT, et al. Multiplexed Detection and Quantification of Human Antibody Response to COVID-19 Infection Using a Plasmon Enhanced Biosensor Platform. Biosens Bioelectron (2021) 171:112679. doi: 10.1016/j.bios.2020.112679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu H, Dai E, Xiao R, Zhou Z, Zhang M, Bai Z, et al. Development of a SERS-Based Lateral Flow Immunoassay for Rapid and Ultra-Sensitive Detection of Anti-SARS-CoV-2 IgM/IgG in Clinical Samples. Sens Actuators B Chem (2020) 329:129196. doi: 10.1016/j.snb.2020.129196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jin YH, Zhan QY, Peng ZY, Ren XQ, Yin XT, Cai L, et al. Chemoprophylaxis, Diagnosis, Treatments, and Discharge Management of COVID-19: An Evidence-Based Clinical Practice Guideline (Updated Version). Mil Med Res (2020) 7:41–74. doi: 10.1186/s40779-020-00270-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lin D, Liu L, Zhang M, Hu Y, Yang Q, Guo J, et al. Evaluations of Serological Test in the Diagnosis of 2019 Novel Coronavirus (SARS-CoV-2) Infections During the COVID-19 Outbreak. medRxiv (2020). doi: 10.1101/2020.03.27.20045153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xu GJ, Kula T, Xu Q, Li MZ, Vernon SD, Ndung’u T, et al. Comprehensive Serological Profiling of Human Populations Using a Synthetic Human Virome. Science (2015) 348:698–707. doi: 10.1126/science.aaa0698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mertens P, De Vos N, Martiny D, Jassoy C, Mirazimi A, Cuypers L, et al. Development and Potential Usefulness of the COVID-19 Ag Respi-Strip Diagnostic Assay in a Pandemic Context. Front Med (2020) 7:225. doi: 10.3389/fmed.2020.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Scohy A, Anantharajah A, Bodéus M, Kabamba-Mukadi B, Verroken A, Rodriguez-Villalobos H. Low Performance of Rapid Antigen Detection Test as Frontline Testing for COVID-19 Diagnosis. J Clin Virol (2020) 129:104455–65. doi: 10.1016/j.jcv.2020.104455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, Araos R, et al. Evaluation of a Novel Antigen-Based Rapid Detection Test for the Diagnosis of SARS-CoV-2 in Respiratory Samples. Int J Infect Dis (2020) 99:328–33. doi: 10.1016/j.ijid.2020.05.098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lambert-Niclot S, Cuffel A, Le Pape S, Vauloup-Fellous C, Morand-Joubert L, Roque-Afonso AM, et al. Evaluation of a Rapid Diagnostic Assay for Detection of Sars-Cov-2 Antigen in Nasopharyngeal Swabs. J Clin Microbiol (2020) 58:1–4. doi: 10.1128/JCM.00977-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lin Q, Wen D, Wu J, Liu L, Wu W, Fang X, et al. Microfluidic Immunoassays for Sensitive and Simultaneous Detection of IgG/IgM/Antigen of SARS-CoV-2 Within 15 Min. Anal Chem (2020) 92:9454–8. doi: 10.1021/acs.analchem.0c01635 [DOI] [PubMed] [Google Scholar]
- 82. Hirotsu Y, Maejima M, Shibusawa M, Nagakubo Y, Hosaka K, Amemiya K, et al. Comparison of Automated SARS-CoV-2 Antigen Test for COVID-19 Infection With Quantitative RT-PCR Using 313 Nasopharyngeal Swabs, Including From Seven Serially Followed Patients. Int J Infect Dis (2020) 99:397–402. doi: 10.1016/j.ijid.2020.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nagura-Ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, Mizuno T, et al. Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription-Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test to Diagnose COVID-19. J Clin Microbiol (2020) 58:1–27. doi: 10.1128/JCM.01438-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Diao B, Wen K, Chen J, Liu Y, Yuan Z, Han C, et al. Diagnosis of Acute Respiratory Syndrome Coronavirus 2 Infection by Detection of Nucleocapsid Protein. medRxiv (2020). doi: 10.1101/2020.03.07.20032524 [DOI] [Google Scholar]
- 85. Zhang P, Gao Q, Wang T, Ke Y, Mo F, Jia R, et al. Evaluation of Recombinant Nucleocapsid and Spike Proteins for Serological Diagnosis of Novel Coronavirus Disease 2019 (COVID-19). medRxiv (2020). doi: 10.1101/2020.03.17.20036954 [DOI] [Google Scholar]
- 86. Ying L, Yue-ping L, Bo D, Feifei R, Yue W, Jinya D, et al. Diagnostic Indexes of a Rapid IgG/IgM Combined Antibody Test for SARS-CoV-2. medRxiv (2020). doi: 10.1101/2020.03.26.20044883 [DOI] [Google Scholar]
- 87. Long QX, Deng HJ, Chen J, Hu JL, Liu BZ, Liao P, et al. Antibody Responses to SARS-CoV-2 in COVID-19 Patients: The Perspective Application of Serological Tests in Clinical Practice. medRxiv (2020). doi: 10.1101/2020.03.18.20038018 [DOI] [Google Scholar]
- 88. Zhang B, Zhou X, Zhu C, Song Y, Feng F, Qiu Y, et al. Immune Phenotyping Based on the Neutrophil-To-Lymphocyte Ratio and IgG Level Predicts Disease Severity and Outcome for Patients With COVID-19. Front Mol Biosci (2020) 7:157. doi: 10.3389/fmolb.2020.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liu L, Liu W, Zheng Y, Jiang X, Kou G, Ding J, et al. A Preliminary Study on Serological Assay for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in 238 Admitted Hospital Patients. Microbes Infect (2020) 22:206–11. doi: 10.1016/j.micinf.2020.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, et al. Evaluation of Nucleocapsid and Spike Protein-Based Enzyme-Linked Immunosorbent Assays for Detecting Antibodies Against SARS-CoV-2. J Clin Microbiol (2020) 58:1–25. doi: 10.1128/JCM.00461-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Okba NMA, Müller MA, Li W, Wang C, Geurtsvankessel CH, Corman VM, et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerg Infect Dis (2020) 26:1478–88. doi: 10.3201/eid2607.200841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19). Clin Infect Dis (2020) 71:778–85. doi: 10.1093/cid/ciaa310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Seo G, Lee G, Kim MJ, Baek S-H, Choi M, Ku KB, et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano (2020) 14:5135–51. doi: 10.1021/acsnano.0c02823 [DOI] [PubMed] [Google Scholar]
- 94. Zhang X, Qi Q, Jing Q, Ao S, Zhang Z, Ding M, et al. Electrical Probing of COVID-19 Spike Protein Receptor Binding Domain via a Graphene Field-Effect Transistor. arXiv (2020). [Google Scholar]
- 95. Hashemi SA, Golab Behbahan NG, Bahrani S, Mousavi SM, Gholami A, Ramakrishna S, et al. Ultra-Sensitive Viral Glycoprotein Detection NanoSystem Toward Accurate Tracing SARS-CoV-2 in Biological/Non-Biological Media. Biosens Bioelectron (2021) 171:112731. doi: 10.1016/j.bios.2020.112731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Roberts A, Mahari S, Shahdeo D, Gandhi S. Label-Free Detection of SARS-CoV-2 Spike S1 Antigen Triggered by Electroactive Gold Nanoparticles on Antibody Coated Fluorine-Doped Tin Oxide (FTO) Electrode. Anal Chim Acta (2021) 10:339207. doi: 10.1016/j.aca.2021.339207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. de Lima LF, Ferreira AL, Torres MDT, de Araujo WR, de la Fuente-Nunez C. Minute-Scale Detection of SARS-CoV-2 Using a Low-Cost Biosensor Composed of Pencil Graphite Electrodes. Proc Natl Acad Sci (2021) 118:e2106724118. doi: 10.1073/pnas.2106724118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Huang L, Ding L, Zhou J, Chen S, Chen F, Zhao C, et al. One-Step Rapid Quantification of SARS-CoV-2 Virus Particles via Low-Cost Nanoplasmonic Sensors in Generic Microplate Reader and Point-of-Care Device. Biosens Bioelectron (2021) 171:112685. doi: 10.1016/j.bios.2020.112685 [DOI] [PMC free article] [PubMed] [Google Scholar]