Abstract
Aims
We investigated the prognostic relevance of serpin peptidase inhibitor, clade A member 3 (SERPINA3) in patients admitted with a de novo or worsened heart failure (HF).
Methods and results
In the first stage, 83 HF‐related left ventricular (LV) transcripts were examined in patients with congestive cardiomyopathy (CCMP, n = 44) who died within 5 years and compared with age‐matched and haemodynamically matched CCMP survivors (n = 39) and controls with normal LV function (n = 17). Among 14 differentially expressed transcripts, myocardial gene and circulating SERPINA3 levels were up‐regulated in non‐survivors vs. survivors (2.40 ± 3.66 vs. 0.36 ± 0.22 units, P < 0.01 and 334.7 ± 138.7 vs. 228.2 ± 83.1 μg/mL, P < 0.01, respectively). While no significant transmyocardial gradient was detected, cytokine stimulation of human endothelial cells induced SERPINA3 secretion. In an independent validation cohort with a de novo or worsened HF (n = 387), circulating SERPINA3 levels > 316 μg/mL were associated with increased all‐cause mortality {hazard ratio [HR] [95% confidence interval (CI)]: 2.4 [1.5–3.9], P = 0.0002} and its composite with unplanned cardiovascular readmission [HR (95% CI): 2.0 (1.2–3.3), P = 0.004]. Patients with elevated SERPINA3 levels and elevated either N‐terminal pro brain natriuretic peptide or ST2 showed worse freedom from both endpoints. In a multivariate analysis, including established clinical risk factors, SERPINA3 remained independent predictor of all‐cause mortality together with age, gender, ST2, glomerular filtration, and pulmonary capillary wedge pressure.
Conclusion
In patients with a de novo or worsened HF, increased SERPINA3 levels > 316 μg/mL are associated with increased mortality or unplanned cardiac readmission. Elevated SERPINA3 levels on top of established clinical predictors appear to identify a subgroup of HF patients at higher mortality risk. Prospective studies should further validate its value in prognostic stratification of HF.
Keywords: Heart failure, Prognosis, Biomarkers, Inflammation, Cardiomyopathy
Introduction
Heart failure is a complex clinical syndrome, and its prognostic stratification remains challenging. 1 Several biomarkers have been introduced to aid the clinical guidance and prognostic stratification of heart failure patients. 2 , 3 , 4 , 5 Nevertheless, the prognostic value of individual biomarkers falls often short to address the prognostic stratification at individual patient level. 6 This may be related to the complex pathophysiology and heterogeneous nature of heart failure syndrome where a single marker may not yield optimal precision at the patient level. 7
Alternative approach in scoping relevant biomarkers could be based on comparative analyses of molecular fingerprints of well‐defined cohorts at index clinical and haemodynamic evaluation and known clinical outcomes. Here, we postulated that using the pre‐defined panel of transcripts involved in myocardial structural remodelling and function, we may identify most relevant heart failure biomarkers. Using this strategy, we identified differential expression of SERPINA3 in a well‐defined cohort of phenotypically matched survivors and non‐survivors with congestive cardiomyopathy. SERPINA3, also known as alpha‐1 antichymotrypsin, acts as an inhibitor of several serine proteases. Insufficient serpin regulation can cause excessive or prolonged cathepsin G activity, ultimately leading to tissue damage. 8 SERPINA3 in heart failure has been suggested by observing a decrease in plasma SERPINA3 levels in end‐stage heart failure patients with favourable remodelling under mechanical haemodynamic support. 9 Accordingly, we further investigated the potential prognostic value of SERPINA3 and report on its association with poor survival in patients with a de novo or worsened heart failure.
Methods
Patients
We studied a total of 487 patients in two separate cohorts. First, we selected 100 patients to address the differential myocardial gene expression related to survival in heart failure due to congestive idiopathic cardiomyopathy. They had no history of coronary artery disease or intervention, were free of significant coronary atherosclerosis at catheterization, and presented with severely reduced left ventricular (LV) ejection fraction. This discovery cohort included 44 patients <75 years who died over 5 years of follow‐up from index admission due to de novo heart failure. They were compared with 39 patients surviving the 5 year follow‐up matched for age, gender, LV ejection fraction, and end‐diastolic pressure at indexed invasive evaluation. In these patients, LV myocardial biopsies were collected as part of the routine diagnostic work‐up. Control myocardial biopsies were also obtained in 17 patients with normal LV function undergoing cardiac bypass surgery due to stable coronary artery disease (controls). In all patients, blood samples were available for laboratory analyses.
Second validation cohort comprised 387 heart failure patients admitted with a de novo or worsened heart failure in whom at least 3 year follow‐up data were obtained unless died. This cohort included consecutive hospitalized patients regardless the aetiology and heart failure phenotype. Blood samples from these patients were used for validation of biomarkers plasma levels. All patients gave informed consent and study protocol has been approved by the local ethical committee.
RNA preparation and quantitative RT‐PCR
Total RNA was extracted from the tissues using RNeasy Fibrous Tissue Mini Kit, from blood using QIAamp RNA Blood Mini Kit and from cells using miRNeasy Mini Kit (Qiagen, Venlo, Netherlands) following the manufacturer's instructions. Reverse transcription was performed with random primers using High‐Capacity cDNA Archive System (Applied Biosystems, Foster City, CA). Quantitative RT‐PCR was subsequently performed with TaqMan Gene Expression Assays using a 7500 real‐time PCR system (Applied Biosystems). Expression data were normalized to the housekeeping gene GAPDH in the same sample and expressed in arbitrary units. We compared gene transcripts of 83 transcripts involved in heart failure progression and implicated in pathophysiology of heart failure (Supporting Information, Table S1 ).
Circulating biomarkers and SERPINA3 plasma levels
Blood samples were centrifuged for 10 min at 4000 rpm within 30 min of collection. Plasma was extracted and stored at ≤ −20°C until analysis. Circulating SERPINA3 and ST2 levels were determined using a commercially available ELISA kit (Human Alpha 1‐Antichymotrypsin ELISA, Immunology Consultants Laboratory, Inc., USA and Presage ST2 Assay, Critical Diagnostics, CA, USA) according to the manufacturer's instructions. Serum N‐terminal pro brain natriuretic peptide (NT‐proBNP) levels were determined with an electrochemiluminescence immunoassay (Elecsys NT‐proBNP, Diagnostics Roche). Baseline sST2 value of >35 ng/mL has been accepted by US Food and Drug Administration as a predictor of worse prognosis [http://www.accessdata.fda.gov/cdrh_docs/reviews/k111452.pdf (Assessed 02 April 2017)] while the reference value for NT‐proBNP is set at 1000 pg/mL 10 to indicate risk of poor clinical outcome. Other biochemical and haematological parameters were measured by standard procedures.
Cultured cells
Human coronary artery endothelial cells and human umbilical vein endothelial cells were obtained from Lonza, and blood outgrowth endothelial cells were isolated in our lab from peripheral blood of heart failure patients. Cells were cultured in endothelial medium at passage 3–5. To examine SERPINA3 production in response to inflammatory cytokines, cells were stimulated with interleukin‐1ß (20 ng/mL) or tumour necrosis factor‐α (30 mg/mL) (R&D Systems, Minneapolis, MN). Culture supernatant was assayed for SERPINA3 1, 3, 5, 17, and 24 h after treatment.
Western blotting
Left ventricular myocardial biopsies or cells were lysed in RIPA protein lysis buffer containing PMSF and protease inhibition cocktail (Roche Diagnostics GmbH, Mannheim, Germany) using the Bead Ruptor 4 Mini Homogenizer (Omni International). Protein concentration was determined with a BCA protein assay kit (Thermo Scientific, Rockford, USA). Protein extracts were separated on a polyacrylamide gel and subsequently transferred to a PVDF membrane. The membrane was blocked in 5% non‐fat dry milk/TBS‐T buffer followed by overnight incubation at 4°C with primary antibody. After rinsing, the membrane was incubated with HRP conjugated secondary antibody and detected by use of the ECL detection kit (Bio‐Rad). Densitometric quantification of protein bands was performed with the Chemidoc Touch Imaging System (Bio‐Rad). Following antibodies were used: anti‐SERPINA3 rabbit monoclonal antibody (ab205197, Abcam, Cambridge), Akt antibody (#9272) (Cell Signaling Technologies, Massachusetts), and HRP conjugated goat anti‐rabbit IgG secondary antibody (TA1300233, Origene). GAPDH was used to normalize for different protein content using GAPDH antibody (sc‐47724) and mIgG κ BP‐HRP (sc‐516102) (Santa Cruz Biotechnology, Inc, Dallas, Texas).
Data and statistical analysis
Statistical analysis was performed using Software R 3.6.2. 11 Data are presented as mean ± SD for continuous variables and as percentages for categorical variables. We compared the median of the three groups with the Kruskal–Wallis test followed by Dunn's multiple comparison testing for differences in pairs. For two groups, data were analysed using Mann–Whitney t‐test. Univariate and multivariate linear regression were used to predict SERPINA3 plasma levels. A log10 transformation was applied to the right skewed outcome variable. Univariate parameter estimates with P < 0.1 significance level was considered further in a multivariate model. In case of high correlation between two possible predictors, two heart failure specialists determined by consensus the parameter to be used for the analysis. Following parameters were included in a univariate model: age, gender, left ventricle ejection fraction, LV end‐diastolic dimension, mean pulmonary artery pressure, mean pulmonary capillary wedge pressure, white blood cells count, haemoglobin, estimated glomerular filtration rate (eGFR), NT‐proBNP, ST2, and C‐reactive protein (CRP) plasma levels. Graphical assessment was used to check the normality assumption of the residuals.
To calculate the cut‐off value of log10 SERPINA3 levels for predicting the mortality, the restricted cubic spline curve of log10 SERPINA3 levels were plotted against the predicted hazard ratios (HRs) of the univariate Cox proportional hazard model. We dichotomized log10 SERPINA3 levels based on the value given by HR > 1.
Prognostic value of this cut‐off was further evaluated in predicting all‐cause mortality, its composite with unplanned cardiac readmission and unplanned cardiac readmission alone in competing risk with mortality. Cumulative survival curves were derived according to the Kaplan–Meier method, and differences between curves were analysed by log‐rank test. The Cox proportional hazard model was used to examine predictive role of SERPINA3 for outcomes. Model assumptions for proportionality of hazard were checked based on the cumulative residual approach. 12 The observed and simulated score processes were plotted assuming the underlying model has proportional hazard. The Kolmogorov–Smirnov test was used to test the proportional hazard.
Finally, the incremental prognostic value of SERPINA3 was evaluated in the multivariate baseline clinical model including clinical, laboratory or structural, and haemodynamic parameters. They included age, gender, ischaemic aetiology of heart failure, presence of diabetes, eGFR, haemoglobin, white blood cell count > 10 000, CRP, LV end‐diastolic diameter and ejection fraction at echocardiography, invasive mean pulmonary artery pressure, mean pulmonary capillary wedge (PCW) pressure together with NT‐proBNP > 1000 pg/mL and ST2 > 35 ng/mL levels. The incremental value was assessed by the net reclassification improvement (NRI) index, together with NRI's for ‘events’ and ‘non‐events’. The integrated discrimination improvement and the change under the receiver operating characteristic curve were evaluated. The nested models were compared based on likelihood ratio test. Concordance and Akaike information criteria measures were calculated. 13 All statistical tests were considered significant at the P < 0.05 level, except in the selection of possible univariate predictors to enter a multivariate model (P < 0.1).
Results
SERPINA3 in the exploratory cohort of CCMP survivors and non‐survivors
Clinical and haemodynamic characteristics of the discovery patient cohort are shown in Table S1 . Haemodynamic indices across the groups were consistent with the study design. Among 83 transcripts related to heart failure (Table S2 ), 14 were up‐regulated in surviving and non‐surviving patients compared with controls (Table S3 ). Only SERPINA3 (Figure 1 A ), ST2 and GDF‐15 were up‐regulated in non‐survivors vs. survivors. Similar to myocardial expression analyses, plasma SERPINA3 levels were significantly higher in CCMP non‐survivors vs. survivors or controls (334.7 ± 138.7 vs. 228.2 ± 83.1 and 175.9 ± 27.8 μg/mL respectively, Figure 1 B ). Myocardial SERPINA3 gene expression showed significant correlation with serum levels (r = 0.28, P = 0.006). Its gene expression could not be detected in peripheral blood cells (data not shown).
Figure 1.
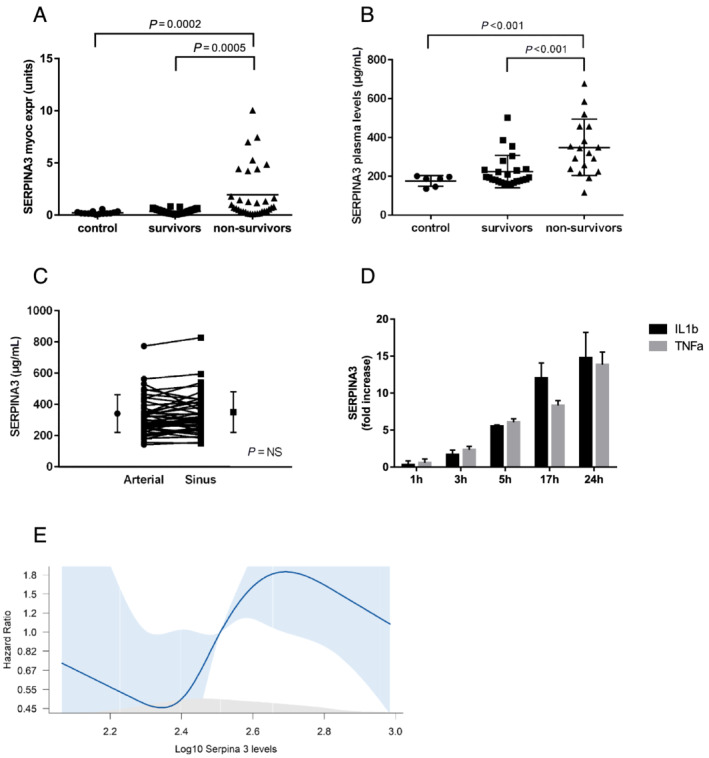
SERPINA3 expression levels. Panel (A) shows SERPINA3 myocardial expression in surviving and non‐surviving heart failure patients compared with control patients. Panel (B) shows SERPINA3 plasma levels in surviving and non‐surviving heart failure patients compared with control patients. Panel (C) shows transmyocardial gradient of SERPINA3 as a difference between coronary sinus and arterial levels. Panel (D) shows fold increase in SERPINA3 secretion by human coronary artery endothelial cells after IL‐1β or TNF‐α stimulation compared with unstimulated cells (data represent four experiments). Panel (E) shows spline curve analysis to determine the optimal SERPINA3 cut‐off value for all‐cause mortality.
To address whether increased plasma SERPINA3 levels are related to myocardial production, we determined SERPINA3 myocardial gradients from a difference between the coronary sinus and arterial blood concentration in 25 non‐survivors and 25 survivors. Arterial and coronary sinus levels of SERPINA3 were similar (313.3 ± 118.8 and 323.3 ± 122.6 μg/mL, P = NS) arguing against significant myocardial gradient (Figure 1 C ).
We examined whether endothelial cells are responsive to inflammatory stimulation and able to secret SERPINA3. Stimulation of endothelial cell subtypes including human coronary artery endothelial cells (Figure 1 D ), human umbilical vein cells or blood outgrowth endothelial cells (Figure S1 A) by interleukin‐1β or tumour necrosis factor‐α led to a significant SERPINA3 release. This secretory response was associated with both SERPINA3 gene and protein expression in the cells (Figure S1 B). SERPINA3 could be blocked by Brefeldin A indicating its secretion through the ER/Golgi secretory pathway (Figure S1 C).
SERPINA3 levels in the validation heart failure cohort
Clinical relevance of elevated SERPINA3 levels was validated in an independent cohort of 387 patients admitted due to de novo or worsened heart failure. In this validation cohort, 80 patients died up to 12 year follow‐up. Characteristics of the study population are in Table 1 . There were no significant differences in their risk factors, prevalence of female gender, or medications except higher use of diuretics in non‐survivors. Both survivors and non‐survivors had similar LV volumes and LV function with similar prevalence of heart failure with preserved ejection fraction. Right‐sided and left‐sided filling pressures were higher in non‐survivors vs. survivors. Plasma levels of ST2, NT‐proBNP, and CRP were also significantly higher in non‐survivors vs. survivors. Serum liver enzymes and ferritin levels were comparable between both groups. Cholesterol levels were lower in non‐survivors.
Table 1.
Baseline characteristics of patients in the validation cohort of all comers with HF
| Validation cohort (N = 387) | |||
|---|---|---|---|
| Total (N = 387) | Survivors (N = 307) | Non‐survivors (N = 80) | |
| Demography and risk factors | |||
| Age (years) at sampling | 59.56 ± 16.01 | 56.93 ± 15.95 | 69.64 ± 11.75**** |
| Male/female (%) | 69/31 | 68/32 | 70/30 |
| BMI (kg/m2) | 26.2 ± 4.5 | 26.2 ± 4.5 | 26.0 ± 4.3 |
| Hyperlipidaemia | 233/387 (60.2%) | 179/307 (58.3%) | 54/80 (67.5%) |
| AHT | 214/387 (55.3%) | 173/307 (56.4%) | 41/80 (51.3%) |
| CAD | 101/387 (26.1%) | 68/307 (22.2%) | 33/80 (41.3%) |
| Diabetes mellitus | 88/387 (22.7%) | 62/307 (20.2%) | 26/80 (32.5%) |
| Echocardiography | |||
| LVEDD (mm) | 58.2 ± 11.3 | 58.4 ± 11.2 | 57.8 ± 11.7 |
| LVESD (mm) | 47.1 ± 12.9 | 47.5 ± 12.9 | 45.7 ± 13.0 |
| LVEF (%) | 39.9 ± 18.0 | 40.2 ± 17.9 | 38.8 ± 18.2 |
| HFpEF (%) | 40.6% | 38.8% | 46.2% |
| LVEDP (mmHg) | 17.3 ± 7.8 | 16.9 ± 7.8 | 18.8 ± 7.5 |
| LVESP (mmHg) | 117.3 ± 24.2 | 117.2 ± 24.2 | 117.7 ± 24.4 |
| LVdevP (mmHg) | 100.1 ± 24.3 | 100.4 ± 24.2 | 98.7 ± 24.6 |
| LVEDVI (mL/m2) | 108.2 ± 42.9 | 107.4 ± 42.2 | 111.4 ± 46.1 |
| LVESVI (mL/m2) | 68.0 ± 41.4 | 67.0 ± 40.5 | 72.6 ± 45.0 |
| LV mass index (g/m2) | 122.7 ± 43.1 | 118.5 ± 38.0 | 137.9 ± 55.6 |
| AP mean (mmHg) | 24.1 ± 9.7 | 23.4 ± 9.8 | 26.7 ± 9.2** |
| RV syst (mmHg) | 38.2 ± 12.4 | 37.1 ± 12.0 | 42.5 ± 13.0** |
| RV diast (mmHg) | 8.4 ± 5.1 | 8.0 ± 4.8 | 9.8 ± 6.0* |
| Mean PCWP (mmHg) | 15.3 ± 8.1 | 14.5 ± 8.0 | 18.0 ± 7.9*** |
| TAPSE (mm) | 18.0 ± 5.0 | 18.0 ± 5.0 | 18.1 ± 5.0 |
| Laboratory parameters | |||
| WBC (n/μL) | 7986 ± 2435 | 7938 ± 2295 | 8166 ± 2918 |
| Haemoglobin (g/dL) | 13.7 ± 1.9 | 13.9 ± 1.9 | 12.8 ± 2.0**** |
| eGFR (mL/min) | 66.9 ± 19.3 | 70.6 ± 16.6 | 53.8 ± 22.3**** |
| SGPT (ALT) (U/L) | 40.1 ± 52.5 | 39.6 ± 47.1 | 42.0 ± 69.3 |
| SGOT (AST) (U/L) | 34.6 ± 46.3 | 33.3 ± 44.5 | 39.2 ± 52.7 |
| Fe (μg/dL) | 81.3 ± 42.1 | 84.1 ± 42.7 | 72.1 ± 39.2 |
| sFerritin (μg/L) | 286.4 ± 322.9 | 285.9 ± 344.0 | 288.3 ± 225.5 |
| sCHOL (mg/dL) | 169.4 ± 43.9 | 173.5 ± 43.4 | 153.8 ± 42.7*** |
| ST2 (ng/mL) | 42.1 ± 38.1 | 39.0 ± 34.9 | 54.2 ± 46.9**** |
| CRP (mg/L) | 16.1 ± 40.3 | 15.8 ± 43.3 | 17.1 ± 25.7**** |
| NT‐proBNP (ng/L) | 4018 ± 5946 | 3205 ± 4920 | 7181 ± 8187**** |
| hsTnT (ng/L) | 92.1 ± 186.6 | 91.6 ± 199.1 | 94.8 ± 96.8**** |
| SERPINA3 (μg/mL) | 375.8 ± 186.8 | 364.0 ± 185.8 | 421.0 ± 184.6*** |
| Medication | |||
| Beta‐blockers | 266/387 (68.7%) | 214/307 (69.7%) | 52/80 (65.0%) |
| ACE/ARB | 249/387 (64.3%) | 199/307 (64.8%) | 50/80 (62.5%) |
| Mineralkortikoid blocker | 263/387 (68.0%) | 203/307 (66.1%) | 60/80 (75.0%) |
| Diuretics | 216/387 (55.8%) | 159/307 (51.8%) | 57/80 (71.2%) |
AHT, arterial hypertension; AP, pulmonary artery pressure; BMI, body mass index; CAD, coronary artery disease; CRP, C‐reactive protein; eGFR, estimated glomerular filtration rate; Fe, iron; HF, heart failure; HFpEF, heart failure with preserved ejection fraction; hsTnT, high sensitivity troponin T; LVdevP, left ventricular developed pressure; LVEDD, left ventricular end‐diastolic diameter; LVEDP, left ventricular end‐diastolic pressure; LVEDVI, left ventricular end‐diastolic volume index; LVEF, left ventricle ejection fraction; LVESD, left ventricular end systolic diameter; LVESP, left ventricular end systolic pressure; LVESVI, left ventricular end systolic volume index; LV Mass Index, left ventricular mass index; NT‐proBNP, N‐terminal pro‐B‐type natriuretic peptide; PCWP, pulmonary capillary wedge pressure; RV diast, right ventricle diastolic pressure; RV syst, right ventricle systolic pressure; sCHOL, serum cholesterol; SGOT, serum glutamic oxaloacetic transaminase; SGPT, serum glutamic pyruvic transaminase; ST2, suppression of tumorigenicity; TAPSE, tricuspid annular plane systolic excursion; WBC, white blood cell count.
P < 0.05,
P < 0.01,
P < 0.001,
P < 0.0001 vs. survivors.
We sought to examine whether laboratory, echocardiographic, and haemodynamic parameters could predict SERPINA3 plasma levels. By using linear regression analysis, none of the echocardiographic or invasive haemodynamic parameters were related to circulating SERPINA3 levels. In contrast, white blood cell counts, haemoglobin, CRP, NT‐proBNP, and ST2 plasma levels emerged as univariate predictors of the log10 transformed SERPINA3 blood levels. All these predictors, except white blood cell counts, remained independent predictors in the multivariate model. Yet this multivariate model yielded only limited prediction precision (adjusted R 2 multivariate model: 0.279) and could not sufficiently account for extent of the circulating SERPINA3 levels.
SERPINA3 and clinical outcomes
Median level of SERPINA3 in the validation cohort was 323 μg/mL (range 116.2–1389.8 μg/mL). In the spline curve analysis, log10 SERPINA3 level of 2.5 corresponding to plasma levels of 316 μg/mL emerged as optimal cut‐off value for mortality (Figure 1 E ).
As shown in Figure 2 A , patients with SERPINA3 levels > 316 μg/mL had a significantly worse survival {HR [95% confidence interval (CI)]: 2.4 [1.5–3.9], P = 0.0002}. As the median follow‐up in the validation cohort was 41 months and number of patients at risk declined under 100 in each group beyond this time point, further analyses describe the relationship between SERPINA3 levels and clinical outcomes by 3 year follow‐up. Survival in patients with SERPINA3 levels > 316 μg/mL was significantly worse as compared with those with levels ≤ 316 μg/mL [Figure 2 B : HR (95% CI): 2.3 (1.1–4.8), P = 0.024]. While freedom from unplanned cardiac readmissions tended to be worse in patients with high SERPINA3 levels [Figure 2 C : HR (95% CI): 1.7 (0.9–3.1), P = 0.078], the freedom from composite of all‐cause mortality and unplanned cardiac readmissions was significantly lower compared with patients with lower levels [Figure 2 D : HR (95% CI): 2 (1.2–3.3), P = 0.004].
Figure 2.
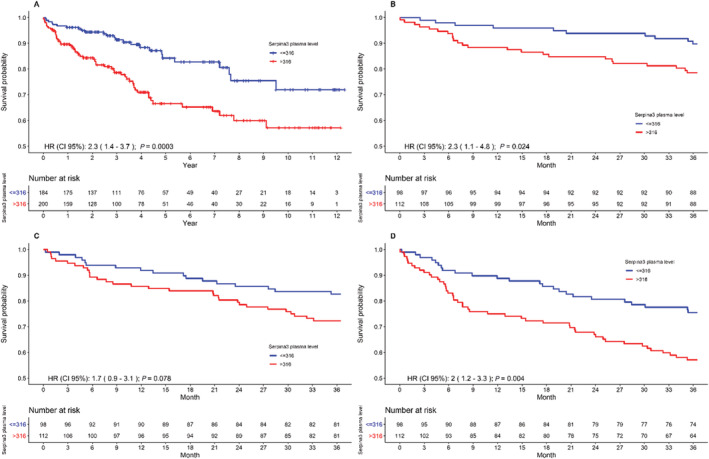
Relationship between SERPINA3 levels and clinical outcome determined from Kaplan–Meier curves. Panel (A) shows Kaplan–Meier curves of all‐cause mortality through the entire follow‐up. Panel (B) shows Kaplan–Meier curves of all‐cause mortality at 3 year follow‐up. Panel (C) shows Kaplan–Meier curves for first unplanned cardiac readmission at 3 year follow‐up. Panel (D) shows Kaplan–Meier curves of the composite endpoint of survival and first unplanned cardiac readmission at 3 year follow‐up.
In further analyses, we explored the added value of SERPINA3 levels to established biomarkers, ST2 and NT‐proBNP on mortality and its composite with unplanned cardiac readmission. The survival rate of patients with elevated SERPINA3 > 316 μg/mL and elevated NT‐proBNP > 1000 pg/mL was lower as compared with other combinations of both biomarkers (Figure 3 A , log‐rank test: P = 0.037). The composite of all‐cause mortality and first unplanned cardiac readmission was also worse in patients with elevated levels of both markers as compared with other combinations (Figure 3 B ). Freedom from this composite endpoint declined to 71% at 1 year follow‐up in these patients compared with 84% in patients with low SERPINA3 levels ≤ 316 μg/mL and elevated NT‐proBNP levels > 1000 pg/mL (risk ratio = 1.87; P = 0.058).
Figure 3.
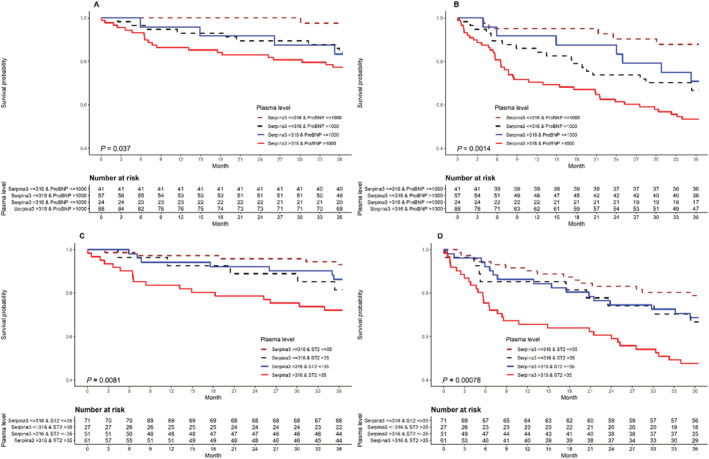
Relationship between SERPINA3 levels and clinical outcome in combination with NT‐proBNP or ST2. All‐cause mortality is shown in panels (A) (SERPINA3 and NT‐proBNP) and (C) (SERPINA3 and ST2) while the composite endpoint of survival and first unplanned cardiac readmission is shown in panels (B) (SERPINA3 and NT‐proBNP) and (D) (SERPINA3 and ST2).
Elevated SERPINA3 with elevated ST2 > 35 ng/mL was also associated with worse survival (Figure 3C, log‐rank test P = 0.0081) as well as its composite with first unplanned cardiac readmission (Figure 3 D , log‐rank test P = 0.00078) as compared with other combinations of both biomarkers. Freedom from both clinical outcomes declined rapidly to 63.6% at 1 year follow‐up in patients with high SERPINA3 and ST2 levels compared with 85.2% in patients with elevated ST2 and low SERPINA3 levels (risk ratio: 1.57; P = 0.059).
A subcohort of patients with elevated SERPINA3 as well as both NT‐proBNP and ST2 showed worse survival (Figure S2 A) and lower freedom from composite of all‐cause mortality and unplanned cardiac readmission at 1 year follow‐up (Figure S2 B) as compared with other combination of all biomarkers.
Prognostic value of SERPINA3 in addition to established risk factors and biomarkers
Cox proportional hazard models were calculated to evaluate the predictive value of the SERPINA3 plasma levels in addition to other clinical parameters for all‐cause mortality, its composite with unplanned cardiac readmission, and unplanned cardiac readmission alone with mortality as competing risk. Of the 387 patients, 81 were deleted due to missing covariates. Analyses were performed using the remaining 306 observations.
Table 2 shows univariate and multivariate predictors of all‐cause mortality by this model. Elevated SERPINA3 emerged as independent predictor of mortality in addition to elevated ST2, eGFR, and mean PCW pressure in a multivariate analysis.
Table 2.
Univariate and multivariate Cox proportional hazard models for overall mortality
| Univariate models | P value | Final multivariate model | P value | |
|---|---|---|---|---|
| Exp. coefficient (95% CI) | Exp. coefficient (95% CI) | |||
| Demographics | ||||
| Age (years) | 1.07 (1.05–1.09) | <0.001 | 1.06 (1.04–1.08) | <0.01 |
| Gender (male) | 1.00 (0.62–1.61) | >0.05 | ||
| Echocardiography | ||||
| LVEDD (mm) | 1.02 (0.99–1.05) | >0.05 | ||
| LVEF (%) | 1,00 (0.99–1.01) | >0.05 | ||
| Invasive haemodynamics | ||||
| AP mean (mmHg) | 1.02 (1.00–1.04) | <0.05 | ||
| Mean PCWP (mmHg) | 1.03 (1.01–1.06) | <0.001 | 1.03 (1.01–1.06) | <0.05 |
| Laboratory | ||||
| WBC count > 10 000/μL | 1.62 (0.94–2.80) | <0.1 | ||
| CRP (mg/L) | 1.00 (0.99–1.01) | >0.05 | ||
| NT‐proBNP >1000 pg/mL | 3.16 (1.56–6.39) | <0.01 | ||
| ST2 > 35 ng/mL | 2.20 (1.41–3.43) | <0.001 | 2.03 (1.26–3.28) | <0.01 |
| SERPINA3 > 316 μg/mL | 2.32 (1.44–3.72) | <0.001 | 1.70 (1.03–2.81) | <0.05 |
| eGFR (mL/min) | 0.96 (0.95–0.97) | <0.001 | 0.98 (0.97–0.99) | <0.01 |
| Haemoglobin (g/dL) | 0.79 (0.71–0.88) | <0.001 | ||
| Other | ||||
| Ischaemic aetiology | 2.13 (1.37–3.33) | <0.001 | ||
| Diabetes | 2.04 (1.22–3.42) | <0.01 | ||
AP, pulmonary artery pressure; CRP, C‐reactive protein; eGFR, estimated glomerular filtration rate; LVEDD, left ventricular end‐diastolic diameter; LVEF, left ventricle ejection fraction; NT‐proBNP, N‐terminal pro‐B‐type natriuretic peptide; PCWP, pulmonary capillary wedge pressure; WBC, white blood cell count.
When comparing the final multivariate model with SERPINA3 (c.statistic: 0.77; AIC: 685) and without SERPINA3 (c.statistic: 0.76; AIC: 688), adding increased SERPINA3 to the risk set significantly improved the model fit (likelihood ratio test: P < 0.05) indicating its incremental value in predicting the mortality hazard. Adding SERPINA3 plasma levels to the model in predicting the 3 year mortality resulted in continuous NRI (>0) of 0.45 (CI: 0.07‐0.77, P < 0.05). The incremental value of NRI (>0) for ‘events’ was 45.2% and NRI (>0) for ‘non‐events’ was 0.1%. The calculated integrated discrimination improvement was 1.9%, and the increase in area under the curve was 0.5%.
In the analysis of predictors of the composite of mortality with unplanned cardiac readmission, SERPINA3 was a significant univariate predictor but was not retained as independent predictor in the multivariate analysis. Here, elevated NT‐proBNP together with ST2, age, eGFR, and PCWP remained independent predictors (c.statistic 0.66). Unplanned cardiac readmission with mortality as competing risk showed only eGFR and ischaemic aetiology as independent predictors (c.statistic: 0.60).
Discussion
In the present study, insights into the potential prognostic role of SERPINA3 in heart failure were enabled by selecting a well‐defined, phenotypically matched cohort of survivors and non‐survivors with heart failure due to congestive cardiomyopathy. While noting up‐regulated myocardial gene expression in non‐survivors, circulating levels were not related to myocardial production or cardiac load. In vitro studies suggested ability of endothelial cell subtypes to secret SERPINA3 in response to inflammatory cytokines. In further validation in an independent cohort of patients with a de novo or worsened heart failure regardless the aetiology and phenotype, circulating SERPINA3 levels were also higher in non‐survivors and predictive of poor survival or unplanned cardiac readmission. In particular, elevated SERPINA3 levels in combination with elevated ST2 and NT‐proBNP levels appeared to identify a vulnerable population of patients at excessive risk for mortality or unplanned cardiac readmission.
Serpins are a large family of protease inhibitors involved in many biological processes. 14 , 15 They are divided into clades based on their phylogenic relationship, 16 termed A to P. Serpins inhibit serine proteinases by an irreversible suicide substrate mechanism using a unique and extensive conformational change. 17 SERPINA3, also known as alpha‐1 antichymotrypsin, is one of the clade A serpins, present with high expression in the retina, kidney, liver, and pancreas. 18 SERPINA3 acts as an inhibitor of several serine proteases, mainly targeting cathepsin G released during the inflammatory response and involved in tissue damage. 8
SERPINA3 is implicated in the pathology of complex human disorders such as Alzheimer disease with vascular dementia 19 , 20 or prion disease. 21 In particular, SERPINA3 overexpression has been documented in several cancer types as a marker of poor prognosis. 22 , 23 , 24 In experimental heart failure, SERPINA3 appeared to accelerate tumour cells growth and thus was implicated to a cancer development in the clinical heart failure setting. 25 In clinical setting, plasma SERPINA3 levels appear to track favourable remodelling under mechanical haemodynamic support in end‐stage heart failure. 9 Increased plasma SERPINA3 concentrations appeared also predictive of adverse outcome in patients with acute myocardial infarction 26 or associated with calcific aortic stenosis. 27
Our study expands these observations by providing the first direct evidence associating SERPINA3 levels with poor clinical outcome in heart failure. In two distinct cohorts of patients, SERPINA3 levels were consistently higher in non‐surviving patients with CCMP or all comers with a de novo or worsened heart failure regardless its phenotype. Our study established a cut‐off value of 316 μg/mL predictive of adverse clinical outcome. Patients with SERPINA3 > 316 μg/mL alone or on top of elevated NT‐proBNP or ST2 showed increased mortality or its composite with first unplanned cardiac readmission. In addition, elevated SERPINA3 provided also additional precision into the prognostic mortality stratification in a model incorporating clinical, laboratory, or haemodynamic risk factors in combination with both biomarkers. Hence, by providing additional prognostic information to either of the established biomarkers, elevated SERPINA3 may help to identify a particularly vulnerable subset of heart failure patients at risk for poor outcomes.
The source of increased circulating plasma SERPINA3 has been primarily attributed to liver. 28 In current heart failure patient cohort, SERPINA3 protein concentrations were similar in coronary sinus and arterial blood despite up‐regulated myocardial transcript levels arguing against myocardial secretion. Yet several members of the Serpin family have a half‐life of several days, 29 which may cloud the interpretation of transmyocardial gradients with difficulties to discern from measured and biologically relevant gradient that might have been induced rapidly in response to inflammatory cytokines. Our findings provide a new experimental evidence that endothelial cells are able to secret SERPINA3 in response to inflammatory cytokines. The underlying molecular mechanisms appear to be related to direct gene and protein SERPINA3 induction followed by secretion via the ER/Golgi secretory pathway. These findings support the hypothesis that vascular endothelium susceptible to inflammatory stimuli may contribute to elevated circulating SERPINA3 levels in heart failure. Nevertheless, in the multivariate model, circulating inflammatory and humoral factors could not fully account for elevated levels and neither of the echocardiographic nor invasive haemodynamic parameters were associated with increased levels. Further precision in defining the mechanism of elevated SERPINA3 in heart failure is to be determined.
Following limitations should be acknowledged. SERPINA3 has been identified as a potential biomarker within pre‐selected gene transcripts. Unbiased genomic or proteomic analyses at myocardial or serum level might have identified other biomarkers of interest. Association between SERPINA3 and clinical outcomes was studied in patients with de novo or worsened heart failure in the limited sample size. Further prospective studies are needed to validate its prognostic value either alone or in the multimarker strategy in addition to NT‐proBNP and ST2 in larger heart failure population. Likewise, male gender was predominant in our population. Although gender did not emerge as independent risk factor, gender differences in SERPINA3 levels in relation to clinical outcomes require further investigation. It is also noteworthy that clinical factors such as renal failure or ischaemic aetiology remained strong predictors of unplanned cardiovascular readmissions alone or in combination with mortality emphasizing their relevance in risk models addressing the prognostic value of circulating biomarkers. Circulating SERPINA3 has been determined at one time point during the admission. The relevance of its dynamic changes in the later stages for prognostic stratification after reaching euvolemic state after discharge remains unknown. The clinical relevance of individual variations in the SERPINA levels within survivors cohort also requires further investigation. Our study attempted primarily to decipher the mechanisms of SERPINA production and its prognostic value. Its potential effects on endothelial cell function or cardiac myocytes as contributing to heart failure pathophysiology are of interest and require further studies.
In conclusion, we identified SERPINA3 as a potential prognostic biomarker in heart failure. Elevated SERPINA3 > 316 μg/mL appears to be associated with higher mortality or its composite with unplanned cardiac readmission. SERPINA3 appears to provide also additional prognostic information in risk models incorporating demographic, clinical, haemodynamic, and laboratory risk including ST2 and NT‐proBNP by identifying a vulnerable subgroup of heart failure patients at increased mortality risk. These findings warrant further validation in prospectively designed studies including a broad population of heart failure patients.
Conflict of interest
There are no conflicts of interest related to this study.
Funding
This study has been supported by VZW Cardiovascular Research Centre Aalst, Belgium.
Supporting information
Table S1. Baseline characteristics of patients in the exploratory cohort of matched survivors and non‐survivors with HF due to idiopathic cardiomyopathy.
Table S2. A total of 83 transcripts involved in heart failure were analysed in surviving and non‐surviving heart failure patients.
Table S3. Differentially myocardial expressed genes (n = 14) in CCMP survivors and non‐survivors vs control patients.
(*p < 0.05;**p < 0.01;***p < 0.001;****p < 0.0001 vs Controls).
Figure S1. Panel A: Fold increase in SERPINA3 protein secretion in human umbilical vein endothelial cells (HUVEC) and circulating precursor blood outgrowth endothelial cells (BOEC) after IL‐1β and TNF‐α stimulation compared to unstimulated cells. Panel B: IL‐1β stimulation of HCAEC induces SERPINA3 secretion and an increase in both SERPINA3 transcripts and protein. Panel C: Brefeldin A, a blocker of protein secretion through the ER/Golgi, is able to block SERPINA3 secretion.
Figure S2. Relationship between SERPINA3 levels and clinical outcome in combination with both NT‐proBNP and ST2. Panel A shows Kaplan‐Meier curves of all‐cause mortality till one year. Panel B shows composite endpoint of survival and first unplanned cardiac readmission.
Delrue, L. , Vanderheyden, M. , Beles, M. , Paolisso, P. , Di Gioia, G. , Dierckx, R. , Verstreken, S. , Goethals, M. , Heggermont, W. , and Bartunek, J. (2021) Circulating SERPINA3 improves prognostic stratification in patients with a de novo or worsened heart failure. ESC Heart Failure, 8: 4780–4790. 10.1002/ehf2.13659.
References
- 1. Kumani S, Squire I. Acute heart failure: definition, classification and epidemiology. Curr Heart Fail Rep 2017; 14: 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gaggin HK, Januzzi JL Jr. Biomarkers and diagnostics in heart failure. Biochim Biophys Acta 2013; 1832: 2442–2450. [DOI] [PubMed] [Google Scholar]
- 3. Wettersten N, Maisel AS. Biomarkers for heart failure: an update for practitioners of internal medicine. Am J Med 2016; 129: 560–567. [DOI] [PubMed] [Google Scholar]
- 4. Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JGF, Coats AJS, Falk V, González‐Juanatey JR, Harjola VP, Jankowska EA, Jessup M, Linde C, Nihoyannopoulos P, Parissis JT, Pieske B, Riley JP, Rosano GMC, Ruilope LM, Ruschitzka F, Rutten FH, van der Meer P, ESC Scientific Document Group . 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC) Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur Heart J 2016; 37: 2129–2200. [DOI] [PubMed] [Google Scholar]
- 5. Chow SL, Maisel AS, Anand I, Bozkurt B, de Boer RA, Felker GM, Fonarow GC, Greenberg B, Januzzi JL Jr, Kiernan MS, Liu PP, Wang TJ, Yancy CW, Zile MR. Role of biomarkers for the prevention, assessment, and managemant of heart failure: a scientific statement from the American Heart Association. Circulation 2017; 135: e1054–e1091. [DOI] [PubMed] [Google Scholar]
- 6. Nadar SK, Shaikh MM. Biomarkers in routine heart failure clinical care. Card Fail Rev 2019; 5: 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Demissei BG, Postmus D, Cleland JG, O'Connor CM, Metra M, Ponikowski P, Teerlink JR, Cotter G, Davison BA, Givertz MM, Bloomfield DM, van Veldhuisen DJ, Dittrich HC, Hillege HL, Voors AA. Plasma biomarkers to predict or rule out early post‐discharge events after hospitalization for acute heart failure. Eur J Heart Fail 2017; 19: 728–738. [DOI] [PubMed] [Google Scholar]
- 8. Baker C, Belbin O, Kalsheker N, Morgan K. SERPINA3 (aka alpha‐1‐antichymotrypsin). Front Biosci 2007; 12: 2821–2835. [DOI] [PubMed] [Google Scholar]
- 9. Lok SJ, van Mil A, Bovenschen N, van der Weide P, van Kuik J, van Wichen D, Peeters T, Siera E, Winkens B, Sluijter JPG, Doevendans PA, da Costa Martins PA, de Jonge N, de Weger RA. Post‐transcriptional regulation of α‐1‐antichymotrypsin by microRNA‐137 in chronic heart failure and mechanical support. Circ Heart Fail 2013; 6: 853–861. [DOI] [PubMed] [Google Scholar]
- 10. Bernstein LH, Zions MY, Alam ME, Haq SA, Heitner JF, Zarich S, Seamonds B, Berger S. What is the best approximation of reference normal for NT‐proBNP? Clinical levels for enhanced assessment of NT‐proBNP (CLEAN). J Med Lab Diagn 2011; 2: 16–21. [Google Scholar]
- 11. Scrucca L, Santucci A, Aversa F. Competing risk analysis using R: an easy guide for clinicans. Bone Marrow Transplant 2007; 40: 381–387. [DOI] [PubMed] [Google Scholar]
- 12. Bellera CA, MacGrogan G, Debled M, Tunon de Lara C, Brouste V, Mathoulin‐Pélissier S. Variables with time‐varying effects and the Cox model: some statistical concepts illustrated with a prognostic factor study in breast cancer. BMC Med Res Methodol 2010; 10: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cook NR. Quantifying the added value of new biomarkers: how and how not. Diagn Progn Res 2018; 2: 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carrell RW, Pemberton PA, Boswell DR. The serpins: evolution and adaptation in a family of protease inhibitors. Cold Spring Harb Symp Quant Biol 1987; 52: 527–535. [DOI] [PubMed] [Google Scholar]
- 15. Gettins PG. Keeping the serpin machine running smoothly. Genome Res 2000; 10: 1833–1835. [DOI] [PubMed] [Google Scholar]
- 16. Irving JA, Pike RN, Lesk AM, Whisstock JC. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res 2000; 10: 1845–1864. [DOI] [PubMed] [Google Scholar]
- 17. Huntington JA, Read RJ, Carrell RW. Structure of a serpin‐protease complex shows inhibition by deformation. Nature 2000; 407: 923–926. [DOI] [PubMed] [Google Scholar]
- 18. Sánchez‐Navarro A, Mejía‐Vilet JM, Pérez‐Villalva R, Carrillo‐Pérez DL, Marquina‐Castillo B, Gamba G, Bobadilla NA. SerpinA3 in the early recognition of acute kidney injury to chronic kidney disease (CKD) transition in the rat and its potentiality in the recognition of patients with CKD. Sci Rep 2019; 9: 10350–13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abraham CR, Selkoe DJ, Potter H. Immunochemical identification of the serine protease inhibitor alpha 1‐antichymotrypsin in the brain amyloid deposits of Alzheimer's disease. Cell 1988; 52: 487–501. [DOI] [PubMed] [Google Scholar]
- 20. Fraser PE, Nguyen JT, McLachlan DR, Abraham CR, Kirschner DA. Alpha 1‐antichymotrypsin binding to alzheimer a beta peptides is sequence specific and induces fibril disaggregation in vitro. J Neurochem 1993; 61: 298–305. [DOI] [PubMed] [Google Scholar]
- 21. Vanni S, Moda F, Zattoni M, Bistaffa E, De Cecco E, Rossi M, Giaccone G, Tagliavini F, Haik S, Deslys JP, Zanusso G, Ironside JW, Ferrer I, Kovacs GG, Legname G. Differential overexpression of SERPINA3 in human prion diseases. Sci Rep 2017; 7: 15637–16650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou J, Cheng Y, Tang L, Martinka M, Kalia S. Up‐regulation of SERPINA3 correlates with high mortality of melanoma patients and increased migration and invasion of cancer cells. Oncotarget 2017; 8: 18712–18725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luo D, Chen W, Tian Y, Li J, Xu X, Chen C, Li F. Serpin peptidase inhibitor, clade a member 3 (SERPINA3) is overexpressed in glioma and associated with poor prognosis in glioma patients. Onco Targets Ther 2017; 10: 2173–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang GD, Yang XM, Lu H, Ren Y, Ma M, Zhu L, Wang J, Song W, Zhan R, Zhang Z. SERPINA3 promotes endometrial cancer cells growth by regulating G2/M cell cycle checkpoint and apoptosis. Int J Clin Exp Pathol 2014; 7: 1348–1358. [PMC free article] [PubMed] [Google Scholar]
- 25. Meijers WC, Maglione M, Bakker SJL, Oberhuber R, Kieneker LM, de Jong S, Haubner BJ, Nagengast WB, Lyon AR, van der Vegt B, van Veldhuisen DJ, Westenbrink BD, van der Meer P, Silljé HHW, de Boer RA. Heart failure stimulates tumor growth by circulating factors. Circulation 2018; 138: 678–691. [DOI] [PubMed] [Google Scholar]
- 26. Zhao L, Zheng M, Guo Z, Li K, Liu Y, Chen M, Yang X. Circulating SERPINA3 levels predict the major adverse cardiac events in patients with myocardial infarction. Int J Cardiol 2020; 300: 34–38. [DOI] [PubMed] [Google Scholar]
- 27. Martin‐Rojas T, Mourino‐Alvarez L, Gil‐Dones F, de la Cuesta F, Rosello‐Lleti E, Laborde CM, Rivera M, Lopez‐Almodovar LF, Lopez JA, Akerstrom F, Padial LR, Barderas MG. A clinical perspective on the utility of alpha 1 antichymotrypsin for the early diagnosis of calcific aortic stenosis. Clin Proteom 2017; 14: 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Travis J, Salvesen GS. Human plasma proteinase inhibitors. Annu Rev Biochem 1983; 52: 655–709. [DOI] [PubMed] [Google Scholar]
- 29. Rau JC, Beaulieu LM, Huntington JA, Church FC. Serpins in thrombosis, hemostasis and fibrinolysis. J Thromb Haemost 2007; 5: 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Baseline characteristics of patients in the exploratory cohort of matched survivors and non‐survivors with HF due to idiopathic cardiomyopathy.
Table S2. A total of 83 transcripts involved in heart failure were analysed in surviving and non‐surviving heart failure patients.
Table S3. Differentially myocardial expressed genes (n = 14) in CCMP survivors and non‐survivors vs control patients.
(*p < 0.05;**p < 0.01;***p < 0.001;****p < 0.0001 vs Controls).
Figure S1. Panel A: Fold increase in SERPINA3 protein secretion in human umbilical vein endothelial cells (HUVEC) and circulating precursor blood outgrowth endothelial cells (BOEC) after IL‐1β and TNF‐α stimulation compared to unstimulated cells. Panel B: IL‐1β stimulation of HCAEC induces SERPINA3 secretion and an increase in both SERPINA3 transcripts and protein. Panel C: Brefeldin A, a blocker of protein secretion through the ER/Golgi, is able to block SERPINA3 secretion.
Figure S2. Relationship between SERPINA3 levels and clinical outcome in combination with both NT‐proBNP and ST2. Panel A shows Kaplan‐Meier curves of all‐cause mortality till one year. Panel B shows composite endpoint of survival and first unplanned cardiac readmission.


